
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 24 May 2019
Sec. Molecular Innate Immunity
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.01084
A correction has been applied to this article in:
Corrigendum: Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS–) vs. Alternatively Activated Macrophages
Macrophages are found in tissues, body cavities, and mucosal surfaces. Most tissue macrophages are seeded in the early embryo before definitive hematopoiesis is established. Others are derived from blood monocytes. The macrophage lineage diversification and plasticity are key aspects of their functionality. Macrophages can also be generated from monocytes in vitro and undergo classical (LPS+IFN-γ) or alternative (IL-4) activation. In vivo, macrophages with different polarization and different activation markers coexist in tissues. Certain mouse strains preferentially promote T-helper-1 (Th1) responses and others Th2 responses. Their macrophages preferentially induce iNOS or arginase and have been called M1 and M2, respectively. In many publications, M1 and classically activated and M2 and alternatively activated are used interchangeably. We tested whether this is justified by comparing the gene lists positively [M1(=LPS+)] or negatively [M2(=LPS–)] correlated with the ratio of IL-12 and arginase 1 in transcriptomes of LPS-treated peritoneal macrophages with in vitro classically (LPS, IFN-γ) vs. alternatively activated (IL-4) bone marrow derived macrophages, both from published datasets. Although there is some overlap between in vivo M1(=LPS+) and in vitro classically activated (LPS+IFN-γ) and in vivo M2(=LPS–) and in vitro alternatively activated macrophages, many more genes are regulated in opposite or unrelated ways. Thus, M1(=LPS+) macrophages are not equivalent to classically activated, and M2(=LPS–) macrophages are not equivalent to alternatively activated macrophages. This fundamental discrepancy explains why most surface markers identified on in vitro generated macrophages do not translate to the in vivo situation. Valid in vivo M1/M2 surface markers remain to be discovered.
Macrophages are critical cells in both tissue homeostasis and inflammation, performing essential tissue-specific functions as well as protecting the organism from infection. Their origin has been debated extensively during the past years (1, 2). It is now clear that most tissue macrophages arrive before definitive hematopoiesis is established. Monocytes derive from a common progenitor called Macrophage Dendritic Cell Precursor (MDP), emphasizing a continuum differentiation potential of monocytes to both inflammatory macrophages and DCs (3). However, although monocytes undoubtedly can become macrophages, data from recent studies have challenged the generalized applicability of this dogma. Sathe et al. also challenged the idea that MDPs are the restricted progenitors of monocytes and macrophages. Thus, they show that MDPs can differentiate into other hematopoietic lineages (4). Other studies nicely summarized by Florent Ginhoux and Martin Guilliams (1) reported that most tissue macrophages in mice are not generated from monocytes in the steady state. Instead, mature tissue macrophages are derived from embryonic precursors that seed the tissues before birth, maintaining their numbers in adults by self-renewal (1). Primitive macrophages first appear within the yolk sac blood islands, then spread into the tissues of the embryo through the blood as soon as the circulatory system is established, giving rise to the fetal macrophage populations. These cells maintain a gene expression signature different from bone marrow-derived macrophages (BMDM). However, whether yolk sac-derived macrophages can persist into adulthood giving rise to specific adult tissue macrophage populations is still debated (5). Of note, all lineage tracing studies of tissue macrophages were completed in mice. While it is reasonable to assume that human tissue macrophages also derive from embryonic precursor cells, this hypothesis is difficult to test rigorously.
Since the discovery of tissue-specific gene signatures in peritoneal macrophages (6), many published transcriptomic data show that each tissue macrophage set has both tissue-specific and shared gene signatures, based on the tissue of origin (7). The tissue-specific gene signatures will not be discussed further. They need to be taken into consideration when comparing macrophages in different parts of the body.
Tissue macrophage transcriptomes dramatically change after transfer to a tissue culture environment (8). Gosselin et al. show that mouse and human microglia transferred to a tissue culture environment gain and lose expression of hundreds of genes. Preferential reduction in expression of microglia-specific genes such as MAF, RUNX, and SMAD3 was documented (8, 9). Thus, there appears to be a specific “in vitro” macrophage transcriptome, reflecting the fact that the cells were removed from their normal tissue environment and placed in a tissue culture dish. Most published macrophage work uses this in vitro setting, most commonly for bone marrow-derived macrophages (BMDM) for mouse macrophages and monocyte-derived macrophages (MDM) for human macrophages. The in vitro environment and the different stimuli used to differentiate the cells such as CSF1 (M-CSF) and CSF2 (GM-CSF) have substantial polarizing effects. These colony-stimulating factors are known to “prime” or “activate” macrophages as well as induce their differentiation (10). Studies comparing M-CSF and GM-CSF in BMDM show that they can activate different pathways: GM-CSF leads to a more pro-inflammatory state (TNF expression), and M-CSF induces a tissue healing state (IL-10 expression) after LPS stimulation (10). In vitro, macrophages change their polarization state based on diverse stimuli such as cytokines, microbes, microbial products, and other modulators (11).
In the literature, there are several terms and definitions to describe the macrophage activation and polarization. Mills defined M1 and M2 in Balb/c and C57BL/6 mice (12). Nathan defined classical macrophage activation by IFN-γ (13), Gordon defined alternative macrophage activation by IL-4 (14), Anderson and Mosser defined regulatory macrophages (Mreg) (15), Kadl defined a macrophage phenotype induced by oxidized lipids (Mox) (16), Gleissner defined the transcriptome of macrophages differentiated in the presence of CXCL4 (PF4, M4) (17), and Mantovani further subdivided alternatively activated macrophages into M2a,b,c (18).
The M1/M2 macrophage polarization nomenclature was introduced in 2000, based on the propensity of C57BL/6J macrophages to be more easily activated to produce NO (M1 polarized) than Balb/c mice (M2 polarized) (12). Different metabolism of arginine after LPS injection elicits different phenotypes of macrophages in C57BL/6J and Balb/c mice. C57BL/6J peritoneal macrophages induced iNOS resulting in nitric oxide and a T-helper 1 (Th1) CD4 T cell response, while Balb/c mice induced arginase to produce ornithine and a Th2 response. In analogy to Th1 and Th2, these macrophages were named M1 and M2. Subsequent work showed that M1 macrophages have a pro-inflammatory phenotype with pathogen-killing abilities and M2 macrophages promote cell proliferation and tissue repair (19). These findings suggested that macrophages from different strains of mice had a different propensity to produce NO and arginase in response to LPS.
We rigorously tested macrophage polarization using the hybrid mouse diversity panel (HMDP) generated by the Lusis group (20, 21). The HMDP is a panel of 83 inbred mouse strains, employed as a surrogate model for human immune diversity. Most bioinformatics tools are not designed to dissect a spectral distribution in a heterogeneous population. Analysis of commonly regulated genes after LPS treatment among all 83 strains yielded an empty set (no genes). Therefore, we established a gene expression-based factor that represents the degree of LPS-induced polarization (polarization factor). Since IL-12 is the known Th1 polarizing cytokine and arginase is the hallmark enzyme of M2 macrophages, we used the ratio of IL-12b/arginase-1 gene expression as a continuous parameter to rank all 83 mouse strains along the M1/M2 axis. Peritoneal macrophage transcriptomes were used to rank the 83 mouse strains based on their response to LPS (22). We found a continuous spectrum of LPS-induced activation: M1-polarized mice responded by upregulating many pro-inflammatory genes that were positively correlated with IL12/arginase. In accordance with Murray's recommendation to list the stimulus in parentheses (11), the M1 signature is synonymous with (=LPS+), i.e., genes whose expression show a positive correlation with the IL-12/arginase ratio in response to LPS. M2-polarized mice responded by upregulating genes that were negatively correlated with IL12/arginase (=LPS–) (22). A similar spectrum of responses was seen in two human macrophage datasets (22), suggesting that the HMDP is a valid way to reflect human macrophage diversity.
Cytokines can polarize macrophages in vitro. The classical activation is induced by LPS and IFN-γ. Classically activated macrophages are often called M1, but this is misleading, as our analysis will show. LPS activates oxidative metabolism and antimicrobial activity of macrophages (23). In the 1990s Gordon's group reported that the Th2 cytokine IL-4 increased the expression of the macrophage mannose receptor (MR, also known as CD206), which is normally down-regulated by IFN-γ, inducing an alternative activation of macrophages that promoted an anti-inflammatory and pro-healing phenotype (14, 24). Other cytokines such as IL-10 and TGF-β were also linked with macrophage polarization in vitro. Mantovani et al. called classically activated macrophages (by IFN-γ combined with LPS or tumor necrosis factor [TNF]) M1. In vitro alternatively activated macrophages (by IL-4) were re-named M2a. Two other M2-like macrophage phenotypes were induced by Fc receptor engagement by immune-complexes (M2b) or by IL-10 and glucocorticoids (M2c) (18). Based on Mantovani's work, many researchers started to use M1 for classical activated and M2 for alternatively activated macrophages in vitro. Unfortunately, that led to more confusion than clarification in the field.
With the final purpose to better understand the differences and similarities between in vivo and in vitro macrophage response, in this review, we compared mouse macrophage transcriptomes in vivo extracted from Buscher et al. (22) (GSE38705) to in vitro transcriptomes of bone marrow derived macrophages BMDMs that were either classically (LPS+IFN-γ, GSE69607) or alternatively (IL-4, GSE69607) activated (25).
The in vitro polarization signatures used in this review as described in Table 1 were obtained from Jablonski et al. (GSE69607) transcriptomic data and defined by comparing the differently expressed (DE) genes of in vitro alternatively activated BMDMs (IL-4, 20 ng/ml for 24 h) vs. the in vitro classically activated BMDMs (LPS, 100 ng/mL + IFN-γ 20 ng/mL for 24 h). All genes with log2 fold change (FC) >1 were considered in the in vitro alternatively activated macrophage (Mφ) signature; all genes with log2 FC < -1 were considered in the in vitro classically activated Mφ signature. An FDR < 0.05 were applied in this analysis (Supplementary Table 1).
To further increase the significance of our analysis we filtered the two in vitro full signatures with the gene sets derived from in vitro classically activated (LPS+IFN-γ) and alternatively activated (IL-4) transcriptomes vs. untreated BMDMs transcriptome, respectively (GSE69607). All the DE genes passing the FDR < 0.05 cutoff were considered and used for the further analysis and discussions (Supplementary Table 2).
The signatures of in vivo bacterial LPS positive responders [M1(=LPS+)] vs. negative responders [M2(=LPS–)] peritoneal macrophages as established in Buscher et al. (22) (GSE38705) were used (Supplementary Table 3). Since the in vitro classically activated (LPS+IFN-γ treated) and alternatively (IL-4-treated) activated Mφ transcriptomes were derived from C57BL/6J mice, we filtered the full in vivo signatures specifically for the C57Bl/6J strain from LPS treated and untreated mice by listing the differently up- and downregulated genes (FDR < 0.05) between control and LPS-treated samples for the C57BL/6J strain only (20, 21) (Supplementary Table 4). The two newly originated in vivo macrophage signatures M1(=LPS+)C57BL/6J, M2(=LPS–)C57BL/6J, and the in vitro classically activated (LPS+IFN-γ treated) and alternatively activated (IL-4-treated) signatures were then analyzed and compared using the Venny 2.1 online tool (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Of the 322 M1 (=LPS+) genes positively correlated with IL-12/arginase, 117 were also upregulated in in vitro classically activated (LPS+IFN-γ) Mφ, but 28 were also upregulated in in vitro alternatively activated (IL-4) Mφ. Conversely, of the 186 genes in the in vivo M2(=LPS–)C57BL/6J signature whose expression was negatively correlated with IL12/arginase, seven each were shared with in vitro classically and alternatively activated (IL-4) Mφ (Figure 1A, Supplementary Table 5). Strikingly, 460 genes were private to in vitro classically activated (LPS+IFN-γ) Mφ and 349 genes were private to in vitro alternatively activated (IL-4) Mφ, respectively. One thousand seven hundred seventy-six genes were private to M1(=LPS+)C57BL/6J and 173 were private to in vivo M2(=LPS–)C57BL/6J. Since in vivo M1 is widely considered synonymous with in vitro classically activated (LPS+IFN-γ) and M2 is considered synonymous with in vitro alternatively activated (IL-4) Mφ, these discrepancies are striking. Only very few genes (15) were downregulated in vitro classically activated (LPS+IFN-γ) Mφ compared to unstimulated Mφ, or in alternatively activated (IL-4) Mφ compared to unstimulated Mφ (36 genes, Figure 1B, Supplementary Table 5). There was almost no overlap (total of five genes) between the gene set genes) between the gene set downregulated by activation in vitro and the in vivo gene set M1/M2(=LPS±) positively or negatively correlated with IL-12/arginase (Figure 1B). Since M1(=LPS+)C57BL/6J is considered the opposite of in vitro alternatively activated (IL-4) and M2(=LPS–)C57BL/6J is considered the opposite of classically activated, we will focus on the differentially expressed private genes that are not shared, because they point out the difference between in vivo and in vitro.
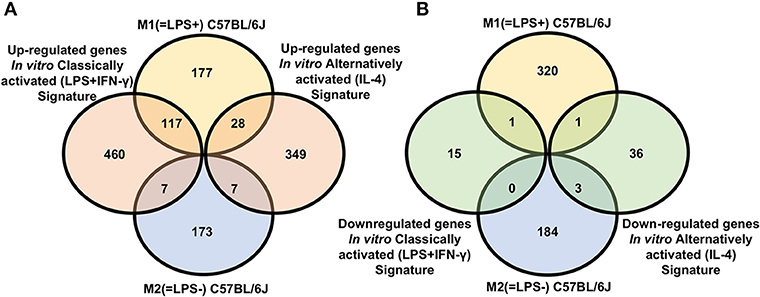
Figure 1. Signature comparison for C57BL/6 macrophages. (A) Venn diagram showing overlap between genes whose expression was positively [M1(=LPS+)C57BL/6J] or negatively [M2(=LPS–)C57BL/6J] correlated with IL12/arginase ratio in vivo with genes upregulated in vitro classically activated (LPS+IFN-γ) macrophages (left) or in vitro alternatively activated (IL-4) (right) macrophages vs. unstimulated. (B) Venn diagram showing overlap between genes whose expression was positively [M1(=LPS+)C57BL/6J] or negatively [M2(=LPS–)C57BL/6J] correlated with IL12/arginase in vivo with genes downregulated in vitro classically activated (LPS+IFN-γ) macrophages (left) or in vitro alternatively activated (IL-4) (right) macrophages vs. unstimulated.
Among the 117 genes upregulated in both in vivo M1(=LPS+)C57BL/6J and in vitro classically activated (LPS+IFN-γ) Mφ (Figure 1A, Supplementary Table 5) are many interferon-induced genes. Among interferon-regulated genes were Irf9, Irf7, Ifi35, Ifnar2, Isg20, Ifit2, and Ifih1. Interferon regulatory factors actively regulate macrophage activation and polarization with the expression of pro-M1 genes such as IL-12 (Il12a and Il12b) (26, 27). The expression of these genes is mediated by the activation of Janus kinase (Jak2) JAK/signal, transducer and activator of transcription signaling pathway guided by signal transducer and activator of transcription (Stat1/Stat2) (28).
Costimulatory molecules such as Cd86 and Cd40 relevant to antigen presentation were also represented among both in vivo M1(=LPS+)C57BL/6J and in vitro classically activated (LPS+IFN-γ) Mφ signatures (29–31). CD86 is a ligand for CD28 and is a known inflammatory Mφ/DC marker (32). CD40 is a member of the TNF receptor superfamily (Tnfrsf5) and involved in antibody isotype switching and vascular inflammation (33). Another interesting gene expressed is Cd38, described by Jablonski et al. (25) to be a good marker for in vitro classical macrophage discrimination. Here we found that Cd38 upregulation is shared among signatures. However, the exact functional role of CD38 in macrophages is still unclear. If Cd38 surface expression is correlated with Cd38 gene expression, CD38 might be a good new marker for M1 polarization (34).
Chemokines and their receptors are the main players in recruiting leukocytes during an active immune response. Cytokine receptors shape their function. Shared between the M1 and classically activated signatures we found Cxcl16, Cxcl9, Il15ra, and Il17ra. The chemokines Cxcl16 and Cxcl9 are known to support M1 polarization mostly primed by IFN stimulation (35). A previous study has shown that IL-15R alpha expression on macrophages supports the early transition of antigen-specific effector CD8(+) T cells to memory cells (36).
The TNF receptor-associated factor (Traf1) expression, together with TNF receptor superfamily members like Tnfrsf1b suggest that both M1 and classically activated macrophages respond strongly to TNF-like signals. Pro-inflammatory cytokine expression is well-known to increase adhesion molecule expression such as ICAM-1 in endothelial cells and macrophages in an NFκB-dependent manner (37). When integrin molecules on the leukocyte surface bind endothelial ICAM-1, this leads to outside-in signaling. In vivo M1(=LPS+)C57BL/6J and in vitro classically activated (LPS+IFN-γ) Mφ share upregulation of VCAM-1 (Vcam1) and ICAM-1 (Icam1). Recently, it was suggested that Icam1 may regulate macrophage polarization by inhibiting the M2 polarization in tumor-associated macrophages (TAM) (38).
Intriguingly, Poly(ADP-Ribose) Polymerase Family Member 9 (Parp9) is also in the list of genes shared between in vivo M1(=LPS+)C57BL/6J and in vitro classically activated (LPS+IFN-γ) Mφ. Parp9 is a critical activator of macrophages (39). Modulating its expression has been shown to inhibit the activation of macrophage and can ameliorate atherosclerosis in mouse models (Patent US20160289685).
The in vivo M1(=LPS+)C57BL/6J signature contains 177 uniquely up-regulated genes (Figure 1A, Supplementary Table 5). Compared to the in vitro signatures we found the expression of many genes correlated with the LPS mediated activation of Toll-like receptor 4 (Tlr4). Downstream of Tlr4 is myeloid differentiation factor 88 (MyD88), Nfkb1, Rela, Rps6ka2, Tank, and Ripk2, (40) all expressed in M1(=LPS+)C57BL/6J. These genes are responsible for an anti-bacterial immune response. The chemokine Ccl2 and the hematopoietic cytokine Csf2 (GM-CSF) are positively correlated with M1 polarization in vivo, but not increased in response to IFN-γ and LPS in vitro. The same is true for the pro-inflammatory cytokines Il15, Il23a, Irf1, and Ifnb1.
Several pro-inflammatory cytokines, such as TNF and IL-12 have been reported to induce Csf2, whereas IL-4 and IL-10 suppress Csf2 (41). Its expression enhances macrophage differentiation toward a pro-inflammatory state by activating the expression of cytokines (i.e., IL-23) and chemokines such as Ccl2, promoting leukocyte recruitment (42). IL-23a is part of the IL-12 family that includes other structurally related, heterodimeric cytokines such as IL-12, IL-27, and IL-35 (43). IL-23a drives inflammation through the induction of IL-17, promoting a highly pro-inflammatory Th17 response (44).
IL-15 is up-regulated in macrophages in response to LPS (45, 46). IL-15 was described to induce Th1 and Natural Killer cell (NK) immune responses by inducing IFN-γ and the transcription factor T-bet (47). Interferon regulatory factors in macrophages like Irf1 actively regulate a macrophage effector activation with the expression of other pro-M1 genes such as IL-12 (Il12a and Il12b) and iNos (Nos2) (27).
Modulation of genes involved in general cellular metabolic activities is a prominent feature of macrophage differentiation and polarization. M1 macrophages mostly activate catabolic processes whereas M2 is skewed toward anabolic metabolism. The in vivo M1(=LPS+)C57BL/6J signature indeed includes genes responsible for an active protein catabolic process such as Psme2. Psme2 and other members of the Psme family are related to proteasome modulation. Proteasome modulation regulates macrophage function (48). The metabolism of proteins and amino acids (such as arginine) is fundamental for the correct activation of macrophages. The LPS stimulation indeed can induce the metabolism of arginine to NO and citrulline (49). This effect is mediated by different genes such as the cationic amino acid transporters Slc7a2, Slc12a4, Slc1a4, Slc39a14, Slc3a2, and Slc4a7 found expressed exclusively in the in vivo M1(=LPS+)C57BL/6J signature.
The in vitro classically activated (LPS+IFN-γ) Mφ signature present after activating BMDMs with LPS and IFN-γ includes many genes involved in chemotaxis and cell migration (Figure 1A, Supplementary Table 5). Chemokines found in the in vitro classically activated (LPS+IFN-γ) Mφ signature include Cxcl1 and Cxcl2, which recruit neutrophils (50). Ccl5 (Rantes), a potent chemoattractant for T Cells, is also present. Other chemokines and chemokine receptors such as Ccl3, Cxcl10, Cxcl11, Ccl25, Cx3cr1, and Ccr7 are in the in vitro signature. It also includes many cytokines well-known in classically activated macrophages: IL-1α, IL1β, IL-6, and TNF (Il1a, Il1b, Il6, and Tnf).
TNFR-associated factors (TRAFs) and other TNF induced proteins (Tnfaip) are important mediators of innate immune receptor signaling. Traf2 and Tnfaip3 are expressed in the classically activated macrophages in vitro, their function mostly related to regulating TLR signaling and mediating the response of other TNFR family members. Traf2 and Tnfaip3 induce proinflammatory cytokines and type I interferons in response to LPS or other stimuli (51).
Fifteen genes are down-regulated in classically activated macrophages but increase with LPS in M1 macrophages (Figure 1B, Supplementary Table 5). Many of them relate to the endocytosis process such as Adrb2, Aif1, Apoc2, Coro1a, Sorl1, and Sirpb1a (51).
Among the in vivo M2(=LPS–)C57BL/6J and in vitro alternatively activated (IL-4) Mφ signatures, seven genes are up-regulated in common (Figure 1A, Supplementary Table 5): Lpxn, Dhrs3, Mical1, Dnmt3a, Jun, Gab1, and P2ry1. Gab1, Jun, and P2ry1 are related to positive regulation of the MAPK cascade.
The Grb2-associated binder (Gab) proteins, which belong to the Gab/DOS family of docking proteins, function as essential elements in signal integration during the assembly of downstream signaling complexes (52). Gab1 and Gab2 (Gab1/2) are widely expressed in various cell types, including immune cells such as T cells, and mast cells. A recent study demonstrated that the deficiency of either Gab1 or Gab2 resulted in impaired M2 polarization (53). Gab1 regulates IL-4-induced macrophage polarization by activating AKT signaling (53). Another interesting gene is Jun Proto-Oncogene (Jun). The transcription factor family AP-1 is composed of homo- and heterodimeric complexes, which consist of Jun, Fos, activating transcription factor, and musculoaponeurotic fibrosarcoma proteins. These dimers are involved in different cellular processes, such as proliferation, apoptosis, and differentiation.
G Protein-Coupled Receptors (GPCRs) are important receptors able to regulate inflammation and immunity. P2Y1 (P2ry1) is an ADP receptor known to be expressed in several macrophages except for microglia, where it does not appear to be expressed (54). P2Y1 function in macrophages is not entirely understood. LPS decreases its expression in vitro (55).
The Glutamine synthetase (Glul) is a gene associated with glutamine metabolism, an essential pathway for the differentiation and function of macrophages (56). Glul indeed is known to be enhanced in M2 macrophages (56).
The in vivo M2(=LPS–)C57BL/6J signature includes 170 private genes not shared by in vitro classically or alternatively activated (IL-4) macrophages (Figures 1A,B, Supplementary Table 5).
As stated before, the M2 polarization exerts a switch on arginine metabolism and other metabolic pathways (57). The majority of genes present in the signature are indeed related to the modulation of the cell metabolism, i.e., lipid catabolic process. M2 polarized macrophages obtain their energy mostly from fatty acid oxidation and oxidative metabolism. The fatty acid oxidation process in in vivo M2(=LPS–)C57BL/6J macrophages is highlighted by the presence of several genes in the signatures such as Acyl-CoA Dehydrogenase (Long chain) and medium-chain (Acadm) (58). Carnitine Palmitoyltransferase 1A (Cpt1a) is also present in the signature, and it is associated with fatty acid beta-oxidation (59, 60).
The nuclear receptor PPARγ plays a dominant role in adipogenesis and storage of fatty acids as triglycerides in adipocytes. In macrophages, its activation enhances CD36 expression and increases the uptake of oxidized LDL and triacylglycerides (61). The nuclear receptor Pparγ activates several changes in the macrophage metabolism enhancing an M2-like response, and IL-4 is responsible for the first induction of its expression (62). The lipid modification and repair are another fundamental property in M2 macrophage function. This mechanism is mediated by the expression of Acadm, and other genes such as B4galnt1, Hadh, Inpp5d, Soat1, Ip6k1, Hacl1, Echs1, Dgkz, and Hadhb, present only in the in vivo M2 signature.
Other important cellular processes are post-translational modifications (PTM) that contribute to cell physiology by regulating protein stability, localization, and functions. Many of these modifications occur in response to LPS and regulate the interaction with environmental cues, being particularly relevant to macrophage functions. Several PTM genes were found in the M2(=LPS–)C57BL/6J signature. The ADP-ribosylating activity is one of them and is associated with the expression of poly(ADP-ribose)-polymerases (PARPs). Parp1 it is present in M2(=LPS–)C57BL/6J macrophages and accounts for the majority of the poly(ADP-ribose) (PAR) polymer synthesis. Although enriched in the M2(=LPS–)C57BL/6J signature, Parp1 has a pro-inflammatory function (63).
Another important PMT gene present in the M2(=LPS−) signature is the protein tyrosine phosphatase Ptpn22. Ptpn22 phosphatase activity is still not fully understood. Ptpn22 in macrophages skews the polarization toward M2 (64).
To define macrophage polarization, the field is in urgent need of good surface markers. At the mRNA level, we found Cd300a, a type I transmembrane protein with a long cytoplasmatic tail, known to regulate immune cells and macrophages (65, 66); and Cd84, a member of the SLAM family as candidates. CD84 is a cell surface receptor already known to be modulated during LPS stimulation (67). It is not known how CD300 or CD84 surface expression correlates with their mRNAs.
The in vitro alternatively activated (IL-4) Mφ signature contains 349 genes solely upregulated by IL-4 treatment in vitro (Figure 1A, Supplementary Table 5).
Among the genes up-regulated by IL-4 treatment in vitro we found the known M2 marker mannose receptor Mrc1 also known as Cd206, as well as the tetraspanin Cd9, the TCR-associated molecule Cd74, Bcl2, Arg1, and the scavenger receptor Cd36.
It has been reported that several types of tissue-resident macrophages express Cd206 in both mouse and humans (68). The depletion of mannose receptors has proven to increase the level of pro-inflammatory proteins (69). Cd206 indeed promotes the expression of several anti-inflammatory cytokines and chemokines such as Tgf β, Il10, and Ccl18 (the last also expressed in the signature), inducing a pro-fibrotic effect (68).
The tetraspanin CD9 is a cell surface glycoprotein, known to modulate cell adhesion and migration. In macrophages it has been found to regulate the LPS induced activation negatively; the loss of CD9 in CD9 knockout macrophages enhances LPS signaling (70). IFN-gamma signaling has been found to reduce CD9 expression in macrophages (71).
CD74 is also a cell-surface receptor for the cytokine macrophage migration inhibitory factor (MIF), mostly expressed and studied in B cells. MIF binding to CD74 induces its intramembrane cleavage and the release of its cytosolic intracellular domain, which regulates cell survival. CD74 with MIF also induces the expression and secretion of the cytokine, midkine. Midkine suppresses apoptosis by elevating the expression of Bcl-2 and inhibiting caspase 3 and 7 activity (72).
B Cell lymphoma 2 (Bcl2) is also expressed among the 350 up-regulated genes. Bcl2 inhibits pro-apoptotic proteins (73). The lack of Bcl2 expression in macrophages has been reported to accelerate the progression of atherosclerotic plaques in Apoe−/− mice (74).
Other well-reported M2 macrophage markers are Arg1 and Cd36. Arginase-1 is an enzyme of the urea cycle; its action catalyzes the hydrolysis of arginine to ornithine. Ornithine is the substrate for ornithine decarboxylase (ODC). This pathway regulates a multitude of cellular processes like DNA replication, protein translation, cell growth, and differentiation (75). Moreover, the absence of L-arginine was proven to reduce T-cell proliferation (76).
CD36 is a class B scavenger receptor for the endocytosis of triacylglycerol-rich lipoprotein particles, such as LDL and VLDL. It is also known to be an essential macrophage receptor for apoptotic cell recognition and phagocytosis (77).
Among the 36 down-regulated genes vs. untreated Mφ (Figure 1B) to be noted is the presence of genes commonly referred for cell cycle modulation such as Ccnd1, Chek1, Cdc25b, and Ncapg2.
In the signature comparison, 28 genes up-regulated in in vivo M1(=LPS+)C57BL/6J and in vitro alternatively activated (IL-4) Mφ signatures were found (Figure 1A, Supplementary Table 5), including four chemokines: Ccl7, Ccl17, Ccl22, and Ccl24.
Ccl7 is known to be expressed by both M1 and M2 macrophages (78). CCL17 and CCL22 are both ligands of the CCR4 receptor. In vitro alternatively activated macrophages have been shown to produce all these chemokines in high amounts in the response of Th2 cytokines such as IL-4, and IL-13 (18, 79). Functionally, CCL17 and CCL22 together with CCL24 are potent chemoattractants, favoring the attraction of immune-inhibitory cells such as regulatory T-cells (Treg) (18). Another gene, type 1 myosin (Myo1c), related to cytoskeleton rearrangement, is fundamental for the migration and phagocytosis process of macrophages. Interestingly, TLR4 stimulation by LPS showed enrichment of cytoskeleton-associated proteins such as Myo1C among the LPS-regulated phosphopeptides (80). The cell surface markers CD44 (Cd44), and CD83 (Cd83) are also regulated in the different signatures. This is a good example illustrating that the cell surface phenotype observed in vitro alternatively activated (IL-4) Mφ cannot be perfectly translated into an M2 in vivo phenotype.
Intriguingly, in vivo M2(=LPS–)C57BL/6J and in vitro classically activated (LPS+IFN-γ) Mφ shared the up-regulation of seven genes; Fos, Tuba4a, Tnfrsf21, Sdc1, Ggta1, Rhov, and Psen2 (Figure 1A, Supplementary Table 5).
Presenilin 2 (Psen2) is the catalytic core component of protease complexes that mediate regulated intramembrane proteolysis of their substrates. The lack of normal Psen2 function has been associated with diminished LPS-induced macrophage responsiveness (81). Another modulated gene is Syndecan1 (Sdc1), that is related to the wound healing process (82, 83). A TNF receptor family gene (Tnfrsf21) is among these seven in the M2(=LPS–)C57BL/6J and classically activated Mφ in vitro (Supplementary Table 5). Activation of TNFRSF receptors induces the apoptotic process through the initiation of specific receptors (so-called Death Receptors). The best-known example is Fas (Tnfrsf6) (84). These genes, indeed, can activate the death effector domain leading to caspases activation and cell death (85).
The gene lists for the different macrophage signatures were further analyzed by Ingenuity Pathways Analysis (IPA). The enriched canonical pathways for each signature with a p-value cutoff of 0.001 are reported in Supplementary Table 6.
The enriched pathways for in vivo macrophage signatures M1(=LPS+)C57BL/6J, M2(=LPS–)C57BL/6J and the in vitro classically activated (LPS+IFN-γ) and alternatively activated (IL-4) signatures were then analyzed and compared using the Venny 2.1 online tool (Figure 2A, Supplementary Table 7). The z-score is a prediction on the state of the pathway based on the expression level of the genes associated with that pathway. A positive z-score implies that the pathway is activated and a negative Z-score implies that the pathway is inhibited. z-score < -2 or z-score >2 was considered significant. The genes enriched for each pathway and the corresponding z-score are fully reported in Supplementary Table 8. Reassuringly, the largest number, 65 pathways are shared between the in vivo M1(=LPS+)C57BL/6J and in vitro classically activated (LPS+IFN-γ) macrophages (Figure 2A). Our analysis shows 31 pathways that are private to in vivo M1(=LPS+)C57BL/6J and two pathways private to in vivo M2(=LPS–)C57BL/6J (Figure 2A). In vitro alternatively and classically activated Mφ show 7 and 11 private pathways, respectively. Interestingly, no pathways are commonly modulated by in vivo M2(=LPS–)C57BL/6J and in vitro alternatively activated (IL-4) Mφ, suggesting that these macrophage phenotypes are very different from each other. Instead, five pathways are shared by the in vivo M1(=LPS+)C57BL/6J and in vitro alternatively activated (IL-4) Mφ. Another five are shared between M1(=LPS+)C57BL/6J, classically and alternatively activated (IL-4) Mφ (Figure 2A).
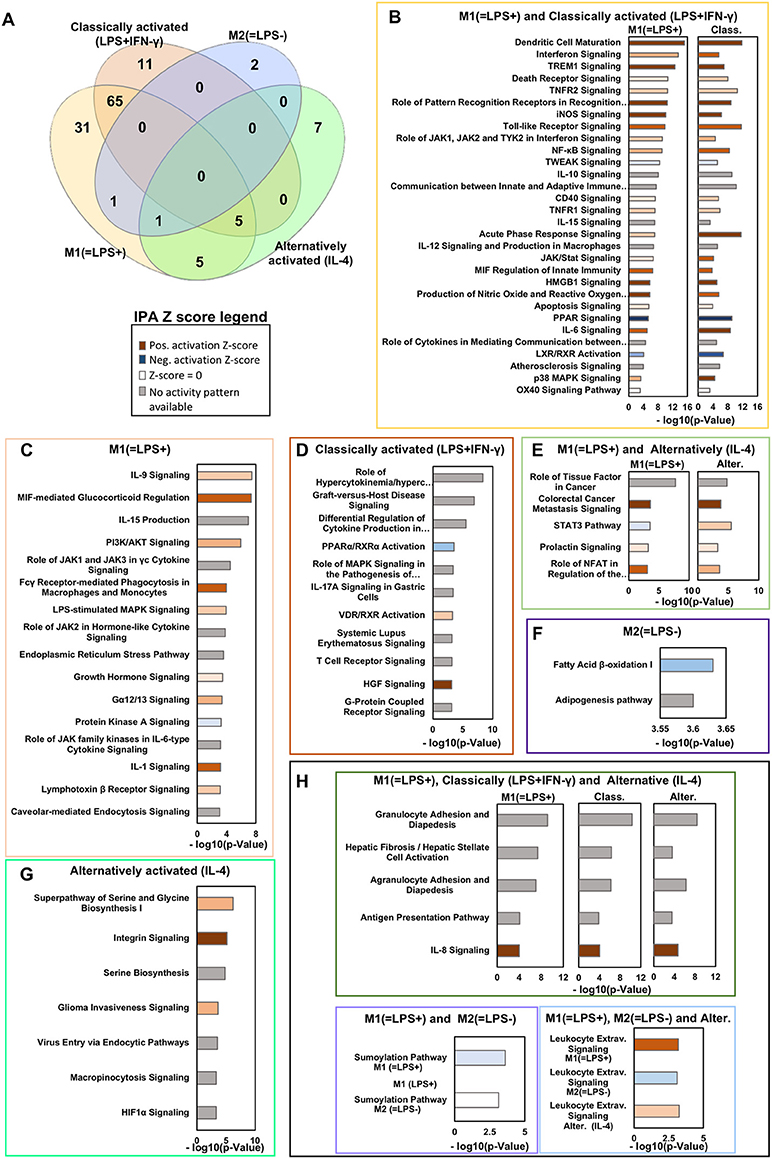
Figure 2. Ingenuity pathways analysis in vivo M1(=LPS+)C57BL/6J, M2(=LPS–)C57BL/6J, and in vitro classically (LPS+IFN-γ) or alternatively activated (IL-4) macrophages. (A) Venn diagram showing the key canonical pathways enriched in vivo M1(=LPS+)C57BL/6J, M2(=LPS–)C57BL/6J, M2(=LPS–)C57BL/6J, classically activated (LPS+IFN-γ) and alternatively activated (IL-4) macrophage signatures as determined by Ingenuity pathway analysis (IPA). The number of pathways is sorted by a P-value cutoff of 0.001. (B–H) Selected canonical pathways ranked based on –log(P-value) divided as reported in the Venn diagram are shown in the boxes. The Z-score of each pathway is reported by the color of the bars (see legend). Light/Dark shades represent smaller/larger absolute values of Z-score.
In vivo M1(=LPS+)C57BL/6J and in vitro classically activated (LPS+IFN-γ) Mφ present 65 pathways in common (Supplementary Table 7). The top 30 are shown in Figure 2B. The top modulated pathways by p-value include Dendritic Cell Maturation, showing a close relationship between M1 macrophages, and mature dendritic cells.
The Dendritic Cell Maturation pathway is a big pathway that includes ≈190 genes. Among these genes many are commonly activated during M1 like macrophage polarization such as Csf2, Il12, Il18, Il15, Il23 etc. (the full list of genes found inside this pathway for M1(=LPS+) and classically activated Mφ is reported in Supplementary Table 8). A prominent inflammatory DC is the TIP-DC, producing TNF and iNOS (86).
Next is Interferon Signaling, driven by the genes Irf1, Irf9, Jak2, Stat1, Ifnar2, Stat2, Ifi35, Ptpn2, Ifitm3, Tap1, Psmb8, and Rela. Trem1 Signaling is also highly significant. Death Receptor Signaling, OX40 Signaling Pathway and TNFR2 Signaling are related and contain many of the TNF receptor superfamily signaling pathways. The pathway Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses contains TLRs (also in the pathway Toll-like Receptor Signaling) and other receptors. iNOS Signaling is prominently enriched; iNOS is the main bacterial killing pathway employed by M1 and classically activated macrophages.
The z-scores given for iNOS signaling in M1(=LPS+) and classically activated (LPS+IFN-γ) Mφ present comparable values with a slightly lower p-value in M1(=LPS+) than classically activated macrophages. Muller et al. previously reported that classically activated Mφ (LPS+IFN-γ) produce more Nos2, than LPS activated macrophages (87, 88). The z-score is a prediction on the state of the pathway based on the expression level of the several genes associated with that pathway but is not reliable for single gene expression. In this particular case the genes modulated inside the iNOS signaling pathway by M1(=LPS+) signature were slightly different compared to the in vitro classically activated (LPS+IFN-γ) macrophage signature as reported in Supplementary Table 8.
Related to this is the Production of Nitric Oxide and Reactive Oxygen Species in Macrophages. Next is the Role of JAK1, JAK2, and TYK2 in Interferon Signaling and NF-κB Signaling. Unexpectedly, TWEAK Signaling ranks highly (p = 3 × 10−9). TWEAK is an often-overlooked member of the TNF receptor superfamily. Two other TNF receptor superfamily pathways are CD40 Signaling and TNFR1 Signaling. The anti-inflammatory IL-10 Signaling pathway is also prominent, showing that an anti-inflammatory program is detectable in the generally pro-inflammatory in vivo M1(=LPS+)C57BL/6J and in vitro classically activated (LPS+IFN-γ) macrophages. Communication between Innate and Adaptive Immune Cells shows that activated Mφ also instruct adaptive immunity. Related to this is the Role of Cytokines in Mediating Communication between Immune Cells pathway. IL-15 Signaling is a pathway related to IL-2. IL-15 signaling stimulates T cell proliferation and inhibits IL-2-mediated activation-induced cell death (89). Acute Phase Response Signaling is normally expected in liver cells, but also active in in vivo M1(=LPS+)C57BL/6J and in vitro classically activated classically activated (LPS+IFN-γ) Mφ. IL-12 Signaling and Production is a defining proinflammatory property of macrophages. JAK/Stat Signaling covers many cytokine signaling pathways. MIF Regulation of Innate Immunity focuses on an atypical chemokine, MIF, with powerful effects in innate immunity. HMGB1 Signaling is also pro-inflammatory and together with CD14 mediates the activation of TLR4 (90).
Significant enrichment of Apoptosis Signaling shows the precarious situation of these highly activated macrophages that teeter on the edge of oblivion, as is necessary for terminating the aggressive phase of inflammation. Related to this is activation of the LXR/RXR Activation pathway, which tends to be anti-inflammatory. PPAR Signaling is related to metabolic effects. IL-6 Signaling is its own module, which is of key importance in cardiovascular inflammation, suggesting that in vitro classically activated (LPS+IFN-γ) and in vivo M1(=LPS+)C57BL/6J macrophages share the ability to promote atherosclerosis. Indeed, the Atherosclerosis Signaling pathway is also significantly enriched.
Some pathways are private to in vivo M1(=LPS+)C57BL/6J (Figure 2C, Supplementary Table 7). They include IL-9 Signaling, IL-15 Production (although a related pathway is found in both in vivo M1(=LPS+)C57BL/6J and classically activated Mφ), and PI3K/AKT Signaling. The Role of JAK1 and JAK3 in common gamma chain (γc) Cytokine Signaling shows than common gamma chain cytokine pathways are more enriched in vivo M1(=LPS+)C57BL/6J macrophages. Interestingly, Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes is only enriched in in vivo M1(=LPS+)C57BL/6J and not in vitro classically activated (LPS+IFN-γ) Mφ, as is LPS-stimulated MAPK Signaling. In vivo M1(=LPS+)C57BL/6J also have a signature for Endoplasmic Reticulum Stress Pathway. Enriched Gα12/13 Signaling suggests activation of chemokine receptor signaling. Protein Kinase A Signaling is an anti-inflammatory pathway. This pathway has a negative z-score, meaning it is likely suppressed in vivo M1(=LPS+)C57BL/6J macrophages. The pathway Role of JAK family kinases in IL-6-type Cytokine Signaling is similar to the IL-6 pathway mentioned above, which is shared between in vitro classically activated (LPS+IFN-γ) Mφ and M1(=LPS+)C57BL/6J. Of great interest for cardiovascular disease, IL-1 Signaling is private to the in vivo M1(=LPS+)C57BL/6J signature and not enriched in in vitro classically activated (LPS+IFN-γ) Mφ. Finally, the Lymphotoxin β Receptor Signaling is present in M1(=LPS+)C57BL/6J; it is a TNF receptor superfamily member involved in organizing lymphoid tissues believed to control pro-inflammatory response reactions (91). Also this pathway suggests that in vivo M1(=LPS+)C57BL/6J macrophages keep control of inflammation avoiding harmful hyper reactivity with a balance of pro and anti-inflammatory stimuli.
In vitro classically activated (LPS+IFN-γ) Mφ show 11 private pathways (Figure 2D). The top modulated pathways by p-value include Role of Hypercytokinemia/Hyperchemokinemia and Graft Vs. Host Disease Signaling, both driven by a similar set of cytokines including Il18, Il1rn, Il1b, Tnf, Il1a, and Il6 (Supplementary Table 3). The third highly significant modulated pathway is Differential Regulation of Cytokine Production in Macrophages and T Helper Cells by IL-17A and IL-17F, driven by the modulation of Il12a, Il1b, Ccl5, Tnf, Il12b, and Il6.
The involvement of IL-17 signaling in in vitro classical Mφ activation is also suggested by the IL-17A Signaling pathway driven by, among other genes, Il17ra. The HGF Signaling pathway is also induced. HGF is a well-known motogenic factor, inducing directional migration, and differentiation of monocytes (92). By contrast, the PPARα/RXRα Activation pathway is negatively modulated (Figure 2D). The negative regulation of this pathway might be linked with switching off lipid metabolism and boosting the glycolytic pathway (93).
Intriguingly, no pathways were regulated in common among the M2(=LPS–)C57BL/6J in vivo and in vitro alternatively activated (IL-4) Mφ signature at the p-value cut-off applied (Figure 2E).
The two pathways privately expressed in vivo M2(=LPS–)C57BL/6J are the adipogenesis pathway and fatty acid synthesis pathway, both involved in anabolic processes (Figure 2F). Opposed to the in vivo M1(=LPS+)C57BL/6J activation, in vivo M2(=LPS–)C57BL/6J regulated gene transcription occurs in conditions favoring mitochondrial metabolism and oxidative glucose metabolism, with an increase of anabolic process that properly corresponds with the in vivo M2 functions (94, 95).
The in vitro alternatively activated (IL-4) Mφ show seven private pathways (Figure 2G). Serine Biosynthesis and Serine and Glycine Biosynthesis I are two of the top three enriched pathways by p-value, confirming that in vitro alternatively activated (IL-4) Mφ polarization involves coordinated metabolic and transcriptional rewiring. The third top modulated pathway by p-value is the Integrin Signaling pathway (Figure 2G), suggesting the activation of the molecular machineries responsible of tissue infiltration. Integrins allow the infiltration of macrophages and other cells that start the removal of necrotic tissue and initiate the tissue regeneration processes (96, 97).
Some pathways are common among opposite signatures. Specifically, we found 11 pathways shared between in vivo M1(=LPS+)C57BL/6J and in vitro alternatively activated (IL-4) Mφ, with five of them also present in vitro classically activated (LPS+IFN-γ) Mφ (Figure 2H). The top two regulated pathways common for all three signatures are the Granulocyte Adhesion and Diapedesis and the Agranulocyte Adhesion and Diapedesis pathways that are driven by several chemokines and adhesion molecules including Ccl2, Cxcl9, Ccl7, Ccl24, Ccl22, Ccl9, Cxcl16, Ccl17, Icam1, and Vcam1 (Supplementary Table 3).
The IL-8 signaling pathway has a high Z score in vivo M1(=LPS+)C57BL/6J, and in vitro classically and alternatively activated (IL-4) Mφ. This pathway is known to support acute inflammation inducing the active neutrophil recruitment (98).
Exclusive for in vivo M1(=LPS+)C57BL/6J and in vitro alternatively activated (IL-4) Mφ, we found the Role of Tissue Factor pathway, driven by the expression of Hbegf, Hck, Csf2, Src, Itgav, Jak2, Plaur, F10, Egr1, Mmp13, Rps6ka2, Stat5a, Csf1, and Rps6ka4 (Supplementary Table 3).
The STAT3 pathway showed an opposite Z score between in vitro alternative Mφ and in vivo M1(=LPS+)C57BL/6J (Figure 2H). The other three pathways instead show similar positively regulation among the two signatures (Figure 2H).
In vivo M1(=LPS+)C57BL/6J, M2(=LPS–)C57BL/6J and in vitro alternatively activated (IL-4) Mφ shared the Leukocyte extravasation signaling pathway.
Among the in vivo M1(=LPS+)C57BL/6J, and M2(=LPS–)C57BL/6J only the sumoylation pathway is shared, but presumably downregulated by the M1(=LPS+)C57BL/6J (Figure 2H).
To address the question whether in vivo M(LPS±) or in vitro classically/alternatively activated (IL-4) gene signatures can predict cancer survival, we used the PRECOG database that ranks genes by overall tumor survival.
As proof of concept, survival data from Buddingh et al. human osteosarcoma (GSE21257) was analyzed for the impact of the original in vivo M1(=LPS+) and in vitro classically activated (LPS+IFN-γ) and in vivo M2(=LPS–) and in vitro alternatively activated (IL-4) gene expression signatures in the tumor biopsy transcriptome from pre-chemotherapy biopsies of osteosarcoma patients. The genes applied in the analysis were extracted based on Cox linear regression analysis from the original signatures. Only the genes with a p-value < 0.01 from the in vitro and in vivo macrophages signatures were subsequently used to analyze survival in the published osteosarcoma datasets retrospectively (GSE21257) (Figure 3, Supplementary Table 9). Kaplan–Meier curves were plotted using ProggeneV2, divided by the median of the mean expression of a tumor-specific gene list.
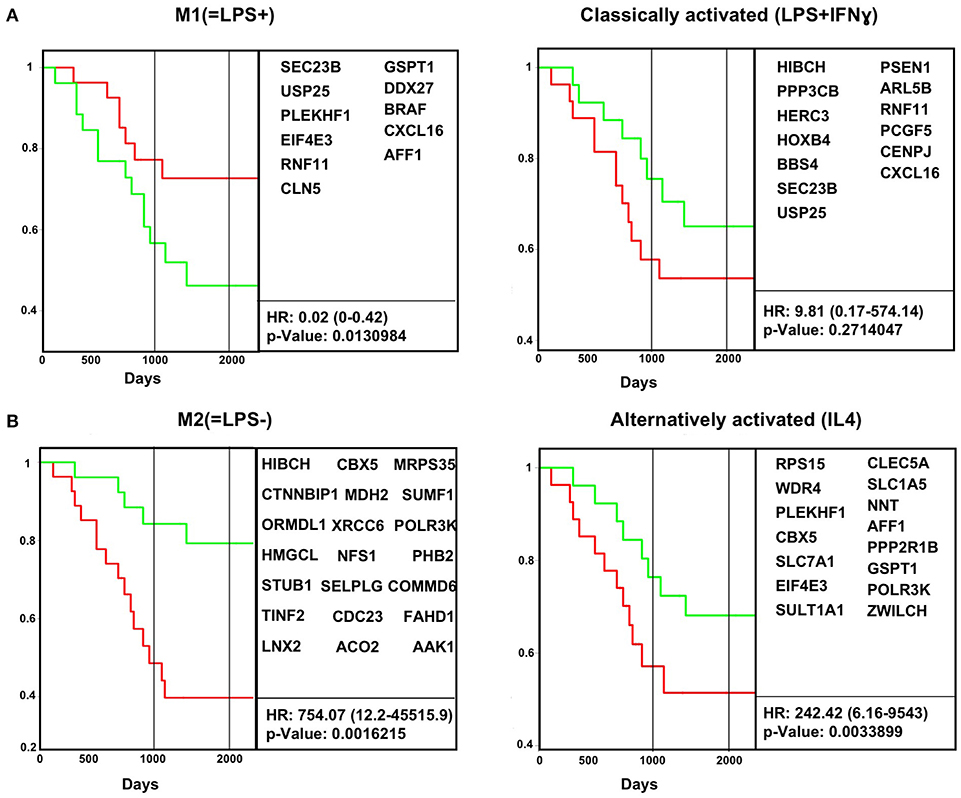
Figure 3. In vivo macrophage signatures predict survival in osteosarcoma cancer biopsy transcriptomes. Survival data for human osteosarcoma cancer biopsies (GSE21257) were analyzed for the impact of M1(=LPS+) and classically activated (LPS+IFN-γ) (A) and M2(=LPS−) and alternatively activated (IL-4) (B) gene expression signatures in the tumor biopsy transcriptome. Kaplan–Meier curves were plotted using ProggeneV2, divided by the median of the mean expression of a tumor-specific gene list (in boxes). Hazard ratio (HR, cox proportional hazard analysis) and significance (log rank P-value) are shown. Red, green curves indicate high, low expression of the respective signature genes. The two vertical black lines indicates 3 and 5 years, respectively.
We find the in vivo M1(=LPS+) signature correlates positively with survival, which means patients with tumors with above median expression of the M1(=LPS+) signature genes survived significantly longer (Figure 3A). Conversely, enrichment of M2(=LPS–) signature genes correlates with cancer death (Figure 3B). The in vitro classical M1 signatures did not correlate with survival. The in vitro alternative M2 gene signature correlates with cancer death, but less than the M2(=LPS–) in vivo signature.
In vitro evidence on BMDMs suggest that activation with two molecular signals from the microenvironment is required for efficient induction of M1 like phenotype in murine macrophages as defined by tumoricidal activity, NO production, and secretion of pro-inflammatory and Th1-polarizing cytokines (87).
However, it is not clear whether cytokines or other cancer therapies can shift macrophages to M1 in an individual with genetic predisposition for M2. M1 polarizers show preclinical evidence of improving cancer outcomes (99, 100). M1/M2 is also important in other diseases like atherosclerosis (101) and autoimmune diseases.
Several efforts have been made trying to define the molecular networks underlying polarized activation of macrophages. This review highlights the difference between cytokine effects on BMDMs in vitro vs. the response of peritoneal macrophages to LPS. Transcriptomes of LPS positive responder [M1(=LPS+)] vs. negative responder [M2(=LPS–)] macrophages show some overlap with in vitro classically (LPS+IFN-γ) vs. alternatively activated (IL-4) BMDMs. However, the majority of regulated genes are different. Thus, in vivo M1(=LPS+) and M2(=LPS–) macrophages and in vitro classically (by LPS, IFN-γ) and alternatively activated (by IL-4) Mφ are not comparable. Some genes and pathways are even shared between the in vivo and the in vitro opposite signature.
Bone marrow contains immature macrophage precursors, which are not found in peripheral tissues like the peritoneum, so BMDMs are a mix of monocyte-derived and precursor-derived macrophages. We show that the in vivo M1(=LPS+) and M2(=LPS–) gene signatures are also applicable to human transcriptomes. Human alveolar macrophages for example expressed about 70% of the mouse-derived gene signatures. Human monocyte-derived macrophages (hMDM) transcriptome (102) in response to LPS stimuli expressed about 57% of the M1(=LPS+) gene signature and only 37% of the in vitro classically activated (LPS+IFN-γ) signature (Supplementary Table 10). We also demonstrated an increased enrichment score for the in vivo M1(=LPS+) signature in isolated human synovial macrophages from rheumatoid arthritis patients (22). The in vivo M2(=LPS–) signature instead presented a high enriched score in isolated human synovial macrophages from healthy donors (22).
In conclusion, classical and alternative macrophage activation in vitro does not match the in vivo M1/M2 polarization. This fundamental discrepancy explains why most surface markers identified on in vitro generated macrophages fail to translate to macrophages in vivo. We propose that a new effort is needed to discover valid M1/M2 markers, which will help understand macrophage behavior in vivo.
KL and MO wrote the paper. MO, YG, and AP performed the analysis, constructed the Venn diagrams, and input to the Supplementary Tables.
This study was funded by the National Institutes of Health R01 HL115232 (to KL), and American Heart Association post-doctoral fellowship AHA18POST34060251 (to MO).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.01084/full#supplementary-material
1. Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. (2016) 44 439–49. doi: 10.1016/j.immuni.2016.02.024
2. van Furth R, Cohn ZA. The origin and kinetics of mononuclear phagocytes. J Exp Med. (1968) 128:415–35. doi: 10.1084/jem.128.3.415
3. Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. (2006) 311:83–7. doi: 10.1126/science.1117729
4. Sathe P, Metcalf D, Vremec D, Naik SH, Langdon WY, Huntington ND, et al. Lymphoid tissue and plasmacytoid dendritic cells and macrophages do not share a common macrophage-dendritic cell-restricted progenitor. Immunity. (2014) 41:104–15. doi: 10.1016/j.immuni.2014.05.020
5. Lavin Y, Mortha A, Rahman A, Merad M. Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol. (2015) 15:731–44. doi: 10.1038/nri3920
6. Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. (2014) 157:832–44. doi: 10.1016/j.cell.2014.04.016
7. Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. (2012) 13:1118–28. doi: 10.1038/ni.2419
8. Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. (2017) 356:eaal3222. doi: 10.1126/science.aal3222
9. Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. (2014) 159:1327–40. doi: 10.1016/j.cell.2014.11.023
10. Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol. (2007) 178:5245–52. doi: 10.4049/jimmunol.178.8.5245
11. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. (2014) 41:14–20. doi: 10.1016/j.immuni.2014.06.008
12. Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. (2000) 164:6166–73. doi: 10.4049/jimmunol.164.12.6166
13. Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med. (1983) 158:670–89. doi: 10.1084/jem.158.3.670
14. Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. (1992) 176:287–92. doi: 10.1084/jem.176.1.287
15. Anderson CF, Mosser DM. A novel phenotype for an activated macrophage: the type 2 activated macrophage. J Leukoc Biol. (2002) 72:101–6. doi: 10.1189/jlb.72.1.101
16. Kadl A, Meher AK, Sharma PR, Lee MY, Doran AC, Johnstone SR, et al. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ Res. (2010) 107:737–46. doi: 10.1161/CIRCRESAHA.109.215715
17. Gleissner CA, Shaked I, Little KM, Ley K. CXC chemokine ligand 4 induces a unique transcriptome in monocyte-derived macrophages. J Immunol. (2010) 184:4810–8. doi: 10.4049/jimmunol.0901368
18. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. (2004) 25:677–86. doi: 10.1016/j.it.2004.09.015
19. Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol. (2014) 5:514. doi: 10.3389/fimmu.2014.00514
20. Lusis AJ, Seldin MM, Allayee H, Bennett BJ, Civelek M, Davis RC, et al. The hybrid mouse diversity panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. J Lipid Res. (2016) 57:925–42. doi: 10.1194/jlr.R066944
21. Orozco LD, Bennett BJ, Farber CR, Ghazalpour A, Pan C, Che N, et al. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell. (2012) 151:658–70. doi: 10.1016/j.cell.2012.08.043
22. Buscher K, Ehinger E, Gupta P, Pramod AB, Wolf D, Tweet G, et al. Natural variation of macrophage activation as disease-relevant phenotype predictive of inflammation and cancer survival. Nat Commun. (2017) 8:16041. doi: 10.1038/ncomms16041
23. Nathan CF. Mechanisms of macrophage antimicrobial activity. Trans R Soc Trop Med Hyg. (1983) 77:620–30. doi: 10.1016/0035-9203(83)90190-6
24. Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. (2010) 32:593–604. doi: 10.1016/j.immuni.2010.05.007
25. Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado DJde, Popovich PG, Partida-Sanchez S, et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS ONE. (2015) 10:e0145342. doi: 10.1371/journal.pone.0145342
26. Gunthner R, Anders HJ. Interferon-regulatory factors determine macrophage phenotype polarization. Mediat Inflamm. (2013) 2013:731023. doi: 10.1155/2013/731023
27. Xie C, Liu C, Wu B, Lin Y, Ma T, Xiong H, et al. Effects of IRF1 and IFN-beta interaction on the M1 polarization of macrophages and its antitumor function. Int J Mol Med. (2016) 38:148–60. doi: 10.3892/ijmm.2016.2583
28. Gough DJ, Levy DE, Johnstone RW, Clarke CJ. IFNgamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev. (2008) 19:383–94. doi: 10.1016/j.cytogfr.2008.08.004
29. Ley K, Gerdes N, Winkels H. ATVB distinguished scientist award: how costimulatory and coinhibitory pathways shape atherosclerosis. Arterioscler Thromb Vasc Biol. (2017) 37:764–77. doi: 10.1161/ATVBAHA.117.308611
30. Subauste CS, de Waal Malefyt R, Fuh F. Role of CD80 (B7.1) and CD86 (B7.2) in the immune response to an intracellular pathogen. J Immunol. (1998) 160:1831–40.
31. Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. (2009) 229:152–72. doi: 10.1111/j.1600-065X.2009.00782.x
32. Sansom DM. CD28, CTLA-4 and their ligands: who does what and to whom? Immunology. (2000) 101:169–77. doi: 10.1046/j.1365-2567.2000.00121.x
33. Zirlik A, Maier C, Gerdes N, MacFarlane L, Soosairajah J, Bavendiek U, et al. CD40 ligand mediates inflammation independently of CD40 by interaction with Mac-1. Circulation. (2007) 115:1571–80. doi: 10.1161/CIRCULATIONAHA.106.683201
34. Amici SA, Young NA, Narvaez-Miranda J, Jablonski KA, Arcos J, Rosas L, et al. CD38 is robustly induced in human macrophages and monocytes in inflammatory conditions. Front Immunol. (2018) 9:1593. doi: 10.3389/fimmu.2018.01593
35. Donlin LT, Jayatilleke A, Giannopoulou EG, Kalliolias GD, Ivashkiv LB. Modulation of TNF-induced macrophage polarization by synovial fibroblasts. J Immunol. (2014) 193:2373–83. doi: 10.4049/jimmunol.1400486
36. Mortier E, Advincula R, Kim L, Chmura S, Barrera J, Reizis B, et al. Macrophage- and dendritic-cell-derived interleukin-15 receptor alpha supports homeostasis of distinct CD8+ T cell subsets. Immunity. (2009) 31:811–22. doi: 10.1016/j.immuni.2009.09.017
37. Hubbard AK, Giardina C. Regulation of ICAM-1 expression in mouse macrophages. Inflammation. (2000) 24:115–25. doi: 10.1023/A:1007029409521
38. Yang M, Liu J, Piao C, Shao J, Du J. ICAM-1 suppresses tumor metastasis by inhibiting macrophage M2 polarization through blockade of efferocytosis. Cell Death Dis. (2015) 6:e1780. doi: 10.1038/cddis.2015.144
39. Iwata H, Goettsch C, Sharma A, Ricchiuto P, Goh WW, Halu A, et al. PARP9 and PARP14 cross-regulate macrophage activation via STAT1 ADP-ribosylation. Nat Commun. (2016) 7:12849. doi: 10.1038/ncomms12849
40. Laird MH, Rhee SH, Perkins DJ, Medvedev AE, Piao W, Fenton MJ, et al. TLR4/MyD88/PI3K interactions regulate TLR4 signaling. J Leukoc Biol. (2009) 85:966–77. doi: 10.1189/jlb.1208763
41. Ushach I, Zlotnik A. Biological role of granulocyte macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF) on cells of the myeloid lineage. J Leukoc Biol. (2016) 100:481–9. doi: 10.1189/jlb.3RU0316-144R
42. Shinohara H, Yano S, Bucana CD, Fidler IJ. Induction of chemokine secretion and enhancement of contact-dependent macrophage cytotoxicity by engineered expression of granulocyte-macrophage colony-stimulating factor in human colon cancer cells. J Immunol. (2000) 164:2728–37. doi: 10.4049/jimmunol.164.5.2728
43. Stow JL, Low PC, Offenhauser C, Sangermani D. Cytokine secretion in macrophages and other cells: pathways and mediators. Immunobiology. (2009) 214:601–12. doi: 10.1016/j.imbio.2008.11.005
44. Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. (2011) 12:231–8. doi: 10.1038/ni.1990
45. Ruckert R, Brandt K, Ernst M, Marienfeld K, Csernok E, Metzler C, et al. Interleukin-15 stimulates macrophages to activate CD4+ T cells: a role in the pathogenesis of rheumatoid arthritis? Immunology. (2009) 126:63–73. doi: 10.1111/j.1365-2567.2008.02878.x
46. Liew FY, McInnes IB. Role of interleukin 15 and interleukin 18 in inflammatory response. Ann Rheum Dis. (2002) 61 (Suppl. 2):ii100-2. doi: 10.1136/ard.61.suppl_2.ii100
47. Strengell M, Matikainen S, Siren J, Lehtonen A, Foster D, Julkunen I, et al. IL-21 in synergy with IL-15 or IL-18 enhances IFN-gamma production in human NK and T cells. J Immunol. (2003) 170:5464–9. doi: 10.4049/jimmunol.170.11.5464
48. Qureshi N, Morrison DC, Reis J. Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim Biophys Acta. (2012) 1823:2087–93. doi: 10.1016/j.bbamcr.2012.06.016
49. Yeramian A, Martin L, Arpa L, Bertran J, Soler C, McLeod C, et al. Macrophages require distinct arginine catabolism and transport systems for proliferation and for activation. Eur J Immunol. (2006) 36:1516–26. doi: 10.1002/eji.200535694
50. Reutershan J, Morris MA, Burcin TL, Smith DF, Chang D, Saprito MS, et al. Critical role of endothelial CXCR2 in LPS-induced neutrophil migration into the lung. J Clin Invest. (2006) 116:695–702. doi: 10.1172/JCI27009
51. Jin J, Xiao Y, Hu H, Zou Q, Li Y, Gao Y, et al. Proinflammatory TLR signalling is regulated by a TRAF2-dependent proteolysis mechanism in macrophages. Nat Commun. (2015) 6:5930. doi: 10.1038/ncomms6930
52. Gu H, Neel BG. The “Gab” in signal transduction. Trends Cell Biol. (2003) 13:122–30. doi: 10.1016/S0962-8924(03)00002-3
53. Guo X, Li T, Xu Y, Xu X, Zhu Z, Zhang Y, et al. Increased levels of Gab1 and Gab2 adaptor proteins skew interleukin-4 (IL-4) signaling toward M2 macrophage-driven pulmonary fibrosis in mice. J Biol Chem. (2017) 292:14003–15. doi: 10.1074/jbc.M117.802066
54. Ley K, Pramod AB, Croft M, Ravichandran KS, Ting JP. How mouse macrophages sense what is going on. Front Immunol. (2016) 7:204. doi: 10.3389/fimmu.2016.00204
55. Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, et al. Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. (2008) 4:5. doi: 10.1186/1745-7580-4-5
56. Liu PS, Wang H, Li X, Chao T, Teav T, Christen S, et al. alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol. (2017) 18:985–994. doi: 10.1038/ni.3796
57. Galvan-Pena S, O'Neill LA. Metabolic reprograming in macrophage polarization. Front Immunol. (2014) 5:420. doi: 10.3389/fimmu.2014.00420
58. Tucci S, Floegel U, Beermann F, Behringer S, Spiekerkoetter U. Triheptanoin: long-term effects in the very long-chain acyl-CoA dehydrogenase-deficient mouse. J Lipid Res. (2017) 58:196–207. doi: 10.1194/jlr.M072033
59. Nomura M, Liu J, Rovira II, Gonzalez-Hurtado E, Lee J, Wolfgang MJ, et al. Fatty acid oxidation in macrophage polarization. Nat Immunol. (2016) 17:216–7. doi: 10.1038/ni.3366
60. Skotte L, Koch A, Yakimov V, Zhou S, Soborg B, Andersson M, et al. CPT1A missense mutation associated with fatty acid metabolism and reduced height in greenlanders. Circ Cardiovasc Genet. (2017) 10:e001618. doi: 10.1161/CIRCGENETICS.116.001618
61. Chawla A. Control of macrophage activation and function by PPARs. Circ Res. (2010) 106:1559–69. doi: 10.1161/CIRCRESAHA.110.216523
62. Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. (2006) 4:13–24. doi: 10.1016/j.cmet.2006.05.011
63. Gongol B, Marin T, Peng IC, Woo B, Martin M, King S, et al. AMPKalpha2 exerts its anti-inflammatory effects through PARP-1 and Bcl-6. Proc Natl Acad Sci USA. (2013) 110:3161–6. doi: 10.1073/pnas.1222051110
64. Chang HH, Miaw SC, Tseng W, Sun YW, Liu CC, Tsao HW, et al. PTPN22 modulates macrophage polarization and susceptibility to dextran sulfate sodium-induced colitis. J Immunol. (2013) 191:2134–43. doi: 10.4049/jimmunol.1203363
65. Borrego F. The CD300 molecules: an emerging family of regulators of the immune system. Blood. (2013) 121:1951–60. doi: 10.1182/blood-2012-09-435057
66. Nakahashi-Oda C, Tahara-Hanaoka S, Shoji M, Okoshi Y, Nakano-Yokomizo T, Ohkohchi N, et al. Apoptotic cells suppress mast cell inflammatory responses via the CD300a immunoreceptor. J Exp Med. (2012) 209:1493–503. doi: 10.1084/jem.20120096
67. Sintes J, Romero X, de Salort J, Terhorst C, Engel P. Mouse CD84 is a pan-leukocyte cell-surface molecule that modulates LPS-induced cytokine secretion by macrophages. J Leukoc Biol. (2010) 88:687–97. doi: 10.1189/jlb.1109756
68. Roszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm. (2015) 2015:816460. doi: 10.1155/2015/816460
69. Lee SJ, Evers S, Roeder D, Parlow AF, Risteli J, Risteli L, et al. Mannose receptor-mediated regulation of serum glycoprotein homeostasis. Science. (2002) 295:1898–901. doi: 10.1126/science.1069540
70. Suzuki M, Tachibana I, Takeda Y, He P, Minami S, Iwasaki T, et al. Tetraspanin CD9 negatively regulates lipopolysaccharide-induced macrophage activation and lung inflammation. J Immunol. (2009) 182:6485–93. doi: 10.4049/jimmunol.0802797
71. Wang XQ, Evans GF, Alfaro ML, Zuckerman SH. Down-regulation of macrophage CD9 expression by interferon-gamma. Biochem Biophys Res Commun. (2002) 290:891–7. doi: 10.1006/bbrc.2001.6293
72. Gil-Yarom N, Radomir L, Sever L, Kramer MP, Lewinsky H, Bornstein C, et al. CD74 is a novel transcription regulator. Proc Natl Acad Sci USA. (2017) 114:562–7. doi: 10.1073/pnas.1612195114
73. Adams JM, Cory S. The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. (2018) 25:27–36. doi: 10.1038/cdd.2017.161
74. Shearn AI, Deswaerte V, Gautier EL, Saint-Charles F, Pirault J, Bouchareychas L, et al. Bcl-x inactivation in macrophages accelerates progression of advanced atherosclerotic lesions in Apoe(-/-) mice. Arterioscler Thromb Vasc Biol. (2012) 32:1142–9. doi: 10.1161/ATVBAHA.111.239111
75. Rath M, Muller I, Kropf P, Closs EI, Munder M. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front Immunol. (2014) 5:532. doi: 10.3389/fimmu.2014.00532
76. Munder M, Schneider H, Luckner C, Giese T, Langhans CD, Fuentes JM, et al. Suppression of T-cell functions by human granulocyte arginase. Blood. (2006) 108:1627–34. doi: 10.1182/blood-2006-11-010389
77. Greenberg ME, Sun M, Zhang R, Febbraio M, Silverstein R, Hazen SL. Oxidized phosphatidylserine-CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J Exp Med. (2006) 203:2613–25. doi: 10.1084/jem.20060370
78. Huang SC, Everts B, Ivanova Y, O'Sullivan D, Nascimento M, Smith AM, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. (2014) 15:846–55. doi: 10.1038/ni.2956
79. Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. (2009) 27:451–83. doi: 10.1146/annurev.immunol.021908.132532
80. Weintz G, Olsen JV, Fruhauf K, Niedzielska M, Amit I, Jantsch J, et al. The phosphoproteome of toll-like receptor-activated macrophages. Mol Syst Biol. (2010) 6:371. doi: 10.1038/msb.2010.29
81. Agrawal V, Sawhney N, Hickey E, McCarthy JV. Loss of presenilin 2 function is associated with defective LPS-mediated innate immune responsiveness. Mol Neurobiol. (2016) 53:3428–38. doi: 10.1007/s12035-015-9285-0
82. Alexopoulou AN, Multhaupt HA, Couchman JR. Syndecans in wound healing, inflammation and vascular biology. Int J Biochem Cell Biol. (2007) 39:505–28. doi: 10.1016/j.biocel.2006.10.014
83. Akilesh S, Suleiman H, Yu H, Stander MC, Lavin P, Gbadegesin R, et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest. (2011) 121:4127–37. doi: 10.1172/JCI46458
84. Marques-Fernandez F, Planells-Ferrer L, Gozzelino R, Galenkamp KM, Reix S, Llecha-Cano N, et al. TNFalpha induces survival through the FLIP-L-dependent activation of the MAPK/ERK pathway. Cell Death Dis. (2013) 4:e493. doi: 10.1038/cddis.2013.25
85. Matsumura H, Shimizu Y, Ohsawa Y, Kawahara A, Uchiyama Y, Nagata S. Necrotic death pathway in Fas receptor signaling. J Cell Biol. (2000) 151:1247–56. doi: 10.1083/jcb.151.6.1247
86. Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. (2003) 19:59–70. doi: 10.1016/S1074-7613(03)00171-7
87. Muller E, Christopoulos PF, Halder S, Lunde A, Beraki K, Speth M, et al. Toll-like receptor ligands and interferon-gamma synergize for induction of antitumor M1 macrophages. Front Immunol. (2017) 8:1383. doi: 10.3389/fimmu.2017.01383
88. Muller E, Speth M, Christopoulos PF, Lunde A, Avdagic A, Oynebraten I, et al. Both type I, and type II interferons can activate antitumor M1 macrophages when combined with TLR stimulation. Front Immunol. (2018) 9:2520. doi: 10.3389/fimmu.2018.02520
89. Marks-Konczalik J, Dubois S, Losi JM, Sabzevari H, Yamada N, Feigenbaum L, et al. IL-2-induced activation-induced cell death is inhibited in IL-15 transgenic mice. Proc Natl Acad Sci USA. (2000) 97:11445–50. doi: 10.1073/pnas.200363097
90. Kim S, Kim SY, Pribis JP, Lotze M, Mollen KP, Shapiro R, et al. Signaling of high mobility group box 1 (HMGB1) through toll-like receptor 4 in macrophages requires CD14. Mol Med. (2013) 19:88–98. doi: 10.2119/molmed.2012.00306
91. Wimmer N, Huber B, Barabas N, Rohrl J, Pfeffer K, Hehlgans T. Lymphotoxin beta receptor activation on macrophages induces cross-tolerance to TLR4 and TLR9 ligands. J Immunol. (2012) 188:3426–33. doi: 10.4049/jimmunol.1103324
92. Galimi F, Cottone E, Vigna E, Arena N, Boccaccio C, Giordano S, et al. Hepatocyte growth factor is a regulator of monocyte-macrophage function. J Immunol. (2001) 166:1241–7. doi: 10.4049/jimmunol.166.2.1241
93. Ide T, Shimano H, Yoshikawa T, Yahagi N, Amemiya-Kudo M, Matsuzaka T, et al. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. II. LXRs suppress lipid degradation gene promoters through inhibition of PPAR signaling. Mol Endocrinol. (2003) 17:1255–67. doi: 10.1210/me.2002-0191
94. Tan HY, Wang N, Li S, Hong M, Wang X, Feng Y. The reactive oxygen species in macrophage polarization: reflecting its dual role in progression and treatment of human diseases. Oxid Med Cell Longev. (2016) 2016:2795090. doi: 10.1155/2016/2795090
95. Izquierdo E, Cuevas VD, Fernandez-Arroyo S, Riera-Borrull M, Orta-Zavalza E, Joven J, et al. Reshaping of human macrophage polarization through modulation of glucose catabolic pathways. J Immunol. (2015) 195:2442–51. doi: 10.4049/jimmunol.1403045
96. Zhang L, Dong Y, Dong Y, Cheng J, Du J. Role of integrin-beta3 protein in macrophage polarization and regeneration of injured muscle. J Biol Chem. (2012) 287:6177–86. doi: 10.1074/jbc.M111.292649
97. Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. (2007) 7:678–89. doi: 10.1038/nri2156
98. Yoshimura T, Matsushima K, Tanaka S, Robinson EA, Appella E, Oppenheim JJ, et al. Purification of a human monocyte-derived neutrophil chemotactic factor that has peptide sequence similarity to other host defense cytokines. Proc Natl Acad Sci USA. (1987) 84:9233–7. doi: 10.1073/pnas.84.24.9233
99. Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature. (2016) 539:437–442. doi: 10.1038/nature19834
100. Najafi M, Hashemi Goradel N, Farhood B, Salehi E, Nashtaei MS, Khanlarkhani N, et al. Macrophage polarity in cancer: a review. J Cell Biochem. (2019) 120:2756–65. doi: 10.1002/jcb.27646
101. Leitinger N, Schulman IG. Phenotypic polarization of macrophages in atherosclerosis. Arterioscler Thromb Vasc Biol. (2013) 33:1120–6. doi: 10.1161/ATVBAHA.112.300173
Keywords: macrophage, innate immunity, M1, M2, cancer
Citation: Orecchioni M, Ghosheh Y, Pramod AB and Ley K (2019) Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS–) vs. Alternatively Activated Macrophages. Front. Immunol. 10:1084. doi: 10.3389/fimmu.2019.01084
Received: 31 January 2019; Accepted: 29 April 2019;
Published: 24 May 2019.
Edited by:
Liwu Li, Virginia Tech, United StatesReviewed by:
Inger Øynebråten, Oslo University Hospital, NorwayCopyright © 2019 Orecchioni, Ghosheh, Pramod and Ley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Klaus Ley, a2xhdXNAbGppLm9yZw==
†Present address: Klaus Ley, Division of Inflammation Biology, La Jolla Institute for Immunology, La Jolla, CA, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.