
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 26 April 2019
Sec. Molecular Innate Immunity
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.00922
This article is part of the Research Topic Transcriptional and Chromatin Regulation in Adaptive and Innate Immune Cells View all 11 articles
Over 100 types of cellular RNA modifications have been identified in both coding and a variety of non-coding RNAs. N6-methyladenosine (m6A) is the most prevalent and abundant post-transcriptional RNA modification on eukaryote mRNA, and its biological functions are mediated by special binding proteins (i.e., methyltransferases, demethylases, and effectors) that recognize this modification. The presence of m6A on transcripts contributes to diverse fundamental cellular functions, such as pre-mRNA splicing, nuclear transport, stability, translation, and microRNA biogenesis, implying an association with numerous human diseases. This review principally summarizes recent progress in the study of m6A methylation mechanisms and relevant roles they play in immunoregulation.
In 1970, m6A was first recognized as an abundant nucleotide modification in eukaryotic messenger RNA (1), by far the most prevalently known of over 100 kinds of RNA modifications identified in various classes of RNAs, including mRNA, rRNA, tRNA, snRNA, microRNA (miRNA), and long non-coding RNA (lncRNA) (2). m6A is present in 0.1–0.4% of all adenosines in global cellular RNAs and accounts for ~50% of all methylated ribonucleotides (3). However, little was known about its potential functional significance and extent transcript identities until very recently. m6A occurs primarily in two consensus sequence motifs, G m6A C (~70%) and A m6A C (~30%) (4, 5). Long internal exons, locations upstream of stop codons, and the 3′-UTR of mRNA are preferred modification sites for m6A, implying roles involving translational control, influencing affinities of RNA binding proteins or unique m6A-derived transcriptome topology (6–9).
The discovery of proteins involved in m6A regulation has been among the most significant achievements in this area of study, elucidating their roles as “writers” (m6A methyltransferases), “erasers” (m6A demethyltransferases), and “readers” (effectors recognizing m6A) (10). Methyltransferase like-3 (METTL3, also known as MT-A70) was among the first of all identified core writer components, responsible for installing m6A on RNA (11), Other core enzyme components such as METTL14 (12, 13), in addition to accessory components including Wilms′ Tumor 1-associating Protein (WTAP) (12, 14), KIAA1429 (15), RNA binding motif protein 15 (RBM15), RBM15 paralog (RBM15B) (16), and zinc finger protein 217 (ZFP217) (17) have also been studied. The METTL3 complex acts at the consensus RRACH motif (R = A or G, H = A, C, or U) (18). METTL3 or METTL14 depletion reduces the ratio of m6A/A, while knockdown of WTAP decreases amounts of METTL3 complex bound to RNA, implying that WTAP may be responsible for the recruitment of RNAs (12, 14). m6A demethylase fat mass and obesity-associated (FTO) protein was the first recognized “eraser” enzyme that reverses RNA modification and controls cellular homeostasis (19). ALKB homolog 5 (ALKBH5) was also recognized as a demethylase involved in alkylated DNA repair (20). Three “reader” proteins that can directly interact with m6A sites via their YTH domains have been discovered to date. YTH domain-containing family (YTHDF) proteins 1 and 3 promote translation of m6A-modified mRNA via interaction with translation initiation factors (21, 22), whereas YTHDF2 promotes RNA degradation via recruitment of m6A modified mRNA to nuclear processing bodies (P bodies) (23).
m6A as an mRNA modification that is abundant in some viruses and nearly all eukaryotes (2). A variety of cytopathologic processes involving nuclear RNA export, splicing, mRNA stability, circRNA translation, miRNA biogenesis, and lncRNA metabolism have recently been linked to aberrant levels of m6A (Figure 1) (24–26). In addition, m6A modification has been associated with numerous physiological and pathological phenomena, including obesity, immunoregulation, yeast meiosis, plant development, and carcinogenesis (2, 27).
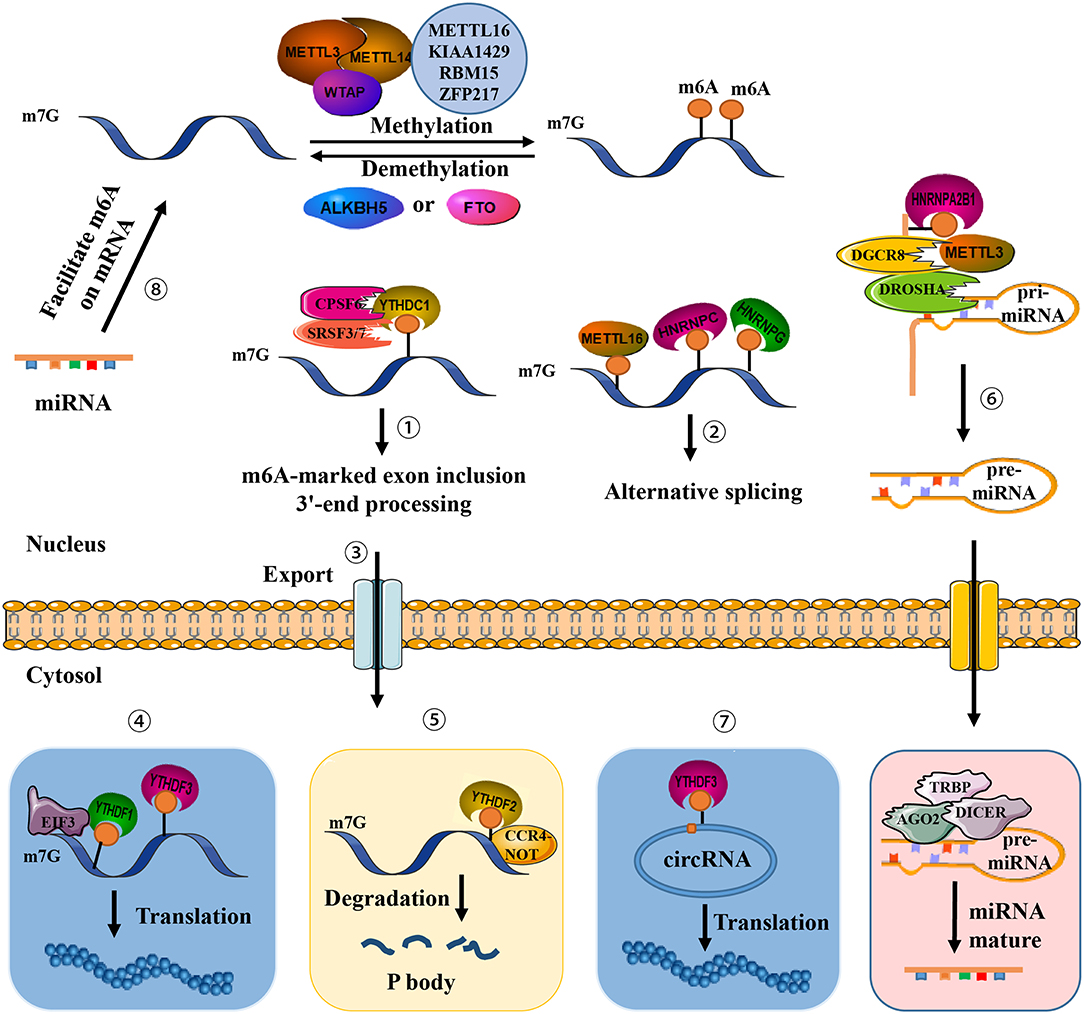
Figure 1. Presence of m6A on transcripts contributes to diverse fundamental cellular functions. ①-⑤ m6A regulates exon inclusion and 3′-end processing, alternative splicing, nuclear export, translation, and degradation. ⑥ m6A regulates miRNA processing. ⑦ m6A regulates circRNA translation. ⑧ miRNA regulates m6A formation.
This review summarizes the most recent progress in research concerning m6A and analyzes newly identified roles this modification plays in the regulation of gene expression and immune responses.
A methyltransferase complex within the nuclear speckle, mainly consisting of METTL3 and METTL14, installs m6A modification on distinct target RNAs via the methyl groups of S-adenosylmethionine (SAM) transferase (11, 12). Two subunits constitute an m6A methyltransferase complex: MT-A (200-kDa) and MT-B (800-kDa). METTLE3 (or MT-A70), a 70-kDa protein, was first identified in 1997 and possesses a SAM-binding domain and a DPPW motif (Asp-Pro-Pro-Trp). It serves as a catalytic subunit and constituent of a 200-kDa methyltransferase complex isolated from the nuclear extract of HeLa cells (11). Knockdown of METTL3 leads to decreased m6A levels and concomitant apoptosis of human HeLa and HepG2 cells (12). The other core writer component, METTL14, harbors two conserved functional domains, a SAM-binding domain and an EPPL motif (Glu-Pro-Pro-Leu) involved in catalyzing the methylation reaction, an additional N-terminal coiled-coil domain for mediating protein–protein interaction, and a G-rich sequence at the C-terminal end (10). METTL3-METTL14, in a ratio of 1:1, form a stable heterodimer (1 MDa) localized at nuclear speckles (28). In addition, METTL14 supports METTL3 in recognizing special RNA substrates. The heterodimer preferentially methylates RNA substrates exhibiting a consensus GGACU domain and contains a moderate preference for substrates with less structure (29). WTAP, a splicing factor, acts as the third crucial component of the writer. Although it does not possess any recognizable domains or motifs, WTAP binds to the METTL3-METTL14 heterodimer and abundantly regulates m6A deposition inside cells (12). WTAP may mediate the position of the heterodimer on nuclear speckles and recruit target RNA for m6A modification (14), thus indirectly enhancing the catalytic capacity of the writer. WTAP may also recruit unknown factors to bind to the methyltransferase complex and modulate methylation.
FTO is the first discovered demethylase that removes methyl groups from m6A (19), indicating that m6A is a dynamically reversible RNA modification. FTO modulates alternative splicing of RUNX1T1, which is involved in adipogenesis (30), as well as the 3′-end mRNA processing in 293T cells (31). ALKBH5 is the second identified demethylase and exhibits distinct physiological functions (20). An ALKB domain is commonly situated in the middle regions of FTO and ALKBH5, consisting of two active motifs termed as HXDXnH and RXXXXXR (X = any amino acid), which binds to Fe(II), as well as α-ketogluterate (α-KG) and substrate, respectively. Compared to ALKBH5, a distinctive fold mediates protein interaction at the C-terminal end of FTO. The N-terminus of ALKBH5 is characterized by an additional A-rich motif responsible for localizing ALKBH5 at nuclear speckles (10). FTO and ALKBH5 have substantial tissue-specificity and diverse intracellular localization. FTO has been reported to be highly abundant in adipose and cerebral tissue, while ALKBH5 has been reported to be primarily expressed in the testes (20). Thus, demethylation in some tissues may be performed solely by either FTO or ALKBH5.
While writer proteins install m6A at a specific domain on target RNA, altering its secondary or tertiary structure (32), another class of proteins, termed readers, recognize, and preferentially bind the RNA to confer its fate and regulate downstream functions. An RNA-pull down assay initially revealed that YTH domain-containing family proteins YTHDF1-3 rich in the P/Q/N motif were discovered in mammalian cells to be m6A readers that recognize the consensus sequence G[G > A]m6ACU (7, 23). Among this protein family, YTHDF2 has the strongest affinity to localize P bodies via its P/Q/N motif where the concentration of mRNA turnover factors in facilitating RNA degradation (23). Notably, YTHDF2 recruits the deadenylase complex CCR4-NOT via the YTHDF2 N-terminus and mediates RNA degradation in mammalian cells (33). YTHDF1 was found to interact with translation initiation factors to evoke m6A-containing mRNA translation (21). YTHDC1, a YTH domain-containing protein, was subsequently validated as a nuclear m6A reader and showed almost completely overlapping sites with m6A in nuclear RNAs. YTHDC1 recruits the serine and arginine-rich splicing factor 3 (SRSF3), restricts exon-skipping factor SRSF10 binding, and promotes exon inclusion (34). YTHDC1 also plays a critical role in pre-mRNA processing in the oocyte nucleus via interaction with the pre-mRNA 3′-end processing factors CPSF6, SRSF3, and SRSF7 (35). A nuclear m6A reader has been identified recently within the pre-mRNA consensus motif RRACH that destabilizes the stem structure, enabling the U-tract motif to become exposed as a single-strand and become more accessible for heterogeneous nuclear ribonucleoprotein (hnRNP) HNRNPC binding, altering alternative splicing of target RNA (36). HNRNPG is another critical protein containing a low-complexity domain at its C-terminus. Arg-Gly-Gly repeat sequences bind directly m6A sites and alter the expression and alternative splicing pattern of target mRNA (37). Additionally, the hnRNP family protein HNRNPA2B1 binds to the m6A RGAC motif in a subset of primary miRNA transcripts, recruits a microprocessor complex to facilitate miRNA processing, and elicits alternative splicing effects similar to those of METTL (37, 38).
A number of m6A readers have been recently identified in addition to the aforementioned proteins. FMR1, a protein that contains a RGG domain, three KH domains, and two Agenet domains at its N-terminus, preferentially recognizes the sequence GGm6ACU via RGG domain binding and represses translation by stalling ribosomal translocation (39, 40). Further data revealed that a class of proteins termed m6A-repelled proteins preferentially bind only to unmodified mRNA. The stress granule proteins G3BP1 and G3BP2 are reported to be the most forceful of the repelled proteins. G3BP1 consistently interacts with the GGACU but not the GGm6ACU motif to stabilize target RNA (39, 41). Interestingly, METTL16 serves as a both m6A writer and reader of U6 snRNA, and is concerned with mRNA splicing (42). METTL16-dependent sites are mainly located in introns or exon-intron boundaries, unlike common m6A sites.
The profile of m6A sites throughout the transcriptome remained unclear until two independent sequencing methods, m6A Seq and MeRIP-Seq (m6A-specific methylated RNA immunoprecipitation (IP) with next-generation sequencing), were established in 2012 (7, 9). In brief, the mRNA is first randomly fragmented into approximately 100 nt prior to IP using m6A-specific antibody. Then, RNA-seq is applied to the RNA pool, in which m6A-tagged RNA fragments are enriched. These two methods revealed m6A to be a pervasive and dynamically reversible modification, particularly enriched in 3′-UTR regions and near mRNA stop codons. It was also reported that m6A sites in some RNAs have very high levels of conservation between human and mouse transcriptomes. Although the aforementioned research methods are easily and effectively applied toward the field of epitranscriptomics, a resolution of approximately 200 nt is impractical to precisely identify m6A positions.
PA-m6A-seq (photo-crosslinking-assisted m6A sequencing) has recently been developed as one of two UV-induced RNA-antibody crosslinking strategies (43). It significantly improves resolution on the basis of m6A-seq/MeRIP-seq. Addition of 4-thiouridine (4SU) into medium results in it embedding into RNA. This is followed by anti-m6A antibody IP with subsequent m6A-containing RNA crosslinking using 365-nm UV light. Afterwards, RNase T1 digests the crosslinked RNA to about 30 nt, allowing for efficient sequencing. Within approximately 23 nt, single consensus methylation sequences can be determined by PA-m6A-seq at single-base resolution. However, m6A sites cannot be detected if the distance between them and those of 4SU incorporation is too great. This UV crosslinking strategy effectively provides insight into m6A-containing RNA and RNA-binding proteins.
Additional UV crosslinking strategies are known as m6A-CLIP/IP and miCLIP (m6A individual-nucleotide-resolution crosslinking and immunoprecipitation) (8, 44). Under 254-nm UV light, crosslinking can be brought about between RNA fragments and anti-m6A antibodies. The crosslinked fragments are subsequently retrieved by applying proteinase K and reverse transcription, thereby leading to highly specific mapping of mutation or truncation profiles to precise m6A sites at single-nucleotide resolution throughout the transcriptome.
To determine the m6A status of a random mRNA or lncRNA from a total RNA pool rather than from purified distinct RNAs and quantify m6A stoichiometry at specific locations, SCARLET (site-specific cleavage and radioactive-labeling followed by ligation-assisted extraction and thin-layer chromatography) was developed in 2013 (45). To achieve site-specific cleavage, RNase H is added to the total RNA sample followed with radiolabeling using 32P. Labeled RNA fragments are then splint-ligated to DNA oligonucleotides by DNA ligase. All RNAs are subsequently digested completely with RNases T1/A, but 32P-labeled sites remain protected from digestion. Finally, after gel purification and nuclease P1 digestion, samples are analyzed using thin-layer chromatography (TLC). Although SCARLET can precisely determine m6A modification sites at single-nucleotide resolution, it is incapable of use in high throughput screening and is time-consuming as a whole (2).
A recently established m6A-LAIC-seq (m6A-level and isoform-characterization sequencing) method can also quantify m6A presence, even on a transcriptome-wide level (46). First, full-length RNA IP is performed employing excess anti m6A antibody. The sample is next treated with External RNA Controls Consortium (ERCC), and the internal standards are supernatant (m6A-negative fraction) and eluate (m6A-positive fraction). In the two pools, an ERCC-normalized RNA enrichment ratio quantifies m6A modification of each gene, followed by library construction and sequencing. m6A-LAIC-seq was found to indicate that under 50% m6A modification levels exist in a majority of genes, and that 3′-ends of RNA molecules containing m6A would become shorter due to proximal alternative polyadenylation sites.
Identifying precise m6A sites in RNA transcripts is a critical step toward comprehending the biological functions of this modification. Although existing methods have been adopted widely in multiple areas and resulted in some achievements, challenges remain at both single-base and quantitative sequencing levels (47). For example, how to relatively distinguish the m6A levels of different sites in a common transcript, and the establish of new methods getting rid of m6A-special antibody will make perfect sense.
Identification of m6A methyltransferases, demethylases, and effectors have revealed that m6A modification is critical throughout the whole RNA life cycle, including pri-mRNA splicing, mRNA nuclear transport, molecular stability, translation, and subcellular localization (48), additionally involved in miRNA biogenesis, lncRNA processing, and circRNA functions. m6A modification is especially abundant in circRNA, and only a single m6A residue is sufficient to drive circRNA translation via recruitment of YTHDF3 and translation initiation factors eIF4G2 and eIF3A. Hundreds of endogenous circRNAs thus possess translation potential (Table 1). Of note, m6A modification is reversely regulated by miRNA. Via sequence pairing with mRNA containing miRNA targeting sites, miRNA modulates METTL3 binding to target RNA, resulting in an increase of m6A modifications. Mouse embryonic fibroblasts are thus reprogrammed into pluripotent stem cells (18).
m6A modification is necessary in the biogenesis and functions of RNA. Modification aberrancies have been associated with various pathophysiologies. Recently, m6A modification has been recognized as crucial regulator in T cell homeostasis and the immune response to bacterial or viral infection. Selectively altered m6A levels along with other types of immunotherapies may be efficient management strategies in a variety of immunological diseases.
The suppressor of cytokine signaling (SOCS) protein family encodes inhibitory proteins involved in JAK-STAT signaling, including SOCS1, SOCS3, and CISH, playing a vital role in T cell proliferation and differentiation (51, 52). In naive T cells, which are induced by “gatekeeper” IL-7 stimulation as well-known immediate-early genes, SOCS genes control IL-7 signal and play critical roles in adaptive immunity (51, 52). All three SOCS genes were found to undergo m6A modification via the RNA-IP assay. The modification induces mRNA degradation of SOCS genes that initiates naïve T cells re-programming for proliferation and differentiation. IL-7/JAK signaling is activated in vitro and in vivo via relieving the inhibition on IL-7-STAT5 signaling (Figure 2A) by an evolutionarily-conserved m6A-dependent mechanism (53). This modification is likely a crucial factor in the regulation of immune homeostasis and the mitigation of various autoimmune diseases.
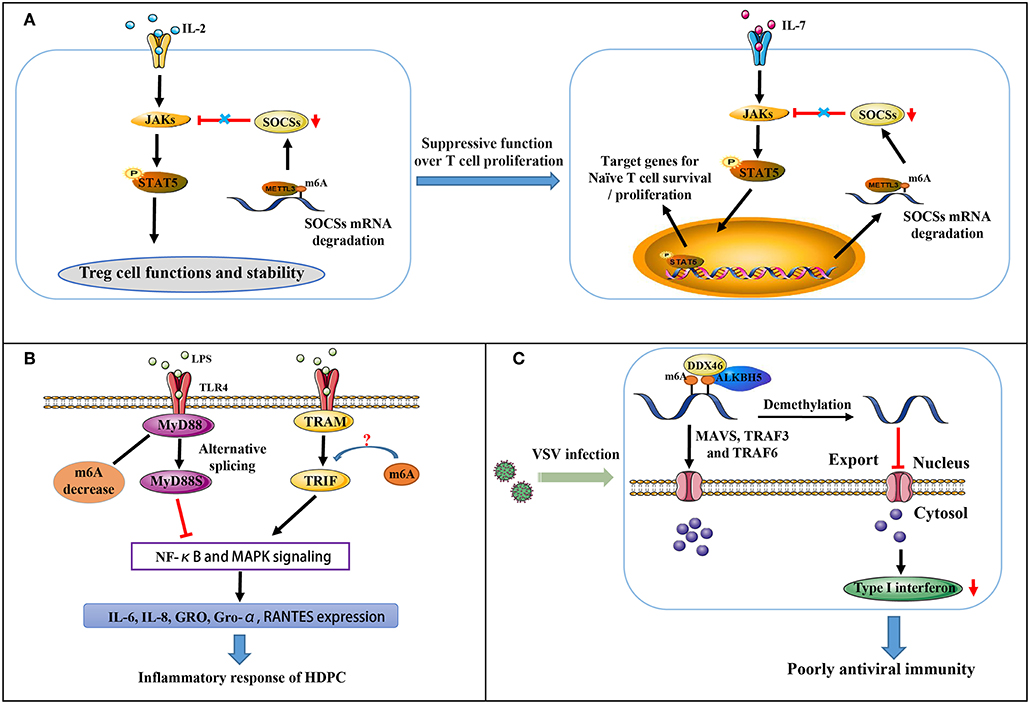
Figure 2. Partial immune mechanisms regulated by m6A modification. (A) m6A methylation regulates the suppressive function of Tregs on naïve T cells. (B) m6A mediates MyD88 alternative splicing that is responsible for LPS-induced inflammatory reactivity in HDPCs. (C) m6A represses type I interferon production in an innate antiviral state.
Regulatory T cells (Tregs) are a crucial specialized T cell lineage, and are involved in reducing inflammation and immunosuppression (54). Chronic intestinal inflammation in METTL3 knockout mice has occurred when the mice reach at least 3 months of age. Co-culture assay of naïve CD4+ T cells and Tregs with m6A KO revealed naïve T cells to exert more rapid proliferative influences due to a complete lack of suppressive function for Tregs (Figure 2A) (55). In CD4+ T cells, m6A modification is indeed enriched at the GG/AACA/U domain at 3′-UTR and at 5′-UTR of SOCS genes. Decreased m6A modification enhances the mRNA stability of SOCS genes, thereby blocking transduction of cytokine signaling in the IL2-STAT5 pathway (55). As this pathway is critically essential for the suppressive function and stability of Tregs (54), m6A levels are considerably responsible in controlling naïve T cells homeostasis.
Dental pulp inflammation, which can progress to pulp necrosis and periapical diseases, is characterized by a partial accumulation of inflammatory mediators and is a typical inflammatory disease (56, 57). In pulpal and periapical diseases, it is recognized that bacterial infection is a major pathogenic factor (58). Recent findings indicate that in LPS-treated human dental pulp cells (HDPCs), METTL3 expression and m6A modification levels are up-regulated instead of METTL14, FTO, and ALKBH5. Moreover, METTL3 knockdown decreases the expression of LPS-induced inflammatory cytokines, including IL-6, IL-8, GRO, Gro-α and RANTES. At the same time, NF-κB and MAPK signaling pathway activation is suppressed (58). MyD88 exists in two forms (MyD88L and MyD88S). MyD88Land TRIF pathway activate the innate immune response by transducing TLR signals, whereas MyD88S inhibits the response (59). Further research revealed that m6A inhibition significantly increases MyD88S mRNA levels, suggesting that m6A mediates alternative splicing of MyD88 and mediates the LPS-induced inflammatory reaction in HDPCs (Figure 2B) (58). Whether or not m6A also regulates TRIF signaling remains unclear.
Influenza virus and Rous sarcoma virus were previously reported to produce viral transcripts with m6A modifications. At an antiviral innate state, RNAs containing m6A modifications is unable to stimulate RIG-I-mediated antiviral signaling and induce interferon expression (60). Further research has suggested that m6A modification is involved in the export and translation of signaling molecules, including MAVS, TRAF3, and TRAF6, thus regulating interferon production in the antiviral innate immune response (Figure 2C) (61). The DEAD-box (DDX) helicase family contains 12 conservative domains, many of which have been identified vital in the recognition of viral nucleic acids and regulation of downstream pathways (62–65). DDX46 was recently found to negatively regulate innate antiviral transcripts via recruitment of ALKBH5. This results in reduced m6A levels on MAVS, TRAF3, and TRAF6 RNA and prevents their transport from nucleus into cytoplasm, thus reducing their translation. Moreover, the target mRNA CCGGUU motif is responsible for the effects DDX46 exerts on the antiviral innate immune response by decreasing the production of type I interferons (61). DDX3 also interacts with ALKBH5, the only protein among the identified methyltransferases and demethylases as partnering with DDX3 via the ATP-binding domain of DDX3 and the DSBH domain of ALKBH5. DDX3 is involved in diverse biological processes via the interactions between its different domains and many distinct proteins (66). Interestingly, recent article revealed a seemingly opposing mechanism of m6A in type I interferon response to the herpesvirus human cytomegalovirus (HCMV) infection. m6A level is dramatically upregulated in primary human foreskin fibroblasts infected by HCMV, and required for viral propagation (67). Following infection in METTL3-depleted cells, decrease of m6A modification results in enhanced mRNA stability of IFNB and sustained IFN-β production, the main type I interferon in human non-immune cells, thus triggering a stronger antiviral response to block HCMV growth. Three putative adenosines proximal to stop codon are mechanistically direct targets of m6A, responsible for IFNB mRNA stability. It is probable that for different viruses, the contribution of m6A machinery for immune response may vary even adverse.
Human CD4+ T cells infected by HIV-1 can trigger a massive m6A increase in both T cells and HIV-1 mRNA (68). In the CDS and UTR sequences of HIV-1 mRNA, additional splice junctions and splicing regulatory domains have been identified to possess 14 methylation peaks. Gene ontology analysis has revealed that 56 host genes with special m6A modification are active in viral infection. Silencing of METTL3/METTL14 or AKBH5 either decreases or increases HIV-1 replication, respectively. Mechanically, the formation of the HIV-1 Rev-RRE (Rev response element) complex is enhanced by the A7883 site methylation in the stem loop II region of RRE RNA. This complex thereby influences HIV-1 replication and nuclear export (68). Furthermore, one third of the 56 genes which encode diverse functional proteins, including MOGS, TRAF2, and HSPA1A, have been linked with HIV-1 replication. It thus seems that altered host genes modified with m6A are involved in the antiviral T cell immune response due to regulation of RNA expression and biological metabolism.
Kaposi′s sarcoma-associated herpesvirus (KSHV) has been reported as the leading cause of cancer in AIDS patients, resulting in both primary effusion lymphoma and the lymphoproliferative disorders multicentric Castleman′s disease (69). In KSHV-infected renal carcinoma cell line iSLK.219 treated with doxycycline, the level of total m6A was found to be markedly increased, and about one third of KSHV transcripts with m6A modification were found to contribute to KSHV gene expression (70). METTL3 and YTHDF2 are involved in triggering production of virion in cells infected by KSHV. ORF50 protein, a major viral transcriptional trans-activator, upon depletion of METTL3 or YTHDF2 is initially significantly reduced and unable to expedite the expression of KSHV lytic genes. However, ORF50 expression is independent of KSHV infection. m6A initially promotes ORF50 mRNA abundance, but knockdown of YTHDF2 or METTL3 leads to a subsequent negative feedback on the ORF50 promoter. Furthermore, in a KSHV-infected iSLK.BAC16 cell line, ORF50 protein expression is inverted following METTL3 or YTHDF2 depletion, suggesting that m6A variably functions in a cell-specific way. Additionally, m6A regulates newly transcribed viral RNA splicing, stability, and protein translation to control viral lytic gene expression and KSHV replication. Blockade of an event regulated by m6A modification decreases viral protein expression and halts virion production (71). Moreover, m6A also plays important roles in latent and lytic KSHV replication as well as KSHV-induced oncogenesis (72). m6A may be a novel, effective target in the management of KSHV infection.
In 1979, m6A residues were identified on the polyomavirus simian virus 40 (SV40) (73). Overexpression of YTHDF2 substantially enhances SV40 replication in the SV40-permissive cell line BSC40 (74). PA-m6A-seq has revealed that SV40 possesses an early region where two m6A clusters are located, and a late region that possesses 11. Instead of detectably affecting SV40 late transcript alternative splicing, m6A present on VP1 ORF principally increases VP1 expression by expediting translation. By decreasing the formation of SAM without affecting mRNA capping, 3-deazaadenosine (DAA), as an inhibitor of methylation, reduces overall m6A modification (75). Subsequent depletion of m6A via DAA treatment inhibits SV40 replication. Drugs that alter m6A modification levels as needed might be in a position to repress replication of different kinds of pathogenic viruses, in particular of those causing acute infections.
Tumor neoantigens are important for generating spontaneous antitumor immunity and for valuating clinical responses to immunotherapies (76, 77). Recent article indicates that m6A-modificated mRNAs encoding lysosomal cathepsins are recognized by YTHDF1 in dendritic cells (DCs), subsequently the binding of YTHDF1 facilitates translation of cathepsins, suppressing the cross-priming ability of DCs. Loss of Ythdf1 inhibits tumors growth and host survival in mice model, owing to elevated infiltration of neoantigen-specific CD8+ T cells in tumors. Consistently, DC-specific Ythdf1 depletion enhances the cross-presentation of tumor antigens and the cross-priming of CD8+ T cells in vivo. Furthermore, given that Ythdf1 depletion promotes IFNγ production, followed by PD-L1 increase in CD8+ T cells (78), combining PD-L1 checkpoint inhibitor with Ythdf1 depletion shows stronger therapeutic efficacy. In combination with burgeoning checkpoint blockade strategy, YTHDF1 could be a potential new therapeutic target for immunotherapy.
Being exquisitely regulated by “writers,” “erasers,” and “readers,” additional repelled proteins or miRNAs, m6A modification relates to nearly any step of mRNA metabolism, as well as ncRNA processing and circRNA translation. There is compelling evidence suggesting that m6A modification is especially critical in a variety of pathologic and physiologic immune responses including T cell homeostasis and differentiation, inflammation, and type I interferon production. Further results have indicated that aberrancies of interferon and Th17 frequencies in systemic lupus erythematosus (SLE) patients may be caused by relevant genes changes for the altered m6A levels (79, 80). Whether functional dysregulations observed in SLE are associated with altered m6A modification warrants future exploration (81). Furthermore, m6A controls stem cell self-renewal, differentiation and pluripotency, additional critical functions in metabolism, as well as metastasis in many cancers (82), including acute myeloid leukemia, glioblastoma, breast cancer and lung carcinoma. Altered m6A modification might thus be an effective therapeutic target in preventing and treating human diseases. m6A exerts a variety of functions that result in alterations of particular functional proteins with m6A on mRNAs.
Though the field of m6A modification has become increasingly attractive, serious challenges in research remain. More advanced technology will be needed for quantification of m6A modification on a transcriptome-wide level and identification of precise m6A sites. How one of the members of writers, readers, and erasers performs its separate functions and interacts with one another remains to be elucidated. It is likely that either FTO or ALKBH5 performs the sole demethylation in different tissues as the presence of one has been found to indicate absence of the other. More biochemical participants in the machinery involved in m6A modification remain to be uncovered, and whether the diverse functions of m6A work in concert or are antagonistic in cellular biological processes remains to be researched in detail. We anticipate that subsequent focus on researching m6A modification in the setting of physiological and pathological processes will enrich our knowledge concerning a variety of conditions, contribute to the advancement of the biological sciences and provide us with novel therapeutic strategies.
CZ the first author, contributed to collection of references and manuscript preparation. JF and YZ contributed to the modification of the manuscript.
This work was supported by grants from the National Key R&D Program of China (2016YFC1305102), National Natural Science Foundation of China (81671561), 1000 Young Talents Plan Program of China, Initial Funding for New PI, Fudan Children's Hospital and Fudan University, the International Joint Laboratory Program of National Children's Medical Center (EK1125180109) and Shanghai Municipal Planning Commission of Science and Research Fund (201740065 to YZ). Shanghai Pujiang Program 16PJ1401600 (to JF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Nat Acad Sci USA. (1974) 71:3971–75. doi: 10.1073/pnas.71.10.3971
2. Wei W, Ji X, Guo X, Ji S. Regulatory role of N(6) -methyladenosine (m(6) A) Methylation in RNA processing and human diseases. J Cel Biochem. (2017) 118:2534–43. doi: 10.1002/jcb.25967
3. Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. (1975) 4:379–86. doi: 10.1016/0092-8674(75)90158-0
4. Wei CM, Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. (1977) 16:1672–6. doi: 10.1021/bi00627a023
5. Wei CM, Gershowitz A, Moss B. 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry. (1976) 15:397–401. doi: 10.1021/bi00647a024
6. Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. (2014) 15:707–19. doi: 10.1016/j.stem.2014.09.019
7. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. (2012) 485:201–06. doi: 10.1038/nature11112
8. Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Gene Dev. (2015) 29:2037–53. doi: 10.1101/gad.269415.115
9. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. (2012). 149:1635–46. doi: 10.1016/j.cell.2012.05.003
10. Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m(6)A and U-tail. Cell. (2014) 158:980–7. doi: 10.1016/j.cell.2014.08.005
11. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. (1997). 3:1233–47.
12. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. (2014) 10:93–5. doi: 10.1038/nchembio.1432
13. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. (2014) 16:191–8. doi: 10.1038/ncb2902
14. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. (2014) 24:177–89. doi: 10.1038/cr.2014.3
15. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. (2014) 8:284–96. doi: 10.1016/j.celrep.2014.05.048
16. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
17. Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, et al. Coordination of m(6)A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell. (2015) 17:689–704. doi: 10.1016/j.stem.2015.09.005
18. Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. (2015) 16:289–301. doi: 10.1016/j.stem.2015.01.016
19. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. (2011) 7:885–7. doi: 10.1038/nchembio.687
20. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015
21. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
22. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. (2017) 27:315–28. doi: 10.1038/cr.2017.15
23. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. (2014) 505:117–20. doi: 10.1038/nature12730
24. Roignant JY, Soller M. m(6)A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. (2017) 33:380–90. doi: 10.1016/j.tig.2017.04.003
25. Shafik A, Schumann U, Evers M, Sibbritt T, Preiss T. The emerging epitranscriptomics of long noncoding RNAs. Biochim et Biophys Acta. (2016) 1859:59–70. doi: 10.1016/j.bbagrm.2015.10.019
26. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. (2017) 27:626–41. doi: 10.1038/cr.2017.31
27. Wang S, Sun C, Li J, Zhang E, Ma Z, Xu W, et al. Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett. (2017) 408:112–20. doi: 10.1016/j.canlet.2017.08.030
28. Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. (2016) 63:306–17. doi: 10.1016/j.molcel.2016.05.041
29. Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. (2015) 29:1343–55. doi: 10.1101/gad.262766.115
30. Merkestein M, Laber S, McMurray F, Andrew D, Sachse G, Sanderson J, et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat Commun. (2015) 6:6792. doi: 10.1038/ncomms7792
31. Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. (2017) 45:11356–70. doi: 10.1093/nar/gkx778
32. Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. (2015) 137:2107–15. doi: 10.1021/ja513080v
33. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. (2016) 7:12626. doi: 10.1038/ncomms12626
34. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. (2016) 61:507–19. doi: 10.1016/j.molcel.2016.01.012
35. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. (2018) 14:e1007412. doi: 10.1371/journal.pgen.1007412
36. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. (2015) 518:560–4. doi: 10.1038/nature14234
37. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. (2017) 45:6051–63. doi: 10.1093/nar/gkx141
38. Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. (2015) 162:1299–308. doi: 10.1016/j.cell.2015.08.011
39. Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. (2017) 24:870–78. doi: 10.1038/nsmb.3462
40. Myrick LK, Hashimoto H, Cheng X, Warren ST. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Human Mol Genet. (2015) 24:1733–40. doi: 10.1093/hmg/ddu586
41. Arguello AE, DeLiberto AN, Kleiner RE. RNA Chemical proteomics reveals the N(6)-methyladenosine (m(6)A)-regulated protein-RNA interactome. J Am Chem Soc. (2017) 139:17249–52. doi: 10.1021/jacs.7b09213
42. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. (2017) 169:824–35 e814. doi: 10.1016/j.cell.2017.05.003
43. Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N, et al. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew Chem Int Ed Engl. (2015) 54:1587–90. doi: 10.1002/anie.201410647
44. Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. (2015) 12:767–72. doi: 10.1038/nmeth.3453
45. Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. (2013) 19:1848–56. doi: 10.1261/rna.041178.113
46. Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Methods. (2016) 13:692–8. doi: 10.1038/nmeth.3898
47. Li X, Xiong X, Yi C. (2016). Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods. 14:23–31. doi: 10.1038/nmeth.4110
48. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. (2017) 18, 31–42. doi: 10.1038/nrm.2016.132
49. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. (2015) 519:482–5. doi: 10.1038/nature14281
50. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. (2017) 20:2262–76. doi: 10.1016/j.celrep.2017.08.027
51. Palmer DC, Restifo NP. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. (2009) 30:592–602. doi: 10.1016/j.it.2009.09.009
52. Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. (2007) 7:454–65. doi: 10.1038/nri2093
53. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. (2017) 548:338–42. doi: 10.1038/nature23450
54. Li MO, Rudensky AY. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol. (2016) 16:220–33. doi: 10.1038/nri.2016.26
55. Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. (2018) 28:253–56. doi: 10.1038/cr.2018.7
56. Song F, Sun H, Wang Y, Yang H, Huang L, Fu D, et al. Pannexin3 inhibits TNF-alpha-induced inflammatory response by suppressing NF-kappaB signalling pathway in human dental pulp cells. J Cell Mol Med. (2017) 21:444–55. doi: 10.1111/jcmm.12988
57. Renard E, Gaudin A, Bienvenu G, Amiaud J, Farges JC, Cuturi MC, et al. Immune cells and molecular networks in experimentally induced pulpitis. J Dental Res. (2016) 95:196–205. doi: 10.1177/0022034515612086
58. Feng Z, Li Q, Meng R, Yi B, Xu Q. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J Cell Mol Med. (2018) 22:2558–68. doi: 10.1111/jcmm.13491
59. De Arras L, Alper S. Limiting of the innate immune response by SF3A-dependent control of MyD88 alternative mRNA splicing. PLoS Genet. (2013) 9:e1003855. doi: 10.1371/journal.pgen.1003855
60. Durbin AF, Wang C, Marcotrigiano J, Gehrke L. RNAs containing modified nucleotides fail To Trigger RIG-I conformational changes for innate immune signaling. mBio. (2016) 7:e00833-16. doi: 10.1128/mBio.00833-16
61. Zheng Q, Hou J, Zhou Y, Li Z, Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nature Immunol. (2017) 18:1094–103. doi: 10.1038/ni.3830
62. Loo YM, Gale M. Immune signaling by RIG-I-like receptors. Immunity. (2011) 34:680–92. doi: 10.1016/j.immuni.2011.05.003
63. Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, et al. (2012). The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 13:1155–161. doi: 10.1038/ni.2460
64. Zhang Z, Yuan B, Lu N, Facchinetti V, Liu YJ. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J Immunol. (2011) 187:4501–08. doi: 10.4049/jimmunol.1101307
65. Schroder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. (2008). 27:2147–57. doi: 10.1038/emboj.2008.143
66. Shah A, Rashid F, Awan HM, Hu S, Wang X, Chen L, et al. (2017). The DEAD-Box RNA helicase DDX3 interacts with m(6)A RNA demethylase ALKBH5. Stem Cells Int. 2017:8596135. doi: 10.1155/2017/8596135
67. Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, et al. Publisher correction: m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol. (2019) 20:243. doi: 10.1038/s41590-019-0314-4
68. Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol. (2016) 1:16011. doi: 10.1038/nmicrobiol.2016.11
69. Myoung J, Ganem D. Infection of lymphoblastoid cell lines by Kaposi's sarcoma-associated herpesvirus: critical role of cell-associated virus. J Virol. (2011) 85:9767–77. doi: 10.1128/JVI.05136-11
70. Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi's sarcoma-associated herpesvirus infection. PLoS Pathog. (2018) 14:e1006995. doi: 10.1371/journal.ppat.1006995
71. Ye F. RNA N(6)-adenosine methylation (m(6)A) steers epitranscriptomic control of herpesvirus replication. Inflam Cell Signal. (2017) 4:e1604. [Epub ahead of print]
72. Tan B, Gao SJ. RNA epitranscriptomics: regulation of infection of RNA and DNA viruses by N(6) -methyladenosine (m(6) A). Rev Med Virol. (2018) 2018:e1983. doi: 10.1002/rmv.1983
73. Canaani D, Kahana C, Lavi S, Groner Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. (1979) 6:2879–99. doi: 10.1093/nar/6.8.2879
74. Tsai K, Courtney DG, Cullen BR. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog. (2018) 14:e1006919. doi: 10.1371/journal.ppat.1006919
75. Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. (2013) 155:793–806. doi: 10.1016/j.cell.2013.10.026
76. Han D, Liu J, Chen C, Dong L, Liu L, Chang R, et al. Author Correction: Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. (2019) 568:E3. doi: 10.1038/s41586-019-1046-1
77. Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. Corrigendum: an immunogenic personal neoantigen vaccine for patients with melanoma. Nature. (2018) 555:402. doi: 10.1038/nature25145
78. Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. (2016) 167:1540–54 e1512. doi: 10.1016/j.cell.2016.11.022
79. Luo S, Wang Y, Zhao M, Lu Q. The important roles of type I interferon and interferon-inducible genes in systemic lupus erythematosus. Int Immunopharmacol. (2016) 40:542–9. doi: 10.1016/j.intimp.2016.10.012
80. Shah K, Lee WW, Lee SH, Kim SH, Kang SW, Craft J, et al. Dysregulated balance of Th17 and Th1 cells in systemic lupus erythematosus. Arthritis Res Therap. (2010) 12:R53. doi: 10.1186/ar2964
81. Li LJ, Fan YG, Leng RX, Pan HF, Ye DQ. Potential link between m(6)A modification and systemic lupus erythematosus. Mol Immunol. (2018) 93:55–63. doi: 10.1016/j.molimm.2017.11.009
Keywords: N6-methyladenosine, binding proteins, mechanisms, immunoregulation, cellular functions
Citation: Zhang C, Fu J and Zhou Y (2019) A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Front. Immunol. 10:922. doi: 10.3389/fimmu.2019.00922
Received: 01 October 2018; Accepted: 10 April 2019;
Published: 26 April 2019.
Edited by:
Keiko Ozato, National Institutes of Health (NIH), United StatesReviewed by:
Eswari Dodagatta-Marri, University of California, San Francisco, United StatesCopyright © 2019 Zhang, Fu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yufeng Zhou, eWZ6aG91MUBmdWRhbi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.