- 1Department of Immunology, IRCSS Bambino Gesù Children's Hospital, Rome, Italy
- 2Department of Pathology, Sacro Cuore Don Calabria Hospital, Negrar, Italy
Innate lymphoid cells (ILCs) belong to a family of immune cells. Recently, ILCs have been classified into five different groups that mirror the function of adaptive T cell subsets counterparts. In particular, NK cells mirror CD8+ cytotoxic T cells while ILC1, ILC2, ILC3, and Lymphoid tissue inducer (LTi)-like cells reflect the function of CD4+T helper (Th) cells (Th1, Th2, and Th17 respectively). ILCs are involved in innate host defenses against pathogens and tumors, in lymphoid organogenesis, and in tissue remodeling/repair. In recent years, important molecular inducible checkpoints (PD-1, TIM3, and TIGIT) were shown to control/inactivate different immune cell types. The expression of many of these receptors has been detected on NK cells and subsets of tissue-resident ILCs in both physiological and pathological conditions, including cancer. In particular, it has been demonstrated that the interaction between PD-1+ immune cells and PD-L1/PD-L2+ tumor cells may compromise the anti-tumor effector function leading to tumor immune escape. However, while the effector function of NK cells in tumor is well-established, limited information exists on the other ILC subsets. We will summarize what is known to date on the expression and function of these checkpoint receptors on NK cells and ILCs, with a particular focus on the recent data that reveal an essential contribution of the blockade of PD-1 and TIGIT on NK cells to the immunotherapy of cancer. A better information regarding the presence and the function of different ILCs and of the inhibitory checkpoints in pathological conditions may offer important clues for the development of new immune therapeutic strategies.
Introduction
Innate Lymphoid Cells (ILCs) represent a heterogeneous group of developmentally related lymphocytes (1). However, distinct aspects differentiate T or B lymphocytes from ILCs. Thus, unlike T and B cells, ILCs are characterized by the lack of expression of recombination activating genes (RAG-1 and RAG-2)-dependent rearranged antigen receptors and rely on a set of germ-line encoded receptors to exert their function (2–5). Thus, while T cells responses require longer time intervals due to antigen-mediated clonal selection and expansion, ILCs can exert a prompt response to recognition of conserved molecular patterns from pathogens and infected or injured tissues (6). For this reason and for their tissue-residency ILCs may represent the leading orchestrator of immune responses. ILCs release effector and regulatory cytokines that play a role in tissue repair and immune defense being also able to coordinate the adaptive immune responses (7–9). Thus, ILCs might also play a primary role in sensing cells that underwent malignant transformation and in initiating anti-tumor immune response, even though, as will be discussed in this review, their actual role in tumor suppression is controversial.
ILC Differentiation and Classification
Recently, five main ILCs groups have been identified according to the transcription factors required for their development and function (6, 10). These groups are represented by natural killer cells (NK), group 1 ILCs (ILC1), group 2 ILCs (ILC2), group 3 ILCs (ILC3), and lymphoid tissue-inducer (LTi) cells. All ILCs derive from common lymphoid progenitors (CLPs) that mainly reside in the bone marrow, even though ILCs progenitors are also found in other tissues, such as fetal liver, tonsils, decidua, and intestinal lamina propria (9, 11–16). ILCs are divided into cytotoxic-ILC and helper-ILC, which resemble cytotoxic and helper T cell subsets (17). In particular, NK cells mirror cytotoxic CD8+ cells while ILC1, ILC2, and ILC3 resemble the T helper cell subsets Th1, Th2, and Th17, respectively. Originally NK and ILC1 represented the group 1 of ILC family because both subsets express the transcription factor T-bet, and secrete interferon (IFN)-γ and tumor necrosis factor (TNF)-α (18, 19) (Figure 1). However, it must be noticed that NK cells, apart from T-bet, rely also on Eomes expression for their development (20) (Figure 1). NK cells, the first ILC subset to be identified, circulate in the bloodstream where they represent about the 15% of peripheral blood lymphocytes (1, 21). However, tissue-resident NK cells have also been found in liver, uterus and decidua (15, 22–24). In humans, two main PB-NK cell subsets can be distinguished based on the level of CD56 surface expression (25). In particular CD56dim, expressing high levels of perforin and granzyme, are characterized by a high cytotoxic activity, while CD56bright cells secrete inflammatory cytokines and are prevalently found in tissues and secondary lymphoid organs (26). NK cells play a major role in innate defenses against viruses and tumors, both by direct cell killing and by promoting the initiation of inflammation (27–30). On the contrary, ILC1 express low level of perforin and are barely found in PB while they mainly reside within tissues, such as intestine, lung, skin and decidua where they are involved in the first line of defense against viruses and bacteria (19, 31–33). ILC2s, which rely on GATA3 expression, are mostly found in lung, intestine, adipose tissue, skin, and gut (34). Upon activation in response to epithelia-derived stimuli (mainly IL-33 and IL-25), they release type-2 cytokines (primarily IL-5, IL-13, and IL-9) and promote defenses against parasites, viral infections, and contribute to metabolic homeostasis (7, 31, 35, 36). In addition, ILC2s produce amphiregulin, an epidermal growth factor family member involved in tissue repair (17). The most heterogeneous ILC subset includes fetal LTi and postnatal ILC3, both depending on the Rorγt transcription factor. They were previously called “group 3 ILCs” but, because of their different developmental trajectories, they are now classified into two distinct subsets (1). LTi cells play a pivotal role in the formation of secondary lymphoid structures, including lymph nodes and Peyer's patches during fetus development (37, 38). After birth, ILC3s can be found in gut, tonsils, and intestine where, through release of IL-22, play an important role in the innate immunity against bacteria and fungi (38, 39). However, LTi and a particular subset of ILC3, namely NCR+ ILC3, have also been found in decidual tissue during early pregnancy (33).
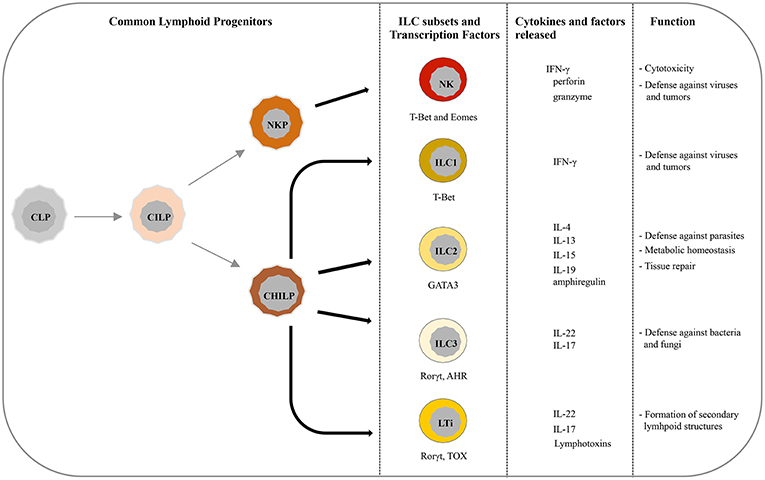
Figure 1. Current view of differentiation and immune function of ILC subsets. ILCs originate from common lymphoid progenitors (CLPs) that give rise to common innate lymphoid progenitors (CILPs). On one side, CILPs can differentiate into NK precursor (NKP) that, through T-bet and Eomes, will originate NK cells, and on the other side into common helper innate lymphoid precursors (CHILP) from which originate both Innate lymphoid precursors (ILP) and lymphoid tissue inducer progenitors (LTiP). ILC1/2/3 subsets and LTi cells differentiate, according to the different transcription factors required, from ILP and LTiP precursors, respectively. As specified in the text, NK/ILC cells release various cytokines and exert different immune functions.
While the role of NK cells in anti-tumor immunity has been widely studied and well-established, ILCs function in the immune defenses against tumors is still controversial (40). Their preferential localization at the mucosal surfaces may even suggest a negative role, as some of their cytokines exacerbate the development of chronic inflammation and potentially favor tumor growth. Indeed, IFN-γ released by ILC1 in inflamed conditions could have detrimental effects by favoring tumor growth. Similarly, type-2 cytokines produced by ILC2s are associated with poor prognosis in cancer patients and can create a pro-tumorigenic environment through the stimulation of myeloid derived suppressor cells (MDSC) or M2 macrophages (2, 41, 42). Moreover, ILC3s have been associated with tumor growth and metastasis in different type of cancers (10). However, available evidence suggests that ILCs function may depend on the tumor microenvironment (43–48). Indeed, the different cytokines, soluble factors, and cell types that characterize the tumor microenvironment can shape the function of different immune cells, dampening antitumor immunity (7). In this context, ILCs plasticity, that allows them to convert from one subset into another depending on the surrounding stimuli, might have a negative role in immune defenses. Therefore, a deeper understanding of NK and ILCs in protective immunity and how tumor cells and the tumor microenvironment can inhibit their functions is of extreme interest especially for the development of new immunotherapies.
NK/ILC Cell Receptors
NK/ILC are able to discriminate between healthy and virus- or tumor- infected cells through an array of inhibitory and activating receptors that recognize specific ligands induced by virus infection or tumor transformation (49–51). Natural Cytotoxicity Receptors (NCR), which include NKp46, NKp44, and NKp30, represent the major NK cells activating receptors (52–57). NCRs can be also expressed by specific ILCs subsets, with ILC1 expressing NKp46, ILC2 NKp30 and tonsil-derived ILC3 and mucosal NCR+ ILC3 bearing NKp30 and NKp46, respectively (58). NKG2D and DNAM-1 represent other important activating receptors able to recognize ligands that are de novo expressed or upregulated upon cell stress or tumor transformation (59–62). Additionally, NK cells express co-activating receptors, such as NTB-A and 2B4, whose function depends on the simultaneous co-engagement of one or more activating receptors (57, 63–65). The function of activating receptors is counterbalanced by inhibitory receptors that are mainly represented by the killer Ig-like receptors (KIR) and the heterodimer CD94/NKG2A which recognize the main type of HLA class-I molecules and function as true checkpoints in NK cell activation (29, 66–68). Indeed, in normal conditions these inhibitory receptors recognize HLA-I ligands expressed on healthy cells preventing their killing. As a consequence, loss of MHC expression on tumor cells is increasing rather than decreasing their susceptibility to NK cell-mediated killing (69). Recently, additional inhibitory checkpoints (such as PD-1, TIGIT, etc.), which under normal conditions maintain immune cell homeostasis, have been shown to facilitate tumor escape. Indeed, different studies demonstrated that, in these pathological conditions, checkpoint regulators, usually absent on resting NK cells, can be induced de novo and contribute to the downregulation of NK cell anti-tumor function upon interaction with their ligands expressed at the tumor cell surface (70).
In the next paragraphs, we will summarize what is known to date about the expression and function of these checkpoint receptors on NK cells and ILCs, with a particular focus on PD-1, TIGIT, and CD96.
PD-1
PD-1, a member of immunoglobulin superfamily, is a cell surface inhibitory receptor, functioning as a major checkpoint of T cell activation. It binds PD-L1 and PD-L2, ligands expressed on many tumors, on infected cells, on antigen-presenting cells in inflammatory foci, and in secondary lymphoid organs. Lack of PD-1 expression results in the suppression of tumor growth and metastasis in mice (71). The efficacy of PD-1 blockade has been mainly correlated with the restoration of a preexisting T cell response. PD-1 expression, initially described on T, B, and myeloid cells, has been recently described also on NK cells (72, 73) (Figure 2). In particular, PD-1 expression was shown on NK cells from some healthy individuals and in most cancer patients, including Kaposi sarcoma, ovarian and lung carcinoma and Hodgkin lymphoma, where it can negatively regulate NK cell function (73–78). The contribution of PD-1 blockade on NK cells in immunotherapy has been demonstrated in several mouse models of cancer, where PD-1 engagement by PD-L1+ tumor cells could strongly suppress NK cell–mediated anti-tumor immunity (79). PD-1 expression was found more abundant on NK cells with an activated and more responsive phenotype rather than on NK cells with an exhausted phenotype (79). However, to date the molecular mechanisms regulating the expression of this inhibitory receptor on NK cells are not clear. It has been demonstrated in a mouse model of cytomegalovirus infection (MCMV) that endogenous glucocorticoids integrate the signals from the microenvironment to induce PD-1 expression at the transcriptional level, highlighting the importance of a tissue-specific cooperation of cytokines and the neuroendocrine system in this regulation (80). Regarding the cancer setting, however, recent data suggest that PD-1 is accumulated inside NK cells and translocated on the cell surface rather than induced at the transcriptional level (81). However, the stimuli required for its surface expression are unknown.
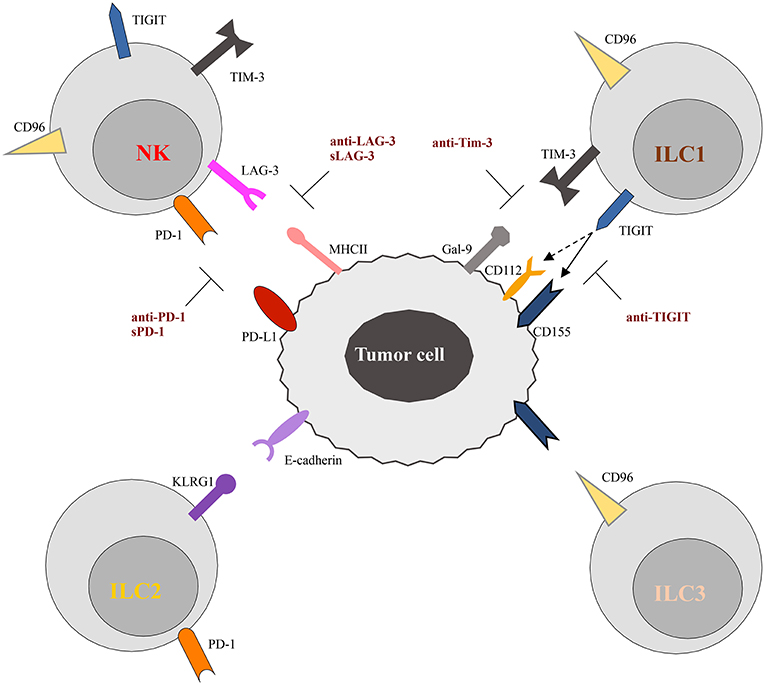
Figure 2. Schematic representation of checkpoint receptors and their ligands expressed by ILC and tumor cells, respectively. NK cells express multiple immune checkpoint receptors, such as PD-1, TIM-3, Lag-3, TIGIT, and CD96. ON the other hand, these checkpoint receptors are instead differentially expressed by ILC subsets. Thus, TIGIT and TIM-3 have been detected only on ILC1 cells, while CD96 is expressed on both ILC1 and ILC2. Surface expression of KLRG1 and PD-1 appears to be restricted to ILC2 cells. The inhibitory ligands expressed by tumor cells, specifically interact with the checkpoint receptors preventing cells activation. However, different therapeutic approaches, aimed to block receptor/ligand interactions, have been demonstrated to restore the anti-tumor activity of immune cells, as illustrated in the Figure. The solid and dotted arrows indicate the strong and weak binding affinity of TIGIT for the different ligands, respectively.
Two recent papers described that, in mice, PD-1 expression identifies ILC committed progenitors, capable of generating ILC1s, ILC2s, ILC3s, and a small number of circulating NK cells (82, 83). High expression of PD-1 is lost upon differentiation, but upregulated on effector tissue resident ILC2s upon lung inflammation (83). In agreement with these findings, it was shown that mouse ILC2s express PD-1 in different percentages depending on their tissue origin and that this expression is enhanced by IL-33 stimulation, reducing their ability to release cytokines (84). This is particularly relevant in type 2 infections, such as helminth infection, but the role of PD-1 expression on ILC2s in the context of cancer remains to be investigated. Nonetheless, the finding that it is possible to modulate ILC2s effector functions by using PD-1 blocking antibodies suggests that targeting this receptor with checkpoint inhibitors could also affect type 2 responses in cancer patients and favor cancer growth by restoring the production of type-2 cytokines. The possible unfavorable effect of this ILC2-mediated response and its contribution to therapy with checkpoint inhibitors should be further explored to further improve the efficacy of cancer treatment.
Recent studies provided the first evidence that also ILC3s can express a functional PD-1 receptor. In particular, PD-1 expression has been detected on both NK cells and ILC3s in malignant pleural effusions of patients with primary and metastatic tumors (85). Moreover, it has been shown that NK cells and ILC3s in human decidua express PD-1 during the first trimester of pregnancy, while the invading trophoblast expresses PD-L1. The PD-1/PD-L1 molecular interaction regulates ILC3 production of cytokines, suggesting that it may play a regulatory role at the feto-maternal interface (16).
TIGIT and CD96
TIGIT (T cell Ig and ITIM domain) and CD96 are co-inhibitory receptors expressed on subsets of T cells, human NK cells, ILC1s, and ILC3s (Figure 2). They belong to a group of immunoglobulin superfamily receptors comprising also the co-stimulatory receptor DNAM-1 (CD226). They recognize nectin and nectin-like ligands, frequently upregulated on tumor cells. CD155 is a ligand shared by the three receptors, with CD96 showing the highest binding affinity (86). These receptors initiate a pathway that is analogous to the CD28/CTLA-4 one. In this pathway, ligands and differential receptor:ligand affinities can fine-tune the immune response. The work of Zhang et al. (87) recently demonstrated that TIGIT constitutes a previously unappreciated checkpoint in NK cells, and that targeting TIGIT alone or in combination with other checkpoint receptors may represent a promising anti-cancer therapeutic strategy. It has been shown that, in patients with colon cancer, TIGIT expression is increased on tumor associated NK cells. In addition, evidences has been provided that, beyond the targeting of effector and regulatory T cells, the mode of action of TIGIT blockade also involves NK cells (87). In particular, genetic KO or mAb-mediated blockade of TIGIT was able to unleash both NK cell and T cell antitumor activity, leading to a substantial improvement in the control of tumor growth in several preclinical mouse models. Moreover, TIGIT blockade prevented NK cell exhaustion in the absence of adaptive immunity, and elicited a potent T cell–mediated memory response to tumor re-challenge through a not yet identified mechanism (87). Increased TIGIT and CD96 expression and lower levels of DNAM1 were also detected on ILC1s induced by TGF-β, contributing to the impairment of their anti-tumor response (88).
Although the role of TIGIT and CD96 as immune checkpoint receptors are just beginning to be uncovered, accumulating data would support the notion that targeting of these receptors for improving anti-tumor immune responses also involves NK cells and ILCs.
Other Checkpoints on NK Cells and ILCs
KLRG1 is another inhibitory receptor expressed by mature NK cells whose expression varies with cell activation. It is a C-type lectin-like receptor containing one ITIM, and it has been used as a marker for distinct NK and T-cell differentiation stages (89). However, KLRG1 knock-out mice showed that it does not play a deterministic role in the generation and functional characteristics of these lymphocyte subsets. KLRG1 is also expressed by mast cells, basophils, eosinophils, and ILC2s, suggesting a role in type 2 immune responses. Experiments in mice showed that in vivo administration of IL-25 elicits the expansion of a subset of ILC2s referred to as “inflammatory” ILC2s that are characterized by high expression of KLRG1 and that participate in the control of helminth infection (90). In the tumor context, KLRG1 expression was found on ILC2 associated to the tumor in NSCLC and CRC (91, 92). While the interaction of KLRG1 and its E-cadherin ligand has been shown to inhibit human ILC2s in vitro, its function in vivo remains to be established (90, 93).
Lag-3 and Tim-3 are inhibitory receptors whose expression has been reported on NK cells and ILC1s (Figure 2). Tim-3 is a type 1 glycoprotein expressed by mature NK cells, and its expression is further increased on NK cells in melanoma and lung adenocarcinoma patients, impairing NK cell effector functions (94, 95). More recently, Tim-3 expression has been reported on human decidual NK cells and also on ILC3s. It was demonstrated that Tim-3 is expressed in higher percentages in CD56+ILC3s compared to LTi-like cells, and that its triggering is able to significantly reduce IL-22 production by CD56+ILC3s (16). Lag-3 is a negative costimulatory receptor that is homologous to CD4 and binds MHC-II molecules with very high affinity. Although its role in downregulating T cell proliferation, activation, and homeostasis is clear, its mechanism of action in NK cells remains to be dissected in detail (96).
NKG2A is a HLA-E-specific inhibitory receptor that plays an important regulatory role in NK cell function. Also antigen- or cytokine-stimulated T cells were shown to express a functional NKG2A that may antagonize T cell function (97, 98). It has recently been reported that NKG2A is expressed on NK and T cells in the tumor bed in many human cancers such as squamous cell carcinoma of the head and neck (SCCHN) and colorectal carcinoma (CRC) (99). In addition, its ligand, HLA-E, is frequently overexpressed in these tumors. NKG2A targeting with monalizumab (a humanized anti-NKG2A antibody) has been shown to enhance the anti-tumor immunity mediated by NK and CD8+ T cells when used as a single agent or in combination with other therapeutic antibodies such as durvalumab (blocking PD-L1), or cetuximab (directed against the epidermal growth factor receptor, EGFR) (99, 100).
In mouse tumor models, it has been shown that TGF-β signaling in the microenvironment induces NK cell conversion to ILC1s. These tumor-associated ILC1s express higher levels of inhibitory receptors (NKG2A, KLRG1, CTLA4, LAG3) as compared to NK cells. While NK cells favored tumor immune surveillance in this setting, the higher expression of immunological checkpoint receptors on ILC1s was associated with a lower ability to control local tumor growth and metastasis (88). These evidences suggest that NK cell conversion to ILC1s displaying a functional impairment could represent an additional mechanism by which tumor escapes immune surveillance.
Cancer Immunotherapies
During the past few years, different strategies have been developed to overcome the immunosuppressive tumor environment and restore antitumor immune activity. The use of blocking antibodies against inhibitory receptors or their ligands, in order to restore the T or NK cell function has been demonstrated to be an efficient and safe cancer immunotherapy in the treatment of several tumors (70). Considering the wide expression of PD-1 on immune cells, most therapies have been developed in order to block PD-1/PD-L1/2 interactions. Indeed, some anti-PD-1 mAbs have already been approved by FDA, showing encouraging results in patients with melanoma or lung cancer (101, 102). Currently, there are ongoing phase I and II clinical trials for anti-KIR, -NKG2A, -Tim3, -LAG3, -TIGIT inhibitory receptors (102). Interestingly, considering that checkpoint inhibitors can act in synergy with each other, combinations of mAbs are also under investigation as a new approach for optimal boost of the immune system. In particular, clinical trials are investigating the combination of anti-PD1 therapy with anti-TIM3 or anti-TIGIT blocking antibodies in different tumors (70, 103). Moreover, encouraging results obtained in phase II clinical trial for SCCHN using a combination of monalizumab and cetuximab suggested that, targeting checkpoint receptors on NK cells, may be an efficient tool to complement first-generation immunotherapies against cancer (99).
Of notice is also the discovery of soluble forms of LAG-3 (sLAG3) and PD-1 (sPD-1) (70, 104). Different studies have been focused on the role of sPD-1 as a putative antitumor agent. Indeed, in mice an increase in anti-tumor activity was observed upon delivery of sPD-1 encoding plasmid at tumor site (105, 106). Moreover, clinical studies have investigated the presence of sPD-1 and its correlation with the overall survival of patients with different cancers (107, 108). It has been shown that sLAG3 is able to induce NK cytokines (IFN-γ and TNF-α) production in ex vivo assay (109). Moreover, a phase II clinical trial is investigating the role of sLAG3 in stimulating the immune system in combination with anti-PD-1 therapy (70).
Conclusions and Future Perspectives
It is now evident that NK/ILC family plays a pivotal role in the immune defenses. Recent studies in murine and human settings demonstrated that the expression of several inhibitory checkpoints, that may be detrimental in the tumor context, is not restricted to T lymphocytes, revealing an important, yet poorly appreciated, contribution of their expression on innate immune cells. Thus, in the recent years different immunotherapy approaches, based on the blockade of inhibitory NK cell receptors, have been developed in order to unleash NK cell cytotoxicity. This is particularly important in the context of tumors that downregulate HLA-I expression and become invisible to T cells. However, it must be considered that most inhibitory checkpoints, targeted by mAbs therapies, are shared by T, NK and ILCs. Therefore, further studies are required in order to identify all the receptors regulating NK/ILC cells function for the development of new mAbs-based immunotherapies. In addition, considering the role exerted by the tumor microenvironment on ILCs plasticity and functions, it is necessary to better clarify the role of tumor infiltrating innate immune cells to improve the selectivity of cancer therapies. Therefore, also accurate patient analysis and deeper examinations of tumor biopsies will become key aspects to consider in order to construct personalized protocols. In this context, studies have been performed to determine the exact number of biopsies required to have a more precise PD-L1 expression profile that would more closely resemble to whole tumor section (110–112). Thus, despite the great improvement reached in the last years, further studies are required to investigate the expression of these checkpoints both in NK cells and on the other subsets of ILCs, and their precise role in human pathologies in order to improve the efficacy of immunotherapies thanks to a more personalized approach.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
The present study has been supported by the following grants: Associazione Italiana per la Ricerca sul Cancro (AIRC) IG 2014 Id. 15283 (LM), IG 2017 Id. 19920 (LM), Special Project 5X1000 no.21147 (LM), Ricerca Corrente OPBG (LM), and Ministero della Salute GR-2013-02356568 (PV).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Montaldo E, Vacca P, Vitale C, Moretta F, Locatelli F, Mingari MC, et al. Human innate lymphoid cells. Immunol Lett. (2016) 179:2–8. doi: 10.1016/j.imlet.2016.01.007
2. Carrega P, Campana S, Bonaccorsi I, Ferlazzo G. The yin and yang of innate lymphoid cells in cancer. Immunol Lett. (2016) 179:29–35. doi: 10.1016/j.imlet.2016.06.003
3. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol. (2013) 13:145–9. doi: 10.1038/nri3365
4. Cording S, Medvedovic J, Aychek T, Eberl G. Innate lymphoid cells in defense, immunopathology and immunotherapy. Nat Immunol. (2016) 17:755–7. doi: 10.1038/ni.3448
5. Medzhitov R, Janeway CA Jr. Innate immune recognition and control of adaptive immune responses. Semin Immunol. (1998) 10:351–3.
6. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell. (2018) 174:1054–66. doi: 10.1016/j.cell.2018.07.017
7. Mattner J, Wirtz S. Friend or Foe? The ambiguous role of innate lymphoid cells in cancer development. Trends Immunol. (2017) 38:29–38. doi: 10.1016/j.it.2016.10.004
8. von Burg N, Chappaz S, Baerenwaldt A, Horvath E, Bose Dasgupta S, Ashok D, et al. Activated group 3 innate lymphoid cells promote T-cell-mediated immune responses. Proc Natl Acad Sci USA. (2014) 111:12835–40. doi: 10.1073/pnas.1406908111
9. Vacca P, Munari E, Tumino N, Moretta F, Pietra G, Vitale M, et al. Human natural killer cells and other innate lymphoid cells in cancer: friends or foes? Immunol Lett. (2018). 201:14–9. doi: 10.1016/j.imlet.2018.11.004
10. Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol. (2018) 18:671–88. doi: 10.1038/s41577-018-0061-z
11. Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell. (2017) 168:1086–100 e10. doi: 10.1016/j.cell.2017.02.021
12. Scoville SD, Mundy-Bosse BL, Zhang MH, Chen L, Zhang X, Keller KA, et al. A progenitor cell expressing transcription factor RORgammat generates all human innate lymphoid cell subsets. Immunity. (2016) 44:1140–50. doi: 10.1016/j.immuni.2016.04.007
13. Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol. (2016) 17:765–74. doi: 10.1038/ni.3489
14. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. (2014) 508:397–401. doi: 10.1038/nature13047
15. Vacca P, Vitale C, Montaldo E, Conte R, Cantoni C, Fulcheri E, et al. CD34+ hematopoietic precursors are present in human decidua and differentiate into natural killer cells upon interaction with stromal cells. Proc Natl Acad Sci USA. (2011) 108:2402–7. doi: 10.1073/pnas.1016257108
16. Vacca P, Pesce S, Greppi M, Fulcheri E, Munari E, Olive D, et al. PD-1 is expressed by and regulates human group 3 innate lymphoid cells in human decidua. Mucosal Immunol. (2019). 12:624–31. doi: 10.1038/s41385-019-0141-9
17. Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. (2014) 41:354–65. doi: 10.1016/j.immuni.2014.09.005
18. Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. (2013) 38:769–81. doi: 10.1016/j.immuni.2013.02.010
19. Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. (2013) 14:221–9. doi: 10.1038/ni.2534
20. Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. (2012) 36:55–67. doi: 10.1016/j.immuni.2011.11.016
21. Kiessling R, Klein E, Pross H, Wigzell H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. (1975) 5:117–21. doi: 10.1002/eji.1830050209
22. Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. (2014) 211:563–77. doi: 10.1084/jem.20131560
23. Doisne JM, Balmas E, Boulenouar S, Gaynor LM, Kieckbusch J, Gardner L, et al. Composition, development, and function of uterine innate lymphoid cells. J Immunol. (2015) 195:3937–45. doi: 10.4049/jimmunol.1500689
24. Yu J, Freud AG, Caligiuri MA. Location and cellular stages of natural killer cell development. Trends Immunol. (2013) 34:573–82. doi: 10.1016/j.it.2013.07.005
25. Caligiuri MA. Human natural killer cells. Blood. (2008) 112:461–9. doi: 10.1182/blood-2007-09-077438
26. Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. (2001) 97:3146–51. doi: 10.1182/blood.V97.10.3146
27. Moretta L, Pietra G, Vacca P, Pende D, Moretta F, Bertaina A, et al. Human NK cells: from surface receptors to clinical applications. Immunol Lett. (2016) 178:15–9. doi: 10.1016/j.imlet.2016.05.007
28. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. (2011) 331:44–9. doi: 10.1126/science.1198687
29. Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, et al. Receptors for HLA class-I molecules in human natural killer cells. Annu Rev Immunol. (1996) 14:619–48. doi: 10.1146/annurev.immunol.14.1.619
30. Moretta L, Bottino C, Pende D, Mingari MC, Biassoni R, Moretta A. Human natural killer cells: their origin, receptors and function. Eur J Immunol. (2002) 32:1205–11. doi: 10.1002/1521-4141(200205)32:5<1205::AID-IMMU1205>3.0.CO;2-Y
31. Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. (2011) 12:1045–54. doi: 10.1031/ni.2131
32. Klose CSN, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. (2014) 157:340–56. doi: 10.1016/j.cell.2014.03.030
33. Vacca P, Montaldo E, Croxatto D, Loiacono F, Canegallo F, Venturini PL, et al. Identification of diverse innate lymphoid cells in human decidua. Mucosal Immunol. (2015) 8:254–64. doi: 10.1038/mi.2014.63
34. Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. (2012) 37:634–48. doi: 10.1016/j.immuni.2012.06.020
35. Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. (2010) 463:540–4. doi: 10.1038/nature08636
36. Brestoff JR, Kim BS, Saenz SA, Stine RR, Monticelli LA, Sonnenberg GF, et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. (2015) 519:242–6. doi: 10.1038/nature14115
37. Cherrier M, Eberl G. The development of LTi cells. Curr Opin Immunol. (2012) 24:178–83. doi: 10.1016/j.coi.2012.02.003
38. Montaldo E, Juelke K, Romagnani C. Group 3 innate lymphoid cells (ILC3s): origin, differentiation, and plasticity in humans and mice. Eur J Immunol. (2015) 45:2171–82. doi: 10.1002/eji.201545598
39. Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. (2008) 14:282–9. doi: 10.1038/nm1720
40. Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. (2016) 16:7–19. doi: 10.1038/nrc.2015.5
41. Zhou R, Qian S, Gu X, Chen Z, Xiang J. Interleukin-13 and its receptors in colorectal cancer (Review). Biomed Rep. (2013) 1:687–90. doi: 10.3892/br.2013.132
42. Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. (2010) 22:231–7. doi: 10.1016/j.coi.2010.01.009
43. Bie Q, Zhang P, Su Z, Zheng D, Ying X, Wu Y, et al. Polarization of ILC2s in peripheral blood might contribute to immunosuppressive microenvironment in patients with gastric cancer. J Immunol Res. (2014) 2014:923135. doi: 10.1155/2014/923135
44. Jovanovic IP, Pejnovic NN, Radosavljevic GD, Pantic JM, Milovanovic MZ, Arsenijevic NN, et al. Interleukin-33/ST2 axis promotes breast cancer growth and metastases by facilitating intratumoral accumulation of immunosuppressive and innate lymphoid cells. Int J Cancer. (2014) 134:1669–82. doi: 10.1002/ijc.28481
45. Ikutani M, Yanagibashi T, Ogasawara M, Tsuneyama K, Yamamoto S, Hattori Y, et al. Identification of innate IL-5-producing cells and their role in lung eosinophil regulation and antitumor immunity. J Immunol. (2012) 188:703–13. doi: 10.4049/jimmunol.1101270
46. Eisenring M, vom Berg J, Kristiansen G, Saller E, Becher B. IL-12 initiates tumor rejection via lymphoid tissue-inducer cells bearing the natural cytotoxicity receptor NKp46. Nat Immunol. (2010) 11:1030–8. doi: 10.1038/ni.1947
47. Chan IH, Jain R, Tessmer MS, Gorman D, Mangadu R, Sathe M, et al. Interleukin-23 is sufficient to induce rapid de novo gut tumorigenesis, independent of carcinogens, through activation of innate lymphoid cells. Mucosal Immunol. (2014) 7:842–56. doi: 10.1038/mi.2013.101
48. Crome SQ, Nguyen LT, Lopez-Verges S, Yang SY, Martin B, Yam JY, et al. A distinct innate lymphoid cell population regulates tumor-associated T cells. Nat Med. (2017) 23:368–75. doi: 10.1038/nm.4278
49. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. (2008) 9:495–502. doi: 10.1038/ni1581
50. Sivori S, Carlomagno S, Pesce S, Moretta A, Vitale M, Marcenaro E. TLR/NCR/KIR: which one to use and when? Front Immunol. (2014) 5:105. doi: 10.3389/fimmu.2014.00105
51. Barrow AD, Colonna M. Tailoring Natural Killer cell immunotherapy to the tumour microenvironment. Semin Immunol. (2017) 31:30–6. doi: 10.1016/j.smim.2017.09.001
52. Sivori S, Vitale M, Morelli L, Sanseverino L, Augugliaro R, Bottino C, et al. p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J Exp Med. (1997) 186:1129–36.
53. Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, et al. Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J Exp Med. (1998) 188:953–60.
54. Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, et al. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med. (1999) 189:787–96.
55. Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E, et al. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med. (1998) 187:2065–72.
56. Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. (1999) 190:1505–16.
57. Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. (2001) 19:197–223. doi: 10.1146/annurev.immunol.19.1.197
58. Guia S, Fenis A, Vivier E, Narni-Mancinelli E. Activating and inhibitory receptors expressed on innate lymphoid cells. Semin Immunopathol. (2018) 40:331–41. doi: 10.1007/s00281-018-0685-x
59. Shibuya A, Campbell D, Hannum C, Yssel H, Franz-Bacon K, McClanahan T, et al. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. (1996) 4:573–81.
60. Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. (2003) 198:557–67. doi: 10.1084/jem.20030788
61. Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. (1999) 285:727–9.
62. Morisaki T, Onishi H, Katano M. Cancer immunotherapy using NKG2D and DNAM-1 systems. Anticancer Res. (2012) 32:2241–7.
63. Bottino C, Falco M, Parolini S, Marcenaro E, Augugliaro R, Sivori S, et al. NTB-A [correction of GNTB-A], a novel SH2D1A-associated surface molecule contributing to the inability of natural killer cells to kill Epstein-Barr virus-infected B cells in X-linked lymphoproliferative disease. J Exp Med. (2001) 194:235–46. doi: 10.1084/jem.194.3.235
64. Parolini S, Bottino C, Falco M, Augugliaro R, Giliani S, Franceschini R, et al. X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J Exp Med. (2000) 192:337–46. doi: 10.1084/jem.192.3.337
65. Bryceson YT, March ME, Ljunggren HG, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. (2006) 107:159–66. doi: 10.1182/blood-2005-04-1351
66. Moretta A, Bottino C, Pende D, Tripodi G, Tambussi G, Viale O, et al. Identification of four subsets of human CD3-CD16+ natural killer (NK) cells by the expression of clonally distributed functional surface molecules: correlation between subset assignment of NK clones and ability to mediate specific alloantigen recognition. J Exp Med. (1990) 172:1589–98.
67. Ciccone E, Pende D, Viale O, Di Donato C, Tripodi G, Orengo AM, et al. Evidence of a natural killer (NK) cell repertoire for (allo) antigen recognition: definition of five distinct NK-determined allospecificities in humans. J Exp Med. (1992) 175:709–18.
68. Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, Augugliaro R, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. (1993) 178:597–604.
69. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. (2008) 9:503–10. doi: 10.1038/ni1582
70. Chiossone L, Vienne M, Kerdiles YM, Vivier E. Natural killer cell immunotherapies against cancer: checkpoint inhibitors and more. Semin Immunol. (2017) 31:55–63. doi: 10.1016/j.smim.2017.08.003
71. Okazaki T, Chikuma S, Iwai Y, Fagarasan S, Honjo T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat Immunol. (2013) 14:1212–8. doi: 10.1038/ni.2762
72. Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. (2017) 545:60–5. doi: 10.1038/nature22079
73. Pesce S, Greppi M, Tabellini G, Rampinelli F, Parolini S, Olive D, et al. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: a phenotypic and functional characterization. J Allergy Clin Immunol. (2017) 139:335–46 e3. doi: 10.1016/j.jaci.2016.04.025
74. Beldi-Ferchiou A, Lambert M, Dogniaux S, Vely F, Vivier E, Olive D, et al. PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget. (2016) 7:72961–77. doi: 10.18632/oncotarget.12150
75. Della Chiesa M, Pesce S, Muccio L, Carlomagno S, Sivori S, Moretta A, et al. Features of memory-like and PD-1(+) human NK cell subsets. Front Immunol. (2016) 7:351. doi: 10.3389/fimmu.2016.00351
76. Iraolagoitia XL, Spallanzani RG, Torres NI, Araya RE, Ziblat A, Domaica CI, et al. NK cells restrain spontaneous antitumor CD8+ T cell priming through PD-1/PD-L1 interactions with dendritic cells. J Immunol. (2016) 197:953–61. doi: 10.4049/jimmunol.1502291
77. Liu Y, Cheng Y, Xu Y, Wang Z, Du X, Li C, et al. Increased expression of programmed cell death protein 1 on NK cells inhibits NK-cell-mediated anti-tumor function and indicates poor prognosis in digestive cancers. Oncogene. (2017) 36:6143–53. doi: 10.1038/onc.2017.209
78. Vari F, Arpon D, Keane C, Hertzberg MS, Talaulikar D, Jain S, et al. Immune evasion via PD-1/PD-L1 on NK cells and monocyte/macrophages is more prominent in Hodgkin lymphoma than DLBCL. Blood. (2018) 131:1809–19. doi: 10.1182/blood-2017-07-796342
79. Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois-Daigneault MC, Trevino TN, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest. (2018) 128:4654–68. doi: 10.1172/JCI99317
80. Quatrini L, Wieduwild E, Escaliere B, Filtjens J, Chasson L, Laprie C, et al. Endogenous glucocorticoids control host resistance to viral infection through the tissue-specific regulation of PD-1 expression on NK cells. Nat Immunol. (2018) 19:954–62. doi: 10.1038/s41590-018-0185-0
81. Mariotti FR, Petrini S, Ingegnere T, Tumino N, Besi F, Scordamaglia F, et al. PD-1 in human NK cells: evidence of cytoplasmic mRNA and protein expression. Oncoimmunology. (2019) 8:1557030. doi: 10.1080/2162402X.2018.1557030
82. Seillet C, Mielke LA, Amann-Zalcenstein DB, Su S, Gao J, Almeida FF, et al. Deciphering the innate lymphoid cell transcriptional program. Cell Rep. (2016) 17:436–47. doi: 10.1016/j.celrep.2016.09.025
83. Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, et al. Single-cell RNA-seq identifies a PD-1(hi) ILC progenitor and defines its development pathway. Nature. (2016) 539:102–6. doi: 10.1038/nature20105
84. Taylor S, Huang Y, Mallett G, Stathopoulou C, Felizardo TC, Sun MA, et al. PD-1 regulates KLRG1(+) group 2 innate lymphoid cells. J Exp Med. (2017) 214:1663–78. doi: 10.1084/jem.20161653
85. Tumino N, Martini S, Munari E, Scordamaglia F, Besi F, Mariotti FR, et al. Presence of innate lymphoid cells in pleural effusions of primary and metastatic tumors: functional analysis and expression of PD-1 receptor. Int J Cancer. (2019). doi: 10.1002/ijc.32262 [Epub ahead of print]
86. Dougall WC, Kurtulus S, Smyth MJ, Anderson AC. TIGIT and CD96: new checkpoint receptor targets for cancer immunotherapy. Immunol Rev. (2017) 276:112–20. doi: 10.1111/imr.12518
87. Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat Immunol. (2018) 19:723–32. doi: 10.1038/s41590-018-0132-0
88. Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol. (2017) 18:1004–15. doi: 10.1038/ni.3800
89. Huntington ND, Tabarias H, Fairfax K, Brady J, Hayakawa Y, Degli-Esposti MA, et al. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J Immunol. (2007) 178:4764–70. doi: 10.4049/jimmunol.178.8.4764
90. Huang Y, Guo L, Qiu J, Chen X, Hu-Li J, Siebenlist U, et al. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory' type 2 innate lymphoid cells. Nat Immunol. (2015) 16:161–9. doi: 10.1038/ni.3078
91. Simoni Y, Fehlings M, Kloverpris HN, McGovern N, Koo SL, Loh CY, et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. (2017) 46:148–61. doi: 10.1016/j.immuni.2016.11.005
92. Carrega P, Loiacono F, Di Carlo E, Scaramuccia A, Mora M, Conte R, et al. NCR(+)ILC3 concentrate in human lung cancer and associate with intratumoral lymphoid structures. Nat Commun. (2015) 6:8280. doi: 10.1038/ncomms9280
93. Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska-Owsiak D, Wang X, et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med. (2013) 210:2939–50. doi: 10.1084/jem.20130351
94. da Silva IP, Gallois A, Jimenez-Baranda S, Khan S, Anderson AC, Kuchroo VK, et al. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade. Cancer Immunol Res. (2014) 2:410–22. doi: 10.1158/2326-6066.CIR-13-0171
95. Xu L, Huang Y, Tan L, Yu W, Chen D, Lu C, et al. Increased Tim-3 expression in peripheral NK cells predicts a poorer prognosis and Tim-3 blockade improves NK cell-mediated cytotoxicity in human lung adenocarcinoma. Int Immunopharmacol. (2015) 29:635–41. doi: 10.1016/j.intimp.2015.09.017
96. Miyazaki T, Dierich A, Benoist C, Mathis D. LAG-3 is not responsible for selecting T helper cells in CD4-deficient mice. Int Immunol. (1996) 8:725–9.
97. Mingari MC, Ponte M, Bertone S, Schiavetti F, Vitale C, Bellomo R, et al. HLA class I-specific inhibitory receptors in human T lymphocytes: interleukin 15-induced expression of CD94/NKG2A in superantigen- or alloantigen-activated CD8+ T cells. Proc Natl Acad Sci USA. (1998) 95:1172–7.
98. Bertone S, Schiavetti F, Bellomo R, Vitale C, Ponte M, Moretta L, et al. Transforming growth factor-beta-induced expression of CD94/NKG2A inhibitory receptors in human T lymphocytes. Eur J Immunol. (1999) 29:23–9. doi: 10.1002/(SICI)1521-4141(199901)29:01<23::AID-IMMU23>3.0.CO;2-Y
99. Andre P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, et al. Anti-NKG2A mAb Is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. (2018) 175:1731–43 e13. doi: 10.1016/j.cell.2018.10.014
100. Mingari MC, Pietra G, Moretta L. Immune checkpoint inhibitors: anti-NKG2A antibodies on board. Trends Immunol. (2019) 40:83–5. doi: 10.1016/j.it.2018.12.009
101. Mahoney KM, Freeman GJ, McDermott DF. The next immune-checkpoint inhibitors: PD-1/PD-L1 blockade in melanoma. Clin Ther. (2015) 37:764–82. doi: 10.1016/j.clinthera.2015.02.018
102. Davis ZB, Vallera DA, Miller JS, Felices M. Natural killer cells unleashed: checkpoint receptor blockade and BiKE/TriKE utilization in NK-mediated anti-tumor immunotherapy. Semin Immunol. (2017) 31:64–75. doi: 10.1016/j.smim.2017.07.011
103. Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. (2016) 7:10501. doi: 10.1038/ncomms10501
104. Nielsen C, Ohm-Laursen L, Barington T, Husby S, Lillevang ST. Alternative splice variants of the human PD-1 gene. Cell Immunol. (2005) 235:109–16. doi: 10.1016/j.cellimm.2005.07.007
105. Zhu X, Lang J. Soluble PD-1 and PD-L1: predictive and prognostic significance in cancer. Oncotarget. (2017) 8:97671–82. doi: 10.18632/oncotarget.18311
106. Elhag OA, Hu XJ, Wen-Ying Z, Li X, Yuan YZ, Deng LF, et al. Reconstructed adeno-associated virus with the extracellular domain of murine PD-1 induces antitumor immunity. Asian Pac J Cancer Prev. (2012) 13:4031–6. doi: 10.7314/APJCP.2012.13.8.4031
107. Kruger S, Legenstein ML, Rosgen V, Haas M, Modest DP, Westphalen CB, et al. Serum levels of soluble programmed death protein 1 (sPD-1) and soluble programmed death ligand 1 (sPD-L1) in advanced pancreatic cancer. Oncoimmunology. (2017) 6:e1310358. doi: 10.1080/2162402X.2017.1310358
108. Sorensen SF, Demuth C, Weber B, Sorensen BS, Meldgaard P. Increase in soluble PD-1 is associated with prolonged survival in patients with advanced EGFR-mutated non-small cell lung cancer treated with erlotinib. Lung Cancer. (2016) 100:77–84. doi: 10.1016/j.lungcan.2016.08.001
109. Brignone C, Grygar C, Marcu M, Schakel K, Triebel F. A soluble form of lymphocyte activation gene-3 (IMP321) induces activation of a large range of human effector cytotoxic cells. J Immunol. (2007) 179:4202–11. doi: 10.4049/jimmunol.179.6.4202
110. Munari E, Zamboni G, Lunardi G, Marchionni L, Marconi M, Sommaggio M, et al. PD-L1 Expression heterogeneity in non-small cell lung cancer: defining criteria for harmonization between biopsy specimens and whole sections. J Thorac Oncol. (2018) 13:1113–20. doi: 10.1016/j.jtho.2018.04.017
111. Munari E, Zamboni G, Lunardi G, Marconi M, Sommaggio M, Brunelli M, et al. PD-L1 expression comparison between primary and relapsed non-small cell lung carcinoma using whole sections and clone SP263. Oncotarget. (2018) 9:30465–71. doi: 10.18632/oncotarget.25770
112. Munari E, Rossi G, Zamboni G, Lunardi G, Marconi M, Sommaggio M, et al. PD-L1 Assays 22C3 and SP263 are not interchangeable in non-small cell lung cancer when considering clinically relevant cutoffs: an interclone evaluation by differently trained pathologists. Am J Surg Pathol. (2018) 42:1384–9. doi: 10.1097/PAS.0000000000001105
Keywords: NK, ILC, PD-1, checkpoint receptors, cancer immunotherapies
Citation: Mariotti FR, Quatrini L, Munari E, Vacca P and Moretta L (2019) Innate Lymphoid Cells: Expression of PD-1 and Other Checkpoints in Normal and Pathological Conditions. Front. Immunol. 10:910. doi: 10.3389/fimmu.2019.00910
Received: 03 December 2018; Accepted: 09 April 2019;
Published: 26 April 2019.
Edited by:
Michael A. Caligiuri, City of Hope National Medical Center, United StatesReviewed by:
Georg Gasteiger, Julius-Maximilians-Universität, GermanyRafael Solana, Universidad de Córdoba, Spain
Copyright © 2019 Mariotti, Quatrini, Munari, Vacca and Moretta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenzo Moretta, bG9yZW56by5tb3JldHRhQG9wYmcubmV0
 Francesca Romana Mariotti
Francesca Romana Mariotti Linda Quatrini
Linda Quatrini Enrico Munari
Enrico Munari Paola Vacca
Paola Vacca Lorenzo Moretta
Lorenzo Moretta