- 1Immunology Program, Memorial Sloan Kettering Cancer Center, Sloan Kettering Institute, New York, NY, United States
- 2Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, United States
- 3Weill Cornell Medical College, New York, NY, United States
Cumulative activating and inhibitory receptor signaling controls the functional output of individual natural killer (NK) cells. Investigation of how competing signals impact response, however, has been hampered by the lack of available antibodies capable of distinguishing inhibitory and activating receptors with highly homologous ectodomains. Utilizing a novel combination of monoclonal antibodies with specificity for discrete inhibitory KIR2DL1 and activating KIR2DS1 allotypes found among 230 healthy donors, we investigated allele-specific receptor expression and function driven by KIR2DL1 and KIR2DS1 alleles. We found that co-expression of the HLA-C2 ligand diminishes KIR2DL1, but not KIR2DS1, cell surface staining, but does not impact the respective frequencies of KIR2DL1- and KIR2DS1-expressing cells within the NK repertoire. We can distinguish by flow cytometry NK cell populations expressing the most common KIR2DL1-C245 allotypes from those expressing the most common KIR2DL1-R245 allotypes, and we show that the informative differential binding anti-KIR2DL1/S1 clone 1127B is determined by amino acid residue T154. Although both KIR2DL1-C245 and KIR2DL1-R245 subtypes can be co-expressed in the same cell, NK cells preferentially express one or the other. Cells expressing KIR2DL1-C245 exhibited a lower KIR2DL1 cell surface density and lower missing-self reactivity in comparison to cells expressing KIR2DL1-R245. We found no difference, however, in sensitivity to inhibition or cell surface stability between the two KIR2DL1 isoforms, and both demonstrated similar expansion among NKG2C+ KIR2DL1+ NK cells in HCMV-seropositive healthy individuals. In addition to cell surface density of KIR2DL1, copy number of cognate HLA-C2 hierarchically impacted the effector capacity of both KIR2DL1+ cells and the tolerization of KIR2DS1+ NK cells. HLA-C2 tolerization of KIR2DS1+ NK cells could be overridden, however, by education via co-expressed self-specific inhibitory receptors, such as the heterodimer CD94/NKG2A. Our results demonstrate that effector function of NK cells expressing KIR2DL1 or KIR2DS1 is highly influenced by genetic variability and is calibrated by co-expression of additional NK receptors and cognate HLA-C2 ligands. These findings define the molecular conditions under which NK cells are activated or inhibited, potentially informing selection of donors for adoptive NK therapies.
Introduction
NK cells are key innate lymphocytes, capable of discriminating malignant or infected cells from their healthy counterparts. They are controlled by a multiplicity of combinations of inhibitory and activating receptors, including the killer immunoglobulin-like receptors (KIR). Inhibitory KIR and the heterodimer CD94/NKG2A (NKG2A) recognize self-human leukocyte antigen (HLA) class I molecules, whose presence on healthy cells protects from NK autoaggression, but whose absence on diseased cells permits NK release of cytotoxic granules, resulting in targeted cell killing (1). The same interaction between the inhibitory receptor and its cognate HLA class I ligand in the tissue environment lowers the threshold for functional responses in a process termed “education,” “licensing,” or “tuning” (2), resulting in an NK repertoire in which cells expressing inhibitory receptors for self-MHC exhibit high functional and killing capacity when encountering MHC-deficient, damaged cells, but are inhibited from killing HLA-sufficient healthy cells. In contrast, NK cells expressing inhibitory receptors for which the individual lacks the cognate HLA class I ligand are relatively hyporesponsive, thereby avoiding potential autoreactivity. Activating KIR are less well-characterized, but can recognize classical or non-classical class-I HLA proteins and drive cellular activation or tolerance to self (3–5). Recent studies have demonstrated peptide specificity for various activating KIR (6–8).
The inhibitory KIR2DL1 and activating KIR2DS1 receptors have highly homologous extracellular domains (9), and both bind HLA-C allotypes with Lys80, collectively referred to as group HLA-C2 alleles. Signaling opposite effects upon engagement with the same HLA ligand, KIR2DL1 endows the NK cell with functional competence but inhibits the NK cell when encountering HLA-C2 on neighboring cells, while KIR2DS1 signals activation and cytotoxicity toward the same cell. In yet another aspect of NK education, KIR2DS1-bearing NK cells in individuals homozygous for HLA-C2/C2 are tolerized to the ligand on surrounding cells, thereby avoiding autoreactivity (4, 5).
Various KIR-HLA interactions influence NK education with known impacts on human health (3). Subtype variability for KIR3DL1 and its ligand HLA-Bw4 diversifies NK cell response, with predictable impacts on the outcome of hematopoietic cell transplantation in patients with leukemia, antibody therapy in patients with neuroblastoma, and killing of HIV-infected autologous CD4+ T cells (10–12). KIR2DS1 in hematopoietic cell donors is beneficial to HLA-matched recipients, but only in the absence of homozygosity for HLA-C2, which renders KIR2DS1+ NK cells hyporesponsive (13). Finally, the KIR2DL1 alleles associated with KIR-A, but not KIR-B haplotypes have been recently shown as positively correlated with the likelihood of developing pre-eclampsia (14). The majority of studies have investigated the impacts of single KIR-HLA partnerships in isolation, but, in reality, the majority of NK cells express more than one receptor that can interact with HLA or other ligands; understanding how this diversity impacts outcomes will therefore be a critical step toward fully understanding NK cell interactions and potential function against diseased cells.
To date, 38 different alleles have been described for KIR2DL1 and nine alleles for KIR2DS1 (15). Previous studies have demonstrated that copy number and allelic variation of inhibitory KIR impact frequency of receptor expression in the NK repertoire and density on the cell surface (16–18). Only dimorphism of the amino acid in position 245 [arginine (R) or cysteine (C)] of KIR2DL1 has been shown to have a functional impact, with KIR2DL1-C245 allotypes demonstrating lower capacity for inhibition against cognate HLA (16). However, this study was completed using cell line transfectants; whether the same dimorphism is relevant in primary human NK cells has not been tested. Conducting these studies has been challenging, due to lack of high throughput technologies to identify KIR alleles routinely (19–21) and access to ethnically diverse populations bearing allelic variability in the KIR genes of interest. Lack of antibodies that can distinguish between the highly homologous inhibitory and activating KIR2DL1/S1 isoforms and their allele subtypes further hampered the ability to discern the contribution of each receptor to NK cells bearing both.
We recently developed a method to distinguish KIR2DL alleles and allele groups and genotyped a bank of PBMC from 230 ethnically diverse healthy individuals (22). In the present study, we investigate how KIR2DL1/S1 allelic diversity, allele copy number, and co-expression of the HLA-C2 ligand impact the NK repertoire and education in individuals without a large human cytomegalovirus (HCMV)-associated adaptive NK cell compartment, which can skew the NK repertoire toward a more educated population (17, 23). Using a novel combination of antibodies with known specificities for KIR2DL1 allotypes, we can now compare allotype-specific differences in KIR2DL1 and KIR2DS1 expression and function on cell populations with discrete combinations of receptor allotypes within the same individual. Our results demonstrate that effector function of NK cells expressing KIR2DL1 or KIR2DS1 is highly influenced by genetic variability and co-expression of HLA-C2 ligands, and demonstrate how multiple inhibitory and activating interactions collaborate for NK cell education at the single cell and repertoire levels. Taken together, these findings endorse study of multiple receptor-ligand partnerships in combination to best predict NK cell function. This understanding of the dynamic and hierarchical contributions of allele and copy number variation, activating and inhibitory input may inform donor selection for hematopoietic stem cell transplant, selection of efficient NK cells in cellular therapy for cancer treatment, and prognosis for patients with infectious disease.
Methods
Cell Sources and Preparation
DNA samples were extracted using blood mini kits according to the manufacturer's instructions (Qiagen). Peripheral blood samples were collected from healthy human donors following approval from the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board, and donors provided informed, written consent. Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll centrifugation. Additional PBMC were isolated from buffy coats obtained from healthy volunteer donors via the New York Blood Center (NYBC, http://nybloodcenter.org/). The MSKCC IRB waived the need for additional research consent for anonymous NYBC samples. PBMC were cryopreserved in fetal bovine serum with 10% DMSO. Human cytomegalovirus (HCMV) serostatus was provided by NYBC.
HLA-Class I and KIR2DL1 and KIR2DS1 Allele Typing and Copy Number Calculation
An intermediate resolution Amplification Refractory Mutation System (ARMS) PCR method for distinguishing KIR2DL1 allele groups and some individual alleles was used to identify KIR2DL1 alleles for 230 donors (22). The majority of donor samples used in the present study (200/230) also had allele-level genotyping confirmed by sequence-based typing in a previous study (21, 22). Copy number for KIR was imputed based on known linkage disequilibrium between alleles permitting KIR haplotype assignment (22, 24). Allele typing for HLA-A, B, and C was performed by Histogenetics (Ossining, NY), and KIR ligand designations were assigned using the IPD-HLA database Version 3.34.0 (available online: https://www.ebi.ac.uk/ipd/imgt/hla/).
Target Cells, Effector Cells, and Culture Conditions
The cell lines K562 (ATCC), 721.221, 721.221 transfected with cDNA encoding HLA-Cw2 or HLA-Cw4, collectively referred to as HLA-C2 (kindly provided by Dr. Peter Parham, Stanford University, Stanford, CA), HL60 [HLA-C2/C2, genotype HLA-C*06:02/C*06:02 (25)] (ATCC) and MONO-MAC-1 (HLA-C1/C2) (kindly provided by Dr. Stephen Nimer, University of Miami, Miami, FL) were used as target cells for in vitro assays and were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 1% sodium pyruvate, and 1% 2-mercaptoethanol and incubated at 37°C with 5% CO2. To upregulate HLA class I expression, recombinant human IFN-γ (PeproTech) was added at 1,000 units/mL daily for 72 h (12, 26, 27). Effector cells were cultured in medium, described above, supplemented with human IL-2 (Proleukin, Prometheus) at 200 U/mL and incubated at 37°C with 5% CO2 for 12 to 16 h prior to assaying function.
Phenotypic Analysis by Flow Cytometry
PBMCs (2 × 105 cells per well) were stained 25 min at room temperature with the following antibodies: anti-CD56, anti-CD3, anti-CD158a with or without anti-CD158h and with anti-NKG2C (Table 1). Dead cells were excluded by staining with DAPI (Invitrogen), and NK cells were defined by CD3−CD56dim-gated cells. HLA-C class I expression was evaluated with the anti-HLA-C antibody produced by the DT9 hybridoma (kindly provided by Dr. Mary Carrington, NCI Frederick) with a secondary staining with goat anti-mouse IgG (Ab specifically adsorbed to minimize cross-reactivity). All FACS analyses were performed on an LSR Fortessa (BD Biosciences) and analyzed using FlowJo software (9.8.5, Treestar). For multicolor compensation and gating, unstained, single-color, and FMO controls were used. Donors with NKG2C+ expanded NK cells were excluded from studies, except where specifically noted, given that cytomegalovirus infection can modify NK repertoire, leading to stable imprints, which may confound our interpretations (17).
NK Activation by Flow Cytometric Analysis
CD107a mobilization was used to determine effector cell activation. PBMCs (2–5 × 105 cells per well) were incubated with target cells at a 1:5 ratio in U-bottom 96 well plates in the presence of anti-CD107a. After co-culture, cells were stained with anti-CD56, anti-CD3, anti-CD158a, anti-CD158h, anti-CD158b1/b2/j, anti-CD158i, anti-CD158e1, anti-NKG2A, and anti-NKG2C (Table 1). NK cells exclusively expressing a single inhibitory KIR receptor were evaluated, excluding cells co-expressing other inhibitory receptors that could contribute to NK education (KIR2DL2, KIR2DL3, KIR3DL1, and NKG2A) or activating receptors (NKG2C and KIR2DS2), and cells co-expressing KIR2DL1 and KIR2DS1 could be discriminated using a combination of antibody clones 1127B and 143211 (Table 1), as described in Results. For activation with soluble antibody, PBMC were incubated with a saturating concentration of conjugated 1127B mAb (0.25 μg/mL) or 11PB6 (2.5 μg/mL) mAb prior to co-incubation with target cells.
DNA Constructs and Transfections
KIR2DL3*005 was used as a target for site-directed mutagenesis to determine the critical amino acid residues for 1127B antibody binding. KIR2DL3*005 cDNA was cloned into pcDNA3.1(+) (Invitrogen), and point mutations were introduced by substitution with the Q5 Site-Directed Mutagenesis Kit (New England BioLabs) according to the manufacturer's instructions. DNA probes were designed with the online software NEBaseChanger. All constructs were prepared as per manufacturer's instructions using the QIAprep Spin Miniprep Kit (QIAGEN) and transformed into SURE 2 Supercompetent Cells (Stratagene) following the manufacturer's instructions. cDNA sequences were confirmed by Sanger sequencing (MSKCC DNA Sequencing Core Facility). HEK293 cells (ATCC) were transfected using jetPRiME (Polyplus-Transfection) according to the manufacturer's instructions and cultured in DMEM medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and incubated at 37°C with 5% CO2. Transgene expression was measured by flow cytometry 48 h after transfection.
Statistics
Unpaired or paired Mann-Whitney tests were used to compute comparisons between the phenotypically different NK populations. A test with a p < 0.05 was considered statistically significant.
Results
The Frequency of NK Cells Expressing KIR2DL1 or KIR2DS1 Is Predicted by the Specific Allele and by Copy Number, but Not Influenced by the Presence of Cognate Ligand
Expression of KIR molecules, including KIR2DL1 and KIR2DS1, has been described as “stochastic” or random (28, 29), but more recently, investigations into allele subtype diversity have demonstrated that expression frequencies and cell surface densities of KIR molecules can be predicted by their allele subtype and copy number (10, 18, 30, 31). To determine if the copy number of KIR2DL1 and KIR2DS1 influences the frequency of NK cells expressing the receptors, we stratified 230 healthy human donors based on their KIR2DL1 and KIR2DS1 allele typing and allele copy number and measured the frequency of NK cells expressing KIR2DL1 or KIR2DS1 (Figures 1A,B). Consistent with published findings (20), we found the most common alleles for KIR2DL1 were *002, *003, and *004; for KIR2DS1, donors most frequently carried the *002 allele. In general, we found that donors exhibiting two copies of KIR2DL1 showed higher frequencies of KIR2DL1+ NK cells in their repertoires. Among KIR2DL1 allotypes, *004 was expressed on the lowest number of NK cells, but KIR2DL1*001, *002 and *006 were similarly expressed. Compared with donors exhibiting a single copy of the KIR2DL1*003 allele, the frequency of KIR2DL1+ NK cells approximately doubled in donors having two copies of the same allele (p < 0.0001) (Figure 1A). In donors with two different KIR2DL1 alleles, the frequency of KIR2DL1+ NK cells reflected the additive frequencies of each allele alone. Similarly, the frequency of KIR2DS1+ NK cells was lower in donors exhibiting one allele copy compared with those encoding two; this was especially pronounced when we limited our analysis to donors encoding one vs. two allele copies of KIR2DS1*002, the most common KIR2DS1 allele (p < 0.01) (Figure 1B).
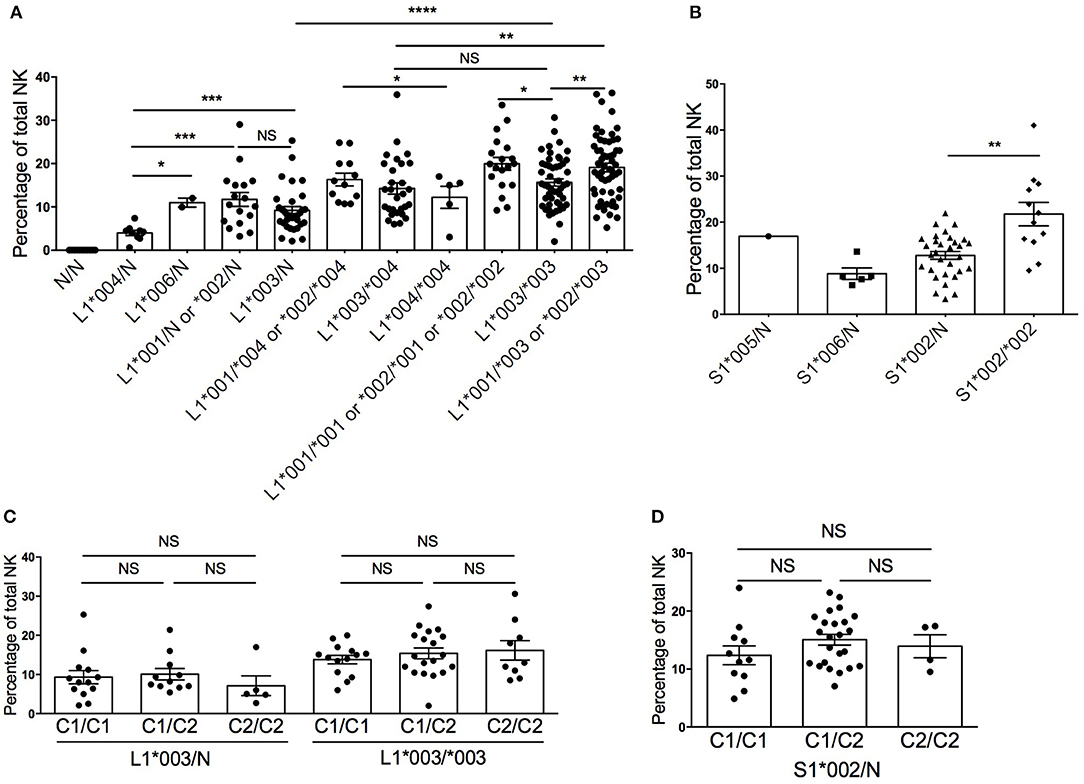
Figure 1. Diversity in frequency of KIR2DL1+ and KIR2DS1+ NK cells according to allele typing in 230 healthy blood donors. (A) KIR2DL1 allele/copy number and frequency of KIR2DL1+ NK cells. (B) KIR2DS1 allele and copy number and frequency of KIR2DS1+ NK cells. (C) HLA-C KIR ligands and frequency of KIR2DL1+ NK cells. (D) HLA-C KIR ligands and frequency of KIR2DS1+ NK cells. N represents absence of a KIR2DL1 allele on the KIR haplotype. Unpaired Mann-Whitney test was used. Symbols represent individual samples (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Previous studies have reported that co-inheritance of HLA ligands can impact the frequency of NK cells exhibiting cognate KIR among human repertoires (18, 32). Our finding that KIR2DL1+ and KIR2DS1+ NK cell frequencies are impacted by allele subtype, however, suggests an alternate explanation for these findings. Donors exhibiting a large HCMV-associated adaptive NKG2Cbright NK cell population were excluded, as these cells have been reported to have modulated functional capacity (33, 34) and skew the NK repertoire toward an educated phenotype (17, 23). Limiting our analysis to the most common alleles for KIR2DL1 (*003) and KIR2DS1 (*002), and stratifying based on donor copy number for HLA-C2, we observed no significant changes in the frequency of KIR2DL1*003+ or KIR2DS1*002+ NK cells (Figures 1C,D). Hence, the frequency of KIR2DL1+ or KIR2DS1+ NK cells among human NK cell repertoires is driven by allele subtype and gene copy number, but not by cognate HLA-C2 ligand.
Cell Surface Density of KIR2DL1 Is Diminished in the Presence of the Cognate HLA-C2 Ligand and Influenced by Allelic Variation, but Unchanged by Copy Number
A previous study has reported a reduction in cell surface density of KIR when cognate ligands are present in a donor's genome (18). To determine whether this remains true even after KIR2DL1 and KIR2DS1 allelic variation are considered, we assessed the median fluorescent intensity (MFI) of KIR2DL1*003, KIR2DL1*002 and KIR2DL1*004 as a measure of receptor cell surface density, stratifying based on the presence and copy number of cognate HLA-C2 molecules. The MFI of KIR2DL1 did not differ based on KIR allele copy number, suggesting that only one copy of each gene is active in a given NK cell, and consistent with previous studies that have demonstrated dominant expression of single KIR allotype in individual NK cells (30). However, in comparison to donors who lack the HLA-C2 ligand (HLA-C1/C1), KIR2DL1*003 MFI was lower on NK cells from individuals expressing one or two copies of HLA-C2 (p < 0.001 and p < 0.01, respectively) (Figure 2A).
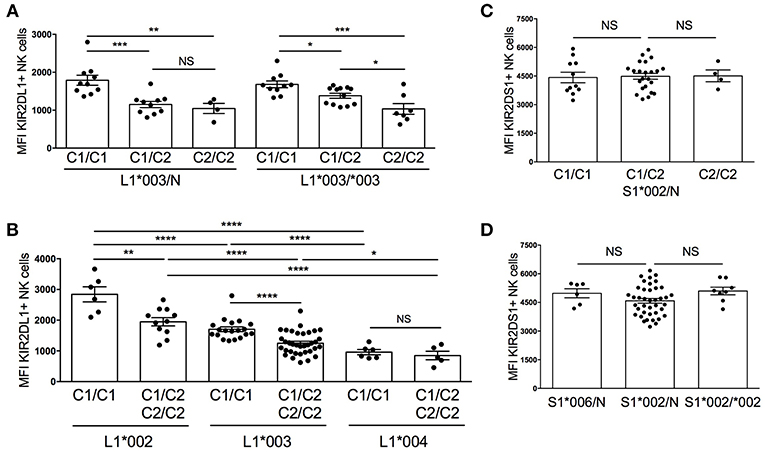
Figure 2. Diversity in cell surface expression of KIR2DL1 and KIR2DS1 alleles. KIR2DL1 and KIR2DS1 cell surface expression was measured by flow cytometry on NK cells from healthy blood donors. (A) KIR2DL1 cell surface expression segregated by KIR allele copy number and HLA-C KIR ligand. (B) Diversity of cell surface expression associated with KIR2DL1 allele typing. (C) KIR2DS1 cell surface expression segregated by KIR ligand. (D) KIR2DS1 allele, copy number, and KIR2DS1 cell surface expression. Two hundred thirty individuals were evaluated. Unpaired Mann-Whitney test was used. Symbols represent individual samples (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Since MFI did not change with KIR2DL1 copy number, we next grouped donors according to KIR2DL1 allele typing and stratified analysis based on donor HLA-C2 copy number. Like for KIR2DL1*003, we found that expression of HLA-C2 ligands reduced the MFI of KIR2DL1*002, measured by the 14321 antibody; however, KIR2DL1*004 was found to exhibit low cell surface density that was not significantly influenced by co-expression of cognate HLA-C2 (Figure 2B). In individuals lacking HLA-C2 ligand (HLA-C1/C1), KIR2DL1*002 expression exhibited the highest MFI in comparison to *003 and *004 (p < 0.0001) and *003 was more densely expressed than *004 (p < 0.0001). Similar results were observed between KIR2DL1 alleles using the anti-CD158a/h EB6B mAb (Supplemental Figure 1), which binds a different epitope than the 14321 antibody, suggesting that expression density differs between isoforms and is not due to differences in the availability of epitopes or binding affinities for each antibody. For KIR2DS1, neither the presence of the cognate ligand HLA-C2, allele copy number nor specific allele impacted MFI (Figures 2C,D). Altogether, these findings suggest that expression of cognate HLA-C2 ligand impacts the availability of high-expressing KIR2DL1 allotypes, with no apparent impact on surface density of the low-expression allotype KIR2DL1*004 or of KIR2DS1.
Antibody Clone 1127B Binds KIR2DL1 Allotypes Exhibiting T154
The anti-KIR2DS1 monoclonal antibody (mAb) clone 1127B is described by the manufacturer to identify expression of KIR2DS1 and some undefined allotypes of KIR2DL1. We therefore sought to identify the KIR2DL1 allotypes that bind clone 1127B (Figure 3A). The gating of the different KIR2DL1 and KIR2DS1 subpopulations is described in Supplemental Figure 2A. We performed phenotyping on PBMC from individuals with different KIR2DL1 alleles and found that four KIR2DL1 allotypes were recognized by clones 1127B and 143211 (*004, *007, *010, *011), and five allotypes were recognized only by 143211 (*001, *002, *003, *006, 008) and not by the 1127B mAb. We noted that all the KIR2DL1 allotypes recognized by the 1127B mAb are KIR2DL1 allotypes with Cys245 (C245), except for KIR2DL1*010. Furthermore, all the KIR2DL1 allotypes not recognized by the 1127B mAb are allotypes with Arg245 (R245), except for KIR2DL1*006 (Figure 3B). KIR2DL1*004 is the most common of the KIR2DL1-C245 allotypes, and compared to KIR2DL1-R245 allotypes, surface expression of KIR2DL1*004 is known to be lower (18). We also tested the 1127B mAb against KIR2DL3*005, previously found to be bound by the anti-KIR2DL1/S1 EB6B mAb (35), and found no specific binding by 1127B (Supplemental Figure 2B).
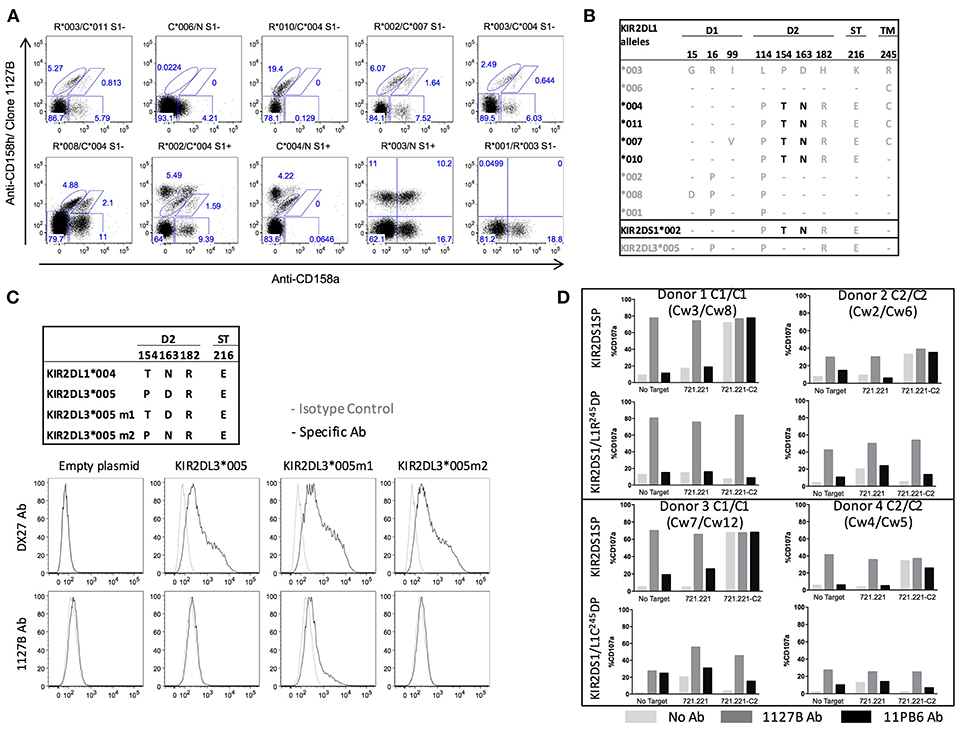
Figure 3. Differential binding and agonist effects of antibody 1127B to KIR2DL1 and KIR2DS1 allotypes. (A) Differential staining of KIR2DL1 allotypes by anti-CD158a and clone 1127B monoclonal antibodies. (B) Alignment of the amino acid sequences of KIR2DL1 allotypic variants. Dashes indicate identity with the consensus KIR2DL1*003 allotype. Structural domains are indicated: Ig like domains (D1 and D2), stem domain (ST) and transmembrane domain (TM). Alleles in black encode receptor variants recognized by both anti-CD158a and clone 1127B antibody staining. Candidate amino acid residues involved in recognition by clone 1127B are indicated in black. (C) Binding specificities by flow cytometry of anti-CD158b1/b2/j (DX27) and clone 1127B on HEK293 cells transfected with cDNA for KIR2DL3*005 or mutation variant exhibiting amino acid substitutions in D2, as indicated. (D) Cytotoxic response, as measured by CD107a mobilization, in KIR2DL1+KIR2DS1+ and KIR2DS1sp cells following co-culture with 721.221 and 721.221-HLA-Cw4 target cells without additional antibody or in the presence of 1127B or 11PB6 antibody. Results are shown from four different donors, representative of ten tested.
Aligning the protein sequences encoded by the different KIR2DL1 alleles, with KIR2DL3*005 and KIR2DS1*002 for comparison, we identified two amino acid residues that segregate the alleles based on receptor binding by the 1127B mAb: T154 and N163 (Figure 3B). Using site-directed mutagenesis, we then altered KIR2DL3*005 to create a knock-in point mutation at either site to introduce the amino acid encoded in the group of alleles bound by 1127B, and tested antibody binding to HEK293 cells transfected with either mutant (Figure 3C). Neither the anti-KIR2DL2/L3/S2 mAb (clone DX27) nor the 1127B mAb bound untransfected cells, but cells transfected with either the KIR2DL3*005P→T154 or KIR2DL3*005D→N163 were bound by DX27. Only cells transfected with KIR2DL3*005P→T154 were bound by 1127B, indicating that it is the T154 amino acid that enables recognition by the 1127B mAb. As relatively uncommon alleles, KIR2DL1*006 is notable for being the only KIR2DL1-C245 allele member lacking T154 and N163, and *010 is notable for being the only KIR2DL1-R245 allele member with T154 and N163, explaining the divergence of their antibody-binding specificities.
KIR2DS1 Signaling Can Override KIR2DL1 Inhibition
That the 1127B antibody binds to only a subset of KIR2DL1 (mostly KIR2DL1-C245) but not to others (mostly KIR2DL1-R245), as well-binding to KIR2DS1, provides an opportunity to study the features of each population and the combined impacts of the same HLA-C2 signal on cells co-expressing KIR2DL1 and KIR2DS1 by flow cytometry (Supplemental Figure 3A). We selected donors positive for both KIR2DL1 (C245 or R245) and KIR2DS1 from HLA-C1/C1 or HLA-C2/C2 individuals and challenged them with the HLA-negative target cell 721.221 or with a transfected derivative that expresses HLA-C2 (721.221-C2). We additionally included during stimulation either the 1127B antibody, or 11PB6, which universally binds KIR2DL1 and KIR2DS1 (Figure 3D).
We found that binding of the soluble 1127B mAb was able to agonize NK cells exclusively expressing KIR2DS1 (KIR2DS1sp) from HLA-C2-negative donors (HLA-C1/C1), activating them to a level equivalent to that induced by 721.221 target cells expressing the HLA-C2 ligand (Figure 3D). The agonist effect appears dose-dependent, reaching saturation at 200 ng/mL (Supplemental Figure 4). Tolerance was maintained in HLA-C2/C2 individuals, however, and KIR2DS1sp NK cells from these donors exhibited blunted activation upon binding to either the 1127B mAb or the 721.221-C2 target cell (Figure 3D). Irrespective of their HLA-C background, cells expressing both KIR2DS1 and KIR2DL1-R245 were not activated upon co-culture with the 721.221-HLA-C2 target, indicating that the inhibitory signal provided by KIR2DL1-R245 could override target cell-induced activation signaling via KIR2DS1 upon binding to HLA-C2 in trans. Despite this, addition of soluble 1127B mAb, which agonizes KIR2DS1 but not KIR2DL1-R245, elicited activation of the double positive NK cells irrespective of donor HLA-C background, indicating that the activating signal driven by soluble 1127B is stronger than the inhibitory signal provided via KIR2DL1-R245 (Figure 3D). Interestingly, for KIR2DS1/L1-R245 double positive cells, while 1127B stimulation could overcome KIR2DL1-mediated inhibition by an HLA-C2-bearing target, the maximum antibody-induced response was still lower than that seen in the uneducated counterpart from HLA-C1/C1 cells, indicating that some measure of tolerance is maintained in KIR2DS1+ cells taken from an HLA-C2/C2 environment.
Using cells expressing both KIR2DS1 and KIR2DL1-C245, we determined that like for KIR2DS1, we could observe an agonist effect on KIR2DL1-C245 receptor upon binding of the 1127B antibody, due to a diminution of overall activation in the KIR2DS1+KIR2DL1-C245+ in comparison to the NK KIR2DS1sp cells (Figure 3D). This opened the opportunity to test the relative importance of inhibitory and activating signals driven by the same KIR ligand in the same NK cell. Addition of the 1127B antibody to HLA-C1/C1 NK cells positive for both KIR2DS1 and KIR2DL1-C245 resulted in overall activation, although the maximum activation was lower in comparison to antibody-stimulated KIR2DS1sp cells from the same individual. The agonist effects of the 1127B mAb were specific to KIR2DS1 and KIR2DL1-C245, as NK cells negative for KIR2DS1 and KIR2DL1 did not demonstrate activation by the 1127B mAb (Supplemental Figure 3B).
NK Cells Educated by KIR2DL1-C245 Exhibit Lower “Missing Self” Reactivity Than KIR2DL1-R245 NK Cells
Since clone 1127B binds only a subset of KIR2DL1 allotypes, we could identify and separately study NK cells expressing different KIR2DL1 alleles within the same individual by counterstaining with the pan-KIR2DL1 antibody 143211 (Figure 3A). This affords the opportunity to compare the function of NK cells educated in the same HLA environment by different KIR2DL1 alleles. Importantly, all staining for KIR2DL1, including with the 1127B antibody, was completed after stimulation; therefore, the antibodies could not impact the reactivity of NK cells.
We first analyzed the frequency of the different subpopulations of NK cells expressing KIR2DL1 in individuals co-expressing either KIR2DL1*004 or *007 (both KIR2DL1-C245 allotypes) and a KIR2DL1-R245 allotype (Figure 4A). The percentage of NK cells that express KIR2DL1*004 or *007 was lower in comparison to the percentage of cells expressing KIR2DL1-R245 (p < 0.0001), confirming our earlier results observed in individuals homozygous and hemizygous for KIR2DL1 (Figure 1A). Co-expression of two KIR2DL1 allotypes was uncommon regardless of KIR2DL1-C245 or -R245 categorization and was observed on an average of 9.11 ± 0.64% of all KIR2DL1+ NK cells.
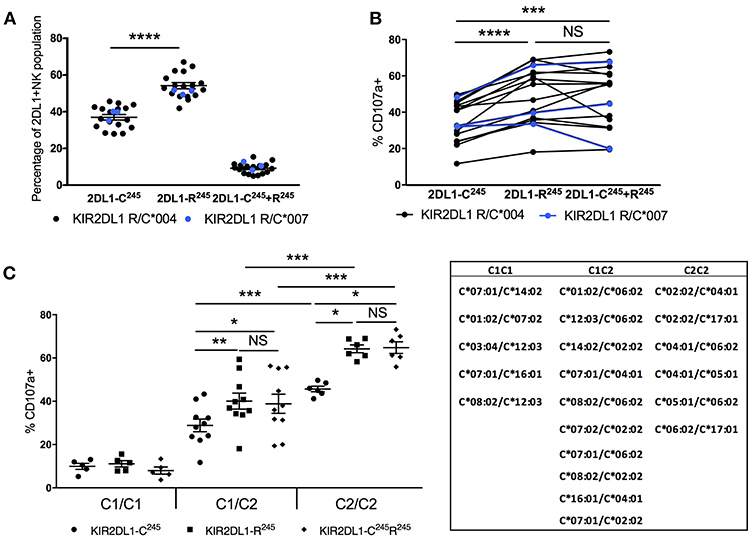
Figure 4. Impact of KIR2DL1 allelic diversity and HLA-C2 copy number on NK education. Cytotoxic response, measured by CD107a mobilization following 5-h co-culture with K562 target cells, was evaluated on NK cells single positive for KIR2DL1 and negative for other inhibitory KIR and NKG2A. (A) Frequencies of NK populations with single positive or double positive for KIR2DL1-C245 and KIR2DL1-R245 allotypes among individuals with both. KIR2DL1-C245 allotypes *004 and *007 display similar expression frequencies. (B) Hierarchical differences in cytotoxic response among NK populations single positive for KIR2DL1-C245 or KIR2DL1-R245 allotypes or double positive for both. (C) Impact of HLA-C2 copy number on the education of NK cells expressing KIR2DL1-R245 and KIR2DL1-C245 allotypes. Donor HLA-C allotypes are listed. Paired and unpaired Mann-Whitney tests were used. Symbols represent individual samples (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
We compared the functional capacity of NK cells expressing different KIR2DL1 allotypes by measuring degranulation response (CD107a) in response to challenge with the HLA-negative target cell line K562 for 5 h. Using donors that carried both KIR2DL1-R245 and –C245 alleles, we could examine within the same donor, functional differences between NK populations expressing each KIR2DL1 receptor allotype (Figure 4B). We analyzed NK cells singly expressing KIR2DL1, excluding cells co-expressing other inhibitory receptors that could contribute to NK education. We found that NK cells expressing KIR2DL1-C245 (KIR2DL1*004 or *007) allotypes displayed a lower frequency of degranulation in comparison to the NK cells expressing KIR2DL1-R245 allotypes within the same donor (p < 0.0001). NK cells co-expressing both alleles had a similar pattern of degranulation to cells expressing only KIR2DL1-R245 allotypes, suggesting a dominant program of education by KIR2DL1-R245.
HLA-C2 Copy Number Calibrates the Education of KIR2DL1-Expressing NK Cells
We next analyzed the same functional data, grouping donors based on genomic HLA-C2 copy number (Figure 4C). Compared with donors exhibiting two copies of HLA-C2 alleles, KIR2DL1+ NK cells from donors having only one copy of HLA-C2 exhibited lower degranulation in response to K562 target cells. This differential function was observed for the NK cells expressing KIR2DL1-C245 (p < 0.001) or KIR2DL1-R245 (p < 0.001) or both (p < 0.001) and suggests that the education of KIR2DL1+ NK cells is instructed by the quantity of available ligand, similar to the partnership of KIR3DL1 and HLA-Bw4 (11).
KIR2DL1-C245 and KIR2DL1-R245 Receptors Are Similarly Inhibited by HLA-C2 and Not Internalized in the Presence of Their Ligand
Using cells transfected to express either the KIR2DL1-C245 or -R245 alleles, a previous study reported that KIR2DL1 isoforms conveyed different sensitivities to inhibition potentially due to differences in receptor internalization after 24 h incubation with HLA class I expressing targets (16). To study the relevance of this finding in primary human NK cells, we performed a similar 24 h target cell co-incubation, comparing degranulation of KIR2DL1+ NK cells from 12 individuals co-expressing the KIR2DL1-C245 allele KIR2DL1*004 and a KIR2DL1-R245 allele, including individuals with or without the cognate HLA-C2 ligand. Target cells included the 721.221 cell line transfected with an HLA-C2 allele and the HLA-C2+ acute myelogenous leukemia cell lines HL-60 (HLA-C2/C2) and MONO-MAC-1 (HLA-C1/C2). HLA expression is high without additional stimulation of the HL-60 cell line, and exposure to IFN-γ induced upregulation of HLA-C on the MONO-MAC-1 leukemia cell lines (Figure 5A). As expected, and reflecting their lack of education, KIR2DL1+ NK cells from HLA-C1/C1 donors were hyporesponsive compared to those from HLA-C2+ donors against 721.221 target cells and MONO-MAC-1 cells unstimulated with IFN-γ. In contrast, KIR2DL1+ NK cells from HLA-C2+ donors were responsive against 721.221 and unstimulated MONO-MAC-1 cells, and inhibited by HLA-C2+ target cells, including the 721.221-C2 transfectant, HL-60 cells and IFN-γ-stimulated MONO-MAC-1 cells to differing degrees (Figure 5B). Using the 24 h co-incubation condition and in contrast to the previous study (16), we observed no difference in degree of inhibition of NK cells expressing the KIR2DL1-C245 allele KIR2DL1*004 compared to those expressing a KIR2DL1-R245 allele (Figure 5B). Differences in degranulation between NK cells expressing the different KIR2DL1 subtypes also decreased after 24 h co-incubation, in contrast to what we observe after the more conventional 5 h co-culture condition, demonstrating that less educated NK cells can achieve the same degree of activation as more educated cells given longer stimulation (Supplemental Figures 5A,B).
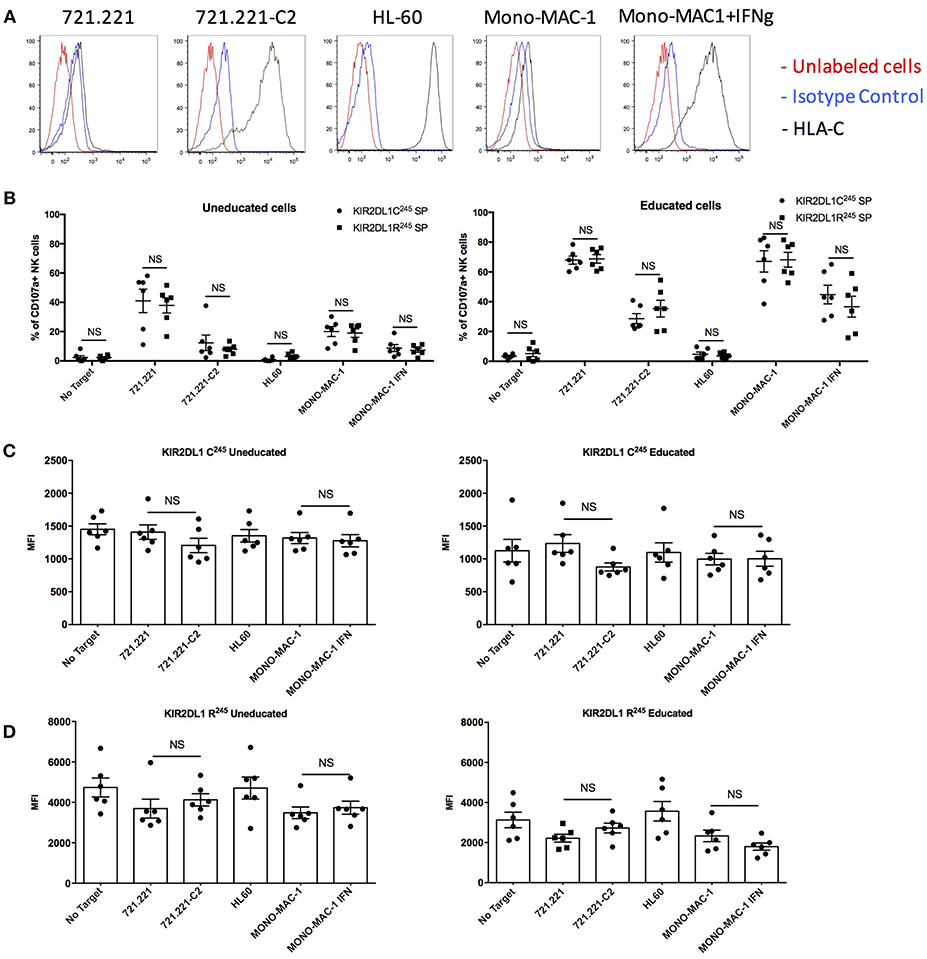
Figure 5. Inhibitability of the KIR2DL1 allotypes following 24 h of co-culture with HLA-C2+ target cells. (A) Cell surface expression of HLA-C on the HLA-class I negative 721.221 target cell, 721.221 transfected with HLA-Cw2 (721.221-C2), and the HLA-C2-bearing AML cell lines HL-60 and Mono-MAC-1 with or without treatment with IFN-γ. (B) Cytotoxic response and inhibition among educated and uneducated NK populations expressing different KIR2DL1 alleles to 721.221 and the HLA-C2 ligand-expressing target cells, as indicated. (C) Surface MFI of KIR2DL1-C245 receptors following incubation with ligand-expressing target cell. (D) Surface MFI of KIR2DL1-R245 receptors following incubation with ligand-expressing target cells. Unpaired and paired Mann-Whitney tests were used. Symbols represent individual samples (mean ± SEM).
We measured the cell surface expression of the KIR2DL1-C245 KIR2DL1*004 allele and of the KIR2DL1-R245 alleles after 24 h of co-culture in the presence or absence of HLA-C2-expressing target cells. The results showed equivalent cell surface expression with or without target cells expressing the HLA-C2 ligand, for cells expressing KIR2DL1-C245 KIR2DL1*004 (Figure 5C and Supplemental Figure 5C) or a KIR2DL1-R245 allele (Figure 5D and Supplemental Figure 5C). These results contrast with a previous study using an NK cell line transfected with each of the two KIR2DL1 isoforms (16), and suggest that both are, in fact, stably present on the cell surface on primary NK cells.
KIR2DS1-Mediated Education Is Titrated by HLA-C2 Copy Number and by NKG2A
The activating KIR2DS1 receptor also impacts the education of NK cells (4, 5). In contrast to inhibitory KIR2DL1, which programs higher education and responsiveness in the presence of autologous ligand HLA-C2, KIR2DS1 renders the cell more tolerant when it is expressed in the presence of the same ligand. We compared the reactivity of KIR2DS1+ NK cells against 721.221 target cells transfected with the activating ligand HLA-C2, stratifying our analysis based on the donor's HLA-C2 copy number (Figure 6A and Supplemental Figure 6A). In HLA-C2/C2 individuals, NK cells exclusively expressing KIR2DS1 were the least responsive cells, consistent with tolerization of this population (4, 5). In contrast, the most responsive KIR2DS1+ NK cells were from donors lacking HLA-C2 (HLA-C1/C1), while HLA-C1/C2 individuals exhibited intermediate reactivity compared to both groups, demonstrating that tolerance or “negative education” is titratable by environmental HLA.
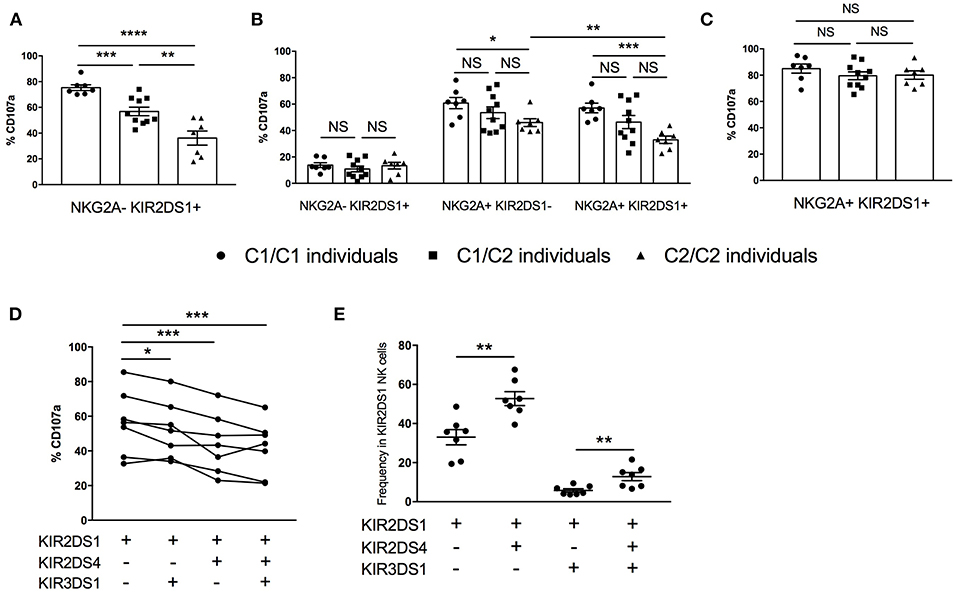
Figure 6. KIR ligand titration of KIR2DS1+ NK tolerance, integration of NK education via NKG2A, and co-expression of other activating KIR. (A) Cytotoxic response among KIR2DS1sp cells following activation by 721.221-Cw4 cells, segregated by autologous HLA-C KIR ligand genotype. (B) Response capacity of KIR2DS1+ and/or NKG2A+ NK cells to 721.221 target cells, segregated by autologous HLA-C KIR ligand genotype. (C) Cytotoxic response of KIR2DS1+NKG2A+ to 721.221-Cw4, segregated by autologous HLA-C KIR ligand genotype. (D) Response capacity of KIR2DS1+ cells co-expressing KIR2DS4 and/or KIR3DS1 to 721.221 target cells (E) Relative frequencies of KIR2DS1 NK cells co-expressing KIR2DS4 and/or KIR3DS1. Unpaired and paired Mann-Whitney test were used. Symbols represent individual samples (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Studies examining the education of NK cells based on expression of a single educating receptor are vital to our understanding of the contribution of individual receptor-ligand pairs to NK education, but the majority of NK cells in the human repertoire co-express multiple KIR and/or NKG2A (17, 36). To better understand the impact of KIR2DS1 and HLA-C2 on NK cell function in a more physiologically relevant context, we analyzed NK cells co-expressing the educating activating receptor KIR2DS1 and inhibitory NKG2A. Against the HLA-negative 721.221 target cell line, NKG2A+ single-positive NK cells from HLA-C2/C2 donors trended toward less degranulation than those collected from HLA-C1/C2 or HLA-C1/C1 donors (Figure 6B). Among the same HLA-C2/C2 individuals, NK cells co-expressing NKG2A, and KIR2DS1 were even less responsive than NKG2A single positive cells, demonstrating that the tolerization of KIR2DS1 by HLA-C2/C2 can diminish NK cell education by NKG2A (p < 0.01) (4).
When the same cells were used to compare reactivity against 721.221-C2 cells, we were surprised to find that where KIR2DS1+ NK cells were previously demonstrated to be tolerized (e.g., in an HLA-C2/C2 individual), we now found equal and strong reactivity among all KIR2DS1+NKG2A+ NK cells, irrespective of the donor HLA-C background, suggesting that education through NKG2A overrides the tolerization driven by KIR2DS1 in HLA-C2/C2 donors (Figure 6C and Supplemental Figure 6B).
Activating KIR molecules other than KIR2DS1 are not known to contribute to NK education, either in a potentiating or tolerizing fashion, but they have not previously been examined systematically. Using KIR2DS1-mediated NK cytotoxic response to HLA-C2 on the target cell for comparison, we examined how, within the same individuals, co-expression with KIR2DS1 of the activating receptors KIR2DS4 or KIR3DS1 on the NK cell impacts responsiveness to the same HLA-C2+ target. Interestingly, co-expression of either KIR2DS4 or KIR3DS1 reduced the likelihood of a KIR2DS1-expressing NK cell to respond, as measured by CD107a staining (Figure 6D). Importantly, there was no reduction in MFI of KIR2DS1 in the setting of a co-expressed activating receptor (data not shown). Notably, contrary to the product rule for co-expression of additional KIR (37), KIR2DS1+ NK cells were unexpectedly more likely to co-express KIR2DS4 than not (Figure 6E). This was not the case, however, for KIR3DS1, which was rarely co-expressed with KIR2DS1. In comparison, there was a higher frequency of cells expressing all three activating KIR, suggesting the preferential co-expression of KIR2DS4+ if KIR2DS1 is expressed, even with KIR3DS1. Therefore, in individuals with KIR2DS1 and KIR2DS4 in their genotype, KIR2DS1+ NK cells are more likely to co-express KIR2DS4, potentially resulting in dampened repertoire response to HLA-C2 activation.
KIR2DL1+ NKG2C+ NK Expansion in HCMV+ Individuals can be Mono- or Bi-allelic
Previous studies have demonstrated expansion of NKG2C-expressing NK cells among some individuals previously exposed to HCMV; in particular, these “adaptive NK” cells most often co-express inhibitory KIR2DL molecules educated by “self” HLA-C molecules (17). Since we found that KIR2DL1-C245 and KIR2DL1-R245 allotypes differently educate NK cells for missing self-reactivity, we explored whether that the magnitude of NK education also impacts selective expansion of NKG2C+ NK cells.
We analyzed seven different individuals with an NKG2C+ population and having at least one copy of HLA-C2 and co-inheriting both KIR2DL1-C245 and KIR2DL1-R245 allotypes to determine whether higher education and stronger “missing self” reactivity was preferentially associated with expansion of NKG2C after HCMV infection (Figure 7A). Within seven samples, we found four with a co-dominant KIR2DL1 allotype NK expansion, two with a preferential KIR2DL1-R245 allotype expansion and one with a preferential KIR2DL1-C245 expansion (Figure 7A). The same observation was made in NKG2C+ CD8+ T cells, but the particular isoform expansion was not necessarily the same in T and NK cells from the same donor (Figure 7A). In HCMV-seropositive individuals, despite the preferential expression of self-specific educating KIR in the adaptive NK cell compartment, there is no apparent preferential expression of the KIR2DL1 allele conferring higher education.
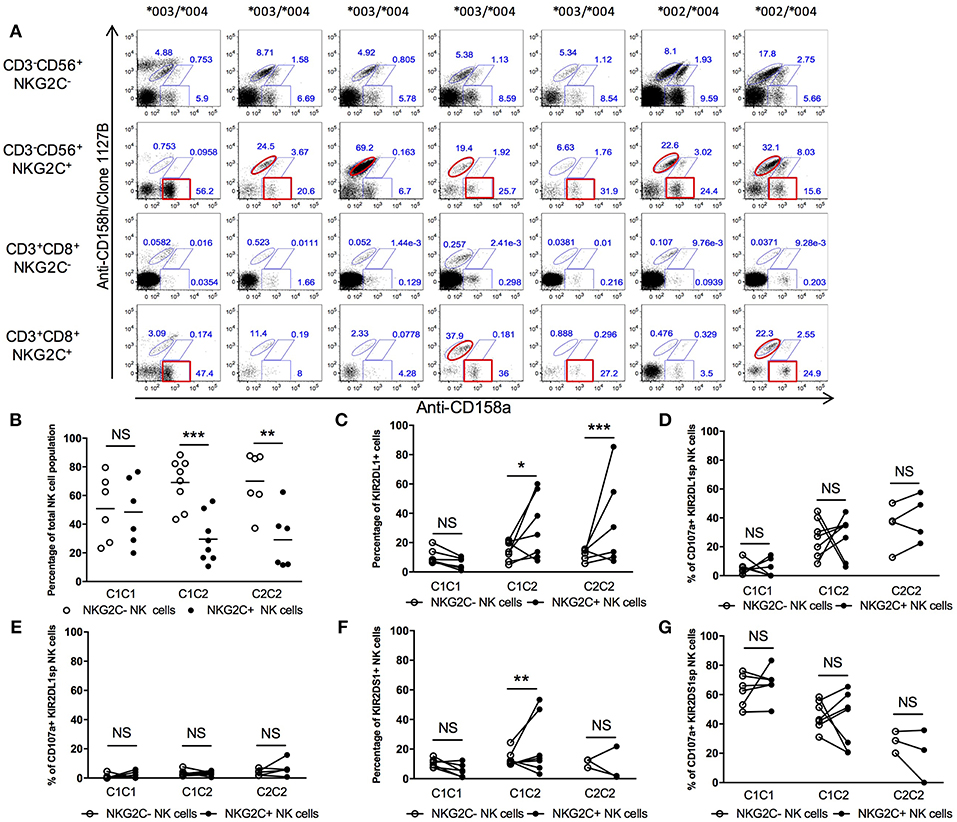
Figure 7. KIR2DL1-S1 expression and function in NKG2C+ compartments. (A) Differential KIR2DL1 allele frequencies in NKG2C+ NK cells. (B) Frequency of NKG2C− and NKG2C+ NK compartments in HCMV+ healthy donors. (C) Frequency of KIR2DL1+ cells within the NKG2C− and NKG2C+ NK cell compartments. (D) Responsiveness of KIR2DL1SP NK cells in NKG2C− and NKG2C+ compartment, against 721.221. (E) Inhibition of KIR2DL1sp cells in NKG2C− and NKG2C+ compartment, with 721.221-Cw4. (F) Frequency of KIR2DS1+ cells in NKG2C− and NKG2C+ NK populations. (G) Cytotoxic responsiveness of KIR2DS1sp with 721.221-Cw4 cells. Unpaired Mann-Whitney test was used. Symbols represent individual samples (mean ± SEM). *p < 0.05, **p < 0.01, and ***p < 0.001.
NKG2C Expression Does Not Modulate the Function of KIR2DL1+ or KIR2DS1+ NK Cells in a Predictive Way
Recent studies characterized phenotypic and functional differences between adaptive and non-adaptive NK cells (17, 34, 38, 39). We examined if the function of KIR2DL1+ and KIR2DS1+ cells differed between the adaptive NK population compared to the non-adaptive NK population.
Cells from 19 HCMV-seropositive individuals, all of whom displayed an adaptive NKG2C+ NK population, were co-cultured with 721.221 or with the 721.221-C2 transfectant (Figure 7B). It should be noted that while in this small cohort, HLA-C2+ donors displayed an overall lower NKG2C+ NK cell population in comparison to HLA-C1 donors, this was not confirmed in a larger cohort (data not shown). We analyzed the frequency of KIR2DL1+ NK cells among the NKG2C+ NK population and, consistent with their educated phenotype, found KIR2DL1+ NKG2C+ NK cells only in individuals having at least one HLA-C2 ligand for KIR2DL1 (Figure 7C). We found that KIR2DL1sp cells had on average the same degranulation against 721.221 in comparison to those co-expressing NKG2C+ but with important variations within individuals (Figure 7D). We did not observe any difference in inhibition by 721.221-C2 between adaptive and non-adaptive KIR2DL1+ NK cells (Figure 7E). We also only observed the KIR2DS1+NKG2C+ NK cell population in HLA-C2+ donors (Figure 7F), the first evidence that the presence of ligand can influence expansion of activating KIR+ cells within the adaptive NK repertoire of HCMV-seropositive individuals and suggesting possible auto-reactivity of KIR2DS1+ NK cells partially driven by HLA-C2. In co-culture with 721.221-C2, KIR2DS1+NKG2C+ NK cells did not behave differently in average from KIR2DS1sp cells but with important variations within individuals (Figure 7G). We conclude that KIR2DS1+ NK cell expansion in HCMV-seropositive individuals is only observed in HLA-C2 individuals, and NKG2C does not impact NK cell function in a predictive way.
Discussion
KIR2DL1 and KIR2DS1 allelic variation and copy number and the availability of their ligand, HLA-C2, combine to impact the phenotype and function of primary human NK cells. Compared with KIR2DL1-C245, KIR2DL1-R245 allotypes are expressed at higher cell surface densities and convey greater missing self-reactivity when educated by HLA-C2 group ligands, consistent with previous findings (14). Despite this, the KIR2DL1 isoforms similarly signal inhibition of NK cells, including those co-expressing the activating KIR2DS1 receptor. While KIR2DL1 is expressed on the expanded NKG2C+ population in HCMV-seropositive, HLA-C2+ donors, neither KIR2DL1-C245 nor KIR2DL1-R245 isoform exhibits dominance among the NKG2C+ adaptive NK population. Together, our findings suggest that KIR2DL1 isoforms co-evolved with HLA-C2 to enable differential detection of and response to target cells missing self HLA, while maintaining a dominant sensitivity for inhibition to avoid autoimmunity.
Using a combination of commercially available monoclonal antibodies including the 1127B clone, which we show to be specific for T154 present in KIR2DL1-C245 allotypes (except for *006), but not KIR2DL1-R245 allotypes (except for *010), we compared function between NK cells expressing KIR2DL1-R245 and cells expressing KIR2DL1-C245 in heterozygous donors. Primary, unmodified NK cells, educated in a shared HLA-C2+ environment, offered the opportunity to directly compare NK cell education imparted by the different allotypes. We found that KIR2DL1 alleles are expressed at predictable cell surface densities and on predictable frequencies of NK cells and that KIR2DL1 is expressed in a primarily monoallelic fashion per cell. Co-expression of both KIR2DL1 isoforms was found on just ~9% of the KIR2DL1+ NK cell population, consistent with the product rule for receptor expression and co-expression (37). Moreover, our results confirm a linear correlation between KIR2DL1 and KIR2DS1 allele copy number and the frequency of NK cells that express these receptors (30), where specific KIR2DL1 allele subtypes are associated with low (KIR2DL1-C245) or high (KIR2DL1-R245) cell surface expression and frequency within the NK repertoire (18, 40). Therefore, allele variation is a major determinant of the likelihood of KIR2DL1 expression on any given NK cell and the extent to which KIR2DL1 is expressed on its cell surface.
We observed an agonistic effect of soluble 1127B mAb on KIR2DL1 and KIR2DS1 receptors. This result is unique to the 1127B mAb and was not recapitulated by 11PB6, which instead binds and blocks KIR2DL1 and KIR2DS1. Previous reports have demonstrated the importance of the D1 domain for epitope-specific recognition of the HLA-C ligand by KIR2D receptors (9, 41). Here we show threonine at position 154 in D2 is critical for the binding of 1127B antibody to the D2 domain. 1127B binding of KIR2DS1 is sufficient to mimic the binding of HLA-C2, maintaining the hyporesponsiveness induced by the expression of HLA-C2 by the donor. This finding is in apparent contrast to a previous study, which showed that KIR2DS1+ cells could be induced for responsiveness by 11PB6-mediated crosslinking, with no hyporesponsiveness among HLA-C2/C2 donors compared to HLA-C1/C1 donors (4). Using the agonist effect of the 11217B mAb, we observed that activation through KIR2DS1 can override KIR2DL1 on NK cells co-expressing KIR2DS1 and KIR2DL1-R245 upon challenge with a HLA-C2+ target, demonstrating how strong activating signaling can overcome inhibitory signaling. The 1127B mAb appears to have some agonistic inhibitory properties upon binding KIR2DL1-C245, as we observe an attenuated response in HLA-C1/C1 individuals to 1127B stimulation of KIR2DS1+KIR2DL1-C245+ NK cells in comparison to KIR2DS1sp from the same individual. Responses to antibody stimulation in KIR2DS1+KIR2DL1-C245+ NK cells from HLA-C2/C2 individuals were even more blunted. Together, these studies indicate that KIR2DS1 and KIR2DL1 signaling can proceed independently and suggest that the strength of incoming signals determines the reactivity of NK cells. Strong activating signals can still be generated despite KIR2DS1+ NK cell tolerization in HLA-C2/C2 hosts, indicating that tolerization does not impact the innate response capacity, but that the degree of response is driven by the nature of the stimulus. Simultaneous engagement of inhibitory and activating receptors by ligand on a target cell leads to inhibition overruling activation; whereas simultaneous agonist antibody engagement of inhibitory and activating receptors leads to activation overruling inhibition.
We find that KIR2DL1 and KIR2DS1 alleles are naturally expressed at different cell surface densities, and by comparing NK cells educated in different HLA-C environments and by the same KIR2DL1 isoforms, we can conclude that HLA-C2 diminishes the brightness of KIR2DL1 but not KIR2DS1 staining on NK cell surfaces in a dose-dependent manner. A diminution of inhibitory receptor staining has been observed in mice, where education of NK cells is associated with lower MFI of the inhibitory Ly49 receptors that engage MHC class I molecules (42, 43) Molecular studies in mice indicate that this phenotypic change results from cis interactions between the cognate receptor-ligand partnerships that interfere with antibody binding (44–46). Functionally, these interactions interfere with engagement of Ly49 by inhibitory ligands available from neighboring cells in trans, increasing the threshold for NK cell inhibition (47, 48). Cis associations between KIR and HLA have not directly been demonstrated, and the KIR proteins are non-orthologous to Ly49 (49). However, KIR3DL1*004, a stable KIR3DL1 allotype that is retained intracellularly, appears to educate NK cells in the presence of its cognate HLA-Bw4 ligand (10), and the KIR ligand in cis has a profound influence on long-term functionality of NK cells (50). Furthermore, HLA-C expression is modulated specifically in NK cells that express its cognate KIR receptor (51). Therefore, KIR-HLA interactions may occur intracellularly and could impact the availability of KIR proteins for antibody staining and for surface interaction with ligands in trans. HLA-C2 polymorphism could differentially impact KIR2DL1 expression, as HLA-C2 alleles have different densities of expression (52) and different affinities for KIR2DL1 alleles (9).
In individuals exhibiting genes encoding KIR2DL1 and its educating HLA-C2 ligand, the extent of NK education, measured as “missing self” responsiveness, correlates with the cell surface density of the inhibitory receptor: KIR2DL1+ NK cells exhibit greater MFI and missing self-responsiveness when they are educated by the high density KIR2DL1-R245 isoforms compared with the low density KIR2DL1-C245 isoforms. We have similarly reported that KIR3DL1 receptors expressed at the highest cell surface densities exhibit the highest response capacity, especially when coupled with high-density isoforms of their cognate HLA-Bw4 ligand (11). These findings in human NK education correspond to observations in mice, in which the density of Ly49 receptors educated by “self” MHC class I also corresponds with increasing education (53, 54).
We found that the copy number of HLA-C2 increases NK cell education in a dose-dependent manner. Currently, there are no antibodies available to distinguish subtypes of HLA-C, or to distinguish the HLA-C2 and HLA-C1 group members which do or do not, respectively, ligate KIR2DL1. Nevertheless, this finding suggests that, as for HLA-Bw4 copy number and KIR3DL1+ NK cells (11), response capacity in KIR2DL1+ cells is tuned to the availability of cognate ligand at steady state. We extend this finding to the activating receptor, KIR2DS1, where the reactivity of KIR2DS1-expressing NK cells is negatively attenuated in a fashion commensurate with the gene “dose” of HLA-C2 ligand. These findings are in contrast to an earlier report, in which clonally-expanded KIR2DS1sp NK cells were only found to be tolerized in donors with two copies of HLA-C2 (5); using primary cells, we observe that while the most profound diminution of response is found among HLA-C2 homozygotes, we can identify an intermediate educational level conferred by a single copy of HLA-C2 ligand, consistent with a ligand-associated titration of effector response. Further, we find that this “tolerization” extends to cells co-expressing other self-sensitive inhibitory receptors, as NKG2A+KIR2DS1+ cells from HLA-C2/C2 individuals exhibit a reactivity lower than NKG2Asp cells or KIR2DS1+ cells from HLA-C1/C1 individuals.
We observed a lower education of NKG2Asp+ NK cells from HLA-C2/C2 donors in comparison to those from HLA-C1/C2 or HLA-C1/C1 donors. The difference in education is presumably due to dimorphism at position−21 in the leader sequence of HLA-B, which has been found to be associated with education of NKG2A-bearing NK cells (55). A threonine residue at−21 (-21T) does not deliver functional peptides to HLA-E, leading to lower education for NKG2A in individuals bearing HLA haplotypes with−21T HLA-B molecules, which, by positive linkage disequilibrium, are enriched for HLA-C2. The pairing of HLA-C2 and poor NKG2A education through−21T likely accounts for the lower education of NKG2Asp cells from HLA-C2C2 donors.
While we observed an impact of HLA-C2 copy number and KIR2DL1 allele subtype on primary NK effector response capacity, we did not observe a similar difference in sensitivity to inhibition by the HLA-C2 targets allotypes tested in our study. These findings lead us to conclude that in contrast to previously published studies using transfected cells (16), primary NK cells exhibiting KIR2DL1-C245 or KIR2DL1-R245 are similarly sensitive to inhibition. Further investigation is necessary to determine if primary NK cells expressing KIR2DL1-C245 or KIR2DL1-R245 exhibit different capacities to kill tumor cells in vivo.
Until recently, it has been challenging to study the function of highly homologous activating and inhibitory KIR receptors and to distinguish between KIR allotypes on primary NK cells due to the lack of reagents and methodologies. In this study, we propose the combination of two antibodies (1127B and 143211 mAb) to separate KIR2DS1 and the two most common KIR2DL1 allele subtypes at the same time. Other studies have proposed alternative antibody strategies to separate KIR2DS1 and KIR2DL1 or KIR2DL1 alleles (4, 14); and still other combinations of commercially available antibodies can separate the KIR2DL2/2DS2 receptor pair (56).
The ability to dissect KIR2DL1 subtypes by flow cytometry is an important facet of this investigation, because it permits assessment of primary NK cells expressing different KIR2DL1 allotypes but educated in the same HLA-C environment. In this way, we can consider whether the expansion of self-specific KIR+ cells in response to HCMV infection that has been previously described (17) is related to the magnitude of reactivity of NK cells. Our results demonstrate instead that there is not preferential expansion of the more educated, more responsive KIR2DL1-R245+ population over the less responsive KIR2DL1-C245+ population. Interestingly, like KIR2DL1, KIR2DS1 is found on NKG2C+ cells only from individuals expressing HLA-C2.
The expansion of NKG2C+ NK subsets in response to HCMV infection is well-documented (17, 57), but how this population interacts with infected cells and impacts viral control is not well-understood. That HMCV-infected cells exhibit self HLA class I molecules and NKG2C+ NK cells exhibit cognate inhibitory KIR suggests that HMCV-infected cells could drive inhibition of educated NK cells. In our present study, we did not observe preference of any KIR2DL1 allele for expansion on NKG2C+ cells in donors genotyped for both KIR2DL1-R245 and KIR2DL1-C245. Whether subtle differences in the capacity for inhibition, which could be mediated by the particular alleles of HLA-C in each person, or input from other co-expressed receptors, and the impact of KIR2DL1 in combination with NKG2C on autologous cells infected with HCMV, remain to be further investigated.
Control of cancer, infection, autoimmunity, and reproduction have all been linked to KIR-HLA partnerships (3), and we have recently demonstrated that allele subtype variation for KIR3DL1 and HLA-Bw4 can have similar impacts (10, 12). That KIR2DL1, KIR2DS1, and HLA-C2 have co-evolved, diversified and enable variable NK cell function is intriguing and may similarly reflect distinctions in the capabilities of NK cells for maintenance of human health.
Ethics Statement
Peripheral blood samples were collected from healthy human donors following approval from the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board, and donors provided informed, written consent. The MSKCC IRB waived the need for additional research consent for anonymous NYBC samples.
Author Contributions
J-BL designed the study. J-BL and JF performed the experiments. J-BL and JB created the bank samples. J-BL, JB, and KH wrote the manuscript. All authors discussed the results and contributed to the final manuscript. KH supervised the project.
Funding
This work was supported by funding from NIH U01 AI069197, R01 AI125651, and P01 CA23766 to KH, the François Wallace Monahan Fellowship to J-BL, and the Cancer Center Core grant (NIH P03 CA008748) to MSKCC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00734/full#supplementary-material
References
1. Karre K. NK cells, MHC class I molecules and the missing self. Scand J Immunol. (2002) 55:221–8. doi: 10.1046/j.1365-3083.2002.01053.x
2. Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. (2005) 436:709–13. doi: 10.1038/nature03847
3. Boudreau JE, Hsu KC. Natural killer cell education in human health and disease. Curr Opin Immunol. (2018) 50:102–111. doi: 10.1016/j.coi.2017.11.003
4. Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaelsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. (2010) 115:1166–74. doi: 10.1182/blood-2009-09-245746
5. Pittari G, Liu XR, Selvakumar A, Zhao Z, Merino E, Huse M, et al. NK cell tolerance of self-specific activating receptor KIR2DS1 in individuals with cognate HLA-C2 ligand. J Immunol. (2013) 190:4650–60. doi: 10.4049/jimmunol.1202120
6. Chapel A, Garcia-Beltran WF, Holzemer A, Ziegler M, Lunemann S, Martrus G, et al. Peptide-specific engagement of the activating NK cell receptor KIR2DS1. Sci Rep. (2017) 7:2414. doi: 10.1038/s41598-017-02449-x
7. Naiyer MM, Cassidy SA, Magri A, Cowton V, Chen K, Mansour S, et al. KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci Immunol. (2017) 2:eaal5296. doi: 10.1126/sciimmunol.aal5296
8. van der Ploeg K, Chang C, Ivarsson MA, Moffett A, Wills MR, Trowsdale J. Modulation of human leukocyte antigen-C by human cytomegalovirus stimulates KIR2DS1 recognition by natural killer cells. Front Immunol. (2017) 8:298. doi: 10.3389/fimmu.2017.00298
9. Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, et al. Polymorphic HLA-C receptors balance the functional characteristics of KIR haplotypes. J Immunol. (2015) 195:3160–70. doi: 10.4049/jimmunol.1501358
10. Boudreau JE, Giglio F, Gooley TA, Stevenson PA, Le Luduec JB, Shaffer BC, et al. KIR3DL1/ HLA-B subtypes govern acute myelogenous leukemia relapse after hematopoietic cell transplantation. J Clin Oncol. (2017) 35:2268–78. doi: 10.1200/JCO.2016.70.7059
11. Boudreau JE, Mulrooney TJ, Le Luduec JB, Barker E, Hsu KC. KIR3DL1 and HLA-B density and binding calibrate NK education and response to HIV. J Immunol. (2016) 196:3398–410. doi: 10.4049/jimmunol.1502469
12. Forlenza CJ, Boudreau JE, Zheng J, Le Luduec JB, Chamberlain E, Heller G, et al. KIR3DL1 allelic polymorphism and HLA-B epitopes modulate response to anti-GD2 monoclonal antibody in patients with neuroblastoma. J Clin Oncol. (2016) 34:2443–51. doi: 10.1200/JCO.2015.64.9558
13. Venstrom JM, Pittari G, Gooley TA, Chewning JH, Spellman S, Haagenson M, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med. (2012) 367:805–16. doi: 10.1056/NEJMoa1200503
14. Huhn O, Chazara O, Ivarsson MA, Retiere C, Venkatesan TC, Norman PJ, et al. High-resolution genetic and phenotypic analysis of KIR2DL1 alleles and their association with pre-eclampsia. J Immunol. (2018) 201:2593–601. doi: 10.4049/jimmunol.1800860
15. Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. IPD–the immuno polymorphism database. Nucleic Acids Res. (2013) 41:D1234–40. doi: 10.1093/nar/gks1140
16. Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. (2009) 114:5182–90. doi: 10.1182/blood-2009-07-231977
17. Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. (2013) 121:2678–88. doi: 10.1182/blood-2012-10-459545
18. Dunphy SE, Guinan KJ, Chorcora CN, Jayaraman J, Traherne JA, Trowsdale J, et al. 2DL1, 2DL2 and 2DL3 all contribute to KIR phenotype variability on human NK cells. Genes Immun. (2015) 16:301–10. doi: 10.1038/gene.2015.15
19. Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E, et al. Defining KIR and HLA class I genotypes at highest resolution via high-throughput sequencing. Am J Hum Genet. (2016) 99:375–91. doi: 10.1016/j.ajhg.2016.06.023
20. Vierra-Green C, Roe D, Hou L, Hurley CK, Rajalingam R, Reed E, et al. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS ONE. (2012) 7:e47491. doi: 10.1371/journal.pone.0047491
21. Wagner I, Schefzyk D, Pruschke J, Schöfl G, Schöne B, Gruber N, et al. Allele-level KIR genotyping of more than a million samples: workflow, algorithm, and observations. Front Immunol. (2018) 9:2843. doi: 10.3389/fimmu.2018.02843
22. Le Luduec JB, Kudva A, Boudreau JE, Hsu KC. Novel multiplex PCR-SSP method for centromeric KIR allele discrimination. Sci Rep. (2018) 8:14853. doi: 10.1038/s41598-018-33135-1
23. Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. (2015) 42:443–56. doi: 10.1016/j.immuni.2015.02.008
24. Hsu KC, Liu XR, Selvakumar A, Mickelson E, O'Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. (2002) 169:5118–29. doi: 10.4049/jimmunol.169.9.5118
25. Adams S, Robbins FM, Chen D, Wagage D, Holbeck SL, Morse HC III, et al. HLA class I and II genotype of the NCI-60 cell lines. J Transl Med. (2005) 3:11. doi: 10.1186/1479-5876-3-11
26. Radford JE Jr, Chen E, Hromas R, Ginder GD. Cell-type specificity of interferon-gamma-mediated HLA class I gene transcription in human hematopoietic tumor cells. Blood. (1991) 77:2008–15.
27. Tarek N, Le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, et al. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest. (2012) 122:3260–70. doi: 10.1172/JCI62749
28. Cichocki F, Miller JS, Anderson SK. Killer immunoglobulin-like receptor transcriptional regulation: a fascinating dance of multiple promoters. J Innate Immun. (2011) 3:242–8. doi: 10.1159/000323929
29. Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. (1997) 7:739–51. doi: 10.1016/S1074-7613(00)80393-3
30. Beziat V, Traherne JA, Liu LL, Jayaraman J, Enqvist M, Larsson S, et al. Influence of KIR gene copy number on natural killer cell education. Blood. (2013) 121:4703–7. doi: 10.1182/blood-2012-10-461442
31. Boudreau JE, Le Luduec JB, Hsu KC. Development of a novel multiplex PCR assay to detect functional subtypes of KIR3DL1 alleles. PLoS ONE. (2014) 9:e99543. doi: 10.1371/journal.pone.0099543
32. Sips M, Liu Q, Draghi M, Ghebremichael M, Berger CT, Suscovich TJ, et al. HLA-C levels impact natural killer cell subset distribution and function. Hum Immunol. (2016) 77:1147–153. doi: 10.1016/j.humimm.2016.08.004
33. Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. (2015) 42:431–42. doi: 10.1016/j.immuni.2015.02.013
34. Wu Z, Sinzger C, Frascaroli G, Reichel J, Bayer C, Wang L, et al. Human cytomegalovirus-induced NKG2C(hi) CD57(hi) natural killer cells are effectors dependent on humoral antiviral immunity. J Virol. (2013) 87:7717–25. doi: 10.1128/JVI.01096-13
35. Falco M, Romeo E, Marcenaro S, Martini S, Vitale M, Bottino C, et al. Combined genotypic and phenotypic killer cell Ig-like receptor analyses reveal KIR2DL3 alleles displaying unexpected monoclonal antibody reactivity: identification of the amino acid residues critical for staining. J Immunol. (2010) 185:433–41. doi: 10.4049/jimmunol.0903632
36. Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. (2013) 5:208ra145. doi: 10.1126/scitranslmed.3006702
37. Brodin P, Lakshmikanth T, Mehr R, Johansson MH, Duru AD, Achour A, et al. Natural killer cell tolerance persists despite significant reduction of self MHC class I on normal target cells in mice. PLoS ONE. (2010) 5:e13174. doi: 10.1371/journal.pone.0013174
38. Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. (2012) 119:2665–74. doi: 10.1182/blood-2011-10-386995
39. Liu LL, Landskron J, Ask EH, Enqvist M, Sohlberg E, Traherne JA, et al. Critical role of CD2 Co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep. (2016) 15:1088–99. doi: 10.1016/j.celrep.2016.04.005
40. Babor F, Manser AR, Fischer JC, Scherenschlich N, Enczmann J, Chazara O, et al. KIR ligand C2 is associated with increased susceptibility to childhood ALL and confers an elevated risk for late relapse. Blood. (2014) 124:2248–51. doi: 10.1182/blood-2014-05-572065
41. Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol. (2013) 190:6198–208. doi: 10.4049/jimmunol.1300464
42. Held W, Raulet DH. Ly49A transgenic mice provide evidence for a major histocompatibility complex-dependent education process in natural killer cell development. J Exp Med. (1997) 185:2079–88. doi: 10.1084/jem.185.12.2079
43. Olsson MY, Karre K, Sentman CL. Altered phenotype and function of natural killer cells expressing the major histocompatibility complex receptor Ly-49 in mice transgenic for its ligand. Proc Natl Acad Sci USA. (1995) 92:1649–53. doi: 10.1073/pnas.92.5.1649
44. Chalifour A, Scarpellino L, Back J, Brodin P, Devevre E, Gros F, et al. A role for cis interaction between the inhibitory Ly49A receptor and MHC class I for natural killer cell education. Immunity. (2009) 30:337–47. doi: 10.1016/j.immuni.2008.12.019
45. Doucey MA, Scarpellino L, Zimmer J, Guillaume P, Luescher IF, Bron C, et al. Cis association of Ly49A with MHC class I restricts natural killer cell inhibition. Nat Immunol. (2004) 5:328–36. doi: 10.1038/ni1043
46. Scarpellino L, Oeschger F, Guillaume P, Coudert JD, Levy F, Leclercq G, et al. Interactions of Ly49 family receptors with MHC class I ligands in trans and cis. J Immunol. (2007) 178:1277–84. doi: 10.4049/jimmunol.178.3.1277
47. Andersson KE, Williams GS, Davis DM, Hoglund P. Quantifying the reduction in accessibility of the inhibitory NK cell receptor Ly49A caused by binding MHC class I proteins in cis. Eur J Immunol. (2007) 37:516–27. doi: 10.1002/eji.200636693
48. Back J, Chalifour A, Scarpellino L, Held W. Stable masking by H-2Dd cis ligand limits Ly49A relocalization to the site of NK cell/target cell contact. Proc Natl Acad Sci USA. (2007) 104:3978–83. doi: 10.1073/pnas.0607418104
49. Lanier LL. NK cell recognition. Annu Rev Immunol. (2005) 23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526
50. Boudreau JE, Liu XR, Zhao Z, Zhang A, Shultz LD, Greiner DL, et al. Cell-extrinsic MHC class I molecule engagement augments human NK cell education programmed by cell-intrinsic MHC class I. Immunity. (2016) 45:280–91. doi: 10.1016/j.immuni.2016.07.005
51. Li H, Ivarsson MA, Walker-Sperling VE, Subleski J, Johnson JK, Wright PW, et al. Identification of an elaborate NK-specific system regulating HLA-C expression. PLoS Genet. (2018) 14:e1007163. doi: 10.1371/journal.pgen.1007163
52. Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, et al. Influence of HLA-C expression level on HIV control. Science. (2013) 340:87–91. doi: 10.1126/science.1232685
53. Brodin P, Lakshmikanth T, Johansson S, Karre K, Hoglund P. The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood. (2009) 113:2434–41. doi: 10.1182/blood-2008-05-156836
54. Brodin P, Lakshmikanth T, Karre K, Hoglund P. Skewing of the NK cell repertoire by MHC class I via quantitatively controlled enrichment and contraction of specific Ly49 subsets. J Immunol. (2012) 188:2218–26. doi: 10.4049/jimmunol.1102801
55. Horowitz A, Djaoud Z, Nemat-Gorgani N, Blokhuis J, Hilton HG, Beziat V, et al. Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci Immunol. (2016) 1:eaag1672. doi: 10.1126/sciimmunol.aag1672
56. Blunt MD, Rettman P, Bastidas-Legarda LY, Fulton R, Capizzuto V, Naiyer MM, et al. A novel antibody combination to identify KIR2DS2(high) natural killer cells in KIR2DL3/L2/S2 heterozygous donors. HLA. (2019) 93:32–5. doi: 10.1111/tan.13413
Keywords: KIR2DL1, KIR2DS1, NK cell, NK cell education, HLA-C2, KIR allele
Citation: Le Luduec JB, Boudreau JE, Freiberg JC and Hsu KC (2019) Novel Approach to Cell Surface Discrimination Between KIR2DL1 Subtypes and KIR2DS1 Identifies Hierarchies in NK Repertoire, Education, and Tolerance. Front. Immunol. 10:734. doi: 10.3389/fimmu.2019.00734
Received: 15 January 2019; Accepted: 19 March 2019;
Published: 05 April 2019.
Edited by:
Eric O. Long, National Institute of Allergy and Infectious Diseases (NIAID), United StatesReviewed by:
Vivien Béziat, Institut National de la Santé et de la Recherche Médicale (INSERM), FrancePaul J. Norman, University of Colorado Denver, United States
Copyright © 2019 Le Luduec, Boudreau, Freiberg and Hsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katharine C. Hsu, aHN1a0Btc2tjYy5vcmc=
†Present Address: Jeanette E. Boudreau, Departments of Pathology, Microbiology and Immunology, Dalhousie University, Halifax Regional Municipality, NS, Canada
 Jean-Benoît Le Luduec
Jean-Benoît Le Luduec Jeanette E. Boudreau
Jeanette E. Boudreau Julian C. Freiberg
Julian C. Freiberg Katharine C. Hsu
Katharine C. Hsu