- 1Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, Taibah University, Madina, Saudi Arabia
- 2Rajiv Gandhi Technical University, Bhopal, India
Leishmaniasis is one of the major neglected tropical diseases, for which no vaccines exist. Chemotherapy is hampered by limited efficacy coupled with development of resistance and other side effects. Leishmania parasites elude the host defensive mechanisms by modulating their surface proteins as well as dampening the host's immune responses. The parasites use the conventional RNA polymerases peculiarly under different environmental cues or pressures such as the host's milieu or the drugs. The mechanisms that restructure post-translational modifications are poorly understood but altered epigenetic histone modifications are believed to be instrumental in influencing the chromatin remodeling in the parasite. Interestingly, the parasite also modulates gene expression of the hosts, thereby hijacking or dampening the host immune response. Epigenetic factor such as DNA methylation of cytosine residues has been incriminated in silencing of macrophage-specific genes responsible for defense against these parasites. Although there is dearth of information regarding the epigenetic alterations-mediated pathogenesis in these parasites and the host, the unique epigenetic marks may represent targets for potential anti-leishmanial drug candidates. This review circumscribes the epigenetic changes during Leishmania infection, and the epigenetic modifications they enforce upon the host cells to ensure a safe haven. The non-coding micro RNAs as post-transcriptional regulators and correlates of wound healing and toll-like receptor signaling, as well as prognostic biomarkers of therapeutic failure and healing time are also explored. Finally, we highlight the recent advances on how the epigenetic perturbations may impact leishmaniasis vaccine development as biomarkers of safety and immunogenicity.
Introduction
Epigenetics encompasses any process that changes gene expression and is inherited without amending the fundamental DNA sequence (1). These variations are highly dynamic which get altered on advent of any external stress or internal cues (2, 3). Epigenetics controls several cellular processes by switching genes on or off, thereby modulating gene expression. Epigenetics is also associated with various diseased states, wherein, it is shaped by host as well as pathogen selection pressures (4, 5). Recently, there is burgeoning interest in epigenetics landscapes during an infection, particularly alterations in DNA methylome, histone marks and non-coding (nc)RNA or micro (mi)RNA profiles. The epigenetic states result in erratic gene expression profiles of host cells, which are responsible for warding off microbial infections (6, 7).
Leishmania belongs to trypanosomatid family, being among the major neglected vector-borne tropical diseases, ranging in severity from self-healing but disfiguring and stigmatizing cutaneous lesions to disseminating muco-cutaneous and fatal visceral manifestations, depending on the species and host characteristics. Globally, 0.7–1.2 million new cases of cutaneous leishmaniasis (CL) occur every year; while for visceral leishmaniasis (VL), 200,000–400,000 new cases and 20,000–40,000 deaths are reported annually, with 95% of fatal cases occurring in only six countries, namely, India, Bangladesh, Sudan, Nepal, Ethiopia, and Brazil (8). The goal of World Health Organization is to eliminate this public health problem in South-east Asia region by 2020 (9).
Leishmania parasites have a digenetic life cycle that may be zoonotic or anthroponotic, depending upon the infecting parasite species. When an infected female sandfly (Phlebotomus or Lutzomyia species) takes a blood meal, the parasites cause dermal lesions as in CL or visceralize as in VL (10). The infection is amplified in the vector's gut with successive blood meals (11). Invasion of host macrophages by Leishmania triggers a multitude of signaling circuits to eliminate the pathogen. However, the parasite tries to subvert these defense mechanisms to create a safe haven for its survival. Leishmania secretes effector molecules to modulate host immune transcriptome resulting in alterations in the host epigenome, to alter cytokine and chemokine levels, their cross talks and downstream signaling hubs. This adversely affects the recruitment and activation of immune cells, respiratory burst and antigen presentation, leading to immune evasion (12).
Though still at infancy, there is a recent surge of information on the epigenetic regulation during Leishmania infection. This review gives an update on Leishmania epigenetic landscapes and epigenome alterations imposed in the host for immune evasion (summarized in Figure 1 and Table 1). Further, evolving evidence on the probable downstream effects of epigenetic regulation such as targeting epigenetic machinery to reset the waning immune response, via vaccine, or drug development and prognostic markers are also discussed.
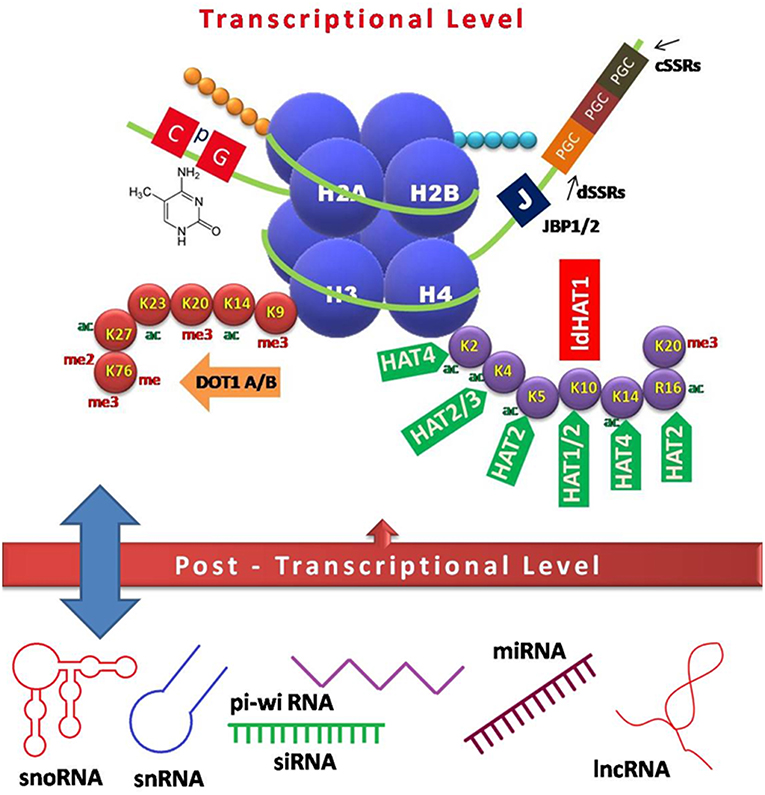
Figure 1. Schematic model of epigenetic regulation during Leishmania infection. Interplay of various factors involved in controlling gene expression at the transcriptional (DNA and histone modifications) and post-transcriptional level (non-coding RNAs) is depicted. CpG-rich regions of repressed genes are usually methylated, which in turn recruit chromatin modifiers to keep the genes in any of the three states, i.e., repressed, expressed, or poised. Heavily expressed genes show neither DNA methylation nor acetylated histones, while repressed genes tend to have both methylated DNA and histones, which inhibit the accessibility of polymerases, and other factors required for transcription. Base J, a DNA modification is crucial for transcriptional control in Leishmania species. Various non-coding RNAs arising mostly from UTRs act as regulatory elements in a feedback loop (13, 14). JBP, Base J binding protein; PGC, polycistronic gene cluster; cSSRs, convergent strand switch regions; dSSRs, divergent strand switch regions; DOT, disrupter of telomere; ac, acetylation; me, methylation; me2, dimethylation; me3, trimethylation; HAT, histone acetyl transferase; sno, small nucleolar; sn, small nuclear; pi-wi, piwi interacting; si, small interfering; mi, micro; lnc, long non-coding.
Epigenetic Changes in Leishmania Parasites
The diverse clinical manifestations of leishmaniasis may be attributed to varying genomic makeup (size, GC content, coding genes, pseudogenes, retrotransposons) of the causative Leishmania species (38). To promote their survival in the host environment, a huge array of epigenetic factors is speculated to interplay in these parasites (39).
DNA Modification
Glycosylated thymine, termed “Base J” or β-D-glucosylhydroxymethyluracil, is present in the telomeric repeat sequence (GGGTTA) of Leishmania (40). J replaces ~1% of T in nuclear DNA and the modified T residue has been implicated in transcriptional regulation and termination. Absence of J is lethal to Leishmania, due to massive read through of transcriptional termination sites (41). However, repression of specific genes has now been identified as an essential role of Base J (15). A thymine base modification, 5-hydroxymethyluracil has recently been mapped to Leishmania genome, but its epigenetic role is yet to be elucidated (42).
Histone Alterations
Acetylation of histone H3 in telomeric divergent strand switch regions, has been reported in L. major promastigotes (43) resulting in chromatin state, with restriction of protein coding genes. Acetylation levels are higher in rapidly growing cells compared to stationary phase cells. Epigenetic marks such as H3K9me3, H3K14ac, H3K23ac, and H3K27ac have also been reported in promoter region of rRNA genes of L. major, favoring transcriptional activation of rRNA genes while H4K20me3 in the coding region is related to transcriptional silencing (44). H3K9me3 is also linked to heterochromatin formation. Histone variants such as H2A.Z and H2B.V have been identified as essential for L. major survival (16).
In L. donovani, histone acetyltransferase (HAT)4 acetylates H4K14, favoring maintenance of euchromatin state (19). H4K4 acetylation by HAT3 (18) and, HAT2-dependant H4K10 acetylation of promoters in L. donovani has been linked with parasite survival (17).
Epigenetic tags are differentially regulated in the promastigote and amastigote stage. Some of the histone deacetylases (HDAC) are preferentially upregulated in L. infantum logarithmic phase promastigotes over intracellular amastigotes, making the amastigotes better able to adapt to intraphagolysosomal environment (20). Sirtuins, NAD-dependent HDAC have been implicated in parasite survival by inhibiting apoptosis (45), and sirtinol, sirtuin inhibitor, selectively induced apoptosis in L. infantum axenic amastigotes but not the promastigotes (46).
Non-coding RNAs
ncRNAs, non-coding sequences of about 22 nucleotides, act as post-transcriptional regulators of RNA encoding proteins (47). A special class of nc RNAs found exclusively in amastigotes of L. infantum and L. donovani, is particularly important for intra-macrophage parasite survival (48). In L. tarentole, ncRNA similar to guide RNA encoded by maxicircles and minicircles has been identified (49). Significant differences in nc RNA repertoire among different Leishmania species and stages has been reported (50).
Leishmania-Induced Host Epigenetic Alterations
Epigenetic mechanisms regulate the interplay of host-pathogen interactions. Although information on epigenetic manipulation of hosts by Leishmania is scarce, the pathogen employs a number of stratagems to manipulate the host epigenome, thereby hijacking its cellular soldiers (23, 51, 52). Genetic heterogeneity among different Leishmania species causes altered gene expression in response to environmental conditions in the host, resulting in varied epigenetic mechanisms.
DNA Modifications in Host
L. donovani has been reported to elicit epigenetic modifications in host macrophages, permanently down-modulating the innate immune defenses (23). These altered epigenetic tags comprise of cytosine methylation at CpG sites of macrophage DNA, upon infection, causing alteration in genes implicated in JAK/STAT, calcium, MAPK, notch, and mTOR signaling pathway as well as in cell adhesion involving integrin β1 and changes in host oxidative phosphorylation. Leishmania-driven epigenomic changes in host macrophages deactivate its innate immune defensive machinery, thereby promoting pathogen survival, and replication.
Epigenetic modification promotes self-healing in CL. Epigenetic repression of wound healing gene, Friend leukemia virus integration 1 (FLI1) via increased methylation of CpG islands in its promoter region, has been found to correlate with up-modulation of pro-fibrotic genes such as collagen type I alpha 1 (Col1α1) and alpha 2 (Col1α2) and, conversely, with down-regulation of matrix metalloproteinase 1 (MMP1) gene, resulting in resolution of lesions caused by L. braziliensis (53, 54). MMP1 cleaves type I collagens to loosen keratinocytes-dermal matrix contact, favoring re-epithelialization or tissue repair. Homocystine-dependent stimulation of IL-6 has been further implicated in epigenetic DNA methylation of CpG-rich promoter of lysyloxidase (LOX) gene, a cross-linker of collagen and elastin, also rendering it transcriptionally inactive (55, 56). These epigenetic regulations of gene expression depend upon the infecting Leishmania species. Contrary to these reports, a recent study showed that increased FLI1 promoter methylation did not translate into low FLI1 gene expression (22).
Histone Modifications in Host
L. amazonensis induces HDAC in infected macrophages, contributing to down regulation of inducible nitric oxide synthase (iNOS) and subsequent parasite survival (25).
ncRNA Induced Gene Silencing
Leishmania infection targets cellular miRNA repertoire and the differential miRNA expression is dependent on infecting species (37). A plethora of studies indicate miRNAs as key regulators of disease phenotype in Leishmania-infected cells (27, 30, 32–34). miRNA-30A-3p mediates survival of intracellular L. donovani and intervention targeting the miRNA resulted in significant reduction in parasite burden by restoring host autophagic machinery (32). miRNA-3620 was found to regulate iron homeostasis and hypoxia in L. donovani infected macrophages while miRNA-3473f was linked with autophagy inhibition (34). Drug resistance due to over expression of efflux pumps such as ABC transporters has also been linked with downregulation of miRNA-763,−1264, and−3473f (34). L. donovani infection causes hypoxic environment within the macrophages by activating hypoxia inducible factor-1α, that in turn up regulates miRNA-210, while down regulating NF-κB mediated pro-inflammatory immune responses, to establish a safe niche for its survival (57).
Leishmania establishes and survives in the host by manipulating its ncRNA network, which includes transcriptional arrest of the major protein coding genes in macrophages (58), downregulating 7SL RNA in SRP complex, knockdown of selected ncRNAs in their host cells by inducing degradation of a specific RNA Pol III transcription factor subunit TFIIIC110 in M2 macrophages (59). Leishmania surface glycoprotein, gp63 and surface glycolipid, LPG have been reported to down modulate ncRNAs in M2 macrophages, thereby promoting infection (60).
Recently, down modulation of 19 miRNAs in L. donovani infected macrophages has been reported (36). The miRNA gene repression correlated with upregulation of host transcription factor, c-myc upon infection, a marker of M2 macrophages, which could possibly be another virulence factor. The expression of c-myc in turn is regulated by several miRNAs, primarily miRNA-34a, which is reciprocally down modulated in Leishmania-infected cells.
miRNA-361-3p and−140-3p have been reported to be more expressed in skin lesions caused by L. braziliensis in localized cutaneous leishmaniasis (LCL) (61). While miR-193b and−671 have been correlated with faster wound healing in L. braziliensis infected patients (31). Autophagy in intramacrophagic L. major has been correlated with miRNA-101c,−129-5p and via inhibiting miRNA-210 (28).
Potential Downstream Effects of Epigenetic Regulation During Leishmania Infection
Epigenetic Reprogramming of Innate Immune Cells
Recent reports shed light on epigenetic reprogramming in monocytes and macrophages via histone trimethylation at H3K4 for innate immune memory or trained immunity (62, 63). Natural killer (NK) cells have also been reported to differentiate into memory NK cells with distinct epigenetic profile (64). However, the epigenetic signatures of innate immune cells during Leishmania infection are limited (65).
Epigenetic Tuning of Cell Signaling Hubs
Epigenetic reprograming at cytokine gene loci is reported to influence its gene expression. A growing body of data suggests that differential cytokine microenvironment modulates T helper (Th) cell polarization, macrophage phenotype differentiation and cytokine-inflammasome crosstalk for optimal immune response (65). Signal transducers and activators of transcription (STAT)-4 and−6 have also been reported to play antagonistic roles in epigenetic tuning for Th cell differentiation (66). The epigenetic marks orchestrating gene regulation in Th cell differentiation (67) and M1/M2 macrophage polarization have been extensively reviewed (68).
Differential expression of miRNAs has been reported to induce T cell differentiation during VL. While miRNA-744 suppresses TGF-β expression and subsequently Treg cell differentiation, miRNA-1272 and−155 downregulate IL-4/IL-13 signaling to mitigate Th2 response during active infection (69). Antimony-resistant L. donovani has been reported to activate miRNA-466 inhibitor to degrade host MyD88 and regulate IL-10/IL-12 axis and establish successful infection (35). Chemokine and chemokine receptor gene expression also contribute to immunopathogenesis of leishmaniasis (70). But the effect of Leishmania–induced epigenetic alterations in regulation of chemokine genes has not been much explored (29).
miRNAs are also known to be involved in activation of monocytes through toll-like receptor (TLR) signaling (31). Following infection with L. major and L. donovani, miRNA expression was down modulated through MAP kinase, JAK-STAT, and TGF-β signaling pathways (31). H3K27 has been found to suppress toll-interacting protein that negatively regulates TLR, thereby promoting TLR-mediated inflammatory cytokine production, and activation of innate immune response against the invading pathogens (65).
Exploiting Epigenetics
Leishmania have evolved stratagems to neutralize macrophage defensive arsenals, the very heart of immune system's defensive machinery, resulting in replication of parasites within phagolysosomal vacuoles of infected macrophages. Unfolding the epigenetic signatures of host-pathogen interactions would help in development of effective drug targets to modulate host immune system and ameliorate the pathogenesis of infection. Some epigenetic marks may serve as putative vaccine candidates. The epigenetic biomarkers may also complement the current diagnostic assays.
Vaccines
An essential hallmark of vaccination is to generate antigen-specific memory T cells for induction of sufficient immune response to protect against re-infection. Epigenetic modifications have been reported to contribute toward memory T cell induction (71, 72). Recombinant histone H1 has been shown to elicit protection in outbred vervet monkeys against CL (73) while histones H2A-2B-3-4 cocktail induced protective immunity against L. donovani challenge in hamsters (74). Sirtuins have been used as vaccine candidate against L. donovani infected hamsters with induction of Th1 immune response (75). Recently, miRNA-21 has been shown to negatively correlate with IL-12 production and priming of protective Th1 response, suggesting declining levels of miRNA-21 as a potential biomarker of safety and immunogenicity in anti-leishmanial vaccines (76). Therapeutic vaccines may be developed to target miRNA-135 and−126 that bias the Th2 response toward protective Th1 type (69).
Epigenomic-Therapeutics
Despite an array of chemotherapeutic arsenal, mostly targeting the parasites directly, treatment failure, and drug resistance are looming large (77). This has been partly attributed to epigenetics-driven evolution of drug resistant phenotypes to override drug pressure (78). Host-directed epigenetic reprogramming may be refractory to resistance and hence offer hope in this regard (79).
Computer-aided drug repurposing for epigenetic targets is revolutionizing drug discovery (80). DNA methylation, particularly of virulence-associated genes, suggests DNA methyl transferases as potential therapeutic targets. An inverse correlation between FLI1 gene expression and MMP1 in cutaneous lesions has also been observed, suggesting MMP1 as a potential therapeutic target in severe forms of leishmaniasis (22). FLI1 and LOX have also been implicated as potent drug targets in L. braziliensis infection (54, 56).
The enzymes effecting histone post-translational modifications, particularly those containing epigenetic reader modules, bromodomains could also be putative therapeutic targets. Sirtuins of L. donovani have been validated as drug targets (81). Crystal structure of L. infantum Sir2 has been elucidated with implications for drug design (82). Sir2 has been suggested as a resistance marker for VL (21). Phenotypic screening of compound libraries against Leishmania has helped in identification of bisnaphthalimidopropyl derivatives as sirtuin inhibitors (83). Imipramine has been found to mediate antileishmanial effect in antimony-resistant Leishmania-infected macrophages via targeting HDAC11, resulting in transcriptional inactivation of IL-10 production (24). KH-TFMDI, a novel sirtuin inhibitor, targets HDAC to promote apoptosis-like cell death in L. amazonensis promastigotes as well as intracellular amastigotes (84). However, none of the clinically approved HDAC inhibitors are effective against L. amazonensis (85).
Studies have deciphered role of miRNA-294 and−721 in Leishmania survival via subversion of macrophage nitric oxide production and hence these may be putative therapeutic targets (26, 32). Recent reports of L. donovani hijacking the host's transcription factor, c-myc and reduction of intramacrophagic parasite burden upon c-myc silencing or inhibition, with consequent miRNA upregulation, implicate c-myc as a potential therapeutic target (36). Epigenetic targets such as miRNAs screened in L. donovani-infected macrophages upon treatment with antileishmanial trans-dibenzalacetone, revealed an imbalance between apoptosis and autophagy (86).
Epigenetic Biomarkers
Leishmania-induced changes in hosts' epigenome may help to predict the clinical outcome of infection and hence complement the existing diagnostics. The state of knowledge regarding epigenetic biomarkers in leishmaniasis is limited. A recent study showed potential of miRNA-361-3p as a prognostic biomarker in CL caused by L. braziliensis (61). miRNA-361-3p expression was upregulated in patients with therapeutic failure to pentavalent antimony and hence required more healing times. miRNA-193b and−671 have also been speculated to be prospective biosignatures for prognosis of LCL but require further validation (31).
Concluding Remarks
The epigenetic mechanisms work in alliance with each other to regulate life cycle of Leishmania parasites and ensure their survival. Pathogens are also capable of eluding cellular defensive machinery by changing the epigenetic states of host gene expression, thereby dampening their immune response. A snapshot of epigenetic imprinting of relevant genes in Th cell polarization, and memory T cell differentiation with triggering of innate immune cell populations may provide a basis for development of improved leishmaniasis vaccines.
Targeting the epigenetic marks could result in drug design with less likelihood of development of resistance, thus extending the pipeline toward disease elimination. Whether Leishmania parasites tailor the epigenetic mechanisms of their vector sandfly to favor their colonization remain to be elucidated. The impact of these pathogens on vector epigenetics could pave a way for development of transmission blocking vaccines. This review may assist to expand our knowledge of epigenetic influences upon host-parasite interplay, and open the doors to investigate epigenetic targets for rapid diagnostics or therapeutic interventions.
Author Contributions
FA: conceptualization, reviewing the studies, writing the mini-review, and critical editing; IK: conceptualization, writing original draft, editing; HH: reviewing the draft; critical editing. All the authors are accountable for all aspects of the review and gave final approval of the version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
2. Poulin R, Thomas F. Epigenetic effects of infection on the phenotype of host offspring: parasites reaching across host generations. Oikos. (2008) 117:331–5. doi: 10.1111/j.2007.0030-1299.16435.x
3. Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. (2012) 13:97–109. doi: 10.1038/nrg3142
4. Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. (2010) 28:1057–68. doi: 10.1038/nbt.1685
5. Gómez-Díaz E, Jordà M, Peinado MA, Rivero A. Epigenetics of host–pathogen interactions: the road ahead and the road behind. PLoS Pathog. (2012) 8:e1003007. doi: 10.1371/journal.ppat.1003007
6. Minarovits J. Microbe-induced epigenetic alterations in host cells: the coming era of patho-epigenetics of microbial infections: a review. Acta Microbiol Immunol Hung. (2009) 56:1–19. doi: 10.1556/AMicr.56.2009.1.1
7. De Monerri NCS, Kim K. Pathogens hijack the epigenome: a new twist on host-pathogen interactions. Am J Pathol. (2014) 184:897–911. doi: 10.1016/j.ajpath.2013.12.022
9. Karunaweera ND, Ferreira MU. Leishmaniasis: current challenges and prospects for elimination with special focus on the South Asian region. Parasitology. (2018) 145:425–9. doi: 10.1017/S0031182018000471
10. Wheeler RJ, Gluenz E, Gull K. The cell cycle of Leishmania: morphogenetic events and their implications for parasite biology. Mol Microbiol. (2011) 79:647–62. doi: 10.1111/j.1365-2958.2010.07479.x
11. Bates PA. Revising Leishmania's life cycle. Nat Microbiol. (2018) 3:529–30. doi: 10.1038/s41564-018-0154-2
12. Martínez-López M, Soto M, Iborra S, Sancho D. Leishmania hijacks myeloid cells for immune escape. Front Microbiol. (2018) 9:883. doi: 10.3389/fmicb.2018.00883
13. Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. (2009) 38:D457–62. doi: 10.1093/nar/gkp851
14. Torres F, Arias-Carrasco R, Caris-Maldonado JC, Barral A, Maracaja-Coutinho V, De Queiroz AT. LeishDB: a database of coding gene annotation and non-coding RNAs in Leishmania braziliensis. Database. (2017) 2017:bax047. doi: 10.1093/database/bax047
15. Reynolds DL, Hofmeister BT, Cliffe L, Siegel TN, Anderson BA, Beverley SM, et al. Base J represses genes at the end of polycistronic gene clusters in Leishmania major by promoting RNAP II termination. Mol Microbiol. (2016) 101:559–74. doi: 10.1111/mmi.13408
16. Anderson BA, Wong IL, Baugh L, Ramasamy G, Myler PJ, Beverley SM. Kinetoplastid-specific histone variant functions are conserved in Leishmania major. Mol Biochem Parasitol. (2013) 191:53–7. doi: 10.1016/j.molbiopara.2013.09.005
17. Chandra U, Yadav A, Kumar D, Saha S. Cell cycle stage-specific transcriptional activation of cyclins mediated by HAT2-dependant H4K10 acetylation of promoters in Leishmania donovani. PLoS Pathog. (2017) 13:e1006615. doi: 10.1371/journal.ppat.1006615
18. Kumar D, Saha S. HAT3-mediated acetylation of PCNA precedes PCNA monoubiquitination following exposure to UV radiation in Leishmania donovani. Nucleic Acids Res. (2015) 43:5423–41. doi: 10.1093/nar/gkv431
19. Yadav A, Chandra U, Saha S. Histone acetyltransferase HAT4 modulates navigation across G2/M and re-entry into G1 in Leishmania donovani. Sci Rep. (2016) 6:27510. doi: 10.1038/srep27510
20. Alcolea PJ, Alonso A, Gómez MJ, Moreno I, Domínguez M, Parro V, et al. Transcriptomics throughout the life cycle of Leishmania infantum: high down-regulation rate in the amastigote stage. Int J Parasitol. (2010) 40:1497–516. doi: 10.1016/j.ijpara.2010.05.013
21. Purkait B, Singh R, Wasnik K, Das S, Kumar A, Paine M, et al. Up-regulation of silent information regulator 2 (Sir2) is associated with amphotericin B resistance in clinical isolates of Leishmania donovani. J Antimicrob Chemother. (2015) 70:1343–56. doi: 10.1093/jac/dku534
22. Almeida L, Silva JA, Andrade VM, Machado P, Jamieson SE, Carvalho EM, et al. Analysis of expression of FLI1 and MMP1 in American cutaneous leishmaniasis caused by Leishmania braziliensis infection. Infect Genet Evol. (2017) 49:212–20. doi: 10.1016/j.meegid.2017.01.018
23. Marr AK, Macisaac JL, Jiang R, Airo AM, Kobor MS, Mcmaster WR. Leishmania donovani infection causes distinct epigenetic DNA methylation changes in host macrophages. PLoS Pathog. (2014) 10:e1004419. doi: 10.1371/journal.ppat.1004419
24. Mukherjee S, Mukherjee B, Mukhopadhyay R, Naskar K, Sundar S, Dujardin J-C, et al. Imipramine exploits histone deacetylase 11 to increase the IL-12/IL-10 ratio in macrophages infected with antimony-resistant Leishmania donovani and clears organ parasites in experimental infection. J Immunol. (2014) 193:4083–94. doi: 10.4049/jimmunol.1400710
25. Calegari-Silva TC, Vivarini ÁC, Pereira RDM, Dias-Teixeira KL, Rath CT, Pacheco AS, et al. Leishmania amazonensis downregulates macrophage iNOS expression via histone: deacetylase 1 (HDAC1): a novel parasite evasion mechanism. Eur J Immunol. (2018) 48:1188–98. doi: 10.1002/eji.201747257
26. Muxel SM, Laranjeira-Silva MF, Zampieri RA, Floeter-Winter LM. Leishmania (Leishmania) amazonensis induces macrophage miR-294 and miR-721 expression and modulates infection by targeting NOS2 and L-arginine metabolism. Sci Rep. (2017) 7:44141. doi: 10.1038/srep44141
27. Lemaire J, Mkannez G, Guerfali FZ, Gustin C, Attia H, Sghaier RM, et al. MicroRNA expression profile in human macrophages in response to Leishmania major infection. PLoS Negl Trop Dis. (2013) 7:e2478. doi: 10.1371/journal.pntd.0002478
28. Frank B, Marcu A, Petersen ALDOA, Weber H, Stigloher C, Mottram JC, et al. Autophagic digestion of Leishmania major by host macrophages is associated with differential expression of BNIP3, CTSE, and the miRNAs miR-101c, miR-129, and miR-210. Parasit Vect. (2015) 8:404. doi: 10.1186/s13071-015-0974-3
29. Mkannez G, Lemaire J, Guerfali F, Gustin C, Attia H, Sghaier R, et al. MicroRNA expression profile in human macrophage in response to Leishmania major infection. Int J Infect Dis. (2014) 21:168. doi: 10.1016/j.ijid.2014.03.772
30. Geraci NS, Tan JC, Mcdowell MA. Characterization of micro RNA expression profiles in Leishmania-infected human phagocytes. Parasite Immunol. (2015) 37:43–51. doi: 10.1111/pim.12156
31. Nunes S, Silva IB, Ampuero MR, Noronha ALLD, Souza LCLD, Correia TC, et al. Integrated analysis reveals that miR-193b, miR-671, and TREM-1 correlate with a good response to treatment of human localized cutaneous leishmaniasis caused by Leishmania braziliensis. Front Immunol. (2018) 9:640. doi: 10.3389/fimmu.2018.00640
32. Singh AK, Pandey RK, Shaha C, Madhubala R. MicroRNA expression profiling of Leishmania donovani-infected host cells uncovers the regulatory role of MIR30A-3p in host autophagy. Autophagy. (2016) 12:1817–31. doi: 10.1080/15548627.2016.1203500
33. Ghosh J, Bose M, Roy S, Bhattacharyya SN. Leishmania donovani targets Dicer1 to downregulate miR-122, lower serum cholesterol, and facilitate murine liver infection. Cell Host Microbe. (2013) 13:277–88. doi: 10.1016/j.chom.2013.02.005
34. Tiwari N, Kumar V, Gedda MR, Singh AK, Singh VK, Singh SP, et al. Identification and characterization of miRNAs in response to Leishmania donovani infection: delineation of their roles in macrophage dysfunction. Front Microbiol. (2017) 8:314. doi: 10.3389/fmicb.2017.00314
35. Mukherjee B, Paul J, Mukherjee S, Mukhopadhyay R, Das S, Naskar K, et al. Antimony-resistant Leishmania donovani exploits miR-466i to deactivate host MyD88 for regulating IL-10/IL-12 levels during early hours of infection. J Immunol. (2015) 1402585. doi: 10.4049/jimmunol.1402585
36. Colineau L, Lambertz U, Fornes O, Wasserman WW, Reiner NE. c-Myc is a novel Leishmania virulence factor by proxy that targets the host miRNA system and is essential for survival in human macrophages. J Biol Chem R A. (2018) 118:002462. doi: 10.1074/jbc.RA118.002462
37. Bragato JP, Melo LM, Venturin GL, Rebech GT, Garcia LE, Lopes FL, et al. Relationship of peripheral blood mononuclear cells miRNA expression and parasitic load in canine visceral Leishmaniasis. PLoS ONE. (2018) 13:e0206876. doi: 10.1371/journal.pone.0206876
38. Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. (2007) 39:839. doi: 10.1038/ng2053
39. Rajabi H, Kufe D. MUC1-C oncoprotein integrates a program of EMT, epigenetic reprogramming and immune evasion in human carcinomas. Biochim Biophys Acta Rev Cancer. (2017) 1868:117–22. doi: 10.1016/j.bbcan.2017.03.003
40. Olinski R, Starczak M, Gackowski D. Enigmatic 5-hydroxymethyluracil: oxidatively modified base, epigenetic mark or both? Mutat Res Rev. (2016) 767:59–66. doi: 10.1016/j.mrrev.2016.02.001
41. Van Luenen HG, Farris C, Jan S, Genest P-A, Tripathi P, Velds A, et al. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. (2012) 150:909–21. doi: 10.1016/j.cell.2012.07.030
42. Kawasaki F, Beraldi D, Hardisty RE, Mcinroy GR, Van Delft P, Balasubramanian S. Genome-wide mapping of 5-hydroxymethyluracil in the eukaryote parasite Leishmania. Genome Biol. (2017) 18:23. doi: 10.1186/s13059-017-1150-1
43. Thomas S, Green A, Sturm NR, Campbell DA, Myler PJ. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genom. (2009) 10:152. doi: 10.1186/1471-2164-10-152
44. Vizuet-De-Rueda JC, Florencio-Martínez LE, Padilla-Mejía NE, Manning-Cela R, Hernández-Rivas R, Martínez-Calvillo S. Ribosomal RNA genes in the protozoan parasite Leishmania major possess a nucleosomal structure. Protist. (2016) 167:121–35. doi: 10.1016/j.protis.2016.02.001
45. Vergnes B, Sereno D, Madjidian-Sereno N, Lemesre J-L, Ouaissi A. Cytoplasmic SIR2 homologue overexpression promotes survival of Leishmania parasites by preventing programmed cell death. Gene. (2002) 296:139–50. doi: 10.1016/S0378-1119(02)00842-9
46. Vergnes B, Vanhille L, Ouaissi A, Sereno D. Stage-specific antileishmanial activity of an inhibitor of SIR2 histone deacetylase. Acta Trop. (2005) 94:107–15. doi: 10.1016/j.actatropica.2005.03.004
47. Bayer-Santos E, Marini MM, Da Silveira JF. Non-coding RNAs in host–pathogen interactions: subversion of mammalian cell functions by protozoan parasites. Front Microbiol. (2017) 8:474. doi: 10.3389/fmicb.2017.00474
48. Dumas C, Chow C, Müller M, Papadopoulou B. A novel class of developmentally regulated noncoding RNAs in Leishmania. Eukaryot Cell. (2006) 5:2033–46. doi: 10.1128/EC.00147-06
49. Madej MJ, Alfonzo JD, Hüttenhofer A. Small ncRNA transcriptome analysis from kinetoplast mitochondria of Leishmania tarentolae. Nucleic Acids Res. (2007) 35:1544–54. doi: 10.1093/nar/gkm004
50. Ruy PDC, Monteiro Teles NM, Miserani Magalhães RD, Freitas De Castro F, Dias L, Aquino Defina TP, et al. Comparative transcriptomics in Leishmania braziliensis: disclosing differential gene expression of coding and putative noncoding RNAs across developmental stages. RNA Biol. (2019) 28:1–22. doi: 10.1080/15476286.2019.1574161
51. Buates S, Matlashewski G. General suppression of macrophage gene expression during Leishmania donovani infection. J Immunol. (2001) 166:3416–22. doi: 10.4049/jimmunol.166.5.3416
52. Gendlina I, De Monerri NS, Kim K. Modification of the host epigenome by parasitic protists. In: Doerfler W, Casadesús J, editors. Epigenetics of Infectious Diseases. New York, NY: Springer (2017). p. 189–220. doi: 10.1007/978-3-319-55021-3_9
53. Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthr Rheumatol. (2006) 54:2271–9. doi: 10.1002/art.21948
54. Almeida L, Oliveira J, Guimarães LH, Carvalho EM, Blackwell JM, Castellucci L. Wound healing genes and susceptibility to cutaneous Leishmaniasis in Brazil: Role of COL1A1. Infect Genet Evol. (2015) 30:225–9. doi: 10.1016/j.meegid.2014.12.034
55. Thaler R, Agsten M, Spitzer S, Paschalis EP, Karlic H, Klaushofer K, et al. Homocysteine suppresses the expression of the collagen cross-linker lysyl oxidase involving IL-6, Fli1, and epigenetic DNA methylation. J Biol Chem. (2011) 286:5578–88. doi: 10.1074/jbc.M110.166181
56. Castellucci LC, Almeida LFD, Jamieson SE, Fakiola M, Carvalho EMD, Blackwell JM. Host genetic factors in American cutaneous leishmaniasis: a critical appraisal of studies conducted in an endemic area of Brazil. Mem Inst Oswaldo Cruz. (2014) 109:279–88. doi: 10.1590/0074-0276140028
57. Kumar V, Kumar A, Das S, Kumar A, Abhishek K, Verma S, et al. Leishmania donovani activates hypoxia inducible factor-1α and miR-210 for survival in macrophages by downregulation of NF-κB mediated pro-inflammatory immune response. Front Microbiol. (2018) 9:385. doi: 10.3389/fmicb.2018.00385
58. Rana T, Misra S, Mittal MK, Farrow AL, Wilson KT, Linton MF, et al. Mechanism of down-regulation of RNA polymerase III-transcribed non-coding RNA genes in macrophages by Leishmania. J Biol Chem. (2011) 286:6614–26. doi: 10.1074/jbc.M110.181735
59. Misra S, Tripathi MK, Chaudhuri G. Down-regulation of 7SL RNA expression and impairment of vesicular protein transport pathways by Leishmania infection of macrophages. J Biol Chem. (2005) 280:29364–73. doi: 10.1074/jbc.M504162200
60. Farrow AL, Rana T, Mittal MK, Misra S, Chaudhuri G. Leishmania-induced repression of selected non-coding RNA genes containing B-box element at their promoters in alternatively polarized M2 macrophages. Mol Cell Biochem. (2011) 350:47–57. doi: 10.1007/s11010-010-0681-5
61. Lago TS, Silva JA, Lago EL, Carvalho EM, Zanette DL, Castellucci LCDC. The miRNA 361-3p, a regulator of GZMB and TNF is associated with therapeutic failure and longer time healing of cutaneous Leishmaniasis caused by L.(viannia) braziliensis. Front Immunol. (2018) 9:2621. doi: 10.3389/fimmu.2018.02621
62. Quintin J, Cheng S-C, Van Der Meer JW, Netea MG. Innate immune memory: towards a better understanding of host defense mechanisms. Curr Opin Immunol. (2014) 29:1–7. doi: 10.1016/j.coi.2014.02.006
63. Van Der Meer JW, Joosten LA, Riksen N, Netea MG. Trained immunity: a smart way to enhance innate immune defence. Mol Immunol. (2015) 68:40–4. doi: 10.1016/j.molimm.2015.06.019
64. Lau CM, Adams NM, Geary CD, Weizman O-E, Rapp M, Pritykin Y, et al. Epigenetic control of innate and adaptive immune memory. Nat Immunol. (2018) 19:963–72. doi: 10.1038/s41590-018-0176-1
65. Mahanta A, Ganguli P, Barah P, Sarkar RR, Sarmah N, Phukan S, et al. Integrative approaches to understand the mastery in manipulation of host cytokine networks by protozoan parasites with emphasis on Plasmodium and Leishmania species. Front Immunol. (2018) 9:296. doi: 10.3389/fimmu.2018.00296
66. Wei L, Vahedi G, Sun H-W, Watford WT, Takatori H, Ramos HL, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. (2010) 32:840–51. doi: 10.1016/j.immuni.2010.06.003
67. Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. (2009) 9:91. doi: 10.1038/nri2487
68. Zhou D, Yang K, Chen L, Zhang W, Xu Z, Zuo J, et al. Promising landscape for regulating macrophage polarization: epigenetic viewpoint. Oncotarget. (2017) 8:57693. doi: 10.18632/oncotarget.17027
69. Pandey RK, Sundar S, Prajapati VK. Differential expression of miRNA regulates T cell differentiation and plasticity during visceral leishmaniasis infection. Front Microbiol. (2016) 7:206. doi: 10.3389/fmicb.2016.00206
70. Navas A, Vargas DA, Freudzon M, Mcmahon-Pratt D, Saravia NG, Gómez MA. Chronicity of dermal leishmaniasis caused by Leishmania panamensis is associated with parasite mediated induction of chemokine gene expression. Infect Immun. (2014) 82:2872–80. doi: 10.1128/IAI.01133-13
71. Youngblood B, Hale JS, Ahmed R. T-cell memory differentiation: insights from transcriptional signatures and epigenetics. Immunology. (2013) 139:277–84. doi: 10.1111/imm.12074
72. Dong J, Chang H-D, Radbruch A. Epigenetic Imprinting of Immunological Memory. In: Doerfler W, Böhm P, editors. Epigenetics-A Different Way of Looking at Genetics. New York, NY: Springer (2016). p. 53–67. doi: 10.1007/978-3-319-27186-6_4
73. Masina SM, Gicheru M, Demotz SO, Fasel NJ. Protection against cutaneous leishmaniasis in outbred vervet monkeys, using a recombinant histone H1 antigen. J Infect Dis. (2003) 188:1250–7. doi: 10.1086/378677
74. Baharia RK, Tandon R, Sahasrabuddhe AA, Sundar S, Dube A. Nucleosomal histone proteins of L. donovani: a combination of recombinant H2A, H2B, H3 and H4 proteins were highly immunogenic and offered optimum prophylactic efficacy against Leishmania challenge in hamsters. PLoS ONE. (2014) 9:e97911. doi: 10.1371/journal.pone.0097911
75. Baharia RK, Tandon R, Sharma T, Suthar MK, Das S, Siddiqi MI, et al. Recombinant NAD-dependent SIR-2 protein of Leishmania donovani: immunobiochemical characterization as a potential vaccine against visceral leishmaniasis. PLoS Negl Trop Dis. (2015) 9:e0003557. doi: 10.1371/journal.pntd.0003557
76. Bhattacharya P, Ismail N, Kaul A, Gannavaram S, Nakhasi HL. Identification of microRNA-21 as a biomarker in live attenuated Leishmania vaccine induced protective immunity. J Immunol. (2017) 198:147.12
77. Sundar S, Singh B. Emerging therapeutic targets for treatment of leishmaniasis. Exp Opin Ther Targets. (2018) 22:467–86. doi: 10.1080/14728222.2018.1472241
78. Lamotte S, Späth GF, Rachidi N, Prina E. The enemy within: targeting host–parasite interaction for antileishmanial drug discovery. PLoS Negl Trop Dis. (2017) 11:e0005480. doi: 10.1371/journal.pntd.0005480
79. Jha PK, Khan MI, Mishra A, Das P, Sinha KK. HAT2 mediates histone H4K4 acetylation and affects micrococcal nuclease sensitivity of chromatin in Leishmania donovani. PLoS ONE. (2017) 12:e0177372. doi: 10.1371/journal.pone.0177372
80. Naveja JJ, Dueñas-González A, Medina-Franco JL. Drug repurposing for epigenetic targets guided by computational methods. In: Medina-Franco JL, editor. Epi-Informatics. Amsterdam: Elsevier (2016). p. 327–57. doi: 10.1016/B978-0-12-802808-7.00012-5
81. Mittal N, Muthuswami R, Madhubala R. The mitochondrial SIR2 related protein 2 (SIR2RP2) impacts Leishmania donovani growth and infectivity. PLoS Negl Trop Dis. (2017) 11:e0005590. doi: 10.1371/journal.pntd.0005590
82. Ronin C, Costa DM, Tavares J, Faria J, Ciesielski F, Ciapetti P, et al. The crystal structure of the Leishmania infantum silent information regulator 2 related protein 1: implications to protein function and drug design. PLoS ONE. (2018) 13:e0193602. doi: 10.1371/journal.pone.0193602
83. Pierce RJ, Khalife J. Parasite epigenetic targets. In: Sippl W, Jung M, editors. Epigenetic Targets in Drug Discovery. New Jersey, NJ: Wiley (2019). doi: 10.1002/9783527809257.ch16
84. Verçoza BRF, Godinho JLP, De Macedo-Silva ST, Huber K, Bracher F, De Souza W, et al. KH-TFMDI, a novel sirtuin inhibitor, alters the cytoskeleton and mitochondrial metabolism promoting cell death in Leishmania amazonensis. Apoptosis. (2017) 22:1169–88. doi: 10.1007/s10495-017-1397-8
85. Chua MJ, Arnold MS, Xu W, Lancelot J, Lamotte S, Späth GF, et al. Effect of clinically approved HDAC inhibitors on Plasmodium, Leishmania and Schistosoma parasite growth. Int J Parasitol. (2017) 7:42–50. doi: 10.1016/j.ijpddr.2016.12.005
Keywords: epigenetics, DNA methylation/demethylation, histone modification, non-coding RNA, leishmaniasis, host-pathogen Interactions, therapeutics, biomarkers
Citation: Afrin F, Khan I and Hemeg HA (2019) Leishmania-Host Interactions—An Epigenetic Paradigm. Front. Immunol. 10:492. doi: 10.3389/fimmu.2019.00492
Received: 31 December 2018; Accepted: 25 February 2019;
Published: 22 March 2019.
Edited by:
Antonio C. R. Vallinoto, Federal University of Pará, BrazilReviewed by:
Theo Araújo-Santos, Universidade Federal do Oeste da Bahia, BrazilRoberta Olmo Pinheiro, Fundação Oswaldo Cruz (Fiocruz), Brazil
Copyright © 2019 Afrin, Khan and Hemeg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Farhat Afrin, ZmFyaGF0YWZyaW5AZ21haWwuY29t; ZmFpc2xhbUB0YWliYWh1LmVkdS5zYQ==
†These authors have contributed equally to this work
 Farhat Afrin
Farhat Afrin Inbesat Khan
Inbesat Khan Hassan A. Hemeg1
Hassan A. Hemeg1