- 1Department of Hematopoiesis, Sanquin Research and Landsteiner Laboratory, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands
- 2Department of Experimental Immunology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands
- 3Department of Molecular and Cellular Hemostasis, Sanquin Research and Landsteiner Laboratory, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands
Tissue-resident memory CD8+ T (TRM) cells that develop in the epithelia at portals of pathogen entry are important for improved protection against re-infection. CD8+ TRM cells within the skin and the small intestine are long-lived and maintained independently of circulating memory CD8+ T cells. In contrast to CD8+ TRM cells at these sites, CD8+ TRM cells that arise after influenza virus infection within the lungs display high turnover and require constant recruitment from the circulating memory pool for long-term persistence. The distinct characteristics of CD8+ TRM cell maintenance within the lungs may suggest a unique program of transcriptional regulation of influenza-specific CD8+ TRM cells. We have previously demonstrated that the transcription factors Hobit and Blimp-1 are essential for the formation of CD8+ TRM cells across several tissues, including skin, liver, kidneys, and the small intestine. Here, we addressed the roles of Hobit and Blimp-1 in CD8+ TRM cell differentiation in the lungs after influenza infection using mice deficient for these transcription factors. Hobit was not required for the formation of influenza-specific CD8+ TRM cells in the lungs. In contrast, Blimp-1 was essential for the differentiation of lung CD8+ TRM cells and inhibited the differentiation of central memory CD8+ T (TCM) cells. We conclude that Blimp-1 rather than Hobit mediates the formation of CD8+ TRM cells in the lungs, potentially through control of the lineage choice between TCM and TRM cells during the differentiation of influenza-specific CD8+ T cells.
Introduction
Long-term memory of previously encountered pathogens is crucial to enable enhanced responses of our body's immune defenses in future encounters with the same pathogen. Antigen-specific memory CD8+ T cells form an essential aspect of immunological memory. Particularly in the context of local infections, tissue-resident memory CD8+ T (TRM) cells situated in the originally infected tissues are important for protective immunity (1). Their position in barrier tissues, including skin, intestine, female reproductive tract and lungs, places non-circulating CD8+ TRM cells at sites of pathogen entry, where they provide efficient early protection against local reinfection (2–5). The superior protective capacity of CD8+ TRM cells is mediated through direct effector functions and by promoting the activation and recruitment of other immune cells. Upon antigen encounter, CD8+ TRM cells rapidly release pro-inflammatory cytokines, including interferon-γ (IFN-γ), which induce a tissue-wide state of alert and initiate the recruitment of circulating B cells and memory T cells (4, 6, 7). In some tissues, CD8+ TRM cells can also directly lyse target cells and limit pathogen spread by employing cytotoxic mechanisms (8, 9). Given their potency in limiting pathogen spread, insights into the mechanisms regulating the development and maintenance of CD8+ TRM cells may contribute to improved strategies to induce protective immunity via vaccination (10, 11).
The localization of CD8+ TRM cells to different organs suggests the existence of tissue-specific adaptions, which, in turn, may influence the local development and maintenance of CD8+ TRM cells. Compared to CD8+ TRM cells in the skin and intestine, lung CD8+ TRM cells are distinct at the transcriptional level (12), indicating specific adaptions to the local microenvironments. Lung CD8+ TRM cells share common features with CD8+ TRM cells from other tissues, including the expression of tissue-retention molecules such as CD69 and the αE integrin CD103 (13, 14). Importantly, lung CD8+ TRM cells provide robust protection against heterosubtypic influenza virus infection (5, 15). However, CD8+ TRM cells in the lungs are divergent from CD8+ TRM cells at other peripheral sites in terms of their maintenance. While CD8+ TRM cells in most tissues are long-lived and self-sustaining (2, 16), the virus-specific CD8+ TRM cell population in the lungs declines over time after pulmonary infection, which coincides with waning of heterosubtypic immunity to influenza virus (5, 14, 17). The mechanisms underlying this limited longevity of lung CD8+ TRM cells are not fully understood. After pulmonary infection, CD8+ TRM cells localize to specific niches at sites of tissue regeneration in the lung, and it has been suggested that the disappearance of these niches over time may account for the limited longevity of lung CD8+ TRM cells (18). The preservation of the resident population in the lungs may additionally require continuous replenishment from a circulating effector memory CD8+ T (TEM) cell pool (17, 19). The gradual decline in the capacity of circulating memory CD8+ T cells to form CD8+ TRM cells may thus contribute to the demise of the lung CD8+ TRM cell population (17). Consequently, tissue residency might be differentially regulated at the transcriptional level for CD8+ T cells in the lung compared to other organs.
We have recently found that the transcription factor Hobit and its homolog Blimp-1 control the generation and/or maintenance of CD8+ TRM cells across several tissues, including skin, small intestine, liver, and kidney (20). These transcription factors instruct a universal program of tissue-residency, in part by directly suppressing the tissue egress receptors CCR7 and S1PR1 (20). Here, we investigated the role of Hobit and Blimp-1 in the development of lung CD8+ TRM cells after pulmonary influenza virus infection. CD8+ TRM cells in the lungs exhibited high expression of Hobit and Blimp-1 at the transcript level. However, we found that Blimp-1, but not Hobit, is essential for the formation of lung CD8+ TRM cells. Blimp-1 also limited the formation of central memory CD8+ T (TCM) cells. These findings highlight the unique transcriptional regulation of CD8+ TRM cells in the lung, which may have implications for future influenza vaccination strategies.
Results
Lung CD8+ TRM Cells Arising After Respiratory Influenza Virus Infection Express Hobit
Infection of mice with influenza virus induces differentiation of virus-specific CD8+ T cells into CD8+ TRM cells, which persist in the lung and provide protection against subsequent reinfection (5, 21, 22). To investigate lung CD8+ TRM cells arising after influenza virus infection, mice were infected intranasally with HKx31 influenza A virus and CD8+ T cells were isolated and analyzed in the memory phase (day 30+ p.i.). Influenza virus infection gave rise to a substantial CD69+ CD8+ TRM cell population in the lung, which partially expressed CD103 (Figure 1A). This population was nearly absent in lungs from naïve mice, indicating that the vast majority of CD8+ TRM cells were a direct result of influenza virus infection. In mice, a core signature of gene-expression has been determined in CD8+ TRM cells (12). Transcriptional profiling of CD69+ and CD69− memory CD8+ T cells isolated from lungs of HKx31-immune mice (day 30+ p.i.) by RNA sequencing confirmed the resident phenotype of the CD69+ CD8+ T cell population arising after influenza virus infection (Figure 1B). When compared to the TRM core signature, the obtained transcriptional profiles of lung CD8+ TRM cells showed a good congruency. Genes associated with tissue-residency, including Cdh1, Itga1, Itgae, Rgs1, and Rgs2, were specifically upregulated in the CD69+ population, while genes associated with circulating memory CD8+ T cells, including tissue-egress factors (e.g., S1pr1 and S1pr5), were substantially downregulated in these cells (Figure 1B). Overall, out of 35 genes of the TRM core signature, 20 were significantly up- or downregulated in the CD69+ CD8+ T cell compartment compared to the CD69− CD8+ T cell pool from the lung. Importantly, the transcription factor Hobit (encoded by Zfp683), which we have recently identified as a key regulator of tissue-residency (20), was also significantly upregulated in lung CD8+ TRM cells, as compared to the circulating memory CD8+ T cell population in the lung (Figures 1B,C). In contrast, expression levels of the related transcription factor Blimp-1 (encoded by Prdm1) were not significantly different between the two memory subsets (Figure 1C). Members of the common γ-chain cytokines, in particular IL-2, IL-7, and IL-15, play an important role in the maintenance of memory CD8+ T cells. In the lung, both circulating and resident memory CD8+ T cells expressed the individual components of the IL-7 (Il7r, Il2rg) and IL-15 receptor (l2rb, Il2rg) (Figure 1D). In contrast, the alpha chain of the IL-2 receptor was upregulated in lung CD8+ TRM cells compared to CD69− memory CD8+ T cells in the lung. Furthermore, in comparison to their circulating counterparts in the lung, the CD69+ lung TRM cells expressed significantly higher levels of pro-inflammatory cytokines, chemokines and cytotoxic mediators, including colony-stimulating factor-1 (Csf1), lymphotactin (Xcl1) granzyme B (Gzmb) and TNF-related apoptosis-inducing ligand (TRAIL, Tnfsf10), indicative of a poised effector state (Figures 1E–G). Taken together, influenza virus infection induced a distinct population of CD69+ CD8+ T cells in the lungs, which were identified as bona fide TRM cells by transcriptional analysis. Importantly, these lung-resident CD8+ T cells exhibited elevated transcript levels of effector molecules and were characterized by high expression of the TRM-associated transcription factor Hobit.
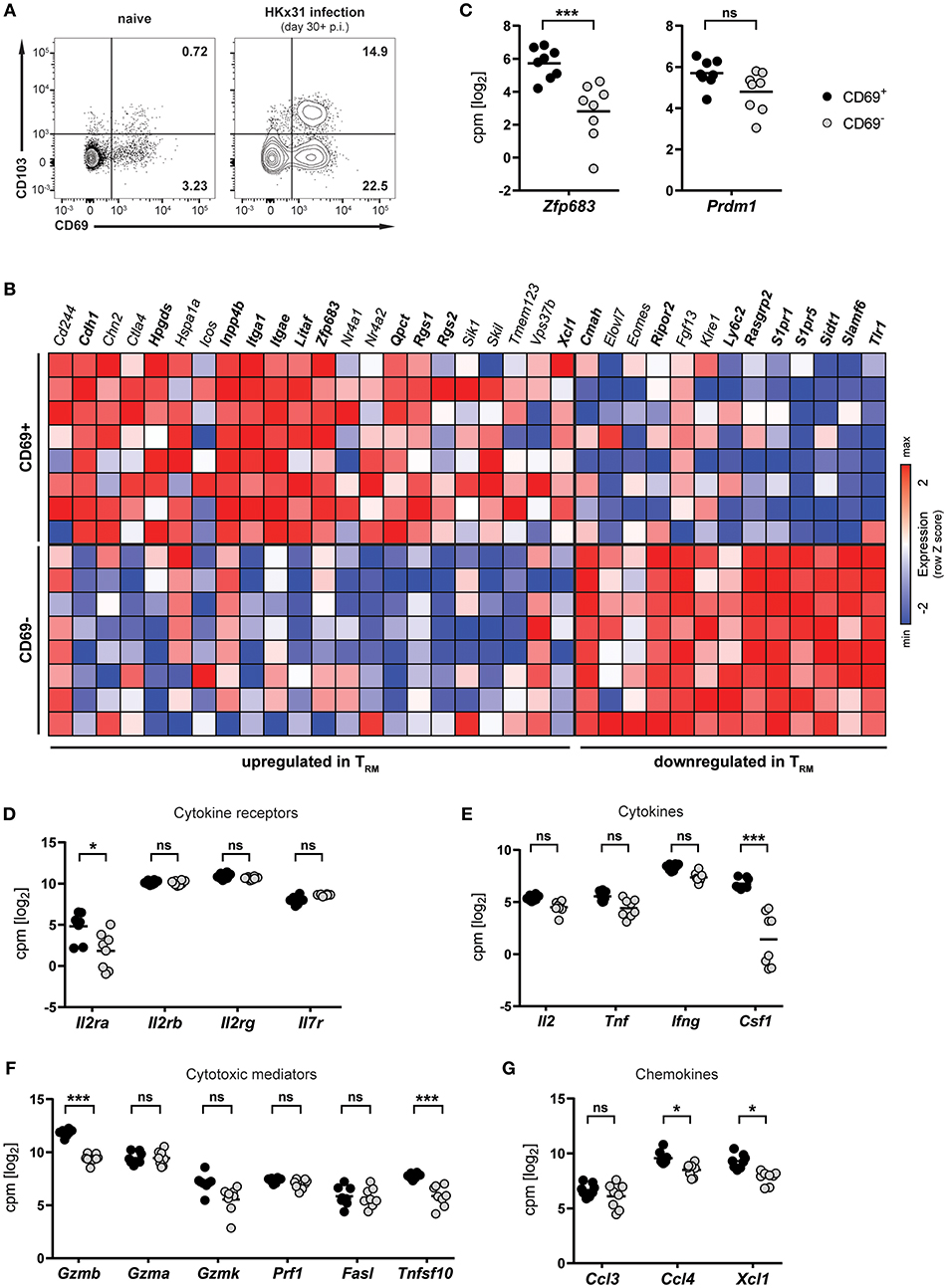
Figure 1. Transcriptional profile of lung CD8+ TRM cells arising after influenza virus infection. (A) Representative flow cytometry plots are shown of CD69 and CD103 expression on memory (CD44high CD62Llo) CD8+ T cells isolated from lungs of naïve mice or lungs of mice at day 30+ (memory phase) after intranasal HKx31 influenza virus infection. (B) Expression (column Z-score) of mRNA from genes belonging to the TRM core signature (12) in CD69+ and CD69− memory (CD44high CD62Llo) CD8+ T cells isolated from murine lungs at day 30+ after HKx31 infection. Genes in bold are differentially expressed in CD69+ vs. CD69− cells. Expression of gene Usp33, part of the TRM core signature, was not detected by our analysis. (C–G) Expression (log2 read counts per million, voom normalized expression) of (C) Hobit (Zfp683) and Blimp-1 (Prdm1), genes encoding for (D) common γ-chain cytokine receptors, (E) cytokines, (F) cytotoxic mediators, and (G) selected chemokines is shown in CD69+ and CD69− memory (CD44high CD62Llo) CD8+ T cells isolated from murine lungs at day 30+ after HKx31 infection. Symbols represent individual mice and line indicates the mean. Data from one experiment (n = 8), taken from Hombrink et al. (23). *FDR adjusted P < 0.05; *** FDR adjusted P < 0.001; ns: not significant.
CD8+ TRM Cell Formation in the Lung Requires Hobit and/or Blimp-1
Given its selective expression in lung CD8+ TRM cells, we hypothesized that Hobit may contribute to the development of these cells. In other tissues, including the skin, liver, kidney, and small intestine, Hobit regulates the generation and/or maintenance of CD8+ TRM cells together with its homolog Blimp-1 (20). In order to investigate the role of these two transcription factors in the development of lung CD8+ TRM cells, mixed bone marrow (BM) chimeric mice were generated, containing a WT compartment and a compartment lacking functional Hobit and Blimp-1 (double knock-out, DKO) (Figure 2A). An approach with mixed BM chimeric mice was chosen to minimize indirect effects on CD8 T cell differentiation through differences in viral clearance. Mice were infected intranasally with HKx31 virus, and the virus-specific (Db NP366+) CD8+ T cell response was analyzed over time. Previous studies have demonstrated a critical role for Blimp-1 in terminal effector cell (TEC) differentiation (24, 25). In line with these findings, analysis of virus-specific Db NP366+ CD8+ T cells in the blood at the peak of the anti-viral effector CD8+ T cell response (day 10 p.i.) revealed a substantial decrease in KLRG1+ CD127− TECs in the DKO compared to the WT compartment (Figures 2B–D). Concomitantly, Db NP366+ cells deficient for both Hobit and Blimp-1 exhibited a sharp increase in CD127+ KLRG1− memory precursor effector cells (MPECs) compared to their WT counterparts (Figures 2C,D). In lung tissue, a distinct CD69+ population was already observed at the effector stage, while CD103 expression was minimal (Figure 2F). Both the WT and the DKO compartment gave rise to similar frequencies of CD69+ CD103− and CD69+ CD103+ cells at this stage, suggesting little impact of Hobit and Blimp-1 deficiency on the formation of these cells (Figures 2E–G). In contrast, Db NP366+ DKO cells generated less TRM cells in the lung at the memory phase than their WT counterparts (Figures 2H,I). This defect was most pronounced for CD69+ CD103+ cells, which were decreased in both frequencies and absolute numbers in the DKO compartment compared to the WT compartment (Figures 2I,J). Interestingly, DKO cells formed CD69+ CD103− TRM cells at near similar frequencies as WT cells, indicating little effect of combined Hobit and Blimp-1 deficiency on the generation of this population (Figures 2I,K). Apart from CD69 and CD103, CD8+ TRM cells across tissues express additional tissue-residency markers, including the chemokine receptor CXCR6 and the integrin CD49a (26–29). Influenza-virus-specific WT CD8+ T cells in the lungs co-expressed CXCR6 and CD49a at similar frequencies as the residency marker CD69, suggesting that both molecules also identify CD8+ TRM cells in this tissue (Figures 2L,M). Interestingly, combined deficiency for Hobit and Blimp-1 impaired the formation of CXCR6+ CD49ahigh cells, which were decreased in both frequencies and absolute numbers in the DKO compartment compared to the WT compartment (Figures 2L,M). In all, these results show that the combined genetic ablation of Hobit and Blimp-1 results in reduced TEC and enhanced MPEC formation during the effector CD8+ T cell response, and impairs the generation of CD103+ lung TRM cells in the memory CD8+ T cell response.
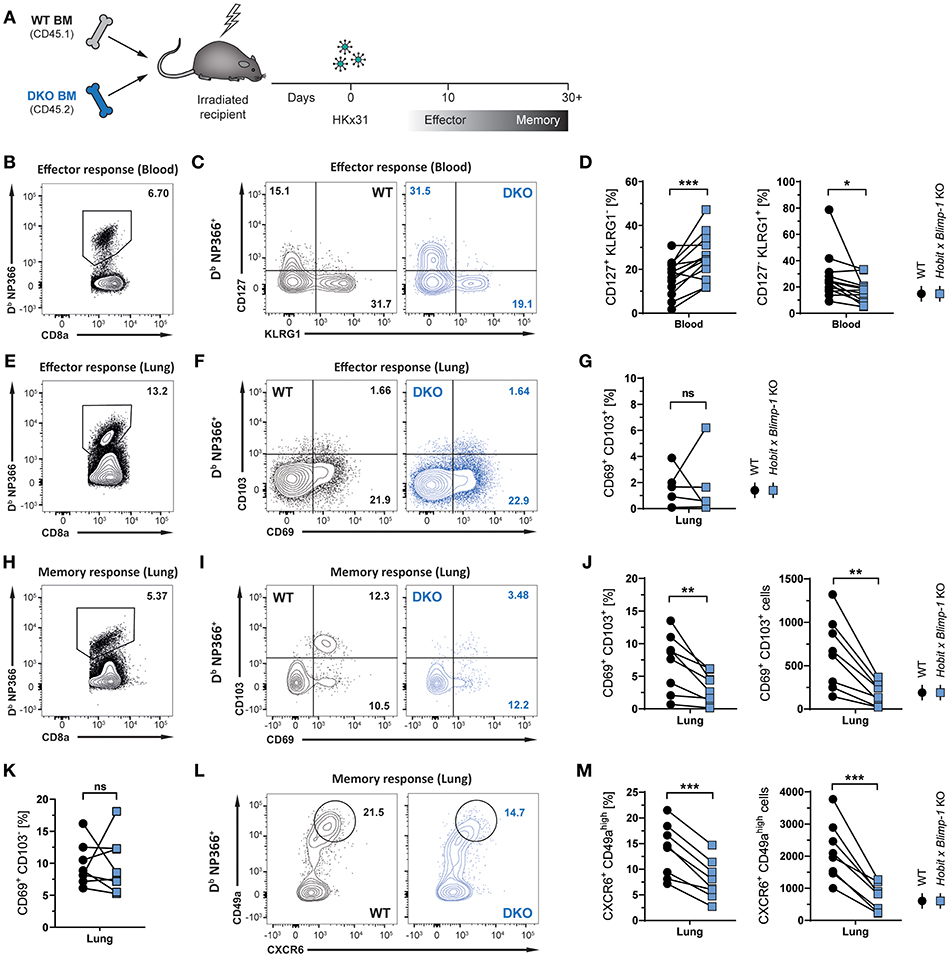
Figure 2. Formation of lung CD8+ TRM cells depends on Hobit and/or Blimp-1. (A) Experimental scheme shows the generation of mixed bone marrow (BM) chimeras from WT and Hobit and Blimp-1 KO (DKO) mice (1:1 ratio) and HKx31 influenza virus infection of these chimeric mice. (B–G) Analysis at the effector time point is shown. (B,E) Representative flow cytometry plot shows frequency of Db NP366+ cells within CD8+ T cell population in (B) blood and (E) lung at day 10 post infection. (C,F) Representative flow cytometry plots show (C) expression of CD127 and KLRG1 and (F) expression of CD69 and CD103 on Db NP366+ donor CD8+ T cells of the WT and DKO compartment from (C) blood and (F) lungs at day 10 post infection. (D,G) The frequencies of (D) CD127+KLRG1− and KLRG1+CD127− and (G) CD69+ CD103+ Db NP366+ donor CD8+ T cells of the WT and DKO compartment from (D) blood and (G) lungs at day 10 post infection were quantified. (H–M) Analysis at the memory time point is shown. (H) Representative flow cytometry plot shows frequency of Db NP366+ cells within CD8+ T cell population in lung at day 30+ post infection. (I,L) Representative flow cytometry plots show (I) expression of CD69 and CD103 and (L) expression of CXCR6 and CD49a on Db NP366+ donor CD8+ T cells of the WT and DKO compartment from lungs at day 30+ post infection. (J,K,M) The frequencies and absolute numbers of (J) CD69+CD103+, (K) CD69+CD103−, and (M) CXCR6+CD49ahigh Db NP366+ donor CD8+ T cells of the WT and DKO compartment from lungs at day 30+ post infection were quantified. Data from (G) one experiment (n = 5) or combined data from (C,J,K,M) two independent experiments (n = 8). Symbols represent individual mice, lines connect paired samples. Paired t-test. *P < 0.05; **P < 0.01; ***P < 0.001, ns: not significant.
Hobit and Blimp-1 Impair Formation of CD8+ TCM Cells
Apart from CD8+ TRM cells in the lung, influenza virus infection also induces the formation of circulating effector memory (TEM) and central memory (TCM) CD8+ T cells (30, 31). To assess the impact of co-deficiency of Hobit and Blimp-1 on these circulating memory subsets, we analyzed virus-specific (Db NP366+) WT and DKO cells isolated from secondary lymphoid organs of mixed BM chimeric mice after HKx31 infection. In both spleen and lung-draining lymph nodes (mediastinal lymph nodes, mLN), virus-specific CD44high CD62L+ CD8+ TCM cells were present at elevated levels in the DKO compartment compared to the WT compartment (Figures 3A,B), both in frequencies and in absolute numbers. In contrast, no effect of Blimp-1 and Hobit deficiency was observed in the CD44high CD62L− CD8+ TEM subset, as these cells were present in both the WT and the DKO compartment in similar numbers (Figure 3C). Taken together, these data suggest that Hobit and/or Blimp-1 not only drive the formation of CD8+ TRM cells, but also suppress the development of CD8+ TCM cells after influenza virus infection.
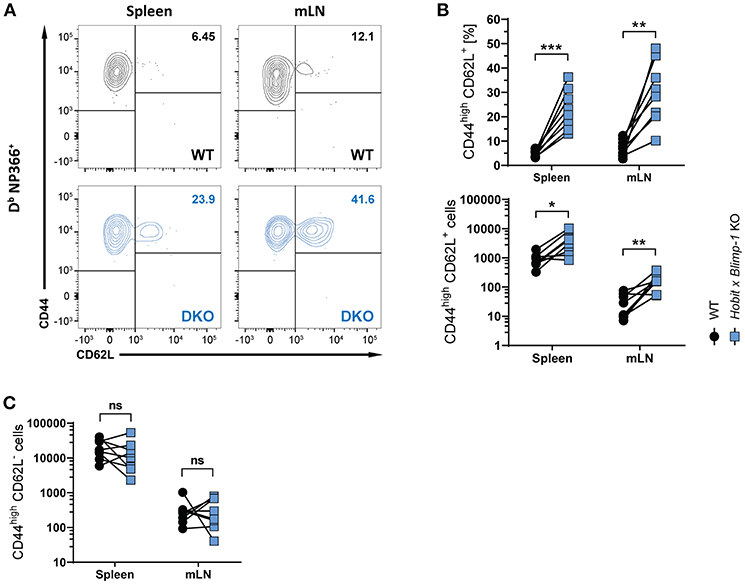
Figure 3. Increased CD8+ TCM formation upon combined loss of Hobit and Blimp-1. (A–C) WT: Hobit × Blimp-1 KO (DKO) mixed bone marrow chimeras were infected intranasally with HKx31 influenza virus. The phenotype of virus-specific (Db NP366+) WT and DKO donor CD8+ T cells was analyzed at day 30+ post infection (memory phase). (A) Representative flow cytometry plots show expression of CD44 and CD62L on Db NP366+ donor CD8+ T cells of the WT and DKO compartment from spleen and mediastinal lymph nodes (mLN). (B,C) The frequencies and absolute numbers of (B) CD44highCD62L+ Db NP366+ donor CD8+ T cells and (C) absolute numbers of CD44highCD62L− Db NP366+ donor CD8+ T cells of the WT and DKO compartment in spleen and mLN were quantified. Combined data from two independent experiments (n = 8). Symbols represent individual mice, lines connect paired samples. Paired t-test. *P < 0.05; **P < 0.01; ***P < 0.001, ns: not significant.
Generation of Lung CD8+ TRM Cells Depends on Blimp-1, but Not Hobit
We next investigated whether Hobit and Blimp-1 collaborated in the development of lung CD8+ TRM cells, as observed in other tissues (20), or whether either one was the dominant transcription factor driving CD8+ TRM formation in the lungs. To this end, three groups of mixed BM chimeric mice were generated, consisting of one control group, containing two WT compartments (CD45.1 and CD45.2) and two experimental groups, containing a WT (CD45.1) compartment next to either a Hobit KO (CD45.2) compartment or a Blimp-1 KO (CD45.2) compartment. The mixed BM chimeric mice were infected intranasally with HKx31 virus, and the virus-specific (Db NP366+) CD8+ T cell response was analyzed over time. As expected, similar to cells lacking both functional Hobit and Blimp-1 (Figures 2C,D), Db NP366+ Blimp-1 KO cells contained elevated frequencies of CD127+ KLRG1− MPECs at the peak of the anti-viral CD8+ T cell effector response (Figures 4A,B). Moreover, Db NP366+ Blimp-1 KO cells nearly lacked KLRG1+ CD127− TECs, which was also observed for cells with combined deficiency for Hobit and Blimp-1 (Figures 2C,D). Neither of these phenotypes was observed for Db NP366+ Hobit-deficient cells, which contained TECs and MPECs at similar frequencies as their WT counterparts (Figures 4A–C). These data indicate a dominant role of Blimp-1 over Hobit in regulating the balance between terminal effector and memory precursor cell differentiation during the effector response. After clearance of the infection, the Db NP366+ Hobit KO compartment exhibited no defects in the formation of CD8+ TRM cells in the lung (Figures 4D,E). In contrast, the Blimp-1 deficient compartment of virus-specific cells was severely impaired in the formation of CD69+ CD103+ CD8+ TRM cells (Figures 4D,E). As previously observed for cells with combined deficiency for Hobit and Blimp-1, Db NP366+ Blimp-1 KO cells did not exhibit a substantial defect in generating CD69+ CD103− cells in the lung (Figure 4F). Thus, Blimp-1, but not Hobit, appears to be essential for the formation of terminal effector cells and CD69+ CD103+ CD8+ TRM cells in the lung.
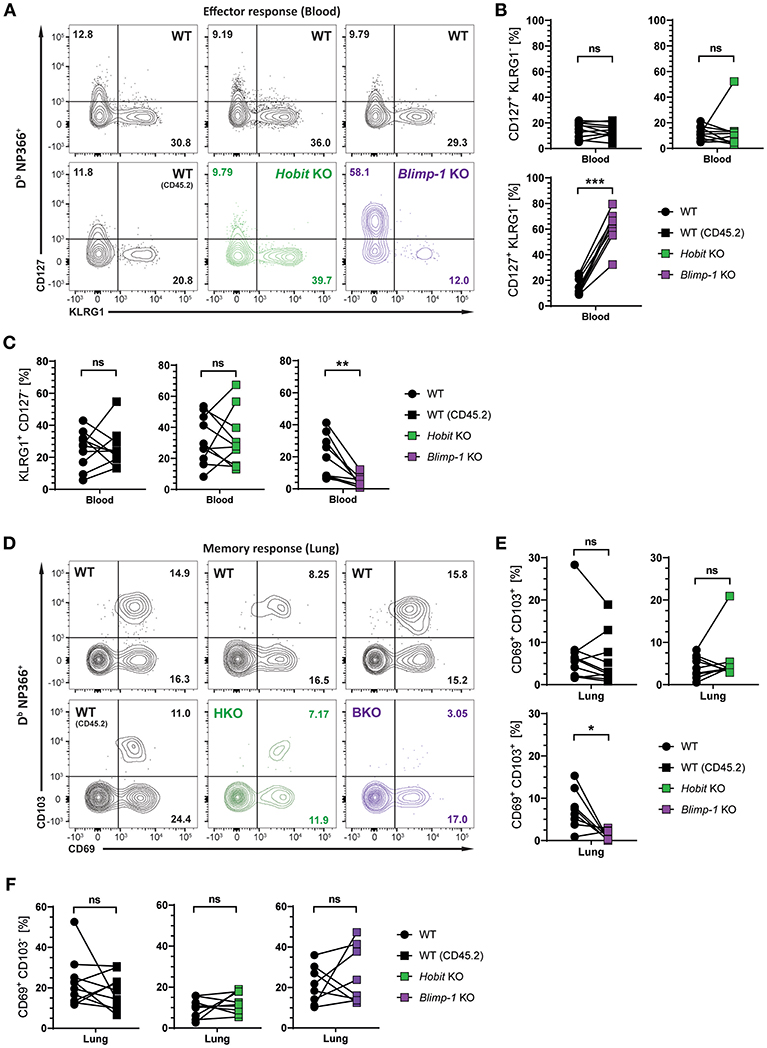
Figure 4. Formation of lung CD8+ TRM cells depends on Blimp-1, but not Hobit. Mixed bone marrow chimeras [WT:WT; WT:Hobit KO (HKO); WT:Blimp-1 KO (BKO)] were infected intranasally with HKx31 influenza virus. The phenotype of virus-specific (Db NP366+) WT (CD45.1+), WT (CD45.2+), HKO (CD45.2+), and BKO (CD45.2+) donor CD8+ T cells was analyzed. (A) Representative flow cytometry plots show expression of CD127 and KLRG1 by Db NP366+ donor CD8+ T cells from the blood of mixed bone marrow chimeras at day 10 post infection. (B,C) Frequencies of (B) CD127+KLRG1− and (C) KLRG1+CD127− Db NP366+ donor CD8+ T cells from the blood of mixed bone marrow chimeras at day 10 post infection were quantified. (D) Representative flow cytometry plots show expression of CD69 and CD103 by Db NP366+ donor CD8+ T cells from the lungs at day 30+ post infection. (E,F) Frequencies of (E) CD69+CD103+ and (F) CD69+CD103− Db NP366+ donor CD8+ T cells from the lungs at day 30+ post infection were quantified. Combined data from two independent experiments (n = 9–10). Symbols represent individual mice, lines connect paired samples. Paired t-test. *P < 0.05; **P < 0.01; ***P < 0.001, ns: not significant.
Blimp-1 Suppresses CD8+ TCM Formation
We next considered the role of Blimp-1 and its homolog Hobit in the formation of other memory CD8+ T cell subsets. Consistent with previous research demonstrating the repressive role of Blimp-1 in the formation of CD8+ TCM cells (24, 25), we observed elevated frequencies of CD44high CD62L+ Db NP366+ cells in the Blimp-1 KO compartment of mixed BM chimeras already during the effector CD8+ T cell response (Figures 5A,B). Importantly, Blimp-1 deficiency resulted in a strong increase in the frequency of Db NP366+ CD8+ TCM cells in the circulation, lung and secondary lymphoid organs during the memory phase (Figures 5C–E). This effect was not observed for cells lacking functional Hobit, neither during the effector nor the memory response (Figures 5B,D). Consequently, these data suggest that Blimp-1 suppresses CD8+ TCM development as early as in the effector response, while Hobit does not appear to have an essential role in this process.
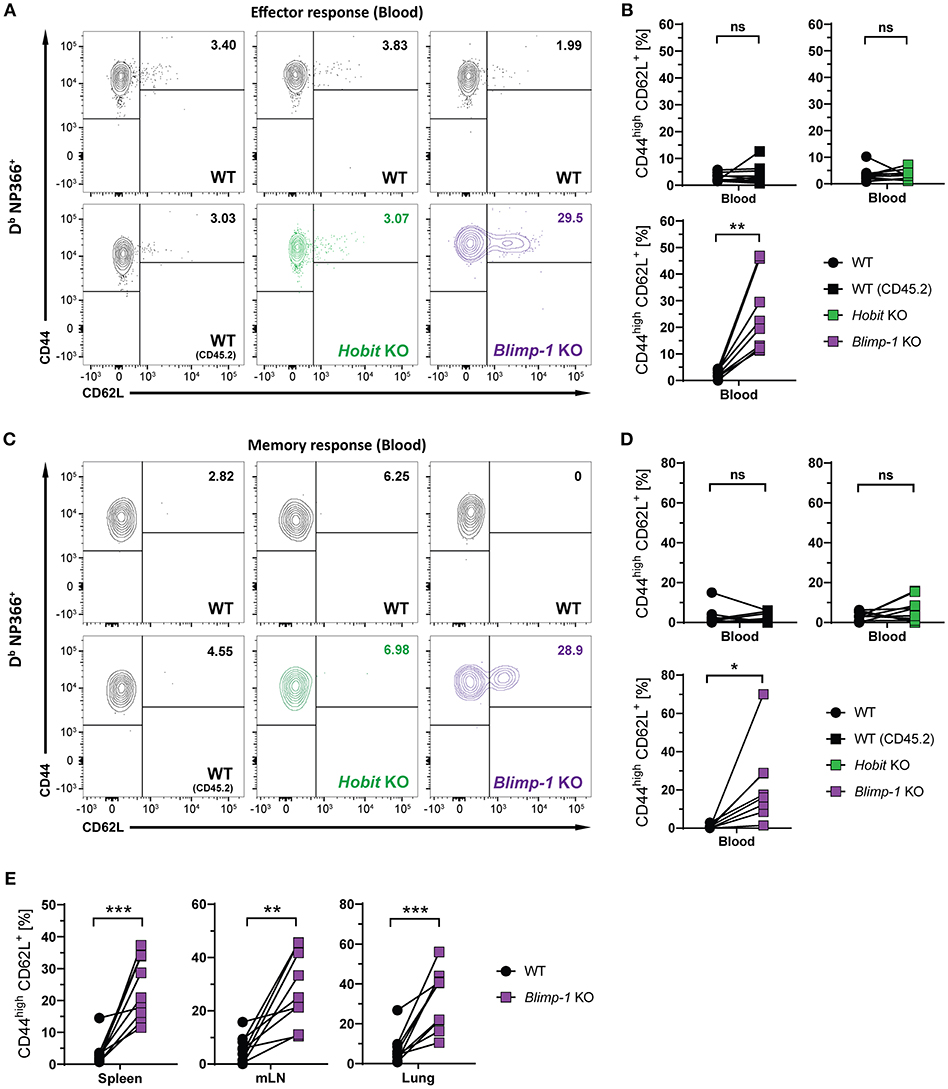
Figure 5. CD8+ TCM formation is suppressed by Blimp-1. Mixed bone marrow chimeras [WT:WT; WT:Hobit KO (HKO); WT:Blimp-1 KO (BKO)] were infected intranasally with HKx31 influenza virus. The phenotype of virus-specific (Db NP366+) WT (CD45.1+), WT (CD45.2+), HKO (CD45.2+) and BKO (CD45.2+) donor CD8+ T cells was analyzed in blood (A–D) and in the indicated tissues (E). (A,C) Representative flow cytometry plots show expression of CD44 and CD62L on Db NP366+ donor CD8+ T cells from the blood at (A) day 10 and (C) day 30+ post infection. (B,D,E) Frequencies of CD44highCD62L+ cells Db NP366+ donor CD8+ T cells from (B) blood at day 10 post infection, and from (D) blood as well as from (E) spleen, mediastinal lymph nodes (mLN) and lung at day 30+ post infection were quantified. Combined data from two independent experiments (n = 9–10). Symbols represent individual mice, lines connect paired samples. Paired t-test. *P < 0.05; **P < 0.01; ***P < 0.001, ns: not significant.
Blimp-1 Suppresses TCF-1 Expression in Lung CD8+ TRM Cells
The transcription factors Hobit and Blimp-1 regulate the development of CD8+ TRM cells in part by suppressing genes, which are incompatible with the establishment of tissue-residency (20). A direct target of both Hobit and Blimp-1 is T-cell factor 1 (TCF-1). TCF-1 is a nuclear effector of the canonical Wingless/Integration 1 (Wnt) signaling pathway and constitutes an essential transcription factor for the development of CD8+ TCM cells (32, 33). Given the enhanced formation of CD8+ TCM cells upon genetic ablation of Blimp-1, we analyzed TCF-1 expression in different memory CD8+ T cells subsets arising after influenza infection. As expected, WT Db NP366+ CD8+ TCM cells exhibited high uniform expression of TCF-1 at the protein level, while virus-specific WT CD8+ TRM cells in the lung showed markedly lower expression of TCF-1 (Figures 6A,B). Hobit deficiency did not significantly impact TCF-1 expression in both CD8+ TCM and TRM populations. Loss of Blimp-1 had no effect on the high levels of TCF-1 expression in CD8+ TCM cells. However, Blimp-1 deficiency resulted in strongly increased TCF-1 protein expression in the remaining virus-specific CD8+ TRM cells present in the lung (Figures 6A,B). These results indicate that Blimp-1 mediates suppression of TCF-1 expression in lung CD8+ TRM cells, which may contribute to the instruction of CD8+ TRM development in the lungs.
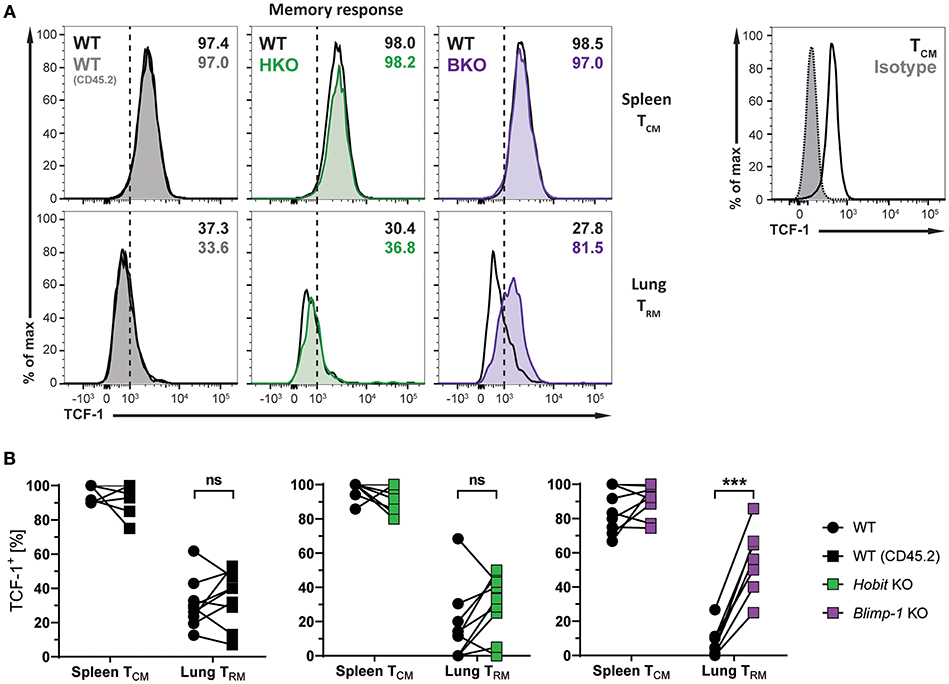
Figure 6. TCF-1 expression in lung CD8+ TRM cells is suppressed by Blimp-1. (A,B) Mixed bone marrow chimeras [WT:WT; WT:Hobit KO (HKO); WT:Blimp-1 KO (BKO)] were infected intranasally with HKx31 influenza virus. At day 30+ post infection (memory phase), the expression of TCF-1 by virus-specific (Db NP366+) WT (CD45.1+), WT (CD45.2+), HKO (CD45.2+), and BKO (CD45.2+) donor CD8+ T cells was analyzed. Expression of TCF-1 was analyzed in donor CD8+ TCM cells (CD44highCD62L+) from spleen and donor CD8+ TRM cells (CD69+CD103+) from lungs. TCF-1 staining was validated using isotype control staining on CD8+ TCM cells (CD44highCD62L+) from spleen. (A) Representative histograms show expression of TCF-1, and (B) graphs display frequencies of TCF-1+ cells. Combined data from two independent experiments (n = 9–10). Symbols represent individual mice, lines connect paired samples. Paired t-test. ***P < 0.001, ns: not significant.
Discussion
The maintenance of lung CD8+ TRM cells is distinct from that of CD8+ TRM cells at other sites. In contrast to CD8+ TRM cells in skin and small intestine, lung-resident CD8+ T cells are not maintained as stable populations, but slowly decline over time. Here, we show that CD8+ TRM cells in the lungs diverge from TRM cells in the liver, intestine and skin in terms of their transcriptional regulation. Unlike their counterparts in other tissues, differentiation and/or maintenance of lung CD8+ TRM cells after influenza virus infection was entirely independent of Hobit. Instead, we found that CD103+ lung CD8+ TRM cells exclusively depended on Blimp-1. Blimp-1 repressed the formation of virus-specific CD8+ TCM cells and the expression of the transcription factor TCF-1 in lung CD8+ TRM cells. These findings may suggest that Blimp-1 controls the lineage choice between CD8+ TCM and TRM cells after influenza infection and highlight a unique aspect of the transcriptional regulation of CD8+ TRM cells in the lung.
In the skin, liver, kidney, and small intestine, CD8+ TRM cell formation and/or maintenance is co-regulated by Hobit and Blimp-1 (20). Our data demonstrate that CD103+ CD8+ TRM cells in the lungs do not require Hobit for their formation, but entirely rely on Blimp-1. These findings may reflect differences in signals from the local microenvironment modulating Hobit and Blimp-1 expression. While influenza virus causes an acute infection of the airways, residual viral antigen can persist in the lung and the draining lymph nodes for extended periods of time after clearance of the infection (34, 35). Presence of remaining viral antigen can induce T cell responses long after the infection and modulates the migration and localization of virus-specific memory CD8+ T cells (34–36). Persistent antigen may thus impact on the local CD8+ TRM cell pool in the airways. Previous research has demonstrated the requirement of local antigen encounter for the establishment of CD8+ TRM cells in the lungs (37). However, it remains unclear whether residual antigen also influences the maintenance of lung CD8+ TRM cells. TCR signaling can induce Blimp-1 expression in T cells (38). Persistent viral antigen in the lungs may thus modulate Blimp-1 expression in CD8+ TRM cells at this site and in turn bias the dependence of lung CD8+ TRM cells toward Blimp-1. In addition, Blimp-1 expression is upregulated by pro-inflammatory cytokines, including IL-2 and IL-12 (39). After influenza virus infection, CD8+ TRM cells localize to sites of tissue regeneration in the lungs (18), which may be characterized by residual inflammation, thus favoring Blimp-1 expression. Our transcriptional analysis revealed selective upregulation of the IL-2 receptor alpha chain (Il2ra) by CD69+ CD8+ TRM cells arising in the lungs after influenza infection, potentially indicating increased responsiveness to IL-2. Furthermore, IL-2 signaling is required for residency of CD4+ TRM cells in the lungs (40). Consequently, IL-2 and other pro-inflammatory cytokines may also act on lung CD8+ TRM cells and enhance Blimp-1 expression, resulting in Blimp-1 dependent maintenance of these memory T cells.
In contrast to Blimp-1, Hobit does not appear to play an essential role in the differentiation and/or maintenance of lung CD8+ TRM cells. Nevertheless, Hobit is specifically expressed by CD69+ CD8+ T cells in the lungs after influenza virus infection, indicating the presence of local cues driving Hobit mRNA expression. The cytokine interleukin-15 (IL-15) mediates Hobit expression in a T-bet-dependent manner (20). Local IL-15 and potential other signals may thus promote Hobit mRNA expression in lung CD8+ TRM cells. Epithelial cells in the lung airways constitutively express IL-15 (41), and the cytokine mediates the recruitment of effector CD8+ T cells into the inflamed lung after influenza virus infection (42). Furthermore, IL-15 is required for persistence of CD103+ CD8+ TRM cells in the lungs (43), as well as for CD8+ TRM maintenance in skin, salivary glands, and kidney (43, 44). Taken together, these findings suggest a prominent role for IL-15 in modulating CD8+ T cell responses in the lungs. Whether IL-15 facilitates Hobit expression in lung CD8+ TRM cells in a similar fashion as in CD8+ TRM cells at other sites, remains to be determined. Despite being differentially expressed, Hobit does not appear to have an essential role in the formation of lung CD8+ TRM cells. Using transcriptional profiling, we have detected Hobit and Blimp-1 expression at the mRNA level. However, this does not necessarily reflect the expression of Hobit and Blimp-1 at the protein level, which may be subject to posttranscriptional regulation. Currently, there are no tools available to study Hobit protein expression in mice. Regarding Blimp-1, we have recently demonstrated a discrepancy between the expression of Blimp-1 at the transcriptional and protein level in memory CD8+ T cells (9), suggesting regulation at the level of translation or degradation. In line with this, Blimp-1 is similarly expressed by both CD69+ and CD69− memory CD8+ T cells in the lungs at the transcript level, while Blimp-1 deficiency only perturbs the formation of lung CD8+ TRM cells. Therefore, differences at the level of posttranscriptional regulation of Hobit and Blimp-1 expression may shape the unique dependency of lung CD8+ TRM cells on Blimp-1.
Our data demonstrate that lung CD8+ TRM cells do not require Hobit for their development, and this may have implications for their effector functions. In contrast to circulating memory CD8+ T cells, CD8+ TRM cells in many tissues maintain high levels of the cytotoxic mediator granzyme B (8, 26, 45). On a transcriptional level, we could show that lung CD8+ TRM cells express elevated levels of granzyme B mRNA. However, lung CD8+ TRM cells do not retain granzyme B protein expression and exhibit little cytotoxicity (46, 47). We recently showed that Hobit is essential for maintenance of granzyme B expression in liver and intestinal CD8+ TRM cells (9). This may indicate a functional link between the poor cytolytic potential of lung CD8+ TRM cells and their independence from Hobit-mediated regulation. Whether Blimp-1 and/or Hobit regulate effector responses of lung CD8+ TRM cells upon reinfection, remains to be determined.
Immunosurveillance of peripheral tissues for reinfection is primarily performed by CD8+ TRM cells (48). These resident populations are characterized by shared phenotypic, transcriptional and functional features, but also exhibit distinct differences across tissues (20, 28). In addition, growing evidence suggests further heterogeneity of CD8+ TRM cells within one tissue. Expression of CD103 and CD49a delineates CD8+ TRM cell populations with discrete functional capacities in the intestine and skin, respectively (29, 49). In the lung, both CD69+ CD103− and CD69+ CD103+ CD8+ TRM cells arise after influenzas virus infection. While genetic ablation of Blimp-1 greatly impaired the formation of CD69+ CD103+ CD8+ TRM cells, the CD69+ CD103− population was less affected, suggesting distinct transcriptional regulation of the two subsets. Moreover, CD103− CD8+ TRM cells may occupy different sites, as they are unable to undergo interaction with E-cadherin expressing epithelial cells, a process requiring CD103 expression (50, 51). In line with this, CD103− CD8+ TRM cells in the intestine display localization and functional properties separate from their CD103+ counterparts (49). Hence, lung tissue may harbor distinct CD8+ TRM cell populations, delineated by CD103 expression and dependency on Blimp-1.
The transcriptional programs governing the differentiation of naïve CD8+ T cells into different subsets of memory CD8+ T cells, namely TCM, TEM and TRM cells, are gradually being uncovered. However, it is still incompletely understood how transcription factors control the fate choice between the memory CD8+ T cell subsets. Previous research has highlighted the importance of the transcription factors Runx3 and Notch in the formation and/or maintenance of CD8+ TRM cells in the lungs. While Runx3 is required for the formation of lung CD8+ TRM cells, Notch regulates the maintenance of these cells (23, 52). Here, we show that the transcription factor Blimp-1 promotes formation of lung CD8+ TRM cells, while suppressing the development of CD8+ TCM cells. These phenotypes were confined to the Blimp-1 deficient, but not the WT compartment of mixed BM chimeras, and are thus likely due to cell-intrinsic effects rather than indirect effects involving competition for shared resources. Interestingly, Blimp-1 had no impact on the formation of CD44high CD62L− CD8+ TEM cells, indicating that this transcription factor primarily regulates the lineage choice between CD8+ TRM and TCM cells. We have previously demonstrated that Blimp-1 suppresses the expression of TCF-1 via direct binding to the TCF-1 encoding Tcf7 locus. Consistently, Tcf7 downregulation is a common feature of resident lymphocyte populations (20). We also observed low expression of TCF-1 protein in lung CD8+ TRM cells, which was strongly increased upon genetic ablation of Blimp-1. TCF-1 is an essential transcription factor driving the development of circulating memory CD8+ T cells, in particular of CD8+ TCM cells (32, 33, 53). The suppression of TCF-1 by Blimp-1 may suggest that Blimp-1, in concert with TCF-1, controls the fate choice between CD8+ TRM and TCM cells. It will be interesting to determine if and how these two transcription factors drive this lineage choice during CD8+ T cell differentiation. This is of particular interest in the context of therapeutic approaches aiming to modulate the balance between different memory CD8+ T cell subsets.
Similar to mice, crossprotection against influenza virus in humans is strongly correlated with the presence of CD8+ T cells specific to conserved viral epitopes (54, 55). Moreover, CD8+ T cells recognizing respiratory viruses, including influenza virus, are enriched in the human lung (56, 57). A substantial fraction of these virus-specific CD8+ T cells express markers of tissue-residency, including CD69 and CD103 (58, 59), indicating the presence of bona fide influenza-specific CD8+ TRM cells within the human lung. Transcriptional profiling of human lung CD8+ TRM cells has demonstrated a strong overlap with gene signatures of CD8+ TRM cells from different murine tissues, including the selective upregulation of tissue retention molecules (e.g., CD49a, CD103, CD69), and the downregulation of tissue egress receptors (e.g., S1PR1, S1PR5) (23, 28). In line with our findings in mice, CD8+ TRM cells in the human lung exhibit no differential expression of Blimp-1 on the transcriptional level in comparison to circulating CD8+ TEM cells (23). In contrast to their murine counterparts, Hobit mRNA is not differentially expressed in human lung CD8+ TRM cells, relative to the expression in CD8+ TEM cells (23, 28), partly because the latter population also expresses Hobit in humans (23, 60). This may suggest that Hobit is not specifically involved in the differentiation of human lung CD8+ TRM cells. It remains to be determined if Blimp-1, on the other hand, promotes CD8+ TRM differentiation in humans.
We show that CD103+ CD8+ TRM cell formation in the lungs after influenza virus infection is dependent on the transcription factor Blimp-1. Blimp-1 driven suppression of TCF-1 expression may instruct the lineage choice between CD8+ TCM and TRM cells during memory CD8+ T cell formation. As there is growing recognition of the clinical importance of tissue-resident T cell memory, we believe that insights into the transcriptional mechanisms governing local CD8+ TRM cell differentiation are an important prerequisite for the development of novel vaccination approaches in the lungs.
Materials and Methods
Mice
C57BL/6JRj (CD45.2+ WT) mice were purchased from Janvier and B6.SJL-Ptprca Pepcb/BoyJ (CD45.1+ WT) mice from the Jackson Laboratory. Both lines were crossed to generate B6.SJL-Ptprca Pepcb/BoyJ x C57BL/6JRj mice. Zfp683−/− (Hobit KO) (61), Prdm1flox/flox × Lck Cre (Blimp-1 KO) mice (24) and Zfp683−/− × Prdm1flox/flox × Lck Cre mice (20) were maintained on a C57BL/6JRj background. For the generation of mixed bone marrow (BM) chimeras, CD45.1 × CD45.2 (B6.SJL-Ptprca Pepcb/BoyJ × C57BL/6JRj) recipient mice were irradiated (2x 5 Gy) and reconstituted with i.v. transfer of 10*106 BM cells per genotype. Mixed BM chimeras were used 12–16 weeks after reconstitution and chimerism of lymphocytes was confirmed prior to usage. All mice were maintained under SPF conditions in the animal facility of the Netherlands Cancer Institute (NKI). Animal experiments were conducted according to institutional and national guidelines.
Influenza Virus Infection
Mice were infected intranasally with 100x TCID50 (median tissue culture infectious dose) of HKx31 influenza A virus in a volume of 50 μl after anesthetization by inhalation of isoflurane. HKx31 virus was kindly provided by Dr. Guus Rimmelzwaan (University of Veterinary Medicine Hannover). At the indicated time points after infection, mice were sacrificed, and tissues were harvested for analysis of CD8+ T cell responses.
Tissue Preparation
Single cell suspensions from spleen and lymph nodes were prepared by mechanical disruption via passing of the tissues over a 70 μm cell strainer. Lung tissue was cut into pieces of 1 mm2 and enzymatically digested for 30 min at 37°C with 750 U ml−1 Collagenase Type I (Worthington) and 0.31 mg ml−1 DNase I (Roche, from bovine pancreas, grade II) in RPMI 1,640 supplemented with 10% (v/v) fetal calf serum (FCS). Single cell suspensions were generated by filtering over a 70 μm cell strainer and the isolated lymphocytes were purified by density centrifugation on a 66/44% Percoll gradient (GE Healthcare). Lung lymphocytes were extracted from the interphase of the Percoll gradient. Contaminating erythrocytes were removed using red blood cell lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA).
Flow Cytometry
Cells were incubated with antibodies and tetramers for 25 min at 4°C and washed with PBS supplemented with 0.5% (v/v) FCS. The following anti-mouse monoclonal antibodies were used: anti-CD3 (17A2), anti-CD4 (RM4-5), anti-CD8a (53–6.7), anti-CD44 (IM7), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD49a (Ha31/8), anti-CD62L (MEL-14), anti-CD69 (H1.2F3), anti-CD103 (M290), anti-CD127 (A7R34), anti-CXCR6 (SA051D1), anti-KLRG1 (2F1), anti-TCF-1 (S33-966), mouse IgG1, κ isotype control (P3.6.2.8.1). Antibodies were purchased from BioLegend, eBiosciences, BD Biosciences, or BD Pharmingen. Influenza-virus-specific CD8+ T cells were detected using H-2 Db ASNENMETM (NP366−374) tetramers (Db NP366), which were a kind gift of Dr. Anja ten Brinke (Sanquin Research). Exclusion of dead cells was performed with live/dead fixable near-IR dead cell stain kit (Thermo Fisher Scientific). For staining of intracellular molecules, the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) was used according to the manufacturer's specifications. Samples were acquired on an LSR Fortessa flow cytometer (BD) and data was analyzed using FlowJo V10 (Tree star) software.
RNA-Seq Analysis
A subset of previously published RNA-seq (CEL-seq) data (geo accession number GSE79774) (23) was reanalysed. DMSO-treated CD69+ and CD69− memory (CD44high CD62Llo) CD8+ T cells isolated from murine lungs at day 30+ after HKx31 infection were analyzed to determine the tissue-resident phenotype of these populations. Reads were first aligned to mm10 genome with STAR, followed by read count quantification with featureCounts using Ensembl's v92 annotation with modification of Zfp683. Since CEL-seq was used, only the 3′ end of the genes is sequenced, and inspecting the alignments manually revealed that the reads that mapped to Zfp683 were outside Ensembl's defined region of the gene. We therefore extended the 3′ end of Zfp683 with 1,057 nucleotides to include all reads of this gene. To find differentially expressed (DE) genes, first low expressed genes were removed (retained genes had more than 1 cpm (counts per million) in at least four samples), followed by limma voom and quantile normalization. A linear model was fitted to each gene using a design that also included mouse (paired) and library (batch) effect, and eBayes moderated t-statistics were used to find DE genes between CD69+ and CD69− memory CD8+ T cells. Benjamini-Hochberg procedure was used for FDR correction, and adjusted P < 0.05 were considered significant.
Statistical Analysis
For pairwise comparisons, a standard two-sided Student's t-test (paired), was applied with GraphPad Prism 6 software. P < 0.05 were considered statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001). The Morpheus software (https://software.broadinstitute.org/morpheus), developed by the Broad Institute, was used to generate heat maps. Values were converted to heat map colors using the mean and maximum values for each row.
Ethics Statement
This study was carried out in accordance with the national guidelines of the Central Commission for Animal Experiments (Centrale Commissie Dierproeven). The experimental protocols were approved by the Animal Welfare Body of the Netherlands Cancer Institute (NKI).
Author Contributions
FB and KvG conceived and designed studies. FB, NK, TW, and RS performed animal experiments and FB analyzed data. BN analyzed RNAseq data provided by PH and DA. FB and KvG wrote the manuscript. All authors provided critical feedback and approved the manuscript.
Funding
FB and KvG were supported by Vidi grant 917.13.338 from the Netherlands Organization for Scientific Research (NWO) and a fellowship of the Landsteiner Foundation for Blood Transfusion Research. RS was supported by a Fellowship from the Alexander von Humboldt Foundation and by Veni grant 016.186.116 from NWO.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Anja ten Brinke (Sanquin Research) for kindly providing H-2 Db ASNENMETM (NP366−374) tetramers and Mark Hoogenboezem for technical support. We thank Dr. Guus Rimmelzwaan (University of Veterinary Medicine Hannover) for providing HKx31 influenza virus.
References
1. Mueller SN, Mackay LK. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol. (2016) 16:79–89. doi: 10.1038/nri.2015.3
2. Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR. Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. (2009) 10:524–530. doi: 10.1038/ni.1718
3. Sheridan BS, Pham QM, Lee YT, Cauley LS, Puddington L, Lefrançois L. Oral infection drives a distinct population of intestinal resident memory CD8+ T cells with enhanced protective function. Immunity. (2014) 40:747–57. doi: 10.1016/j.immuni.2014.03.007
4. Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. (2014) 346:98–101. doi: 10.1126/science.1254536
5. Wu T, Hu Y, Lee YT, Bouchard KR, Benechet A, Khanna K, et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J Leukoc Biol. (2014) 95:215–24. doi: 10.1189/jlb.0313180
6. Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8+ T cells. Nat Immunol. (2013) 14:509–13. doi: 10.1038/ni.2568
7. Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song JY, et al. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science. (2014) 346:101–5. doi: 10.1126/science.1254803
8. Steinbach K, Vincenti I, Kreutzfeldt M, Page N, Muschaweckh A, Wagner I, et al. Brain-resident memory T cells represent an autonomous cytotoxic barrier to viral infection. J Exp Med. (2016) 213:1571–87. doi: 10.1084/jem.20151916
9. Kragten NAM, Behr FM, Vieira Braga FA, Remmerswaal EBM, Wesselink TH, Oja AE, et al. Blimp-1 induces and Hobit maintains the cytotoxic mediator granzyme B in CD8 T cells. Eur J Immunol. (2018) 48:1644–62. doi: 10.1002/eji.201847771
10. Clark RA. Resident memory T cells in human health and disease. Sci Transl Med. (2015) 7:269rv261. doi: 10.1126/scitranslmed.3010641
11. Schenkel JM, Masopust D. Tissue-Resident Memory T Cells. Immunity. (2014) 41:886–97. doi: 10.1016/j.immuni.2014.12.007
12. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The developmental pathway for CD103+ CD8+ tissue-resident memory T cells of skin. Nat Immunol. (2013) 14:1294–301. doi: 10.1038/ni.2744
13. Wakim LM, Gupta N, Mintern JD, Villadangos JA. Enhanced survival of lung tissue-resident memory CD8(+) T cells during infection with influenza virus due to selective expression of IFITM3. Nat Immunol. (2013) 14:238–45. doi: 10.1038/ni.2525
14. Pizzolla A, Nguyen THO, Smith JM, Brooks AG, Kedzieska K, Heath WR, et al. Resident memory CD8+ T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci Immunol. (2017) 2:eaam6970. doi: 10.1126/sciimmunol.aam6970
15. Zens KD, Chen JK, Farber DL. Vaccine-generated lung tissue–resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight. (2016) 1:e85832. doi: 10.1172/jci.insight.85832
16. Mackay LK, Stock AT, Ma JZ, Jones CM, Kent SJ, Mueller SN, et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci USA. (2012) 109:7037–42. doi: 10.1073/pnas.1202288109
17. Slütter B, Van Braeckel-Budimir N, Abboud G, Varga SM, Salek-Ardakani S, Harty JT. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci Immunol. (2017) 2:eaag2031. doi: 10.1126/sciimmunol.aag2031
18. Takamura S, Yagi H, Hakata Y, Motozono C, McMaster SR, Masumoto T, et al. Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J Exp Med. (2016) 213:3057–73. doi: 10.1084/jem.20160938
19. Ely KH, Cookenham T, Roberts AD, Woodland DL. Memory T cell populations in the lung airways are maintained by continual recruitment. J Immunol. (2006) 176:537–43. doi: 10.4049/jimmunol.176.1.537
20. Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. (2016) 352:459–63. doi: 10.1126/science.aad2035
21. Anderson KG, Sung H, Skon CN, Lefrancois L, Deisinger A, Vezys V, et al. Cutting edge: intravascular staining redefines lung CD8 T cell responses. J Immunol. (2012) 189:2702–6. doi: 10.4049/jimmunol.1201682
22. Gilchuk P, Hill TM, Guy C, McMaster SR, Boyd KL, Rabacal WA, et al. A distinct lung-interstitium-resident memory CD8+ T cell subset confers enhanced protection to lower respiratory tract infection. Cell Rep. (2016) 16:1800–9. doi: 10.1016/j.celrep.2016.07.037
23. Hombrink P, Helbig C, Backer RA, Piet B, Oja AE, Stark R, et al. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat Immunol. (2016) 17:1467–78. doi: 10.1038/ni.3589
24. Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 Transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity. (2009) 31:283–95. doi: 10.1016/j.immuni.2009.06.021
25. Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, et al. Transcriptional repressor blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. (2009) 31:296–308. doi: 10.1016/j.immuni.2009.05.014
26. Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, et al. Liver-resident memory CD8(+) T cells form a front-line defense against malaria liver-stage infection. Immunity. (2016) 45:889–902. doi: 10.1016/j.immuni.2016.08.011
27. Zaid A, Hor JL, Christo SN, Groom JR, Heath WR, Mackay LK, et al. Chemokine receptor–dependent control of skin tissue–resident memory T cell formation. J Immunol. (2017) 199:2451–9. doi: 10.4049/jimmunol.1700571
28. Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. (2017) 20:2921–34. doi: 10.1016/j.celrep.2017.08.078
29. Cheuk S, Schlums H, Gallais Sérézal I, Martini E, Chiang SC, Marquardt N, et al. CD49a Expression defines tissue-resident CD8+ T cells poised for cytotoxic function in human skin. Immunity. (2017) 46:287–300. doi: 10.1016/j.immuni.2017.01.009
30. Kedzierska K, Venturi V, Field K, Davenport MP, Turner SJ, Doherty PC. Early establishment of diverse T cell receptor profiles for influenza-specific CD8+ CD62Lhi memory T cells. Proc Natl Acad Sci USA. (2006) 103:9184–9. doi: 10.1073/pnas.0603289103
31. Roberts AD, Woodland DL. Cutting Edge: effector memory CD8+ T cells play a prominent role in recall responses to secondary viral infection in the lung. J Immunol. (2004) 172:6533–7. doi: 10.4049/jimmunol.172.11.6533
32. Jeannet G, Boudousquié C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci USA. (2010) 107:9777–82. doi: 10.1073/pnas.0914127107
33. Danilo M, Chennupati V, Silva JG, Siegert S, Held W. Suppression of Tcf1 by inflammatory cytokines facilitates effector CD8 T cell differentiation. Cell Rep. (2018) 22:2107–17. doi: 10.1016/j.celrep.2018.01.072
34. Jelley-Gibbs DM, Brown DM, Dibble JP, Haynes L, Eaton SM, Swain SL. Unexpected prolonged presentation of influenza antigens promotes CD4 T cell memory generation. J Exp Med. (2005) 202:697–706. doi: 10.1084/jem.20050227
35. Zammit DJ, Turner DL, Klonowski KD, Lefrançois L, Cauley LS. Residual antigen presentation after influenza virus infection affects CD8 T cell activation and migration. Immunity. (2006) 24:439–49. doi: 10.1016/j.immuni.2006.01.015
36. Kim TS, Hufford MM, Sun J, Fu YX, Braciale TJ. Antigen persistence and the control of local T cell memory by migrant respiratory dendritic cells after acute virus infection. J Exp Med. (2010) 207:1161–72. doi: 10.1084/jem.20092017
37. McMaster SR, Wein AN, Dunbar PR, Hayward SL, Cartwright EK, Denning TL, et al. Pulmonary antigen encounter regulates the establishment of tissue-resident CD8 memory T cells in the lung airways and parenchyma. Mucos Immunol. (2018) 11:1071–8. doi: 10.1038/s41385-018-0003-x
38. Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. (2006) 7:457–65. doi: 10.1038/ni1320
39. Xin A, Masson F, Liao Y, Preston S, Guan T, Gloury R, et al. A molecular threshold for effector CD8+ T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat Immunol. (2016) 17:422–32. doi: 10.1038/ni.3410
40. Hondowicz BD, An D, Schenkel JM, Kim KS, Steach HR, Krishnamurty AT, et al. Interleukin-2-dependent allergen-specific tissue-resident memory cells drive asthma. Immunity. (2016) 44:155–166. doi: 10.1016/j.immuni.2015.11.004
41. Hocke AC, Lampe MP, Witzenrath M, Mollenkopf H, Zerrahn J, Schmeck B, et al. Cell-specific Interleukin−15 and Interleukin−15 receptor subunit expression and regulation in pneumococcal pneumonia—Comparison to chlamydial lung infection. Cytokine. (2007) 38:61–73. doi: 10.1016/j.cyto.2007.05.009
42. Verbist KC, Cole CJ, Field MB, Klonowski KD. A Role for IL−15 in the Migration of effector CD8 T cells to the lung airways following influenza infection. J Immunol. (2010) 186:174–82. doi: 10.4049/jimmunol.1002613
43. Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, et al. T-box transcription factors combine with the cytokines TGF-β and IL−15 to control tissue-resident memory T cell fate. Immunity. (2015) 43:1101–11. doi: 10.1016/j.immuni.2015.11.008
44. Schenkel JM, Fraser KA, Casey KA, Beura LK, Pauken KE, Vezys V, et al. IL−15–Independent maintenance of tissue-resident and boosted effector memory CD8 T cells. J Immunol. (2016) 196:3920–6. doi: 10.4049/jimmunol.1502337
45. Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol. (2012) 188:4866–75. doi: 10.4049/jimmunol.1200402
46. Kohlmeier JE, Cookenham T, Roberts AD, Miller SC, Woodland DL. Type I interferons regulate cytolytic activity of memory CD8(+) T cells in the lung airways during respiratory virus challenge. Immunity. (2010) 33:96–105. doi: 10.1016/j.immuni.2010.06.016
47. McMaster SR, Wilson JJ, Wang H, Kohlmeier JE. Airway-resident memory CD8 T cells provide antigen-specific protection against respiratory virus challenge through rapid IFN-γ production. J Immunol. (2015) 195:203–9. doi: 10.4049/jimmunol.1402975
48. Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyártó BZ, et al. Quantifying Memory CD8 T cells reveals regionalization of immunosurveillance. Cell. (2015) 161:737–49. doi: 10.1016/j.cell.2015.03.031
49. Bergsbaken T, Bevan MJ. Proinflammatory microenvironments within the intestine regulate the differentiation of tissue-resident CD8+ T cells responding to infection. Nat Immunol. (2015) 16:406–14. doi: 10.1038/ni.3108
50. Cepek KL, Parker CM, Madara JL, Brenner MB. Integrin alpha E beta 7 mediates adhesion of T lymphocytes to epithelial cells. J Immunol. (1993) 150:3459–70.
51. Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, Rimm DL, et al. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the αEβ7 integrin. Nature. (1994) 372:190–3. doi: 10.1038/372190a0
52. Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. (2017) 552:253–7. doi: 10.1038/nature24993
53. Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity. (2010) 33:229–40. doi: 10.1016/j.immuni.2010.08.002
54. McMichael AJ, Gotch FM, Noble GR, Beare PA. Cytotoxic T-Cell Immunity to Influenza. N Engl J Med. (1983) 309:13–17. doi: 10.1056/NEJM198307073090103
55. Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med. (2013) 19:1305–12. doi: 10.1038/nm.3350
56. de Bree GJ, van Leeuwen EM, Out TA, Jansen HM, Jonkers RE, van Lier RA. Selective accumulation of differentiated CD8+ T cells specific for respiratory viruses in the human lung. J Exp Med. (2005) 202:1433–42. doi: 10.1084/jem.20051365
57. Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, Kupper TS. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS ONE. (2011) 6:e16245. doi: 10.1371/journal.pone.0016245
58. Turner DL, Bickham KL, Thome JJ, Kim CY, D'Ovidio F, Wherry EJ, et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. (2014) 7:501–10. doi: 10.1038/mi.2013.67
59. Pizzolla A, Nguyen TH, Sant S, Jaffar J, Loudovaris T, Mannering SI, et al. Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J Clin Invest. (2018) 128:721–33. doi: 10.1172/JCI96957
60. Vieira Braga FA, Hertoghs KM, Kragten NA, Doody GM, Barnes NA, Remmerswaal EB, et al. Blimp-1 homolog Hobit identifies effector-type lymphocytes in humans. Eur J Immunol. (2015) 45:2945–58. doi: 10.1002/eji.201545650
Keywords: hobit, blimp-1/PRDM1, lung T cell, T cell differentiation, influenza virus infection, central memory CD8(+) T cells, tissue-resident memory CD8(+) T cells, TCF-1
Citation: Behr FM, Kragten NAM, Wesselink TH, Nota B, van Lier RAW, Amsen D, Stark R, Hombrink P and van Gisbergen KPJM (2019) Blimp-1 Rather Than Hobit Drives the Formation of Tissue-Resident Memory CD8+ T Cells in the Lungs. Front. Immunol. 10:400. doi: 10.3389/fimmu.2019.00400
Received: 31 October 2018; Accepted: 15 February 2019;
Published: 07 March 2019.
Edited by:
Francesca Di Rosa, Consiglio Nazionale Delle Ricerche (CNR), ItalyReviewed by:
Kim Klonowski, University of Georgia, United StatesLuigia Pace, Italian Institute for Genomic Medicine (IIGM), Italy
Copyright © 2019 Behr, Kragten, Wesselink, Nota, van Lier, Amsen, Stark, Hombrink and van Gisbergen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Felix M. Behr, Zi5tYW51ZWxiZWhyQHNhbnF1aW4ubmw=
Klaas P. J. M. van Gisbergen, ay52YW5naXNiZXJnZW5Ac2FucXVpbi5ubA==
 Felix M. Behr
Felix M. Behr Natasja A. M. Kragten
Natasja A. M. Kragten Thomas H. Wesselink1
Thomas H. Wesselink1 Derk Amsen
Derk Amsen Regina Stark
Regina Stark Pleun Hombrink
Pleun Hombrink Klaas P. J. M. van Gisbergen
Klaas P. J. M. van Gisbergen