
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 12 March 2019
Sec. Comparative Immunology
Volume 10 - 2019 | https://doi.org/10.3389/fimmu.2019.00310
 Soyi Park1†
Soyi Park1† Yong Hun Jo1†
Yong Hun Jo1† Ki Beom Park1
Ki Beom Park1 Hye Jin Ko1
Hye Jin Ko1 Chang Eun Kim1
Chang Eun Kim1 Young Min Bae1
Young Min Bae1 Bobae Kim1
Bobae Kim1 Sung Ah Jun2
Sung Ah Jun2 In Seok Bang3
In Seok Bang3 Yong Seok Lee4
Yong Seok Lee4 Yu Jung Kim5*
Yu Jung Kim5* Yeon Soo Han1*
Yeon Soo Han1*Although it is known that the Drosophila Toll-7 receptor plays a critical role in antiviral autophagy, its function in other insects has not yet been reported. Here, we have identified a Toll-like receptor 7 gene, TmToll-7, in the coleopteran insect T. molitor and examined its potential role in antibacterial and antifungal immunity. We showed that TmToll-7 expression was significantly induced in larvae 6 h after infection with Escherichia coli and Staphylococcus aureus and 9 h after infection with Candida albicans. However, even though TmToll-7 was induced by all three pathogens, we found that TmToll-7 knockdown significantly reduced larval survival to E. coli, but not to S. aureus, and C. albicans infections. To understand the reasons for this difference, we examined the effects of TmToll-7 knockdown on antimicrobial peptide (AMP) gene expression and found a significant reduction of E. coli-induced expression of AMP genes such as TmTenecin-1, TmDefensin-1, TmDefensin-2, TmColeoptericin-1, and TmAttacin-2. Furthermore, TmToll-7 knockdown larvae infected with E. coli showed significantly higher bacterial growth in the hemolymph compared to control larvae treated with Vermilion dsRNA. Taken together, our results suggest that TmToll-7 plays an important role in regulating the immune response of T. molitor to E. coli.
Toll and Toll-like receptors in Drosophila and mammals play a major role in innate immunity (1). So far, in Drosophila, nine Toll receptors (Toll, 18-Wheeler, and Toll-3 to Toll-9) have been identified (2). Toll, which was the first of these receptors to be discovered, was initially noted for its role in dorsal-ventral patterning in Drosophila embryos (3) and was later shown to participate in the immune response to fungi and Gram-positive bacteria in larvae and adults (4). In addition to its role in salivary gland morphogenesis (5), the 18-wheeler or Toll-2 receptor is required for antibacterial defense in larvae but dispensable for immune responses in adults (6). Among what is known so far of the remaining Toll receptors is that Toll-7 and Toll-8 may have developmental functions that are redundant with 18-wheeler (7), and although they do not induce antibacterial responses in flies, Toll-7 is involved in antiviral defense (8) and Toll-8 acts as a negative regulator of antibacterial responses in respiratory epithelial cells (9). Furthermore, in addition to its role in development and immunity, Toll-7 along with Toll-6 have been shown to have neurotrophic functions and are required for locomotion behavior, motor axon targeting, and neuronal survival (10). More recently, Toll-6 and Toll-7 were also discovered to mediate axon and dendrite targeting during Drosophila olfactory circuit assembly (11). Toll-5 has been shown to activate Drosomycin and Metchnikowin expression in transfected cells (12). However, whether Toll-5 has the same function in vivo still remains to be determined. Finally, overexpression of Toll-9 in S2 cells and larval fat body tissues results in constitutive expression of antimicrobial peptides (13, 14). Based on these findings, it was proposed that Toll-9 may have a constitutive function in activating antimicrobial defense (13, 14). However, a later study showed that Toll-9 loss-of function-mutants could still induce AMP expression under unchallenged conditions, suggesting that Toll-9 is not required to maintain a basal antimicrobial response as initially hypothesized, but may function redundantly with other Toll proteins (15).
Overall, the structure of insect Tolls are similar to mammalian TLRs in that they have an extracellular leucine-rich repeat (LRR) domain, a transmembrane domain, and a cytoplasmic Toll/Interleukin-1 receptor (TIR) domain (16). Despite this similarity, a number of structural and functional differences exist. One notable difference is that the Drosophila Tolls contain cysteine-rich clusters at either, or both, the N- and C-terminal ends of the LRRs, whereas mammalian TLRs contain a single cysteine-rich cluster at the C-terminal end of the LRRs, adjacent to the plasma membrane (16–18). The one exception is Toll-9, which has a single cysteine cluster, and because its TIR domain is more similar to the mammalian TLRs than to the other eight Drosophila Tolls, it falls within the same phylogenetic subfamily as the TLRs rather than with other insect Tolls (16, 17, 19). Thus, it might be expected that it would share functional similarities with mammalian TLRs, which serve to initiate inflammatory pathways following recognition of conserved pathogen associated molecular patterns (PAMPs) on various microbes, including bacteria, viruses, fungi, and protozoa (2, 19–21). However, the significance of Toll-9 and its role in immunity is unclear (13, 14).
Much of what is known about Toll activation in insect immunity comes from studies with Drosophila Toll-1, which unlike mammalian TLRs, does not respond directly to microbial molecules, but is activated by the cytokine ligand Spätzle (Spz) (22). However, for this binding to occur, Spz must be cleaved by the serine protease Spätzle-processing enzyme (SPE), which in turn is activated by three protease cascades. Two of them, which converge at the level of the ModSP-Grass proteases, are initiated when pattern recognition receptors detect cell wall components from Gram-positive bacteria (Lysine-type peptidoglycans) and fungi (β-1,3-glucan), respectively (23). Furthermore, four other serine proteases (Spirit, Spheroide, and Sphinx1/2) are thought to function in this pathway between Grass and SPE (24). The third cascade is triggered when the protease Persephone (Psh) is activated by microbial proteases or endogenous danger signals. With the former, it is believed that Persephone can be targeted for direct cleavage by subtilisin-like serine proteases produced by Gram-positive bacteria or fungi (25, 26). With the latter, although it is unclear what signals are actually sensed by Persephone and how, one possibility is that it is able to detect abnormal proteolytic activities in the hemolymph (26). This was suggested based on the observation that Grass overexpression in transgenic flies results in Persephone-dependent Toll activation, similar to what occurs when pathogen-derived proteases are injected into flies (26). In short, all three modes of detection lead to downstream activation of SPE. However, currently it is unknown what protease directly cleaves and activates SPE in Drosophila, although an SPE-activating enzyme (TmSAE) has been found in T. molitor (27, 28).
The mechanisms leading to Toll receptor activation in T. molitor and Drosophila are similar, but they also have many important differences. In both of these insects, Toll signaling is initiated when the peptidoglycan-recognition protein-SA (PGRP-SA) and Gram-negative binding protein 1 (GNBP1) recognize lysine-type peptidoglycan from Gram-positive bacteria, and GNBP3 recognizes β-1,3 from fungi (29, 30). However, in the case of the Tenebrio PGRP-SA/GNBP1 complex, it also recognizes meso-diaminopimelic acid (DAP)-type peptidoglycan, which is found in most Gram-negative bacteria and certain Gram-positive bacteria (e.g., Bacillus species and L. monocytogenes) (29). This is unlike Drosophila, where DAP-type peptidoglycan recognition depends on two other pattern recognition receptors, PGRP-LC and PGRP-LE, both of which are known to activate the Imd pathway (31). A further difference is that while both systems have a modular serine protease (ModSP) that functions directly downstream of PGRP-SA/GNBP1 and GNBP3, the Tenebrio ModSP proceeds through sequential activation of clip-domain-containing serine proteases, Tm-SAE and Tm-SPE, whereas in Drosophila, it activates the protease Grass, which in turn leads to activation of SPE (27, 32). In recent years, the mechanism for Spätzle activation in T. molitor in response to components of Gram-positive and Gram-negative bacteria and fungi has largely been determined using purified proteases (28). These studies have provided insights into the extracellular recognition mechanisms involved in Toll signaling (27, 29, 33); however, the Toll receptors themselves and their downstream components have not been characterized so far. Here, we have cloned a Toll-like receptor 7 gene from T. molitor (TmToll-7) and examined its role in vivo by RNAi. Our current study reveals that TmToll-7 is important for immune mediating responses to Gram-negative bacteria E. coli.
T. molitor larvae were reared on wheat bran diet at 27 ± 1°C, 60 ± 5% relative humidity, and under dark conditions. All experiments were conducted with 10–12th instar larvae. To investigate the immunological function of TmToll-7 against infections, three microorganisms, including Escherichia coli K12, Staphylococcus aureus RN4220, and Candida albicans were used. Overnight cultures of E. coli, S. aureus, and C. albicans were grown in Luria-Bertani (LB) broth and Sabouraud Dextrose broth at 37°C, respectively. The microorganisms were harvested, washed, and suspended in phosphate-buffered saline (PBS, pH 7.0) by centrifugation at 3,500 rpm for 10 min, and the concentrations were measured at OD600. Finally, 106 cells/μl of E. coli and S. aureus and 5 × 104 cells/μl of C. albicans were used in infection experiments.
The amino acid sequence (EEZ99327.1) of Toll-like receptor 7 (TcToll-7) in Tribolium castaneum was used to identify the TmToll-7 gene from a nucleotide sequence database constructed with T. molitor EST and RNAseq by local-tblastn analysis (34). The TmToll-7 nucleotide sequence was annotated with the Genbank nr database by the blastx program.
To obtain the full-length cDNA sequence of TmToll-7 ORF cloning and 5′- and 3′-RACE, PCR was conducted with the gene specific primers listed in Table 1. All primers were designed by using the Primer 3 plus program (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi). TmToll-7 was amplified by reverse transcription-PCR (RT-PCR) using Ex Taq polymerase (TaKaRa, Japan) under the following conditions: pre-denaturation at 98°C for 3 min, denaturation at 98°C for 10 s, annealing at 53°C for 30 s, and extension at 72°C for 1 min for 35 cycles. To obtain 5′- and 3′- end sequences of TmToll-7, 5′- and 3′-RACE PCR was carried with the SMARTer RACE cDNA amplification kit (Clontech Laboratories, Palo Alto, CA, USA) according to the manufacturer's instructions. Conditions for TmToll-7 RACE PCR were as follows: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s for 35 cycles. The RT-PCR products and nested PCR products were purified with AccuPrep ® PCR Purification Kit (Bioneer, Korea), cloned into TOPO TA cloning vector (Invitrogen Co., Carlsbad, CA), and subsequently transformed into E. coli DH5α competent cells and sequenced. The obtained sequences were assembled by the Cap3.dat program.
Specific domains of TmToll-7 were analyzed with the SignalP 4.1 program (http://www.cbs.dtu.dk/services/), InterProScan 5 program (35–37), and TMHMM 2.0 program (http://www.cbs.dtu.dk/services/TMHMM/). Phylogenetic analysis and percentage identity/distance analysis were conducted by using the Clustal X2 (38) and MEGA 6 programs (39), respectively. Human TLR4 amino acid sequences were used as an outgroup.
Developmental and tissue specific expression patterns of TmToll-7 were investigated by relative quantitative PCR (qRT-PCR) method using the Exicycler Real-Time PCR Quantification System (Bioneer Co., Daejon, South Korea). To investigate developmental and tissue specific expression patterns of TmToll-7, samples were collected from various developmental stages including late instar larval, pre-pupal, 1–7 day old pupal, and 1–2 day old adult stages and tissues dissected from late instar larvae (integument, gut, fat body, Malpighian tubules, and hemocytes) and 5 day old adult (integument, gut, fat body, Malpighian tubules, hemocytes, ovary, and testis) individuals. To examine induction patterns of TmToll-7 challenged by microorganisms, E. coli (106 cells/μl), S. aureus (106 cells/μl), and/or C. albicans (5 × 104 cells/μl) were injected into T. molitor larvae and samples were collected at different time points, including 3, 6, 9, and 12 h post-injection of microorganisms. PBS-injected T. molitor larvae were used as a negative control.
Total RNAs were extracted by using FavorPrep™ Tri-RNA Reagent (Favorgen biotech corp., Ping-Tung, Taiwan), following which 2 μg of total RNAs were used to synthesize cDNA using AccuPower® RT PreMix (Bioneer, Korea) with Oligo (dT)12−18 primer on a MyGenie 96 thermal block (Bioneer, Korea). To investigate the expression levels of TmToll-7 transcripts, relative quantitative PCR was examined by using AccuPower® 2X GreenStar qPCR Master Mix (Bioneer) with synthesized cDNAs and specific primers, TmToll-7_qPCR_Fw and TmT7_qPCR_Rv (Table 1) at an initial denaturation of 95°C for 20 s, followed by 45 cycles at 95°C for 5 s, and 60°C for 20 s. T. molitor ribosomal protein (TmL27a) was used as an internal control, and the results were analyzed by using ΔΔCt methods.
The PCR product (510 bp sequence) containing the T7 promotor sequences was amplified by Ex Taq polymerase with TmToll-7_T7_Fw and Rv primers (Table 1) using the same PCR condition mentioned above. dsRNA for TmToll-7 was synthesized by using AmpliScribe T7-Flash Transcription Kit (Epicentre, Madison, Wisconsin, USA) and was purified by PCI (Phenol: Chloroform: Isopropyl alcohol mixture), ammonium acetate purification and ethanol precipitation. 2 μg of synthesized dsTmToll-7 was injected into 10–11th instar larvae for gene silencing and the dsTmVer was used as a control.
To investigate the effect of TmToll-7 knockdown in response to microorganisms, 106 cells/μl of E. coli and S. aureus and 5 × 104 cells/μl of C. albicans were injected into dsTmToll-7-treated T. molitor larvae, respectively. The dead larvae were counted up to 10 days post-injection of microorganisms. Ten insects per group were used for this assay and the experiments were replicated three times.
To characterize the function of TmToll-7 on humoral innate immune response, TmToll-7 RNAi technique was applied and microorganisms, including E. coli, S. aureus and C. albicans, were injected subsequently. Samples were collected at 24 h post-injection of microorganisms. PBS was used as an injection control and dsTmVer-treated T. molitor was used as a negative control. Temporal expression patterns of 14 antimicrobial peptide (AMP) genes, including TmTene-1 (Figure 6A: TmTenecin-1), TmTene-2 (Figure 6B: TmTenecin-2), TmTene-3 (Figure 6C: TmTenecin-3), TmTene-4 (Figure 6D: TmTenecin-4), TmDef-1 (Figure 6E: TmDefensin-1), TmDef-2 (Figure 6F: TmDefensin-2), TmCole-1 (Figure 6G: TmColeoptericin-1), TmCole-2 (Figure 6H: TmColeoptericin-2), TmAtt-1a (Figure 6I: TmAttacin-1a), TmAtt-1b (Figure 6J: TmAttacin-1b), TmAtt-2 (Figure 6K: TmAttacin-2), TmCec-2 (Figure 6L: TmCecropin-2), TmTLP-1 (Figure 6M: TmThaumatin-like protein-1), and TmTLP-2 (Figure 6N: TmThaumatin-like protein-2) were investigated by relative quantitative-PCR with AMP gene specific primers detailed in Table 1.
The effect of TmToll-7 RNAi on antimicrobial activity against E. coli was investigated by using a colony formation assay (40). E. coli (106 cells) was injected into TmToll-7 dsRNA-treated T. molitor larvae and hemolymph was isolated in 100 μl 1X PBS at 24 h after E. coli injection. PBS-injected T. molitor hemolymph samples were used as an uninfected control, and dsTmVer-treated hemolymph was used as a dsRNA control. Hemolymph samples were centrifuged at 15,000 rpm at 4°C for 5 min and then the supernatants were boiled at 100°C for 5 min and centrifuged again at 15,000 rpm at 4°C for 5 min. Fifty nanogram of hemolymph samples were assayed with 106 cells of E. coli in 1X PBS at 37°C for 2 h. 2,000-fold diluted samples on LB-agar plates were incubated at 37°C for 16 h. The colony numbers of assayed plates were then counted.
All experiments were performed in triplicate and all data are shown as means ± S.E. The one-way analysis of variance (ANOVA) and Tukey's multiple range tests were used to evaluate the difference between groups (p < 0.05).
In this study, a Toll-7 homolog from Tenebrio molitor (TmToll-7, Accession number: MK234903) was identified by an EST and RNA-seq search using the T. castaneum protein sequence as a query. Based on this sequence information, we cloned the corresponding full-length cDNA from Tenebrio larval RNA by RT-PCR and 5′- and 3′-RACE. The 6,097-bp cDNA contains an 847-bp 5′-untranslated region (UTR), a 231-bp 3′-UTR (excluding the poly-A tail), and a 3,939-bp open reading frame (ORF) encoding a protein of 1,311 amino acids (Figure 1). Domain analysis of the deduced amino acid sequence shows that TmToll-7 has an N-terminal signal peptide from amino acids 1 to 18, seven leucine-rich repeats between amino acids 135 and 889, a transmembrane domain from amino acids 980 to 1,004, and a C-terminal Toll/interleukin-1 receptor homology (TIR) domain at positions 1,036 to 1,174 (Figure 1).

Figure 1. Nucleotide and deduced amino acid sequences of TmToll-7. Amino acid sequences of TmToll-7 were deduced by using the BLAST program and “DNA to protein translation program” provided from biophp (http://biophp.org/). Domains were analyzed by using the SignalP 4.1 program (http://www.cbs.dtu.dk/services/SignalP/), InterProScan program (http://www.ebi.ac.uk/Tools/pfa/iprscan5/), and TMHMM Server v. 2.0 program (http://www.cbs.dtu.dk/services/TMHMM-2.0/). The arrow, yellow box, black box, and green box indicate the signal peptide region (one), leucine rich repeat region (seven), transmembrane domain (one), and Toll/interleukin-1 receptor homology (TIR) domain (one), respectively.
Multiple alignment of Toll-7 homologs indicates that the TIR domain is highly conserved in insects (Figure 2A). Further, phylogenetic analysis based on the full-length amino acid sequences of TmToll-7 and other insect Toll receptors (which include D. melanogaster Toll-1 through Toll-9) indicates that it clusters together with the Toll-7 proteins from T. castaneum, Nilaparvata lugens, D. melanogaster, and Aedes aegypti, and with the 18W proteins from Bombyx mori, Operophtera brumata, Apis mellifera, and Drosophila melanogaster (Figure 2B). Furthermore, within this branch, TmToll-7 appears to be most closely related to TcToll-7 (93% aa identity) from T. Castaneum, which belongs to the same insect order as T. molitor (Coleoptera), with AmToll-7 (68% aa identity) from Apis mellifera (Hymenoptera) and Bm18W (61% aa identity) from Bombyx mori (Lepidoptera) being the next two closely related sequences, followed by Toll-7 (55% aa identity) and 18W (53% aa identity) from D. melanogaster (Diptera).
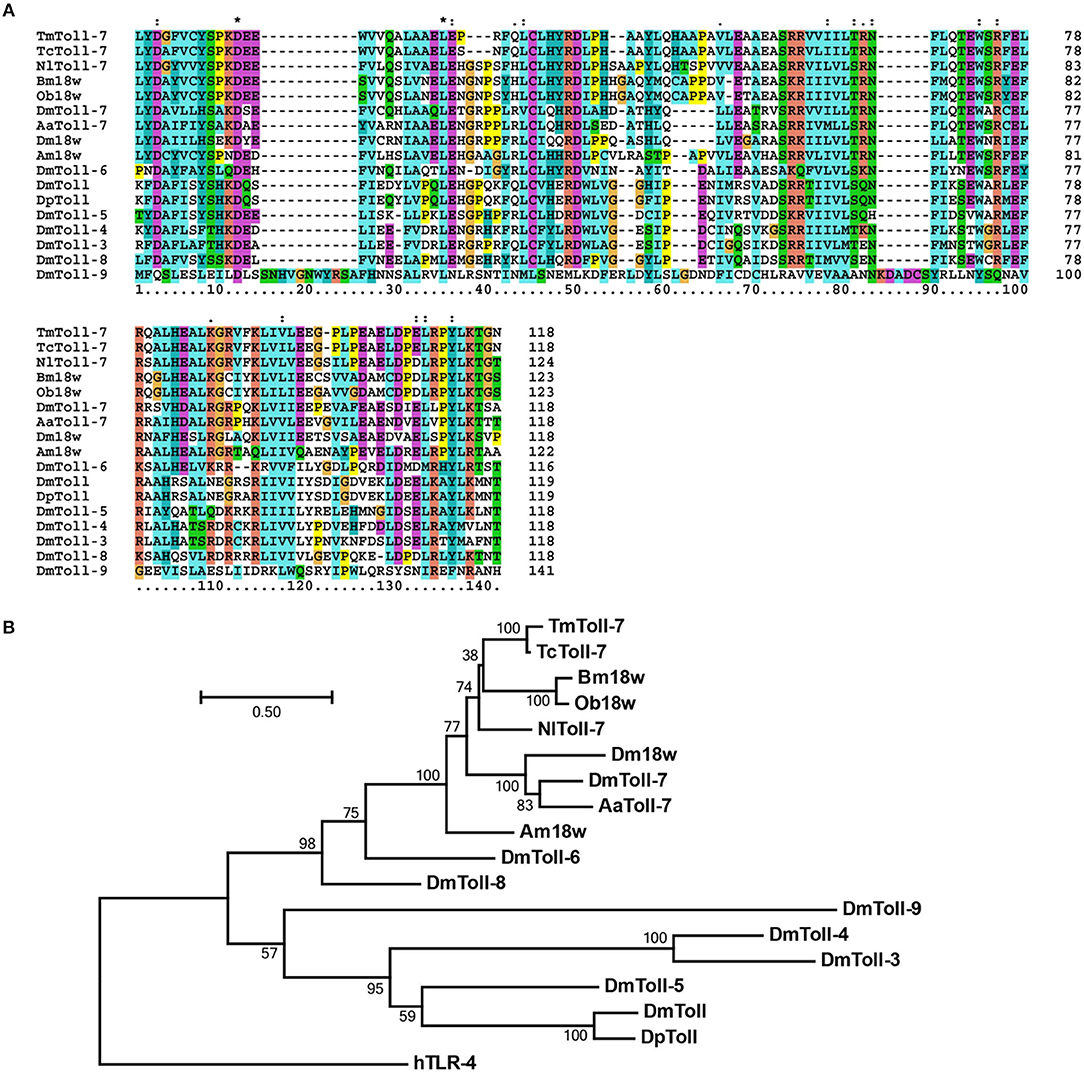
Figure 2. Multiple alignment and molecular phylogenetic analysis of insect TLR7s. (A) The highly conserved TIR domain was aligned by using ClustalX 2.1 software and the result was visualized by Gendoc software. (B) Molecular phylogenetic analysis was performed with whole amino acid sequences of insect TLR7s. ClustalX 2.1 software was used to perform multiple amino acid alignments before phylogenetic analysis. The phylogenic tree was constructed by MEGA 6.06 software. Bootstrap analysis of 1,000 replications are shown. TmToll-7, T. molitor Toll-like receptor 7 (MK234903); TcToll-7, T. castaneum Toll-like-7 protein (EEZ99327.1); NlTollR-7, Nilaparvata lugens Toll-like receptor 7 (AGK40936.1); Bm18w, Bombyx mori 18 wheeler (BAB85498.1); DmToll-7, Drosophila melanogaster Toll-7 (AAF57514.1); Am18w, Apis mellifera 18-wheeler precursor (NP_001013379.1); Ob18w, Operophtera brumata 18 wheeler (KOB69977.1); AaToll-7, Aedes aegypti Toll-like receptor 7 (EAT46215.1); DmToll, Drosophila melanogaster Toll protein (AAA28941.1); DpToll, Drosophila pseudoobscura Toll protein (L25390); DmToll-3, Drosophila melanogaster Toll-3 (AAF54021.3); DmToll-4, Drosophila melanogaster Toll-4 (AAF52747.3); DmToll-5, Drosophila melanogaster Toll-5 (AAF86227.1); DmToll-6, Drosophila melanogaster Toll-6 (AAF86226.1); DmToll-8, Drosophila melanogaster Toll-8 (AAF86224.1); DmToll-9, Drosophila melanogaster Toll-9 (NP_649214.1); and HsTLR4, Homo sapiens toll-like receptor 4 precursor (AAY82270.1).
We investigated expression pattern of TmToll-7 by qRT-PCR at four different developmental stages (late-instar larvae, pre-pupae, 1 to 7-day-old pupae, and 1 to 5-day-old adults). Our results showed that TmToll-7 expression level was high in the late-instar larval stage, but then gradually decreased between the pre-pupal and adult stages (Figure 3A). We further examined TmToll-7 transcript levels in different tissues of late-instar larvae and adults. As shown in Figure 3B, TmToll-7 expression was relatively high in the Malpighian tubules of late-instar larvae and low in the fat body, integument, hemocytes, and gut. In comparison, in adult tissues, TmToll-7 was predominantly expressed in the integument (albeit at low levels similar to those found in late-instar larvae) and to a lesser extent in other tissues, including testes and hemocytes (Figure 3C).
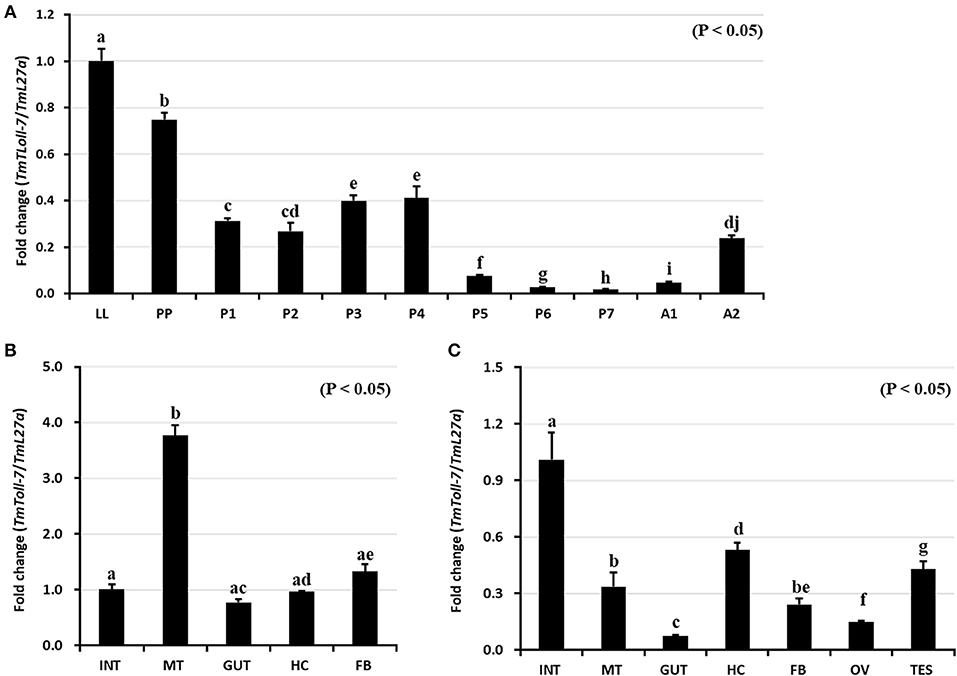
Figure 3. Developmental and tissue specific expression patterns of TmToll-7. (A) For developmental expression analysis of TmToll-7, total RNAs were isolated from late instar larvae (LL), pre-pupae (PP), 1~7 day old pupae (P1~P7), and 1~2 day old adults (A1~A2). T. molitor 60S ribosomal protein 27a (TmL27a) primers were used as an internal control. TmToll-7 was highly expressed in late-instar larval and prepupal stage (N = 3). Tissue specific expression patterns of TmToll-7 in late instar larvae (B), and adults (C). Total RNAs were isolated from various tissues, including integument (INT), Malpighian tubule (MT), gut (GUT), hemocytes (HC), and fat body (FB) in late instar larvae and integument (INT), Malpighian tubule (MT), gut (GUT), hemocytes (HC), fat body (FB), ovary (OV) and testis (TES) from 5th day old adults. Signals were detected by using relative quantitative PCR and ΔΔCT method was used to analyze tissue specificity (N = 3).
Next, to determine whether TmToll-7 expression is regulated in response to immune challenge, we further examined temporal changes in TmToll-7 mRNA expression in T. molitor larvae after infection with either Gram-negative (E. coli) or Gram-positive (S. aureus) bacteria, or fungus (C. albicans). Briefly, this was done by isolating total RNA from control and immune-challenged larvae (tenth or eleventh instar) at 3, 6, 9, and 12 h post-infection, followed by reverse-transcription and qRT-PCR using TmToll-7-specific primers. As shown in Figures 4A,B, challenging larvae with E. coli or S. aureus resulted in similar time-course changes in TmToll-7 mRNA levels, which became significantly upregulated at 6 h post-infection, and then decreased, but remained higher than control levels at 9 and 12 h post-infection. However, in the case of C. albicans infection, TmToll-7 mRNA levels reached a maximum at 9 h post-infection, and then declined to control levels at 12 h post-infection (Figure 4C).
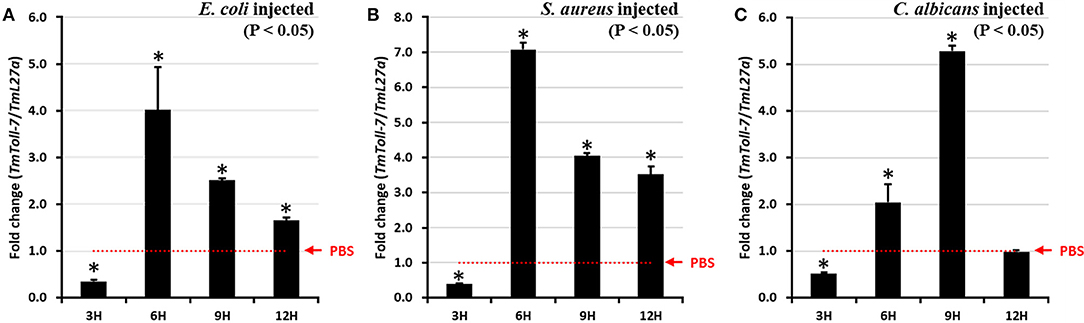
Figure 4. Temporal expression patterns of TmToll-7 in late instar larvae whole body. E.coli (A), S. aureus (B), and C. albicans (C) were injected into 10–11th instar larvae and total RNAs were isolated from whole-body at 3, 6, 9, and 12 h post injection of microorganisms. PBS was used as an injection control. The results show that TmToll-7 was highly induced at 6 h post injection of E. coli and S. aureus, and 9 h after injection of C. albicans (n = 3).
Since we found TmToll-7 expression to be induced after infection by E. coli, S. aureus, or C. albicans, we next wanted to determine if TmToll-7 plays a role in resistance to bacteria and fungi by monitoring the survival rates of infected T. molitor larvae after treatment with either control dsRNA (TmVer) or TmToll-7 dsRNA. Figure 5A shows that 7 days after injection, TmToll-7 mRNA levels decreased by 70% in larvae treated with TmToll-7 dsRNA compared to those treated with control dsRNA. After confirming efficient knockdown, we then challenged dsTmToll-7-treated and control larvae by injecting them with 1 μl of bacterial (E. coli or S. aureus, 1 × 106/μl) or fungal suspension (C. albicans, 5 × 104/μl) and followed their survival for 10 days. We found that compared to dsTmVer (control) larvae, dsTmToll-7 larvae were significantly more susceptible to E. coli infection (93 vs. 49% mortality) (Figure 5B), whereas their survival rates after infection with S. aureus or C. albicans were not significantly different from the controls (Figures 5C,D).
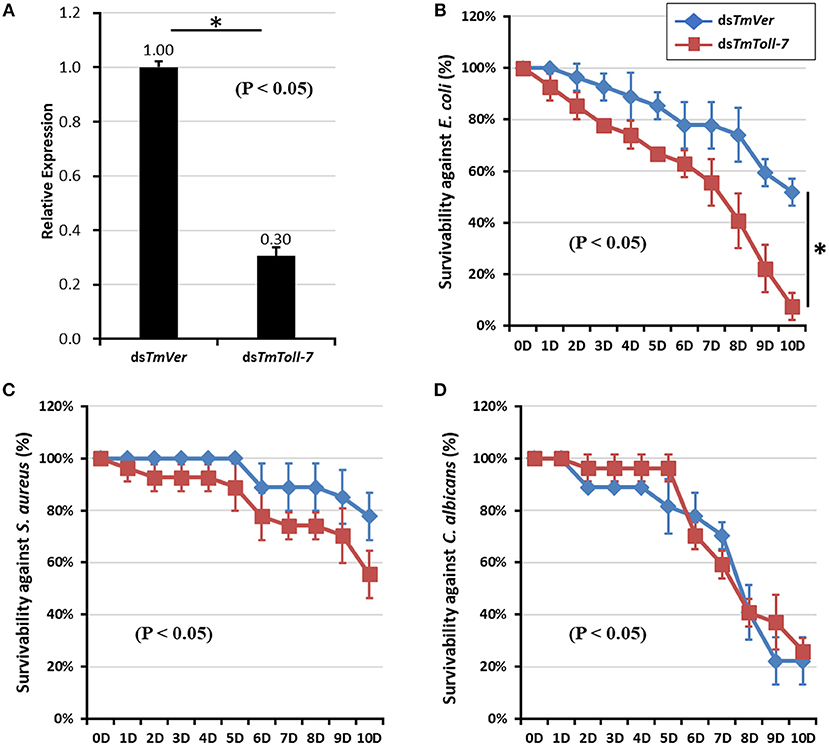
Figure 5. Effect of TmToll-7 gene silencing on survivability of T. molitor larvae. (A) Down-regulated TmToll-7 transcripts by RNAi. dsRNA for TmToll-7 (2 μg per insect) was injected into 10~11th instar larvae. To determine knockdown levels of TmToll-7 transcripts, quantitative PCR was performed by using total RNA isolated from 10~11th instar larvae (n = 3). The transcripts levels of TmToll-7 was reduced at 7-days after TmToll-7 RNAi. “*” indicates a significant (p < 0.05) difference between control treatments. Survival curves of T. molitor larvae infected with E.coli (B), S. aureus (C), C. albicans (D) following down-regulation of TmToll-7 mRNA transcripts. At the end of the experimental period (10 days post-infection), the survival rates of larvae were 7% at 10 h post-E. coli infection and 52% in TmToll-7-depleted larvae compared to the dsTmVer control group.
Since we found that TmToll-7 knockdown rendered T. molitor larvae significantly more susceptible to E. coli infection, but had no significant effect on survival rates after S. aureus or C. albicans infection, our data suggested that TmToll-7 is important for defense against Gram-negative bacteria. Thus, we next wanted to determine if TmToll-7 mediates this protective effect through AMP production. To do so, we knocked down TmToll-7 once again and measured the expression levels of 14 different AMP genes following challenge with E. coli, S. aureus, or C. albicans. What we hoped to find in this experiment were AMPs that were significantly induced upon infection by E. coli, but reversed by TmToll-7 knockdown. If so, this would suggest that TmToll-7, at least in part, mediates the activation of these AMPs in response to E. coli infection. Our data shows that among the 14 AMPs tested, TmTenecin-1, TmDefensin-1, TmDefensin-2, TmColeroptericin-1, and TmAttacin-2 gene expression was induced in response to E. coli infection, but pretreatment with TmToll-7 dsRNA suppressed their upregulation (Figures 6A,E–G,K). Interestingly, and in contrast to these effects, TmToll-7 knockdown increased the mRNA levels of TmTenecin-2, TmTenecin-4, TmColeoptericin-2, TmAttacin-1a, and TmAttacin-1b in S. aureus-challenged larvae (Figures 6B,D,H–J). Finally, none of the 14 AMPs showed significant responses to C. albicans infection, regardless of whether or not TmToll-7 was knocked down prior to infection.
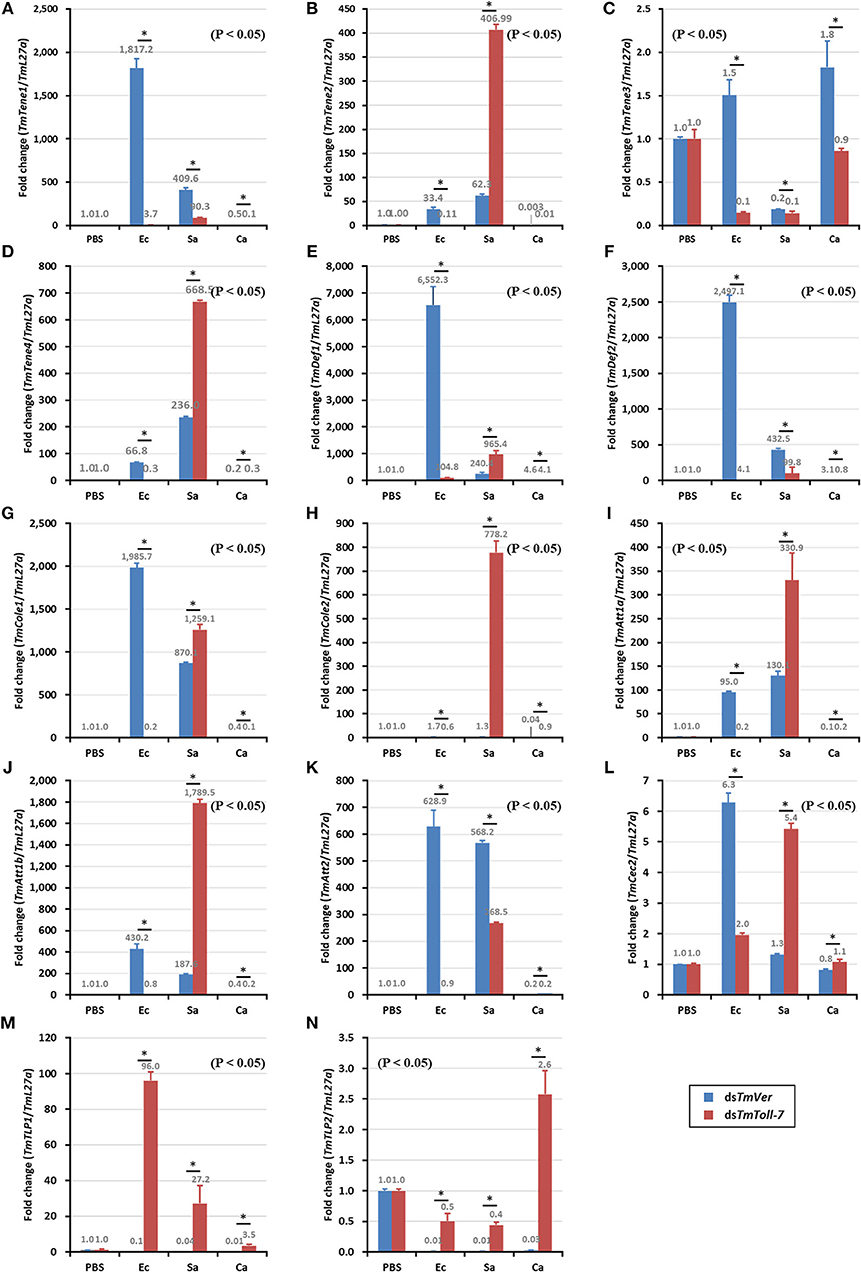
Figure 6. Induction patterns of 14 AMP genes in response to pathogenic microbial injection in TmToll-7 silenced T. molitor larvae. E. coli, C. albicans, and S. aureus were injected into dsTmToll-7-teated T. molitor larvae and whole body samples were collected at 24 h post injection. Expression of AMP genes, including TmTene-1 (A, TmTenecin-1); TmTene-2 (B, TmTenecin-2); TmTene-3 (C, TmTenecin-3); TmTene-4 (D, TmTenecin-4); TmDef-1 (E, TmDefensin-1); TmDef-2 (F, TmDefensin-2); TmCole-1 (G, TmColeoptericin-1); TmCole-2 (H, TmColeoptericin-2); TmAtt-1a (I, TmAttacin-1a); TmAtt-1b (J, TmAttacin-1b); TmAtt-2 (K, TmAttacin-2); TmCec-2 (L, TmCecropin-2); TmTLP-1 (M, TmThaumatin-like protein-1); and TmTLP-2 (N, TmThaumatin-like protein-2) were investigated by using qPCR. TmVer dsRNA was used as a knock down control and T. molitor ribosomal protein (TmL27a) was used as an internal control. PBS, PBS-injected control; Ec, E. coli-injected; Sa, S. aureus injected; Ca, C. albicans injected.
To further examine the role of TmToll-7 in regulating the immune response against E. coli, we investigated how TmToll-7 RNAi might affect the expression of six Toll pathway-related genes (TmMyD88, TmTRAF, TmCactin, TmCactus, TmDorsal-1, and TmDorsal-2) and one Imd-related gene (TmRelish) using specific primers listed in Supplementary Table 1. Our results showed that, except for TmTRAF and TmDorsal-2, the expression of the other four Toll pathway-related genes were significantly reduced by TmToll-7 RNAi (p < 0.05), while the level of TmRelish was not strongly affected (Supplementary Figure 2; Supplementary Table 1). This suggests that TmToll-7 can positively regulate downstream genes of the Toll signaling pathway, and moreover, that TmToll-7 functions through the Toll signaling pathway to regulate AMP expression.
Since knockdown of TmToll-7 by RNAi clearly suppressed E. coli-induced AMP expression, we wanted to determine whether this suppression would result in greater bacterial proliferation in the insect's hemolymph. To test this, we performed colony-forming unit assays. First, we pretreated larvae with TmToll-7 dsRNA or TmVer dsRNA (negative control) and infected them with E. coli to induce their immune system. After 24 h post-infection, cell-free hemolymph samples were prepared from these larvae and re-incubated with fresh E. coli for 2 h before being diluted and plated on LB agar for CFU counting. We found that pre-exposure to E. coli inhibited bacterial growth in the hemolymph of uninjected and TmVer dsRNA-injected control larvae, but not in the hemolymph of TmToll-7 dsRNA-injected larvae since it gave high E. coli counts, equal to those obtained from control larvae that had been pre-exposed to PBS instead (Figures 7A,B). By contrast, when we screened the hemolymph for antimicrobial activity against S. aureus (Gram-positive) and C. albicans (fungi), we found that RNAi of TmToll-7 did not significantly change the CFU numbers for both pathogens, i.e., the CFUs were low and comparable to the TmVermilion dsRNA-infected controls (~300 and 35 CFU, respectively) (Supplementary Figure 1). These results indicate that TmToll-7 knockdown suppresses anti-Gram negative activity and the ability to clear E. coli from the hemolymph.
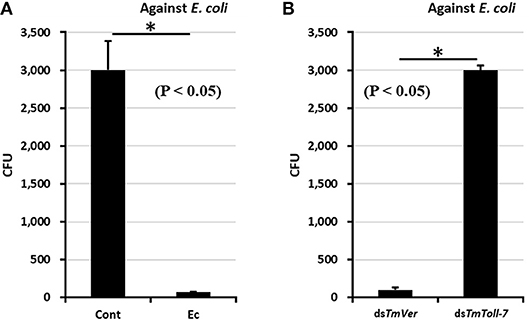
Figure 7. Antimicrobial activity assay against E. coli with TmToll-7-silenced T. molitor hemolymph by CFU method. (A) Antimicrobial activity was induced by E. coli (106 cells/μl) injection. PBS-injected T. molitor hemolymph was used as a negative control. E. coli-injected T. molitor hemolymph had high antimicrobial activity compared with control hemolymph. Cont; assayed with PBS-injected T. molitor hemolymph, Ec, assayed with E. coli-injected T. molitor hemolymph (B) E. coli (106 cells/μl) was injected into TmToll7-depleted T. molitor larvae. dsTmVer-treated T. molitor larvae was used as a negative control. The result indicates that the antimicrobial activity was dramatically decreased by treatment of dsTmToll-7 compared with the dsTmVer-treated group. dsTmVer, assayed with E. coli-injected T. molitor hemolymph after treatment with dsTmVer; dsTmToll-7, assayed with E. coli-injected T. molitor hemolymph after treatment with dsTmToll-7.
Using BLASTP with TmToll7 sequence as a query, we found more than 200 additional homologs in other insect species, with identities ranging from 48 to 93% (data not shown), suggesting that Toll-7 is widely conserved and essential in insects. This includes Toll-7, 18-wheeler, Toll-6 from Drosophila, which so far are the only 3 homologs (out of the 200) for which functional data are available (6–8, 10, 16). Overall, TmToll-7 shares 53, 51, and 43% sequence identity, respectively, with these three proteins, and like them, it has an N-terminal leucine-rich repeat (LRR) domain that is longer (~700 residues with 28 repeats) compared to other Drosophila Tolls (17 repeats in Toll-1, 2 in Toll-3, 17 in Toll-4, and 12 in Toll-5).
Although it is known, at least for Toll-1, that its LRR domain (specifically the first 13 LRRs) is required for formation of an active signaling complex (i.e., two Spz molecules bind to the LRR domains of two Toll receptor molecules, inducing a conformational change that enables dimer formation) (41, 42), the roles of the remaining Toll receptors (18-wheeler/Toll-2 to Toll-9, or any other insect TLR for that matter), are currently unclear. For example, it is unclear if and how they function in immunity or what ligands are required for their activation. However, it was recently shown in one study that Toll-7 and Toll-6 bind to and function together with Spätzle paralogs DNT-1/Spz-2 and DNT-2/Spz-5 in the Drosophila CNS to regulate motor-axon targeting and neuronal survival (10). Interestingly, the authors of this study also showed that in S2 cells, when either Toll-6 or 7 were stimulated with DNT-2 ligand, this activated the expression of a drosomycin reporter gene, which led them to suggest that Toll-6 and 7 could induce NF-kb signaling, possibly by using downstream signaling components, such as dMyD88, which are required by Toll-1 during embryogenesis and immunity (10). However, they ruled out the possibility that both receptors might have functional roles in immunity similar to Toll-1, since studies by Tauszig et al. and McIlroy et al. showed that Toll-6 and 7 do not activate the drosomycin reporter gene after immune challenge (10, 16).
It should be noted, however, that in the study by Tauszig et al. they examined the induction of the drosomycin reporter using chimeric constructs of Toll-2 to -8, in which the transmembrane and cytoplasmic domains of each receptor (devoid of their extracellular LRR domain) were fused to a truncated Toll-1 ectodomain. This was done for the purpose of determining if the cytoplasmic TIR domains of these receptors might be capable of activating antimicrobial peptide promoters and exhibit the same signaling specificity as Toll-1 (16). While their study showed that none of the chimeric proteins significantly induced expression of antimicrobial peptide genes, except for Toll-5, which activated drosomycin expression (16), we cannot help but wonder if one of the reasons why there is a discrepancy between Tauszig et al.'s (16) results and McIlroy et al.'s (10) is because Toll-6 and Toll-7 rely on their own extracellular LRR domain for ligand binding and signaling (the corresponding domain from Toll-1 cannot be substituted). If indeed they are required, this might explain why McIlroy et al. saw an induction in drosomycin promoter activity when they added purified DNT-2/Spz-5 protein to Toll-6 and Toll-7 expressing S2 cells.
We must add that, while writing this section, a preprint paper by Yu et al. showed that Toll-7, in its full-length (i.e., non-chimeric) form, is also capable of activating the drosomycin promoter when co-expressed with Spz-1 or Spz-2 (43). Importantly, they used co-immunoprecipitation assays to show that this activation may be mediated, in part, by binding of Toll-7, through its extracellular domain, to Spz-1, Spz-2, or Spz-5 (43). Given this evidence, and the fact that TmToll-7 shares 65% amino acid identity in its extracellular domain with Toll-7, there is reason to suggest that TmToll-7 binds to similar ligands as Toll-7.
Incidentally, a previous RNA-sequencing study conducted with bacterial-induced transcripts from T. molitor adults identified 9,570 putative orthologs of previously annotated T. castaneum genes, 213 of which were classified as immune-related (44). Interestingly, among these, an ortholog of Toll-7, one of only two Toll genes, the other being an ortholog of Toll-3, was identified along with seven Spätzle-like protein encoding genes (Spz 1–7). In addition to spz-3, 4, 6, and 7, our group has also recently identified five more spätzle-like genes: four are homologous to Tribolium spz-1b, spz-like, spz-7a, and spz-7b, the latter two of which encode 141- and 185-amino acid isoforms of Spz-7, and one is homologous to Drosophila Spz-5. We are now in the process of determining which of these ligands are required for AMP expression through knockdown studies. Of the nine ligands listed above, we have so far looked at three (TmSpz-7, -7a, and -7b), and our preliminary studies indicate that RNAi of TmSpz-7 and -7b reduces the induction of several AMPs, including the five aforementioned AMPs (Cho et al., unpublished results), which we have demonstrated here to be suppressed by TmToll-7 RNAi after E. coli challenge. It will be interesting to see, in future studies, whether either of these ligands binds directly to TmToll-7 (or other receptors) and, if they do, whether their interaction is relevant to immunity and/or development in T. molitor.
Although generally recognized as being responsive to Gram-positive bacteria and fungi, the Toll pathway in Drosophila has recently been demonstrated by Duneau et al. (45) and Yu et al. (43) to be sexually dimorphic in response to Gram-negative bacteria infection (43, 45). In the Duneau et al. study, they found that the induction of Drosomycin was much stronger in wild-type males than in females in response to Gram-negative Providencia rettgeri infection; however, this dimorphism was lost in spätzle and persephone mutants, but not in modSP mutants. Based on these results, Duneau et al. hypothesized that, in the case of P. rettgeri infection, Toll pathway activation specifically occurring via the Persephone branch leads to higher expression of AMP genes in males. In the study by Yu et al. upon observing that Toll-1 is expressed in both adult males and females while Toll-7 is expressed in males only, they looked at the susceptibility of Toll-7 mutant males to Gram-negative Pseudomonas aeruginosa infection and found that they were significantly more susceptible than Toll-1 mutant males. Together, these differential effects suggest that Toll-7 plays a major role in defense against P. aeruginosa infection in adult males only (43). However, it should be noted in our study, although we found that TmToll-7 was induced in Tenebrio adults (males and females combined) in response to Gram-negative E. coli infection, it was to a lesser extent than that observed in infected larvae. Thus, for the remainder of our studies, we focused on the effects of TmToll-7 silencing in larvae, which rendered larvae more susceptible to E. coli infection and suppressed certain AMP genes induced by E. coli challenge (including TmDefensin-1, TmDefensin-2, TmColeoptericin-1, and TmAttacin-2, all four of which belong to AMP families characterized as having antibacterial activity against Gram-negative bacteria) (46–48). In addition to the above studies in Drosophila, in vitro reconstitution experiments showing that E. coli DAP-type peptidoglycans can induce cleavage of pro-ModSP and pro-Spätzle after incubation with purified Tenebrio PGRP-SA and GNBP1 (signaling components upstream of Toll) provide further evidence that E. coli is able to activate Toll signaling in T. molitor larvae, in this case, we suggest can occur via TmToll-7 (29). Moreover, these in vitro findings suggest that unlike in Drosophila, the Tenebrio Toll pathway can be activated by recognition of Gram-negative bacteria through cleavage of ModSP, although at this point we cannot rule out the possibility that Toll activation may also occur via the Persephone branch.
One general question that arises from this study is what makes TmToll-7 different from Drosophila Toll-1 and other insect Tolls that mediate immune responses to Gram-positive bacteria and fungi? To try and answer this question, we have further compared the TIR sequence of TmToll-7 with those of Drosophila 18W and Toll-7, given that they are closely related phylogenetically. Protein sequence alignments show that the TmToll-7 TIR domain shares 53% identity and 70% similarity with that of Toll-7 and 48% identity and 68% similarity with that of 18W. These similarities were higher than when compared with the TIR domain of Toll-1 (35% identity and 55% similarity), suggesting that TmToll-7 may activate similar intracellular signaling components as Toll-7 and 18W; but given that Toll-1 also shares a certain level of identity with TmToll-7, we cannot rule out the possibility that they too may share some conserved signaling components.
While so far, none of the intracellular proteins that interact with and/or function downstream of Toll-7 and 18W have been identified, it is known, at least for 18W, that it is required for the nuclear import of the NF-kB transcription factor, Dif, and for the induction of the antibacterial peptide gene attacin (49). Moreover, studies have shown that the induction of attacin involves the Imd transcription factor Relish, in addition to Dif (i.e., Dif and Relish form a heterodimer that binds to the promoter of this antibacterial gene in order to regulate its expression) (50). Given that orthologs for most of the intracellular components of the Toll-1 and Imd pathways have been identified in T. molitor, including two Dif orthologs and a Relish ortholog (44), and based on our data above showing that TmToll-7 RNAi inhibits Tmattacin-2 expression, we hypothesize that similar to 18W, TmToll-7 in T. molitor plays a role in mediating nuclear translocation of Dif and that it also uses a Dif-Relish heterodimer for activating AMP gene expression. However, one major difference between these receptors may be that unlike 18W, TmToll-7 can be activated by Gram-negative bacteria (Figure 8) given that it has been demonstrated through in vitro experiments that DAP-type peptidoglycan of Gram-negative bacteria binds to the Tenebrio recognition protein PGRP-SA and induces proteolytic activation of Spätzle (29). In addition to Tmattacin-2, we also found that Tmdefensin-1 and Tmdefensin-2 were among the five AMPs that resulted in suppressed expression after E. coli challenge and TmToll-7 RNAi. Earlier studies in Drosophila S2 cells have shown that Dif by itself is sufficient to activate defensin expression (51), and when in combination with Relish upregulates defensin activity (50), but whether this also occurs in vivo in T. molitor remains to be determined. Now that several NF-kB protein sequences (Dif1, Dif2, Rel1, and Rel2) from T. molitor have been identified, we can begin to perform further studies to determine which combinations of these Rel proteins are formed and which AMP promoters they directly bind to and regulate.
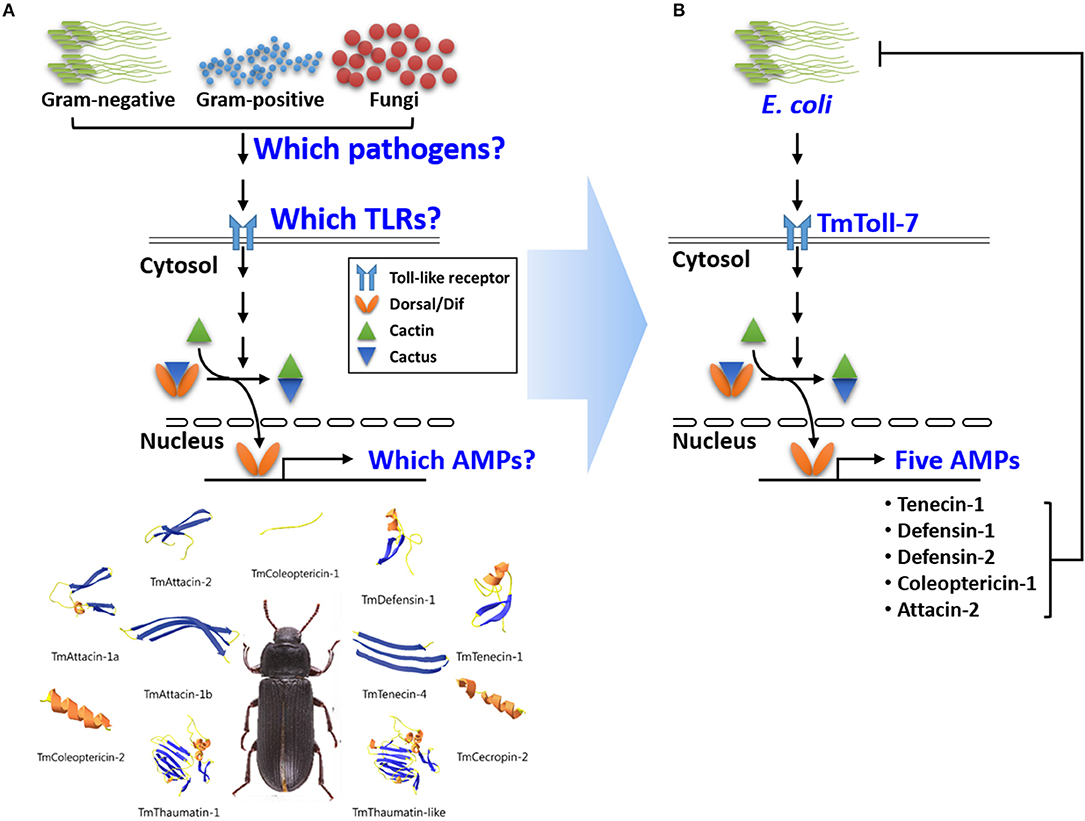
Figure 8. Graphical illustration of TmToll-7 on Toll-signaling pathway in T. molitor. (A) The questions and (B) answers for our study. Our results suggest that the Gram-negative bacterium, E. coli, stimulates TmToll7, resulting in the induction of five AMP genes that kill E. coli.
YH and YJ: conceived and designed the experiments; SP, KP, HK, CK, YB, and BK: performed the experiments; YJ and SP: analyzed the data; YH and YL: contributed reagents, materials, analysis tools; YH, YJ, SP, and YK: wrote the manuscript; SJ, IB, and YL: revised the manuscript.
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and future Planning (Grant No. 2018R1A2A2A05023367).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00310/full#supplementary-material
1. Uematsu S, Akira S. Toll-Like receptors (TLRs) and their ligands. Handb Exp Pharmacol. (2008) 1–20. doi: 10.1007/978-3-540-72167-3_10
2. Bilak H, Tauszig-Delamasure S, Imler JL. Toll and Toll-like receptors in Drosophila. Biochem Soc Trans. (2003) 31 (Pt 3):648–51. doi: 10.1042/bst0310648
3. Belvin MP, Anderson KV. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol. (1996) 12:393–416. doi: 10.1146/annurev.cellbio.12.1.393
4. Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
5. Kolesnikov T, Beckendorf SK. 18 wheeler regulates apical constriction of salivary gland cells via the Rho-GTPase-signaling pathway. Dev Biol. (2007) 307:53–61. doi: 10.1016/j.ydbio.2007.04.014
6. Ligoxygakis P, Bulet P, Reichhart JM. Critical evaluation of the role of the Toll-like receptor 18-Wheeler in the host defense of Drosophila. EMBO Rep. (2002) 3:666–73. doi: 10.1093/embo-reports/kvf130
7. Yagi Y, Nishida Y, Ip YT. Functional analysis of Toll-related genes in Drosophila. Dev Growth Differ. (2010) 52:771–83. doi: 10.1111/j.1440-169X.2010.01213.x
8. Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. (2012) 36:658–67. doi: 10.1016/j.immuni.2012.03.003
9. Akhouayri I, Turc C, Royet J, Charroux B. Toll-8/Tollo negatively regulates antimicrobial response in the Drosophila respiratory epithelium. PLoS Pathog. (2011) 7:e1002319. doi: 10.1371/journal.ppat.1002319
10. McIlroy G, Foldi I, Aurikko J, Wentzell JS, Lim MA, Fenton JC, et al. Toll-6 and Toll-7 function as neurotrophin receptors in the Drosophila melanogaster CNS. Nat Neurosci. (2013) 16:1248–56. doi: 10.1038/nn.3474
11. Ward A, Hong W, Favaloro V, Luo L. Toll receptors instruct axon and dendrite targeting and participate in synaptic partner matching in a Drosophila olfactory circuit. Neuron. (2015) 85:1013–28. doi: 10.1016/j.neuron.2015.02.003
12. Luo C, Shen B, Manley JL, Zheng L. Tehao functions in the Toll pathway in Drosophila melanogaster: possible roles in development and innate immunity. Insect Mol Biol. (2001) 10:457–64. doi: 10.1046/j.0962-1075.2001.00284.x
13. Ooi JY, Yagi Y, Hu X, Ip YT. The Drosophila Toll-9 activates a constitutive antimicrobial defense. EMBO Rep. (2002) 3:82–7. doi: 10.1093/embo-reports/kvf004
14. Bettencourt R, Tanji T, Yagi Y, Ip YT. Toll and Toll-9 in Drosophila innate immune response. J Endotoxin Res. (2004) 10:261–8. doi: 10.1179/096805104225004897
15. Narbonne-Reveau K, Charroux B, Royet J. Lack of an antibacterial response defect in Drosophila Toll-9 mutant. PLoS ONE. (2011) 6:e17470. doi: 10.1371/journal.pone.0017470
16. Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci USA. (2000) 97:10520–5. doi: 10.1073/pnas.180130797
17. Imler JL, Zheng L. Biology of Toll receptors: lessons from insects and mammals. J Leukoc Biol. (2004) 75:18–26. doi: 10.1189/jlb.0403160
18. Leulier F, Lemaitre B. Toll-like receptors–taking an evolutionary approach. Nat Rev Genet. (2008) 9:165–78. doi: 10.1038/nrg2303
19. Beutler B, Poltorak A. The sole gateway to endotoxin response: how LPS was identified as Tlr4, and its role in innate immunity. Drug Metab Dispos. (2001) 29:474–8.
20. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006) 124:783–801. doi: 10.1016/j.cell.2006.02.015
21. Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. (2009) 22:240–73. doi: 10.1128/CMR.00046-08
22. Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelievre E, Gascan H, Ray KP, et al. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat Immunol. (2003) 4:794–800. doi: 10.1038/ni955
23. Valanne S, Wang JH, Ramet M. The Drosophila Toll signaling pathway. J Immunol. (2011) 186:649–56. doi: 10.4049/jimmunol.1002302
24. Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, et al. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. (2006) 16:808–13. doi: 10.1016/j.cub.2006.03.020
25. Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang CS, Butt TM, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. (2006) 127:1425–37. doi: 10.1016/j.cell.2006.10.046
26. El Chamy L, Leclerc V, Caldelari I, Reichhart JM. Sensing of “danger signals” and pathogen-associated molecular patterns defines binary signaling pathways “upstream” of Toll. Nat Immunol. (2008) 9:1165–70. doi: 10.1038/ni.1643
27. Kim CH, Kim SJ, Kan H, Kwon HM, Roh KB, Jiang R, et al. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J Biol Chem. (2008) 283:7599–607. doi: 10.1074/jbc.M710216200
28. Rolff J, Reynolds SE. Insect Infection and Immunity: Evolution Ecology and Mechanisms. New York, NY: Oxford University Press (2009).
29. Yu Y, Park JW, Kwon HM, Hwang HO, Jang IH, Masuda A, et al. Diversity of innate immune recognition mechanism for bacterial polymeric meso-diaminopimelic acid-type peptidoglycan in insects. J Biol Chem. (2010) 285:32937–45. doi: 10.1074/jbc.M110.144014
30. Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster-from microbial recognition to whole-organism physiology. Nat Rev Immunol. (2014) 14:796–810. doi: 10.1038/nri3763
31. Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. (2006) 7:715–23. doi: 10.1038/ni1356
32. Ganesan S, Aggarwal K, Paquette N, Silverman N. NF-kappaB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr Top Microbiol Immunol. (2011) 349:25–60. doi: 10.1007/82_2010_107
33. Roh KB, Kim CH, Lee H, Kwon HM, Park JW, Ryu JH, et al. Proteolytic cascade for the activation of the insect toll pathway induced by the fungal cell wall component. J Biol Chem. (2009) 284:19474–81. doi: 10.1074/jbc.M109.007419
34. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
35. Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. (2001) 17:847–8. doi: 10.1093/bioinformatics/17.9.847
36. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, et al. InterProScan: protein domains identifier. Nucleic Acids Res. (2005) 33:W116–20. doi: 10.1093/Nar/Gki442
37. Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. (2014) 30:1236–40. doi: 10.1093/bioinformatics/btu031
38. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. (2007) 23:2947–8. doi: 10.1093/bioinformatics/btm404
39. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. (2013) 30:2725–9. doi: 10.1093/molbev/mst197
40. Moon HJ, Lee SY, Kurata S, Natori S, Lee BL. Purification and molecular cloning of cDNA for an inducible antibacterial protein from larvae of the coleopteran, Tenebrio molitor. J Biochem. (1994) 116:53–8. doi: 10.1093/oxfordjournals.jbchem.a124502
41. Gangloff M, Arnot CJ, Lewis M, Gay NJ. Functional insights from the crystal structure of the N-terminal domain of the prototypical toll receptor. Structure. (2013) 21:143–53. doi: 10.1016/j.str.2012.11.003
42. Lewis M, Arnot CJ, Beeston H, McCoy A, Ashcroft AE, Gay NJ, et al. Cytokine Spatzle binds to the Drosophila immunoreceptor Toll with a neurotrophin-like specificity and couples receptor activation. Proc Natl Acad Sci USA. (2013) 110:20461–6. doi: 10.1073/pnas.1317002110
43. Yu X-Q, Chowdhury M, Li C-F, He Z, Lu Y, Liu X, et al. Multiple toll-spätzle pathways in Drosophila melanogaster immunity. bioRxiv [preprint]. (2018). doi: 10.1101/420679
44. Johnston PR, Makarova O, Rolff J. Inducible defenses stay up late: temporal patterns of immune gene expression in Tenebrio molitor. G3. (2014) 4:947–55. doi: 10.1534/g3.113.008516
45. Duneau DF, Kondolf HC, Im JH, Ortiz GA, Chow C, Fox MA, et al. The Toll pathway underlies host sexual dimorphism in resistance to both Gram-negative and Gram-positive bacteria in mated Drosophila. BMC Biol. (2017) 15:124. doi: 10.1186/s12915-017-0466-3
46. Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, et al. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest. (1985) 76:1427–35. doi: 10.1172/JCI112120
47. Bulet P, Cociancich S, Dimarcq JL, Lambert J, Reichhart JM, Hoffmann D, et al. Insect immunity. Isolation from a coleopteran insect of a novel inducible antibacterial peptide and of new members of the insect defensin family. J Biol Chem. (1991) 266:24520–5.
48. Carlsson A, Nystrom T, de Cock H, Bennich H. Attacin–an insect immune protein–binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology. (1998) 144 (Pt 8):2179–88. doi: 10.1099/00221287-144-8-2179
49. Williams MJ, Rodriguez A, Kimbrell DA, Eldon ED. The 18-wheeler mutation reveals complex antibacterial gene regulation in Drosophila host defense. EMBO J. (1997) 16:6120–30. doi: 10.1093/emboj/16.20.6120
50. Han ZS, Ip YT. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J Biol Chem. (1999) 274:21355–61. doi: 10.1074/jbc.274.30.21355
Keywords: Toll-7, Tenebrio molitor, microbial infection, RNAi, antimicrobial peptides, AMP assay
Citation: Park S, Jo YH, Park KB, Ko HJ, Kim CE, Bae YM, Kim B, Jun SA, Bang IS, Lee YS, Kim YJ and Han YS (2019) TmToll-7 Plays a Crucial Role in Innate Immune Responses Against Gram-Negative Bacteria by Regulating 5 AMP Genes in Tenebrio molitor. Front. Immunol. 10:310. doi: 10.3389/fimmu.2019.00310
Received: 27 September 2018; Accepted: 06 February 2019;
Published: 12 March 2019.
Edited by:
Jun-ichi Hikima, University of Miyazaki, JapanReviewed by:
Anchalee-Tassanakajon, Chulalongkorn University, ThailandCopyright © 2019 Park, Jo, Park, Ko, Kim, Bae, Kim, Jun, Bang, Lee, Kim and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Jung Kim, eWpraW1AY3N1c2IuZWR1
Yeon Soo Han, aGFueXNAam51LmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.