
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 22 November 2018
Sec. B Cell Biology
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.02715
 David Benhamou1
David Benhamou1 Verena Labi2,3
Verena Labi2,3 Andrew Getahun4
Andrew Getahun4 Eli Benchetrit1
Eli Benchetrit1 Reem Dowery1
Reem Dowery1 Klaus Rajewsky2
Klaus Rajewsky2 John C. Cambier4
John C. Cambier4 Doron Melamed1*
Doron Melamed1*Appropriate PI3K signals generated by the antigen receptor are essential to promote B cell development. Regulation of recombination activating gene (RAG)-1 and RAG-2 expression is one key process that is mediated by PI3K to ensure developmental progression and selection. When PI3K signals are too high or too low, expression of RAGs does not turn off and B cell development is impaired or blocked. Yet, the mechanism which tunes PI3K activity to control RAG expression during B cell development in the bone marrow is unknown. Recently we showed that a c-Myc/miR17-92/PTEN axis regulates PI3K activity for positive and negative selection of immature B cells. Here, we show that the c-Myc/miR17-92/PTEN axis tunes PI3K activity to control the expression of RAGs in proB cells. Using different genetically engineered mouse models we show that impaired function of the c-Myc/miR17-92/PTEN axis alters the PI3K/Akt/Foxo1 pathway to result in dis-regulated expression of RAG and a block in B cell development. Studies using 38c-13 B lymphoma cells, where RAGs are constitutively expressed, suggest that this regulatory effect is mediated post-translationally through Foxo1.
The development of B lymphocytes in the bone marrow (BM) is a highly regulated process guided by the successive attempts to recombine immunoglobulin (Ig) genes and to assemble the B cell antigen receptor (BCR). Recombination activating gene (RAG)-1 and RAG-2 are key enzymes in this process and their expression is tightly regulated by signals generated by the antigen receptor to promote developmental progression through negative and positive selection check-points. Binding self-antigens activates tolerance mechanisms in immature and transitional B cells (1–3), whereas appropriate ligand-independent (tonic) signals are necessary for positive selection, developmental progression and maturation (4, 5). Thus, tonic signals, which are transmitted by the precursor BCR (preBCR), stimulate proliferative expansion of proB cells (6, 7), and those transmitted by the BCR promote developmental progression and maturation of immature B cells expressing non-harmful receptors (8–10). Failure to express preBCR or BCR or their signaling or surface regulatory components impairs or blocks B cell development (4, 11, 12).
The phosphoinositide 3-kinase (PI3K) activity is critical for B cell development and survival (12–16). Tonic PI3K signaling regulates expression of RAGs and is therefore essential for positive selection at the proB and immature B cell stages. Foxo1 is a major transcription factor necessary to induce the transcription of both RAGs. In developing B cells the expression and function of Foxo1 are directly regulated through phosphorylation and inactivation by the PI3K/Akt pathway (17, 18). When PI3K signals are too low, such as in CD19-deficient (CD19KO) B cells or when antigen receptor is ablated, continues RAG expression results in ongoing rearrangements (9, 14, 19–21). On the other hand, PI3K signals that are too high, such as in PTEN-ablated B cells, suppress RAG expression and impose a severe developmental block at the proB stage (22). These studies show that generation of appropriate PI3K activity is crucial to control RAG levels, thus allowing for proper developmental progression. However, the exact physiological mechanism and/or biochemical circuit which tunes PI3K activity to control RAG expression at these early stages of B cell development are unknown.
MicroRNAs were shown to regulate B lymphocyte fate at multiple levels, affecting physiological processes such as development, differentiation, and function (23–26). Recently, we showed that miRNAs are essential to regulate the PI3K/Akt/Foxo1 pathway and RAG expression in peripheral B cells (27). In another work, we reported an autostimulatory axis by which the PI3K regulates expression of its main antagonistic phosphatase, phosphatase and tensin homolog (PTEN). This autostimulatory axis is composed of a transcription factor c-Myc, a microRNA miR17-92, and a signaling molecule PTEN (c-Myc/miR17-92/PTEN axis) and controls PI3K activity for positive and negative selection of immature B cells (28). According this axis, c-Myc, whose expression levels are regulated by PI3K, directly controls expression of miR17-92, which post-transcriptionally promotes degradation of PTEN mRNA to dampen PTEN protein levels and to enhance PI3K activity. The resulting enhanced PI3K further promotes expression of high levels of c-Myc and survival of B cells (see also Figure 8). In the present study, we explored the role of the c-Myc/miR17-92/PTEN axis in regulating RAG expression at early stages of B cell development. We show that in proB cells the c-Myc/miR17-92/PTEN axis tunes expression of RAG through controlling the activity of PI3K. Using different conditional mutated mouse models we show that impaired function of the c-Myc/miR17-92/PTEN axis alters the PI3K/Akt/Foxo1 pathway to result in dis-regulated expression of RAG and a block in B cell development. In further studies we used 38c-13 B lymphoma cells, where RAG genes are constitutively expressed, to suggest that this regulatory effect is mediated post-translationally through Foxo1.
miR17-92fl/fl (29), ROSA26STOPflox PTEN-2AYFP (PTENover) (30), ROSA26STOPflox-P110* (16), CD19KO (31), miR17-92Tg mice (32), and mb1-cre (33) mice have been described. All mice are C57BL/6 or have been backcrossed to the C57BL/6 background for more than 10 generations. Mice were housed and bred at the animal facility of the Technion, Faculty of Medicine (Israel) or at the Max Delbrück Center for Molecular Medicine (Berlin, Germany) or at the University of Colorado, School of Medicine (Aurora CO, United States) and used for experiments at 10–12 weeks of age. Mouse studies at the Technion were approved by the local committee for the supervision of animal experiments. Animal care in Berlin followed guidelines of the Max Delbrück Center for Molecular Medicine and the governmental review board (Landesamt für Gesundheit und Soziales Berlin, LaGeSo). Animal studies at University of Colorado were performed in accordance with the regulations and with approval of the University of Colorado Denver Institutional Animal Care and Use Committee.
38c13 cells (34) were cultured in standard RPMI. Cells were infected with different viral vectors to express a sponge system to miR-19b or to the complete miR17-92 cluster, overexpress PTEN or knockdown PTEN (shPTEN) (see supplemental details). Stable transfectants were selected and used for the experiments. Cells were lysed (see Supplemental Details in methodology) for western blotting and quantification of mRNAs. In some experiments a specific inhibitor for Foxo1 transcription activity AS1842856 (Calbiochem) was added to the cultured cells at 14 μM for 2 h. In some experiments HEK cells were transfected with vectors encoding luciferase reporter bound to 3′UTR of PTEN or Foxo1 as detailed in the Supplementary Section. IL-7-driven BM cultures for B cell precursors were prepared as previously described (35). Cells grown in these primary cultures were harvested after 5 days (>95% B220+) and were used as detailed. In some experiments B cell precursors were grown in IL-7 driven BM cultures (36) and cells were treated with 1 μM miR19b antagomirs (Exiqon).
Single cell suspensions from mouse BM or spleen were stained with fluorescently labeled antibodies for surface and intracellular proteins to identify specific B cell subsets. Antibodies for the following surface markers were used: αB220-BV785 (RA3-6B2), αCD19-BV605 (6D5), αCD93-APC (AA4.1), αB220-PB (RA3-6B2), αCD25-PE (3C7), αc-kit-BV605 (ACK2) from BioLegend, αCD1d-PE (1B1) from eBioscience. Antibodies for intracellular staining used were anti pAkt (4060) and anti PTEN (p559) from Cell Signaling using the BrdU fixation kit (552598) from BD. Data were acquired on LSRFortessa (BD Pharmingen) and analyzed using FlowJo software (Tree Star). In some experiments cells of specific populations based on surface markers expression were sorted using FACSAria (BD).
Western blots were performed as we have previously described (37). Briefly, cells were lysed in RIPA buffer containing complete protease inhibitor cocktail (Roche Diagnostics), 1 mM sodium orthovanadate, 5 mM sodium fluoride and 0.5 mM phenylmethylsulfonyl fluoride. Proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membrane (Millipore, Bedford, MA, United States). Blots were probed with rabbit anti-phospho-Akt (Ser473) (Cat 4060), rabbit anti-Akt (Cat 9272), rabbit anti-PTEN (Cat 9188), (all from Cell Signaling), rabbit anti-Foxo1(Santa Cruz), mouse anti-actin (C4) (from MP biomediacls). To reveal bound Abs we used HRP conjugated secondary Abs. Blots were developed with ECL reagent (Pierce).
To suppress miR19b or the entire miR17-92 cluster in 38c13 cells we used pMSCV-PIG-sp19 or pMSCV-PIG-sp17-92 encoding a sponge system to miR19b or to the entire miR17-92 cluster as has been described (38). To overexpress human PTEN we used an inducible Tet-on expression system that has been described (28). PTEN expression was induced upon induction with 1 μg/ml doxycycline. To knockdown PTEN expression we used a viral vector encoding shPTEN [LENG.ShPten 1523, (39)]. In some experiments, 38c13 cells were first infected with LENG.ShPten1523 vector and after selection the cells were infected with pMSCV-PIG-sp19 to generate cells where PTEN is knockdown and miR19b is suppressed.
Retroviral infections, virus production infection and selection of cells were performed as we have described (28, 40).
Quantitative PCR analysis for RAG-1 RAG-2, PTEN and Hprt were performed as we have previously described (40). Briefly, RNA was extracted by using Tri-Reagent (Sigma) and reverse transcribed to cDNA (Promega). Real-time PCR was performed using Syber green mix (Tiangen) using BioRad CFX connect Real-Time PCR. Samples were normalized with Hprt gene amplification. PCR product specificity was confirmed by melting curve analysis where the crossover (Ct) values were used to calculate gene specific input mRNA amount for each sample according to the calibration curve method. DATA were analyzed using the CFX Manager Software.
To perform the luciferase reporter assay, 105 HEK cells per well in 1 ml of medium were seeded in 24-well plates for 24 h. Cells were then transfected with the modified firefly luciferase vector PmiRGLO- PTEN3'UTR or the PmirGLO-Foxo13′UTR together with a vector encoding miR17-92 or GFP using Calfectin according to the manufacturer's instruction. The 3'UTRs of PTEN and Foxo1 were amplified from DNA isolated from C57BL/6 mice by pcr and cloned into the pmirGLO vector. After 48 h the firefly and renilla luciferase activities were measured with the dual-luciferase reporter assay system (Promega). To control the transfection efficiency, firefly activity was normalized to renilla activity.
The statistical significance of the differences between experimental groups was determined using unpaired two-tailed Student's t test with differences considered significant at p < 0.05.
We have shown that a c-Myc/miR17-92/PTEN axis regulates PI3K activity for positive selection of immature B cells (28). As PI3K signals are necessary to turn off RAG (17, 18, 20), we tested whether the c-Myc/miR17-92/PTEN functions to tune PI3K activity to control RAG expression during B cell development. To do so in vivo, we used conditional mice enabling altering the expression and/or activity of c-Myc/miR17-92/PTEN axis in B lineage cells. We first generated mb1-Cre/miR17-92f/f mice, where the entire miR17-92 cluster is conditionally ablated in early proB cells (33). Analysis of BM cells in these mice revealed a mild block at the proB stage as reflected by a 2-fold increase in their frequency (Figure 1A) and by the changed proB/preB cell ratio (Figure 1B). In agreement with Lai et al (41), we found that ablation of miR17-92 resulted in 15–20% increase in expression of PTEN mRNA in proB and preB cells (Figure 1C) and PTEN protein (Figure 1D), and reduced PI3K activity measured by Akt phosphorylation (Figure 1D). These changes were also validated in pro/pre B cells grown in BM cultures that were treated with miR19b antagomirs (Figure 1E). To further validate these findings in a reciprocal experiment we analyzed hCD2Cre/R26miR17-92stopflox mice overexpressing miR17-92 in all lymphocytes and found that PTEN expression is reduced in pro/pre B cells whereas pAkt is increased (Supplementary Figure 1B). Consistent with this and with our hypothesis, we found that expression levels of both RAG-1 and RAG-2 were elevated in preB cells from mb1-Cre/miR17-92f/f mice relative to the controls (Figure 1F). Despite of this mild pro-to-pre B cell block we found no significant differences in splenic B cells (total number and specific subsets, Supplementary Figure 1). These findings suggest that intrinsic deletion of miR17-92 in proB cells impairs the regulatory activity of the c-Myc/miR17-92/PTEN axis to result in enhanced RAG expression and a partial pro-to-preB block.
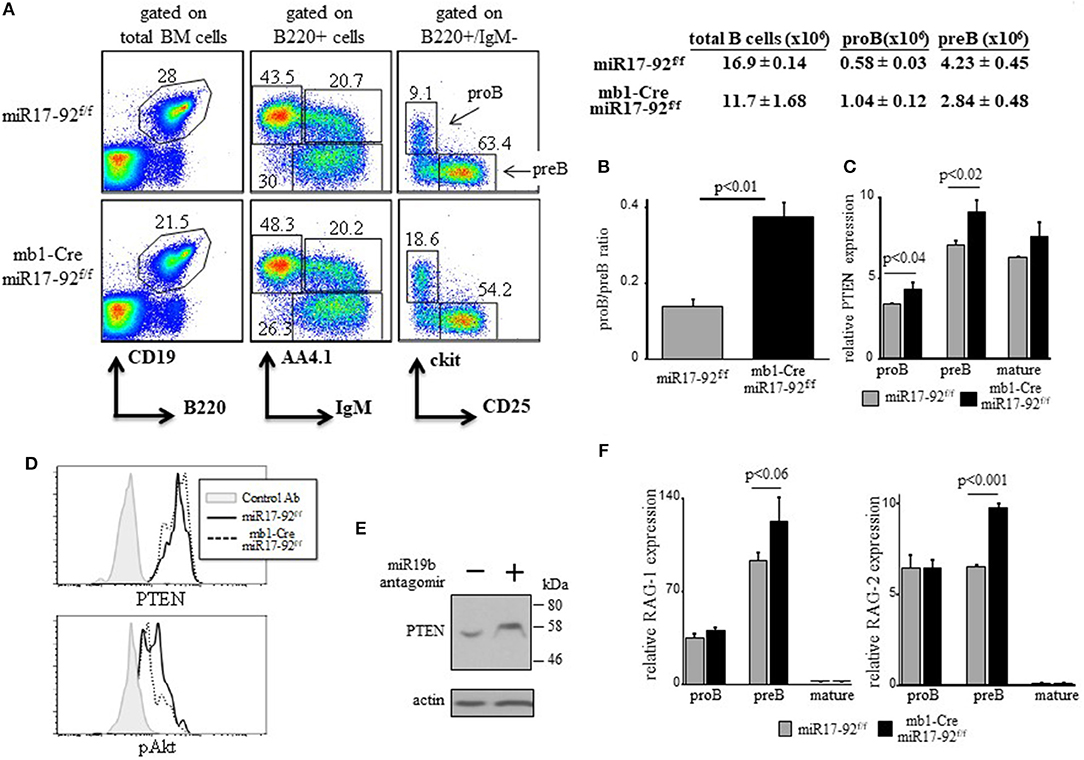
Figure 1. Ablation of miR17-92 in proB cells imposes a mild pro-to-pre B cell block and elevates expression of RAGs. (A) Representative flow cytometry analysis of BM cells from mice with the indicated genotypes (3 mice from each genotype). Initial forward and side scatter gates were set to exclude dead cells and debris. Numbers adjacent to outlined areas indicate % cells amongst total BM cells in each gate. The proB (B220 + IgM- AA4.1 + CD25- ckit+) and preB (B220 + IgM- AA4.1 + CD25 + ckit-) populations are marked with arrows. Also shown are absolute cell counts. (B) The proB and preB cells were quantified for each individual mouse and are expressed as proB/preB ratio. Plot depicts mean from 3 individual mice ± SE. (C) The proB, preB and mature B (B220 + IgM + AA4.1-) cells were sorted from the gates shown in (A) and analyzed for relative expression of PTEN mRNA by qPCR and normalized to Hprt. Results are presented as mean from 3 individual mice ± SE. (D) Intracellular stain for PTEN and pAKT of BM cells gated on B220+/IgM- pro/pre B cells. Graph represents 2 mice in each group. (E) BM culture wild-type pro/preB cells were treated with or without miR19b antagomirs for 48 h and analyzed for the indicated proteins by western blotting. (F) Sorted proB, preB and mature B cells were analyzed for relative expression of RAG-1 (top) and RAG-2 (bottom) by qPCR normalized to Hprt. Graph depicts mean from 3 individual mice ± SE.
To confirm the function of the c-Myc/miR17-92/PTEN axis in tuning RAG expression in early B cell development we generated mb1-Cre/ROSA26STOPflox PTEN-2AYFP compound mice (30), where selective over-expression of PTEN by about 15–20% is obtained in the B lineage starting from the proB stage (Supplementary Figure 1C). Over-expression of PTEN in these mice is encoded by PTEN cDNA and is therefore not subjected to regulation by the miR17-92. Analysis of B cell development and RAG expression in these compound mice recapitulated our findings obtained in the miR17-92 ablated mice (Figure 1). Thus, overexpression of PTEN imposed a partial block at the proB stage as revealed by about 2-fold increase in proB cell frequency (Figure 2A) and a changed proB/preB cell ratio (Figure 2B). Similarly, overexpression of PTEN, obtained by transduction of hematopoietic stem cells (HSCs) with a lentivirus vector encoding human PTEN, impaired B cell development as reflected by reduced frequencies of total B cells in BM and spleen, and accumulation of B220+/CD43 + cells (mostly proB cells). However, when the transducing vector encoded a human PTEN construct ligated to a miR19b binding site [the major miRNA regulating PTEN (32)] PTEN expression was suppressed and B cell development was rescued (Supplementary Figure 2). Quantification of RAG expression in sorted cells revealed that RAG-1 and RAG-2 were significantly increased in proB and preB cells that over-express PTEN (Figure 2C). These results suggest that the autostimulatory c-Myc/miR17-92/PTEN axis tunes RAG expression in B cell development through balancing PI3K activity. Alteration of this axis, by either ablation of the miR17-92 cluster or over-expression of PTEN, enhances expression of RAG and partially blocks B cell development at the proB stage.
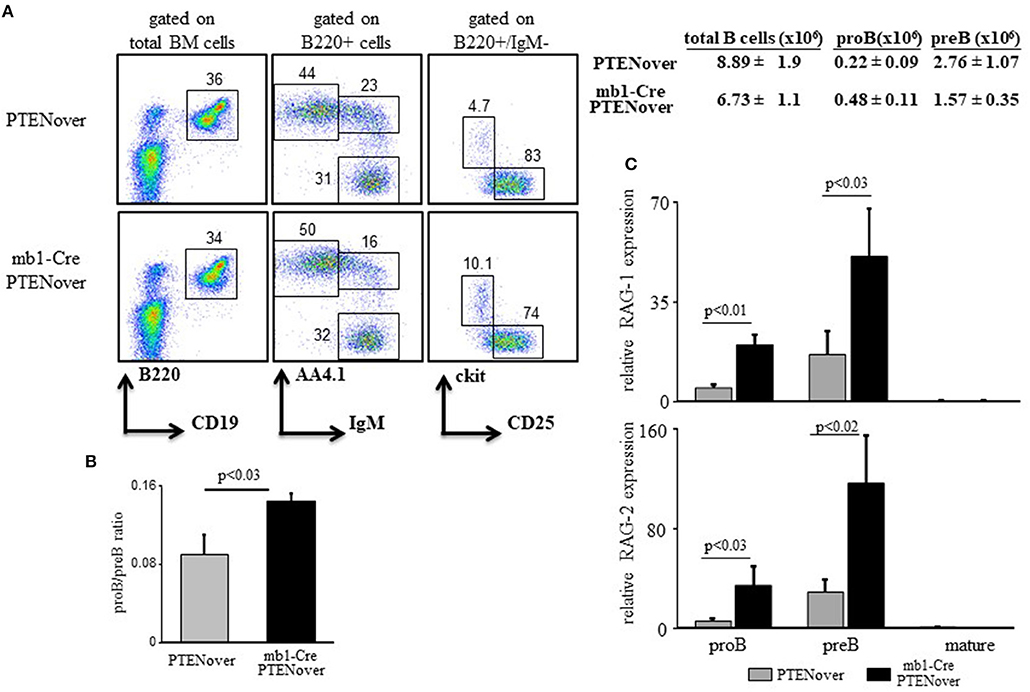
Figure 2. PTEN overexpression partially blocks pro-to-pre B cell transition and elevates expression of RAGs. (A) Representative flow cytometry analysis of BM cells from mice with the indicated genotypes (3–5 mice from each genotype). Initial forward and side scatter gates were set to exclude dead cells and debris. Numbers adjacent to outlined areas indicate % cells amongst total splenocytes in each gate. Also shown are absolute cell counts. Total numbers of B cells in the BM were calculated as total number of nucleated cells purified from one femur and one tibia multiplied by percentage of B220+ cells. (B) The proB and preB cells (gated as in Figure 1 legend) were quantified for each individual mouse and are expressed as proB/preB ratio. Plot depicts mean from 3 to 5 individual mice ± SE. (C) The proB, preB, and mature B cells were sorted using the indicated gates and analyzed for relative expression of RAG-1 (top) and RAG-2 (bottom) by qPCR normalized to Hprt. Graph depicts mean from 3 to 5 individual mice ± SE.
To probe for the mechanism by which the c-Myc/miR17-92/PTEN axis controls RAG expression we used the 38c13 immature B lymphoma cells which constitutively express RAG (34). To alter c-Myc/miR17-92/PTEN axis in these cells we used a sponge system specific to miR19b (38) to suppress expression of this miRNA. We found that suppression of miR19b resulted in a significant increase in expression of PTEN protein and a consequential reduction in PI3K activity as revealed by Akt phosphorylation (Figure 3A). Consistent with the reduced PI3K activity, expression of RAG-1 and RAG-2 were significantly enhanced by ~2-fold relative to the control (Figure 3B). Reciprocally, transfection with a vector encoding PTEN specific shRNA [shPTEN (39)] enhanced Akt phosphorylation significantly and suppressed expression of RAG-1 and RAG-2 by ~2-fold relative to the control (Figures 3A,B). Furthermore, in cells co-transfected with miR19b-sponge and shPTEN, activity of PI3K was enhanced to effectively suppress RAG expression (Figures 3A,B), suggesting that the increased expression of RAG induced by the miR19b-sponge system is mediated via alteration of PTEN expression. Consistent with the regulatory feedback of the c-Myc/miR17-92/PTEN axis we show that reduced PI3K activity obtained by suppression of miR19b reduced expression of MYC whereas enhanced PI3K activity obtained by suppression of PTEN enhanced MYC expression (Figure 3A).
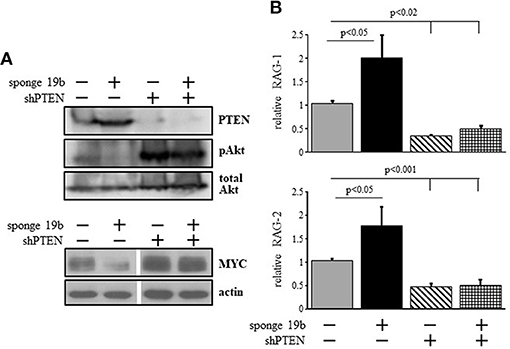
Figure 3. The c-Myc/miR17-92/PTEN axis regulates RAG expression in 38c13 cells through balancing PI3K activity. 38c13 cells were infected with vector encoding miR19b sponge or shPTEN (to suppress miR19b or PTEN, respectively) or with both, and stable transfectants were selected. (A) Representative western blot for the indicated proteins (4 different experiments). (B) The indicated stably transfected 38c13 cells were lyzed and analyzed for relative RAG-1 and RAG-2 expression by qPCR normalized to Hprt. Graph depicts mean of 4 experiments ± SE.
To further validate this, we infected 38c13 cells with a vector encoding an inducible system for human PTEN expression [PTEN-OE (42)]. Over expression of PTEN in these cells was evident by PTEN protein level and a suppressed PI3K activity (revealed by Akt phosphorylation) in the selected subclones (Figure 4A). In agreement with the results obtained upon introducing the miR19b-sponge system (Figure 3), we found that RAG-1 and RAG-2 were significantly elevated in 38c13 cells over-expressing PTEN (Figure 4B). Collectively, these results confirmed that in 38c13 cells the regulation of RAG expression is mediated by the c-Myc/miR17-92/PTEN axis.
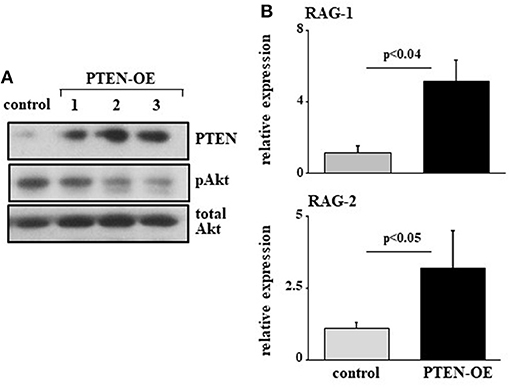
Figure 4. Over expression of PTEN suppresses PI3K and enhances RAG expression in 38c13 cells. 38c13 cells were stably transfected with a Tet-on inducible system to over-express human PTEN (PTEN-OE). Cells were treated with doxycycline and analyzed after 24 h. (A) Representative western blot for the indicated proteins (shown are three different clones for PTEN-OE). (B) The stably PTEN-OE and control 38c13 cells were lyzed and analyzed for relative RAG-1 and RAG-2 expression by qPCR normalized to Hprt. Graph depicts mean of 3–6 experiments ± SE.
Previous studies have shown that regulation of RAG by PI3K is mediated post-translationally through phosphorylation and inactivation of Foxo1 (17, 18). In agreement with this we find elevated expression of Foxo1 in 38c13 cells transfected with miR17-92 or the miR19b sponge (Figure 5A), or over-expressing PTEN (Figure 5). Moreover, we show that the induction of RAG expression obtained upon transfection with the miR19b sponge system is blocked by a Foxo inhibitor (Figure 5C), suggesting that the regulatory effects of the c-Myc/miR17-92/PTEN axis on RAG expression are mediated by Foxo1. Yet, bioinformatic analysis of the Foxo1 3′UTR (using the TargetScanHuman program; http://www.targetscan.org) revealed a potential binding site for miR19b, proposing a direct post-transcriptional regulatory effect of miR19b on expression of Foxo1. To test this we transfected human embryonic kidney (HEK) cells with a vector encoding luciferase reporter ligated to Foxo1 3′UTR or PTEN 3'UTR, together with a vector encoding miR17-92 or GFP control. The results in Figure 5D show that miR17-92 significantly reduced luciferase activity in HEK cells transfected with a luciferase reporter ligated to PTEN 3'UTR, whereas no change was recorded in HEK cells transfected with luciferase reporter ligated to Foxo1 3′UTR. Moreover, quantification of Foxo1 mRNA in 38c13 cells transfected with miR19b sponge revealed no significant change relative to control cells (Figure 5E). These results suggest that regulation of Foxo1 expression by the c-Myc/miR17-92/PTEN axis is mediated post-translationally through balancing PI3K activity.
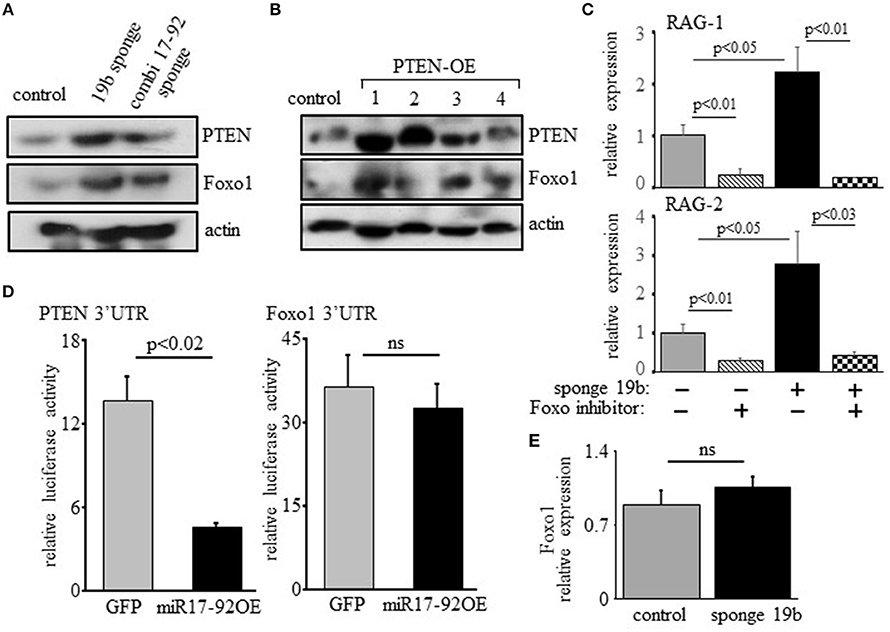
Figure 5. Regulation of RAG expression by the c-Myc/miR17-92/PTEN axis is mediated through Foxo1, but not through direct binding of miR17-92 to the Foxo1 3'UTR. (A) 38c13 cells were transfected with a sponge system to miR19b or to the entire miR17–92 cluster. Shown is a representative western blot for the indicated proteins (from 3 experiments). (B) 38c13 cells were stably transfected with a Tet-on inducible system to over-express human PTEN (PTEN-OE). Cells were treated with doxycycline and analyzed after 24 h lysed and analyzed by western blotting for the indicated proteins (shown are four different clones for PTEN-OE). (C) 38c13 cells that are wild-type or transfected with the miR19b sponge system were cultured for 2 h in the presence of Foxo1 inhibitor. Cells were then lyzed and analyzed for relative RAG-1 and RAG-2 expression by qPCR normalized to Hprt. Graph depicts mean of 3 experiments ± SE. (D) HEK cells were transfected with vector encoding luciferase reporting system that is ligated to PTEN3′UTR or to Foxo1 3'UTR, together with a vector encoding miR17-92 or GFP. After 48 h luciferase activity was determined. Graph depicts mean of 3 experiments ± SE. (E) 38-c13 cells that are wild-type control or transfected with the miR19b sponge system were lyzed and relative Foxo1 mRNA was quantified by qPCR normalized to Hprt. Graph depicts mean of 3 experiments ± SE.
To address physiological roles of the c-Myc/miR17-92/PTEN axis in controlling RAG expression during B cell development we used CD19KO mice, where RAG expression in developing B cells does not turn off (9) owing to impaired PI3K signaling (20, 43). Consequentially, B cell maturation in CD19KO mice is impaired with evidence for ongoing Ig gene rearrangements (9, 19, 44). Overexpression of miR17-92 (28) or loss of PTEN (45) in developing CD19KO B cells enhanced PI3K activity, to effectively reconstitute maturation and differentiation of CD19KO B cell (28). The results in Figure 6 show that enhancing PI3K activity through overexpression of miR17-92 effectively suppressed RAG expression in developing CD19KO miR17-92Tg B cells to promote their differentiation in BM cultures. Thus, while RAG-1 and RAG-2 expression were significantly elevated in developing CD19KO B cells (~18 and ~5-fold, respectively), expression of both RAGs in developing CD19KO miR17-92Tg B cells was suppressed to a level that is not different from that found in developing control B cells. These results suggest that reconstitution of B cell development and maturation in CD19KO miR17-92Tg mice is mediated by the suppression of RAG expression, which is in turn mediated through tuning PI3K activity by the c-Myc/miR17-92/PTEN axis.
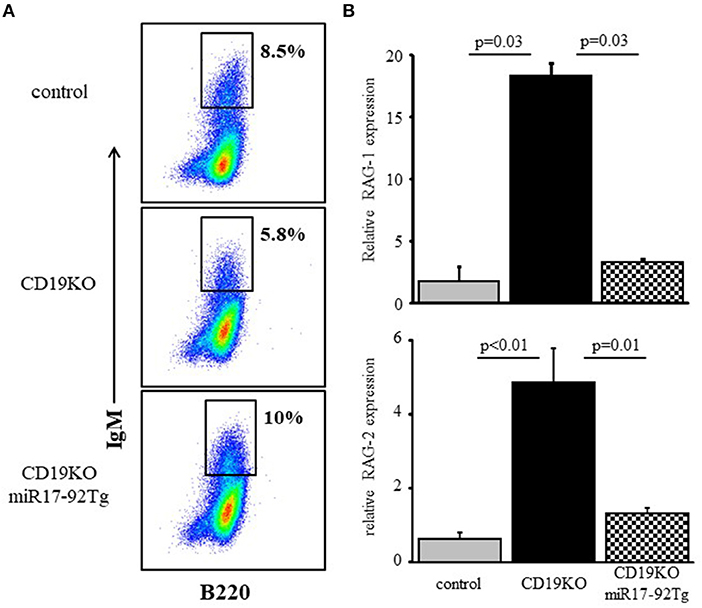
Figure 6. Overexpression of miR17-92 suppresses RAG in developing CD19KO B cells. IL-7-driven BM cultures from mice of the indicated genotypes were prepared. (A) Cells grown were harvested on day 5, stained for the indicated surface markers and analyzed by FACS. Initial forward and side scatter gates were set to exclude dead cells and debris. Shown are representative plots from 3 to 4 mice from each group. (B) Cells were lyzed and relative RAG-1 and RAG-2 mRNA was quantified by qPCR normalized to Hprt. Graph depicts mean of 3–4 mice from each group ± SE.
In all mouse models detailed here earlier (miR17-92 ablated, PTEN-over expression and CD19KO) B cell development is perturbed due to a low PI3K activity and enhanced RAG expression, resulting from reduced activity of the c-Myc/miR17-92/PTEN axis. To further confirm the importance of this axis we performed a reciprocal experiment and tested whether constitutively active PI3K would suppress RAG gene expression in developing B cells to impair their development. To do so, we generated mice carrying a conditional allele encoding constitutively active PI3K subunit P110α [P110* (16)] specifically expressed in the B lineage starting at the early proB stage (mb1-Cre/ROSA26STOPflox-P110* compound mice). As shown in Figure 7, the enhanced PI3K activity blocked B cell development at the early proB stage (ckit-/CD25-) with only few proB (cKit+/CD25-) and preB cells (cKit-/CD25+) detected (Figure 7A), explaining the nearly complete absence of B220+/IgM+ cells in the BM (Figure 7A). Analysis of RAG in sorted proB and preB cells from mb1-Cre/ROSA26STOPflox-P110* mice revealed a significantly suppressed expression in both B cell subsets relative to control (Figure 7B). These results suggest that enhanced PI3K activity suppresses RAG expression to block B lymphopoiesis.
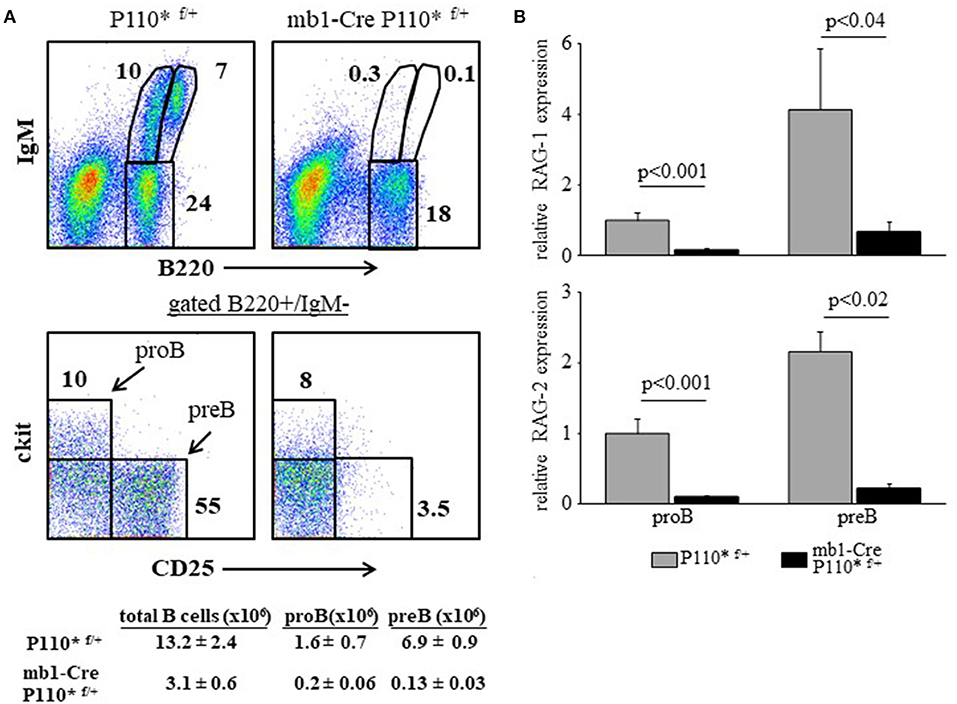
Figure 7. Enhanced PI3K activity abolishes RAG expression and blocks B cell development at the early proB stage. (A) Representative flow cytometry analysis of BM cells from mice with the indicated genotypes (3 mice from each genotype). Initial forward and side scatter gates were set to exclude dead cells and debris. Also shown are absolute cell counts. (B) The proB and preB cells were sorted using the indicated gates and analyzed for relative expression of RAG-1 (top) and RAG-2 (bottom) by qPCR normalized to Hprt. Graph depicts mean from 3 individual mice ± SE.
This study suggests that the autostimulatory c-Myc/miR17-92/PTEN axis functions as a post-translational mechanism balancing the PI3K/Foxo1/RAG pathway during early stages of B cell development. We show that modification of this axis altered expression of RAGs and impacted B cell development. Hence, while BCR-mediated PI3K signaling is most important for B cell development and survival, the c-Myc/miR17-92/PTEN axis functions to properly tune it for regulation of RAGs and to promote developmental progression.
The mechanism by which the c-Myc/miR17-92/PTEN axis balances the PI3K/Foxo1/RAG pathway in B cell development is illustrated in Figure 8. In developing B cells the PI3K/Akt/Foxo1 is most important to control RAG expression. Both, enhanced or suppressed PI3K significantly impact RAG expression and consequentially alter B cell development. Thus, enhanced PI3K activity obtained upon ablation of PTEN suppresses RAG expression and blocks B cell development at the proB stage (22), whereas deficiency of CD19 (9), or lack of the p85(alpha) regulatory subunit of PI3K (20) suppresses PI3K activity to result in ongoing RAG expression and Ig gene rearrangements (9, 19, 44). This study shows that the PI3K activity in developing B cells is balanced by the c-Myc/miR17-92/PTEN axis for appropriately controlled RAG expression (Figure 8). Accordingly, we show that altering the c-Myc/miR17-92/PTEN axis, by deletion of miR17-92 or by overexpressing PTEN, suppressed PI3K activity, led to enhanced RAG expression in early developing B cells, and imposed a mild pre-to-pro B cell block (Figures 1, 2). A similar mild pro-to-pre B cell block is also evident in CD19-/- mice, although RAG expression levels in pro and pre B cells were not measured in this study (46). Yet, in all these mutated mouse models significant B cell development is still observed, as revealed by the number of mature B cells in BM and spleen, probably suggesting the existence of compensatory mechanisms to enhance PI3K activity (9, 44). In agreement with this, Sandoval et al reported that cycling large preB cells express MYC and high level of inactivated (phosphorylated) Foxo1, whereas non-cycling preB cells do not have MYC protein but have abundant expression of RAG and DNA rearrangements (47).
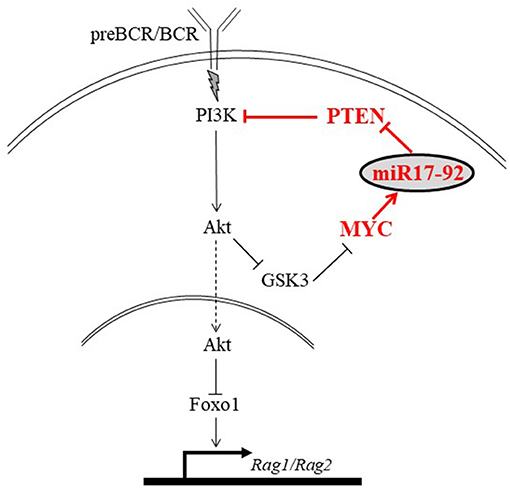
Figure 8. An illustration proposing a mechanism by which the MYC/miR17-92/PTEN axis balances the PI3K/Foxo1/RAG pathway in developing B cells. Signals generated by the antigen receptor activate PI3K/Akt to phosphorylate and inactivate Foxo1, leading to suppression of RAG. We propose that in developing B cells PI3K activation is balanced by the autostimulatory MYC/miR17-92/PTEN axis (shown in red). Accordingly, c-Myc regulates expression of miR-17-92 which reduces expression of PTEN to enhance PI3K activity and promote suppression of RAG.
On the other hand, it is more difficult to compensate for enhanced PI3K activity as B cell development in PTEN-ablated mice or transgenic for P110* is blocked at the proB or early proB stage [respectively, (22) and Figure 7]. Owing to the increased PI3K activity and the resulting enhanced phosphorylation and degradation of Foxo1, RAG expression in pro and pre B cells from these mice was too low and probably insufficient to promote Ig gene rearrangements. This loss of Foxo1 also impairs expression of the transcription factor Ikaros that is necessary for Ig gene rearrangements (22). Remarkably, in the P110* mice B cell development is blocked at an earlier stage relative to that observed in RAG-deficient mice (pre-proB in P110 relative to proB in RAG-/-), suggesting that in addition to the suppression of RAG other mechanisms may be involved. Studies have shown that developmental progression from pre-proB to the preB stage is mediated through transcriptional regulation by EBF or E2A (48), as well as by expression and signaling of the proBCR (8, 49). Hence, it is possible that the overly activated PI3K in the P110* mice impairs expression and/or function of EBF and E2A, or alters expression and signaling of the proBCR to abort B cell development at the pre-proB stage. In addition, transcription from the RAG locus is required for transition into the earliest lymphoid progenitor stage (50). In RAG–/– mice this transcription allows differentiation to proB cells but not beyond due to the non-functional RAG mutation. It is possible that the overly activated PI3K blocks this transcription and prevents development of pre-proB to a proB stage in the P110* mice.
In mature B cells a PI3K/Akt/Foxo1 pathway has been shown to control survival (16). Interestingly, suppressing PI3K by chemical inhibitors or by ablating biogenesis of miRNAs stimulates expression of RAG in mature splenic B cells (27). As tonic PI3K activity increases with B cell maturation to promote survival (3), it is possible that the increased PI3K activity is also important to prevent re-expression of RAGs in peripheral B cells. Thus, during B cell development in the BM the c-Myc/miR17-92/PTEN axis regulates PI3K for RAG expression (this study) and for positive and negative selection of immature B cells (28), but the regulatory circuits balancing tonic PI3K activity for survival and to seal-off RAG expression are less known.
The enhanced expression of RAG in developing B cells was delayed to some extent in miR17-92-ablated mice relative to PTEN over expressing mice. Thus, while differences in RAGs were detected only in preB cells in miR17-92-ablated mice, significant elevation in RAGs was readily seen already in proB cells in PTEN over expressing mice. We attribute this to the elapsing time from genomic deletion until actual affecting PI3K activity. Thus, in PTEN over expressing mice PI3K activity was immediately suppressed due to the enhanced PTEN activity, whereas genomic ablation of the miR17-92 cluster did not immediately abolish all mature miRNAs of this cluster, which continue to function until degradation based on their turnover rate (51). A similar delayed effect was evident when PI3K activity was enhanced in PTEN-ablated mice (22) relative to P110* Tg mice (Figure 7). Thus, genomic ablation of PTEN does not abolish the function of residual PTEN mRNA and protein in balancing PI3K for development until they are degraded, allowing development up to proB stage. In the P110*Tg mice though, we observed a block earlier at the early pro B stage which is driven by the enhanced PI3K activity in these cells.
Interestingly, in mb1-Cre miR17-92fl/fl mice we observed a mild block in B cell development compared to that obtained when miR17-92 is ablated early in hematopoietic progenitors (29), or when the miR17-92 is deleted together with its paralog clusters miR106a-363 and miR106b-25 (41). It is possible that an early deletion of the miR17-92 cluster in the hematopoietic lineage may affect commitment and differentiation into the B lineage, and loss of all paralog clusters aborts all regulatory and compensatory functions by this group of miRNAs. Nevertheless, the block at the proB cell stage that is readily observed in Ventura et al (29), in Nojima et al (52) and here indicates the cell-autonomous important function of the miR17-92 cluster in promoting B cell development at this developmental check-point.
In addition to PTEN, several studies have shown that the miR17-92 controls other important genes regulating the pro-to-preB cell transition such as E2F family of transcription factors (53), as well as the pro-apoptotic Bim (29). Moreover, Lai et al have shown that loss of one PTEN allele fails to rescue B cell development in mice where the miR17-92 is deleted together with its paralog clusters miR106a-363 and miR106b-25 (41). Thus, the miR17-92 cluster and its paralogs are critical for B cell development by regulating multiple target genes controlling developmental transition. The mild pro-to-pre B cell block we show here in miR17-92-deleted or PTENover mice supports this.
Finally, PI3K is most critical for HSCs maintenance and lineage differentiation (54, 55). High level of PI3K (obtained by constitutive active Akt or by PTEN heterozygosity) promotes leukemia and aplastic BM, whereas low level of PI3K prevents self-renewal and differentiation (56, 57). The regulation of PI3K activity in HSCs is unknown. Studies indicate that multipotent HSCs depend on Dicer and miRNA biogenesis for persistence (58), and that PTEN expression regulates HSCs activity by repressing the PI3K/Akt/mTor signaling (59). It is possible that the c-Myc/miR17-92/PTEN axis may also function to regulate PI3K in HSCs. Alternatively, the PTEN-PI3K relationship in balancing PI3K activity may be regulated in the hematopoietic compartment by a different miRNA. Thus, while balancing PI3K is most critical for hematopoiesis in general, the present work proposes that in the B lineage lymphopoiesis PI3K activity is strictly balanced by the c-Myc/miR17-92/PTEN axis to regulate RAG expression and to promote developmental progression.
DB and DM: conceived of the study; DB, VL, AG, EB, and RD: performed the experiments and analyzed the data. KR, JC, and DM: wrote and edited the manuscript and supervised the experiments.
This work was supported by grants from the Israel Science Foundation (1408/13), The Wolens Gerontology Research Fund and the Colleck Research Fund to DM, and The Advanced ERC grant 268921 to KR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge Veronicca Andrews and Johannes Zuber for providing the shPTEN vector, Joost Kluiver for providing the miR19b-sponge vector and Andrea Ventura for providing the miR17-92 vector. We are grateful to Soojin Kim, Janine Woehlecke and Claudia Grosse for technical support and to Roi Gazit for establishing transduced chimeric mice.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02715/full#supplementary-material
1. Pogue SL, Kurosaki T, Bolen J, Herbst R. B cell antigen receptor-induced activation of Akt promotes B cell survival and is dependent on Syk kinase. J Immunol. (2000) 165:1300–6. doi: 10.4049/jimmunol.165.3.1300
2. Clayton E, Bardi G, Bell SE, Chantry D, Downes CP, Gray A, et al. A crucial role for the p110δ subunit of phosphatidylinositol 3-Kinase in B cell development and activation. J Exp Med. (2002) 196:753–63. doi: 10.1084/jem.20020805
3. Cheng S, Hsia CY, Feng B, Liou ML, Fang X, Pandolfi PP, et al. BCR-mediated apoptosis associated with negative selection of immature B cells is selectively dependent on Pten. Cell Res. (2009) 19:196–207. doi: 10.1038/cr.2008.284
4. Monroe JG. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat Rev Immunol. (2006) 6:283–94. doi: 10.1038/nri1808
5. Rowland SL, Tuttle K, Torres RM, Pelanda R. Antigen and cytokine receptor signals guide the development of the naive mature B cell repertoire. Immunol Res. (2013) 55:231–40. doi: 10.1007/s12026-012-8366-7
6. Karasuyama H, Nakamura T, Nagata K, Kuramochi T, Kitamura F, Kuida K. The roles of preB cell receptor in early B cell development and its signal transduction. Immunol Cell Biol. (1997) 75:209–16. doi: 10.1038/icb.1997.32
7. Melchers F, Ten Boekel E, Yamagami T, Andersson J, Rolink A. The roles of preB and B cell receptors in the stepwise allelic exclusion of mouse IgH and L chain gene loci. Semin Immunol. (1999) 11:307–17. doi: 10.1006/smim.1999.0187
8. Bannish G, Fuentes-Panana EM, Cambier JC, Pear WS, Monroe JG. Ligand-independent signaling functions for the B lymphocyte antigen receptor and their role in positive selection during B lymphopoiesis. J Exp Med. (2001) 194:1583–96. doi: 10.1084/jem.194.11.1583
9. Shivtiel S, Leider N, Sadeh O, Kraiem Z, Melamed D. Impaired light chain allelic exclusion and lack of positive selection in immature B cells expressing incompetent receptor deficient of CD19. J Immunol. (2002) 168:5596–604. doi: 10.4049/jimmunol.168.11.5596
10. Von Muenchow L, Engdahl C, Karjalainen K, Rolink AG. The selection of mature B cells is critically dependent on the expression level of the co-receptor CD19. Immunol Lett. (2014) 160:113–9. doi: 10.1016/j.imlet.2014.01.011
11. Edry E, Melamed D. Receptor editing in positive and negative selection of B lymphopoiesis. J Immunol. (2004) 173:4265–71. doi: 10.4049/jimmunol.173.7.4265
12. Werner M, Hobeika E, Jumaa H. Role of PI3K in the generation and survival of B cells. Immunol Rev. (2010) 237:55–71. doi: 10.1111/j.1600-065X.2010.00934.x
13. Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. (2003) 3:317–30. doi: 10.1038/nri1056
14. Tze LE, Schram BR, Lam KP, Hogquist KA, Hippen KL, Liu J, et al. Basal immunoglobulin signaling actively maintains developmental stage in immature B cells. PLoS Biol. (2005) 3:e82. doi: 10.1371/journal.pbio.0030082
15. Herzog S, Hug E, Meixlsperger S, Paik JH, Depinho RA, Reth M, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol. (2008) 9:623–31. doi: 10.1038/ni.1616
16. Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, Depinho RA, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell (2009) 139:573–86. doi: 10.1016/j.cell.2009.08.041
17. Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. (2008) 9:613–22. doi: 10.1038/ni.1612
18. Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. (2008) 9:1388–98. doi: 10.1038/ni.1667
19. Diamant E, Keren Z, Melamed D. CD19 regulates positive selection and maturation in B lymphopoiesis: Lack of CD19 imposes developmental arrest of immature B cells and consequential stimulation of receptor editing. Blood (2005) 105:3247–54. doi: 10.1182/blood-2004-08-3165
20. Verkoczy L, Duong B, Skog P, Ait-Azzouzene D, Puri K, Vela JL, et al. Basal B cell receptor-directed phosphatidylinositol 3-Kinase signaling turns off RAGs and promotes B cell-positive selection. J Immunol. (2007) 178:6332–41. doi: 10.4049/jimmunol.178.10.6332
21. Ramadani F, Bolland DJ, Garcon F, Emery JL, Vanhaesebroeck B, Corcoran AE, et al. The PI3K Isoforms p110α and p110δ are essential for Pre–B cell receptor signaling and B cell development. Sci Signal. (2010) 3:ra60. doi: 10.1126/scisignal.2001104
22. Alkhatib A, Werner M, Hug E, Herzog S, Eschbach C, Faraidun H, et al. FoxO1 induces Ikaros splicing to promote immunoglobulin gene recombination. J Exp Med. (2012) 209:395–406. doi: 10.1084/jem.20110216
23. Simpson LJ, Ansel KM. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J Clin Invest. (2015) 125:2242–9. doi: 10.1172/JCI78090
24. Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell (2009) 136:26–36. doi: 10.1016/j.cell.2008.12.027
25. De Yébenes VG, Bartolomé-Izquierdo N, Ramiro AR. Regulation of B-cell development and function by microRNAs. Immunol Rev. (2013) 253:25–39. doi: 10.1111/imr.12046
26. Coffre M, Koralov SB. miRNAs in B cell development and lymphomagenesis. Trends Mol Med. (2017) 23:721–36. doi: 10.1016/j.molmed.2017.06.001
27. Coffre M, Benhamou D, Rieß D, Blumenberg L, Snetkova V, Hines MJ, et al. miRNAs are essential for the regulation of the PI3K/AKT/FOXO pathway and receptor editing during B cell maturation. Cell Rep. (2016) 17:2271–85. doi: 10.1016/j.celrep.2016.11.006
28. Benhamou D, Labi V, Novak R, Dai I, Shafir-Alon S, Weiss A, et al. A c-Myc/miR17-92/Pten axis controls PI3K-Mediated positive and negative selection in B cell development and reconstitutes CD19 deficiency. Cell Rep. (2016) 16:419–31. doi: 10.1016/j.celrep.2016.05.084
29. Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 Through 92 family of miRNA clusters. Cell (2008) 132:875–86. doi: 10.1016/j.cell.2008.02.019
30. Getahun A, Wemlinger SM, Rudra P, Santiago ML, Van Dyk LF, Cambier JC. Impaired B cell function during viral infections due to PTEN-mediated inhibition of the PI3K pathway. J Exp Med.(2017) 214:931–41. doi: 10.1084/jem.20160972
31. Rickert RC, Rajewsky K, Roes J. Impairment of T-cell-dependent B-cell responses and B-1 cell development in CD19-deficient mice. Nature (1995) 376:352–5. doi: 10.1038/376352a0
32. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. (2008) 9:405–14. doi: 10.1038/ni1575
33. Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. (2006) 103:13789–94. doi: 10.1073/pnas.0605944103
34. Maes J, Caspi Y, Rougeon F, Haimovich J, Goodhardt M. Secondary V(D)J rearrangements and B cell receptor-mediated down-regulation of recombination activating gene-2 expression in a murine B cell line. J Immunol. (2000) 165:703–9. doi: 10.4049/jimmunol.165.2.703
35. Leider N, Melamed D. Differential c-Myc responsiveness to B cell receptor ligation in B cell-negative selection. J Immunol. (2003) 171:2446–52. doi: 10.4049/jimmunol.171.5.2446
36. Melamed D, Benschop RJ, Cambier JC, Nemazee D. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor selection. Cell (1998) 92:173–82. doi: 10.1016/S0092-8674(00)80912-5
37. Keren Z, Diamant E, Ostrovsky O, Bengal E, Melamed D. Modification of ligand-independent B cell receptor tonic signals activates receptor editing in immature B lymphocytes. J Biol Chem. (2004) 279:13418–24. doi: 10.1074/jbc.M311970200
38. Kluiver J, Gibcus JH, Hettinga C, Adema A, Richter MKS, Halsema N, et al. Rapid generation of MicroRNA sponges for MicroRNA inhibition. PLoS ONE (2012) 7:e29275. doi: 10.1371/journal.pone.0029275
39. Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. (2013) 5:1704–13. doi: 10.1016/j.celrep.2013.11.020
40. Novak R, Jacob E, Haimovich J, Avni O, Melamed D. The MAPK/ERK and PI3K pathways additively coordinate the transcription of recombination-activating genes in B lineage cells. J Immunol. (2010) 185:3239–47. doi: 10.4049/jimmunol.1001430
41. Lai M, Gonzalez-Martin A, Cooper AB, Oda H, Jin HY, Shepherd J, et al. Regulation of B-cell development and tolerance by different members of the miR-17~92 family microRNAs. Nat Commun. (2016) 7:12207. doi: 10.1038/ncomms12207
42. Furnari FB, Lin H, Huang H-JS, Cavenee WK. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci USA. (1997) 94:12479–84. doi: 10.1073/pnas.94.23.12479
43. Buhl AM, Pleiman CM, Rickert RC, Cambier JC. Qualitative regulation of B cell antigen receptor signaling by CD19: selective requirement for PI3-kinase activation, inositol-1,4,5-trisphosphate production and Ca2+ mobilization. J Exp Med. (1997) 186:1897–910. doi: 10.1084/jem.186.11.1897
44. Buhl AM, Nemazee D, Cambier JC, Rickert R, Hertz M. B-cell antigen receptor competence regulates B-lymphocyte selection and survival. Immunol Rev. (2000) 176:141–53. doi: 10.1034/j.1600-065X.2000.00613.x
45. Anzelon AN, Wu H, Rickert RC. Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat Immunol. (2003) 4:287–94. doi: 10.1038/ni892
46. Otero DC, Rickert RC. CD19 function in early and late B cell development. II. CD19 facilitates the pro-B/pre-B transition. J Immunol. (2003) 171:5921–30. doi: 10.4049/jimmunol.171.11.5921
47. Sandoval GJ, Graham DB, Bhattacharya D, Sleckman BP, Xavier RJ, Swat W. Cutting edge: cell-autonomous control of IL-7 response revealed in a novel stage of precursor B cells. J Immunol. (2013) 190:2485–9. doi: 10.4049/jimmunol.1203208
48. Sigvardsson M, O'riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity (1997) 7:25–36. doi: 10.1016/S1074-7613(00)80507-5
49. Meffre E, Casellas R, Nussenzweig MC. Antibody regulation of B cell development. Nat Immunol. (2000) 1:379–85. doi: 10.1038/80816
50. Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity (2002) 17:117–30. doi: 10.1016/S1074-7613(02)00366-7
51. Gantier MP, Mccoy CE, Rusinova I, Saulep D, Wang D, Xu D, et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. (2011) 39:5692–703. doi: 10.1093/nar/gkr148
52. Nojima T, Haniuda K, Moutai T, Matsudaira M, Mizokawa S, Shiratori I, et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. (2011) 2:465. doi: 10.1038/ncomms1475
53. Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. (2013) 20:1603. doi: 10.1038/cdd.2013.125
54. Perry JM, He XC, Sugimura R, Grindley JC, Haug JS, Ding S, et al. Cooperation between both Wnt/β-catenin and PTEN/PI3K/Akt signaling promotes primitive hematopoietic stem cell self-renewal and expansion. Genes Dev. (2011) 25:1928–42. doi: 10.1101/gad.17421911
55. Polak R, Buitenhuis M. The PI3K/PKB signaling module as key regulator of hematopoiesis: implications for therapeutic strategies in leukemia. Blood (2012) 119:911–23. doi: 10.1182/blood-2011-07-366203
56. Yilmaz ÖH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature (2006) 441:475–82. doi: 10.1038/nature04703
57. Kharas MG, Okabe R, Ganis JJ, Gozo M, Khandan T, Paktinat M, et al. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood (2010) 115:1406–15. doi: 10.1182/blood-2009-06-229443
58. Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA. (2010) 107:14229–34. doi: 10.1073/pnas.0913574107
Keywords: B cell development, recombination activating gene (RAG), PI3K–AKT pathway, microRNA, PTEN (phosphatase and tensin homolog)
Citation: Benhamou D, Labi V, Getahun A, Benchetrit E, Dowery R, Rajewsky K, Cambier JC and Melamed D (2018) The c-Myc/miR17-92/PTEN Axis Tunes PI3K Activity to Control Expression of Recombination Activating Genes in Early B Cell Development. Front. Immunol. 9:2715. doi: 10.3389/fimmu.2018.02715
Received: 16 August 2018; Accepted: 05 November 2018;
Published: 22 November 2018.
Edited by:
Amy L. Kenter, University of Illinois at Chicago, United StatesReviewed by:
Rachel Maurie Gerstein, University of Massachusetts Medical School, United StatesCopyright © 2018 Benhamou, Labi, Getahun, Benchetrit, Dowery, Rajewsky, Cambier and Melamed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Doron Melamed, bWVsYW1lZGRAdGVjaG5pb24uYWMuaWw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.