
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 02 October 2018
Sec. Molecular Innate Immunity
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.02256
This article is part of the Research TopicOdyssey of Surfactant Proteins SP-A and SP-D: Innate Immune Surveillance MoleculesView all 15 articles
 Zhenwu Lin1†
Zhenwu Lin1† Nithyananda Thorenoor2†
Nithyananda Thorenoor2† Rongling Wu3†
Rongling Wu3† Susan L. DiAngelo2
Susan L. DiAngelo2 Meixia Ye3,4
Meixia Ye3,4 Neal J. Thomas2
Neal J. Thomas2 Xiaojie Liao2
Xiaojie Liao2 Tony R. Lin2
Tony R. Lin2 Stuart Warren2
Stuart Warren2 Joanna Floros2,5*
Joanna Floros2,5*Surfactant proteins (SP) are involved in surfactant function and innate immunity in the human lung. Both lung function and innate immunity are altered in CF, and altered SP levels and genetic association are observed in Cystic Fibrosis (CF). We hypothesized that single nucleotide polymorphisms (SNPs) within the SP genes associate with CF or severity subgroups, either through single SNP or via SNP-SNP interactions between two SNPs of a given gene (intragenic) and/or between two genes (intergenic). We genotyped a total of 17 SP SNPs from 72 case-trio pedigree (SFTPA1 (5), SFTPA2 (4), SFTPB (4), SFTPC (2), and SFTPD (2)), and identified SP SNP associations by applying quantitative genetic principles. The results showed (a) Two SNPs, SFTPB rs7316 (p = 0.0083) and SFTPC rs1124 (p = 0.0154), each associated with CF. (b) Three intragenic SNP-SNP interactions, SFTPB (rs2077079, rs3024798), and SFTPA1 (rs1136451, rs1059057 and rs4253527), associated with CF. (c) A total of 34 intergenic SNP-SNP interactions among the 4 SP genes to be associated with CF. (d) No SNP-SNP interaction was observed between SFTPA1 or SFTPA2 and SFTPD. (e) Equal number of SNP-SNP interactions were observed between SFTPB and SFTPA1/SFTPA2 (n = 7) and SP-B and SFTPD (n = 7). (f) SFTPC exhibited significant SNP-SNP interactions with SFTPA1/SFTPA2 (n = 11), SFTPB (n = 4) and SFTPD (n = 3). (g) A single SFTPB SNP was associated with mild CF after Bonferroni correction, and several intergenic interactions that are associated (p < 0.01) with either mild or moderate/severe CF were observed. These collectively indicate that complex SNP-SNP interactions of the SP genes may contribute to the pulmonary disease in CF patients. We speculate that SPs may serve as modifiers for the varied progression of pulmonary disease in CF and/or its severity.
Cystic fibrosis (CF) is an autosomal multi-organ recessive inherited disease. Mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) protein are key in CF pathogenesis (1). CFTR is activated via cAMP through β2 adrenoceptor stimulation; coding sequence polymorphisms in the CFTR result in CF (2). CFTR functions as a chloride channel on the surface of airway epithelial cells. In patients with CF, loss of CFTR channel function at the cell surface results in impermeability and increased sodium absorption (3, 4). Depletion of the airway surface liquid causes reduced mucus clearance resulting in bacterial colonization, recurrent infections, chronic inflammation, and irreversible damage to the airway epithelium.
Pulmonary function deterioration is one of the primary complications of CF and pulmonary surfactant is essential for normal lung function. Pulmonary surfactant, a surface-active lipoprotein complex, is composed of 90% lipids and 10% surfactant proteins. The latter includes plasma proteins and surfactant proteins SP-A1, SP-A2, SP-B, and SP-C. The surfactant proteins comprise a hydrophobic group of proteins (SP-B and SP-C) and a hydrophilic group of proteins (SP-A1 and SP-A2). The SP-D, although co-isolates with surfactant, is not an integral part of the surfactant complex, but it is grouped with SP-A1/A2 based on its structural similarity and function (5, 6). Pulmonary surfactant is synthesized and secreted by the alveolar epithelial Type II cells of the lung and maintains the stability of the pulmonary tissue by reducing the surface tension of fluids that coat the lung. Broadly speaking the hydrophobic surfactant proteins (SP-B and SP-C) are primarily involved in surface properties of surfactant and are important for normal lung function (7); the hydrophilic proteins (SP-A1, SP-A2, and SP-D) are primarily involved in host defense (6, 8, 9), although SP-A1 and SP-A2 exhibit differential effects on the surfactant structural reorganization (10), on the organization of phospholipid monolayers containing SP-B (11), and lung mechanics (12). Moreover, lipid-mediated interactions of SP-A/SP-B may contribute to normal lung surfactant function (13).
Unlike in rodents that have a single gene encoding surfactant protein A (SP-A), in humans SP-A consists of SP-A1 and SP-A2 encoded by SFTPA1 and SFTPA2, respectively; each gene has been identified with several genetic variants (14), and these have been shown to associate with several pulmonary diseases (15, 16). The human SP-D locus is linked to the SP-A locus and is located proximal to the centromere at approximately 80–100 kb from the SFTPA2 gene (14). Genetic associations between SFTPD SNPs and lung disease have also been identified (17, 18). Although SP-A1, SP-A2, and SP-D are molecules of the innate immunity of the lung, there may be differences in the mechanisms via which host defense is achieved (19, 20). Surfactant proteins SP-B and SP-C play a key role in lung function in normal lung. SP-B is essential for life (21), and both SP-B and SP-C via their role in surfactant function may contribute to CF.
Lung surfactant proteins may contribute to the outcome in CF (22). Bronchoalveolar lavage levels of SP-A have been shown to be increased early in the course of the CF (23), but decrease as disease progresses. Lower levels are correlated with more inflammation and diminished lung function (22, 24–27). Although, the level of SP-B (encoded by SFTPB) was unchanged in BAL from CF patients (24, 28, 29), in CF patients with mild lung disease (30) SP-B was found to be increased, but SP-A did not change. No changes were observed in the levels of SP-C and SP-D, encoded by the SFTPC and SFTPD genes, respectively. However, increased levels of SP-D were observed in serum of CF patients (31).
Moreover, in CF patients with well-conserved lung function (26), SP-C was increased, SP-A was decreased but SP-B and SP-D were not changed. Recently, it was reported that in CF patients, complex forms of SP-A were associated with better lung function. This indicates that the structural organization of SP-A affects its functional activity and this is linked to disease severity (32). SP-A1, shown previously to form higher size oligomers compared to SP-A2 (33), was shown recently to affect more efficiently (than SP-A2) the structural organization of surfactant, which in turn may affect lung function (10). Furthermore, genetic associations of SFTPA1 and SFTPA2 with CF have been observed (34). Moreover, different size SP-D oligomers have been associated with functional differences in patients with chronic lung disease such as CF (35).
Because individuals with identical CFTR mutations may differ in their pulmonary disease, it has been postulated that other genetic factors (i.e. gene modifiers) as well as the microenvironment may contribute to the variable outcome of pulmonary disease (36–41). Surfactant proteins play an important role in surfactant function (7, 34, 42), pulmonary mechanics (12), and innate immunity (6, 8, 9). Furthermore, disruption of these functions can compromise normal lung health. Therefore, we postulated that the surfactant proteins contribute to the progression of the pulmonary disease in CF. We hypothesized that multiple genetic variants of the surfactant protein genes, SFTPA1, SFTPA2, SFTPB, SFTPC, and/or SFTPD, are associated with CF or disease severity subgroups (mild and moderate/severe) through single genetic variations within a gene, and through intragenic or intergenic interactions between variants of a single gene or variants of two different genes. Allele frequencies and linkage disequilibrium of these loci in races and ethnic groups has been previously studied (43). We further hypothesized that some of the associations are unique to patients with mild CF and moderate/severe CF. The observations made indicate that complex SNP-SNP interactions of the surfactant protein genes may contribute to the pulmonary disease in CF patients, and the SPs could serve as modifier genes in lung CF.
The patient samples were collected with informed and written consent from patients and/or parents under an approved protocol by the institutional review board from the Human Subject Protection Office of the Pennsylvania State University College of Medicine. The clinical data of the study samples are given in Supplementary Table 1 and summarized below.
Seventy-two pedigrees (family trees) were studied, of which, 43 pedigrees had one case with two parents, 21 pedigrees had one case with a single parent, five pedigrees had 2 cases with 1 or 2 parents, and 3 pedigrees had 1 or 2 cases in 3 generations. There were a total of 205 study samples in the 72 pedigrees and of these 79 were CF cases. Their ethnicity was as follows: 198-White, 7-Hispanic; 196-American, 6-Mexican, and 3- Unknown (Supplementary Table 1). A correction for ethnicity and sex was performed in the analysis but no correction was made for age. The lung function was assessed by standard spirometry, and for children, <5 years of age, assessment of disease severity was done by the clinical scoring of CXR by the Wisconsin Scoring System (44, 45). CF disease severity was classified as mild, moderate, and severe by lung function impairment based on percent predicted forced expired volume in 1 sec (FEV1). FEV1/FVC: mild = 70–89%; moderate = 40–69%; severe ≤ 40% (Cystic Fibrosis Foundation). The number of patients in the present study under this classification is as follows: severe in 4 cases, moderate in 11 cases, and mild in 64 cases.
Genomic DNAs were prepared from blood samples using QIAamp Blood kit following the manufacturer's instructions (Qiagen, Valencia, CA, United States).
The target surfactant protein genes, SFTPA1, SFTPA2, SFTPB, SFTPC, and SFTPD, were selected based on gene function and association with lung diseases (especially with CF) from our findings and other published data as described above. A total of 17 genetic SNPs were selected. These SNPs were previously shown to associate with various lung diseases, be important in function or structure, or other: 5 SNPs from SFTPA1, rs1059047, rs1136450, rs1136451, rs1059057, and rs4253527; 4 SNPs from SFTPA2, rs1059046, rs17886395, rs1965707, and 1965708; 4 SNPs from SFTPB, rs7316, rs2077079, rs3024798, and rs1130866; 2 SNPs from SFTPC, rs4715 and rs1124; and 2 SNPs from SFTPD, rs721917 and rs2243639. The SNP ID, other used name, nucleotide change, and association with human disease as well as related references are given in Table 1. The genotype frequencies of these SNPs in mild and moderate CF compared to controls are given in Supplementary Table 2.
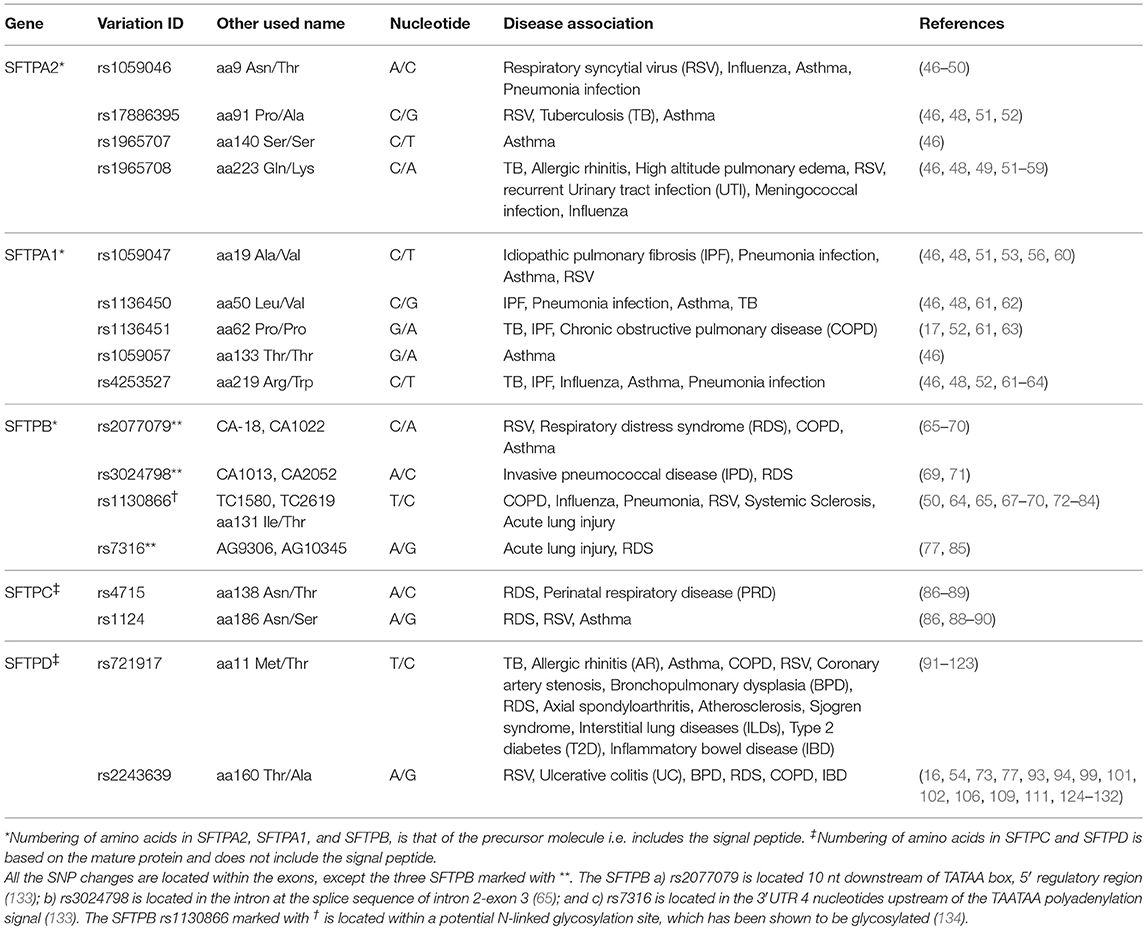
Table 1. Genetic variations of surfactant proteins SFTPA1, SFTPA2, SFTPB, SFTPC, and SFTPD association with disease.
The PCR-based RFLP or cRFLP (135) method was used for genotyping as described in previous publications for SFTPA1, SFTPA2, and SFTPD (136, 137), SFTPB (133, 137), and SFTPC (61). The PCR primer sequences and restriction enzymes used are given in Table 2. Briefly, PCRs were performed at 95°C for 2 min, 5 cycles of 95°C for 30 seconds, 50°C for 1 min, and 70°C for 1 min, then 25 cycles of 95°C for 30 s, 55°C for 1 min, and 70°C for 1 min, followed by an extension at 70°C for 4 min. Five microliter of each PCR products were used for appropriate restriction enzyme digestion (Table 2). The digested PCR product was separated by 8 or 10% of PAGE gel (based on the length of digested PCR fragments). The genotyping was done blindly. As samples (CF and Controls) were received, each was given a sequential laboratory number with no other identifiers and were genotyped all together without knowledge as to which sample is CF or Control. Therefore we believe that there was no bias in the genotyping. The genotype was scored based on the pattern of the digested PCR products for each sample.
We used the Wang et al.'s (138) approach (provides computer code (written in R) for public use), which is a more efficient approach compared to more traditional methods (139) to test and estimate genetic effects of each pair of the 17 SNPs. This approach integrates the principle of quantitative genetics, enabling the decomposition of the overall genetic effect into different components: the additive (a) and dominant genetic effects (d) of each SNP and additive x additive (aa), additive x dominant (ad), dominant x additive (da), and dominant x dominant epistatic effects (dd) between the two SNPs. By estimating the role of each of these components, this approach can provide a better understanding of inheritance mode by which SNPs impact the disease. By analyzing each SNP pair, we calculated p-values for each genetic component. A Bonferroni correction was used to adjust for multiple comparisons.
Associations of single SNPs or SNP-SNP interactions with CF discussed below are shown in Tables 3, 4. The SNPs studied here are not rare alleles (136). The column “interaction type,” in Table 3 as well as in subsequent relevant tables, is the type of the SNP-SNP interaction (interaction between two SNPs within a given gene - intragenic, or interaction between SNPs of two genes-intergenic). The letter a or d is for additive effect or dominant effect. The number 1 or 2 is for the SNP1 or SNP2. In Table 3 the d1 stands for a dominant effect for SNP1, and d2 stands for a dominant effect for SNP2. If it is a1d2 (Table 4), the interaction type is additive effect for SNP1 and dominant effect for SNP2. For example in Table 4, (1) SFTPC rs1124 has a significant dominant effect (d2) (P = 0.0053). This means that the heterozygote is beyond the mean of two homozygotes in the degree of severity at this SNP. (2) SFTPA1 rs1059047 × SFTPC rs1124 has a significant additive x dominant epistatic effect (a1d2) (P = 0.0014). This means that the combination of the homozygote at the first SNP and the heterozygote at the second SNP is significantly different from other combinations. (3) SFTPA1 rs1059047 × SFTPC rs1124 has a significant dominant x dominant epistatic effect (d1d2) (P = 0.0021). This means that the combination of the heterozygote at the first SNP and the heterozygote at the second SNP is significantly different from other combinations. In general, it looks like SFTPB and SFTPC have a dominant effect in most of the interactions, whereas the genes encoding the hydrophilic proteins exhibit primarily an additive effect (Table 4). We speculate that the surfactant related functions imparted by SP-B and SP-C variants play a critical differential role in pulmonary CF and that functions imparted by SP-A1/SP-A2 and SP-D variants further enhance the CF disease progression.
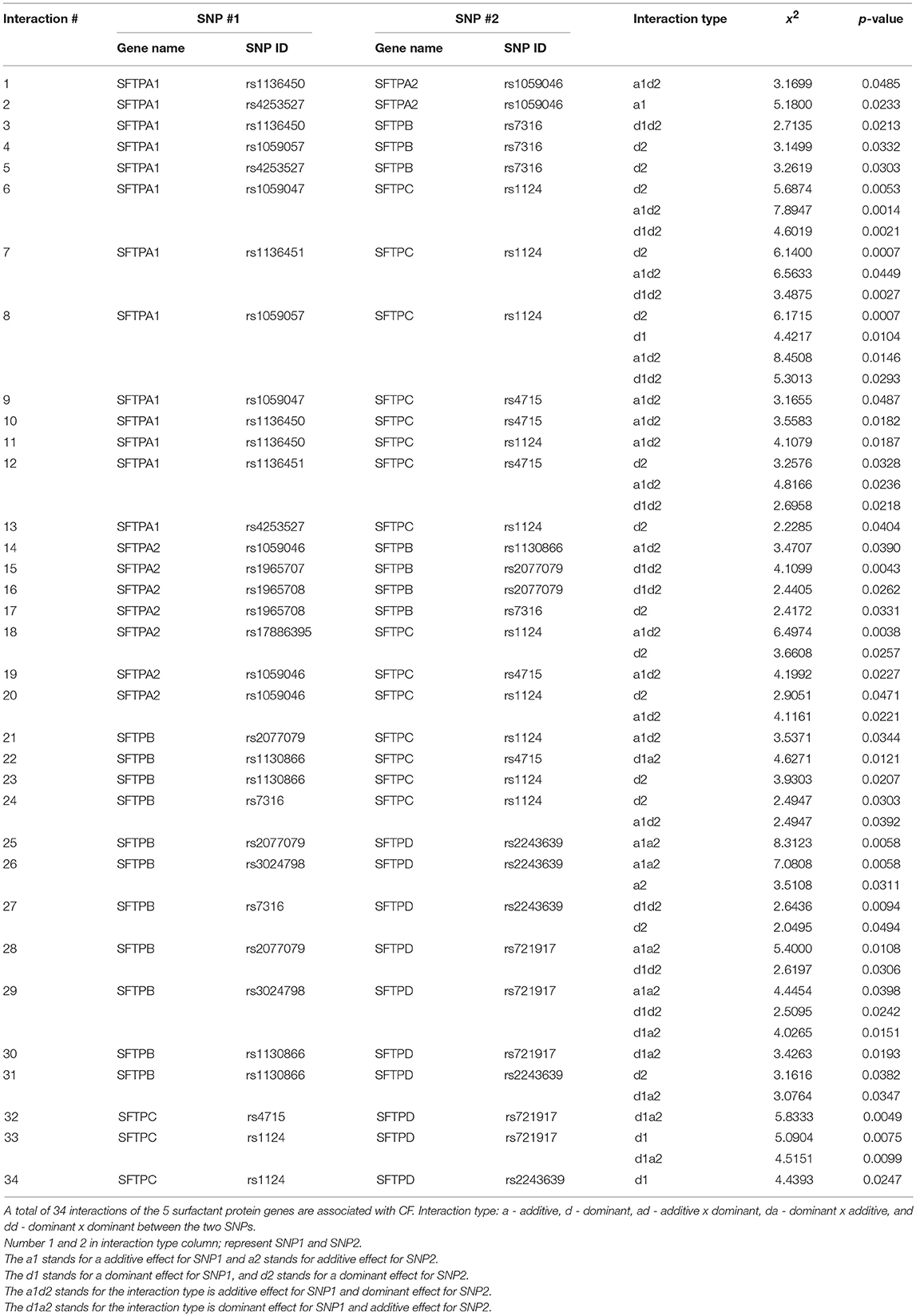
Table 4. Genetic association of surfactant protein genes SFTPA1, SFTPA2, SFTPB, SFTPC, and SFTPD with CF by intergenic interactions.
The data in Table 3 showed that (1) SFTPB SNP rs7316 is associated with CF (X2 = 6.9689, p = 0.0083); (2) another two SFTPB SNPs rs2077079 and rs3024798 are associated with CF through an intragenic interaction (X2 = 3.2688, p = 0.0325). (3) SFTPC SNP rs1124 is associated with CF (X2 = 5.8674, p = 0.0154); (4). Although no single SFTPA1 SNP by itself is associated with CF, the SFTPA1 rs1136451 SNP in an intragenic interaction with either SNP rs1059057 or rs4253527 is shown to associate with CF (X2 = 2.7329, 9 = 0.0469 or X2 = 3.5625, p = 0.0238, respectively) (Table 3).
A total of 34 intergenic interactions of different combinations between SNPs of the studied genes were observed to associate with CF after Bonferroni correction. Significant interactions that include each of the studied genes are as follows: 19 interactions for SFTPB (X2 is 2.045–8.3123, p = 0.0398–0.0043); 18 interactions for SFTPC (X2 is 2.2285–8.4508, p = 0.0487–0.0007); 13 interactions for SFTPA1 (X2 is 2.2285–7.8947, p = 0.0487–0.0007), 9 interactions for SFTPA2 (X2 is 2.4172–6.4974, p = 0.0485–0.0038), and 10 interactions for SFTPD (X2 is 2.2285–8.4508, p = 0.0487–0.0007). Below, we present significant interactions where each interaction contains a SNP from a given gene.
All of the 4 studied SFTPB SNPs, rs7316, rs2077679, rs3024798, and rs1130866, are associated with CF through 18 intergenic SNP-SNP interactions with 3 SNPs in SFTPA1, 4 SNPs in SFTPA2, 4 SNPs in SFTPC, and 7 SNPs in SFTPD (X2 is 2.045–8.3123, p = 0.0398–0.0043) (Table 4). Of the 18 intergenic SNP-SNP interactions that included SFTPB SNPs, only 4 contain SNPs from SFTPC encoding the other hydrophobic surfactant protein, whereas 14 interactions are with genes encoding the hydrophilic surfactant proteins, seven are with SFTPA (3 for SFTPA1 and 4 for SFTPA2) (Figure 1A) and seven with SFTPD (Figure 1B).
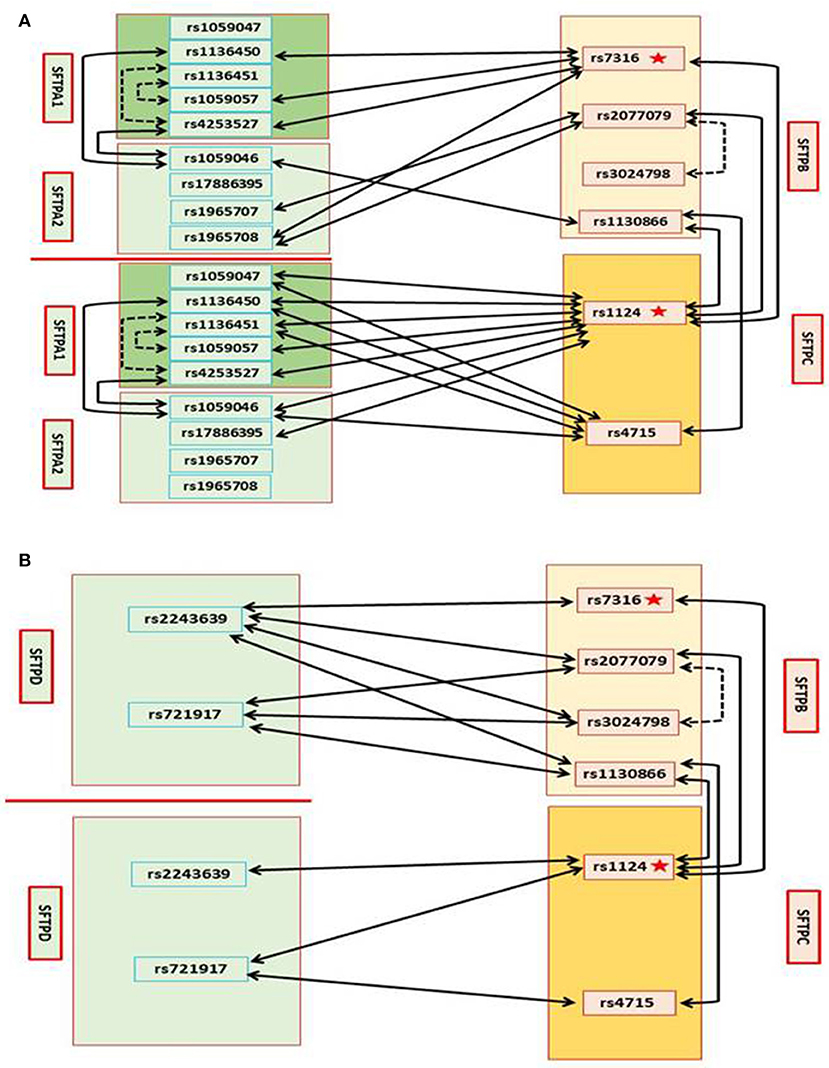
Figure 1. Genetic interactions between SP genes and associations with CF (A). Genetic association of SFTPB and SFTPC with CF by single SNP, and intragenic and intergenic SNP-SNP interactions between SFTPB and SFTPC, and with SFTPA1 and SFTPA2. On the left SNPs of the surfactant genes SFTPA1 and SFTPA2 encoding the hydrophilic surfactant proteins, SP-A1 and SP-A2 and on the right SNPs of the surfactant genes SFTPB and SFTPC, encoding the hydrophobic proteins, SP-B and SP-C, are shown (B). Genetic association of SFTPB and SFTPC with CF by single SNP, and intragenic and intergenic SNP-SNP interactions between SFTPB and SFTPC, and with SFTPD. On the left SNPs of SFTPD and on the right SNPs of SFTPB and SFTPC are shown: In Figure, the star depicts a single SNP association with CF, the black dash line depicts intragenic interactions associated with CF, and the black solid line depicts intergenic interactions associated with CF.
SFTPB SNP rs7316 is associated with CF by itself as noted above (Table 3), and by intergenic interactions with 3 SNPs in SFTPA1 (n = 3), one SNP in SFTPA2, one SNP in SFTPC, and one SNP in SFTPD. SFTPB SNP rs2077079 is associated with CF by intragenic interaction with rs3024798, and by intergenic interaction with SNPs in SFTPA2 (n = 2), SFTPC (n = 1), and SFTPD (n = 2). SFTPB SNP rs3024798 is associated with CF, in addition to the intragenic interaction with rs2077079 (noted above), by intergenic interactions with SFTPD (n = 2). SFTPB SNP rs1130866 is associated with CF by intergenic interaction with SFTPA2 (n = 1; Figure 1A), SFTPC (n = 2), and SFTPD (n = 2; Figure 1B).
Both of the SFTPC SNPs studied, rs4715 and rs1124, are associated with CF through 18 intergenic SNP-SNP interactions with SNPs in SFTPA1, SFTPA2, SFTPC, and SFTPD (X2 is 2.2285–8.4508, p = 0.0487–0.0007) (Table 4). Of the 18 intergenic SNP-SNP interactions associated with CF, eight are with SFTPA1, three are with SFTPA2, four are with SFTPB (Figure 1A), and three are with SFTPD (Figure 1B).
Of the 18 intergenic SFTPC SNP-SNP interactions, only 4 are with SFTPB the gene that encodes the other hydrophobic surfactant protein, whereas 14 interactions are with genes encoding the hydrophilic surfactant proteins (8 for SFTPA1, 3 for SFTPA2, and 3 for SFTPD). SNP rs1124 as noted above (Table 3) is associated with CF by itself, and by 11 intergenic SNP-SNP interactions of the surfactant protein genes, SFTPA1, SFTPA2, SFTPB (Figure 1A), and SFTPD (Figure 1B). SNP rs4715 is associated with CF by 7 intergenic SNP-SNP interactions with SFTPA1, SFTPA2, SFTPB (Figure 1A), and SFTPD (Figure 1B).
All of the 5 studied SFTPA1 SNPs are associated with CF through 13 intergenic SNP-SNP interactions with SNPs in SFTPA2 (n = 2), SFTPB (n = 3), and SFTPC (n = 8) (X2 is 2.2285–7.8947, p = 0.0487–0.0007) (Table 4, Figure 1A). Of interest, no SNP-SNP interaction was observed with SFTPD. Each SFTPA1 SNP is shown to have 1–3 intergenic SNP interactions with the other surfactant protein genes. 1) SNP rs1136451 exhibits a significant association with CF through interactions with another two SFTPA1 SNPs (rs1059057 and rs4253527), as well as with both SFTPC SNPs (rs1124 and rs4715). 2) SNPs rs1136450 and rs4253527 are associated with CF through interaction with SNP rs1059046 of the SFTPA2.
Of the 13 intergenic SFTPA1 SNP-SNP interactions, 11 are with SFTPB and SFTPC encoding the hydrophobic surfactant proteins, and only two interactions are with SFTPA2 that encodes the hydrophilic surfactant protein A2 (Figure 1A), whereas no interactions are observed with SFTPD.
Each SFTPA2 SNP is shown to have 1–3 intergenic SNP interactions with other surfactant protein genes. Table 4 shows that the SFTPA2 gene is associated with CF through 9 intergenic SNP-SNP interactions with SFTPA1 (n = 2), SFTPB (n = 4), and SFTPC (n = 3) (X2 is 2.4172–6.4974, p = 0.0485–0.0038, Figure 1A). Similarly to SFTPA1, no interaction of SFTPA2 with SFTPD was found to be associated with CF. SFTPA2 SNP rs1059046 appears to stand out from the other SFTPA2 SNPs studied, as this SNP (1) is associated with CF via five of the total nine interactions observed with the other SP genes, (2) is the only SNP that shows interactions with two SFTPA1 SNPs, and (3) shows interactions with both hydrophobic surfactant proteins SFTPB and SFTPC (Figure 1A). Of the 9 intergenic interactions, 7 are with the SFTPB and SFTPC hydrophobic surfactant proteins genes, and only two interactions are with the hydrophilic surfactant protein gene, SFTPA1, and no interactions were observed with SFTPD.
Table 4 shows that SFTPD is associated with CF through 10 intergenic SNP-SNP interactions with SNPs in SFTPB and SFTPC, but as noted above no interactions were observed with SFTPA1 or SFTPA2. Of the 10 intergenic SFTPD interactions associated with CF, seven of these are with SFTPB and three with SFTPC (X2 is 2.2285–8.4508, p = 0.0487–0.0007) (Figure 1B). SFTPD SNP rs721917 is associated with CF through 5 intergenic interactions with SFTPB (n = 3) and SFTPC (n = 2); rs2243639 is associated with CF also through 5 intergenic interactions but four of these are with SFTPB and only one with SFTPC (Figure 1B).
In summary, when we studied the entire CF cohort, we observed a) Two SNPs (one from SFTPB and the other from SFTPC) to be individually associated with CF; b) Three intragenic interactions (2 of SFTPA1 and one of SFTPB) to associate with CF; c) A total of 34 intergenic interactions of different combinations between the various genes studied, except between SFTPD and SFTPA1 or SFTPA2, to associate with CF; A summary of all the significant intragenic and intergenic interactions is shown in Supplementary Table 3, and Figure 2. Moreover, our results have a potential clinical impact. For example, since SFTPA1 rs1059047 x SFTPC rs1124 has a significant additive x dominant epistatic effect (a1d2) (P = 0.0014), the combination of the homozygote at the first SNP and the heterozygote at the second SNP is significantly different from other combinations. Thus, we can make a prediction of patients' severity based on their genotypes at these two SNPs.
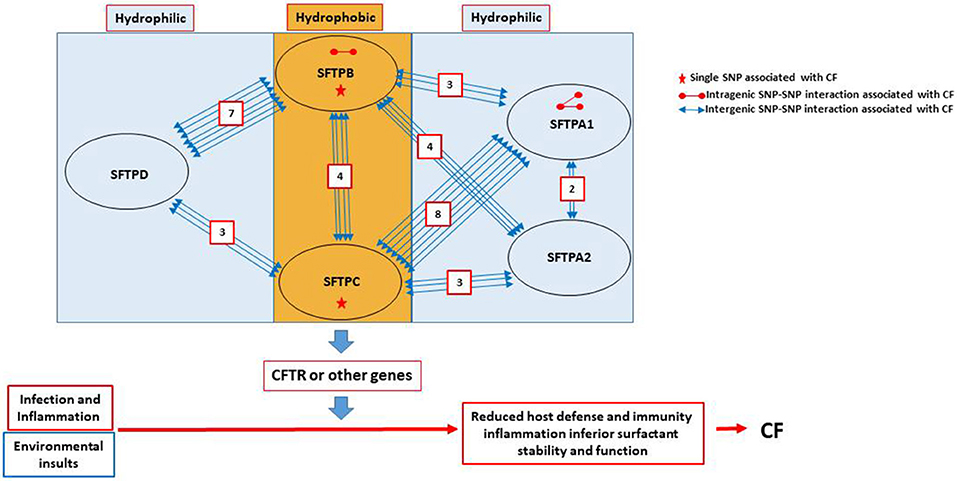
Figure 2. Summary of SNP interactions between genes encoding the surfactant proteins. The cartoon in this figure depicts the observed associations and reveals the following: (a) No association between SFTPD and SFTPA1 or SFTPA2. (b) Out of the 34 intergenic interactions, 28 are between genes encoding the hydrophobic and the hydrophilic surfactant proteins. Only four are between genes (SFTPB and SFTPC) encoding the hydrophobic surfactant proteins and two are between genes (SFTPA1 and SFTPA2) encoding the hydrophilic SP-A1 and SP-A2. (c) SFTPC has the highest number of interactions with SFTPA1 (n = 8), while SFTPB has the highest number of interactions with SFTPD (n = 7). (d) SFTPB has a total of 14 intergenic interactions with genes encoding hydrophilic proteins, seven with SFTPD, four with SFTPA2, and three with SFTPA1. SFTPC also has 14 intergenic interactions, eight with SFTPA1, three with SFTPA2 and only 3 with SFTPD. (e) SFTPA1 has two intergenic interactions with SFTPA2, as well as intragenic interactions. (f) SFTPB is the only other gene (other than SFTPA1) that has an intragenic interaction. (g) Only SFTPB and SFTPC have each a single SNP association with CF.
To gain insight into the contribution of the SP genes to CF disease severity, we separated the CF cohort into two subgroups, mild (n = 64) and moderate/severe (n = 15). The data showed that after Bonferroni correction a single SFTPB SNP (rs7316) to be associated with mild CF and no other SNPs were found to associate with either CF subgroup. However, there are a number of intergenic SNP interactions (p < 0.01 prior to Bonferroni correction) that associated with each CF subgroups (Supplementary Table 4).
Given the smaller number of subjects in each CF subgroup, and as we wished to gain further insight into the interactions observed, we focused our attention on significant associations (p < 0.01) observed in each subgroup that were also significant after Bonferroni correction in the entire CF group. These are shown in Table 5 and Figure 3. Eight intergenic interactions were observed for the mild subgroup and only one for the moderate/severe subgroup. In the mild group, SNPs of the SFTPB exhibited the same number of interactions with SFTPD (n = 4) as they did with SFTPA1+SFTPA2 (n = 4). More interactions were observed between SFTPB and SFTPA1 (n = 3) than SFTPA2 (n = 1) in the mild subgroup. No significant associations with SFTPC were observed with either disease severity group. In addition similar to the entire CF group, no associations were found between SFTPD and SFTPA1 or SFTPA2 in either subgroup.
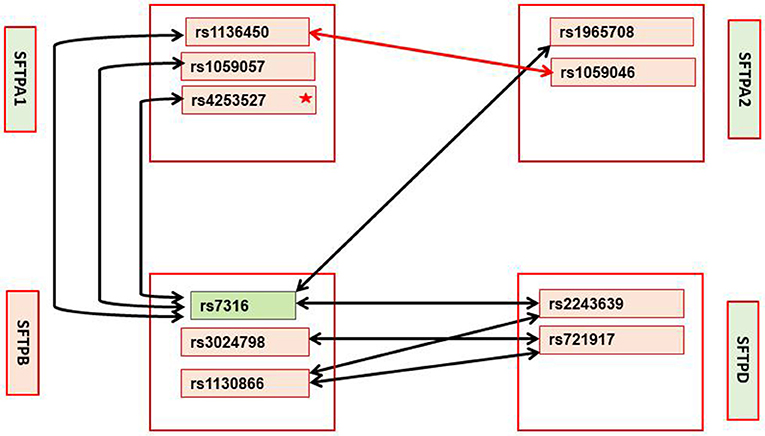
Figure 3. Interactions in mild and moderate/severe CF subgroups with p-value < 0.01; these interactions were significant in the entire CF group after Bonferroni correction. Mild CF: Significant interactions between SFTPA1, SFTPA2, SFTPB, and SFTPD are shown with black arrows. Green box indicates the only SNP that remained significant after Bonferroni correction. Star (*) indicates that this SNP is significant at p < 0.03 by itself. Moderate/severe CF: Red arrow shows the only significant interaction at p < 0.01 that is also significant in the entire CF groups after Bonferroni correction. No significant interactions that include SNPs of the SFTPC are observed in either severity group.
In this study, we investigated the genetic contribution of the surfactant protein genes, SFTPA1, SFTPA2, SFTPB, SFTPC, and SFTPD to CF and disease severity subgroups by genetic association analysis of single SNP and intragenic and intergenic SNP-SNP interactions. (a) For the entire CF group, we observed that all 5 surfactant protein genes are associated with CF by single SNP association, intragenic (Table 3), and/or intergenic SNP-SNP interactions (Table 4).( b) For the CF severity subgroups, we observed after Bonferroni correction a single SFTPB SNP to be associated with mild CF and several interactions (p < 0.01) to associate with mild or moderate/severe CF subgroups (Supplementary Table 4 and Table 5).
Human diseases are complex and determined by environmental factors and genes. Single genetic mutations or multiple mutations in a single gene, constitute a part or a small part of disease mechanisms. To study the full spectrum of gene(s) contributing to a disease either by being the primary cause or by modifying disease expression, an integrated genetic approach is needed to understand genetic control and clinical therapy. By integrating quantitative genetic principles (138) a statistical method was developed to test associations of pairwise SNPs and disease in a case-control setting. This method can decompose the overall genetic effect into its underlying components and test the significance of each component, gaining insight into the mechanisms of how SNPs affect disease. It has been used in our previous studies in human inflammatory bowel diseases (IBD) that included both case-control and case-trios studies, and targeting SNPs in one gene and in multiple genes of a metabolic pathway (140, 141). Because this method is powerful and helps understand genetic contribution to a disease via interactions of genes in a metabolic pathway or gene network, we used it in the present study.
Surfactant proteins SP-B and SP-C are hydrophobic membrane proteins that increase the rate that surfactant spreads over the alveolar surface, and are required for proper biophysical function of surfactant and lung function. SP-B is also important for SP-C processing, as indicated in SP-B deficient states where an aberrant SP-C was observed (7, 142, 143).
All four of the SFTPB SNPs studied showed significant interactions with SNPs of one or more SP genes. The rs7316 is located in the 3′-UTR and could affect regulation of polyadenylation (133) and has previously been associated with acute lung injury (77). This SFTPB SNP is not only significant by itself but interacts with SNPs of all 4 SP genes. The rs2077079 interacts with SNPs of all SP genes, except SFTPA1. It is located at 11 nt downstream of the TATA box and could affect gene transcription. The rs1130866, which also interacts with SNPs of SP genes, except SFTPA1, is a missense mutation that changes the encoded amino acid (Ile/Thr) and an N-linked glycosylation site of the protein, shown previously to be indeed glycosylated in the Thr-containing variant (1580_C) (134). This is a significant change and may be an important contributing factor in various diseases and/or in response to environmental oxidative stress. Moreover, the SFTPB 1580_C (rs1130866) genetic variant has been observed to be a risk factor in several lung disease, such as idiopathic pulmonary fibrosis (IPF) (61), chronic obstructive pulmonary disease (COPD) (17), acute respiratory distress syndrome (ARDS) (137), septic shock and those with risk of respiratory failure after community acquired pneumonia (144), as well as increases mortality, apoptosis, and lung injury in mice carrying the human SP-B 1580_C variant compared to those with the 1580-T variant (145). On the other hand, the SP-B1580-T/T (rs1130866) is associated with protection against interstitial lung disease (ILD) with systemic sclerosis (64). The rs3024798 is located within the splicing sequence of intron 2–exon 3, and although its effect on splicing is unknown (65) it has previously been associated with invasive pneumococcal disease (IPD) (71). This SNP, in addition to an intragenic interaction, is the only SNP that interacts with a single SP gene, the SFTPD. Together these SNP variations in SFTPB could affect SP-B function by altering N-linked glycosylation and/or affect regulation at several levels including transcription and splicing.
SP-C is a hydrophobic surfactant protein and plays an important role in surfactant function. Mutations in SP-C have also been shown to associate with a number of pulmonary diseases (15), such as ILD and pulmonary alveolar proteinosis (PAP). Both SFTPC variants are missense, where amino acids 186 and 138 are changed, Ser/Asn in rs1124, and Thr/Asn in rs4715, respectively. While both SNPs associate with CF through numerous intergenic interactions with the other SPs, the rs1124 is also associated with CF by itself. The potential mechanisms via which these may affect function are not known. The two SP-C variants (rs1124, Ser/Asn and rs4715, Thr/Asn) have previously been associated with RDS (86, 88, 89), and children infected with respiratory syncytial virus (RSV) (90).
In summary, the observations made indicate that the hydrophobic proteins, shown previously to be key in surfactant function and homeostasis and consequently in lung function may play a central role in CF. The only two individual SNPs to associate with CF by themselves were SNPs of SFTPB rs7316 (p = 0.0083) and SFTPC rs1124 (p = 0.0154) (indicated by star in Figure 1A). Moreover, of the 37 interactions shown to associate with CF, SFTPB or SFTPC was part of the 35 interactions. Given the importance of SP-B and SP-C in surfactant function and lung function, and the fact that lung function deterioration is a major issue in CF, it is likely that the SFTPB and SFTPC genes are modifier genes for CF lung function, by modulating surfactant structural organization and stability.
The SP-A1, SP-A2, and SP-D mediate innate immunity in the lung and via interactions with the alveolar macrophage the sentinel cell of innate lung host defense, promote, among others, bacterial and viral phagocytosis, and cytokine production, as well as affect lung inflammatory processes. The mechanisms implicated in these processes may differ among these molecules (19). Of interest in the present study, there were no significant interactions between SNPs of SFTPD and SFTPA1 or SFTPA2 and only two significant intergenic interactions were observed between SFTPA1 and SFTPA2 (Figure 1A). A single SNP in SFTPA2 (rs1059046) interacted with two different SNPs in SFTPA1 (rs1136450, aa 50 Leu/Val and rs4253527, aa 219 Arg/Trp). The SFTPA2 SNP (rs1059046) changes the amino acid (Thr/Asn) at codon 9 of the precursor molecule which is part of the signal peptide, having the potential to affect processing of the SP-A2. This SNP was previously shown to associate with increased risk in RSV (47). The two SFTPA1 SNPs (rs1136450, rs4253527) that interact with this SFTPA2 SNP are located in the collagen-like domain and in the carbohydrate recognition domain (CRD), respectively, of the SP-A2 and change the encoded amino acid (rs1136450, aa 50 Leu/Val, rs4253527, aa 219 Arg/Trp). Moreover, higher- or lower-order of oligomerization of SP-A and SP-D is known to affect their functional capabilities (146–149), and has been observed that naturally occurring SP-A and SP-D oligomers have functional relevance in patients with chronic lung diseases such as CF (32, 35). Thus, each of the surfactant protein variants may differentially affect innate immune functions and in CF may each differentially modify lung host defense.
Multiple interactions between the hydrophobic and the hydrophilic protein genes were observed indicating that these two groups of proteins may co-operatively or synergistically contribute to the expression of pulmonary CF. As depicted in Figure 2, SFTPD SNPs are primarily found in interaction with SNPs of SFTPB (n = 7), whereas SNPs of SFTPA1 are primarily found in interactions with SNPs of SFTPC (n = 8). The latter is of interest because SP-A1 has been shown to affect more efficiently (compared to SP-A2) the structural organization of surfactant (10), indicating that SP-A1 and SP-C may cooperatively affect surfactant structure, which in turn may affect surfactant function and lung function. Moreover, the large number of SNP interactions between SFTPD and SFTPB is puzzling and not intuitively understood as far as surfactant structure or function is concerned. This is because, although SP-D is generally found in the bronchoalveolar lavage fluid and significantly less in alveolar epithelia, and grouped with SP-A1/A2 based on its structural similarity and function with these proteins (5, 6), it is not found in the surfactant lipoprotein complex.
When we studied disease severity subgroups (mild and moderate/severe), we found after Bonferroni correction a single SFTPB SNP (rs7316) to associate with mild CF. This SNP has also been shown to associate with acute lung injury (77). Lack of SP-B is not compatible with life, and SP-B deficiency affects SP-C processing (7, 142, 143). This SFTPB SNP is located within the 3′UTR and may affect regulation of polyadenylation (133). Thus, this SFTPB SNP may contribute to CF either by affecting its regulation (133) and/or by affecting SP-C processing (7, 142, 143). In the CF subgroups of the eight significant intergenic interactions (p < 0.01) that were also significant in the entire CF group after Bonferroni correction, seven were for the mild group and one for the moderate/severe subgroup. The ones for the mild group all were between SFTPB SNPs and SNPs of the genes encoding the hydrophilic surfactant proteins.
The SFTPB SNP (rs7316) that is significant by itself in mild CF after Bonferroni correction was the only SFTPB SNP that interacted with SNPs of SFTPA1 or SFTPA2, whereas the SNP-SNP intergenic interactions between SFTPB and SFTPD include three of the four SFTPB SNPs studied. These indicate the importance of SFTPB in mild CF which may provide protection in the sense of enabling a milder form of pulmonary CF through its surfactant-associated function or perhaps other currently unknown function. Based on the differences of SNP-SNP interactions; it is likely that the mechanisms via which the hydrophilic proteins contribute to mild CF may differ. The SFTPA1 SNP (rs4253527) was also significant by itself (p < 0.03) (Supplementary Table 4) in the mild CF group. SP-A and SP-D bind via their carbohydrate recognition domains (CRD) to the carbohydrate motifs on bacteria, viruses, fungi, etc. (150–152). Thus, SNPs in CRD may differentially affect binding to various ligands to trigger the host's innate immune response. The SFTPA1 SNP (rs4253527) is located in the CRD and changes the encoded amino acid (Arg/Trp) holding the potential to differentially affect innate immunity under various conditions including oxidative stress, since Trp is more sensitive to oxidation than Arg (33). In fact SP-A1 variants that differ in CRD at rs4253527 have been shown to differ in their ability to enhance phagocytosis (153) and cytokine production (154). Moreover, SP-A1 has been shown to more efficiently affect surfactant reorganization (than SP-A2) in the alveolar space (10). Whether this SNP provides protection in CF via its role in surfactant function or innate immunity or both remains to be determined. Recently, SP-A1 and SP-A2 have been shown to differentially affect lung mechanics (12) indicating another role of the SP-As beyond innate immunity.
For the moderate/severe group the only significant interaction is between the SNP rs1136450, (aa 50 Leu/Val) of SFTPA1 and SNP rs1059046 (aa 9 Thr/Asn) of SFTPA2. This interaction may be unique to moderate/severe disease group because it was not identified in the mild group even in interactions with p < 0.05 (Supplementary Table 4). Similarly, interactions 1, 3, 4, 7, and 8 in the mild group (Table 5) were not identified in the moderate/severe disease group even in interactions with p < 0.05 (Supplementary Table 4), indicating that these may be unique to the mild CF group.
In summary, a single SNP of the SFTPB is a marker for mild pulmonary disease in CF. A number of intergenic interactions that all include SNPs of SFTPB as well as a single intragenic SFTPA1 and SFTPA2 interaction are likely to be markers for mild and moderate/severe pulmonary disease in CF, respectively.
Limitations of the study include: (a) The small number of subjects especially after the CF group was divided in the mild and moderate/severe subgroups. This also precluded analysis of the individual components of FEV1 and FVC; (b) The limited clinical information; the samples were collected at an earlier time using only FEV1 as a discriminator for the severity subgroups. However, this is still the main biomarker to assess disease severity. (c) The lack of associations with bacterial strain and correction for age. (d) The CFTR mutation is not known although most of the subjects are expected to carry the ΔF508, which is approximately found in 70% of the CF patients.
However, in spite of these limitations the present findings indicate that both groups of surfactant proteins, those involved in innate immunity, and those affecting surfactant functions, associate with CF via complex interactions. These may contribute to pulmonary disease progression in CF, by affecting surfactant structure and function and/or by affecting innate immunity functions. Altered surfactant leads to a compromised lung function, and lung function deterioration is a major setback in CF pulmonary disease. Similarly host defense and inflammatory processes are partially affected in CF and SPs may play a role in these. Thus, based on the collective information with regards to their function and the observations made here, the SP genes are likely to be significant gene modifiers of CF and must be studied further. As gene modifiers, SPs may explain the varied progression of the pulmonary disease in CF in terms of lung function and host defense.
All the data are presented as supplementary files.
All protocols used in this study were evaluated and approved by the institutional review board from the Human Subject Protection Office of the Pennsylvania State University College of Medicine.
ZL analyzed and synthesized the data, contributed to the manuscript writing. NT analyzed and synthesized data for CF subgroups and contributed to manuscript writing. RW performed statistical analysis and contributed to manuscript writing. SD performed all the genotyping. MY assisted with statistical analysis. NJT attended to human subjects issues, sample collection and contributed to manuscript writing. XL checked genotype data sheets after multiple transfers. TL reviewed literature and made Table 1. SW helped with clinical assessment. JF designed the study and provided oversight to the entire project, involved in data analysis, integration, and writing of the manuscript.
This work was supported by NIH HL68947 of JF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AT and handling Editor declared their shared affiliation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02256/full#supplementary-material
CF, Cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; FEV1, Forced expired volume in 1 sec; SP-A, surfactant protein A; SFTPA1, gene encoding SP-A1; SFTPA2, gene encoding SP-A2; SFTPB, gene encoding SP-B; SFTPC, gene encoding SP-C; SFTPD, gene encoding SP-D; SNPs, single-nucleotide polymorphism.
1. O'Sullivan BP, Freedman SD. Cystic fibrosis. Lancet (2009) 373:1891–904. doi: 10.1016/s0140-6736(09)60327-5
2. Buscher R, Eilmes KJ, Grasemann H, Torres B, Knauer N, Sroka K, et al. beta2 adrenoceptor gene polymorphisms in cystic fibrosis lung disease. Pharmacogenetics (2002) 12:347–53.
3. Boucher RC, Cotton CU, Gatzy JT, Knowles MR, Yankaskas JR. Evidence for reduced Cl- and increased Na+ permeability in cystic fibrosis human primary cell cultures. J Physiol. (1988) 405:77–103.
5. Wright JR. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol. (2005) 5:58–68. doi: 10.1038/nri1528
6. Kishore U, Greenhough TJ, Waters P, Shrive AK, Ghai R, Kamran MF, et al. Surfactant proteins SP-A and SP-D: structure, function and receptors. Mol Immunol. (2006) 43:1293–315. doi: 10.1016/j.molimm.2005.08.004
7. Serrano AG, Perez-Gil J. Protein-lipid interactions and surface activity in the pulmonary surfactant system. Chem Phys Lipids (2006) 141:105–18. doi: 10.1016/j.chemphyslip.2006.02.017
8. Phelps DS. Surfactant regulation of host defense function in the lung: a question of balance. Pediatr Pathol Mol Med. (2001) 20:269–92. doi: 10.1080/15513810109168822
9. Floros J, Wang G, Mikerov AN. Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2–impact on function. Crit Rev Eukaryot Gene Expr. (2009) 19:125–37. doi: 10.1615/CritRevEukarGeneExpr.v19.i2.30
10. Lopez-Rodriguez E, Pascual A, Arroyo R, Floros J, Perez-Gil J. Human pulmonary surfactant protein SP-A1 provides maximal efficiency of lung interfacial films. Biophys J. (2016) 111:524–36. doi: 10.1016/j.bpj.2016.06.025
11. Wang G, Taneva S, Keough KM, Floros J. Differential effects of human SP-A1 and SP-A2 variants on phospholipid monolayers containing surfactant protein B. Biochim Biophys Acta (2007) 1768:2060–9. doi: 10.1016/j.bbamem.2007.06.025
12. Thorenoor N, Zhang X, Umstead TM, Scott Halstead E, Phelps DS, Floros J. Differential effects of innate immune variants of surfactant protein-A1 (SFTPA1) and SP-A2 (SFTPA2) in airway function after Klebsiella pneumoniae infection and sex differences. Respir Res. (2018) 19:23. doi: 10.1186/s12931-018-0723-1
13. Sarker M, Jackman D, Booth V. Lung surfactant protein A (SP-A) interactions with model lung surfactant lipids and an SP-B fragment. Biochemistry (2011) 50:4867–76. doi: 10.1021/bi200167d
14. Floros J, Hoover RR. Genetics of the hydrophilic surfactant proteins A and D. Biochim Biophys Acta (1998) 1408:312–22.
15. Floros J, Thomas N. Genetic variation of surfactant protiens and lung injury, In: Nakos G and Papathanasiou A editors. Surfactant in Pathogenesis and Treatment of Lung Disease. India: Research Signpost (2009). pp. 25–48.
16. Silveyra P, Floros J. Genetic variant associations of human SP-A and SP-D with acute and chronic lung injury. Front Biosci. (2012) 17:407–29. doi: 10.2741/3935
17. Guo X, Lin HM, Lin Z, Montano M, Sansores R, Wang G, et al. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J. (2001) 18:482–90.
18. Sorensen GL. Surfactant protein D in respiratory and non-respiratory diseases. Front Med. (2018) 5:18. doi: 10.3389/fmed.2018.00018
19. Nayak A, Dodagatta-Marri E, Tsolaki AG, Kishore U. An Insight into the diverse roles of surfactant proteins, SP-A and SP-D in innate and adaptive immunity. Front Immunol. (2012) 3:131. doi: 10.3389/fimmu.2012.00131
20. Carreto-Binaghi LE, Aliouat el M, Taylor ML. Surfactant proteins, SP-A and SP-D, in respiratory fungal infections: their role in the inflammatory response. Respir Res. (2016) 17:66. doi: 10.1186/s12931-016-0385-9
21. Nogee LM, Wert SE, Proffit SA, Hull WM, Whitsett JA. Allelic heterogeneity in hereditary surfactant protein B (SP-B) deficiency. Am J Respir Crit Care Med. (2000) 161(3 Pt 1):973–981. doi: 10.1164/ajrccm.161.3.9903153
22. Noah TL, Murphy PC, Alink JJ, Leigh MW, Hull WM, Stahlman MT, et al. Bronchoalveolar lavage fluid surfactant protein-A and surfactant protein-D are inversely related to inflammation in early cystic fibrosis. Am J Respir Crit Care Med. (2003) 168:685–91. doi: 10.1164/rccm.200301-005OC
23. Hull J, South M, Phelan P, Grimwood K. Surfactant composition in infants and young children with cystic fibrosis. Am J Respir Crit Care Med. (1997) 156:161–5. doi: 10.1164/ajrccm.156.1.9609090
24. Griese M, Birrer P, Demirsoy A. Pulmonary surfactant in cystic fibrosis. Eur Respir J. (1997) 10:1983–8.
25. Meyer KC, Sharma A, Brown R, Weatherly M, Moya FR, Lewandoski J, et al. Function and composition of pulmonary surfactant and surfactant-derived fatty acid profiles are altered in young adults with cystic fibrosis. Chest (2000) 118:164–74. doi: 10.1378/chest.118.1.164
26. Griese M, Essl R, Schmidt R, Rietschel E, Ratjen F, Ballmann M, et al. Pulmonary surfactant, lung function, and endobronchial inflammation in cystic fibrosis. Am J Respir Crit Care Med. (2004) 170:1000–5. doi: 10.1164/rccm.200405-575OC
27. Peterson-Carmichael SL, Harris WT, Goel R, Noah TL, Johnson R, Leigh MW, et al. Association of lower airway inflammation with physiologic findings in young children with cystic fibrosis. Pediatr Pulmonol. (2009) 44:503–11. doi: 10.1002/ppul.21044
28. Bernhard W, Haagsman HP, Tschernig T, Poets CF, Postle AD, van Eijk ME, et al. Conductive airway surfactant: surface-tension function, biochemical composition, and possible alveolar origin. Am J Respir Cell Mol Biol. (1997) 17:41–50. doi: 10.1165/ajrcmb.17.1.2594
29. Postle AD, Mander A, Reid KB, Wang JY, Wright SM, Moustaki M, et al. Deficient hydrophilic lung surfactant proteins A and D with normal surfactant phospholipid molecular species in cystic fibrosis. Am J Respir Cell Mol Biol. (1999) 20:90–8. doi: 10.1165/ajrcmb.20.1.3253
30. Griese M, Essl R, Schmidt R, Ballmann M, Paul K, Rietschel E, et al. Sequential analysis of surfactant, lung function and inflammation in cystic fibrosis patients. Respir Res. (2005) 6:133. doi: 10.1186/1465-9921-6-133
31. Olesen HV, Holmskov U, Schiotz PO, Sorensen GL. Serum-surfactant SP-D correlates inversely to lung function in cystic fibrosis. J Cyst Fibros (2010) 9:257–62. doi: 10.1016/j.jcf.2010.03.011
32. Griese M, Heinrich S, Ratjen F, Kabesch M, Paul K, Ballmann M, et al. Surfactant protein a in cystic fibrosis: supratrimeric structure and pulmonary outcome. PLoS ONE (2012) 7:e51050. doi: 10.1371/journal.pone.0051050
33. Wang G, Bates-Kenney SR, Tao JQ, Phelps DS, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry (2004) 43:4227–39. doi: 10.1021/bi036023i
34. Choi EH, Ehrmantraut M, Foster CB, Moss J, Chanock SJ. Association of common haplotypes of surfactant protein A1 and A2 (SFTPA1 and SFTPA2) genes with severity of lung disease in cystic fibrosis. Pediatr Pulmonol. (2006) 41:255–62. doi: 10.1002/ppul.20361
35. Kotecha S, Doull I, Davies P, McKenzie Z, Madsen J, Clark HW, et al. Functional heterogeneity of pulmonary surfactant protein-D in cystic fibrosis. Biochim Biophys Acta (2013) 1832:2391–400. doi: 10.1016/j.bbadis.2013.10.002
36. Parad RB, Gerard CJ, Zurakowski D, Nichols DP, Pier GB. Pulmonary outcome in cystic fibrosis is influenced primarily by mucoid Pseudomonas aeruginosa infection and immune status and only modestly by genotype. Infect Immun. (1999) 67:4744–50.
37. Buscher R, Grasemann H. Disease modifying genes in cystic fibrosis: therapeutic option or one-way road? Naunyn Schmiedebergs Arch Pharmacol. (2006) 374:65–77. doi: 10.1007/s00210-006-0101-2
38. Guillot L, Beucher J, Tabary O, Le Rouzic P, Clement A, Corvol H. Lung disease modifier genes in cystic fibrosis. Int J Biochem Cell Biol. (2014) 52:83–93. doi: 10.1016/j.biocel.2014.02.011
39. de Lima Marson FA, Bertuzzo CS, Ribeiro JD. Personalized drug therapy in cystic fibrosis: from fiction to reality. Curr Drug Targets (2015) 16:1007–17. doi: 10.2174/1389450115666141128121118
40. De Boeck K, Amaral MD. Progress in therapies for cystic fibrosis. Lancet Respir Med. (2016) 4:662–74. doi: 10.1016/s2213-2600(16)00023-0
41. Marson FAL. Disease-modifying genetic factors in cystic fibrosis. Curr Opin Pulm Med. (2018) 24:296–308. doi: 10.1097/mcp.0000000000000479
42. Floros J, Kala P. Surfactant proteins: molecular genetics of neonatal pulmonary diseases. Annu Rev Physiol. (1998) 60:365–84. doi: 10.1146/annurev.physiol.60.1.365
43. Liu W, Bentley CM, Floros J. Study of human SP-A, SP-B and SP-D loci: allele frequencies, linkage disequilibrium and heterozygosity in different races and ethnic groups. BMC Genet. (2003) 4:13. doi: 10.1186/1471-2156-4-13
44. Weatherly MR, Palmer CG, Peters ME, Green CG, Fryback D, Langhough R, et al. Wisconsin cystic fibrosis chest radiograph scoring system. Pediatrics (1993) 91:488–95.
45. Koscik RE, Kosorok MR, Farrell PM, Collins J, Peters ME, Laxova A, et al. Wisconsin cystic fibrosis chest radiograph scoring system: validation and standardization for application to longitudinal studies. Pediatr Pulmonol. (2000) 29:457–67. doi: doi: 10.1002/(SICI)1099-0496(200006)29:6<457::AID-PPUL8>3.0.CO;2-9
46. Pettigrew MM, Gent JF, Zhu Y, Triche EW, Belanger KD, Holford TR, et al. Respiratory symptoms among infants at risk for asthma: association with surfactant protein A haplotypes. BMC Med Genet. (2007) 8:15. doi: 10.1186/1471-2350-8-15
47. El Saleeby CM, Li R, Somes GW, Dahmer MK, Quasney MW, DeVincenzo JP. Surfactant protein A2 polymorphisms and disease severity in a respiratory syncytial virus-infected population. J Pediatr. (2010) 156:409–14. doi: 10.1016/j.jpeds.2009.09.043
48. Garcia-Laorden MI, Rodriguez de Castro F, Sole-Violan J, Rajas O, Blanquer J, Borderias L, et al. Influence of genetic variability at the surfactant proteins A and D in community-acquired pneumonia: a prospective, observational, genetic study. Crit Care (2011) 15:R57. doi: 10.1186/cc10030
49. Herrera-Ramos E, Lopez-Rodriguez M, Ruiz-Hernandez JJ, Horcajada JP, Borderias L, Lerma E, et al. Surfactant protein A genetic variants associate with severe respiratory insufficiency in pandemic influenza A virus infection. Crit Care (2014) 18:R127. doi: 10.1186/cc13934
50. Dudina KR, Kutateladze MM, Bokova NO, Znoiko OO, Abramov DD, Kelly EI, et al. [ASSOCIATION OF POLYMORPHISM GENES OF SURFACTANT PROTEINS IN PATIENTS WITH INFLUENZA]. Zh Mikrobiol Epidemiol Immunobiol (2015) 6:71–7.
51. Lofgren J, Ramet M, Renko M, Marttila R, Hallman M. Association between surfactant protein A gene locus and severe respiratory syncytial virus infection in infants. J Infect Dis. (2002) 185:283–9. doi: 10.1086/338473
52. Malik S, Greenwood CM, Eguale T, Kifle A, Beyene J, Habte A, et al. Variants of the SFTPA1 and SFTPA2 genes and susceptibility to tuberculosis in Ethiopia. Hum Genet. (2006) 118:752–9. doi: 10.1007/s00439-005-0092-y
53. Saxena S, Kumar R, Madan T, Gupta V, Muralidhar K, Sarma PU. Association of polymorphisms in pulmonary surfactant protein A1 and A2 genes with high-altitude pulmonary edema. Chest (2005) 128:1611–9. doi: 10.1378/chest.128.3.1611
54. Grupe A, Li Y, Rowland C, Nowotny P, Hinrichs AL, Smemo S, et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. (2006) 78:78–88. doi: 10.1086/498851
55. Jack DL, Cole J, Naylor SC, Borrow R, Kaczmarski EB, Klein NJ, et al. Genetic polymorphism of the binding domain of surfactant protein-A2 increases susceptibility to meningococcal disease. Clin Infect Dis. (2006) 43:1426–33. doi: 10.1086/508775
56. Liu J, Hu F, Liang W, Wang G, Singhal PC, Ding G. Polymorphisms in the surfactant protein a gene are associated with the susceptibility to recurrent urinary tract infection in chinese women. Tohoku J Exp Med. (2010) 221:35–42. doi: 10.1620/tjem.221.35
57. Deng Y, Chen S, Chen J, Tao Z, Kong Y, Xu Y, et al. Relationship between surfactant protein A polymorphisms and allergic rhinitis in a Chinese Han population. Mol Biol Rep. (2011) 38:1475–82. doi: 10.1007/s11033-010-0254-4
58. Foster MW, Thompson JW, Ledford JG, Dubois LG, Hollingsworth JW, Francisco D, et al. Identification and quantitation of coding variants and isoforms of pulmonary surfactant protein a. J Proteome Res. (2014) 13:3722–32. doi: 10.1021/pr500307f
59. Wu AL, Xiong YS, Li ZQ, Liu YG, Quan Q, Wu LJ. Correlation between single nucleotide polymorphisms in hypoxia-related genes and susceptibility to acute high-altitude pulmonary edema. Genet Mol Res. (2015) 14:11562–72. doi: 10.4238/2015.September.28.8
60. Pettigrew MM, Gent JF, Zhu Y, Triche EW, Belanger KD, Holford TR, et al. Association of surfactant protein A polymorphisms with otitis media in infants at risk for asthma. BMC Med Genet. (2006) 7:68. doi: 10.1186/1471-2350-7-68
61. Selman M, Lin HM, Montano M, Jenkins AL, Estrada A, Lin Z, et al. Surfactant protein A and B genetic variants predispose to idiopathic pulmonary fibrosis. Hum Genet. (2003) 113:542–50. doi: 10.1007/s00439-003-1015-4
62. Cooper DN, Kehrer-Sawatzki H. Exploring the potential relevance of human-specific genes to complex disease. Hum Genomics (2011) 5:99–107. doi: 10.1186/1479-7364-5-2-99
63. Guan J, Liu X, Xie J, Xu X, Luo S, Wang R, et al. Surfactant protein a polymorphism is associated with susceptibility to chronic obstructive pulmonary disease in Chinese Uighur population. J Huazhong Univ Sci Technolog Med Sci. (2012) 32:186–9. doi: 10.1007/s11596-012-0033-7
64. Sumita Y, Sugiura T, Kawaguchi Y, Baba S, Soejima M, Murakawa Y, et al. Genetic polymorphisms in the surfactant proteins in systemic sclerosis in Japanese: T/T genotype at 1580 C/T (Thr131Ile) in the SP-B gene reduces the risk of interstitial lung disease. Rheumatology (Oxford) (2008) 47:289–91. doi: 10.1093/rheumatology/kem355
65. Lin Z, deMello DE, Wallot M, Floros J. An SP-B gene mutation responsible for SP-B deficiency in fatal congenital alveolar proteinosis: evidence for a mutation hotspot in exon 4. Mol Genet Metab. (1998) 64:25–35. doi: 10.1006/mgme.1998.2702
66. Floros J, Fan R, Diangelo S, Guo X, Wert J, Luo J. Surfactant protein (SP) B associations and interactions with SP-A in white and black subjects with respiratory distress syndrome. Pediatr Int. (2001) 43:567–76. doi: 10.1046/j.1442-200X.2001.01474.x
67. Puthothu B, Forster J, Heinze J, Heinzmann A, Krueger M. Surfactant protein B polymorphisms are associated with severe respiratory syncytial virus infection, but not with asthma. BMC Pulm Med. (2007) 7:6. doi: 10.1186/1471-2466-7-6
68. Foreman MG, DeMeo DL, Hersh CP, Carey VJ, Fan VS, Reilly JJ, et al. Polymorphic variation in surfactant protein B is associated with COPD exacerbations. Eur Respir J. (2008) 32:938–44. doi: 10.1183/09031936.00040208
69. Hamvas A, Heins HB, Guttentag SH, Wegner DJ, Trusgnich MA, Bennet KW, et al. Developmental and genetic regulation of human surfactant protein B in vivo. Neonatology (2009) 95:117–24. doi: 10.1159/000153095
70. Baekvad-Hansen M, Nordestgaard BG, Dahl M. Surfactant protein B polymorphisms, pulmonary function and COPD in 10,231 individuals. Eur Respir J. (2011) 37:791–9. doi: 10.1183/09031936.00026410
71. Lingappa JR, Dumitrescu L, Zimmer SM, Lynfield R, McNicholl JM, Messonnier NE, et al. Identifying host genetic risk factors in the context of public health surveillance for invasive pneumococcal disease. PLoS ONE (2011) 6:e23413. doi: 10.1371/journal.pone.0023413
72. Lin Z, Wang G, Demello DE, Floros J. An alternatively spliced surfactant protein B mRNA in normal human lung: disease implication. Biochem J. (1999) 343(Pt 1):145–149.
73. Hersh CP, Demeo DL, Lange C, Litonjua AA, Reilly JJ, Kwiatkowski D, et al. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am J Respir Cell Mol Biol. (2005) 33:71–8. doi: 10.1165/rcmb.2005-0073OC
74. Wood AM, Stockley RA. The genetics of chronic obstructive pulmonary disease. Respir Res. (2006) 7:130. doi: 10.1186/1465-9921-7-130
75. Raleigh SM, Davies BM, Cleal D, Ribbans WJ. No association between coding polymorphism within Exon 4 of the human surfactant protein B gene and pulmonary function in healthy men. J Physiol Sci. (2007) 57:199–202. doi: 10.2170/physiolsci.SC002607
76. Wood AM, Tan SL, Stockley RA. Chronic obstructive pulmonary disease: towards pharmacogenetics. Genome Med. (2009) 1:112. doi: 10.1186/gm112
77. Dahmer MK, O'Cain P, Patwari PP, Simpson P, Li SH, Halligan N, et al. The influence of genetic variation in surfactant protein B on severe lung injury in African American children. Crit Care Med. (2011) 39:1138–44. doi: 10.1097/CCM.0b013e31820a9416
78. Christie JD, Wurfel MM, Feng R, O'Keefe GE, Bradfield J, Ware LB, et al. Genome wide association identifies PPFIA1 as a candidate gene for acute lung injury risk following major trauma. PLoS ONE (2012) 7:e28268. doi: 10.1371/journal.pone.0028268
79. Guo Y, Gong Y, Pan C, Qian Y, Shi G, Cheng Q, et al. Association of genetic polymorphisms with chronic obstructive pulmonary disease in the Chinese Han population: a case-control study. BMC Med Genomics (2012) 5:64. doi: 10.1186/1755-8794-5-64
80. O'Mahony DS, Glavan BJ, Holden TD, Fong C, Black RA, Rona G, et al. Inflammation and immune-related candidate gene associations with acute lung injury susceptibility and severity: a validation study. PLoS ONE (2012) 7:e51104. doi: 10.1371/journal.pone.0051104
81. To KKW, Zhou J, Song YQ, Hung IFN, Ip WCT, Cheng ZS, et al. Surfactant protein B gene polymorphism is associated with severe influenza. Chest (2014) 145:1237–43. doi: 10.1378/chest.13-1651
82. Yang J, Wang B, Zhou HX, Liang BM, Chen H, Ma CL, et al. Association of surfactant protein B gene with chronic obstructive pulmonary disease susceptibility. Int J Tuberc Lung Dis. (2014) 18:1378–84. doi: 10.5588/ijtld.13.0569
83. Tochimoto A, Kawaguchi Y, Yamanaka H. Genetic susceptibility to interstitial lung disease associated with systemic sclerosis. Clin Med Insights Circ Respir Pulm Med. (2015) 9(Suppl. 1):135–140. doi: 10.4137/ccrpm.S23312
84. Ge L, Liu X, Chen R, Xu Y, Zuo YY, Cooney RN, et al. Differential susceptibility of transgenic mice expressing human surfactant protein B genetic variants to Pseudomonas aeruginosa induced pneumonia. Biochem Biophys Res Commun. (2016) 469:171–5. doi: 10.1016/j.bbrc.2015.11.089
85. Fatahi N, Niknafs N, Kalani M, Dalili H, Shariat M, Amini E, et al. Association of SP-B gene 9306 A/G polymorphism (rs7316) and risk of RDS. J Matern Fetal Neonatal Med. 31:2965–70. doi: 10.1080/14767058.2017.1359829
86. Lahti M, Marttila R, Hallman M. Surfactant protein C gene variation in the Finnish population - association with perinatal respiratory disease. Eur J Hum Genet. (2004) 12:312–20. doi: 10.1038/sj.ejhg.5201137
87. Salminen A, Paananen R, Karjalainen MK, Tuohimaa A, Luukkonen A, Ojaniemi M, et al. Genetic association of SP-C with duration of preterm premature rupture of fetal membranes and expression in gestational tissues. Ann Med. (2009) 41:629–42. doi: 10.1080/07853890903186176
88. Wambach JA, Yang P, Wegner DJ, An P, Hackett BP, Cole FS, et al. Surfactant protein-C promoter variants associated with neonatal respiratory distress syndrome reduce transcription. Pediatr Res. (2010) 68:216–20. doi: 10.1203/00006450-201011001-00421
89. Fatahi N, Dalili H, Kalani M, Niknafs N, Shariat M, Tavakkoly-Bazzaz J, et al. Association of SP-C gene codon 186 polymorphism (rs1124) and risk of RDS. J Matern Fetal Neonatal Med. (2017) 30:2585–9. doi: 10.1080/14767058.2016.1256994
90. Puthothu B, Krueger M, Heinze J, Forster J, Heinzmann A. Haplotypes of surfactant protein C are associated with common paediatric lung diseases. Pediatr Allergy Immunol. (2006) 17:572–7. doi: 10.1111/j.1399-3038.2006.00467.x
91. Floros J, Lin HM, Garcia A, Salazar MA, Guo X, DiAngelo S, et al. Surfactant protein genetic marker alleles identify a subgroup of tuberculosis in a Mexican population. J Infect Dis. (2000) 182:1473–8. doi: 10.1086/315866
92. Lahti M, Lofgren J, Marttila R, Renko M, Klaavuniemi T, Haataja R, et al. Surfactant protein D gene polymorphism associated with severe respiratory syncytial virus infection. Pediatr Res. (2002) 51:696–9. doi: 10.1203/00006450-200206000-00006
93. Pavlovic J, Papagaroufalis C, Xanthou M, Liu W, Fan R, Thomas NJ, et al. Genetic variants of surfactant proteins A, B, C, and D in bronchopulmonary dysplasia. Dis Markers (2006) 22:277–91. doi: 10.1155/2006/817805
94. Thomas NJ, Fan R, Diangelo S, Hess JC, Floros J. Haplotypes of the surfactant protein genes A and D as susceptibility factors for the development of respiratory distress syndrome. Acta Paediatr. (2007) 96:985–9. doi: 10.1111/j.1651-2227.2007.00319.x
95. Brandt EB, Mingler MK, Stevenson MD, Wang N, Khurana Hershey GK, Whitsett JA, et al. Surfactant protein D alters allergic lung responses in mice and human subjects. J Allergy Clin Immunol. (2008) 121:1140–7.e1142. doi: 10.1016/j.jaci.2008.02.011
96. Berg KK, Madsen HO, Garred P, Wiseth R, Gunnes S, Videm V. The additive contribution from inflammatory genetic markers on the severity of cardiovascular disease. Scand J Immunol. (2009) 69:36–42. doi: 10.1111/j.1365-3083.2008.02187.x
97. Deng YQ, Tao ZZ, Kong YG, Xiao BK, Chen SM, Xu Y, et al. Association between single nucleotide polymorphisms of surfactant protein D and allergic rhinitis in Chinese patients. Tissue Antigens (2009) 73:546–52. doi: 10.1111/j.1399-0039.2009.01232.x
98. Hilgendorff A, Heidinger K, Bohnert A, Kleinsteiber A, Konig IR, Ziegler A, et al. Association of polymorphisms in the human surfactant protein-D (SFTPD) gene and postnatal pulmonary adaptation in the preterm infant. Acta Paediatr. (2009) 98:112–7. doi: 10.1111/j.1651-2227.2008.01014.x
99. Thomas NJ, DiAngelo S, Hess JC, Fan R, Ball MW, Geskey JM, et al. Transmission of surfactant protein variants and haplotypes in children hospitalized with respiratory syncytial virus. Pediatr Res. (2009) 66:70–3. doi: 10.1203/PDR.0b013e3181a1d768
100. van Diemen CC, Postma DS, Aulchenko YS, Snijders PJ, Oostra BA, van Duijn CM, et al. Novel strategy to identify genetic risk factors for COPD severity: a genetic isolate. Eur Respir J. (2010) 35:768–75. doi: 10.1183/09031936.00054408
101. Foreman MG, Kong X, DeMeo DL, Pillai SG, Hersh CP, Bakke P, et al. Polymorphisms in surfactant protein-D are associated with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. (2011) 44:316–22. doi: 10.1165/rcmb.2009-0360OC
102. Lin Z, John G, Hegarty JP, Berg A, Yu W, Wang Y, et al. Genetic variants and monoallelic expression of surfactant protein-D in inflammatory bowel disease. Ann Hum Genet. (2011) 75:559–68. doi: 10.1111/j.1469-1809.2011.00662.x
103. Ishii T, Hagiwara K, Ikeda S, Arai T, Mieno MN, Kumasaka T, et al. Association between genetic variations in surfactant protein d and emphysema, interstitial pneumonia, and lung cancer in a Japanese population. Copd (2012) 9:409–16. doi: 10.3109/15412555.2012.676110
104. Ishii T, Hagiwara K, Kamio K, Ikeda S, Arai T, Mieno MN, et al. Involvement of surfactant protein D in emphysema revealed by genetic association study. Eur J Hum Genet. (2012) 20:230–5. doi: 10.1038/ejhg.2011.183
105. Kim DK, Cho MH, Hersh CP, Lomas DA, Miller BE, Kong X, et al. Genome-wide association analysis of blood biomarkers in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2012) 186:1238–47. doi: 10.1164/rccm.201206-1013OC
106. Shakoori TA, Sin DD, Bokhari SN, Ghafoor F, Shakoori AR. SP-D polymorphisms and the risk of COPD. Dis Markers (2012) 33:91–100. doi: 10.3233/dma-2012-0909
107. Pueyo N, Ortega FJ, Mercader JM, Moreno-Navarrete JM, Sabater M, Bonas S, et al. Common genetic variants of surfactant protein-D (SP-D) are associated with type 2 diabetes. PLoS ONE (2013) 8:e60468. doi: 10.1371/journal.pone.0060468
108. Horimasu Y, Hattori N, Ishikawa N, Tanaka S, Bonella F, Ohshimo S, et al. Differences in serum SP-D levels between German and Japanese subjects are associated with SFTPD gene polymorphisms. BMC Med Genet. (2014) 15:4. doi: 10.1186/1471-2350-15-4
109. Issac MS, Ashur W, Mousa H. Genetic polymorphisms of surfactant protein D rs2243639, Interleukin (IL)-1beta rs16944 and IL-1RN rs2234663 in chronic obstructive pulmonary disease, healthy smokers, and non-smokers. Mol Diagn Ther. (2014) 18:343–54. doi: 10.1007/s40291-014-0084-5
110. Johansson SL, Tan Q, Holst R, Christiansen L, Hansen NC, Hojland AT, et al. Surfactant protein D is a candidate biomarker for subclinical tobacco smoke-induced lung damage. Am J Physiol Lung Cell Mol Physiol. (2014) 306:L887–895. doi: 10.1152/ajplung.00340.2013
111. Sorensen GL, Dahl M, Tan Q, Bendixen C, Holmskov U, Husby S. Surfactant protein-D-encoding gene variant polymorphisms are linked to respiratory outcome in premature infants. J Pediatr. (2014) 165:683–9. doi: 10.1016/j.jpeds.2014.05.042
112. Ou CY, Chen CZ, Hsiue TR, Lin SH, Wang JY. Genetic variants of pulmonary SP-D predict disease outcome of COPD in a Chinese population. Respirology (2015) 20:296–303. doi: 10.1111/resp.12427
113. Soto-Cardenas MJ, Gandia M, Brito-Zeron P, Arias MT, Armiger N, Bove A, et al. Etiopathogenic role of surfactant protein d in the clinical and immunological expression of primary Sjogren syndrome. J Rheumatol. (2015) 42:111–8. doi: 10.3899/jrheum.140394
114. Chang HY, Li F, Li FS, Zheng CZ, Lei YZ, Wang J. Genetic polymorphisms of SP-A, SP-B, and SP-D and risk of respiratory distress syndrome in preterm neonates. Med Sci Monit. (2016) 22:5091–100. doi: 10.12659/MSM.898553
115. Perez-Rubio G, Silva-Zolezzi I, Fernandez-Lopez JC, Camarena A, Velazquez-Uncal M, Morales-Mandujano F, et al. Genetic variants in IL6R and ADAM19 are associated with COPD severity in a mexican mestizo population. Copd (2016) 13:610–5. doi: 10.3109/15412555.2016.1161017
116. Shakoori TA, Sin DD. SNP rs3088308 is a risk factor for poor lung function in healthy smokers. J Pak Med Assoc. (2016) 66:1137–41.
117. Sorensen GL, Bladbjerg EM, Steffensen R, Tan Q, Madsen J, Drivsholm T, et al. Association between the surfactant protein D (SFTPD) gene and subclinical carotid artery atherosclerosis. Atherosclerosis (2016) 246:7–12. doi: 10.1016/j.atherosclerosis.2015.12.037
118. Wu CW, Zhang XF, Liu W, Wang HL, Hao XH, Guo ZY, et al. [Association of surfactant protein D gene polymorphisms at rs3088308 and rs721917 with susceptibility to silicosis]. Nan Fang Yi Ke Da Xue Xue Bao (2016) 36:1004–7.
119. Obeidat M, Li X, Burgess S, Zhou G, Fishbane N, Hansel NN, et al. Surfactant protein D is a causal risk factor for COPD: results of Mendelian randomisation. Eur Respir J. (2017) 50:1700657. doi: 10.1183/13993003.00657-2017
120. Choi Y, Lee DH, Tu THK, Ban GY, Park H, Shin YS, et al. Surfactant protein D alleviates eosinophil-mediated airway inflammation and remodeling in patients with aspirin-exacerbated respiratory disease. Allergy (2018). doi: 10.1111/all.13458. [Epub ahead of print].
121. Fakih D, Akiki Z, Junker K, Medlej-Hashim M, Waked M, Salameh P, et al. Surfactant protein D multimerization and gene polymorphism in COPD and asthma. Respirology (2018) 23:298–305. doi: 10.1111/resp.13193
122. Hsieh MH, Ou CY, Hsieh WY, Kao HF, Lee SW, Wang JY, et al. Functional analysis of genetic variations in surfactant protein d in mycobacterial infection and their association with tuberculosis. Front Immunol. (2018) 9:1543. doi: 10.3389/fimmu.2018.01543
123. Munk HL, Fakih D, Christiansen L, Tan Q, Christensen AF, Ejstrup L, et al. Surfactant protein-D, a potential mediator of inflammation in axial spondyloarthritis. Rheumatology (2018) doi: 10.1093/rheumatology/key187. [Epub ahead of print].
124. Hoffjan S, Nicolae D, Ostrovnaya I, Roberg K, Evans M, Mirel DB, et al. Gene-environment interaction effects on the development of immune responses in the 1st year of life. Am J Hum Genet. (2005) 76:696–704. doi: 10.1086/429418
125. Constantin JM, Mira JP, Guerin R, Cayot-Constantin S, Lesens O, Gourdon F, et al. Lemierre's syndrome and genetic polymorphisms: a case report. BMC Infect Dis. (2006) 6:115. doi: 10.1186/1471-2334-6-115
126. Tanaka M, Arimura Y, Goto A, Hosokawa M, Nagaishi K, Yamashita K, et al. Genetic variants in surfactant, pulmonary-associated protein D (SFTPD) and Japanese susceptibility to ulcerative colitis. Inflamm Bowel Dis. (2009) 15:918–25. doi: 10.1002/ibd.20936
127. Byers HM, Dagle JM, Klein JM, Ryckman KK, McDonald EL, Murray JC, et al. Variations in CRHR1 are associated with persistent pulmonary hypertension of the newborn. Pediatr Res. (2012) 71:162–7. doi: 10.1038/pr.2011.24
128. Ryckman KK, Dagle JM, Kelsey K, Momany AM, Murray JC. Genetic associations of surfactant protein D and angiotensin-converting enzyme with lung disease in preterm neonates. J Perinatol. (2012) 32:349–55. doi: 10.1038/jp.2011.104
129. Xie ZK, Huang QP, Huang J, Xie ZF. Association between the IL1B, IL1RN polymorphisms and COPD risk: a meta-analysis. Sci Rep. (2014) 4:6202. doi: 10.1038/srep06202
130. Valverde G, Zhou H, Lippold S, de Filippo C, Tang K, Lopez Herraez D, et al. A novel candidate region for genetic adaptation to high altitude in Andean populations. PLoS ONE (2015) 10:e0125444. doi: 10.1371/journal.pone.0125444
131. Liu Y, Yan S, Poh K, Liu S, Iyioriobhe E, Sterling DA. Impact of air quality guidelines on COPD sufferers. Int J Chron Obstruct Pulmon Dis. (2016) 11:839–72. doi: 10.2147/copd.S49378
132. Rivera L, Siddaiah R, Oji-Mmuo C, Silveyra GR, Silveyra P. Biomarkers for Bronchopulmonary Dysplasia in the preterm infant. Front Pediatr. (2016) 4:33. doi: 10.3389/fped.2016.00033
133. Lin Z, deMello DE, Batanian JR, Khammash HM, DiAngelo S, Luo J, et al. Aberrant SP-B mRNA in lung tissue of patients with congenital alveolar proteinosis (CAP). Clin Genet. (2000) 57:359–369. doi: 10.1034/j.1399-0004.2000.570506.x
134. Wang G, Christensen ND, Wigdahl B, Guttentag SH, Floros J. Differences in N-linked glycosylation between human surfactant protein-B variants of the C or T allele at the single-nucleotide polymorphism at position 1580: implications for disease. Biochem J. (2003) 369(Pt 1):179–184. doi: 10.1042/bj20021376
135. Lin Z, Cui X, Li H. Multiplex genotype determination at a large number of gene loci. Proc Natl Acad Sci USA. (1996) 93:2582–7.
136. DiAngelo S, Lin Z, Wang G, Phillips S, Ramet M, Luo J, et al. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis Markers (1999) 15:269–81.
137. Lin Z, Pearson C, Chinchilli V, Pietschmann SM, Luo J, Pison U, et al. Polymorphisms of human SP-A, SP-B, and SP-D genes: association of SP-B Thr131Ile with ARDS. Clin Genet. (2000) 58:181–91. doi: 10.1034/j.1399-0004.2000.580305.x
138. Wang Z, Liu T, Lin Z, Hegarty J, Koltun WA, Wu R. A general model for multilocus epistatic interactions in case-control studies. PLoS ONE (2010) 5:e11384. doi: 10.1371/journal.pone.0011384
139. Liu T, Thalamuthu A, Liu JJ, Chen C, Wang Z, Wu R. Asymptotic distribution for epistatic tests in case-control studies. Genomics (2011) 98:145–51. doi: 10.1016/j.ygeno.2011.05.001
140. Lin Z, Hao H, Hegarty JP, Lin TR, Wang Y, Harris LR, et al. Association of the haem oxygenase-1 gene with inflammatory bowel disease. Swiss Med Wkly. (2017) 147:w14456. doi: 10.4414/smw.2017.14456
141. Lin Z, Wang Z, Hegarty JP, Lin TR, Wang Y, Deiling S, et al. Genetic association and epistatic interaction of the interleukin-10 signaling pathway in pediatric inflammatory bowel disease. World J Gastroenterol. (2017) 23:4897–909. doi: 10.3748/wjg.v23.i27.4897
142. Vorbroker DK, Profitt SA, Nogee LM, Whitsett JA. Aberrant processing of surfactant protein C in hereditary SP-B deficiency. Am J Physiol (1995) 268(4 Pt 1):L647–656. doi: 10.1152/ajplung.1995.268.4.L647
143. Akinbi HT, Breslin JS, Ikegami M, Iwamoto HS, Clark JC, Whitsett JA, et al. Rescue of SP-B knockout mice with a truncated SP-B proprotein. Function of the C-terminal propeptide J Biol Chem. (1997) 272:9640–7.
144. Quasney MW, Waterer GW, Dahmer MK, Kron GK, Zhang Q, Kessler LA, et al. Association between surfactant protein B + 1580 polymorphism and the risk of respiratory failure in adults with community-acquired pneumonia. Crit Care Med. (2004) 32:1115–9. doi: 10.1097/01.CCM.0000124872.55243.5A
145. Xu Y, Ge L, Abdel-Razek O, Jain S, Liu Z, Hong Y, et al. Differential susceptiblity of human SP-B genetic variation on lung injury caused by bacterial pneumonia and the effect of a chemically mdified curcumin. Shock (2016) 45:375–84. doi: 10.1097/shk.0000000000000535
146. Crouch EC. Collectins and pulmonary host defense. Am J Respir Cell Mol Biol. (1998) 19:177–201. doi: 10.1165/ajrcmb.19.2.140
147. Hickling TP, Malhotra R, Sim RB. Human lung surfactant protein A exists in several different oligomeric states: oligomer size distribution varies between patient groups. Mol Med. (1998) 4:266–75.
148. Griese M, Maderlechner N, Ahrens P, Kitz R. Surfactant proteins A and D in children with pulmonary disease due to gastroesophageal reflux. Am J Respir Crit Care Med. (2002) 165:1546–50. doi: 10.1164/rccm.2107147
149. Sanchez-Barbero F, Rivas G, Steinhilber W, Casals C. Structural and functional differences among human surfactant proteins SP-A1, SP-A2 and co-expressed SP-A1/SP-A2: role of supratrimeric oligomerization. Biochem J. (2007) 406:479–89. doi: 10.1042/bj20070275
150. Sano H, Kuronuma K, Kudo K, Mitsuzawa H, Sato M, Murakami S, et al. Regulation of inflammation and bacterial clearance by lung collectins. Respirology (2006) 11(Suppl.):S46–50. doi: 10.1111/j.1440-1843.2006.00808.x
151. Brummer E, Stevens DA. Collectins and fungal pathogens: roles of surfactant proteins and mannose binding lectin in host resistance. Med Mycol. (2010) 48:16–28. doi: 10.3109/13693780903117473
152. Silveyra P, Floros J. Air pollution and epigenetics: effects on SP-A and innate host defence in the lung. Swiss Med Wkly. (2012) 142:w13579. doi: 10.4414/smw.2012.13579
153. Mikerov AN, Wang G, Umstead TM, Zacharatos M, Thomas NJ, Phelps DS, et al. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun. (2007) 75:1403–12. doi: 10.1128/iai.01341-06
Keywords: surfactant protein, SP-A, SP-B, SP-C, SP-D, cystic fibrosis
Citation: Lin Z, Thorenoor N, Wu R, DiAngelo SL, Ye M, Thomas NJ, Liao X, Lin TR, Warren S and Floros J (2018) Genetic Association of Pulmonary Surfactant Protein Genes, SFTPA1, SFTPA2, SFTPB, SFTPC, and SFTPD With Cystic Fibrosis. Front. Immunol. 9:2256. doi: 10.3389/fimmu.2018.02256
Received: 01 June 2018; Accepted: 11 September 2018;
Published: 02 October 2018.
Edited by:
Uday Kishore, Brunel University London, United KingdomReviewed by:
Anthony George Tsolaki, Brunel University London, United KingdomCopyright © 2018 Lin, Thorenoor, Wu, DiAngelo, Ye, Thomas, Liao, Lin, Warren and Floros. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanna Floros, anhmMTlAcHN1LmVkdQ==; amZsb3Jvc0BwZW5uc3RhdGVoZWFsdGgucHN1LmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.