- 1Biological Resource Center, Taizhou Hospital of Zhejiang Province, Wenzhou Medical University, Linhai, China
- 2Medical Research Center, Taizhou Hospital of Zhejiang Province, Wenzhou Medical University, Linhai, China
Phenotypic heterogeneity has been observed in most malignancies, which represents a considerable challenge for tumor therapy. In recent decades, the biological function and clinical significance of the human leukocyte antigen (HLA)-G have been intensively explored. It is now widely accepted that HLA-G is a critical marker of immunotolerance in cancer cell immune evasion and is strongly associated with disease progress and prognosis for cancer patients. Moreover, it has recently been emphasized that the signaling pathway linking HLA-G and immunoglobulin-like transcripts (ILTs) is considered an immune checkpoint. In addition, HLA-G itself can generate at least seven distinct isoforms, and intertumor and intratumor heterogeneity of HLA-G expression is common across different tumor types. Furthermore, HLA-G heterogeneity in cancers has been related to disease stage and outcomes, metastatic status and response to different therapies. This review focuses on the heterogeneity of HLA-G expression in malignant lesions, and clinical implications of this heterogeneity that might be relevant to personalized treatments are also discussed.
Introduction
Cancer is a very complex and heterogeneous disease that involves a broad range of mixed cells with distinct features. Tumor cells not only vary in morphology and phenotype but also, in genomes, transcriptomes, epigenomes and proteomes (1). Diversity among tumor cells termed heterogeneity can be observed between tumor cells within a tumor (intratumor heterogeneity). However, tumor heterogeneity is also common between the primary tumor and metastatic tumors, between metastases from the same patient, and between tumors of the same histotype from different patients (intertumor heterogeneity) (2). Tumor heterogeneity is now widely acknowledged to influence tumor cell characteristics such as growth, survival, metastasis and response to various therapies and immune evasion. Consequently, tumor heterogeneity presents a formidable obstacle in cancer treatment (3).
Human leukocyte antigen (HLA)-G, a non-classical HLA class I molecule, encompasses at least four membrane-bound (mHLA-G, HLA-G1~HLA-G4) and three soluble (sHLA-G, HLA-G5~HLA-G7) isoforms resulting from alternative splicing of in its primary mRNA. HLA-G expression was initially observed in extravillous cytotrophoblasts and is considered to play important roles in maintenance of fetal-maternal immune tolerance (4, 5). In addition to extravillous cytotrophoblasts, HLA-G expression is restricted to a few healthy adult tissues, including the cornea, thymic medulla and pancreatic islets (6–8). However, HLA-G expression can be switched on in various pathological conditions such as cancers, viral infection, organ transplantation, and autoimmune and inflammatory diseases (9).
Though the functions of HLA-G were first explored in reproductive immune regulation, its pre-clinical significance in tumor biology has been intensively investigated (10). In the context of tumor biology, Paul et al. (11) first reported that HLA-G expression was specifically observed in melanoma lesions but, absent in the adjacent non-tumorous tissues. This finding has been solidified by numerous subsequent studies with thousands of samples from more than thirty different types of tumors. It is now well established that HLA-G expression in cancers is highly related to immune suppressive microenvironments, advanced tumor stage, and poor therapeutic responses and prognosis (12, 13).
In the context of heterogeneity of HLA-G expression in cancers, the degree of HLA-G expression and the isoform profiles vary dramatically among tumor types and patients, within tumors of the same type, and between the primary tumor and metastases (10). In clinical settings, the relevance of HLA-G heterogeneity in therapeutic and immune responses, tumor progression and prognosis has been documented in a body of previous studies (14, 15). Herein, we reviewed the literature on HLA-G heterogeneity in cancers, and the clinical implications of this heterogeneity that might be relevant to personalized treatments were also discussed.
Diversity of the HLA-G Isoforms
Unlike the case for classical HLA class I counterparts, at least seven HLA-G isoforms, including four membrane-bound (HLA-G1, -G2, -G3, and -G4) and three soluble (HLA-G5, -G6, and -G7) isoforms, can be generated by alternative splicing of HLA-G primary transcripts (16). Moreover, proteolytic cleavage of cell surface HLA-G1 by metalloproteinases (MMPs) such as MMP-2 results in another soluble isoform called shedding HLA-G1 (17). Different HLA-G isoforms are distinguished by the number of extracellular immunoglobulin-like domains and whether intronic sequence encoded residues are included or not; however, all seven reported HLA-G isoforms have the extracellular α1 domain (18). The conformation of HLA-G1 and -G5 is similar to that of the classical HLA class I antigens, which contain three extracellular domains including α1, α2, and α3 non-covalently bound to β2-macroglobulin (β2m). The antigen presenting peptide-binding cleft is formed by the α1 and α2 domains in HLA-G1 and HLA-G5 molecules (19).
Other HLA-G isoforms lack one or two extracellular domains (α2, or α3, or both) and are smaller than HLA-G1. The extracellular structure of HLA-G2 contains α1 and α3 domains but lacks the α 2 domain; HLA-G3 contains only an α1 domain, and both α 2 and α 3 are deleted; HLA-G4 contains α1 and α2 domains, while the α 3 domain is deleted. The C terminals of soluble HLA-G isoforms including HLA-G5 (counterpart of HLA-G1) and HLA-G6 (counterpart of HLA-G2) are encoded by intron 4. HLA-G7 consists of only an α1 domain linked to two amino acid residues encoded by intron 2 (10). Notably, a study by Tronik-Le Roux et al. (20) recently reported that previously undescribed novel HLA-G isoforms were predicted by transcriptome analysis in renal cancer lesions. However, more efforts should be carried out in the development of new antibodies to identify these new isoforms and in evaluating their biological functions and clinical relevance (Figure 1).
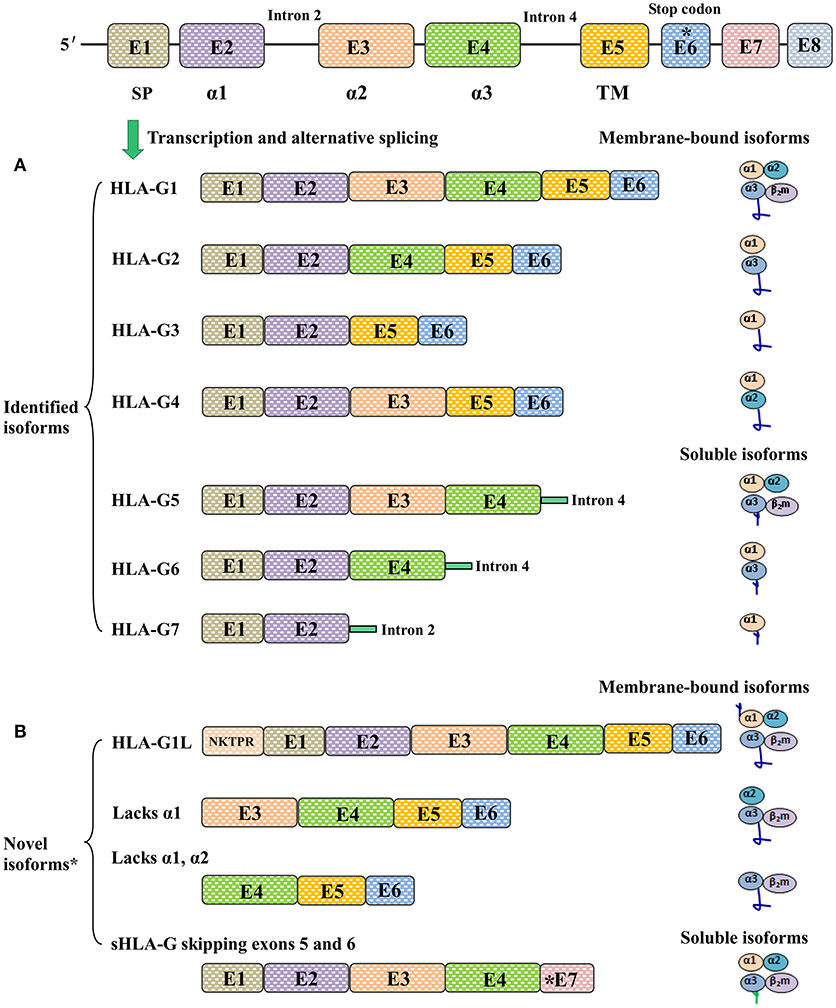
Figure 1. Different HLA-G isoforms generated by alternative splicing of HLA-G mRNA. (A) Seven identified HLA-G isoforms including four membrane-bound (HLA-G1, -G2, -G3, -G4) and three soluble (HLA-G5, -G6, -G7) molecules. The extracellular structures of HLA-G1 and HLA-G5 contain α1, α2, and α3 domains; HLA-G2 and HLA-G6 contain α1 and α3 domains; HLA-G3 contains α1 domains; HLA-G4 contains α1 and α2 domains; HLA-G7 contains an α1 domain linked to two amino acids encoded by intron 2. (B) Novel HLA-G isoforms predicted by Tronik-Le Roux et al. (20)*. N-terminal ends including the additional five amino acids (NKTPR) in HLA-G1L, and potential isoforms contain α2 and α3 domains or only the α3 domain. Novel soluble HLA-G isoforms generated by skipping exons 5 and 6, and with distinct C-terminal ends.
Among the HLA-G isoforms reported to date, HLA-G1 and HLA-G5 have been studied more extensively than the others due to the available antibodies (12). However, available antibodies for HLA-G detection have a limited ability to discriminate different isoforms, which has made it difficult to test the functional importance of a specific isoform. Currently available antibodies for HLA-G detection and their specificities are detailed in Supplementary Table 1.
In addition to the different isoforms, the structure of HLA-G is even more complex, since these isoforms can be presented as homo- and hetero-multimers resulting from intermolecular disulphide bonds by Cys42 or Cys147 in the α1 or α2 domain, respectively. Furthermore, HLA-G molecules can be ubiquitinated, glycosylated and nitrated by post-translational modifications (21–23). Finally, HLA-G has been reported as a part of exosomes (24).
HLA-G-Mediated Immune Suppression
HLA-G has comprehensive suppressive functions exerted in multiple steps to impair anti-tumor immune responses by interacting with receptors expressed on immune cells. To date, several receptors for HLA-G have been identified, such as CD85j/immunoglobulin-like transcript 2 (ILT2), CD85d/ILT4, and CD158d/killer cell immunoglobulin-like receptor 2DL4 (KIR2DL4). Moreover, CD8 and CD160 have also been reported to bind HLA-G (25). Among these receptors, ILT2 is present on all monocytes and B lymphocytes, and on subsets of dendritic cells (DCs), myeloid derived suppressive cells (MDSCs), natural-killer (NK) cells and T cells (26). ILT4 is mainly expressed on DCs and monocytes, neutrophils and MDSCs (27–29); KIR2DL4 has been found predominately in decidual NK cells (30). Other receptors such as CD160 are expressed by subsets of CD8+, CD4+, Tγ/δ and CD56dim NK cells, and by activated endothelial cells and intestinal intraepithelial cells. CD8 is a hallmark of cytotoxic T cells and is also expressed by some NK cells (26).
The mechanisms involved in HLA-G/receptor (particularly ILT2 and ILT4) mediated immune suppression have been documented in previous studies and include impairment of immune cell proliferation, differentiation, cytotoxicity, cytokine secretion and chemotaxis; and induction of regulatory cells and MDSCs or M2 type macrophages (31–33) (Figure 2). ILT2 and ILT4 have four tandem immunoglobulin-like extracellular domains (D1~D4), a transmembrane region of 23 amino acids and three immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in their cytoplasmic tails (34). The extracellular domains (D1~D2) of ILTs bind to the α3 domain in the HLA I molecule. Among HLA I family members, HLA-G binds ILTs with the highest affinity. Structural analysis showed that ILT4 could recognize both HLA-G associated with β2m and free HLA-G heavy chains, whereas ILT2 only recognized HLA-G associated with β2m. The HLA-G binding affinities of the ILT2 and ILT4 are also different; ILT4 binds HLA-G with higher affinity than ILT2 due to the different recognition specificities. Previous studies revealed that residues Tyr36 and Arg38 in ILT4 recognize the 195–197 loop, whereas the Tyr38 and Tyr76 locations of ILT2 bind Phe195 in the α3 domain of the HLA-G molecule (35). Interaction of HLA-G with ILTs is a critical step for HLA-G-mediated immune regulation. This HLA-G/ILTs interaction causes phosphorylation of ITIMs and recruits protein tyrosine phosphatase Src homology 2 (SH2) domain-containing proteins such as SHP-1 and SHP-2, which initiates the inhibitory signaling cascade (36, 37).
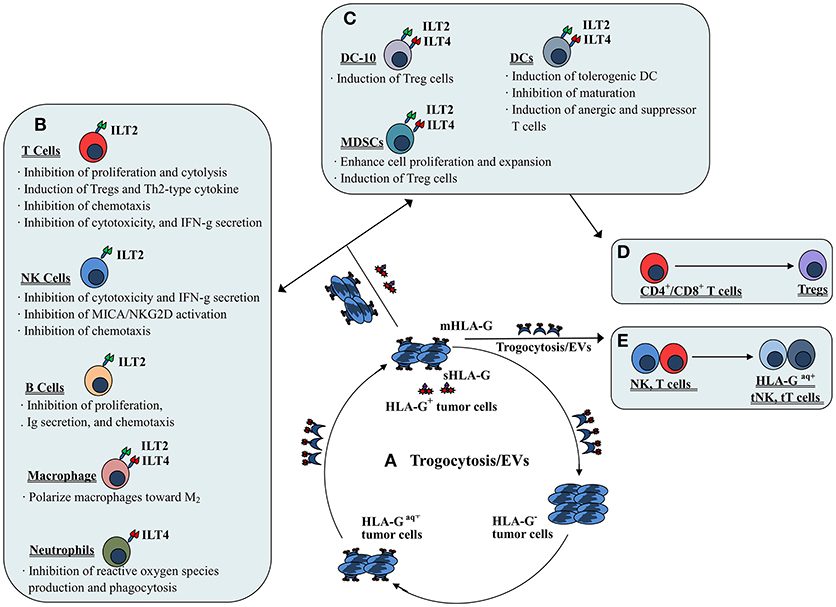
Figure 2. Mechanisms of both membrane-bound and soluble HLA-G-mediated immune suppression in tumor immune evasion. (A) Dynamic transferring of HLA-G by trogocytosis (membrane-bound HLA-G) and/or extracellular vesicles (EVs, both membrane-bound and soluble HLA-G) between HLA-G+ and HLA-G− tumor cells. (B) Direct HLA-G-mediated immunosuppressive effects through engagement of inhibitory receptors (ILT2 and/or ILT4) expressed by immune cells such as T cells, NK cells, B cells, macrophages and neutrophils. (C) Indirect HLA-G-mediated immunosuppressive effects by induction of immune suppressive or regulatory cells such as tolerogenic DCs and MDSCs, which induce (D) CD4+/CD8+ T cells to become regulatory T cells (Tregs). (E) Immune effectors such as NK cells and T cells efficiently act as suppressor cells when they acquire HLA-G from HLA-G+ tumor cells or HLA-G+ immune cells via the process of trogocytosis and/or EVs.
Moreover, it has also been reported that HLA-G can up-regulate tumor-promoting agents such as MMPs (38). Because of these functions, neoexpression of the HLA-G molecule on tumors favors carcinogenesis and tumor progression. Fortunately, HLA-G down-regulation with RNA interference or antibody blockade can recover the functions of immune effectors and prevent tumor reoccurrence, raising the possibility that HLA-G/ILTs interaction blockade could be a potential immuno-therapeutic strategy for cancer treatment (32, 39, 40).
Direct HLA-G-Mediated Immune Suppression Through Inhibitory Receptor Engagement
Direct immunosuppressive effects induced by HLA-G could occur through the engagement of inhibitory receptors expressed by various immune cells such as NK cells, T cells, B cells, neutrophils, macrophages, and DCs.
By interaction with the receptor ILT2, HLA-G could directly inhibit the immune function of NK cells, T cells and B cells. Inhibition of NK cell lysis against targets by HLA-G expression has been observed for isolated decidual NK cells, peripheral blood and NK cell lines (41, 42). HLA-G expression was also found to inhibit xenogeneic cytotoxicity and rolling adhesion of activated human NK cells to porcine endothelial cells (43, 44). Furthermore, HLA-G/ILT2 mediated inhibitory signals have been shown to mitigate major histocompatibility complex class I-related chain A (MICA)/ natural-killer group 2 member D (NKG2D)-induced activation (45). Our previous studies revealed that both the HLA-G1 and HLA-G5 isoforms could suppress NK cell cytolysis in a manner dependent on the HLA-G expression, and HLA-G1 and HLA-G5 isoforms had an additive effect on NK cytolysis suppression (46, 47). Other studies also reported that HLA-G expression in tumor cell lines such as ovarian carcinoma, hepatocellular carcinoma (HCC), glioma, and renal cell carcinoma could be protective against NK cytolysis. These studies also revealed that the inhibition could be reversed by blocking HLA-G antigens or its receptor with respective specific antibodies (48–51). For T cells, HLA-G could inhibit the CD4+ T cell alloproliferative response and cytolytic functions of CD8+ and Vγ9Vδ2 T cells through ILT2 (52–54). Regarding B cells, Naji et al. (55) demonstrated that the sHLA-G/ILT2 pathway could induce B cell G0/G1 cycle arrest. Consequently, the proliferation, differentiation, and immunoglobulin (Ig) secretion of B cells was suppressed.
Moreover, sHLA-G/ILT2 interaction impairs expression and functions of chemokine receptors in T cells, NK cells and B cells (31). sHLA-G could down-regulate the expression of CCR2, CXCR3, and CXCR5 on CD4+ T cells, and CXCR3 on CD8+ T cells and Vγ9Vδ2 T cells. Additionally, sHLA-G could impair the CD4+ T cell chemotaxis response to CCL2, CCL8, CXCL10, the CXCL11, CD8+ T cells response to CXCL10 and CXCL11, and the Vγ9Vδ2 T cells response to CXCL10 and CXCL11 (56). For NK cells, sHLA-G/ILT2 binding could down-modulate CXCR3, CX3CR1, and CCR2 expression and binding to their specific ligands such as CCL2 and CXCL10 (26). Moreover, HLA-G could decrease germinal center B cell CXCR5 expression and dampen the chemotaxis of those cells to their chemokines (57).
The immune suppressive functions of HLA-G on neutrophils have been demonstrated in a study by Baudhuin et al. (27) who found that HLA-G5/ILT4 engagement impairs neutrophil phagocytosis and reactive oxygen species production. For DCs, the expression of HLA-G has been observed to inhibit DC maturation and differentiation, and to disturb their cross-talk with NK cells. A study by Liang et al. (58) revealed that HLA-G/ILT4 interaction plays a critical role in DC differentiation modulation through the IL-6-STAT3 signaling pathway.
When HLA-G binds ILT2 and ILT4 expressed on macrophages, it not only induces macrophage differentiation toward an M2 phenotype but also enhances macrophage IL-6 and CXCL1 secretion, which could inhibit the production of interferon-γ (IFN-γ) by T cells (33). Notably, Barkal et al. (59) recently released important findings that the MHC class I component β2m expressed by cancer cells could directly prevent malignant cells from performing phagocytosis through ILT2. However, knowledge of the biological relevance of HLA-G expression for the regulation of macrophage functions is very limited. Therefore, more studies to explore the significance of interactions between HLA-G and macrophage receptors such as ILTs are needed.
Indirect HLA-G-Mediated Immune Suppression Through Suppressor/Regulatory Cells
Indirect immunosuppressive effects of HLA-G could be mediated by the induction of immune suppressive or regulatory cells such as regulatory T cells (Tregs), tolerogenic DCs and MDSCs.
HLA-G1-expressing antigen presenting cells (APCs) have been found to induce CD4+ T cell anergy and differentiation into suppressive cells. Furthermore, HLA-G-induced tolerogenic DCs can induce the generation of CD4+CD25+CTLA-4+ and CD8+CD28+ regulatory T cells (25, 60). The tolerogenic DC sub-population DC-10s, expresses high levels of HLA-G and can potently induce adaptive type 1 regulatory T cells (Tr1) through the HLA-G/ILT4 signaling pathway (28).
The evidence for a role of HLA-G in MDSC proliferation and function modulation has been obtained mainly from experiments with murine models. The HLA-G/ILT2 signaling pathway is considered to play an important role in the expansion of CD11b+Gr1+ MDSCs in ILT2 transgenic C57BL/6 mice and is directly related to the long-term survival of skin allografts (61). HLA-G5 expression has been observed to favor the expansion of CD11b+Ly6G+ MDSCs in a tumor-bearing Balb/c murine model with the murine mammary carcinoma cell line 4T1 (62). Similar findings, such as the fact that HLA-G and paired immunoglobulin-like receptor B (PIR-B, murine homolog of human ILT receptors) engagement expanded the population of CD11b+Gr1+PIR-B+ MDSCs in an M8-HLA-G1 (human melanoma cell line) tumor-bearing mouse model, have also been reported (32). In addition to the murine models, a recent study by Köstlin et al. (29) revealed that HLA-G/ILT4 interaction could promote MDSC accumulation and suppressive activity during human pregnancy.
HLA-G-Mediated Immune Suppression Through Trogocytosis/Extracellular Vesicles
In addition to HLA-G expression and interactions with receptor, HLA-G mediated immune suppression by intercellular transfer mechanisms such as trogocytosis or extracellular vesicles has gained considerable attention in recent years.
Trogocytosis is a rapid process of transferring cell membrane fragments containing molecules from one cell to another during cell-to-cell contact (63). Immune cells such as activated NK cells, T cells and monocytes can rapidly acquire membrane fragments containing functional HLA-G from other cells (i.e., HLA-G+ immune or tumor cells) in their vicinity by the process of trogocytosis. The acquired HLA-G molecule can then immediately reverse the functional phenotype of the recipient from effector to regulatory cells (64). In this scenario, when activated NK cells acquired HLA-G1 from M8-HLA-G1 tumor cells, NK-HLA-G1acq+ cells lost their cytolytic functions and ceased cell proliferation. Moreover, NK-HLA-G1acq+ cells behaved as suppressor cells capable of suppressing the cytolytic functions other NK cells (65). In other immune cells such as T cells and monocytes, acquisition of HLA-G from tumor cells or APCs can also rapidly reverse the phenotype from effector to regulatory (66). In clinical settings, CD25–FoxP3– T cells can acquire HLA-G from HLA-G+ malignant plasma cells in multiple myeloma patients. Functionally, these HLA-G+ T cells act as Tregs with inhibitory functions similar to those of natural Tregs (67). Furthermore, intercellular transfer of HLA-G from allogeneic as well as from autologous HLA-G positive tumor cells to HLA-G negative tumor cells via trogocytosis has been demonstrated (68).
Extracellular vesicles (EVs) are highly heterogeneous small phospholipid bilayer vesicles that are released by most normal and malignant cells (69). EVs harbor different types of genetic materials and proteins. HLA-G-bearing EVs were first observed in culture supernatants of an HLA-G+ melanoma cell line (Fon), and later in ascites and pleural exudates from cancer patients (21, 70). Moreover, high levels of HLA-G-bearing EVs were observed to be positively associated with disease progression in advanced breast cancer patients undergoing neoadjuvant chemotherapy (71). A recent study by Grange et al. (72) reported that HLA-G-bearing EVs derived from renal cancer cells could impair monocyte-derived DC differentiation and T cell immune responses, indicating the potential of HLA-G-bearing EVs to modulate functions of both the innate and adaptive immune systems. These findings highlighted that the effects of HLA-G-mediated tumor immune evasion can be extended to HLA-G-negative tumor cells by the pathways of trogocytosis and/or extracellular vesicles.
Given the abovementioned immune suppressive functions of HLA-G in cancer immunology, the HLA-G/ILTs signaling pathway has been recently recognized as a new immune checkpoint in addition to other immune checkpoints such as cytotoxic T lymphocyte-associated protein 4 (CTLA-4)/B7 and programmed cell death protein-1 (PD-1)/PD-L1 (40). Functionally, CTLA4/B7 interactions specifically inhibit T cell responses during T cell priming by competing with CD28 for the B7 receptor and thereby preventing CD4+ T cell activation. PD1/PDL1 interactions have prominent roles in interfering with T cell receptor (TCR) signaling and result in dysfunction and exhaustion of activated T cells (73). Thus, blocking either CTLA-4 or PD-1 has emerged as a promising anti-cancer strategy. However, it is evident from large clinical trials that only a fraction of patients respond with durable remission or are cured, while many patients will relapse (74). One can hypothesize that different responses to checkpoint inhibitor therapy could be a consequence of heterogeneous intra- and inter-tumor expression of different kinds of checkpoints, though data on the expression status of ILTs and CTLA-4 and PD-L1 in cancers are rather limited. Indeed, recent findings by Rouas-Freiss et al. (75) revealed that intra- and inter-tumor heterogeneity of PD-1/PDL-1 and HLA-G/ILT2/ILT4 expression does exist in various areas of the same lesion or among different renal-cell carcinoma lesions and in tumor infiltrating immune cells. In this regard, blocking multiple checkpoints is urgently needed to target the entire tumor.
In addition to the complexity of the tumor itself, heterogeneous tumor immune microenvironments including varying profiles of immune checkpoints molecule expression can lead to different responses to immune therapy. In this context, MHC I and II molecule expression pattern in melanoma patients were reported to confer differential sensitivity to CTLA-4 and PD-1 blockade therapy. Data revealed that an anti-CTLA-4 therapy response requires MHC class I, while an anti-PD-1 response is associated with MHC class II expression (76). However, the significance of HLA-G expression for anti-CTLA-4 and/or anti-PD-1 therapy needs to be evaluated. Given this evidence, the proposed multiple checkpoint blockade to overcome resistance to therapy and relapse should take MHC expression status into consideration (75, 77).
HLA-G Heterogeneity in Cancers
Genetic and proteomic heterogeneity regarding markers such as HLA-G is a common phenomenon in tumors. The inter- and intra-tumor heterogeneity of HLA-G expression can be increased by the complexity of mechanisms involved in the regulation of HLA-G expression (20, 75). In addition to the genetic background of HLA-G, multiple transcriptional, epigenetic, post-transcriptional and environmental mechanisms and contexts are involved in modulating HLA-G mRNA and/or protein expression (13, 78). In this context, a number of HLA-G specific microRNAs, including the miR-152 family (miR-148a, miR-148b, miR-152) and miR-133 have been demonstrated to control HLA-G expression (39, 79). Furthermore, cytokines such as IFN-γ and/or TNF-α differentially regulated HLA-G isoform expression in a retinal pigment epithelial cell line (80). In pathological settings, fewer functional HLA-G molecules and differential expression of HLA-G isoforms were observed in asthma and prostatic adenocarcinoma patients, respectively (81, 82). All these mechanisms of regulation and disease status alone or in combination can contribute to the heterogeneity of HLA-G expression.
Intratumor HLA-G Heterogeneity
Intratumor heterogeneity is very common in cancers; however, information on intratumor HLA-G heterogeneity is rather limited. In a cohort of nineteen clear cell renal-cell carcinoma patients (ccRCC), normal adjacent tissues and tumor lesions were collected for each patient. Among these samples, different tumor areas including 3–4 zones per tumor were obtained, and HLA-G expression was evaluated with immunohistochemistry. The authors found that intratumor HLA-G heterogeneity was present in all the ccRCC patients; levels of HLA-G expression varied markedly among different areas. In some patients, a high degree of HLA-G expression was observed in all selected areas; however, in other patients, HLA-G expression was only found in some but not all areas among CA9+ ccRCC tumor cells (75).
In another study, expression patterns of HLA-G isoforms in ccRCC lesions were analyzed with next-generation sequencing technologies and immunohistochemical labeling (20). In this in-depth study, expression levels of HLA-G isoforms such as HLA-G1, -G5, and -G6 were highly variable among tumors from different patients (intertumor heterogeneity) and among distinct areas or subcellular locations in the same tumor (intratumor heterogeneity). Importantly, the authors also identified novel HLA-G isoforms that had not been recognized before, such as the predicted isoforms without a transmembrane region and α1 domain. However, mechanisms underlying the high diversity of HLA-G expression and whether they involve a manner of spatial separation or selection of subclones remain to be explored.
Intertumor HLA-G Heterogeneity
Interpatient Intertumor HLA-G Heterogeneity
Aberrant HLA-G expression in cancers was first reported in 1998 (11), and studies later documented that up-regulated HLA-G expression could only be found in primary and metastatic melanoma cells; but not in tumor regression sites or in normal adjacent skin tissues from a melanoma patient (83, 84). Based on these pioneering studies, the hypothesis that HLA-G expression is associated with malignant transformation and tumor cell immune evasion was proposed.
Since Paul's first report (11), HLA-G expression has been analyzed and evaluated worldwide in thousands of malignant samples of various types of cancers (12). Aberrant HLA-G expression in cancers has been found to be associated with advanced tumor stage, metastasis status and poor disease outcome. However, among and within different tumor types, discrepancies regarding HLA-G expression profiles have been observed in various types of cancers, such as breast cancer (85–94), colorectal cancer (CRC) (95–100), cervical cancer (101–103), endometrial cancer (104, 105), oesophageal squamous cell carcinoma (ESCC) (106–108), Ewing sarcoma (109), gastric cancer (110–112), glioblastoma (113), HCC (49, 114, 115), lung cancer (116–118), lymphoma (119–122), nasopharyngeal carcinoma (123), oral squamous cell carcinoma (124), ovarian cancer (125–127), pancreatic adenocarcinoma (128–130), and thyroid cancer (131, 132). The details of the methods applied and main conclusions drawn from these studies are summarized in Supplementary Table 2.
Notably, the percentage of HLA-G expressing malignant cells depends on the type of cancers, and expression levels vary from negative to totally positive among different types of cancers (133, 134). However, the degree of HLA-G expression also varies dramatically among different laboratories for almost every histological type of tumor studied. For example, HLA-G expression was assessed by immunohistochemistry with the same HLA-G monoclonal antibody (mAb), 4H84, in seven previous studies on breast cancer, in which the percentages of HLA-G expression ranged from 24 to 94.1% (91, 94). In four other studies using the HLA-G mAbs MEM-G1 (92), MEM-G2 (90), HGY (87) and 5A6G7 (89), the percentages of HLA-G positive staining cells were 43.8, 62.2, 66, and 59.6%, respectively. Similarly, rather low interlaboratory concordance for HLA-G expression has been observed for other malignancies.
The low concordance between different laboratories for the same type of tumor lesions may reflect differences between assessment methods and tumor heterogeneity. Indeed, among these studies, there is a wide variation of immunohistochemistry protocols, including variations in the antibodies used and their dilution, the incubation time and staining evaluation criteria. Different cohort sizes and clinicopathological parameters such as treatment history, tumor subhistological type and tumor immune microenvironment could all affect the evaluation of the status of HLA-G expression among cancer patients.
Furthermore, genetic backgrounds of HLA-G may also influence the profile of HLA-G expression; for example, in the case of HLA-G molecules generated by the primary transcripts from two HLA-G null alleles. One null allele is the HLA-G*0105N (135) with a cytosine deletion at position 1597 (ΔC) in exon 3, which causes an open reading frame shift mutation and generates a premature stop at either codon 189 (TGA) in exon 4 or at codon 297 (TAG) in exon 5. As a result of this mutation, the translation and expression of the α2 domain-containing HLA-G isoforms, including HLA-G1, -G4, and HLA-G5, are disrupted, while the functional isoforms HLA-G2, -G3, -G6, and HLA-G7 can be still normally expressed (136, 137). The other null allele, HLA-G*01:21N (138), is a nucleotide mutation at codon 226 (CAG) that results in an early stop codon (TAG) in exon 4 and, is predicted to translate a truncated and presumably non-functional protein.
Intrapatient Intertumor HLA-G Heterogeneity
This heterogeneity is noticeable in a patient with multiple tumors of the same type. In cervical cancers, Ferns et al. (139) analyzed HLA-A, -B, -C, HLA-E and HLA-G expression on primary tumors and case-matched lymph node (LN) metastases by immunohistochemistry. In a cohort of 136 patients, HLA-G expression (probing with mAb 4H84) was detected in 25% of the primary cervical cancers and in 11% of paired metastatic LNs. In squamous cell carcinoma (SCC) subtypes, HLA-G positive staining was found in 22% (20/90) of the primary tumors and 20% (18/90) of their LN metastases. In adenocarcinoma (AC), positive HLA-G was staining observed in 31% (10/32) of the primary tumors and in 28% (9/32) of paired LN metastases. These findings revealed that primary tumors and paired LN metastases in both histology types of cervical cancers had similar HLA-G expression patterns. Similarly, a study by Guimarães et al. (103) revealed that, in invasive cervical cancer (ICC) patients, HLA-G5 expression was positive in 31.6% (25/79) of the patients. Among these patients, HLA-G5 expression was observed in 29.6% (8/27) of the patients with LN metastasis and in 32.7% (17/52) patients without LN metastasis.
An evaluation of HLA-G expression in primary CRC and liver metastases using different HLA-G mAbs including 4H84, MEM-G/1 and MEM-G/2, was performed by Swets et al. (95). In this study, HLA-G expression was observed in 29, 6, and 10% of primary CRC lesions, and 30, 4, and 0% of paired liver metastases when using the mAbs 4H84, MEM-G/1 and MEM-G/2, respectively. These findings indicated that HLA-G molecule probing with different antibodies with different specificities could dramatically impact the interpretation of the relevance of HLA-G in clinical settings.
HLA-G Heterogeneity and Cancer Therapy
Alterations in HLA expression are frequent and early events during tumor development and progression as a result of immune editing (140). Consequently, tumor cells that fail to present surface neoantigens due to an altered classical HLA class I complex can avoid killing by T cells, and aberrant induction of HLA-G expression can suppress the functions of various immune cells for immune escape (141).
As with other molecules, mechanisms underlying the regulation of HLA-G expression involve in multiple levels such as epigenetic modification, transcriptional regulation and post-transcriptional modifications (78, 79). In addition, microenvironmental factors such cytokine profiles and even therapeutics can enhance HLA-G expression. In this context, previous studies showed that HLA-G expression could be induced by 5-aza-2′-deoxycytidine and IFN-γ treatment in vitro in glioblastoma, and an enhanced peripheral sHLA-G level was observed in melanoma patients treated with IFN-α (113, 142). Given the immune suppressive functions of HLA-G in favoring tumor immune evasion and progression, induction of HLA-G expression during immune therapy which may impair the therapeutic effects, should be considered.
Moreover, the fact that HLA-G expression status can dramatically affect therapeutic responses has been addressed in tumor patients and patients with other diseases. In 2000, Wagner et al. (15) highlighted the importance of classic and non-classic HLA molecule expression status in melanoma patients treated with postsurgery adjuvant IFN-α-2b. In that study, they found that the status of classic HLA I molecules and HLA-G expression were significantly related to the outcome of melanoma patients receiving IFN-α-2b treatment. Disease relapse occurred in patients with HLA-G expression that had lost classic HLA I molecules. However, with or without HLA-G co-expression, no relapse was observed in patients with classic HLA I molecule expression. This study indicated that HLA-G expression can enable classic HLA I deficient melanoma cells to lose responsiveness to IFN-α treatment. Thus, evaluation of the status HLA antigen expression before immunotherapy may have important practical implications for treatment.
Conclusion
Tumor heterogeneity has gained attention in the field of individualized cancer therapy due to its implications for oncogenesis, and metastasis, and its potential to affect responses to therapies and clinical outcomes (3). Tumor heterogeneity can be caused by genetic mutations during clonal evolution, and/or by tumor microenvironment selective pressure during tumor development (143).
Alteration of HLA antigen expression is a well-recognized mechanism employed by malignant cells to avoid innate and adaptive immune surveillance and responses (144). Many efforts have been made by multiple laboratories worldwide to assess the expression and clinical significance of HLA-G expression in cancers. From these studies, a strong relationship of HLA-G expression with tumor progression and patient outcomes has been established.
Based on numerous previous studies, HLA-G has been promoted as a new checkpoint molecule and a promising immunotherapy target, such as via application of HLA-G antagonists or anti-HLA-G or anti-ILT antibodies to block the interaction between ILT receptors and HLA-G. Additionally, HLA-G as an effective target for drug delivery was proposed by Zhang et al. (145). In that study, authors successfully developed HLA-G antibody and methotrexate (MTX)-loaded nanobubbles (mAbHLA−G/MTX/PLGA NBs) as HLA-G-targeted drug delivery molecule. The results showed that mAbHLA−G/MTX/PLGA NBs could specifically transport to the HLA-G positive tumor cells in vitro or tumor tissues in vivo in a murine model, and the released MTX from the NBs could kill the residual tumor cells and inhibit the reoccurrence of tumors.
However, available results from the literature reveal that intertumor and intratumor heterogeneity of HLA-G expression varies dramatically. In addition to the heterogeneity that inherently occurs in tumors, the low concordance in HLA-G detection and evaluation results among different laboratories remains a major obstacle for the interpretation of the clinical significance of HLA-G. These controversies might be caused by the use of different HLA-G monoclonal antibodies, technical procedures, immunostaining evaluation criteria, or different clinicopathological and HLA-G genetic backgrounds of the included cohorts. Therefore, international recommended standardization protocols, larger cohorts and prospective studies are required to confirm and validate HLA-G as a target before routine clinical application. Furthermore, in addition to the seven well-known HLA-G isoforms, novel unrecognized HLA-G isoforms do exist, as presented by Dr. Carosella and colleagues (20). In this context, more specific antibodies for a particular HLA-G isoform and new technologies such as next generation sequencing are needed. With these tools, we can further discriminate different isoforms and evaluate their respective functions in tumor biology to finally improve and optimize personalized medicine.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (31370920, 81372247), and by Zhejiang Provincial program for the cultivation of high-level innovative health talents.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02164/full#supplementary-material
References
1. Gambara G, Gaebler M, Keilholz U, Regenbrecht CRA, Silvestri A. From chemotherapy to combined targeted therapeutics: in vitro and in vivo models to decipher intra-tumor heterogeneity. Front Pharmacol. (2018) 9:77. doi: 10.3389/fphar.2018.00077
2. Grzywa TM, Paskal W, Wlodarski PK. Intratumor and intertumor heterogeneity in melanoma. Transl Oncol. (2017) 10:956–75. doi: 10.1016/j.tranon.2017.09.007
3. Stanta G, Bonin S. Overview on clinical relevance of intra-tumor heterogeneity. Front Med. (2018) 5:85. doi: 10.3389/fmed.2018.00085
4. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science (1990) 248:220–3. doi: 10.1126/science.2326636
5. Ferreira LMR, Meissner TB, Tilburgs T, Strominger JL. HLA-G: at the interface of maternal-fetal tolerance. Trends Immunol. (2017) 38:272–86. doi: 10.1016/j.it.2017.01.009
6. Le Discorde M, Moreau P, Sabatier P, Legeais JM, Carosella ED. Expression of HLA-G in human cornea, an immune-privileged tissue. Hum Immunol. (2003) 64:1039–44. doi: 10.1016/j.humimm.2003.08.346
7. Mallet V, Blaschitz A, Crisa L, Schmitt C, Fournel S, King A, et al. HLA-G in the human thymus: a subpopulation of medullary epithelial but not CD83(+) dendritic cells expresses HLA-G as a membrane-bound and soluble protein. Int Immunol. (1999) 11:889–98.
8. Cirulli V, Zalatan J, McMaster M, Prinsen R, Salomon DR, Ricordi C, et al. The class I HLA repertoire of pancreatic islets comprises the nonclassical class Ib antigen HLA-G. Diabetes (2006) 55:1214–22. doi: 10.2337/db05-0731
9. Gonzalez A, Rebmann V, LeMaoult J, Horn PA, Carosella ED, Alegre E. The immunosuppressive molecule HLA-G and its clinical implications. Crit Rev Clin Lab Sci. (2012) 49:63–84. doi: 10.3109/10408363.2012.677947
10. Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: an immune checkpoint molecule. Adv Immunol. (2015) 127:33–144. doi: 10.1016/bs.ai.2015.04.001
11. Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, et al. HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc Natl Acad Sci USA. (1998) 95:4510–5.
12. Lin A, Yan WH. HLA-G expression in cancers: roles in immune evasion, metastasis and target for therapy. Mol Med. (2015) 21:782–91. doi: 10.2119/molmed.2015.00083
13. Alegre E, Rizzo R, Bortolotti D, Fernandez-Landazuri S, Fainardi E, Gonzalez A. Some basic aspects of HLA-G biology. J Immunol Res. (2014) 2014:657625. doi: 10.1155/2014/657625
14. Yan WH. Human leukocyte antigen-G in cancer: are they clinically relevant? Cancer Lett. 311:123–30. doi: 10.1016/j.canlet.2011.07.019
15. Wagner SN, Rebmann V, Willers CP, Grosse-Wilde H, Goos M. Expression analysis of classic and non-classic HLA molecules before interferon alfa-2b treatment of melanoma. Lancet (2000) 356:220–1. doi: 10.1016/S0140-6736(00)02486-7
16. Paul P, Cabestre FA, Ibrahim EC, Lefebvre S, Khalil-Daher I, Vazeux G, et al. Identification of HLA-G7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6, and -G7 transcripts in human transfected cells. Hum Immunol. (2000) 61:1138–49. doi: 10.1016/S0198-8859(00)00197-X
17. Rizzo R, Trentini A, Bortolotti D, Manfrinato MC, Rotola A, Castellazzi M, et al. Matrix metalloproteinase-2 (MMP-2) generates soluble HLA-G1 by cell surface proteolytic shedding. Mol Cell Biochem. (2013) 381:243–55. doi: 10.1007/s11010-013-1708-5
18. Dahl M, Djurisic S, Hviid TV. The many faces of human leukocyte antigen-G: relevance to the fate of pregnancy. J Immunol Res. (2014) 2014:591489. doi: 10.1155/2014/591489
19. Yan WH. HLA-G expression in cancers: potential role in diagnosis, prognosis and therapy. Endocr Metab Immune Disord Drug Targets (2011) 11:76–89. doi: 10.2174/187153011794982059
20. Tronik-Le Roux D, Renard J, Verine J, Renault V, Tubacher E, LeMaoult J, et al. Novel landscape of HLA-G isoforms expressed in clear cell renal cell carcinoma patients. Mol Oncol. (2017) 11:1561–1578. doi: 10.1002/1878-0261.12119
21. Alegre E, Rebmann V, Lemaoult J, Rodriguez C, Horn PA, Diaz-Lagares A, et al. In vivo identification of an HLA-G complex as ubiquitinated protein circulating in exosomes. Eur J Immunol. (2013) 43:1933–9. doi: 10.1002/eji.201343318
22. Nardi Fda S, Konig L, Wagner B, Giebel B, Santos Manvailer LF, Rebmann V. Soluble monomers, dimers and HLA-G-expressing extracellular vesicles: the three dimensions of structural complexity to use HLA-G as a clinical biomarker. HLA (2016) 88:77–86. doi: 10.1111/tan.12844
23. Diaz-Lagares A, Alegre E, Gonzalez A. Detection of 3-nitrotyrosine-modified human leukocyte antigen-G in biological fluids. Hum Immunol. (2009) 70:976–80. doi: 10.1016/j.humimm.2009.07.018
24. Rebmann V, Konig L, Nardi Fda S, Wagner B, Manvailer LF, Horn PA. The potential of HLA-G-bearing extracellular vesicles as a future element in HLA-G immune biology. Front Immunol. (2016) 7:173. doi: 10.3389/fimmu.2016.00173
25. Fainardi E, Castellazzi M, Stignani M, Morandi F, Sana G, Gonzalez R, et al. Emerging topics and new perspectives on HLA-G. Cell Mol Life Sci. (2011) 68:433–51. doi: 10.1007/s00018-010-0584-3
26. Rouas-Freiss N, Moreau P, LeMaoult J, Carosella ED. The dual role of HLA-G in cancer. J Immunol Res. (2014) 2014:359748. doi: 10.1155/2014/359748
27. Baudhuin J, Migraine J, Faivre V, Loumagne L, Lukaszewicz AC, Payen D, et al. Exocytosis acts as a modulator of the ILT4-mediated inhibition of neutrophil functions. Proc Natl Acad Sci USA. (2013) 110:17957–62. doi: 10.1073/pnas.1221535110
28. Gregori S, Tomasoni D, Pacciani V, Scirpoli M, Battaglia M, Magnani CF, et al. Differentiation of type 1 T regulatory cells (Tr1) by tolerogenic DC-10 requires the IL-10-dependent ILT4/HLA-G pathway. Blood (2010) 116:935–44. doi: 10.1182/blood-2009−07-234872
29. Kostlin N, Ostermeir AL, Spring B, Schwarz J, Marme A, Walter CB, et al. HLA-G promotes myeloid-derived suppressor cell accumulation and suppressive activity during human pregnancy through engagement of the receptor ILT4. Eur J Immunol. (2017) 47:374–84. doi: 10.1002/eji.201646564
30. Rajagopalan S, Long EO. KIR2DL4 (CD158d): an activation receptor for HLA-G. Front Immunol. (2012) 3:258. doi: 10.3389/fimmu.2012.00258
31. Morandi F, Rouas-Freiss N, Pistoia V. The emerging role of soluble HLA-G in the control of chemotaxis. Cytokine Growth Factor Rev. (2014) 25:327–35. doi: 10.1016/j.cytogfr.2014.04.004
32. Agaugue S, Carosella ED, Rouas-Freiss N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood (2011) 117:7021–31. doi: 10.1182/blood-2010-07-294389
33. Lee CL, Guo Y, So KH, Vijayan M, Wong VH, Yao Y, et al. Soluble human leukocyte antigen G5 polarizes differentiation of macrophages toward a decidual macrophage-like phenotype. Hum Reprod. (2015) 30:2263–74. doi: 10.1093/humrep/dev196
34. Zhang F, Zheng J, Kang X, Deng M, Lu Z, Kim J, et al. Inhibitory leukocyte immunoglobulin-like receptors in cancer development. Sci China Life Sci. (2015) 58:1216–25. doi: 10.1007/s11427-015-4925-1
35. Shiroishi M, Kuroki K, Rasubala L, Tsumoto K, Kumagai I, Kurimoto E, et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d). Proc Natl Acad Sci USA. (2006) 103:16412–7. doi: 10.1073/pnas.0605228103
36. Yu Y, Wang Y, Feng M. Human leukocyte antigen-G1 inhibits natural killer cytotoxicity through blocking the activating signal transduction pathway and formation of activating immunologic synapse. Hum Immunol. (2008) 69:16–23. doi: 10.1016/j.humimm.2007.11.005
37. Ketroussi F, Giuliani M, Bahri R, Azzarone B, Charpentier B, Durrbach A. Lymphocyte cell-cycle inhibition by HLA-G is mediated by phosphatase SHP-2 and acts on the mTOR pathway. PLoS ONE (2011) 6:e22776. doi: 10.1371/journal.pone.0022776
38. Lin A, Xu HH, Xu DP, Zhang X, Wang Q, Yan WH. Multiple steps of HLA-G in ovarian carcinoma metastasis: alter NK cytotoxicity and induce matrix metalloproteinase-15 (MMP-15) expression. Hum Immunol. (2013) 74:439–46. doi: 10.1016/j.humimm.2012.11.021
39. Porto IO, Mendes-Junior CT, Felicio LP, Georg RC, Moreau P, Donadi EA, et al. MicroRNAs targeting the immunomodulatory HLA-G gene: a new survey searching for microRNAs with potential to regulate HLA-G. Mol Immunol. (2015) 65:230–41. doi: 10.1016/j.molimm.2015.01.030
40. Carosella ED, Ploussard G, LeMaoult J, Desgrandchamps F. A systematic review of immunotherapy in urologic cancer: evolving roles for targeting of CTLA-4, PD-1/PD-L1, and HLA-G. Eur Urol. (2015) 68:267–79. doi: 10.1016/j.eururo.2015.02.032
41. Chumbley G, King A, Robertson K, Holmes N, Loke YW. Resistance of HLA-G and HLA-A2 transfectants to lysis by decidual NK cells. Cell Immunol. (1994) 155:312–22. doi: 10.1006/cimm.1994.1125
42. Pazmany L, Mandelboim O, Vales-Gomez M, Davis DM, Reyburn HT, Strominger JL. Protection from natural killer cell-mediated lysis by HLA-G expression on target cells. Science (1996) 274:792–5. doi: 10.1126/science.274.5288.792
43. Zeng MH, Fang CY, Wang SS, Zhu M, Xie L, Li R, et al. A study of soluble HLA-G1 protecting porcine endothelial cells against human natural killer cell-mediated cytotoxicity. Transplant Proc. (2006) 38:3312–4. doi: 10.1016/j.transproceed.2006.10.179
44. Forte P, Pazmany L, Matter-Reissmann UB, Stussi G, Schneider MK, Seebach JD. HLA-G inhibits rolling adhesion of activated human NK cells on porcine endothelial cells. J Immunol. (2001) 167:6002–8. doi: 10.4049/jimmunol.167.10.6002
45. Menier C, Riteau B, Carosella ED, Rouas-Freiss N. MICA triggering signal for NK cell tumor lysis is counteracted by HLA-G1-mediated inhibitory signal. Int J Cancer (2002) 100:63–70. doi: 10.1002/ijc.10460
46. Zhang WQ, Xu DP, Liu D, Li YY, Ruan YY, Lin A, et al. HLA-G1 and HLA-G5 isoforms have an additive effect on NK cytolysis. Hum Immunol. (2014) 75:182–9. doi: 10.1016/j.humimm.2013.11.001
47. Chen BG, Xu DP, Lin A, Yan WH. NK cytolysis is dependent on the proportion of HLA-G expression. Hum Immunol. (2013) 74:286–9. doi: 10.1016/j.humimm.2012.12.005
48. Lin A, Yan WH, Xu HH, Gan MF, Cai JF, Zhu M, et al. HLA-G expression in human ovarian carcinoma counteracts NK cell function. Ann Oncol. (2007) 18:1804–9. doi: 10.1093/annonc/mdm356
49. Lin A, Chen HX, Zhu CC, Zhang X, Xu HH, Zhang JG, et al. Aberrant human leucocyte antigen-G expression and its clinical relevance in hepatocellular carcinoma. J Cell Mol Med. (2010) 14:2162–71. doi: 10.1111/j.1582-4934.2009.00917.x
50. Wiendl H, Mitsdoerffer M, Hofmeister V, Wischhusen J, Bornemann A, Meyermann R, et al. A functional role of HLA-G expression in human gliomas: an alternative strategy of immune escape. J Immunol. (2002) 168:4772–80. doi: 10.4049/jimmunol.168.9.4772
51. Bukur J, Rebmann V, Grosse-Wilde H, Luboldt H, Ruebben H, Drexler I, et al. Functional role of human leukocyte antigen-G up-regulation in renal cell carcinoma. Cancer Res. (2003) 63:4107–11.
52. Lesport E, Baudhuin J, Sousa S, LeMaoult J, Zamborlini A, Rouas-Freiss N, et al. Inhibition of human gamma delta [corrected] T-cell antitumoral activity through HLA-G: implications for immunotherapy of cancer. Cell Mol Life Sci. (2011) 68:3385–99. doi: 10.1007/s00018−011-0632-7
53. Le Gal FA, Riteau B, Sedlik C, Khalil-Daher I, Menier C, Dausset J, et al. HLA-G-mediated inhibition of antigen-specific cytotoxic T lymphocytes. Int Immunol. (1999) 11:1351–6.
54. LeMaoult J, Krawice-Radanne I, Dausset J, Carosella ED. HLA-G1-expressing antigen-presenting cells induce immunosuppressive CD4+ T cells. Proc Natl Acad Sci USA. (2004) 101:7064–9. doi: 10.1073/pnas.0401922101
55. Naji A, Menier C, Morandi F, Agaugue S, Maki G, Ferretti E, et al. Binding of HLA-G to ITIM-bearing Ig-like transcript 2 receptor suppresses B cell responses. J Immunol. (2014) 192:1536–46. doi: 10.4049/jimmunol.1300438
56. Morandi F, Ferretti E, Bocca P, Prigione I, Raffaghello L, Pistoia V. A novel mechanism of soluble HLA-G mediated immune modulation: downregulation of T cell chemokine receptor expression and impairment of chemotaxis. PLoS ONE (2010) 5:e11763. doi: 10.1371/journal.pone.0011763
57. Morandi F, Ferretti E, Castriconi R, Dondero A, Petretto A, Bottino C, et al. Soluble HLA-G dampens CD94/NKG2A expression and function and differentially modulates chemotaxis and cytokine and chemokine secretion in CD56bright and CD56dim NK cells. Blood (2011) 118:5840–50. doi: 10.1182/blood-2011-05-352393
58. Gregori S, Amodio G, Quattrone F, Panina-Bordignon P. HLA-G orchestrates the early interaction of human trophoblasts with the maternal niche. Front Immunol. (2015) 6:128. doi: 10.3389/fimmu.2015.00128
59. Barkal AA, Weiskopf K, Kao KS, Gordon SR, Rosental B, Yiu YY, et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat Immunol. (2018) 19:76–84. doi: 10.1038/s41590-017-0004-z
60. Ristich V, Liang S, Zhang W, Wu J, Horuzsko A. Tolerization of dendritic cells by HLA-G. Eur J Immunol (2005) 35: 1133-42. doi: 10.1002/eji.200425741
61. Zhang W, Liang S, Wu J, Horuzsko A. Human inhibitory receptor immunoglobulin-like transcript 2 amplifies CD11b+Gr1+ myeloid-derived suppressor cells that promote long-term survival of allografts. Transplantation (2008) 86:1125–34. doi: 10.1097/TP.0b013e318186fccd
62. Loumagne L, Baudhuin J, Favier B, Montespan F, Carosella ED, Rouas-Freiss N. In vivo evidence that secretion of HLA-G by immunogenic tumor cells allows their evasion from immunosurveillance. Int J Cancer (2014) 135:2107–17. doi: 10.1002/ijc.28845
63. Ahmed KA, Munegowda MA, Xie Y, Xiang J. Intercellular trogocytosis plays an important role in modulation of immune responses. Cell Mol Immunol. (2008) 5:261–9. doi: 10.1038/cmi.2008.32
64. Carosella ED, Gregori S, Rouas-Freiss N, LeMaoult J, Menier C, Favier B. The role of HLA-G in immunity and hematopoiesis. Cell Mol Life Sci. (2011) 68:353–68. doi: 10.1007/s00018-010-0579-0
65. Caumartin J, Favier B, Daouya M, Guillard C, Moreau P, Carosella ED, LeMaoult J. Trogocytosis-based generation of suppressive NK cells. Embo J. (2007) 26:1423–33. doi: 10.1038/sj.emboj.7601570
66. HoWangYin KY, Alegre E, Daouya M, Favier B, Carosella ED, LeMaoult J. Different functional outcomes of intercellular membrane transfers to monocytes and T cells. Cell Mol Life Sci. (2010) 67:1133–45. doi: 10.1007/s00018-009-0239-4
67. Brown R, Kabani K, Favaloro J, Yang S, Ho PJ, Gibson J, et al. CD86+ or HLA-G+ can be transferred via trogocytosis from myeloma cells to T cells and are associated with poor prognosis. Blood (2012) 120:2055–63. doi: 10.1182/blood-2012-03-416792
68. LeMaoult J, Caumartin J, Daouya M, Switala M, Rebmann V, Arnulf B, et al. Trogocytic intercellular membrane exchanges among hematological tumors. J Hematol Oncol. (2015) 8:24. doi: 10.1186/s13045-015-0114-8
69. Chulpanova DS, Kitaeva KV, James V, Rizvanov AA, Solovyeva VV. Therapeutic prospects of extracellular vesicles in cancer treatment. Front Immunol. (2018) 9:1534. doi: 10.3389/fimmu.2018.01534
70. Riteau B, Faure F, Menier C, Viel S, Carosella ED, Amigorena S, Rouas-Freiss N. Exosomes bearing HLA-G are released by melanoma cells. Hum Immunol. (2003) 64:1064–72. doi: 10.1016/j.humimm.2003.08.344
71. Konig L, Kasimir-Bauer S, Hoffmann O, Bittner AK, Wagner B, Manvailer LF, et al. The prognostic impact of soluble and vesicular HLA-G and its relationship to circulating tumor cells in neoadjuvant treated breast cancer patients. Hum Immunol. (2016) 77:791–9. doi: 10.1016/j.humimm.2016.01.002
72. Grange C, Tapparo M, Tritta S, Deregibus MC, Battaglia A, Gontero P, et al. Role of HLA-G and extracellular vesicles in renal cancer stem cell-induced inhibition of dendritic cell differentiation. BMC Cancer (2015) 15:1009. doi: 10.1186/s12885-015-2025-z
73. Menter T, Tzankov A. Mechanisms of immune evasion and immune modulation by lymphoma cells. Front Oncol. (2018) 8:54. doi: 10.3389/fonc.2018.00054
74. Chowdhury PS, Chamoto K, Honjo T. Combination therapy strategies for improving PD-1 blockade efficacy: a new era in cancer immunotherapy. J Intern Med. (2018) 283:110–120. doi: 10.1111/joim.12708
75. Rouas-Freiss N, LeMaoult J, Verine J, Tronik-Le Roux D, Culine S, et al. Intratumor heterogeneity of immune checkpoints in primary renal cell cancer: focus on HLA-G/ILT2/ILT4. Oncoimmunology (2017) 6:e1342023. doi: 10.1080/2162402X.2017.1342023
76. Rodig SJ, Gusenleitner D, Jackson DG, Gjini E, Giobbie-Hurder A, Jin C, et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med. (2018) 10:eaar3342. doi: 10.1126/scitranslmed.aar3342
77. Patel SA, Minn AJ. Combination cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity (2018) 48:417–33. doi: 10.1016/j.immuni.2018.03.007
78. Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res. (2014) 2014:734068. doi: 10.1155/2014/734068
79. Seliger B. Role of microRNAs on HLA-G expression in human tumors. Hum Immunol. (2016) 77:760–3. doi: 10.1016/j.humimm.2016.04.006
80. Svendsen SG, Udsen MS, Daouya M, Funck T, Wu CL, Carosella ED, et al. Expression and differential regulation of HLA-G isoforms in the retinal pigment epithelial cell line, ARPE-19. Hum Immunol. (2017) 78:414–20. doi: 10.1016/j.humimm.2017.04.007
81. Carlini F, Picard C, Garulli C, Piquemal D, Roubertoux P, Chiaroni J, et al. Bronchial epithelial cells from asthmatic patients display less functional HLA-G isoform expression. Front Immunol. (2017) 8:6. doi: 10.3389/fimmu.2017.00006
82. Langat DK, Sue Platt J, Tawfik O, Fazleabas AT, Hunt JS. Differential expression of human leukocyte antigen-G (HLA-G) messenger RNAs and proteins in normal human prostate and prostatic adenocarcinoma. J Reprod Immunol. (2006) 71:75–86. doi: 10.1016/j.jri.2006.01.006
83. Ibrahim EC, Aractingi S, Allory Y, Borrini F, Dupuy A, Duvillard P, et al. Analysis of HLA antigen expression in benign and malignant melanocytic lesions reveals that upregulation of HLA-G expression correlates with malignant transformation, high inflammatory infiltration and HLA-A1 genotype. Int J Cancer (2004) 108:243–50. doi: 10.1002/ijc.11456
84. Paul P, Cabestre FA, Le Gal FA, Khalil-Daher I, Le Danff C, Schmid M, et al. Heterogeneity of HLA-G gene transcription and protein expression in malignant melanoma biopsies. Cancer Res. (1999) 59:1954–60.
85. Kleinberg L, Florenes VA, Skrede M, Dong HP, Nielsen S, McMaster MT, et al. Expression of HLA-G in malignant mesothelioma and clinically aggressive breast carcinoma. Virchows Arch. (2006) 449:31–9. doi: 10.1007/s00428-005-0144-7
86. Chen HX, Lin A, Shen CJ, Zhen R, Chen BG, Zhang X, et al. Upregulation of human leukocyte antigen-G expression and its clinical significance in ductal breast cancer. Hum Immunol. (2010) 71:892–8. doi: 10.1016/j.humimm.2010.06.009
87. He X, Dong DD, Yie SM, Yang H, Cao M, Ye SR, et al. HLA-G expression in human breast cancer: implications for diagnosis and prognosis, and effect on allocytotoxic lymphocyte response after hormone treatment in vitro. Ann Surg Oncol. (2010) 17:1459–69. doi: 10.1245/s10434-009-0891-9
88. de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, et al. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol. (2010) 185:7452–9. doi: 10.4049/jimmunol.1002629
89. da Silva GB, Silva TG, Duarte RA, Neto NL, Carrara HH, Donadi EA, et al. Expression of the classical and nonclassical HLA molecules in breast cancer. Int J Breast Cancer (2013) 2013:250435. doi: 10.1155/2013/250435
90. Ramos CS, Goncalves AS, Marinho LC, Gomes Avelino MA, Saddi VA, Lopes AC, et al. Analysis of HLA-G gene polymorphism and protein expression in invasive breast ductal carcinoma. Hum Immunol. (2014) 75:667–72. doi: 10.1016/j.humimm.2014.04.005
91. Ishibashi K, Kumai T, Ohkuri T, Kosaka A, Nagato T, Hirata Y, et al. Epigenetic modification augments the immunogenicity of human leukocyte antigen G serving as a tumor antigen for T cell-based immunotherapy. Oncoimmunology (2016) 5:e1169356. doi: 10.1080/2162402X.2016.1169356
92. Jeong S, Park S, Park BW, Park Y, Kwon OJ, Kim HS. Human leukocyte antigen-G (HLA-G) polymorphism and expression in breast cancer patients. PLoS ONE (2014) 9:e98284. doi: 10.1371/journal.pone.0098284
93. Singer G, Rebmann V, Chen YC, Liu HT, Ali SZ, Reinsberg J, et al. HLA-G is a potential tumor marker in malignant ascites. Clin Cancer Res. (2003) 9:4460–4.
94. Engels CC, Charehbili A, van de Velde CJ, Bastiaannet E, Sajet A, Putter H, et al. The prognostic and predictive value of Tregs and tumor immune subtypes in postmenopausal, hormone receptor-positive breast cancer patients treated with adjuvant endocrine therapy: a Dutch TEAM study analysis. Breast Cancer Res Treat. (2015) 149:587–96. doi: 10.1007/s10549-015-3269-7
95. Swets M, Konig MH, Zaalberg A, Dekker-Ensink NG, Gelderblom H, van de Velde CJ, et al. HLA-G and classical HLA class I expression in primary colorectal cancer and associated liver metastases. Hum Immunol. (2016) 77:773–9. doi: 10.1016/j.humimm.2016.03.001
96. Ye SR, Yang H, Li K, Dong DD, Lin XM, Yie SM. Human leukocyte antigen G expression: as a significant prognostic indicator for patients with colorectal cancer. Mod Pathol. (2007) 20:375–83. doi: 10.1038/modpathol.3800751
97. Guo ZY, Lv YG, Wang L, Shi SJ, Yang F, Zheng GX, et al. Predictive value of HLA-G and HLA-E in the prognosis of colorectal cancer patients. Cell Immunol. (2015) 293:10–6. doi: 10.1016/j.cellimm.2014.10.003
98. Zhang RL, Zhang X, Dong SS, Hu B, Han QY, Zhang JG, et al. Predictive value of different proportion of lesion HLA-G expression in colorectal cancer. Oncotarget (2017) 8:107441–51. doi: 10.18632/oncotarget.22487
99. Zeestraten EC, Reimers MS, Saadatmand S, Goossens-Beumer IJ, Dekker JW, Liefers GJ, et al. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br J Cancer (2014) 110:459–68. doi: 10.1038/bjc.2013.696
100. Reimers MS, Engels CC, Putter H, Morreau H, Liefers GJ, van de Velde CJ, et al. Prognostic value of HLA class I, HLA-E, HLA-G and Tregs in rectal cancer: a retrospective cohort study. BMC Cancer (2014) 14:486. doi: 10.1186/1471-2407-14-486
101. Miranda LN, Reginaldo FP, Souza DM, Soares CP, Silva TG, Rocha KB, et al. Greater expression of the human leukocyte antigen-G (HLA-G) and interleukin-17 (IL-17) in cervical intraepithelial neoplasia: analytical cross-sectional study. Sao Paulo Med J (2015) 133:336–42. doi: 10.1590/1516-3180.2013.7170009
102. Li XJ, Zhang X, Lin A, Ruan YY, Yan WH. Human leukocyte antigen-G (HLA-G) expression in cervical cancer lesions is associated with disease progression. Hum Immunol. (2012) 73:946–9. doi: 10.1016/j.humimm.2012.07.041
103. Guimaraes MC, Soares CP, Donadi EA, Derchain SF, Andrade LA, Silva TG, et al. Low expression of human histocompatibility soluble leukocyte antigen-G (HLA-G5) in invasive cervical cancer with and without metastasis, associated with papilloma virus (HPV). J Histochem Cytochem. (2010) 58:405–11. doi: 10.1369/jhc.2009.954131
104. Barrier BF, Kendall BS, Sharpe-Timms KL, Kost ER. Characterization of human leukocyte antigen-G (HLA-G) expression in endometrial adenocarcinoma. Gynecol Oncol. (2006) 103:25–30. doi: 10.1016/j.ygyno.2006.01.045
105. Bijen CB, Bantema-Joppe EJ, de Jong RA, Leffers N, Mourits MJ, Eggink HF, et al. The prognostic role of classical and nonclassical MHC class I expression in endometrial cancer. Int J Cancer (2010) 126:1417–27. doi: 10.1002/ijc.24852
106. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of HLA-G is associated with prognosis in esophageal squamous cell carcinoma. Am J Clin Pathol. (2007) 128:1002–9. doi: 10.1309/JNCW1QLDFB6AM9WE
107. Lin A, Zhang X, Zhou WJ, Ruan YY, Xu DP, Wang Q, et al. Human leukocyte antigen-G expression is associated with a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Cancer (2011) 129:1382–90. doi: 10.1002/ijc.25807
108. Zheng J, Xu C, Chu D, Zhang X, Li J, Ji G, et al. Human leukocyte antigen G is associated with esophageal squamous cell carcinoma progression and poor prognosis. Immunol Lett. (2014) 161:13–9. doi: 10.1016/j.imlet.2014.04.007
109. Spurny C, Kailayangiri S, Altvater B, Jamitzky S, Hartmann W, Wardelmann E, et al. T cell infiltration into Ewing sarcomas is associated with local expression of immune-inhibitory HLA-G. Oncotarget (2017) 9:6536–6549. doi: 10.18632/oncotarget.23815
110. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of human leukocyte antigen G (HLA-G) correlates with poor prognosis in gastric carcinoma. Ann Surg Oncol. (2007) 14:2721–9. doi: 10.1245/s10434-007-9464-y
111. Tuncel T, Karagoz B, Haholu A, Ozgun A, Emirzeoglu L, Bilgi O, et al. Immunoregulatory function of HLA-G in gastric cancer. Asian Pac J Cancer Prev. (2013) 14:7681–4. doi: 10.7314/APJCP.2013.14.12.7681
112. Du L, Xiao X, Wang C, Zhang X, Zheng N, Wang L, et al. Human leukocyte antigen-G is closely associated with tumor immune escape in gastric cancer by increasing local regulatory T cells. Cancer Sci. (2011) 102:1272–80. doi: 10.1111/j.1349-7006.2011.01951.x
113. Wastowski IJ, Simoes RT, Yaghi L, Donadi EA, Pancoto JT, Poras I, et al. Human leukocyte antigen-G is frequently expressed in glioblastoma and may be induced in vitro by combined 5-aza-2'-deoxycytidine and interferon-gamma treatments: results from a multicentric study. Am J Pathol. (2013) 182:540–52. doi: 10.1016/j.ajpath.2012.10.021
114. Cai MY, Xu YF, Qiu SJ, Ju MJ, Gao Q, Li YW, et al. Human leukocyte antigen-G protein expression is an unfavorable prognostic predictor of hepatocellular carcinoma following curative resection. Clin Cancer Res. (2009) 15:4686–93. doi: 10.1158/1078-0432.CCR-09-0463
115. Wang Y, Ye Z, Meng XQ, Zheng SS. Expression of HLA-G in patients with hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. (2011) 10:158–63. doi: 10.1016/S1499-3872(11)60025-8
116. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of human leucocyte antigen G (HLA-G) is associated with prognosis in non-small cell lung cancer. Lung Cancer (2007) 58:267–74. doi: 10.1016/j.lungcan.2007.06.011
117. Lin A, Zhu CC, Chen HX, Chen BF, Zhang X, Zhang JG, et al. Clinical relevance and functional implications for human leucocyte antigen-g expression in non-small-cell lung cancer. J Cell Mol Med. (2010) 14:2318–29. doi: 10.1111/j.1582-4934.2009.00858.x
118. Yan WH, Liu D, Lu HY, Li YY, Zhang X, Lin A. Significance of tumour cell HLA-G5/-G6 isoform expression in discrimination for adenocarcinoma from squamous cell carcinoma in lung cancer patients. J Cell Mol Med. (2015) 19:778–85. doi: 10.1111/jcmm.12400
119. Diepstra A, Poppema S, Boot M, Visser L, Nolte IM, Niens M, et al. HLA-G protein expression as a potential immune escape mechanism in classical Hodgkin's lymphoma. Tissue Antigens (2008) 71:219–26. doi: 10.1111/j.1399-0039.2008.01005.x
120. Caocci G, Greco M, Fanni D, Senes G, Littera R, Lai S, et al. HLA-G expression and role in advanced-stage classical Hodgkin lymphoma. Eur J Histochem. (2016) 60:2606. doi: 10.4081/ejh.2016.2606
121. Jesionek-Kupnicka D, Bojo M, Prochorec-Sobieszek M, Szumera-Cieckiewicz A, Jablonska J, Kalinka-Warzocha E, et al. HLA-G and MHC class II protein expression in diffuse large B-cell lymphoma. Arch Immunol Ther Exp (Warsz). (2016) 64:225–40. doi: 10.1007/s00005-015-0372-8
122. Urosevic M, Willers J, Mueller B, Kempf W, Burg G, Dummer R. HLA-G protein up-regulation in primary cutaneous lymphomas is associated with interleukin-10 expression in large cell T-cell lymphomas and indolent B-cell lymphomas. Blood (2002) 99:609–17. doi: 10.1182/blood.V99.2.609
123. Cai MB, Han HQ, Bei JX, Liu CC, Lei JJ, Cui Q, et al. Expression of human leukocyte antigen G is associated with prognosis in nasopharyngeal carcinoma. Int J Biol Sci. (2012) 8:891–900. doi: 10.7150/ijbs.4383
124. Goncalves AS, Wastowski IJ, Capeletti LR, Sacono NT, Cortez AP, Valadares MC, et al. The clinicopathologic significance of the expression of HLA-G in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. (2014) 117:361–8. doi: 10.1016/j.oooo.2013.12.001
125. Jung YW, Kim YT, Kim SW, Kim S, Kim JH, Cho NH, et al. Correlation of human leukocyte antigen-G (HLA-G) expression and disease progression in epithelial ovarian cancer. Reprod Sci. (2009) 16:1103–11. doi: 10.1177/1933719109342131
126. Menier C, Prevot S, Carosella ED, Rouas-Freiss N. Human leukocyte antigen-G is expressed in advanced-stage ovarian carcinoma of high-grade histology. Hum Immunol. (2009) 70:1006–9. doi: 10.1016/j.humimm.2009.07.021
127. Zhang X, Han QY, Li JB, Ruan YY, Yan WH, Lin A. Lesion HLA-G5/-G6 isoforms expression in patients with ovarian cancer. Hum Immunol. (2016) 77:780–4. doi: 10.1016/j.humimm.2015.12.003
128. Xu YF, Lu Y, Cheng H, Jiang J, Xu J, Long J, et al. High expression of human leukocyte antigen-g is associated with a poor prognosis in patients with PDAC. Curr Mol Med. (2015) 15:360–7. doi: 10.2174/1566524015666150401102218
129. Khodabandeh Shahraki P, Zare Y, Azarpira N, Hosseinzadeh M, Farjadian S. Prognostic value of HLA-G in malignant liver and pancreas lesions. Iran J Immunol. (2018) 15:28–37.
130. Zhou L, Niu ZY, Liang ZY, Zhou WX, You L, Wang MY, et al. HLA-G impairs host immune response and predicts poor prognosis in pancreatic cancer. Am J Transl Res. (2015) 7:2036–44.
131. de Figueiredo Feitosa NL, Crispim JC, Zanetti BR, Magalhaes PK, Soares CP, Soares EG, et al. HLA-G is differentially expressed in thyroid tissues. Thyroid (2014) 24:585–92. doi: 10.1089/thy.2013.0246
132. Nunes LM, Ayres FM, Francescantonio IC, Saddi VA, Avelino MA, Alencar Rde C, et al. Association between the HLA-G molecule and lymph node metastasis in papillary thyroid cancer. Hum Immunol. (2013) 74:447–51. doi: 10.1016/j.humimm.2012.12.012
133. Anastassiou G, Rebmann V, Wagner S, Bornfeld N, Grosse-Wilde H. Expression of classic and nonclassic HLA class I antigens in uveal melanoma. Invest Ophthalmol Vis Sci. (2003) 44:2016–9. doi: 10.1167/iovs.02-0810
134. Goldman-Wohl D, Ariel I, Greenfield C, Hochner-Celnikier D, Lavy Y, Yagel S. A study of human leukocyte antigen G expression in hydatidiform moles. Am J Obstet Gynecol. (2001) 185:476–80. doi: 10.1067/mob.2001.115994
135. Moreau P, Dausset J, Carosella ED, Rouas-Freiss N. Viewpoint on the functionality of the human leukocyte antigen-G null allele at the fetal-maternal interface. Biol Reprod. (2002) 67:1375–8. doi: 10.1095/biolreprod.102.005439
136. Le Discorde M, Le Danff C, Moreau P, Rouas-Freiss N, Carosella ED. HLA-G*0105N null allele encodes functional HLA-G isoforms. Biol Reprod. (2005) 73:280–8. doi: 10.1095/biolreprod.104.037986
137. Sala FG, Del Moral PM, Pizzato N, Legrand-Abravanel F, Le Bouteiller P, Lenfant F. The HLA-G*0105N null allele induces cell surface expression of HLA-E molecule and promotes CD94/NKG2A-mediated recognition in JAR choriocarcinoma cell line. Immunogenetics (2004) 56:617–24. doi: 10.1007/s00251-004-0733-7
138. Faucher MC, Louvanto K, Syrjanen S, Roger M. Characterisation of the novel HLA-G null allele, HLA-G*01:21N, in Finnish individuals. HLA (2018) 91:146–7. doi: 10.1111/tan.13177
139. Ferns DM, Heeren AM, Samuels S, Bleeker MCG, de Gruijl TD, Kenter GG, et al. Classical and non-classical HLA class I aberrations in primary cervical squamous- and adenocarcinomas and paired lymph node metastases. J Immunother Cancer (2016) 4:78. doi: 10.1186/s40425-016-0184-3
140. Concha-Benavente F, Srivastava R, Ferrone S, Ferris RL. Immunological and clinical significance of HLA class I antigen processing machinery component defects in malignant cells. Oral Oncol. (2016) 58:52–8. doi: 10.1016/j.oraloncology.2016.05.008
141. Bukur J, Jasinski S, Seliger B. The role of classical and non-classical HLA class I antigens in human tumors. Semin Cancer Biol. (2012) 22:350–8. doi: 10.1016/j.semcancer.2012.03.003
142. Ugurel S, Rebmann V, Ferrone S, Tilgen W, Grosse-Wilde H, Reinhold U. Soluble human leukocyte antigen–G serum level is elevated in melanoma patients and is further increased by interferon-α immunotherapy. Cancer (2001) 92:369–76. doi: 10.1002/1097-0142(20010715)92:2<369:AID-CNCR1332>3.0.CO;2-U
143. Testa U, Pelosi E, Castelli G. Colorectal cancer: genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Med Sci. (2018) 6:31. doi: 10.3390/medsci6020031
144. Aptsiauri N, Ruiz-Cabello F, Garrido F. The transition from HLA-I positive to HLA-I negative primary tumors: the road to escape from T-cell responses. Curr Opin Immunol. (2018) 51:123–32. doi: 10.1016/j.coi.2018.03.006
Keywords: HLA-G, heterogeneity, cancer, immune evasion, therapy target
Citation: Lin A and Yan W-H (2018) Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front. Immunol. 9:2164. doi: 10.3389/fimmu.2018.02164
Received: 10 May 2018; Accepted: 03 September 2018;
Published: 27 September 2018.
Edited by:
Fang-Ping Huang, University of Hong Kong, Hong KongReviewed by:
Alessandro Poggi, Ospedale Policlinico San Martino, ItalyManel Juan, Hospital Clínic de Barcelona, Spain
Copyright © 2018 Lin and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Hua Yan, eWFud2hjb21AeWFob28uY29t
 Aifen Lin1
Aifen Lin1 Wei-Hua Yan
Wei-Hua Yan