- 1Department of Basic Veterinary, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Wuhan, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Sichuan Agricultural University, Chengdu, China
- 4Department of Aquaculture, College of Animal Science & Technology, Sichuan Agricultural University, Chengdu, China
Outer membrane porins, as the major components of Gram-negative bacterial membrane proteins, have been proven to be involved in interactions with the host immune system and potent protective antigen candidates against bacterial infection in fish. Outer membrane porin F (OmpF) is one of the major porins of Yersinia ruckeri (Y. ruckeri), the causative agent of enteric red mouth disease of salmonid and non-salmonid fish. In the present study, the molecular characterization and phylogenetic analysis of OmpF gene was studied, heterogenous expression, immunogenicity and protective immunity of OmpF were systemically evaluated as a subunit vaccine for channel catfish against Y. ruckeri infection. The results showed that OmpF gene was highly conserved among 15 known Yersinia species based on the analysis of conserved motifs, sequences alignment and phylogenetic tree, and was subjected to negative/purifying selection with global dN/dS ratios value of 0.649 throughout the evolution. Besides, OmpF was also identified to have immunogenicity by western blotting and was verified to be located on the surface of Y. ruckeri using cell surface staining and indirect immunofluorescence assays. Moreover, recombinant OmpF (rtOmpF) as a subunit vaccine was injected with commercial adjuvant ISA763, significantly enhanced the immune response by increasing serum antibody levels, lysozyme activity, complement C3 activity, total protein content, SOD activity, immune-related genes expression in the head kidney and spleen, and survival percent of channel catfish against Y. ruckeri infection. Thus, our present results not only enriched the information of molecular characterization and phylogenetics of OmpF, but also demonstrated that OmpF holds promise to be used as a potential antigen against Y. ruckeri infection in fish.
Introduction
Yersinia ruckeri (Y. ruckeri) is a Gram-negative rod-shaped enterobacterium and the causative agent of enteric red mouth disease (ERM), one of the most serious septicemic bacterial disease of salmonid fish species (1). It has been reported that Y. ruckeri has been increasingly widespread and been detected as an important pathogen of salmonid fish in many other countries (2–6) since its isolation in North American (7–10). Apart from salmonids, Y. ruckeri can also infect other non-salmonid fish species including common carp (11), whitefish (12), sturgeon (13–15), and channel catfish (16, 17). Alternative approaches to traditional control strategies include probiotics and vaccines, which may play greater significance in disease control due to the increasing antibiotic resistance of bacteria (18). Although vaccines against ERM have been widely used for more than 30 years, most of these vaccines are generally inactivated whole-cell vaccines (19–22) and live-attenuated vaccines (23), which have led to selective pressure leading to emergence of other serotypes (18). Moreover, concerns about the environmental safety restricted the commercial use of such live attenuated vaccines (18). Thus, genetically engineered vaccines based on conserved and potent protective antigen genes, are increasingly urgent and need to be developed.
Outer membrane proteins (OMPs) are the major components of Gram-negative bacterial membranes and essential in maintaining the integrity and selective permeability of the outer membrane (24). As one of the membrane surface molecules, OMPs are considered as the major targets of the membrane-environment interaction and easily recognized by the infected host compared with intracellular proteins (25). Bacterial porins, one of the most abundant OMPs (26), are the main channels for many hydrophilic nutrients and antibiotics (27), and are also involved in interactions with the host immune system due to their exposed antigen epitopes on bacterial surface (28). Many studies have reported that OMPs hold promise to serve as vaccine candidate and offer significant protection against bacterial infection in fish (29–39), including OmpA (31, 32), OmpC (33), OmpK (34), OmpN (35), OmpTS (36), OmpU (37), and OmpW (38, 39). OmpF is one of the major porins of Enterobacteriaceae, and has been reported to be the protective antigen and to provide desirable immunoprotection against pathogenic Escherichia coli (40) and Salmonella enterica (41). Besides, based on the perspective of structure and evolution, OmpF porin gene in genus Yersinia was comparably conserved in structure and homology and had putative antigenic epitopes located on several loops (42), indicating that it could be used as candidate protective antigen against Y. ruckeri infection.
Thus, in the present study, the molecular characterization and phylogenetic analysis of Y. ruckeri OmpF gene was studied, heterogenous expression was conducted to serve as a candidate immunogen, the immunogenicity and protective immunity of OmpF were also systemically evaluated as a subunit vaccine against Y. ruckeri infection in channel catfish, which was an excellent biological model for comparative immunology research in teleosts (43–45). Based on the results of this study, OmpF gene was inferred to be a novel protective antigen of Y. ruckeri and recombinant OmpF (rtOmpF) was a promising vaccine candidate for channel catfish against Y. ruckeri infection.
Materials and Methods
Ethics Statement
The biosafety procedures of recombinant DNA technology and the use of laboratory animals in this study were carried out in strict accordance with the guidelines and recommendations of Chinese National Institute of Health. All the procedures of recombinant DNA technology and animal experiments were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University (No. XF201418).
Bacterial Strains, Plasmids, Reagents, and Growth Conditions
Y. ruckeri YRWEL01, a fish pathogen isolated from dying channel catfish in Sichuan province of China, was cultured in Brain-Heart Infusion (BHI) medium at 28°C and stored at our laboratory (17). Escherichia coli strains DH5α and BL21 (DE3) competent cells (Takara; Dalian, China) served as cloning and protein expression host, respectively. Both strains were grown in Luria-Bertani medium containing 100 μg/ml of ampicillin (Amp) at 37°C. Plasmids pMD19-T (Takara) and pET32a (+) (Merck, Germany) served as cloning and expression vectors, respectively. Montanide™ ISA763 A VG (Seppic, France) was selected for use as an adjuvant for the experiment.
PCR Amplification and Molecular Cloning of OmpF
Y. ruckeri genomic DNA was extracted using a TIANamp Bacteria DNA extraction Kit (Tiangen, Beijing, China). The primers OmpF-F1/ OmpF-R1 of the target gene were designed using the Primer Premier 5.0 software based on the Y. ruckeri strain Nr34/85 OmpF gene sequence deposited in GenBank database (HM142671.1, corresponding OmpF protein accession no.: ADK27779.1). OmpF gene was amplified by PCR under the following conditions: 1 cycle of 94°C for 5 min, 30 cycles of 94°C for 1 min, 56°C for 30 s, and 72°C for 90 s, followed by a final extension of 72°C for 10 min. The product of PCR amplification was expected to be about 1098 bp and purified using the Agarose Gel DNA Extraction Kit (TaKaRa), ligated with the pMD19-T using T4 DNA ligase (TaKaRa) and transformed into E. coli DH5α competent cells. The positive recombinant clones were selected on the Amp/LB plate. The recombinant plasmid was identified by PCR under the aforementioned conditions, digested with restriction enzymes NcoI and SacI, and fractionated on 1% agarose gels. DNA sequencing was conducted by TaKaRa Bio Inc. and the sequence was deposited in the NCBI GenBank to obtain accession number. The correct recombinant cloning plasmids were named as T-OmpF.
Sequence and Phylogenetic Analysis of OmpF
The opening reading frame of OmpF nucleotide sequence was analyzed using ORF Finder (46). The OmpF amino acid sequences were derived from the nucleotide sequence. The signal peptide was predicted by SignalP 4.1 Server (47). To delineate the evolutionary dynamics of bacteria OmpF gene, the conserved domains and conserved motifs of OmpF in this study and other 25 reference bacteria OmpFs were searched using the Conserved Domain Database (CDD) in NCBI (48) and MEME software (49), respectively, their amino acid sequences identity were calculated using MegAlign program (DNASTAR, Madison, WI) (50), multiple sequences alignment was performed using MUSCLE (51), the phylogenetic tree was constructed using neighbor-joining method in MEGA 5 (52) with bootstrap test of 1000 replicates. To measure the selection pressures imposed on OmpF gene, the natural selection analysis was conducted based on the dN/dS ratios (the relative rates of non-synonymous (dN) and synonymous (dS) substitutions) which was calculated using Datamonkey (53).
Cloning, Expression, Purification, and Refolding of Truncate OmpF (tOmpF)
Based on the sequence analysis, the signal peptide was removed to obtain the mature OmpF by prokaryotic expression with E. coli BL21 (DE3). The truncated OmpF (tOmpF) gene was amplified using primers OmpF-F2/ OmpF-R2 (Table 1), which contained NcoI and SacI restriction enzyme sites. The resultant amplicons were purified, ligated, transformed, and sequenced as described above, and the positive recombinant cloning plasmid was named as T-tOmpF.
The expression, purification, and refolding of recombinant tOmpF protein were performed as described in our previous studies (54, 55). Briefly, the cloned plasmid T-tOmpF was digested with NcoI and SacI, and the resultant products and the NcoI /SacI -digested pET32a (+) were ligated to construct the recombinant expression plasmid, named as P-tOmpF. Then, the plasmid P-tOmpF was transformed into E. coli BL21 and induced using 1.0 mM IPTG at 37°C for 4 h. Bacterial cells were harvested and resuspended with sterile phosphate buffer saline (PBS), followed by ultrasonication, and detection using 12.5% SDS-PAGE. The purification of recombinant tOmpF (rtOmpF), which was expressed in the form of inclusion bodies in the sediment was conducted using Ni-NTA-Sefinose Column (Sangon Biotech, Shanghai, China). The refolded tOmpF protein was obtained by gradient dialysis and analyzed using 12.5% SDS-PAGE. To rule out the potential bystander effects of LPS/or other impurities, endotoxin in recombinant protein was removed using ToxinEraserTM Endotoxin Removal kit (GenScript Corp. Nanjing, China), and the remaining endotoxin levels were measured using the Chromogenic End-point Endotoxin Assay kit (Limulus reagent biotechnology, Xiamen, China). Less than 0.1 EU/ml was detected in the final protein preparations. The protein was quantified using a NanoDrop spectrophotometer (Thermo Scientific) according to the manufacturer's instructions. Purified protein rtOmpF was stored at −20°C until further use.
Preparation of Rabbit Anti-Y. ruckeri and Anti-rtOmpF Antisera
Rabbit anti-Y. ruckeri and anti-rtOmpF antisera were prepared according to the method described previously (56) using formaldehyde-killed Y. ruckeri (3.0 × 109 CFU/ml) (57) and purified protein rtOmpF as the antigens, respectively. Briefly, New Zealand white rabbits were divided into three groups, one control group and two experimental groups. The purified rtOmpF (2 mg/ml) used as the antigen was emulsified with an equal volume of Freund's Complete Adjuvant (FCA, Sigma, USA) and injected intravenously into rabbits, followed by three intravenous booster injections of rtOmpF-FIA (Freund's Incomplete Adjuvant, Sigma) at 1-week intervals. Similarly, the rabbits in other two groups were immunized on the same day with PBS (control group) and formaldehyde-killed Y. ruckeri, respectively. After the last injection, blood was sampled from the rabbits in all three groups and centrifuged to obtain the antisera at 3,000 × g for 15 min. The antisera (Immunoglobulin G, IgG) were purified by the ammonium sulfate precipitation method (58) and stored at −20°C until required.
Western Blotting
The western blotting analysis of recombinant proteins was carried out as previously described with slight modifications (54, 55). Briefly, the purified proteins were separated using 12.5% SDS-PAGE and transferred onto two PVDF membranes at 150 V for 2 h. The membranes were pre-blocked with TBST containing 3% Bovine Serum Albumin (BSA, Sangon Biotech) for 1 h at 37°C, then incubated with rabbit anti-6 × His antibody (1:1000, Sangon Biotech) and rabbit anti-Y. ruckeri antisera (1:200) respectively for 12 h at 4°C. After washing three times with TBST, the membranes were incubated with goat-anti-rabbit IgG-HRP (1:5000, Sangon Biotech) for 1h at 37°C. The reaction was visualized using DAB (Sigma) for 5 to 15 min, and terminated by rinsing with distilled water.
To verify the cross-protection of OmpF in Yersiniaceae species, Y. ruckeri YRWEL01 (used in this study), Yersinia enterocolitica, and Yersinia pestis were cultured overnight and homogenized as protein sources for Western blotting. They were incubated with rabbit anti-rtOmpF sera.
Surface Display of Y. ruckeri OmpF
Cell Surface Staining of Bacteria
The OmpF protein on the surface of Y. ruckeri was detected and verified using the cell surface staining method described previously (56). Briefly, Y. ruckeri was distributed uniformly on poly-L-lysine-treated slides (Boster, Wuhan, China) after culturing overnight on BHI agar plates. After air drying, flame fixation, and fixation in 100% methanol for 10 min at −20°C, Y. ruckeri coated on the slides were incubated with rabbit anti-rtOmpF sera (1:200), anti-Y. ruckeri sera (positive control), and rabbit negative sera (PBS group, negative control) respectively for 1 h at 37°C. After washing three times with PBST, goat anti-rabbit IgG-HRP (1:5000) was applied and incubated for 1 h at 37°C. The reaction wase visualized using DAB for 5 to 15 min and stopped by rinsing with distilled water. Then the slides were covered by cover glasses and observed using a Nikon microscope (Japan) at a × 1000 magnification.
Indirect Immunofluorescence
To verify the surface localization of OmpF protein on Y. ruckeri, indirect immunofluorescence assay was also carried out as described previously with minor modification (59). Briefly, Y. ruckeri was distributed uniformly on poly-L-lysine-treated slides (Boster, Wuhan, China) after culturing overnight on BHI agar plates. After air drying, flame fixation, and fixation in 100% methanol for 10 min at −20°C, Y. ruckeri coated on the slides were incubated with rabbit anti-rtOmpF sera (1:200), anti-Y. ruckeri sera (positive control), and rabbit negative sera (negative control) respectively for 1 h at 37°C. After washing three times with PBS, each slide was incubated with fluorescein isothiocyanate (FITC) conjugated goat anti rabbit IgG-FITC (1:64, Sangon Biotech) for 1 h at 37°C. The reaction was stopped by rinsing with PBS and cover glasses were covered in a solution containing glycerol and PBS (1:1). The slides were observed using a fluorescence microscope (Nikon Eclipse 80i).
Preparation of Fish and Vaccines
Channel catfish (50.0 ± 5.0 g) were purchased from a fish farm in Chengdu (Sichuan, China) and acclimatized in the laboratory for 2 weeks at 28 ± 1°C before any experimental manipulation. Fish were fed a commercial diet daily and water was partially replaced every day. Before the experiments, blood and tissues including liver, kidney, and spleen were sampled to detect the bacteria. No bacteria was recovered and agglutination tests showed no reaction between the serum and Y. ruckeri. Fish were anesthetized using MS-222 (Sigma) before any experimental manipulation. The recombinant antigen rtOmpF was diluted in PBS to obtain appropriate concentration. To obtain PBS+ ISA763 and rtOmpF+ISA763, the PBS and rtOmpF were emulsified respectively with commercial adjuvant Montanide™ ISA763 at a ratio of 3:7 by ultrasonic disruption (JY92-IIDN, Ningbo Scientz, China). The effective concentration of rtOmpF injected into fish was set as 1.0 mg/ml, which was determined using BCA Protein Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), according to the manufacturer's description.
Vaccination and Bacterial Challenge
Healthy channel catfish were divided randomly into four groups (100 fish /group) including one control group and three test groups. The vaccination protocol was carried out as described previously with some modification (55). Briefly, fish were injected intraperitoneally (i.p.) with 0.2 ml of PBS (control group), PBS+ ISA763, rtOmpF, and rtOmpF+ISA763 respectively, i.e., effective dose of rtOmpF was 4 μg/g fish, which was determined to be an optimal dose in our preliminary experiments. To obtain an optimal immune response, booster vaccination were conducted with the same method and dosage of first vaccination at 2 weeks later. At 4 weeks post-secondary vaccination (psv), 30 fish from each group were randomly selected and challenged by i.p. injection with 0.2 ml of Y. ruckeri YRWEL01 which was resuspended in PBS to 3.5 × 108 CFU/ml (57). Mortality was monitored over a period of 14 days after the challenge, and dying fish were randomly selected for the examination of bacterial recovery from liver, kidney, and spleen. Relative percent of survival (RPS) was calculated according to the following formula: RPS = [1– (% mortality of vaccinated fish/% mortality of control fish)] × 100 (60). Serum samples of five fish in each group were collected for the detection of immune related indexes at 1-8 week psv. Head-kidney and spleen tissues of five fish were taken for qRT-PCR analysis at 24 h post-challenge. Vaccination experiments were performed in duplicate.
Enzyme-Linked Immunosorbent Assay (ELISA)
Sera were collected from the caudal vein of vaccinated fish at 1–8 weeks psv to detect the specific antibody against rtOmpF by ELISA as described perviously (54). Briefly, rtOmpF was diluted to 50 μg/ml in a carbonate buffer (pH = 9.6). Each well of 96-well microplate was coated using 100 μL diluted rtOmpF overnight at 4°C, followed by washing three times with PBST (0.1% Tween-20 in PBS), and then blocking with 3% BSA in PBST for 2 h at 37°C. The sera were added into the wells in triplicate and subsequently incubated for 2 h at 37°C. Rabbit anti-channel catfish IgM antiserum (1:200, prepared in our laboratory) and goat-anti-rabbit IgG-HRP (1:2000) were used as primary and secondary antibodies, respectively. The reaction was visualized using the TMB kit (Tiangen, Beijing, China) and terminated with 2 M H2SO4. The absorbance was measured at 450 nm with a microplate reader (Bio-Rad, Hercules, USA).
Measurement of Innate Immune Parameters
The serum lysozyme activity, complement C3 activity, total protein content, and superoxide dismutase (SOD) activity were measured at 1–8 week psv to evaluate the innate immune responses using the commercial kits (lysozyme kit Cat. No: A050-01; complement C3 kit Cat. No: E032; total protein kit Cat. No: A045-3; SOD kit Cat. No: A001-01) according to the manufacturer's instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The absorbance of lysozyme, complement C3, total protein, and SOD were measured at 530, 340, 562, and 550 nm, respectively, under a microtiter plate reader (Thermo, Varioskan Flash, USA).
qRT-PCR Analysis of Immune-Related Genes Expression
Head kidney and spleen were taken at 24 h post-challenge. Total RNA extraction and cDNA synthesis were performed as described in our previous study (54). qRT-PCR was carried out using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa) in an ABI StepOnePlus™ System (Applied Biosystems, USA) as described previously (54). Each assay was performed in triplicate, two housekeeping genes 18S ribosomal RNA (18S) and elongation factor-1 alpha (EF1α) were used as internal control genes (reference genes). The primers used to amplify reference genes and immune-related genes were shown in Table 1. The relative expression levels of these genes were analyzed by the 2−ΔΔCT method with the geometric mean of the expression levels of 18S and EF1α. All data are presented as relative mRNA expression.
Statistical Analysis
Statistical analysis was performed using SPSS 19.0 software (SPSS Inc., USA). Survival data of bacterial challenge experiment were analyzed by the Kaplan-Meier methods and log-rank tests. The data differences among groups were detected using a one-way analysis of variance (ANOVA). In all cases, the significance level was defined as P < 0.05 and the results were presented as mean ± SE (standard error).
Results
Molecular Characterization and Sequence Analysis of OmpF
The coding sequence length of Y. ruckeri YRWEL01 OmpF gene was 1095 bp (GenBank accession No: KP159420) (Figures 1A,B), which was 3 bp shorter than reference OmpF gene (Accession No.: HM142671.1). The OmpF gene in this study contained a complete open reading frame and encoded 364 amino acid (a.a.), which was predicted to contain a 21 a.a. signal peptide (1-21 a.a.) and a 351 a.a. OM_channels superfamily conserved domain (14-364 a.a.) (Figures 1C,D). Comparing with reference OmpF amino acid sequence (Accession No.: ADK27779.1), although there were 3 a.a. differences between them, both possessed 39 trimer interface polypeptide binding sites and 5 channel eyelet sites (Figure 1C).
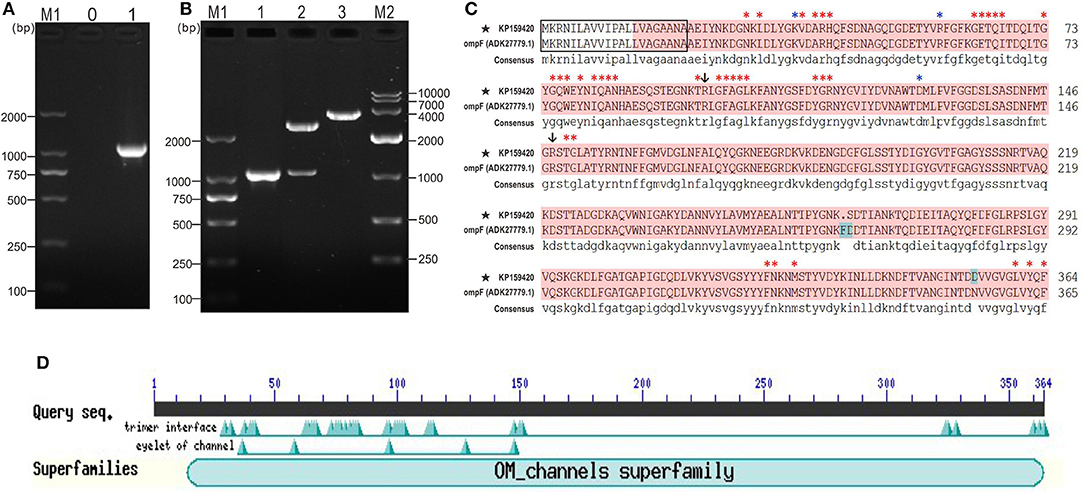
Figure 1. PCR, cloning, alignment and conserved domains analyses of OmpF. (A) PCR amplification of OmpF gene. M1: DNA marker (DL2000); lane 0: negative control; lane 1: PCR product of OmpF gene with 1095 bp. (B) Identification of cloning plasmid T-OmpF. M1: DNA marker (DL2000); lane 1: PCR identification of T-OmpF; lane 2: digestion of T-OmpF with NcoI and SacI; lane 3: digestion of T-OmpF with SacI [A and B are Figures 1A,B from Wang et al. (61) reproduced with permission from South China Fisheries Science]. (C) Amino acid sequence alignment of OmpF in this study with reference Y. ruckeri OmpF deposited in NCBI (ADK27779.1), black box presented the signal peptide sequences (1–22 a.a.), red shade regions presented the OM_channels superfamily conserved domain, red “*” indicated the trimer interface polypeptide binding sites, blue “*” indicated the eyelet of channel, “↓” indicated both trimer interface polypeptide binding sites and eyelet of channel. (D) Conserved domains of OmpF.
Phylogenetic Analysis of OmpF
Figure 2 presented the logos of top five conserved motifs with length ranging from five to fifty (Figure 2A), as well as their corresponding locations in bacteria OmpF proteins (Figure 2C). All these reference bacteria OmpFs possessed all these five conserved motifs (Figure 2C) which constituted about 60% of the OmpF length, indicating OmpF remained comparatively conserved in different bacteria species. Besides, the two-dimensional topology structures of Y. ruckeri OmpF showed that OmpF located extracellular without any transmembrane regions and the signal peptide sequences located into the Motif 1 domain (Figure 2B).

Figure 2. Conserved motifs analysis of OmpF with other 25 reference OmpFs. (A) The logos of top five conserved motifs with length ranging from five to fifty in OmpF amino acid sequences. (B) The distribution of these five motifs in the two-dimensional topology structures of OmpF (used OmpF in this study as an example). (C) The location of these five motifs in bacteria OmpFs. Red shades presented Y. ruckeri, blue shades presented other Yersiniaceae species, dotted box presented other Enterobacteriaceae species.
The results of amino acid sequences identity indicated that OmpF in this study shared 99.2% identity with Y. ruckeri strain Nr34/85 OmpF, shared relative high identity (80.8–86.8%) with other 14 Yersiniaceae species OmpFs and relative low identity (58.8–76.4%) with other Enterobacteriaceae species OmpFs (Figure 3A). The phylogenetic analysis showed that OmpF in this study clustered one clade with Y. ruckeri OmpF with the bootstrap values 100, displayed a closer relationship with other bacteria of Yersiniaceae family and a distant relationship with other Enterobacteriaceae bacteria (Figure 3B). In addition, the result of natural selection pressure analysis indicated that the global dN/dS ratios of bacteria OmpFs was 0.649 with 13 positive/diversifying selection sites and 53 negative/purifying selection sites, which was well below 1.0, a theoretical boundary for positive and negative selection.
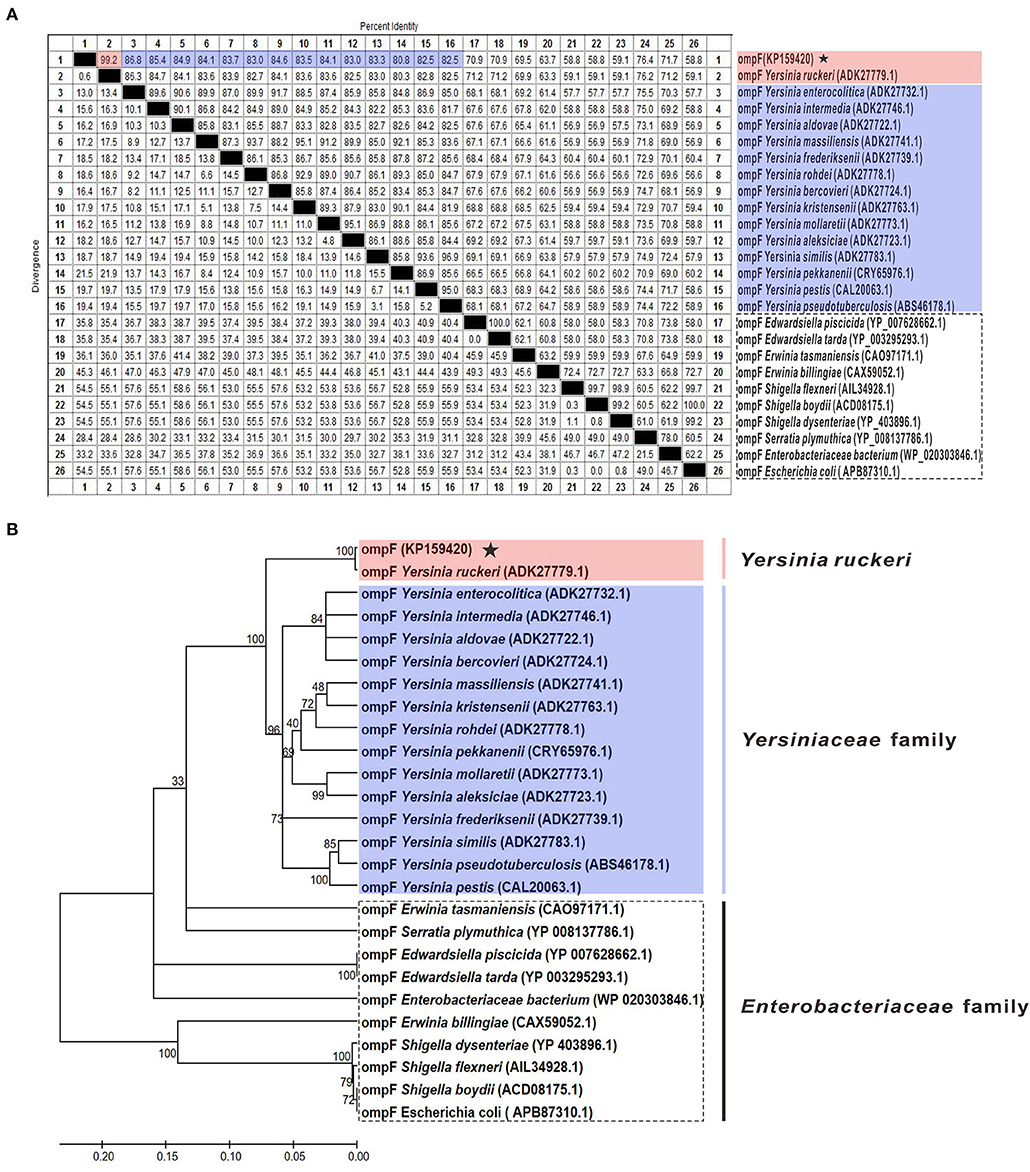
Figure 3. Multiple sequences alignment and phylogenetic tree of OmpF with other 25 reference OmpFs. (A) Multiple sequences alignment of OmpF with other 25 reference OmpFs. Red shades indicated OmpF in this study was consistent with Y. ruckeri OmpF with 99.2% identities, blue shades indicated other Yersiniaceae species OmpFs, dotted box presented other Enterobacteriaceae species. (B) Phylogenetic tree of OmpF with other 25 reference OmpFs. The numbers at each branch represent the bootstrap values obtained with 1000 replicates. “KP159420” represent the OmpF in this study.
Expression, Purification and Western Blotting Analysis of Recombinant tOmpF
To obtain the mature peptide of Y. ruckeri OmpF, the truncated OmpF (~1032 bp) removing the signal peptide sequence was successfully amplified using PCR and cloned into pMD19-T to obtain recombinant cloned plasmid T-tOmpF (Figures 4A, B), followed by constructing recombinant expression plasmid P-tOmpF (Figure 4C) using vector pET32a, expectedly expressed in induced E. coli BL21 (P-tOmpF) sediment and purifying using Ni-NTA metal affinity chromatography. After expression, purification, and refolding, recombinant proteins rtOmpF with about 55 kDa were observed using SDS-PAGE (Figure 4D). Western blotting analysis indicated that the 55 kDa band of rtOmpF specifically reacted with the rabbit anti-6 × His antisera (Figure 4E) and rabbit anti-Y. ruckeri antisera, respectively (Figure 4F). Furthermore, the proteins from Y. ruckeri YRWEL01, Y. enterocolitica, and Y. pestis were also detected by anti-rtOmpF sera with about 38 kDa (Figure 4G). Thus, we infer that rtOmpF may confer cross-protection in Yersiniaceae species.
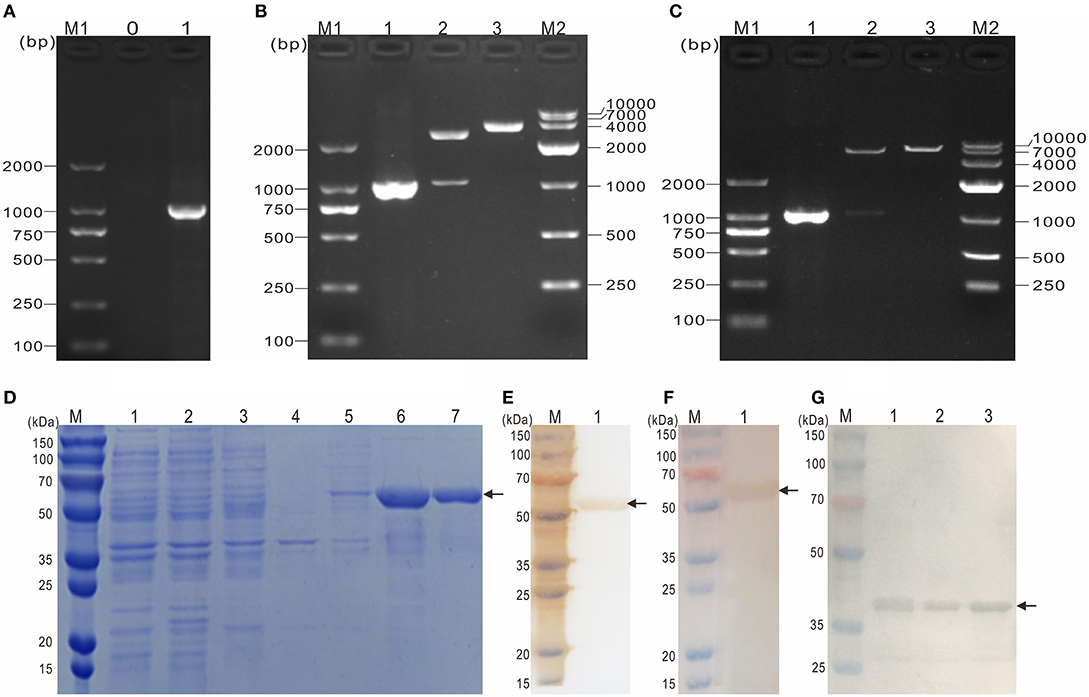
Figure 4. Molecular cloning, expression, purification and western blotting analysis of rtOmpF. (A) PCR amplification of tOmpF gene. M1: DNA Marker (DL2000); lane 0: negative control; lane 1: PCR product of tOmpF gene with 1032 bp. (B) Identification of recombinant cloning plasmid T-tOmpF. M1: DNA Marker (DL2000); lane 1: PCR product of T-tOmpF; lane 2: digestion of T-tOmpF with NcoI and SacI; lane 3: digestion of T-tOmpF with SacI; M2: DNA Marker (DL10000). (C) Identification of recombinant expression plasmid P-tOmpF. M1: DNA Marker (DL2000); lane 1: PCR product of P-tOmpF; lane 2: digestion of P-tOmpF with NcoI and SacI; lane 3: digestion of P-tOmpF with SacI; M2: DNA Marker (DL10000) [C is Figure 1C from Wang et al. (61) reproduced with permission from South China Fisheries Science]. (D) SDS-PAGE analysis of recombinant protein rtOmpF. M: protein marker; lane 1~6: uninduced BL21 (pET32a), induced BL21 (pET32a), uninduced BL21 (P-tOmpF) supernatant, uninduced BL21 (P-tOmpF) sediment, induced BL21 (P-tOmpF) supernatant, induced BL21 (P-tOmpF) sediment, lane 7: purified rtOmpF. (E) Western blotting analysis of rtOmpF with rabbit anti-6 × His antisera. M: protein marker; lane 1: specific binding between rtOmpF and rabbit anti-6 × His antisera. (F) Western blotting analysis of rtOmpF with rabbit anti-Y. ruckeri antisera. M: protein marker; lane 1: specific binding between rtOmpF and rabbit anti-Y. ruckeri antisera [D, E and F are Figure 10 from Wang et al. (61) reproduced with permission from South China Fisheries Science]. (G) The cross-protection of OmpF in Yersiniaceae species was analyzed by Western blotting with rabbit anti-rtOmpF sera. M: protein marker; lane 1~3: Y. ruckeri YRWEL01, Y. enterocolitica and Y. pestis, respectively.
Surface Display of Y. ruckeri OmpF
The rabbit anti-Y. ruckeri antisera and rabbit negative antisera were employed as positive and negative controls, respectively. After staining with DAB, the target bacteria was recognized by rabbit anti-rtOmpF antisera and showed in brown color, which was consistent with the result of positive control, while the negative control showed no color (Figure 5A). Similarly, in indirect immunofluorescence assay, the target bacteria incubated with anti-rtOmpF antisera showed fluorescence signal, although the signal was a little weak compared with that in positive control, and the negative control showed dark without fluorescence signal (Figure 5B).
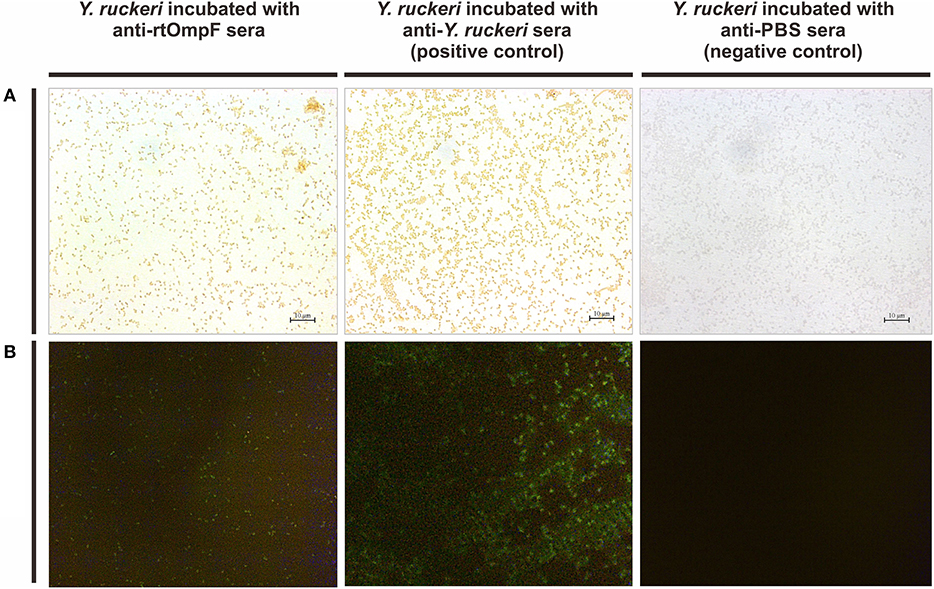
Figure 5. Detection of OmpF localization using bacteria cell surface staining and indirect immunofluorescence. (A) Cell surface staining of Y. ruckeri. Y. ruckeri incubated with rabbit negative antisera (negative control) showed no color. (B) Indirect immunofluorescence assay. Y. ruckeri incubated with rabbit negative antisera (negative control) showed no fluorescence signal.
Serum Antibody Production
Serum specific-antibody was detected continuously using ELISA from 1st to 8th week psv (Figure 6). The results showed that antibody levels in fish vaccinated with rtOmpF and rtOmpF+ISA763 were both significantly higher (P < 0.05) than that of fish vaccinated with PBS and PBS+ ISA763 at 1st−8th weeks psv. Compared with vaccine rtOmpF alone, rtOmpF+ISA763 induced significantly higher (P < 0.05) antibody levels at 4th, 5th and 6th weeks psv, and slightly higher antibody levels at other time points. The antibody peaks of fish vaccinated with rtOmpF and rtOmpF+ISA763 were located at 3rd and 4th week psv, respectively. During whole experimental period, the absorbance values of fish in group PBS+ ISA763 were just slight higher than that in PBS group.
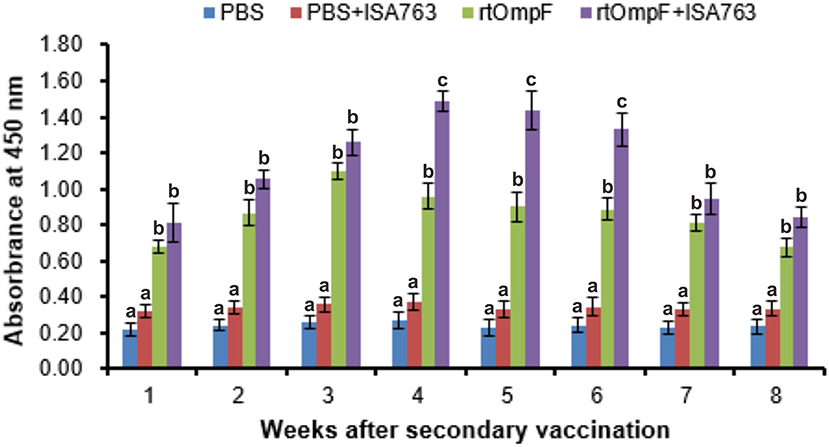
Figure 6. The detection of specific serum antibody in vaccinated fish using ELISA. Channel catfish were vaccinated twice at 2-week intervals, with PBS, PBS+ ISA763, rtOmpF and rtOmpF+ISA763 respectively. Sera were collected from 1st to 8th week psv. Data are presented as means ± SE (n = 5). Different letters above a bar denoted significant difference (P < 0.05).
Measurement of Innate Immune Parameters
Serum Lysozyme Activity
The serum lysozyme activity of fish vaccinated with rtOmpF and rtOmpF+ISA763 increased significantly (P < 0.05) compared with that of fish in PBS and PBS+ ISA763 groups from 1st to 8th week psv (Figure 7A). During the whole experimental period, rtOmpF+ISA763 induced significantly higher (P < 0.05) lysozyme activity than rtOmpF alone except at 5th week psv with slight higher. The highest lysozyme activities of rtOmpF and rtOmpF+ISA763 groups were both detected at 4th week psv. Moreover, the lysozyme activity of fish in PBS+ ISA763 group was just slight higher than that in PBS group except 2nd week psv with significantly higher (P < 0.05).
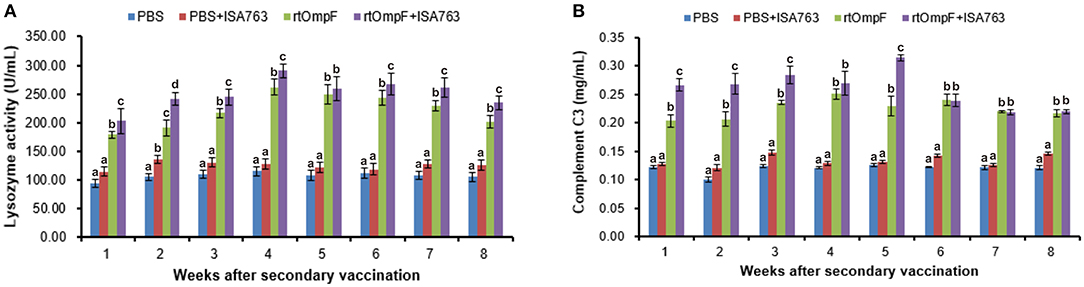
Figure 7. Serum lysozyme activity (A) and complement C3 (B) of vaccinated fish. Channel catfish were vaccinated twice at 2-week intervals, with PBS, PBS+ ISA763, rtOmpF and rtOmpF+ISA763 respectively. Sera were collected from 1st to 8th week psv. Data are presented as means ± SE (n = 5). Different letters above a bar denoted significant difference (P < 0.05).
Serum Complement C3
Figure 7B showed the complement C3 content in serum of fish vaccinated with PBS and vaccines. The results suggested that compared with complement C3 content of fish in PBS and PBS+ ISA763 groups, the complement C3 content of fish vaccinated with rtOmpF and rtOmpF+ISA763 were significantly (P < 0.05) higher throughout whole detection period. Compared with rtOmpF alone, rtOmpF+ISA763 enhanced significantly higher (P < 0.05) complement C3 at 1st, 2nd, 3rd, and 5th weeks psv. The highest complement C3 content of fish vaccinated with rtOmpF and rtOmpF+ISA763 were detected at 4th and 5th weeks psv, respectively. Besides, the differences of complement C3 content of fish in PBS and PBS+ ISA763 were not significant from 1st to 8th week psv.
Serum Total Protein
As shown in Figure 8A, the vaccines rtOmpF and rtOmpF+ISA763 both induced significantly higher (P < 0.05) serum total protein than PBS and PBS+ ISA763 from 1st to 8th week psv. Throughout whole detection period, the total protein levels induced by rtOmpF+ISA763 was significantly higher (P < 0.05) than rtOmpF alone, and the total protein levels of fish in PBS+ ISA763 group were significantly higher than that in PBS group except 1st week psv. Furthermore, the total protein peaks of fish in rtOmpF and rtOmpF+ISA763 were both observed at 4th week psv.
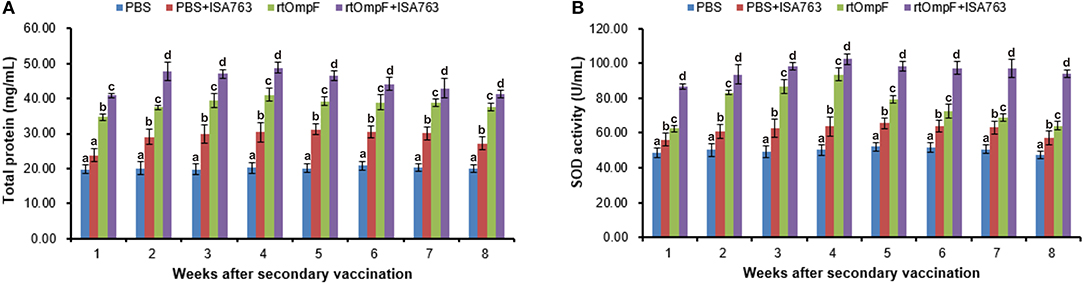
Figure 8. Serum total protein content (A) and SOD activity (B) of vaccinated fish. Channel catfish were vaccinated twice at 2-week intervals, with PBS, PBS+ ISA763, rtOmpF and rtOmpF+ISA763 respectively. Sera were collected from 1st to 8th week psv. Data are presented as means ± SE (n = 5). Different letters above a bar denoted significant difference (P < 0.05).
Serum SOD Activity
Compared with PBS and PBS+ ISA763 groups, a significant (P < 0.05) increase of serum SOD activity was measured in rtOmpF and rtOmpF+ISA763 groups from 1st to 8th week psv (Figure 8B). During whole detection period, the SOD activity of fish in rtOmpF+ISA763 group was significantly (P < 0.05) higher than that in rtOmpF group, and the SOD activity of fish in PBS+ ISA763 group increased significantly (P < 0.05) compared with that in PBS group. Moreover, the highest SOD activity of vaccine groups (rtOmpF and rtOmpF+ISA763) were both detected at 4th week psv.
Expression of the Immune-Related Genes
The immune-related genes expression in the head kidney and spleen at 24 h post-challenge were detected by qRT-PCR with two housekeeping genes 18S and EF1α. The results of melting curve analysis indicated that there was only one peak for the PCR product of each gene (Figure 9A). The results (Figure 9B) of relative expression analysis showed that in the head kidney, the mRNA expression levels of all detected immune-related genes in rtOmpF and rtOmpF+ISA763 groups were significantly increased (P < 0.05) compared with those in PBS and PBS+ ISA763 groups, especially CD8α gene (more than 4.2 fold change) in rtOmpF group, CD4-L2, CD8α and MHC Iα genes (more than 4.8 fold change) in rtOmpF+ISA763 groups. Moreover, the fold changes of all these genes in rtOmpF+ISA763 group were significantly higher (P < 0.05) than that in rtOmpF group. In addition, similar trends were observed in spleen. Compared with PBS group, rtOmpF and rtOmpF+ISA763 induced notably higher (P < 0.05) relative expression of these genes, especially CD8α and MHC IIβ genes (more than 4 fold change) in rtOmpF+ISA763 group. Interestingly, the fold change of each gene in spleen was lower than that in head kidney at the same time.
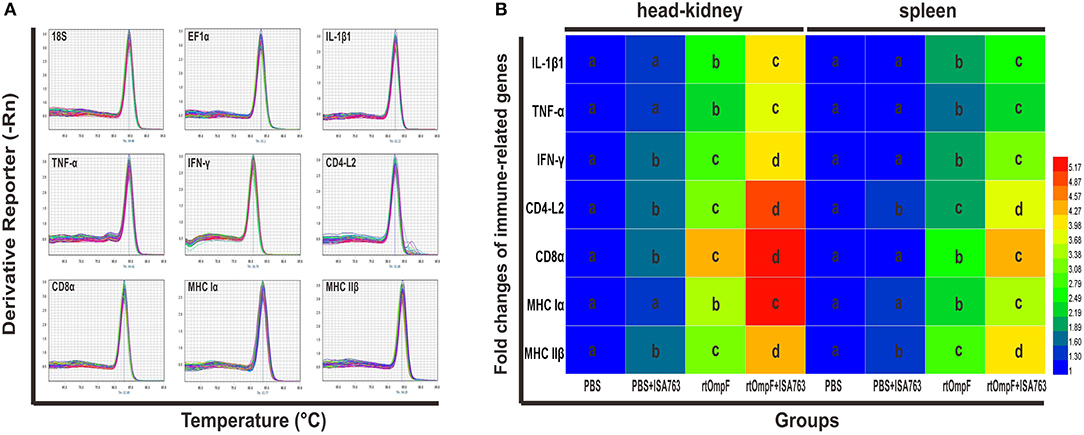
Figure 9. The melt curves and relative expression levels of immune-related genes in the head kidney and spleen of vaccinated fish. (A) The melt curves analysis of two reference genes and seven immune-related genes. (B) Heatmap analysis of the fold changes of immune-related genes determined by qRT-PCR in the head kidney and spleen. For each gene, the mRNA level of the PBS-vaccinated fish was set as 1. Data were presented as means (n = 5). Different letters in the same tissues denoted significant difference (P < 0.05) of the same gene in different groups. The color scale was shown at right of the figure, with blue color indicating low fold changes and red color indicating high fold changes.
Immunoprotection Efficacy Against Y. ruckeri
The results of survival data analyzed using the Kaplan-Meier methods indicated that the percent survivals of fish were 3.33% in PBS group, 10.0% in PBS+ISA763, 66.67% in rtOmpF group, and 76.67% in rtOmpF+ISA763 group during the challenge test with pathogenic Y. ruckeri YRWEL01 at 4th week psv (Figure 10). The results of log-rank analysis suggested that the survivals of fish in rtOmpF and rtOmpF+ISA763 groups were both significantly higher (P < 0.05) than that in PBS and PBS+ ISA763 groups. Moreover, the survival of fish vaccinated with rtOmpF+ISA763 was remarkably higher (P < 0.05) than that of fish vaccinated with rtOmpF alone, and the survival of fish vaccinated with PBS+ ISA763 was significantly higher (P < 0.05) than that of fish vaccinated with PBS alone. Besides, compared with PBS group, the immunoprotective efficacy (in terms of RPS) of PBS+ ISA763, rtOmpF and rtOmpF+ISA763 were 6.90, 65.52, 75.86%, respectively. Furthermore, Y. ruckeri YRWEL01 was the only type of bacterial strain detected in the liver, and kidney of moribund fish, suggesting that mortality was indeed caused by Y. ruckeri YRWEL01 infection.
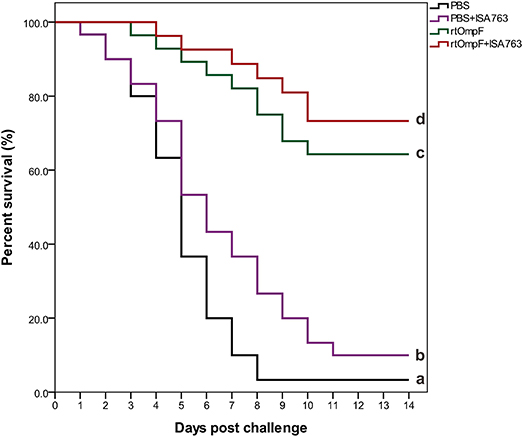
Figure 10. Percent survival analysis of vaccinated fish using Kaplan-Meier method. The differences among groups were analyzed by log-rank test. Different letters denoted significant difference (P < 0.05).
Discussion
With the continuous expansions of aquaculture and yield and the increasing of fish breeding density, many aquatic diseases have appeared and seriously damaged the economic productivity of aquaculture, especially bacterial diseases (62–64). Nowadays, based on the consideration of safety and antimicrobial resistance, traditional control strategies including antibiotics and chemicals are more and more questioned. By contrast, vaccines have become a more effective, safe and green intervention to control bacterial infection in aquaculture. As one of the most promising vaccines, genetically engineered vaccines such as subunit vaccines and DNA vaccines are more safe and serotype-independent due to the basis of protective immunogens (65–67). Therefore, the identification of conserved and protective immunogens is vital for the development of effective genetically engineered vaccines. As the major components and one of the most abundant proteins in the outer membrane, bacterial porin proteins play a critical role in bacterial pathogenesis and interactions with the host immune system (68, 69). OmpF, as one of the best-studied bacterial porins on structural and functional characteristics (70–74), has been reported to be a protective antigen against some bacterial infections (40, 74–77) and been predicted to be a conserved porin located on the surface of Yersinia (42), which suggests it is possible to use as an immunogen candidate providing immunoprotection against Yersinia infection.
In this study, the molecular characteristic results of Y. ruckeri OmpF a.a. sequence suggested OmpF was a member of OM_channels porin superfamily, a β-barrel nonspecific channels consisting of 16 antiparallel β-strands for the transportation of small hydrophillic molecules (78, 79). There were 39 trimer interface polypeptide binding sites located in OM_channels domain, which was related with the typical homotrimer structure of bacterial porins (80, 81). Conserved motifs analysis of Y. ruckeri OmpF with 25 reference OmpFs revealed that even though the sequence divergences existed among different bacterial OmpFs, the components and positions of conserved motifs of bacterial OmpFs remained highly conserved. Besides, multiple sequences alignment and phylogenetic analysis also indicated that OmpF remained comparatively conserved in different Enterobacteriaceae species, especially in Yersiniaceae species (sharing 80.8–86.8% identity in 14 Yersiniaceae species OmpFs). Moreover, the dN/dS ratio was calculated to determine the natural selection pressure imposed on OmpF gene throughout the evolution process. If a ratio dN/dS = 1, a theoretical boundary for positive/diversifying and negative/purifying selection, indicated the absence of selection. A ratio dN/dS > 1 indicated the positive/diversifying selection had occurred. By contrast, negative/purifying selection occurring on the gene should generate dN/dS <1. In the present study, the global dN/dS ratios of bacterial OmpFs was 0.649, implying that negative/purifying selection played a critical role to remove nonsynonymous substitutions from OmpF genes and OmpFs remained comparatively conserved during the evolution process and the interactions with the host immune system, which agreed with the conclusion of Stenkova et al. (42).
To verify the surface location of OmpF protein on Y. ruckeri, bacteria cell surface staining and indirect immunofluorescence assays were conducted with specific anti-rtOmpF antibody. Rabbit anti-Y. ruckeri antisera and rabbit negative antisera were employed as positive and negative controls respectively. The results suggested that positive signals were observed on the surface of Y. ruckeri incubated with rabbit anti-rtOmpF sera, which was consistent with the results of positive control groups, indicating that OmpF was located on the surface of Y. ruckeri and the polyclonal antibody against rtOmpF was successfully generated and had the desirable affinity to Y. ruckeri. Besides, the results of western blotting showed rtOmpF specifically reacted with the rabbit anti-6 × His antiserum and rabbit anti-Y. ruckeri antiserum respectively, and displayed a single predicted band with 55 kDa, which consisted of the OmpF sequence, the His-tag sequences and some sequences of expression plasmid pET32a. Furthermore, the proteins from Y. ruckeri YRWEL01, Y. enterocolitica, and Y. pestis can also be detected by anti-rtOmpF sera with about 38 kDa (Figure 4G), which only consisted of the OmpF sequence in strains. Taken together, these results suggested that recombinant proteins rtOmpF was expressed correctly in vitro and able to confer cross-protection in Yersiniaceae species, and harbored antigenicity properties to serve as candidate immunogen for vaccine development.
Recently, increasing studies haveshown that reference genes might not be stably expressed in different host tissues and one reference gene is not reliable enough for the accurate normalization of target genes expression (82–88). Thus, in the present study, 18S and EF1α genes, two most suitable and stably expressed genes observed in the head kidney and spleen tissues of channel catfish (89), was employed as reference genes. The geometric mean of the expression levels of 18S and EF1α was used for the accurate normalization of immune-related genes expression in qRT-PCR analysis, which was better than arithmetic mean to control the possible outlying values and abundance differences between the different genes (82). The results indicated that compared with control group, rtOmpF significantly enhanced the expression of immune-related genes involved in inflammatory response (IL-1β1, TNF-α), humoral immunity (MHC II β and CD4-L2) and cellular immunity (MHC Iα, CD8α, and IFN-γ) in the head kidney and spleen, especially co-injection of rtOmpF+ISA763, implying rtOmpF had potential as the antigen to induce a series of immune responses for channel catfish against bacterial infection, and ISA763 as the adjuvant improved the immune response. Moreover, we found that the immune-related gene expression levels in head kidney were higher than that in spleen, which may be due to their different roles and functions in fish immune response (90, 91), since head kidney served as not only the site of hematopoiesis, but also both a primary and secondary lymphoid organ, while spleen just served as a secondary peripheral lymphoid organ in fish (45).
In addition, the measurement of immune parameters including adaptive and innate immunity is the direct method to evaluate the vaccine effect. In the present study, serum specific antibody levels, lysozyme activity, complement C3 activity, total protein content, and SOD activity were detected in the control group and treatment groups. The results indicated that rtOmpF significantly enhanced the levels of above five immune parameters compared with PBS. Moreover, commercial adjuvant ISA 763 notably enhanced the immune effects induced by rtOmpF. Furthermore, survival percent and PRS were also calculated to assess the vaccine efficacy against Y. ruckeri infection. It was shown that the percent survival of fish in rtOmpF+ISA763 group was significantly higher than that of rtOmpF group which was remarkably higher than that of control group, and the PRS of rtOmpF and rtOmpF+ISA763 groups was 65.52 and 75.86% respectively, both were higher than that of inactive Y. ruckeri vaccine (RPS with 13.8%) shown in our previous study (57), mainly because of difference types, functions and mechanisms of different antigens.
In conclusion, OmpF gene was shown to be highly conserved among 15 known Yersinia species even in Enterobacteriaceae species based on the analysis of conserved motifs, sequences alignment and phylogenetic tree, and was subjected to negative/purifying selection with global dN/dS ratios value of 0.649 throughout the evolution. Besides, rtOmpF served as a candidate antigen could enhance the immune response by increasing antibody levels, lysozyme activity, complement C3 activity, total protein content, SOD activity, immune-related genes expression, and survival percent against Y. ruckeri infection. Moreover, co-injection of rtOmpF+ISA763 significantly improved the immune effect and immunoprotection induced by rtOmpF. Thus, our present results enriched the information of Y. ruckeri OmpF molecular characterization and phylogenetics, and demonstrated that OmpF holds promise to be used as a potential antigen against Y. ruckeri infection in fish. However, further studies are still required to understand the detailed mechanisms of OmpF used as immunogen and seek for more effective vaccine types like DNA vaccines to enhance and extend the immunoprotection.
Author Contributions
EW, ZQ, ZY, and KW designed the experiment; EW, ZQ, ZY, and XA performed the experimental work. EW, ZQ, and XA analyzed the data, EW and ZY prepared all figures, and EW drafted the paper. QY, TL, DC, and YG participated in fish vaccination and sample collection. XH, PO, and WL contributed to the discussion and revision. All authors reviewed the manuscript.
Funding
This work was supported by the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT0848), Sichuan Technology Support Planning (No. 2014JY0143).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Furones MD, Rodgers CJ, Munn CB. Yersinia ruckeri, the causal agent of enteric redmouth disease (ERM) in fish. Ann Rev Fish Dis. (1993) 3:105–25. doi: 10.1016/0959-8030(93)90031-6
2. Horne MT, Barnes AC. Enteric Redmouth Disease (Yersinia ruckeri). In: Woo PTK, Bruno DW, editors. Fish Diseases and Disorders, Vol. 3: Viral, Bacterial and Fungal Infections. Oxfordshire: CABI Publishing (1999). p. 455–77.
3. Carson J, Wilson T. Yersiniosis in Fish. Australian New Zealand Standard Diagnostic Procedures. Canberra, ACT: Sub-Committee on Animal Health Laboratory Standards, Commonwealth of Australia (2009). p. 1–19.
4. Bastardo A, Bohle H, Ravelo C, Toranzo AE, Romalde JL. Serological and molecular heterogeneity among Yersinia ruckeri strains isolated from farmed Atlantic salmon Salmo salar in Chile. Dis Aquat Organ. (2011) 93:207–14. doi: 10.3354/dao02296
5. Shah SQA, Karatas S, Nilsen H, Steinum TM, Colquhoun DJ, Sorum H. Characterization and expression of the gyrA gene from quinolone resistant Yersinia ruckeri strains isolated from Atlantic salmon (Salmo salar L.) in Norway. Aquaculture (2012) 350:37–41. doi: 10.1016/j.aquaculture.2012.04.022
6. Ormsby MJ, Caws T, Burchmore R, Wallis T, Verner-Jeffreys DW, Davies RL. Yersinia ruckeri isolates recovered from diseased atlantic salmon (Salmo salar) in scotland are more diverse than those from rainbow trout (Oncorhynchus mykiss) and represent distinct subpopulations. Appl Environ Microbiol. (2016) 82:5785–94. doi: 10.1128/AEM.01173-16
7. Ross AJ, Rucker RR, Ewing WH. Description of a bacterium associated with redmouth disease of rainbow trout (Salmo Gairdneri). Can J Microbiol. (1966) 12:763. doi: 10.1139/m66-103
8. Bullock GL, Stuckey HM, Shotts EB. Enteric redmouth bacterium: comparison of isolates from different geographic areas. J Fish Dis. (1978) 1:351–6. doi: 10.1111/j.1365-2761.1978.tb00039.x
10. Stevenson R, Daly JG. Biochemical and serological characteristics of Ontario isolates of Yersinia ruckeri. Can J Fish Aquatic Sci. (1982) 39:870–6. doi: 10.1139/f82-118
11. Berc A, Petrinec Z, Matasin Z, Kozaric Z. Yersinia ruckeri septicaemia in experimentally infected carp (Cyprinus carpio L.) fingerlings. Acta Vet Hung (1999) 47:161–72. doi: 10.1556/004.47.1999.2.2
12. Rintamaki P, Valtonen ET, Frerichs GN. Occurrence of Yersinia ruckeri infection in farmed whitefish, Coregonus peled Gmelin and Coregonus muksun Pallas, and Atlantic salmon, Salmo salar L., in northern Finland. J Fish Dis. (1986) 9:137–40. doi: 10.1111/j.1365-2761.1986.tb00993.x
13. Vuillaume A, Brun R, Chene P, Sochon E, Lesel R. First isolation of Yersinia ruckeri from sturgeon, Acipenser baeri Brandt, in south west of France. B Eur Assoc Fish Pat. (1987) 7:18–9.
14. Shaowu L, Di W, Hongbai L, Tongyan L. Isolation of Yersinia ruckeri Strain H01 from Farm-Raised Amur Sturgeon Acipenser schrencki in China. J Aquat Anim Health (2013) 25:9–14. doi: 10.1080/08997659.2012.728169
15. Li S, Zhang Y, Cao Y, Wang D, Liu H, Lu T. Trancriptome profiles of Amur sturgeon spleen in response to Yersinia ruckeri infection. Fish Shellfish Immunol. (2017) 70:451–60. doi: 10.1016/j.fsi.2017.09.033
16. Danley ML, Goodwin AE, Killian HS. Epizootics in farm-raised channel catfish, Ictalurus punctatus (Rafinesque), caused by the enteric redmouth bacterium Yersinia ruckeri. J Fish Dis. (1999) 22:451–6. doi: 10.1046/j.1365-2761.1999.00196.x
17. Fan F, Wang K, Geng Y, Huang X, Chen D. Isolation, identification and phylogenetic analysis of Yersinia ruckeri in channel catfish Ictalunes punctatus. Oceanol Limnol Sin. (2010) 41:862–8. doi: 10.11693/hyhz201006010010
18. Ormsby M, Davies R. Yersinia ruckeri In: Woo PTK, Cipriano RC, editors. Fish Viruses and Bacteria: Pathobiology and Protection. Oxfordshire: CABI Publishing (2017). p. 339–51.
19. Tinsley JW, Austin DA, Lyndon AR, Austin B. Novel non-motile phenotypes of Yersinia ruckeri suggest expansion of the current clonal complex theory. J Fish Dis. (2011) 34:311–7. doi: 10.1111/j.1365-2761.2011.01237.x
20. Costa AA, Leef MJ, Bridle AR, Carson J, Nowak BF. Effect of vaccination against yersiniosis on the relative percent survival, bactericidal and lysozyme response of Atlantic salmon, Salmo salar. Aquaculture (2011) 315:201–6. doi: 10.1016/j.aquaculture.2011.02.031
21. Deshmukh S, Raida MK, Dalsgaard I, Chettri JK, Kania PW, Buchmann K. Comparative protection of two different commercial vaccines against Yersinia ruckeri serotype O1 and biotype 2 in rainbow trout (Oncorhynchus mykiss). Vet Immunol Immunop. (2012) 145:379–85. doi: 10.1016/j.vetimm.2011.12.014
22. Kumar G, Menanteau-Ledouble S, Saleh M, El-Matbouli M. Yersinia ruckeri, the causative agent of enteric redmouth disease in fish. Vet Res. (2015) 46:103. doi: 10.1186/s13567-015-0238-4
23. Temprano A, Riano J, Yugueros J, Gonzalez P, de Castro L, Villena A, et al. Potential use of a Yersinia ruckeri O1 auxotrophic aroA mutant as a live attenuated vaccine. J Fish Dis. (2005) 28:419–27. doi: 10.1111/j.1365-2761.2005.00646.x
24. Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol R (2003) 67:593. doi: 10.1128/MMBR.67.4.593-656.2003
25. Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. (2010) 74:81–94. doi: 10.1128/MMBR.00031-09
26. Nikaido H, Nakae T. The outer membrane of Gram-negative bacteria. Adv Microb Physiol. (1980) 20:163–250. doi: 10.1016/S0065-2911(08)60208-8
27. Pages JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. (2008) 6:893–903. doi: 10.1038/nrmicro1994
28. Benz R. Structure and function of porins from gram-negative bacteria. Annu Rev Microbiol. (1988) 42:359–93. doi: 10.1146/annurev.mi.42.100188.002043
29. Tang X, Zhan W, Sheng X, Chi H. Immune response of Japanese flounder Paralichthys olivaceus to outer membrane protein of Edwardsiella tarda. Fish Shellfish Immun. (2010) 28:333–43. doi: 10.1016/j.fsi.2009.11.015
30. Phuong PTM. Expression of Recombinant Outer Membrane Protein 48 (OMP48) Developing Vaccine Against Aeromonas hydrophila Infection. Dissertation thesis, HCMC International University, Vietnam (2014).
31. Maiti B, Shetty M, Shekar M, Karunasagar I, Karunasagar I. Recombinant outer membrane protein A (OmpA) of Edwardsiella tarda, a potential vaccine candidate for fish, common carp. Microbiol Res. (2011) 167:1–7. doi: 10.1016/j.micres.2011.02.002
32. Abdelhamed H, Ibrahim I, Nho SW, Banes MM, Wills RW, Karsi A, et al. Evaluation of three recombinant outer membrane proteins, OmpA1, Tdr, and TbpA, as potential vaccine antigens against virulent Aeromonas hydrophila infection in channel catfish (Ictalurus punctatus). Fish Shellfish Immunol. (2017) 66:480–6. doi: 10.1016/j.fsi.2017.05.043
33. Yadav SK, Meena JK, Sharma M, Dixit A. Recombinant outer membrane protein C of Aeromonas hydrophila elicits mixed immune response and generates agglutinating antibodies. Immunol Res. (2016) 64:1087–99. doi: 10.1007/s12026-016-8807-9
34. Ningqiu L, Junjie B, Shuqin W, Xiaozhe F, Haihua L, Xing Y, et al. An outer membrane protein, OmpK, is an effective vaccine candidate for Vibrio harveyi in Orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immun. (2008) 25:829–33. doi: 10.1016/j.fsi.2008.09.007
35. Yang Q, Pan YL, Wang KY, Wang J, He Y, Wang EL, et al. OmpN, outer membrane proteins of Edwardsiella ictaluri are potential vaccine candidates for channel catfish (Ictalurus punctatus). Mol Immunol. (2016) 78:1–8. doi: 10.1016/j.molimm.2016.08.011
36. Khushiramani R, Girisha SK, Karunasagar I, Karunasagar I. Cloning and expression of an outer membrane protein ompTS of Aeromonas hydrophila and study of immunogenicity in fish. Protein Expr Purif. (2007) 51:303–7. doi: 10.1016/j.pep.2006.07.021
37. Wang Q, Chen J, Liu R, Jia J. Identification and evaluation of an outer membrane protein OmpU from a pathogenic Vibrio harveyi isolate as vaccine candidate in turbot (Scophthalmus maximus). Lett Appl Microbiol. (2011) 53:22–9. doi: 10.1111/j.1472-765X.2011.03062.x
38. Maiti B, Shetty M, Shekar M, Karunasagar I, Karunasagar I. Evaluation of two outer membrane proteins, Aha1 and OmpW of Aeromonas hydrophila as vaccine candidate for common carp. Vet Immunol Immunopathol. (2012) 149:298–301. doi: 10.1016/j.vetimm.2012.07.013
39. Neema M, Karunasagar I. In silico homology modeling and epitope prediction of outer membrane protein OMP W, a potential vaccine candidate against Edwardsiella tarda. Int. J. Curr. Microbiol. App. Sci. (2018) 7:2762–73. doi: 10.20546/ijcmas.2018.703.319
40. Liu C, Chen Z, Tan C, Liu W, Xu Z, Zhou R, et al. Immunogenic characterization of outer membrane porins OmpC and OmpF of porcine extraintestinal pathogenic Escherichia coli. Fems Microbiol Lett. (2012) 337:104–11. doi: 10.1111/1574-6968.12013
41. Verma SK, Gautam V, Balakrishna K, Kumar S. Overexpression, purification, and immunogenicity of recombinant porin proteins of Salmonella enterica Serovar Typhi (S. Typhi). J Microbiol Biotechn. (2009) 19:1034–40. doi: 10.4014/jmb.0812.675
42. Stenkova AM, Isaeva MP, Shubin FN, Rasskazov VA, Rakin AV. Trends of the major porin gene (OmpF) evolution: insight from the genus Yersinia. Plos ONE (2011) 6:e20546. doi: 10.1371/journal.pone.0020546
43. Clem LW, Bly JE, Ellsaesser CF, Lobb CJ, Miller NW. Channel catfish as an unconventional model for immunological studies. J Exp Zool. (1990) 4:123–5. doi: 10.1002/jez.1402560420
44. Liu Z, Liu S, Yao J, Bao L, Zhang J, Li Y, et al. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat Commun. (2016) 7:11757. doi: 10.1038/ncomms11757
45. Dickerson HW, Findly RC. Vertebrate adaptive immunity-comparative insights from a teleost model. Front Immunol. (2017) 8:1379. doi: 10.3389/fimmu.2017.01379
46. Rombel IT, Sykes KF, Rayner S, Johnston SA. ORF-FINDER: a vector for high-throughput gene identification. Gene (2002) 282:33–41. doi: 10.1016/S0378-1119(01)00819-8
47. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods (2011) 8:785–6. doi: 10.1038/nmeth.1701
48. Marchlerbauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. (2015) 43:D222. doi: 10.1093/nar/gku1221
49. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. (2009) 37:202–8. doi: 10.1093/nar/gkp335
50. Clewley JP, Arnold C. MEGALIGN. The multiple alignment module of LASERGENE. Methods Mol Biol. (1997) 70:119–29. doi: 10.1385/0-89603-358-9:119
51. Edgar RC. MUSCLE: multiple sequence alignment with improved accuracy and speed. In: Proceedings of the 2004 IEEE Computational Systems Bioinformatics Conference (CSB 2004). Washington, DC: The IEEE Computer Society (2004). p. 728–9.
52. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. (2011) 28:2731. doi: 10.1093/molbev/msr121
53. Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics (2010) 26:2455–7. doi: 10.1093/bioinformatics/btq429
54. Wang E, Wang J, Long B, Wang K, Yang H, Yang Q, et al. Molecular cloning, expression and the adjuvant effects of interleukin-8 of channel catfish (Ictalurus Punctatus) against Streptococcus iniae. Sci Rep. (2016) 6:29310. doi: 10.1038/srep29310
55. Wang Y, Wang E, He Y, Wang K, Yang Q, Wang J, et al. Identification and screening of effective protective antigens for channel catfish against Streptococcus iniae. Oncotarget (2017) 8:30793. doi: 10.18632/oncotarget.16475
56. He Y, Wang K, Xiao D, Chen D, Huang L, Liu T, et al. A recombinant truncated surface immunogenic protein (tSip) plus adjuvant FIA confers active protection against Group B streptococcus infection in tilapia. Vaccine (2014) 32:7025–32. doi: 10.1016/j.vaccine.2014.08.017
57. Yang L, Wang KY, Zhou Y, Wang EL, Wang J, Chen DF, et al. The characteristics and immune efficacy of an oral microspheres vaccine of Yersinia ruckeri. Acta Hydrobiologica Sinica (2015) 39:1149–56. doi: 10.7541/2015.151
58. Page M, Thorpe R. Purification of IgG by precipitation with sodium sulfate or ammonium sulfate. In: Walker JM, editor. The Protein Protocols Handbook. Totowa, NJ: Humana Press (2009). p. 1749–51.
59. Wang J, Wang K, Chen D, Geng Y, Huang X, He Y, et al. Cloning and characterization of surface-localized α-Enolase of Streptococcus iniae, an effective protective antigen in mice. Int J Mol Sci. (2015) 16:14490–510. doi: 10.3390/ijms160714490
61. Wang E, Qin Z, Wang K, Chen D, Wang J, He Y. Molecular cloning, bioinformatics and immunogenicity analyses of outer membrane protein ompF gene of Yersinia ruckeri. South China Fish Sci. (2016) 12:24–34. doi: 10.3969/j.issn.2095-0780.2016.03.004
62. Austin B, Austin DA. Bacterial Fish Pathogens: Disease of Farmed and Wild Fish. Cham: Springer.(2012)
63. Kumagai A. Bacterial Cold-water Disease in Salmonid Fish and Ayu. Fish Pathol. (2016) 51:153–7. doi: 10.3147/jsfp.51.153
64. Haditomo AHC, Ariyati RW, Prayitno SB. The diversity of causative agent associated with bacterial diseases on catfish (Clarias gariepinus) with molecular based from Kendal, Indonesia. Adv Sci Lett. (2017) 23:6479–82. doi: 10.1166/asl.2017.9659
65. Moyle PM, Toth I. Modern subunit vaccines: development, components, and research opportunities. Chemmedchem. (2013) 8:360–76. doi: 10.1002/cmdc.201200487
66. Karch CP, Burkhard P. Vaccine technologies: From whole organisms to rationally designed protein assemblies. Biochem Pharmacol. (2016) 120:1–14. doi: 10.1016/j.bcp.2016.05.001
67. Moyle PM. Biotechnology approaches to produce potent, self-adjuvanting antigen-adjuvant fusion protein subunit vaccines. Biotechnol Adv. (2017) 35:375–89. doi: 10.1016/j.biotechadv.2017.03.005
68. Achouak W, Heulin T, Pages JM. Multiple facets of bacterial porins. Fems Microbiol Lett. (2001) 199:1–7. doi: 10.1111/j.1574-6968.2001.tb10642.x
69. Massari P, Ram S, Macleod H, Wetzler LM. The role of porins in neisserial pathogenesis and immunity. Trends Microbiol. (2003) 11:87–93. doi: 10.1016/S0966-842X(02)00037-9
70. Williams KM, Bigley EC, Raybourne RB. Identification of murine B-cell and T-cell epitopes of Escherichia coli outer membrane protein F with synthetic polypeptides. Infect Immun. (2000) 68:2535–45. doi: 10.1128/IAI.68.5.2535-2545.2000
71. Guzev KV, Isaeva MP, Novikova OD, Solov'Eva TF, Rasskazov VA. Molecular characteristics of OmpF-Like porins from pathogenic Yersinia. Biochemistry (2005) 70:1104–10. doi: 10.1007/s10541-005-0231-z
72. Novikova OD, Vakorina TI, Khomenko VA, Likhatskaya GN, Kim NY, Emelyanenko VI, et al. Influence of cultivation conditions on spatial structure and functional activity of OmpF-like porin from outer membrane of Yersinia pseudotuberculosis. Biochemistry (2008) 73:139–48. doi: 10.1134/S0006297908020041
73. Chistyulin DK, Novikova OD, Portnyagina OY, Khomenko VA, Vakorina TI, Kim NY, et al. Isolation and characterization of OmpF-like Porin from Yersinia ruckeri. Biochemistry (2012) 6:235–42. doi: 10.1134/S1990747812030038
74. Sidorova OV, Khomenko VA, Portnyagina OY, Likhatskaya GN, Vakorina TI, Kim NY, et al. Mutant OmpF porins of Yersinia pseudotuberculosis with deletions of external loops: structure–functional and immunochemical properties. Biochem Bioph Res Co. (2014) 445:428–32. doi: 10.1016/j.bbrc.2014.02.018
75. Hughes EE, Gilleland HE. Ability of synthetic peptides representing epitopes of outer membrane protein F of Pseudomonas aeruginosa to afford protection against P. aeruginosa infection in a murine acute pneumonia model. Vaccine (1995) 13:1750–3. doi: 10.1016/0264-410X(95)00166-X
76. Portnyagina O, Sidorova O, Khomenko V, Novikova O, Issaeva M, Solovev T. Immunochemical properties of recombinant Ompf Porin from outer membrane of Yersinia pseudotuberculosis. In: Abuelzein E, editor. Trends in Immunolabelled and Related Techniques. Rijeka: InTech Open Access Publisher (2012). p. 243–58.
77. Sharma M, Dixit A. Identification and immunogenic potential of B cell epitopes of outer membrane protein OmpF of Aeromonas hydrophila in translational fusion with a carrier protein. Appl Microbiol Biotechnol. (2015) 99:6277–91. doi: 10.1007/s00253-015-6398-3
78. Gromiha MM, Majumdar R, Ponnuswamy PK. Identification of membrane spanning beta strands in bacterial porins. Protein Eng. (1997) 10:497–500. doi: 10.1093/protein/10.5.497
79. Schulz GE. The structure of bacterial outer membrane proteins. Biochimica et Biophysica Acta Biomembranes (2002) 1565:308–17. doi: 10.1016/S0005-2736(02)00577-1
80. Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, et al. Crystal structures explain functional properties of two E. coli porins. Nature (1992) 358:727–33. doi: 10.1038/358727a0
81. Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J Mol Biol. (2006) 362:933–42. doi: 10.1016/j.jmb.2006.08.002
82. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. (2002) 3:H34. doi: 10.1186/gb-2002-3-7-research0034
83. Peters IR, Peeters D, Helps CR, Day MJ. Development and application of multiple internal reference (housekeeper) gene assays for accurate normalisation of canine gene expression studies. Vet Immunol Immunopathol. (2007) 117:55–66. doi: 10.1016/j.vetimm.2007.01.011
84. Chen H, Yang Z, Hu Y, Tan J, Jia J, Xu H, et al. Reference genes selection for quantitative gene expression studies in Pinus massoniana L. Trees-Struct Funct. (2016) 30:685–96. doi: 10.1007/s00468-015-1311-3
85. Jiang M, Lee JN, Bionaz M, Deng XY, Wang Y. Evaluation of suitable internal control genes for RT-qPCR in yak mammary tissue during the lactation cycle. PLoS ONE (2016) 11:e147705. doi: 10.1371/journal.pone.0147705
86. Gütling H, Bionaz M, Sloboda DM, Ehrlich L, Braun F, Gramzow AK, et al. The importance of selecting the right internal control gene to study the effects of antenatal glucocorticoid administration in human placenta. Placenta (2016) 44:19–22. doi: 10.1016/j.placenta.2016.05.011
87. Marcial-Quino J, Fierro F, De la Mora-De LMI, Enriquez-Flores S, Gomez-Manzo S, Vanoye-Carlo A, et al. Validation of housekeeping genes as an internal control for gene expression studies in Giardia lamblia using quantitative real-time PCR. Gene (2016) 581:21–30. doi: 10.1016/j.gene.2016.01.018
88. Palve V, Pareek M, Krishnan NM, Siddappa G, Suresh A, Kuriakose MA, et al. A minimal set of internal control genes for gene expression studies in head and neck squamous cell carcinoma. bioRxiv [Preprint] (2017). doi: 10.1101/108381
89. Small BC, Murdock CA, Bilodeau-Bourgeois AL, Peterson BC, Waldbieser GC. Stability of reference genes for real-time PCR analyses in channel catfish (Ictalurus punctatus) tissues under varying physiological conditions. Comp Biochem Physiol B Biochem Mol Biol. (2008) 151:296–304. doi: 10.1016/j.cbpb.2008.07.010
90. Dickerson H, Clark T. Ichthyophthirius multifiliis: a model of cutaneous infection and immunity in fishes. Immunol Rev. (1998) 166:377–84. doi: 10.1111/j.1600-065X.1998.tb01277.x
Keywords: ompF, molecular characterization, phylogenetic analysis, immunogenicity, immune effect, Yersinia ruckeri
Citation: Wang E, Qin Z, Yu Z, Ai X, Wang K, Yang Q, Liu T, Chen D, Geng Y, Huang X, Ouyang P and Lai W (2018) Molecular Characterization, Phylogenetic, Expression, and Protective Immunity Analysis of OmpF, a Promising Candidate Immunogen Against Yersinia ruckeri Infection in Channel Catfish. Front. Immunol. 9:2003. doi: 10.3389/fimmu.2018.02003
Received: 01 June 2018; Accepted: 14 August 2018;
Published: 13 September 2018.
Edited by:
Hetron Mweemba Munang'andu, Norwegian University of Life Sciences, NorwayReviewed by:
Wenbin Zhan, Ocean University of China, ChinaBiswajit Maiti, Nitte University, India
Shih-Chu Chen, National Pingtung University of Science and Technology, Taiwan
Copyright © 2018 Wang, Qin, Yu, Ai, Wang, Yang, Liu, Chen, Geng, Huang, Ouyang and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaiyu Wang, a3l3YW5nMTk1NUAxMjYuY29t
† These authors have contributed equally to this work
 Erlong Wang
Erlong Wang Zhenyang Qin1†
Zhenyang Qin1† Kaiyu Wang
Kaiyu Wang Yi Geng
Yi Geng