- 1Guangdong Provincial Key Laboratory of Animal Nutrition Control, College of Animal Science, Institute of Subtropical Animal Nutrition and Feed, South China Agricultural University, Guangzhou, China
- 2Scientific Observing and Experimental Station of Animal Nutrition and Feed Science in South-Central, Ministry of Agriculture, Hunan Provincial Engineering Research Center of Healthy Livestock, Key Laboratory of Agro-Ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Department of Animal Science, Hunan Agriculture University, Changsha, Hunan, China
Unrestricted cell proliferation is a hallmark of cancer. Purines are basic components of nucleotides in cell proliferation, thus impaired purine metabolism is associated with the progression of cancer. The de novo biosynthesis of purine depends on six enzymes to catalyze the conversion of phosphoribosylpyrophosphate to inosine 5′-monophosphate. These enzymes cluster around mitochondria and microtubules to form purinosome, which is a multi-enzyme complex involved in de novo purine biosynthesis and purine nucleotides requirement. In this review, we highlighted the purine metabolism and purinosome biology with emphasis on the therapeutic potential of manipulating of purine metabolism or purinosome in cancers. We also reviewed current advances in our understanding of mammalian target of rapamycin for regulating purinosome formation or purine metabolism in cancers and discussed the future prospects for targeting purinosome to treat cancers.
Introduction
Normal cells undergo a series of highly regulated physiological responses to provide necessary substrates for the basic cellular processes, while cancer cells are involved in a complex metabolic rearrangement characterized by an increase in energy production and biosynthetic processes to sustain cell growth and proliferation (1–8). Purines are the most abundant metabolic substrates for all living organisms by providing essential components for DNA and RNA. Besides as building blocks for DNA and RNA, purines provide the necessary energy and cofactors to promote cell survival and proliferation. Thus, purines and their derivatives widely participate in biological processes, including immune responses and host–tumor interaction (9). Notably, high concentrations of purine metabolites have been indicated in tumor cells, and this discovery favors to the development of the earliest antitumor drugs (purine antimetabolites) to treat cancers by blocking DNA synthesis and halting cell growth. Purinosome has been recently identified within purine metabolism, and the formation of purinosome is closely related to the cell cycle (10, 11). These results provide a novel therapeutic strategy for cancers by targeting purinosome formation and purine metabolism.
In this review, we discuss the purine metabolism, including the complementary salvage pathway and de novo biosynthetic pathway. We then discussed the purinosome with emphasis on purinosome formation and composition, and its interaction with mitochondria. We also described the potential therapeutic strategies for cancers by targeting purine metabolism and purinosome, which may be used to reprogram cancer metabolism. Finally, the mechanism of mammalian target of rapamycin (mTOR) regulating the formation of purinosome is discussed.
Purine Metabolism
Purine metabolism maintains cellular pools of adenylate and guanylate via synthesis and degradation of purine nucleotides. In mammalian cells, purine nucleotides are synthesized in two different pathways: the complementary salvage pathway and de novo biosynthetic pathway (Figure 1). Generally, the complementary salvage pathway accounts for most of the cellular requirements for purine by recycling the degraded bases with help of hypoxanthine-guanine phosphoribosyltransferase (HPRT) and adenine phosphoribosyltransferase (Figure 1). HPRT is an Mg2+-dependent enzyme and recycles hypoxanthine and guanine via transferring phosphoribosyl group from phosphoribosylpyrophosphate (PRPP) to generate inosine monophosphate (IMP) and guanine monophosphate (GMP), respectively.
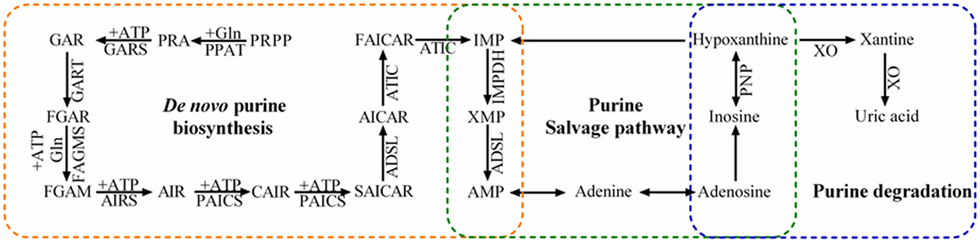
Figure 1. Purine metabolism pathways. Purine metabolism includes de novo purine biosynthetic pathway, purine salvage pathway, and degradation. The de novo purine biosynthetic pathway uses six enzymes to catalyze the transformation of phosphoribosylpyrophosphate (PRPP) into inosine 5′-monophosphate (IMP) via 10 highly conserved steps (orange). Purine salvage (green) recycles hypoxanthine, inosine, and adenine as substrates to generate purine nucleotides. Inosine and hypoxanthine can be further oxidized into xanthine and uric acid in the purine degradation pathway (blue).
Under the conditions with higher requirement for purine nucleotides, such as dividing cells and tumor cells, the de novo biosynthetic pathway is fundamental to replenish the purine pool. In humans, six enzymes catalyze PRPP to form IMP in 10 highly regulated and conserved steps. These enzymes include one trifunctional enzyme (TrifGART: GARS, GART, and AIRS domains), two bifunctional enzymes (PAICS: CAIRS and SAICARS domains; ATIC: AICART and IMPCH domains), and three monofunctional enzymes (PPAT, FGAMS, and ASL). The generated IMP contributes to the production of various intermediates, such as AMP, GMP, adenosine, and inosine. Inosine is further converted to hypoxanthine by purine nucleoside phosphorylase (PNP), and xanthine-oxidase (XO) catalyzes hypoxanthine oxidation to form xanthine (12).
The de novo biosynthetic pathway is energy intensive, and numerous amino acid substrates and one-carbon units contribute to the 10-step enzymatic processes, such as glutamine, ATP, and formate. In this pathway, five molecules of ATP, two molecules of glutamine and formate, and one molecule of glycine, aspartate, and carbon dioxide are necessary for generation of one molecule of IMP (13). Therefore, these substrates play a critical role in purine metabolism especially in rapid proliferating cancer cells. Indeed, metabolic dependencies on glutamine and aspartate are increased to fuel anabolic processes to support cancer growth (14), and glycine metabolism contributes to biosynthetic requirement of purines, ATP, and NADPH in cancer cells (15).
Purinosome
Metabolic pathway is generally assembled with several sequential enzymes into a higher order protein structure to facilitate metabolic flux, and the formation of the protein complex of enzymes is known as metabolon (16). Metabolon has been found in various metabolic pathways, such as the glycosome in the glycolytic pathway (17). Metabolon improves the efficiency of metabolic pathway by increasing the local concentration of intermediates and metabolic substrates, decreasing the concentration of enzymes needed to maintain a given flux, directing the products to a specific subcellular location, or minimizing the escape of reactive intermediates (16, 18). For example, compared to the effector T cells, which have more “fissed” mitochondria, memory T cells have more “fused” mitochondria to densely pack the electron transport chain complexes to form respirasomes, resulting in efficient transfer of electrons and minimizing proton leak during ATP production (19). Purine metabolism has been investigated for decades and there is a wide speculation that enzymes in the de novo purine biosynthesis are organized into a metabolon to maintain cellular purine pool. In 2008, purinosome was first confirmed in living cells through the discovery that all six enzymes in the de novo biosynthetic pathway are recruited to form punctate bodies in the cellular cytoplasm (10). The assembly and disassembly of purinosome can be regulated dynamically by cellular level of purine (10, 20), which directly activates the de novo purine biosynthesis (21, 22).
During cell growth or division, the de novo purine biosynthesis is significantly activated to provide purine nucleotides for growth (G1), duplication of genetic materials (S), division (G2), and divide (M) (23–25). Through the time-lapse fluorescence microscopy assay, high percent of purinosome-positive cells are identified in the G1 phase of cell cycle when purine demand is the highest (11). The requirement of purine decreases with progression of cells into the S and G2/M phases, which accounts for the drop in the relative amount of purinosome-positive cells throughout the cell cycle (11).
Through mapping protein–protein interactions within the purinosome, Deng et al. found that the first three enzymes in the pathway constitute the core scaffolding structure, including PPAT, GART, and FGAMS, whereas PAICS, ADSL, and ATIC appear to interact peripherally (26) (Figure 2). Deficiency or mutation of any enzymes within purinosome impairs purinosome formation and complex stability (27, 28). Purine supplementation or deficient in specific enzymes results either in a complete loss or a significant reduction of purinosomes (13). Various factors have been identified to affect purinosome function. For example, mitochondrial tetrahydrofolate (THF), an essential substrate for de novo purine synthesis, enhances purine biosynthesis by delivering 10-formyl THF to purinosome (29). Interestingly, although purinosome-positive cells vary from G1 to G2/M phases, the expressions of enzymes in the de novo purine biosynthetic pathway are not altered throughout the cell cycle (11). Therefore, we anticipate that the assembly and disassembly of purinosome in cells are not totally governed by protein abundances.
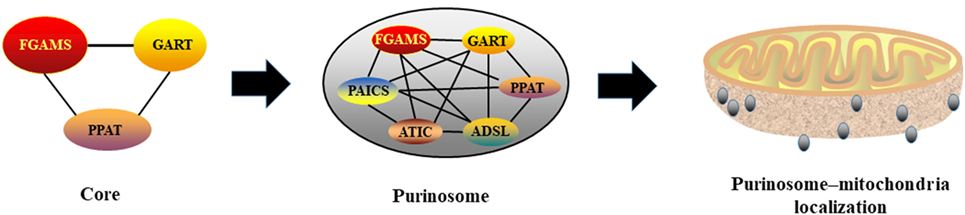
Figure 2. Purinosome formation and its interaction with mitochondria. PPAT, GART, and FGAMS constitute the core scaffolding structure of purionsome, then, PAICS, ADSL, and ATIC appear to interact peripherally. Assembled purionsomes are mainly localized with mitochondria and interact mutually.
Considering that five molecules of ATP are needed for generating one molecule of IMP, mitochondrial function may be associated with purinosome biology and de novo purine biosynthesis. Indeed, mitochondrial THF cycle contributes to non-essential amino acids and one-carbon formyl units to mediate production of purine nucleotides (30). Using two-color 3D STORM imaging, a substantial larger fraction of purinosomes is localized with mitochondria rather than randomly distributed within the cytoplasm in purinosome-positive cells (31), indicating a high possibility of ordered distribution of purinosomes and mitochondria (Figure 2). This is further supported by the discovery that purinosome proteins (i.e., ASL and FGAMS) are co-precipitated with the purified mitochondria, indicating a potential possibility of interaction between mitochondria and purinosome. Mitochondria dysregulation (i.e., defect in electron transport or oxidative phosphorylation) enhances purinosome formation in cells and inhibition of purinosome formation also impairs mitochondrial metabolism in purine-rich conditions (31). Although the molecular mechanisms of the purinosome and mitochondria interaction still need to be uncovered, mitochondria provides enough ATP, amino acid substrates, and one carbon formyl units for the de novo purine biosynthesis, which partially helps to explain the potential mechanism (13).
Purine Metabolism in Cancers
Aberrant cell cycle with uncontrolled cell proliferation is a hallmark of cancers, thus targeting cell cycle has been considered as an attractive strategy for cancer therapy (32–38). Purines and enzymes for de novo purine biosynthetic pathway are enhanced in tumor cells because purine nucleotides are fundamental and necessary for tumor cell proliferation (39–42). For example, inosine strongly enhances proliferation of human melanoma cells (43), and altered ratio of adenosine to inosine has been widely noticed in cancer cells, affecting growth, invasiveness, and metastasis (44, 45). Meanwhile, purines serve as potent modulators in the response of immune cells and cytokine release via various receptor subtypes, such as P2X ligand-gated ion channels and G protein-coupled P2Y receptors (46, 47), which is substantially involved in the development of oncogenesis and tumorigenesis (48–51). For example, adenosine plays a role in the regulation of neutrophil function and modulates the interaction of neutrophils with pathogens (52). Also, mutation or deficiency of adenosine deaminase, which is a key enzyme for purine metabolite degradation or salvage into the nucleotide pool, increases susceptibility to infections and autoimmunity, and adenosine deaminase activity has been used as a diagnostic marker for cancers (53, 54). More specific details about the interaction between purine metabolism and immune system in tumor microenvironment can be found in reviews by Antonioli et al. (55), Kumar (56), and Muller-Haegele et al. (57). Therefore, targeting purine metabolism may serve as a potential therapy in cancers.
Aberrant metabolisms of amino acids and one-carbon units for purine metabolism are also presented in cancer cells, which can be used to predict the subtype of cancer and disease progression (58). For example, glutamine is a key substrate for catalytic activity of PPAT and FGAMS in the purinosome and plays important roles in anabolic processes and physiological responses in cancer cells, including sustaining proliferative signaling, enabling replicative immortality, resisting cell death, and invasion and metastasis (14, 59). Moreover, other substrates of purine metabolism, such as glycine, aspartate, and precursor of one-carbon unit, have also been reported to anticipate in the rapid cancer cell proliferation (60–64), thus, targeting amino acid substrates or one-carbon unit metabolism may serve as a potential therapeutic power of manipulating cell proliferation for treating cancers. Indeed, targeting glutamine metabolism and uptake in cancer cells reduces tumor weight, nodules, and metastasis (65, 66). One-carbon unit also support the high proliferative rate of cancer cells, thus, antifolate drugs that target one-carbon metabolism have long been used in the treatment of cancers. For example, cancer cells are particularly susceptible to deprivation of one-carbon units by serine restriction or inhibition of de novo serine synthesis (67). Also, dietary starvation of serine or glycine reduces tumor growth and improves survival in different cancer models through antioxidant responses and mitochondrial oxidative phosphorylation (68, 69).
Purine antimetabolites are the one of earliest developed chemotherapy drugs and have been widely used in the clinic to treat cancers (70, 71). Currently, more than 10 purine antimetabolites have been approved by the Food and Drug Administration for treatment of cancers, such as 6-mercaptopurine, 6-thioguanine, and methotrexate. Purine antimetabolites are chemical analogs sharing with similar structure to the metabolites in the purine metabolism and can compete to incorporate into purine nucleotides and DNA during the S phase of the cell cycle to inhibit rapid division and proliferation (72, 73). Unfortunately, resistance of antimetabolites often occurs, and various genes are involved in the intolerance (74, 75). In addition, purine antimetabolites also affect the proliferation of healthy cells and thereby cause potential toxicity to normal cells. Therefore, it is urgent to identify new regulatory targets within purine metabolism, which would inhibit tumorigenesis without drug resistance and hurting normal cells.
Targeting purine degradation has also been used to treat cancers as enhanced purine degradation limits the available purines for nucleotides synthesis, resulting in inhibition of cell proliferation in cancers. For example, an intratumoral injection of adenoviral vector expressing E. coli PNP to accelerate inosine degradation shows safety and antitumor activity in the first-in-human clinical trial (76). Activation of XO suppresses disulfide bond formation of breast cancer resistance protein (77), and reactive oxygen species derived from XO further interrupt dimerization of breast cancer resistance protein (78, 79).
Considering that purinosome formation plays an important role in de novo purine biosynthesis, one strategy may arise by targeting purinosome assembly/disassembly in cancers. For example, G protein-coupled receptors (GPCRs) regulate a myriad of biological responses via multiple signaling pathways in both normal and cancer cells (80–82). GPCRs have also been demonstrated to affect purinosome assembly/disassembly to control metabolic flux via de novo purine biosynthesis in human cancer cells (83, 84), which may contribute to the regulatory mechanism of GPCRs in cancers. Heat shock protein 90 (Hsp90) and Hsp70 functionally colocalize with purinosomes (85), thus inhibitors of Hsp90 and Hsp70 reversibly disrupt purinosome formation, and have a synergistic effect with methotrexate (86) to treat cancers (87, 88). Most cancer cells abundantly express Hsp90/Hsp70 (89), and its chaperone machinery in the assembly of the purinosome may provide a novel strategy for the development of advanced anticancer therapies via disrupting purine biosynthesis. Upregulations of specific pathway enzymes (i.e., PPAT, PAICS, and ATIC) in numerous cancers indicate an importance of purine metabolism and purinosomes in tumorigenesis (41, 90), while mutations of these enzymes affect purinosome assembly in cultured skin fibroblasts from patients with AICA-ribosiduria and ADSL deficiency (27). In conclusion, disruption of purinosome formation is likely to mediate cell cycle and to enhance sensitivity to cancer chemotherapeutics (86).
mTOR-Mediated-Purinosome and Purine Metabolism in Cancers
Purinosomes and mitochondria interaction may improve the efficiency of de novo purine biosynthesis, thus disruption in the purinosome juxtaposition to the mitochondria may serve as a novel therapeutic potential for cancers. Using human kinome screen, mTOR has been identified as a putative kinase network associated with the translation of chemical signals into purinosome and mitochondria interaction (31). In sporadic cancers, mTOR activation is the result of amplification/activation mutations in genes encoding upstream tumor signal transduction cascades or deletion/inactivation of tumor suppressors (91–93). In response to proliferating signal in cancer cells, mTOR activates ATF4, which stimulates the expressions of MTHFD2 and other enzymes for serine synthesis and THF cycle, providing amino acid substrates and one-carbon units required for the de novo purine synthesis (30). Rapamycin (a mTOR inhibitor) inhibits the interaction between purinosomes and mitochondria in a dose-dependent manner (31). Mitochondria dysregulation enhances the formation of purinosomes and the percentage of purinosome-positive cells, while these increases are abrogated by rapamycin (31). Meanwhile, mTOR is also suppressed by a de novo purine synthesis antagonist (AG2037) that reduce intracellular purine nucleotide pools via reducing the level of GTP-bound Rheb, an obligate upstream activator of mTOR complex 1 (mTORC1) (94). Meanwhile, AG2037 treatment markedly inhibits mTORC1 activation and robust tumor growth in mice bearing non-small-cell lung cancer xenografts (94). These results confirm the mutual interaction among mTOR, mitochondria, and the de novo purine biosynthetic pathway, and further studies are needed to explore the therapeutic effects of inhibitors for purine biosynthesis and the underlying mechanism of mTOR-mediated purinosome-mitochondria localization and purine metabolism in cancers.
Recent work has shown that alterations of amino acid metabolism are common observed in cancers, and glutamine is an abundant and versatile nutrient that participates in the growth and metabolism of cancer cells (95–97). Glutamine also serves as a substrate of PPAT and FGAMS for catalyzing PRA and FGAM formation in the de novo purine biosynthesis, which may partially explain why glutamine is one of the most highly consumed nutrient by cancer cells. mTOR also serve as a master regulator that senses amino acid availability to regulate cell growth in normal and cancer cells (98–104). For example, glutamine transportation plays a key role in controlling cellular metabolism, growth, and survival via mTOR signal in lung cancer and breast cancer (105, 106). Also, mTOR-mediated alteration of amino acid substrates (glycine and aspartate) for the de novo purine biosynthesis has been reported in a subset of tumors (107–109). Collectively, these studies suggest that mTOR is a critical regulatory signal in the de novo purine biosynthetic pathway by influencing purinosome and mitochondria interaction and amino acid substrates, which further reprogram metabolic responses in cancer cells (Figure 3).
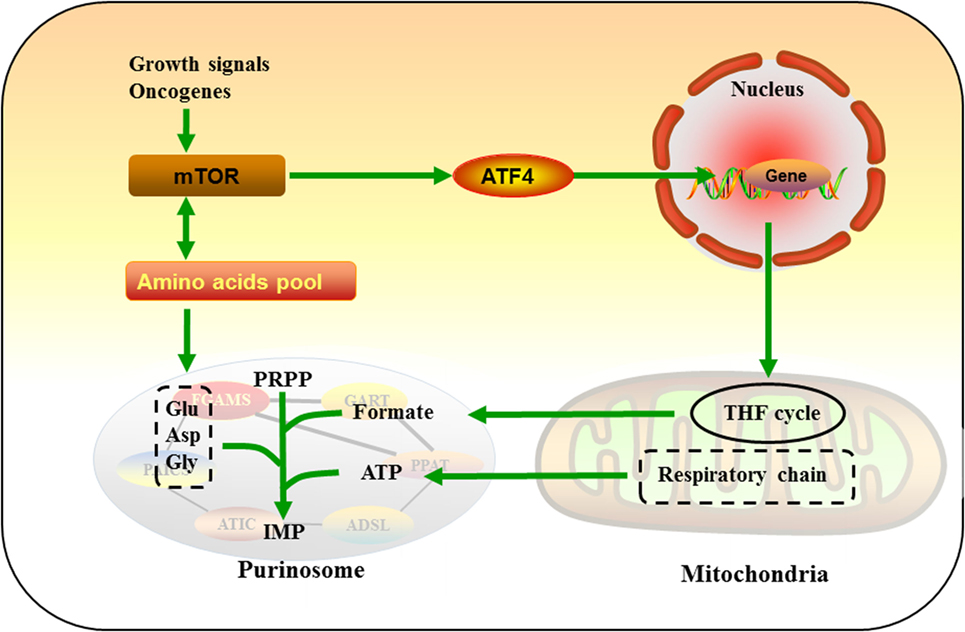
Figure 3. Mammalian target of rapamycin (mTOR)-mediated purine metabolism. In response to growth signal in cancer cells, mTOR activates ATF4, which upregulates enzymes of tetrahydrofolate cycle and provides one-carbon unit formate for de novo purine synthesis. mTOR also serve as a master regulator that senses and mediates amino acids pool, which may further regulate glutamine (Gln), glycine (Gly), and aspartate (Asp) flux into purinosomes.
Conclusion and Future Perspectives
Purines are the scaffold substrates of nucleic acids, coenzymes, allosteric modulators, and energy intermediates for cells. Thus, purine metabolism is associated with several of biochemical reactions, including metabolism, cell cycle, immune function, and signal transduction. Recently, scientists have identified purinosomes, which formed from the de novo purine biosynthesis enzymes to improve the efficiency of metabolic flux in the cells when purines are highly required. In addition to providing ATP for the de novo purine biosynthesis, there may be a potential interaction between mitochondria and purinosome, which further accelerates purine synthesis. Several decades ago, targeting purine metabolism has been used to design the antimetabolite drugs to treat cancers, and purinosome represents a novel target to pharmacologically control cellular metabolism.
Although the current results are inspiring, various questions have been raised after the discovery and characterization of the purinosome. For example, purinosome-mitochondria colocalization plays an important role in purine metabolism, how does purinosome sense and locate mitochondria? Besides mTOR-mediated link between purinosomes and mitochondria, is there any other signaling events involving purinosome formation? Protein-coupled receptors and p38MAPK have been reported to activate purine metabolism to initiate cell cycle in response to stress (25, 83, 110), the mediatory roles of these signals in purinosome need to be further studied.
Author Contributions
JY wrote the manuscript. XH, JD, TL, and YY discussed the topic. WR and XH revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the Public Service Technology Center, Institute of Subtropical Agriculture, Chinese Academy of Sciences for technical support. Many thanks also to the editors and reviewers for the painstaking care taken in helping improve the clarity of the manuscript.
Funding
This study was supported by the National Basic Research Program of China (973) (2013CB127301), National Natural Science Foundation of China (No. 31472106), Hunan Key Research Program (2017NK2320), and China Agriculture Research System (CARS-35).
Abbreviations
mTOR, mammalian target of rapamycin; HPRT, hypoxanthine-guanine phosphoribosyltransferase; APRT, adenine phosphoribosyltransferase; PRPP, phosphoribosylpyrophosphate; IMP, inosine monophosphate; GMP, guanine monophosphate; PNP, purine nucleoside phosphorylase; XO, xanthine-oxidase; THF, tetrahydrofolate; FDA, Food and Drug Administration; GPCRs, G protein-coupled receptors; Hsp90, Heat shock protein 90; NSCLC, non-small-cell lung cancer.
References
1. Heiden MGV, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell (2017) 168(4):657–69. doi:10.1016/j.cell.2016.12.039
2. Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol (2017) 14:11–31. doi:10.1038/nrclinonc.2016.60
3. Vernieri C, Casola S, Foiani M, Pietrantonio F, de Braud F, Longo V. Targeting cancer metabolism: dietary and pharmacologic interventions. Cancer Discov (2016) 6:1315–33. doi:10.1158/2159-8290.CD-16-0615
4. Zaimenko I, Lisec J, Stein U, Brenner W. Approaches and techniques to characterize cancer metabolism in vitro and in vivo. Biochim Biophys Acta (2017) 1868:412–9. doi:10.1016/j.bbcan.2017.08.004
5. Morandi A, Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochim Biophys Acta (2017) 1868:1–6. doi:10.1016/j.bbcan.2016.12.004
6. Corbet C, Feron O. Cancer cell metabolism and mitochondria: nutrient plasticity for TCA cycle fueling. Biochim Biophys Acta (2017) 1868:7–15. doi:10.1016/j.bbcan.2017.01.002
7. Yin J, Ren W, Huang X, Li T, Yin Y. Protein restriction and cancer. Biochim Biophys Acta (2018) 1869:256–62. doi:10.1016/j.bbcan.2018.03.004
8. Li CM, Jeong Y, Kim M. Mammea longifolia Planch. and Triana fruit extract induces cell death in the human colon cancer cell line, SW480, via mitochondria- related apoptosis and activation of p53. J Med Food (2017) 20:485–90. doi:10.1089/jmf.2016.3865
9. Di Virgilio F, Adinolfi E. Extracellular purines, purinergic receptors and tumor growth. Oncogene (2017) 36:293–303. doi:10.1038/onc.2016.206
10. An SG, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science (2008) 320:103–6. doi:10.1126/science.1152241
11. Chan CY, Zhao H, Pugh RJ, Pedley AM, French J, Jones SA, et al. Purinosome formation as a function of the cell cycle. Proc Natl Acad Sci U S A (2015) 112:1368–73. doi:10.1073/pnas.1423009112
12. Maiuolo J, Oppedisano F, Gratteri S, Muscoli C, Mollace V. Regulation of uric acid metabolism and excretion. Int J Cardiol (2016) 213:8–14. doi:10.1016/j.ijcard.2015.08.109
13. Pedley AM, Benkovic SJ. A new view into the regulation of purine metabolism: the purinosome. Trends Biochem Sci (2017) 42:141–54. doi:10.1016/j.tibs.2016.09.009
14. Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature (2013) 496:101–5. doi:10.1038/nature12040
15. Tedeschi PM, Markert EK, Gounder M, Lin HX, Dolfi SC, Chan LLY, et al. Contribution of serine, folate, and glycine metabolism to the ATP, NADPH, and purine requirements of cancer cells. Cell Death Dis (2013) 4:e877. doi:10.1038/cddis.2013.393
16. Laursen T, Borch J, Knudsen C, Bavishi K, Torta F, Martens HJ, et al. Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science (2016) 354:890–3. doi:10.1126/science.aag2347
17. Cull B, Godinho JLP, Rodrigues JCF, Frank B, Schurigt U, Williams RAM, et al. Glycosome turnover in Leishmania major is mediated by autophagy. Autophagy (2014) 10:2143–57. doi:10.4161/auto.36438
18. Hines PJ. Metabolite channeling by a dynamic metabolon. Science (2016) 354:843. doi:10.1126/science.354.6314.843-c
19. Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell (2013) 155:160–71. doi:10.1016/j.cell.2013.08.032
20. Kyoung M, Russell SJ, Kohnhorst CL, Esemoto NN, An S. Dynamic architecture of the purinosome involved in human de novo purine biosynthesis. Biochemistry (2015) 54:870–80. doi:10.1021/bi501480d
21. Zhao H, Chiaro CR, Zhang LM, Smith PB, Chan CY, Pedley AM, et al. Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J Biol Chem (2015) 290:6705–13. doi:10.1074/jbc.M114.628701
22. Pedley AM, Benkovic SJ. Detecting purinosome metabolon formation with fluorescence microscopy. Methods Mol Biol (2018) 1764:279–89. doi:10.1007/978-1-4939-7759-8_17
23. Fridman A, Saha A, Chan A, Casteel DE, Pilz RB, Boss GR. Cell cycle regulation of purine synthesis by phosphoribosyl pyrophosphate and inorganic phosphate. Biocheml J (2013) 454:91–9. doi:10.1042/BJ20130153
24. Law V, Dong S, Rosaless JL, Jeong MY, Zochodne D, Lee KY. Enhancement of peripheral nerve regrowth by the purine nucleoside analog and cell cycle inhibitor, roscovitine. Front Cell Neurosci (2016) 10:238. doi:10.3389/fncel.2016.00238
25. Karigane D, Kobayashi H, Morikawa T, Ootomo Y, Sakai M, Nagamatsu G, et al. p38 alpha activates purine metabolism to initiate hematopoietic stem/progenitor cell cycling in response to stress. Cell Stem Cell (2016) 19:192–204. doi:10.1016/j.stem.2016.05.013
26. Deng YJ, Gam J, French JB, Zhao H, An S, Benkovic SJ. Mapping protein-protein proximity in the purinosome. J Biol Chem (2012) 287:36201–7. doi:10.1074/jbc.M112.407056
27. Baresova V, Skopova V, Sikora J, Patterson D, Sovova J, Zikanova M, et al. Mutations of ATIC and ADSL affect purinosome assembly in cultured skin fibroblasts from patients with AICA-ribosiduria and ADSL deficiency. Hum Mol Genet (2012) 21:1534–43. doi:10.1093/hmg/ddr591
28. Baresova V, Krijt M, Skopova V, Souckova O, Kmoch S, Zikanova M. CRISPR-Cas9 induced mutations along de novo purine synthesis in HeLa cells result in accumulation of individual enzyme substrates and affect purinosome formation. Mol Genet Metab (2016) 119:270–7. doi:10.1016/j.ymgme.2016.08.004
29. Field MS, Anderson DD, Stover PJ. Mthfs is an essential gene in mice and a component of the purinosome. Front Genet (2011) 2:36. doi:10.3389/fgene.2011.00036
30. Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science (2016) 351:728–33. doi:10.1126/science.aad0489
31. French JB, Jones SA, Deng HY, Pedley AM, Kim D, Chan CY, et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science (2016) 351:733–7. doi:10.1126/science.aac6054
32. Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer (2017) 17:93–115. doi:10.1038/nrc.2016.138
33. Sherr CJ. Focus on research a new cell-cycle target in cancer – inhibiting cyclin D-dependent kinases 4 and 6. N Engl J Med (2016) 375:1920–3. doi:10.1056/NEJMp1612343
34. Liang C, Qin Y, Zhang B, Ji SR, Shi S, Xu WY, et al. Metabolic plasticity in heterogeneous pancreatic ductal adenocarcinoma. Biochim Biophys Acta (2016) 1866:177–88. doi:10.1016/j.bbcan.2016.09.001
35. Gill KS, Fernandes P, O’Donovan TR, McKenna SL, Doddakula KK, Power DG, et al. Glycolysis inhibition as a cancer treatment and its role in an anti-tumour immune response. Biochim Biophys Acta (2016) 1866:87–105. doi:10.1016/j.bbcan.2016.06.005
36. Dhillon H, Mamidi S, McClean P, Reindl KM. Transcriptome analysis of piperlongumine-treated human pancreatic cancer cells reveals involvement of oxidative stress and endoplasmic reticulum stress pathways. J Med Food (2016) 19:578–85. doi:10.1089/jmf.2015.0152
37. Ismail A, Noolu B, Gogulothu R, Perugu S, Rajanna A, Babu SK. Cytotoxicity and proteasome inhibition by alkaloid extract from Murraya koenigii leaves in breast cancer cells—molecular docking studies. J Med Food (2016) 19:1155–65. doi:10.1089/jmf.2016.3767
38. Tiloke C, Phulukdaree A, Chuturgoon AA. The antiproliferative effect of Moringa oleifera crude aqueous leaf extract on human esophageal cancer cells. J Med Food (2016) 19:398–403. doi:10.1089/jmf.2015.0113
39. Barfeld SJ, Fazli L, Persson M, Marjavaara L, Urbanucci A, Kaukoniemi KM, et al. Myc-dependent purine biosynthesis affects nucleolar stress and therapy response in prostate cancer. Oncotarget (2015) 6:12587–602. doi:10.18632/oncotarget.3494
40. Di Virgilio F. Purines, purinergic receptors, and cancer. Cancer Res (2012) 72:5441–7. doi:10.1158/0008-5472.CAN-12-1600
41. Goswami MT, Chen GA, Chakravarthi BVSK, Pathi SS, Anand SK, Carskadon SL, et al. Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer. Oncotarget (2015) 6:23445–61. doi:10.18632/oncotarget.4352
42. Bahreyni A, Samani SS, Rahmani F, Behnam-Rassouli R, Khazaei M, Ryzhikov M, et al. Role of adenosine signaling in the pathogenesis of breast cancer. J Cell Physiol (2018) 233:1836–43. doi:10.1002/jcp.25944
43. Soares AS, Costa VM, Diniz C, Fresco P. Inosine strongly enhances proliferation of human C32 melanoma cells through PLC-PKC-MEK1/2-ERK1/2 and PI3K pathways. Basic Clin Pharmacol Toxicol (2015) 116:25–36. doi:10.1111/bcpt.12280
44. Shoshan E, Mobley AK, Braeuer RR, Kamiya T, Huang L, Vasquez ME, et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol (2015) 17:311–21. doi:10.1038/ncb3110
45. Dominissini D, Moshitch-Moshkovitz S, Amariglio N, Rechavi G. Adenosine-to-inosine RNA editing meets cancer. Carcinogenesis (2011) 32:1569–77. doi:10.1093/carcin/bgr124
46. Salinas I, Esteban MA, Meseguer J. Purine nucleosides downregulate innate immune function and arrest apoptosis in teleost fish leucocytes. J Immunol (2007) 178.
48. Alvarado AG, Thiagarajan PS, Mulkearns-Hubert EE, Silver DJ, Hale JS, Alban TJ, et al. Glioblastoma cancer stem cells evade innate immune suppression of self-renewal through reduced TLR4 expression. Cell Stem Cell (2017) 20:450–61. doi:10.1016/j.stem.2016.12.001
49. Byun DJ, Wolchok JD, Rosenberg LM, Girotra M. Cancer immunotherapy – immune checkpoint blockade and associated endocrinopathies. Nat Rev Endocrinol (2017) 13:195–207. doi:10.1038/nrendo.2016.205
50. Grivas P, Koshkin VS, Pal SK. Cancer vaccines at the age of immune checkpoint inhibitors: reasonable approach as combination therapy in advanced urothelial carcinoma? Ann Oncol (2017) 28:680–2. doi:10.1093/annonc/mdx063
51. Rocha KC, Vieira MLD, Beltrame RL, Cartum J, Alves SIPMD, Azzalis LA, et al. Impact of selenium supplementation in neutropenia and immunoglobulin production in childhood cancer patients. J Med Food (2016) 19:560–8. doi:10.1089/jmf.2015.0145
52. Burnstock G, Boeynaems JM. Purinergic signalling and immune cells. Purinergic Signal (2014) 10:529–64. doi:10.1007/s11302-014-9427-2
53. Rai B, Kaur J, Jacobs R, Anand SC. Adenosine deaminase in saliva as a diagnostic marker of squamous cell carcinoma of tongue. Clin Oral Investig (2011) 15:347–9. doi:10.1007/s00784-010-0404-z
54. Dhankhar R, Dahiya K, Sharma TK, Ghalaut VS, Atri R, Kaushal V. Diagnostic significance of adenosine deaminase, uric acid and C-reactive protein levels in patients of head and neck carcinoma. Clin Lab (2011) 57:795–8.
55. Antonioli L, Blandizzi C, Pacher P, Hasko G. Immunity, inflammation and cancer: a leading role for adenosine. Nat Rev Cancer (2013) 13:842–57. doi:10.1038/nrc3613
56. Kumar V. Adenosine as an endogenous immunoregulator in cancer pathogenesis: where to go? Purinergic Signal (2013) 9:145–65. doi:10.1007/s11302-012-9349-9
57. Muller-Haegele S, Muller L, Whiteside TL. Immunoregulatory activity of adenosine and its role in human cancer progression. Expert Rev Clin Immunol (2014) 10:897–914. doi:10.1586/1744666X.2014.915739
58. Naushad SM, Pavani A, Rupasree Y, Divyya S, Deepti S, Digumarti RR, et al. Association of aberrations in one-carbon metabolism with molecular phenotype and grade of breast cancer. Mol Carcinog (2012) 51:E32–41. doi:10.1002/mc.21830
59. Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest (2013) 123:3678–84. doi:10.1172/JCI69600
60. Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science (2012) 336:1040–4. doi:10.1126/science.1218595
61. Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature (2011) 476:346–50. doi:10.1038/nature10350
62. Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci (2014) 39:191–8. doi:10.1016/j.tibs.2014.02.004
63. Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer (2016) 16:650–62. doi:10.1038/nrc.2016.81
64. Wong CC, Qian Y, Li XN, Xu JY, Kang W, Tong JH, et al. SLC25A22 promotes proliferation and survival of colorectal cancer cells with KRAS mutations and xenograft tumor progression in mice via intracellular synthesis of aspartate. Gastroenterology (2016) 151:945–60. doi:10.1053/j.gastro.2016.07.011
65. Yang LF, Achreja A, Yeung TL, Mangala LS, Jiang DH, Han C, et al. Targeting stromal glutamine synthetase in tumors disrupts tumor microenvironment-regulated cancer cell growth. Cell Metab (2016) 24:685–700. doi:10.1016/j.cmet.2016.10.011
66. van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D, et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene (2016) 35:3201–8. doi:10.1038/onc.2015.381
67. Newman AC, Maddocks ODK. One-carbon metabolism in cancer. Br J Cancer (2017) 116:1499–504. doi:10.1038/bjc.2017.118
68. Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature (2017) 544:372–6. doi:10.1038/nature22056
69. Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature (2013) 493:542–6. doi:10.1038/nature11743
70. Fridley BL, Batzler A, Li L, Li F, Matimba A, Jenkins GD, et al. Gene set analysis of purine and pyrimidine antimetabolites cancer therapies. Pharmacogenet Genomics (2011) 21:701–12. doi:10.1097/FPC.0b013e32834a48a9
71. Parker WB. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer. Chem Rev (2009) 109:2880–93. doi:10.1021/cr900028p
72. Peters GJ. Novel developments in the use of antimetabolites. Nucleosides Nucleotides Nucleic Acids (2014) 33:358–74. doi:10.1080/15257770.2014.894197
73. Tiwari M. Antimetabolites: established cancer therapy. J Cancer Res Ther (2012) 8:510–9. doi:10.4103/0973-1482.106526
74. Bennett LL, Brockman RW, Schnebli HP, Chumley S, Dixon GJ, Schabel FM, et al. Activity and mechanism of action of 6-methylthiopurine ribonucleoside in cancer cells resistant to 6-mercaptopurine. Nature (1965) 205:1276. doi:10.1038/2051276a0
75. Xie T, Geng J, Wang Y, Wang LY, Huang MX, Chen J, et al. FOXM1 evokes 5-fluorouracil resistance in colorectal cancer depending on ABCC10. Oncotarget (2017) 8:8574–89. doi:10.18632/oncotarget.14351
76. Rosenthal EL, Chung TK, Parker WB, Allan PW, Clemons L, Lowman D, et al. Phase I dose-escalating trial of Escherichia coli purine nucleoside phosphorylase and fludarabine gene therapy for advanced solid tumors. Ann Oncol (2015) 26:1481–7. doi:10.1093/annonc/mdv196
77. Ogura J, Sasaki S, Kuwayama K, Kaneko C, Koizumi T, Yabe K, et al. Activation of xanthine oxidase suppresses disulfide bond formation of breast cancer resistance protein in Caco-2 cells. Drug Metab Rev (2014) 45:265.
78. Ogura J, Kuwayama K, Sasaki S, Kaneko C, Koizumi T, Yabe K, et al. Reactive oxygen species derived from xanthine oxidase interrupt dimerization of breast cancer resistance protein, resulting in suppression of uric acid excretion to the intestinal lumen. Biochem Pharmacol (2015) 97:89–98. doi:10.1016/j.bcp.2015.06.021
79. Sakuma S, Abe M, Kohda T, Fujimoto Y. Hydrogen peroxide generated by xanthine/xanthine oxidase system represses the proliferation of colorectal cancer cell line Caco-2. J Clin Biochem Nutr (2015) 56:15–9. doi:10.3164/jcbn.14-34
80. Guixa-Gonzalez R, Albasanz JL, Rodriguez-Espigares I, Pastor M, Sanz F, Marti-Solano M, et al. Membrane cholesterol access into a G-protein-coupled receptor. Nat Commun (2017) 8. doi:10.1038/ncomms14505
81. Hasenoehrl C, Feuersinger D, Graf R, Heitzer E, Haybaeck J, Schuligoi R, et al. Differential roles of G protein-coupled receptors GPR55 and CB1 in colon cancer. Gastroenterology (2016) 150:S82–3. doi:10.1016/S0016-5085(16)30394-8
82. Ristic B, Bhutia YD, Ganapathy V. Cell-surface G-protein-coupled receptors for tumor-associated metabolites: a direct link to mitochondrial dysfunction in cancer. Biochim Biophys Acta (2017) 1868:246–57. doi:10.1016/j.bbcan.2017.05.003
83. Verrier F, An SO, Ferrie AM, Sun HY, Kyoung M, Deng HY, et al. GPCRs regulate the assembly of a multienzyme complex for purine biosynthesis. Nat Chem Biol (2011) 7:909–15. doi:10.1038/nchembio.690
84. Fang Y, French J, Zhao H, Benkovic S. G-protein-coupled receptor regulation of de novo purine biosynthesis: a novel druggable mechanism. Biotechnol Genet Eng Rev (2013) 29:31–48. doi:10.1080/02648725.2013.801237
85. Pedley AM, Karras GI, Zhang X, Lindquist S, Benkovic SJ. Role of HSP90 in the regulation of de novo purine biosynthesis. Biochemistry (2018) 57:3217–21. doi:10.1021/acs.biochem.8b00140
86. French JB, Zhao H, An SO, Niessen S, Deng YJ, Cravatt BF, et al. Hsp70/Hsp90 chaperone machinery is involved in the assembly of the purinosome. Proc Natl Acad Sci U S A (2013) 110:2528–33. doi:10.1073/pnas.1300173110
87. Singh R, Fouladi-Nashta AA, Li D, Halliday N, Barrett DA, Sinclair KD. Methotrexate induced differentiation in colon cancer cells is primarily due to purine deprivation. J Cell Biochem (2006) 99:146–55. doi:10.1002/jcb.20908
88. Rinaldi G, Galanti D, Stagno A, Bronte E, Marchese A, Badalamenti G, et al. Methotrexate chemotherapy in elderly patients with locally advanced head and neck cancer: preliminary results. Ann Oncol (2016) 27.
89. Schilling D, Garrido C, Combs SE, Multhoff G. The Hsp70 inhibiting peptide aptamer A17 potentiates radiosensitization of tumor cells by Hsp90 inhibition. Cancer Lett (2017) 390:146–52. doi:10.1016/j.canlet.2017.01.015
90. Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun (2014) 5. doi:10.1038/ncomms4128
91. Karki R, Man SM, Malireddi RKS, Kesavardhana S, Zhu QF, Burton AR, et al. NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature (2016) 540:583. doi:10.1038/nature20597
92. Chiarini F, Evangelisti C, McCubrey JA, Martelli AM. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol Sci (2015) 36:124–35. doi:10.1016/j.tips.2014.11.004
93. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell (2017) 168:960–76. doi:10.1016/j.cell.2017.02.004
94. Emmanuel N, Ragunathan S, Shan Q, Wang F, Giannakou A, Huser N, et al. Purine nucleotide availability regulates mTORC1 activity through the Rheb GTPase. Cell Rep (2017) 19:2665–80. doi:10.1016/j.celrep.2017.05.043
95. Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science (2016) 353:1161–5. doi:10.1126/science.aaf5171
96. Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer (2016) 16:619–34. doi:10.1038/nrc.2016.71
97. Kim JH, Lee KJ, Kwon JH, Lee HJ, Park SJ, Cheon JH, et al. The mechanism of differential effect of metformin on colorectal cancer cells: metabolic alteration via glutamine pathway. Gastroenterology (2016) 150:S1016–1016. doi:10.1016/S0016-5085(16)33441-2
98. Pezze PD, Ruf S, Sonntag AG, Langelaar-Makkinje M, Hall P, Heberle AM, et al. A systems study reveals concurrent activation of AMPK and mTOR by amino acids. Nat Commun (2016) 7. doi:10.1038/ncomms13254
99. Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, et al. A tumor suppressor complex with GAP activity for the rag GTPases that signal amino acid sufficiency to mTORC1. Science (2013) 340:1100–6. doi:10.1126/science.1232044
100. Ren WK, Yin J, Duan JL, Liu G, Tan B, Yang G, et al. mTORC1 signaling and IL-17 expression: defining pathways and possible therapeutic targets. Eur J Immunol (2016) 46:291–9. doi:10.1002/eji.201545886
101. Yin J, Li YY, Zhu XT, Han H, Ren WK, Chen S, et al. Effects of long-term protein restriction on meat quality, muscle amino acids, and amino acid transporters in pigs. J Agric Food Chem (2017) 65:9297–304. doi:10.1021/acs.jafc.7b02746
102. Ren W, Liu G, Yin J, Tan B, Wu G, Bazer FW, et al. Amino-acid transporters in T-cell activation and differentiation. Cell Death Dis (2017) 8. doi:10.1038/cddis.2016.222
103. Ren W, Yin J, Xiao H, Chen S, Liu G, Tan B, et al. Intestinal microbiota-derived GABA mediates interleukin-17 expression during enterotoxigenic Escherichia coli infection. Front Immunol (2016) 7:685. doi:10.3389/fimmu.2016.00685
104. Yin J, Li Y, Han H, Zheng J, Wang L, Ren W, et al. Effects of Lysine deficiency and Lys-Lys dipeptide on cellular apoptosis and amino acids metabolism. Mol Nutr Food Res (2017) 61. doi:10.1002/mnfr.201600754
105. Moses MA, Neckers L. The GLU that holds cancer together: targeting GLUtamine transporters in breast cancer. Cancer Cell (2015) 27:317–9. doi:10.1016/j.ccell.2015.02.010
106. Hassanein M, Hoeksema MD, Shiota M, Qian J, Harris BK, Chen HD, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res (2013) 19:560–70. doi:10.1158/1078-0432.CCR-12-2334
107. Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer (2013) 13:572–83. doi:10.1038/nrc3557
108. Gui DY, Sullivan LB, Luengo A, Hosios AM, Bush LN, Gitego N, et al. Environment dictates dependence on mitochondrial complex I for NAD plus and aspartate production and determines cancer cell sensitivity to metformin. Cell Metab (2016) 24:716–27. doi:10.1016/j.cmet.2016.09.006
109. Allen EL, Ulanet DB, Pirman D, Mahoney CE, Coco J, Si YG, et al. Differential aspartate usage identifies a subset of cancer cells particularly dependent on OGDH. Cell Rep (2016) 17:876–90. doi:10.1016/j.celrep.2016.09.052
Keywords: purine, purinosome, metabolism, cancers, mammalian target of rapamycin
Citation: Yin J, Ren W, Huang X, Deng J, Li T and Yin Y (2018) Potential Mechanisms Connecting Purine Metabolism and Cancer Therapy. Front. Immunol. 9:1697. doi: 10.3389/fimmu.2018.01697
Received: 01 May 2018; Accepted: 10 July 2018;
Published: 30 July 2018
Edited by:
Pinyi Lu, Biotechnology HPC Software Applications Institute (BHSAI), United StatesReviewed by:
Stefania Ceruti, Università degli Studi di Milano, ItalyTingyu Liu, Novartis Institutes for BioMedical Research, United States
Copyright: © 2018 Yin, Ren, Huang, Deng, Li and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenkai Ren, cmVud2Vua2FpMTlAMTI2LmNvbQ==;
Tiejun Li, dGpsaUBpc2EuYWMuY24=;
Yulong Yin, eWlueXVsb25nQGlzYS5hYy5jbg==
 Jie Yin
Jie Yin Wenkai Ren
Wenkai Ren Xingguo Huang3,4
Xingguo Huang3,4