Commentary: Group 3 innate lymphoid cells mediate early protective immunity against tuberculosis
- 1Department of Immunobiology, Institute of Tissue Transplantation and Immunology, Jinan University, Guangzhou, China
- 2Key Laboratory of Functional Protein Research of Guangdong, Higher Education Institutes, Jinan University, Guangzhou, China
- 3School of Stomatology, Jinan University, Guangzhou, China
A commentary on
Regulatory Innate Lymphoid Cells Control Innate Intestinal Inflammation
by Wang S, Xia P, Chen Y, Qu Y, Xiong Z, Ye B, et al. Cell (2017) 171:201–16.e18. doi: 10.1016/j.cell.2017.07.027
Over the past decade, innate lymphoid cells (ILCs) have been identified as a subpopulation of innate immune cells. These cells exist mostly in intestinal mucosal tissue and express no lymphoid differentiation lineage negative (Lin−) markers and antigen-specific receptors that make them distinct from T cells or B cells (1). They were originally classified as ILC1, ILC2, and ILC3 based on their phenotypic features and functions (2). ILC1 cells secrete interferon-γ (IFN-γ) after interleukin-12 (IL-12), IL-15, and IL-18 stimulation and express the transcription factor T-bet (3). ILC2 cells mainly produce the type 2 T helper cells like cytokines IL-5, IL-9, and IL-13 in response to the stimulation of IL-25 and IL-33 in the presence of the transcriptional factor GATA3 (4). ILC3 cells largely generate IFN-γ, IL-17, and IL-22 under the stimulation of IL-1 and IL-23 in the presence of the transcription factors RORγt and aryl hydrocarbon receptor (AhR) (5). Hence, the ILCs participate in the formation of mucosal immunity, development of lymphoid cells, rehabilitation of tissue injury, and the protection of epithelial barriers.
However, in a recent study published in Cell (6), Lin−CD45+CD127+IL-10+ cells were, for the first time, recognized to represent a novel subset of the IL-10-producing ILCs in mice, called regulatory ILCs (ILCreg). These are mostly present in the gut tissue and differentiate and mature in response to pathogen stimulation. In comparison with other ILCs or regulatory T cells (Treg), the ILCreg possess an exclusive gene identity. They express neither CD4 nor FoxP3, the signature marker of Treg cells (6), but are morphologically similar to lymphoid cells with a high nuclear to cytoplasmic ratio. In addition, the authors reported that the ILCreg expressed various phenotypic ILC markers, including CD25 (IL-2Rα), Sca-1, and CD90 (Thy1), but not ILC1 (NK1.1 and NKp46), ILC2 (ST2 and KLRG1), or ILC3 markers (NKp46, CD4, and RORγt) in inflamed intestinal tissue. The ILCreg inhibited the activation of ILC1 and ILC3 via secretion of IL-10 but did not affect ILC2 with undetectable Il10rb expression, showing that it is a novel ILC subpopulation discriminated from other ILC subsets.
Interestingly, Seehus et al. (7) described a distinct subpopulation of IL-10-producing lung ILC2 cells, termed ILC210, that can suppress pathogenic inflammatory immune responses. Unlike most lung ILC2 with the Thy1.1-negative phenotype, the ILC210 are Thy1.1-positive, generate IL10 to exert modulating roles under the stimulation of IL-33 and downregulate proinflammatory genes, such as Tnf, Lta, Il2, and Ccl1. The ILC210 produce higher levels of transcription factors Id3, Foxf1, Atf3, and Klf2 than ILC2. IL-33 and IL7 instruct ILC2 to produce IL-13, but have no effect on the ILC210 in vitro. Analogous to the ILCreg, the ILC210 have no expression of Foxp3, and IL-2 signaling promotes IL-10 production from ILC210. However, they also express the anti-inflammatory gene Retnla, suggesting that the ILC210 may possess anti-inflammatory properties. Interestingly, the ILC210 exhibit memory-like properties in vivo. The ILC210 are in a resting status after in vivo stimulation is discontinued, but are swiftly reactivated and expanded once again when re-stimulated by a minimal dose of IL-33, similar to memory T cells (7). In addition, a third regulatory subset, CD56+CD3− ILCs were recently described to have regulatory functions in human and mice. They simultaneously express NK cell- and ILC-associated molecules, including CD56, CD94, KLRK1 (NKG2D), killer cell Ig-like receptors (KIRs), NCR3 (NKP30), and NCR1 (NKP46). Consistent with the ILCreg, the CD56+CD3− ILCs express no transcription factor Foxp3, but produce high levels of CD94, various KIRS, EOMES, TBX21, GATA3, RORA, and AhR, suggesting the existence of transcriptional factor profiles overlapping with NK cells, ILC2, and ILC3 (8).
A restricted common helper-like innate lymphoid progenitor (CHILP) express the transcriptional factor PLZF and differentiate into the downstream common ILC precursor (ILCP) that give rise to formation of ILC1, ILC2, and ILC3 subsets (9–11), in which ILC2 are a major ILC population in the lungs to play an important role in protecting airway and lung tissue from virus infection (12). Wang et al. (6) demonstrated that the ILCreg originate from CHILP and not ILCPs within the gut tissue (Figure 1). Dissimilarly, the ILC210 are derived from ILCP, but CD56+CD3− ILCs may originate from NK cells (8). Of note, although all these three subsets are considered to display regulatory roles, they appear to be distinct roles as the ILCreg can relieve intestinal inflammation, ILC210 regulate chronic inflammatory conditions, including allergies, whereas CD56+CD3− ILCs inhibit tumor-infiltrating lymphocytes. Neither the ILCreg, ILC210, nor the CD56+CD3− ILCs express the transcription factor Foxp3. However, both the ILCreg and the ILC210 express the transcription factor ID3, which contains a HLH dimerization domain but lacks the basic DNA binding region, to inhibit inflammation (13). The CD56+CD3− ILCs and the ILC210 express the transcription factor GATA3 and RORα. Unlike ILCreg and ILC210, the CD56+CD3− ILCs produce IL-22 but not IL10, with expressions of ILC3 transcription factor AhR and NK cell transcription factor EOMES. It is likely that the different origins and distributions of these three subsets are responsible for their distinct constitutive expressions of transcription factors and cytokines, and hence their functional diversity.
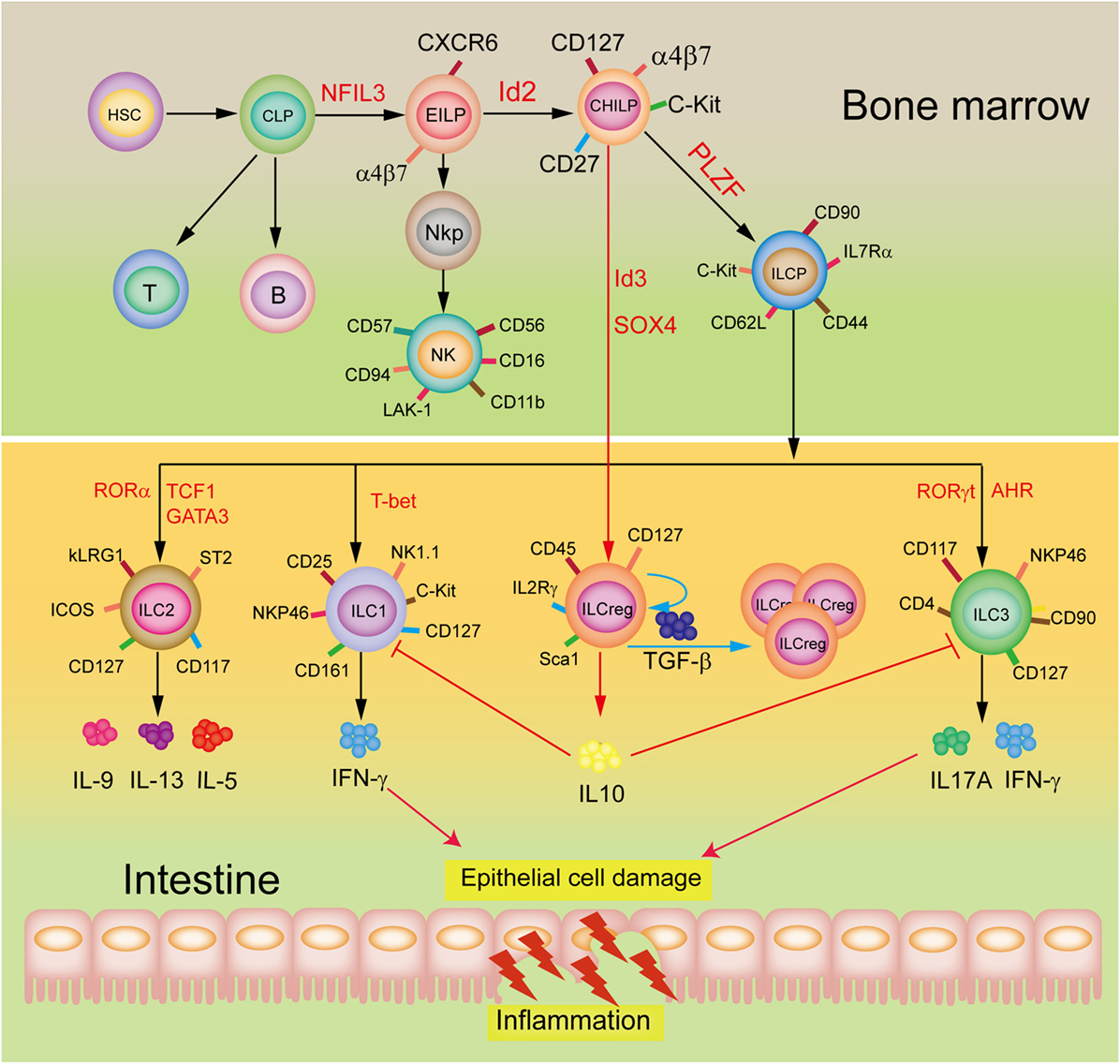
Figure 1. Development and potential functions of regulatory innate lymphoid cells (ILCreg). The ILCreg originate from CHILP not ILCPs and depend on the transcriptional factors Id3 and SOX4. The ILCreg can inhibit the activation of ILC1 and ILC3 to relieve innate intestinal inflammation via IL-10 secretion, and promote self-expansion by secreting TGF-β during inflammation. Abbreviations: CLP, common lymphoid progenitor; CHILP, common helper-like innate lymphoid precursor; EILP, early innate lymphoid precursor; PLZF, promyelocytic leukemia zinc finger; NKP, NK cell progenitor; AhR, aryl hydrocarbon receptor; NFIL3, nuclear factor interleukin 3; GATA3, GATA-binding protein 3; ID2, inhibitor of DNA binding 2; ILCP, ILC precursor; ROR, retinoic acid receptor-related orphan receptor; IFN-γ, interferon-γ; ID3, inhibitor of DNA binding 3; SOX4, sex-determining region Y-box 4; TCF1, T cell factor-1; T-bet, T-box expressed in T cells; TGF-β, transforming growth factor-β.
These new findings not only advances our understanding of intestinal mucosal microenvironment balance but also diseases such as the inflammatory bowel disease, and may contribute significantly toward developing therapeutic applications. Emergence of ILCreg as a novel ILC subset also raises new interesting questions. How do Id3 and SOX4 affect the ILCreg development and fate? Since Treg can generate IL-10 in responses to IL-2 and TGF-β stimulation, is it possible that the ILCreg may be activated by both IL-2 and TGF-β signaling. A mechanistic process by which the ILCreg secrete IL-10 also needs further exploration. Additionally, the populations of ILC1 and ILC3 are obviously increased in the inflamed intestinal mucosa (14). The ILCreg suppress the activation of ILC1 and ILC3 by secreting a high level of IL-10 but why does it not impact ILC2 during intestinal inflammation? An exploration of these will provide a novel insight into the fundamental roles and mechanisms of maintenance of gut innate immune homeostasis and defense against inflamed intestinal mucosa injury.
Author Contributions
FX conceived of this commentary. FX, BZ, and SS drafted it. JL and FX finalized the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers: 81172824, 30971465) and Guangzhou City Science and Technology Program Synergistic Innovation Major Project (grant number: 201604020146) to FX.
References
1. Morita H, Moro K, Koyasu S. Innate lymphoid cells in allergic and nonallergic inflammation. J Allergy Clin Immunol (2016) 138(5):1253–64. doi:10.1016/j.jaci.2016.09.011
2. Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat Immunol (2015) 16(11):1124–33. doi:10.1038/ni.3272
3. Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, et al. Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity (2015) 43(1):146–60. doi:10.1016/j.immuni.2015.06.019
4. Brestoff JR, Kim BS, Saenz SA, Stine RR, Monticelli LA, Sonnenberg GF, et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature (2015) 519(7542):242–6. doi:10.1038/nature14115
5. Magri G, Cerutti A. Role of group 3 innate lymphoid cells in antibody production. Curr Opin Immunol (2015) 33:36–42. doi:10.1016/j.coi.2015.01.008
6. Wang S, Xia P, Chen Y, Qu Y, Xiong Z, Ye B, et al. Regulatory innate lymphoid cells control innate intestinal inflammation. Cell (2017) 171(1):201–16.e18. doi:10.1016/j.cell.2017.07.027
7. Seehus CR, Kadavallore A, Torre B, Yeckes AR, Wang Y, Tang J, et al. Alternative activation generates IL-10 producing type 2 innate lymphoid cells. Nat Commun (2017) 8(1):1900. doi:10.1038/s41467-017-02023-z
8. Crome SQ, Nguyen LT, Lopez-Verges S, Yang SY, Martin B, Yam JY, et al. A distinct innate lymphoid cell population regulates tumor-associated T cells. Nat Med (2017) 23(3):368–75. doi:10.1038/nm.4278
9. Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity (2014) 41(3):354–65. doi:10.1016/j.immuni.2014.09.005
10. Liu M, Zhang C. The role of innate lymphoid cells in immune-mediated liver diseases. Front Immunol (2017) 8:695. doi:10.3389/fimmu.2017.00695
11. Yang Q, Bhandoola A. The development of adult innate lymphoid cells. Curr Opin Immunol (2016) 39:114–20. doi:10.1016/j.coi.2016.01.006
12. Lai D, Tang J, Chen L, Fan EK, Scott MJ, Li Y, et al. Group 2 innate lymphoid cells protect lung endothelial cells from pyroptosis in sepsis. Cell Death Dis (2018) 9(3):369. doi:10.1038/s41419-018-0412-5
13. Miyazaki M, Miyazaki K, Chen S, Itoi M, Miller M, Lu LF, et al. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat Immunol (2014) 15(8):767–76. doi:10.1038/ni.2928
Keywords: regulatory innate lymphoid cells, innate lymphoid cell, development, regulation, inflammatory bowel disease
Citation: Zeng B, Shi S, Liu J and Xing F (2018) Commentary: Regulatory Innate Lymphoid Cells Control Innate Intestinal Inflammation. Front. Immunol. 9:1522. doi: 10.3389/fimmu.2018.01522
Received: 09 March 2018; Accepted: 19 June 2018;
Published: 02 July 2018
Edited by:
Mats Bemark, University of Gothenburg, SwedenReviewed by:
Timotheus You Fu Halim, University of Cambridge, United KingdomCopyright: © 2018 Zeng, Shi, Liu and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Liu, dGpsaXUmI3gwMDA0MDtqbnUuZWR1LmNu;
Feiyue Xing, dGZ5eGluZyYjeDAwMDQwO2pudS5lZHUuY24=
 Boning Zeng
Boning Zeng Shengnan Shi1
Shengnan Shi1 Feiyue Xing
Feiyue Xing