- 1Department of HIV/AIDS, National Institute for Research in Tuberculosis, Chennai, India
- 2Molecular Virology Laboratory, Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore, India
- 3ART Center, Kilpauk Medical College and Hospital, Chennai, India
- 4Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India
- 5Advanced Bioscience Laboratories Inc., Rockville, MD, United States
Strain-specific neutralizing antibodies develop in all human immunodeficiency virus type 1 (HIV-1)-infected individuals. However, only 10–30% of infected individuals produce broadly neutralizing antibodies (bNAbs). Identification and characterization of these bNAbs and understanding their evolution dynamics are critical for obtaining useful clues for the development of an effective HIV vaccine. Very recently, we published a study in which we identified 12 HIV-1 subtype C-infected individuals from India whose plasma showed potent and broad cross-clade neutralization (BCN) ability (1). In the present study, we report our findings on the evolution of host bNAb response over a period of 4 years in a subset of these individuals. Three of the five individuals (NAB033, NAB059, and NAB065) demonstrated a significant increase (p < 0.05) in potency. Interestingly, two of the three samples also showed a significant increase in CD4 binding site-specific antibody response, maintained stable CD4+ T cell counts (>350 cells/mm3) and continued to remain ART-naïve for more than 10 years after initial diagnosis, implying a strong clinical correlation with the development and evolution of broadly neutralizing antibody response against HIV-1.
Introduction
One of the major goals of an HIV-1 vaccine is to induce broadly neutralizing antibodies (bNAbs) that can prevent HIV infection (2). The bNAbs target conserved regions in the HIV-1 envelope (Env) including conformational glycans at positions N160 and N332, the CD4-binding site (CD4BS), gp120-gp41 interface, membrane external proximal region (MPER), and fusion domain in gp41 (2–11). HIV-1 infection in the host is thought to be initiated by one or few transmitted/founder (T/F) viruses (12, 13). Shortly after infection, a strain-specific neutralizing antibody (NAb) response appears against the T/F virus (14–16). Earlier studies have reported that in adults infected with HIV-1, bNAbs usually develop after 2–4 years of infection (17). Interestingly, over a period of time, half of the chronically HIV-1-infected individuals may develop plasma neutralization breadth against 50% of HIV-1 variants (18). The autologous neutralizing response tends to be immunodominant, but when mutations occur in the variable epitopes (V1–V5) in the viral envelope due to error prone reverse transcriptase activity, it leads to virus escape from neutralization (19). Broadly cross-clade neutralizing antibodies (bNAbs) are known to develop in 10–30% of HIV-1 infected individuals. These antibodies bind to the Env trimer and neutralize a broad spectrum of globally circulating HIV-1 strains (20–23). The antibody response is thought to mature with CD4+ T cell help during the course of infection (6). Some recent studies have reported that while non-neutralizing antibodies (n-NAbs) trigger Fc-mediated ADCC, the Fc gamma receptors (FcgRs) play an important role in increasing the neutralizing activity of gp41-specific antibodies (2F5) but not gp120-specific antibodies. Therefore, modifications in the Fc domain to modulate its selective interaction with FcgRs can greatly influence the protective activity of bNAbs (24–27).
The vast majority of HIV-1 infections in India and more than 50% of HIV-1 infections globally is caused by HIV-1 subtype C [UNAIDS 2016 and NACO 2016]. Though a number of recent studies have described the identification and characterization of broadly neutralizing antibodies from HIV-1 subtype C-infected individuals (3, 7, 28–30), very few studies have examined the evolution of the broadly NAb response over time (31, 32). Recently, Patil et al. (33) studied the evolution of broadly NAb response in an elite neutralizer from India and observed an increase in the neutralization activity during follow-up. Improved neutralization activities of bNAbs during the course of evolution have also been reported from individuals infected with other clades of HIV-1 (34–36).
In a previous study, we screened a cohort of (n = 101) HIV-1 subtype C-infected individuals and identified 12 individuals whose plasma exhibited broad cross-clade neutralization (BCN) activity against HIV (1). In the present study, we describe the evolution of the bNAb response in a subset of these individuals in terms of breadth, potency, and neutralization specificity.
Materials and Methods
Ethics Statement
The study was approved by the Institutional Ethics Committee of the National Institute for Research in Tuberculosis (NIRT IEC No: 2011001), and all experiments were performed in accordance with relevant guidelines and regulations. Sample collection was done after obtaining written informed consent from the study participants.
Sample Collection
A follow-up blood sample was obtained from 5 of the 12 individuals identified to have broad cross-clade neutralization property in our previous study (1); follow-up sample could not be obtained from the remaining 7 individuals. The follow-up time point was 4 years after the initial screening, which was within 3 years from the time of initial diagnosis of HIV infection. Blood samples were collected in tri potassium-EDTA (K3-EDTA) treated blood collection tubes (Ref#455036, Greiner Bio-One GmbH, Austria). Plasma was separated by centrifugation at 300 g and stored as 1 mL aliquots at −80°C. The plasma samples were heat inactivated at 56°C for 30 min before use.
Estimation of CD4 + and CD8+ T Cell Counts and CD4/CD8 Ratio
CD4+ and CD8+ T-lymphocyte counts were determined on the day of sample collection, by flow cytometry as described earlier using a FACS count flow cytometer (Becton-Dickinson) (1).
Measurement of HIV-1 Viral Load
Total HIV-1 viral load in cryopreserved plasma was estimated using the fully automated COBAS TaqMan 48 version 2.0 kit (Roche), and expressed as copies of HIV-1 RNA/ml of plasma.
Cell Lines
293 T cells, which are human embryonic kidney cells engineered with the T-antigen from SV40 to support constant growth, were kindly gifted by Dr. Udaykumar Ranga (JNCASR, Bangalore). TZM-bl cells (Catalog Number 8129), also called JC53-BL, were obtained from the NIH AIDS Reagents Repository. This is a genetically engineered HeLa cell clone that expresses CD4, CXCR4, and CCR5 and contains Tat-responsive reporter genes for firefly luciferase and Escherichia coli β-galactosidase under the regulatory control of an HIV-1 Long Terminal Repeat (37, 38). Both cell lines were maintained in DMEM (Lonza, Cat# 12-604 F) containing 10% heat-inactivated fetal bovine serum (Lonza) and 50 µg gentamicin/mL (Sigma) in vented T-75 tissue culture flasks (NUNC). Cultures were incubated at 37°C in a humidified 5% CO2–95% air environment. Cell monolayers were split 1:10 at confluence by treatment with 0.25% trypsin, 1 mM EDTA (Sigma Cat# T4049).
Pseudoviruses
Pseudoviruses were obtained from the NIH AIDS Reagent Repository (www.aidsreagent.org). The pseudovirus panel used in this study is provided in Table S1 in Supplementary Material. Murine leukemia virus (MuLV) env plasmid (env-pseudotyped virus containing the Env of MuLV in the same backbone vector as all HIV-1 env-pseudotyped viruses, MuLV/pSG3Δenv) was kindly gifted by Dr. V. Raghavan (IISc, Bangalore). The MuLV Env-pseudotyped virus was used as negative control in the virus neutralization assays.
Pseudovirus Production and Titration
HEK293T cells were transiently transfected with two plasmids, one carrying the HIV-1 envelope (pEnv), and the other carrying the entire HIV-1 genome except the envelope (pSGΔenv), using the standard calcium phosphate protocol (39). Briefly, exponentially dividing 293 T cells were seeded at a concentration of 0.7 × 106 cells in a T-25 tissue culture flask and incubated at 37°C in a humidified 5% CO2–95% air environment overnight. On the following day, old medium was replaced with fresh medium, and cells were co-transfected with the rev/env expression plasmid (5 µg) and env-deficient HIV-1 backbone vector (pSG3ΔEnv) (10 µg) using the calcium phosphate method, and incubated for 5 h at 37°C in a humidified 5% CO2–95% air environment. After 5 h, the entire medium was removed from the flask and fresh DMEM was added. Pseudovirus-containing culture supernatant was harvested 2 days later, filtered using a 0.45 µm pore size filter, and stored as 1 mL stocks with 20% FBS at −80°C.
The 50% tissue culture infectious dose (TCID50) was determined in TZM-bl cells as described by Montefiori (40). After 2 days of infection, the culture medium was removed from the wells. 100 µL of Britelite Plus reagent (Perkin Elmer) was added to each well, and the plate was incubated at room temperature (RT) for 2 min to allow complete cell lysis. The contents of the wells were mixed by pipettor action (at least two strokes) and 100 µL of cell suspension was transferred to corresponding wells of a 96-well black (opaque) plate. The plate was read immediately in a luminometer and TCID50 was calculated using “TCID” macro (https://www.hiv.lanl.gov/content/nab-reference-strains/htmL/home.htm).
Neutralization Assay
The neutralization potential of the plasma samples was evaluated using the standard neutralization assay with 200 TCID50 pseudoviruses as described earlier (1). In order to determine the neutralization potency (ID50 value), serial dilutions of the plasma samples ranging from 1:20 to 1:43,740 were used. Anti-glycan neutralization activity was evaluated using Du156WT, Du156N160K, Du156N332A, JR-FLWT, and JR-FL E168K pseudoviruses. Further, polyclonal IgG was eluted from the plasma samples using tosylactivated MyOne Dynabeads coupled with 93IN101-gp145 trimer, and neutralization (IC50) and cell-surface binding (EC50) ability of the samples were determined against a tier-2 pseudovirus, JR-FL WT, and its mutant, JR-FL E168K.
Coupling, Adsorption, and Elution of pIndie gp145-Specific Antibodies From BCN Samples
Paramagnetic polystyrene, tosylactivated MyOne Dynabeads (Life Technologies) were coupled with recombinant gp145 protein of Indie-C1 which is an Indian HIV-1 reference strain. The gp145 of Indie-C1 is codon optimized and well-studied (41). Briefly, 0.5 mg of protein was coupled with 2.50 mg of tosylactivated magnetic, i.e., MyOne Dynabeads. Coupling was done at room temperature in a total volume of 1.25 ml in coupling buffer (0.1 M sodium borate buffer [pH 9.5] containing 1 M ammonium sulfate) with gentle rocking for 48 h. The Dynabeads and bound protein were separated from the coupling buffer using a magnet and resuspended in 5 ml of blocking buffer (PBS [pH 7.4] containing 0.5% [wt/vol] bovine serum albumin [BSA] and 0.05% Tween 20) for an additional 48 h. The blocking buffer was removed by aspiration, and the protein-coupled Dynabeads were washed two times with 5 ml of wash buffer (PBS [pH 7.4] containing 0.1% BSA and 0.05% Tween 20). The beads were then resuspended in 0.5 ml of storage buffer (PBS [pH 7.4] supplemented with 0.1% BSA, 0.05% Tween 20, and 0.02% sodium azide + protease inhibitors) and stored at 4°C. On the following day, the protein-coupled beads were washed three times with DMEM containing 10% fetal bovine serum, and incubated in fresh DMEM containing 10% FBS at RT for 30 min to block nonspecific binding to the beads. All the follow-up samples were diluted 1:5 in DMEM containing 10% FBS, and 1 ml of the diluted plasma was incubated with 0.5 ml of beads at RT for 1 h. This step was repeated three times for the better removal of antibodies. Beads coupled with gp145-specific antibodies were stored in PBS containing 0.2% BSA and 0.02% sodium azide at 4°C. The protein-coupled beads were washed three times with PBS containing 500 mM NaCl and once with PBS, and the antibodies were eluted by decreasing the pH in a stepwise manner. First, the beads were mixed with 100 mM glycine-HCl elution buffer (pH 2.7) for 30 s. The acid-eluted solution containing IgG was quickly removed and placed in a separate tube containing 0.5 ml of 1 M Tris (pH 9.0) buffer so as to attain a pH of 7.0 to 7.4. This process was repeated three times. The eluted IgG was diluted in DMEM and concentrated over a 30-kDa Centricon plus filter (Millipore Corp.). Subsequently, the same procedure was performed on the beads at an elution pH of 2.2 to recover any IgG resistant to elution at pH 2.7. The IgG fractions recovered by both acid elution steps were combined and the concentration of the combined IgG fractions was measured using nanodrop technology (Thermofisher). The eluted IgG was characterized using cell-surface binding assay and virus neutralization assay.
Cell-Surface Binding Assay
Human immunodeficiency virus type 1 envelope glycoprotein (Env) expression on the cell surface was achieved as previously described (42). Briefly, 293 T cells were transiently co-transfected with codon-optimized JR-FL wild-type or JR-FL E168K mutant Env-encoding pSVIII plasmid with a pctat plasmid at 3:1 ratio using the calcium phosphate method (1). Cells were harvested 48 h post-transfection, washed thrice with FACS buffer (PBS with 2% FBS), stained with monoclonal neutralizing antibodies or polyclonal IgG eluted from BCN samples, and incubated for 1 h at 37°C at concentrations ranging from 10 µg/ml to 0.0001 µg/ml. After washing, the cells were stained with a PE-conjugated mouse anti-human IgG secondary antibody (1:100 dilutions, BD Bioscience) for 30 min. Cells were again washed and resuspended in 0.5% paraformaldehyde (in PBS). Cell-surface binding of HIV-1 Env-specific antibodies was analyzed by flow cytometry (FACS Verse/Canto analyzer II, BD Bioscience and FlowJo software version 10.0.6, Tree Star Inc.), and binding curves were generated by plotting the mean fluorescence intensity of antigen binding as a function of antibody concentration. Known monoclonal neutralizing antibodies (MAbs) VRC01, PG9, 2G12, and 4E10 that bind specifically to the CD4BS, N160-glycan, N332-glycan, and MPER regions of the HIV-1 envelope, respectively, were used as positive controls.
Enzyme Linked Immunosorbent Assays
Linear PepScan ELISA
Linear gp160 PepScan ELISA was performed as per the procedure described previously (1). Briefly, fifteen amino acid long linear peptides with 11 amino acids overlap, spanning the entire length of gp160 of the Indian subtype C virus, 93IN101 (Table S2 in Supplementary Material), were synthesized commercially (Infinity Biotech and Resource Inc., PA, USA). The peptides were adsorbed onto 96-well ELISA immunomaxisorp plates (Thermo Fisher) at a concentration of 5 µg/mL in 100 mM NaHCO3, pH 9.6, by overnight incubation at 4°C. The plates were washed four times with PBST (1XPBS containing 0.05% Tween20) using a plate washer (ELx50-BioTek) and blocked for 2 h at RT in PBS containing 1% BSA and 0.05% Tween 20. The plates were again washed four times, and heat-inactivated plasma samples diluted 1:50 with the diluent (ABL Inc., MD, USA) were added and incubated at 37°C for 1 h. Following another four washes, goat anti-human IgG horse radish peroxidase conjugated secondary antibody (ThermoFisher) at 1:120 K dilution was added and incubated for 45 min at 37°C. Plates were washed four times with wash buffer and developed using the One-STEP TMB (Thermo Fisher) substrate. After 30 min, the reaction was stopped with 1 N H2SO4, and the plates were read using a microplate reader (ELx808-BioTek). This experiment was performed two times for each sample and mean values were determined to represent the binding reactivity. Healthy human plasma pool (HHP) was used as the negative plasma control.
ELISA With Conformational Proteins
ELISA was performed with 93IN101 gp145 recombinant [cytoplasmic tail (CT) of 93IN101 gp160 completely deleted for the expression of gp145 protein] trimer and dimer proteins that express intact conformational epitopes targeted by the major classes of bNAbs, namely, CD4BS, N160-V1/V2 glycan, N332- V3 glycan and MPER (41), as well as HXB2 gp120 monomer and NL4-3 gp41 monomer proteins (kindly gifted by Dr. V. S. Kalyanaraman, ABL Inc). BCN samples were tested at a dilution of 1:200 following the protocol described earlier (1). HHP was used as the negative control.
ELISA With RSC3 and CD4 Outer Domain (CD4OD) Proteins
ELISA was performed with recombinant re-stabilized core 3 WT (RSC3) protein that is employed widely for the identification of CD4BS antibodies, a double mutant of the RSC3 protein which contains a deletion at position 371 and a substitution at position 363 (Proline substituted with Asparagine), resulting in addition of an N-linked glycan on the β15 strand near the CD4-binding loop (RSC3Δ371I/P363N), and CD4OD protein which comprises of the outer domain of the CD4 molecule that is based on the HIV-1 HXB2 sequence but with better exposure of the highly conserved CD4BS leading to better identification of CD4BS-specific antibodies that could not be identified using RSC3 protein (43–45). BCN samples were tested at a dilution of 1:200 as described previously (1). HHP was used as the negative control.
Healthy Human Plasma Pool (HHP)
Blood samples were collected from 23 healthy HIV negative individuals in tri potassium-EDTA (K3-EDTA) treated blood collection tubes (Ref#455036, Greiner Bio-One GmbH, Austria). Plasma was separated by centrifugation at 300 g for 10 min. Plasma were pooled and stored as 1 mL aliquots at −80°C until use. This plasma was used as the negative control for all our experiments. The plasma was heat inactivated at 56°C for 30 min before use.
Statistical Analysis
All statistical analyses were performed using Graphpad prism 5.0. Experiments were performed two or more times and values obtained from three replicate samples were averaged in each experiment. Statistical significance was tested using t-test. Differences were considered significant at p < 0.05.
Results
Clinical Profile of Study Subjects
Follow-up samples were obtained from five broad cross-clade neutralizers (four males and one female, aged between 35 and 45 years). Four of these individuals had CD4+ T cell counts >350 cells/mm3 (median 489 cells/mm3) at the time of follow-up. Only one had a lower CD4+ T cell count of 204 cells/mm3. Two of the five individuals (NAB033 and NAB059) continued to be ART-naïve till the time of the present study, while the remaining three were started on antiretroviral therapy during the intervening period. Demographic details and clinical profile of the study subjects are provided in Table 1.
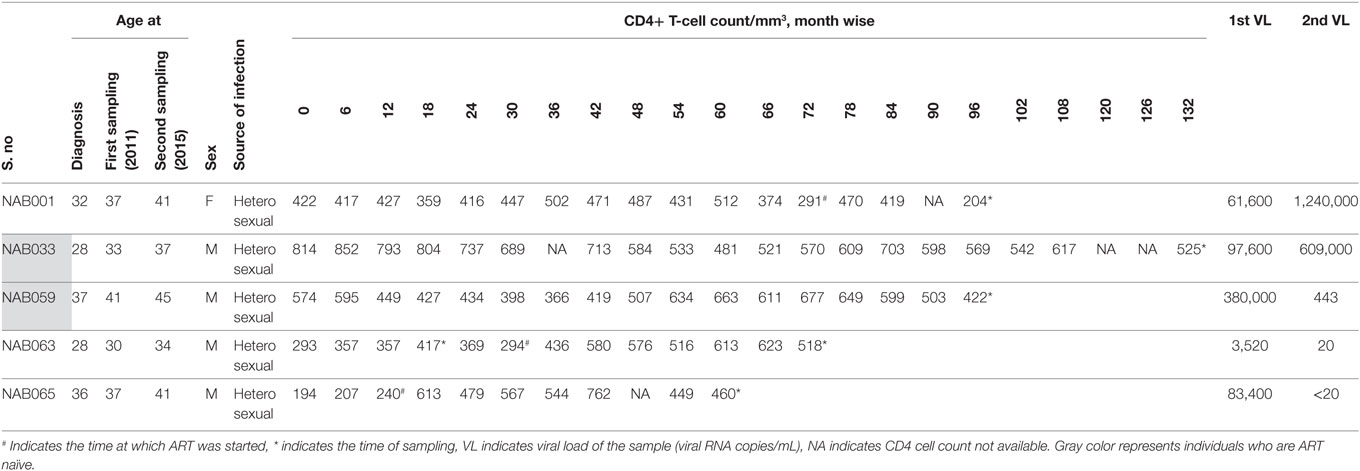
Table 1. Clinical and demographic details of the BCN samples at the time of follow-up sample collection.
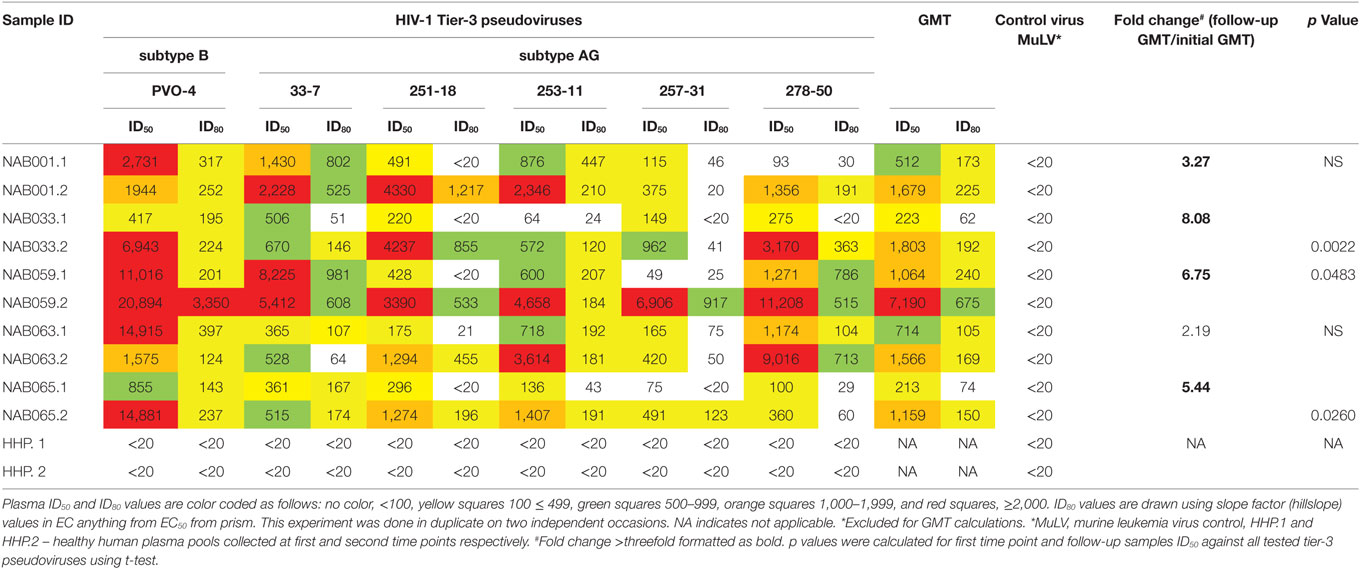
Increase in Neutralization Potency Against Tier-3 Pseudoviruses
Since the plasma samples of the study subjects were tested previously for neutralization against tier-1, tier-2, and tier-3 pseudoviruses (1), the follow-up samples were directly tested with a panel of 6 tier-3 pseudoviruses at varying dilutions. All five samples exhibited strong neutralization potency against the tested viruses. A heat map of the log neutralization titer of the 5 samples is shown in Figure 1; the dendrogram at the top of the figure shows three clusters of two, three and one viruses, with the one on the right (PVO-4) being most sensitive to neutralization, those in the middle (251-18, 253-11, and 278-50) being moderately sensitive to neutralization and those on the left (257-31 and 33-7) being less sensitive to neutralization with the tested samples (Table 2). Results of the neutralization titer analysis of the plasma samples are represented in graphical format in Figure 2. The 50% inhibitory dilution (ID50) values were calculated using a dose-response curve fit with non-linear function in Graph Pad Prism, v. 5.0. The Geometric mean titer (GMT) was calculated for each sample against all the tier-3 pseudoviruses (Figure 2; Table 2). Four of the 5 samples (NAB001, NAB033, NAB063, and NAB065) neutralized all the tier-3 pseudoviruses with a GMT of 1,678; 1,803; 1,565; and 1,159, respectively. The fifth sample (NAB059) showed an extraordinarily high neutralization potency with a GMT of 7190, prompting us to categorize this individual as an elite neutralizer. Further, we compared first time point neutralization potency (1) with that seen during follow-up, and found that the neutralization potency of 3 of the 5 follow-up samples had increased significantly over time (Table 2; Figure S1 in Supplementary Material). Interestingly, two of these individuals (NAB033 and NAB059) continued to maintain stable CD4 counts and remained ART-naïve.
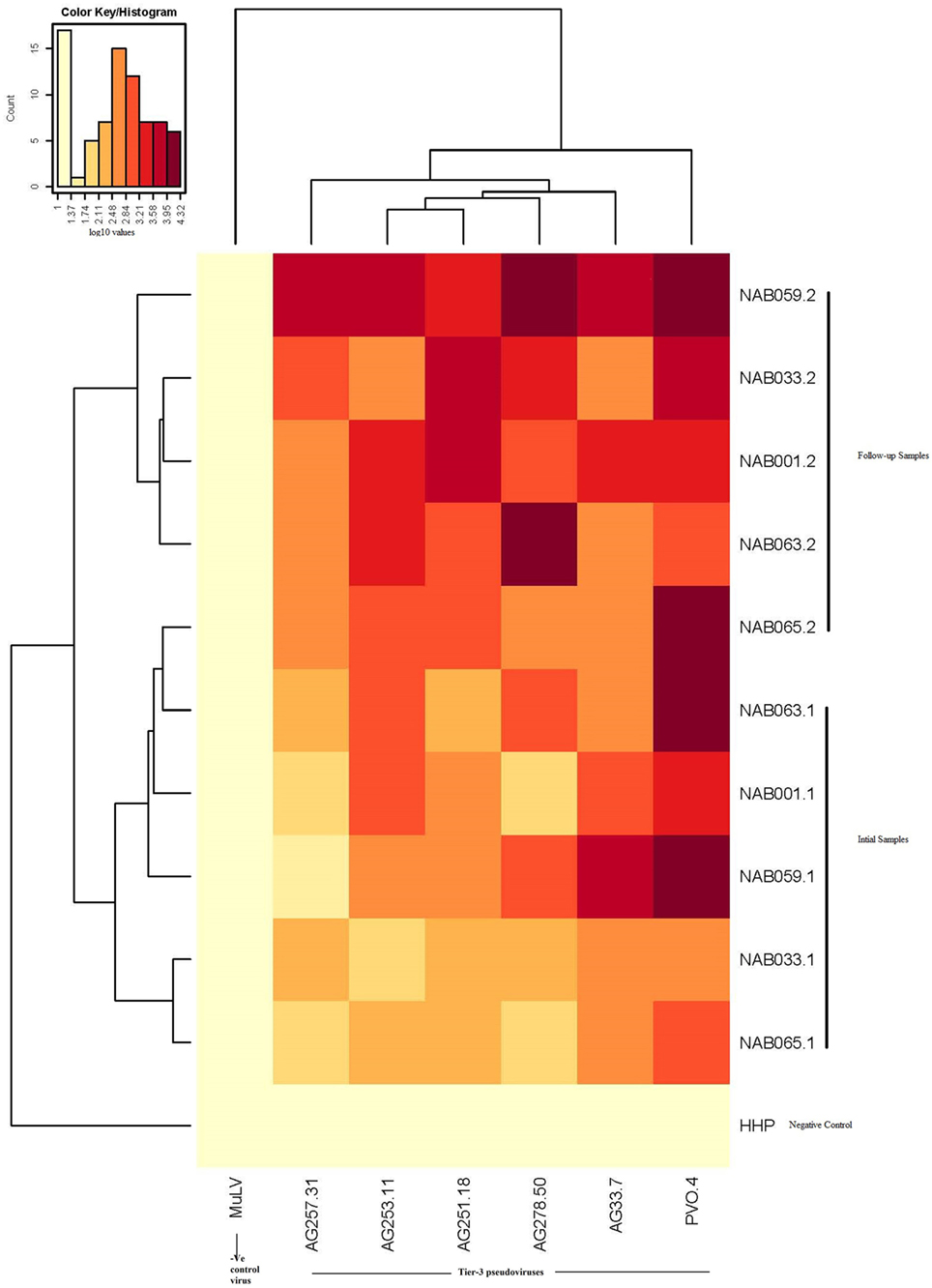
Figure 1. Heat map of the results of the neutralization assay of the follow-up samples tested against tier-3 pseudoviruses, and hierarchical clustering of plasma and viruses based on neutralization titers.
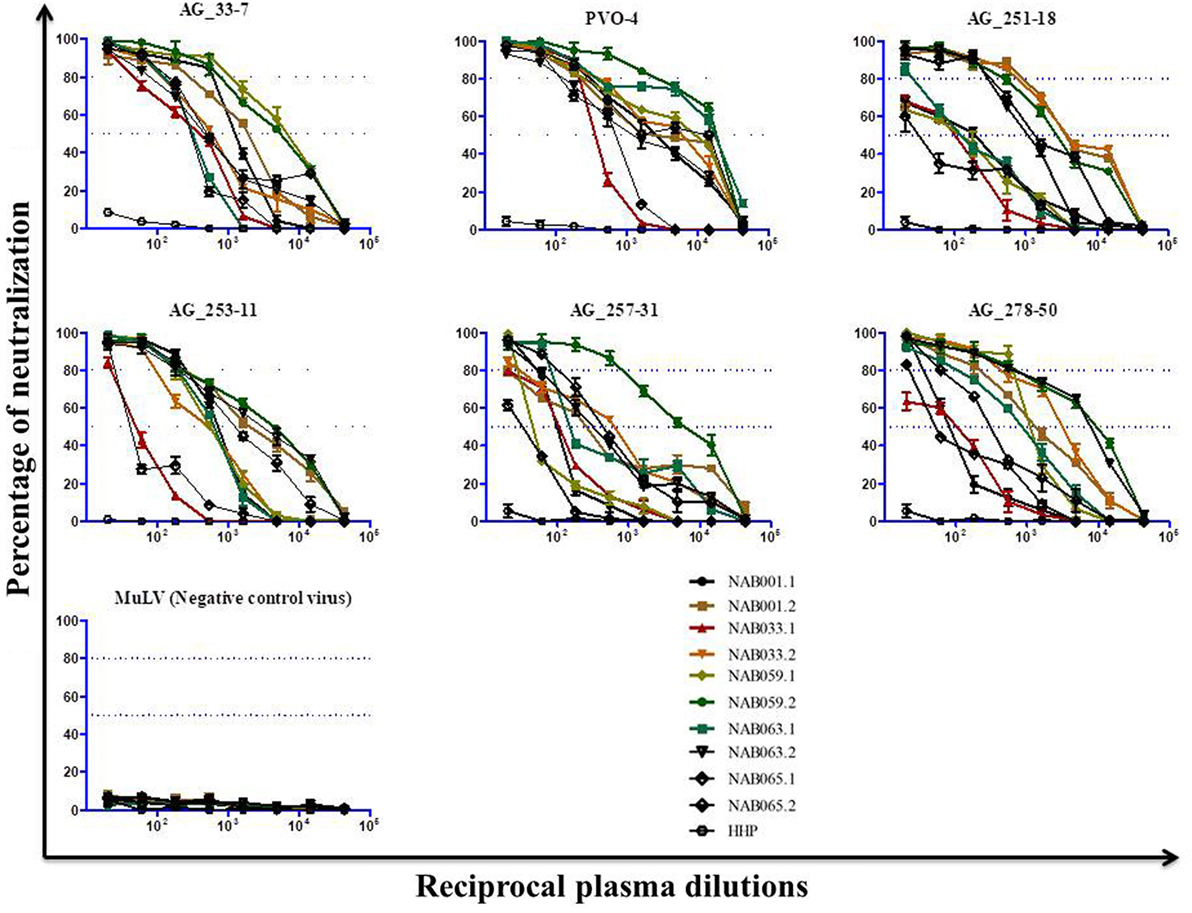
Figure 2. Neutralization activity of the follow-up plasma samples (n = 5) against tier-3 pseudoviruses. Neutralization titration (dilutions 1:20 to 1:43,740) analysis was performed against 6 tier-3 pseudoviruses in TZM-bl cells.
Increase in the Breadth of Reactivity to Env Peptides
PepScan ELISA was performed with linear overlapping HIV-1 gp160 peptides to identify if newer antibody responses were generated in any of the BCN samples. Samples were considered reactive if the sample absorbance was two-fold more than that of the negative control (HHP) (Figure 3; Table S3 in Supplementary Material). Interestingly, samples NAB033 and NAB059, showed newer antibody responses with binding reactivity spread across the regions C2, V4, C4, C5, HR2, and CT of the envelope protein. We also found that samples NAB063 and NAB065 acquired additional binding antibody response to V4, V5, C5 and C3, C4, V4 regions of HIV-1 Env protein. As in the earlier time point, all the second time point samples also bound strongly to the V3 loop (peptide nos. 68 to 70) and Immuno Dominant region of gp41 (peptide nos.140 to 143) (Figure 3). Sample NAB059 showed strong binding reactivity to the C2 region which comprises a part of the CD4BS, and also to the HR1 and HR2 regions. NAB033 showed binding reactivity to the HR2 region. The HR1 and HR2 regions are known to play a major role in virus-host cell fusion.
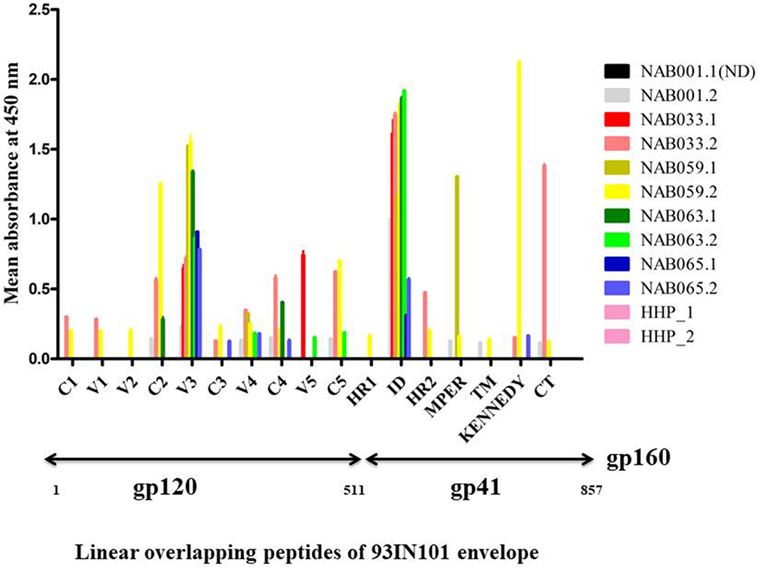
Figure 3. Antibody binding to linear overlapping peptides of 93IN101 Env gp160. ELISA was performed with a total of 192 peptides, 15 amino acids long with 11 amino acids overlap, covering the entire 93IN101 envelope (gp160) protein. Plasma samples from BCN samples (n = 5) were tested at a dilution of 1:50 for determining binding reactivity. Healthy Human Plasma pool (HHP) was used as the negative control. This experiment was performed in duplicate on two independent occasions. Mean absorbance of HHP was 0.054; twofold of HHP absorbance (0.162) was considered as positive reactivity. Normalized absorbance values are used for the graphical representation. ND indicates not done.
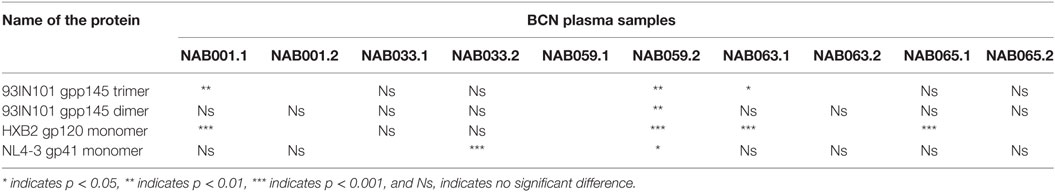
Enhanced Reactivity to Conformational Env Proteins
The plasma samples were tested in an ELISA with different forms of the HIV-1 envelope protein, viz. 93IN101 gp145 trimer and dimer, HXB2 gp120 monomer and NL4-3 gp41 monomer, and the results were compared with that obtained at the first time point (Figure 4; Table 3). Interestingly, the second time point sample of NAB059 showed a significant increase in the binding reactivity to all four conformational proteins as compared to the previous time point (p < 0.05). On the other hand, NAB033 showed a significant increase in binding to the NL4-3 gp41 monomer (p < 0.001) but not to the other forms of the envelope protein. There was no significant increase in the binding reactivity to the conformational envelope proteins over time in the remaining samples.
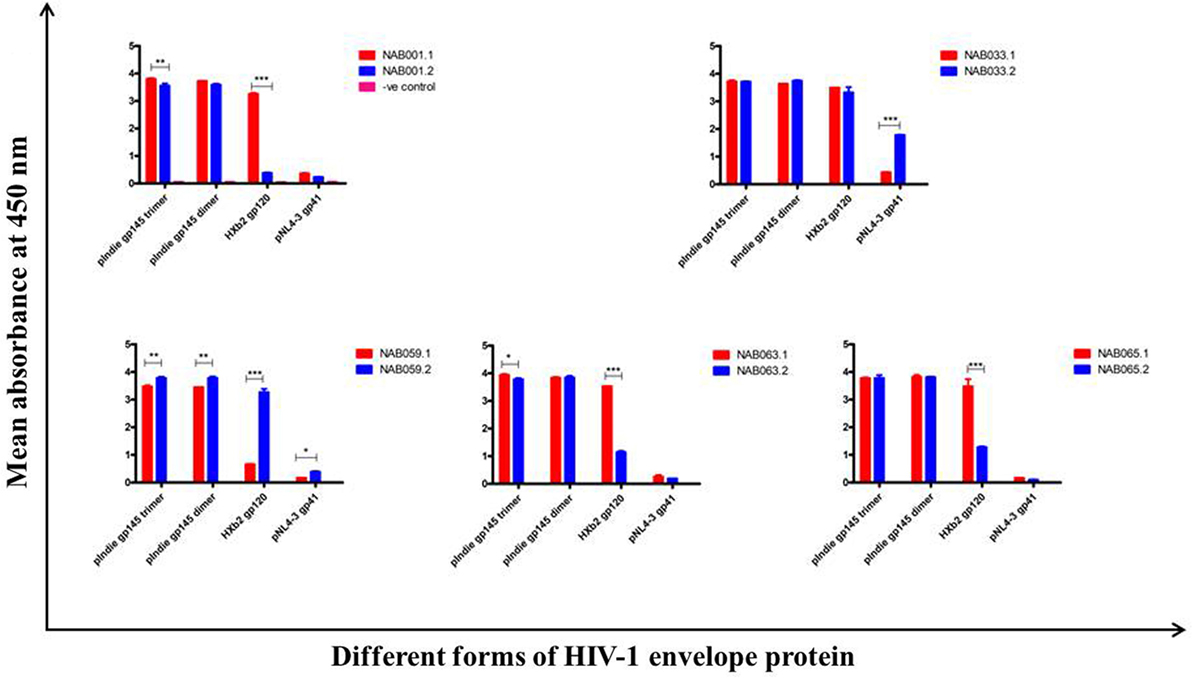
Figure 4. ELISA with recombinant HIV-1 envelope proteins. ELISA was performed with 93IN101 gp145 trimer, dimer, HXB2 gp120 monomer, and NL4-3 gp41 monomer proteins. All the BCN plasma samples were tested at 1:200 dilution, and HHP was used as the negative control. Each experiment was performed in triplicate on two independent occasions. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001.
Increase in CD4BS-Specific Antibody Response Over Time
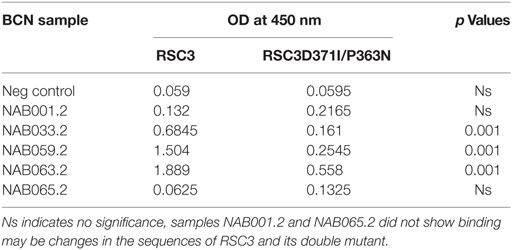
The samples were tested in an ELISA with the wild-type RSC3 and double mutant RSC3Δ371I/P363N CD4BS recombinant proteins, at a dilution of 1:200 to investigate changes in CD4BS specific antibody responses in the BCN samples. Three of the five samples which showed CD4BS specificity at the initial time point again showed CD4BS-specific antibody response as revealed by greater binding response to the RSC3 wild-type protein than to the RSC3 mutant (RSC3Δ371I/P363N) protein (Table 4). When these results were compared with the corresponding results of the first time point samples, it was found that the level of RSC3 binding antibodies had significantly increased with time in two of the three samples (NAB033 and NAB059) (p < 0.001) (Figure 5A).
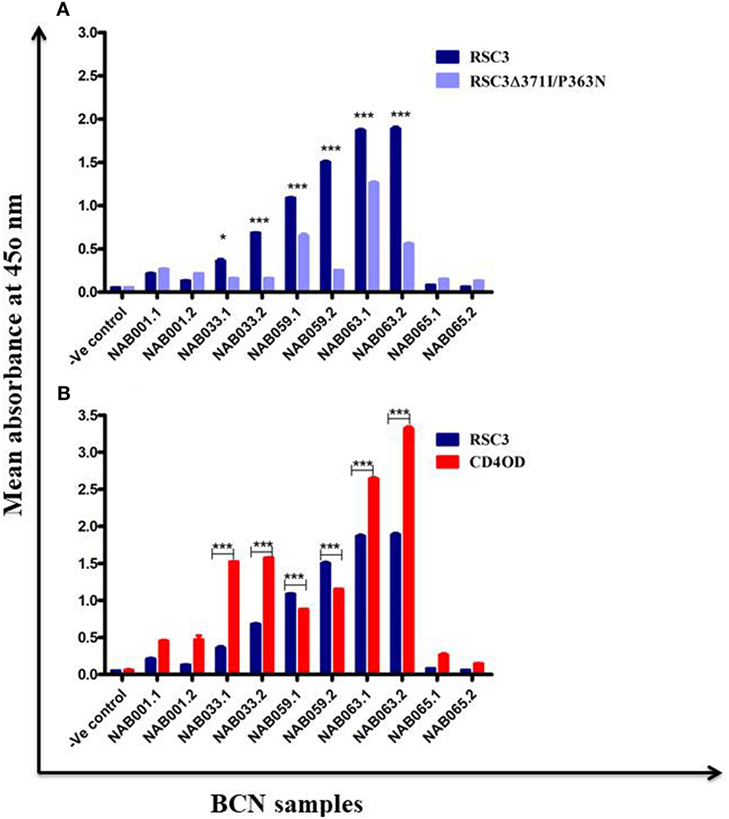
Figure 5. ELISA with recombinant RSC3 WT, RSC3 mutant and CD4 outer domain (CD4OD) proteins. (A) ELISA was performed with recombinant wild type RSC3 and double mutant RSC3Δ371I/P363N proteins. (B) ELISA was performed with HIV-1 subtype B-CD4OD protein and RSC3 recombinant protein. Each experiment was performed in triplicate on two different occasions. Difference was considered significant if p < 0.05.
The recombinant RSC3 protein is designed based on a HIV-1 subtype B strain (8) and has been widely used for the identification and isolation of several well-characterized CD4BS neutralizing antibodies like VRC01, VRC02, VRC03, etc. However, there is sufficient evidence to show that the CD4BS of Indian subtype C HIV-1 Env varies from that of subtype B due to differences at the molecular level (46, 47). Since all our samples were obtained from HIV-1 subtype C-infected individuals, we decided to use an additional HIV-1 envelope protein for identification of CD4BS antibodies. Saha et al. (43) designed the CD4OD protein (CD4OD) for the identification of CD4BS antibodies from HIV-1 subtype C-infected individuals. We performed ELISA with the CD4OD protein and found that four of the five BCN samples showed significantly higher (p < 0.01) binding reactivity to the CD4OD protein than to RSC3 (Figure 5B). In contrast, one sample (NAB059) showed significantly higher (p < 0.001) binding to the RSC3 protein than to the CD4OD protein.
Increase in PG9/PG16 Like NAb Response Over Time
Previously, we had identified four BCN plasma samples with neutralization specificities similar to the PG9/PG16 class of bNAbs (1). However, second time point sample could be collected only from one of the four individuals. This sample (NAB033) was tested in a neutralization assay with pseudoviruses DU156WT and DU156 N160K. Strong neutralization of the DU156 WT pseudovirus was observed (Figure 6A; Table 5) with a 6.7-fold lesser neutralization of the DU156 N160K mutant as compared to the wild type, indicating the persistence of significant amount of antibodies that target the N160 glycan of the HIV-1 Env in this sample.
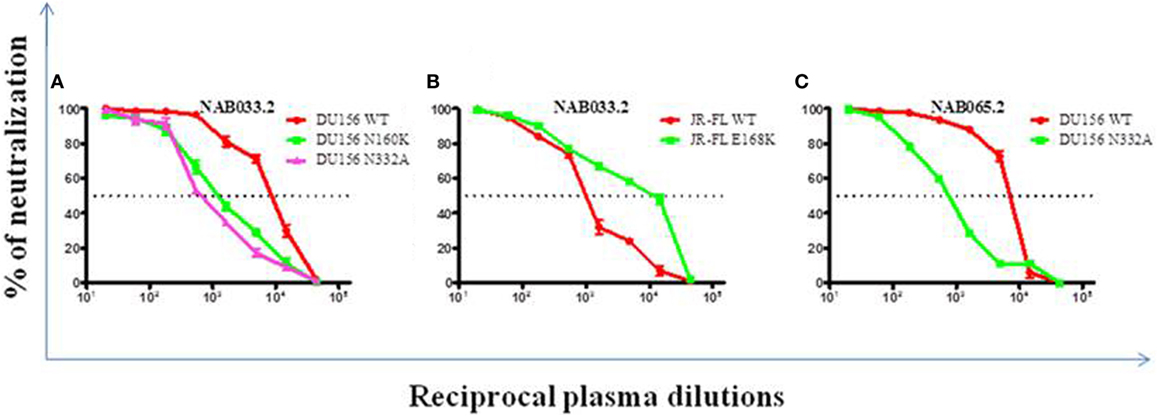
Figure 6. Neutralization analysis to identify glycan-dependent bNAbs. (A) Neutralization assay was performed against DU156WT, DU156N160K, and DU156N332A. (B) Presence of PG9/PG16 like bNAb specificity in sample NAB033 was confirmed by performing neutralization assay with JR-FL WT and JR-FL E168K. (C) Presence of PGT series like bNAbs in the sample NAB065 was confirmed by performing neutralization assay with DU156WT and DU156N332A. Each sample was tested in duplicate (1:20 to 1:43,740 dilutions), and the experiment was repeated on two independent occasions. ID50 values were calculated using non-linear regression with variable loops using Graph pad prism 5.0. software.
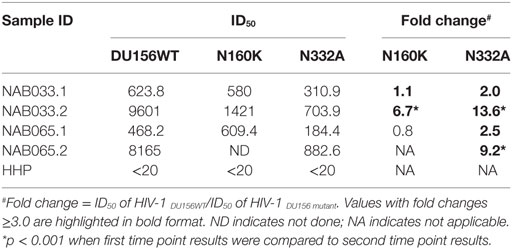
Table 5. Neutralization activity of BCN samples against DU156 pseudovirus with mutations in peptidoglycans targeted by the PG9/PG16 and PGT series of bNAbs.
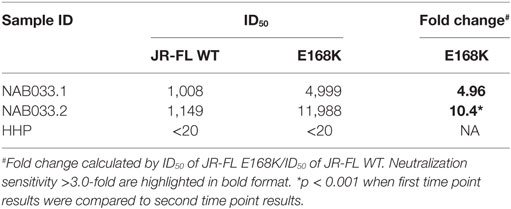
Neutralization assay was also performed with JR-FL WT and JR-FL E168K mutant viruses. The PG9/PG16 series of bNAbs are unable to neutralize the JR-FL WT HIV-1 strain because the glycan at position N160 is hidden by the glutamic acid (E) present at position 168 in this virus. The PG9/PG16 bNAbs show strong neutralization of the JR-FL pseudovirus when the glutamic acid at position 168 is replaced with lysine (K) in the JR-FL back bone (JR-FL E168K) (3). It was observed that the plasma of NAB033 showed >10-fold stronger neutralization of the JR-FL E168K mutant than the JR-FL WT pseudovirus, providing further evidence to support the presence of significant amounts of PG9/PG16-like antibodies in this sample (Figure 6B; Table 6). Interestingly, we observed significant increase in the neutralization property specifically targeting the N160 glycan of the HIV-1 Env at the second time point in NAB033 as compared to the first time point (p < 0.001) (Figure S2 in Supplementary Material).
Increase in PGT Series—Like Neutralization Specificity Over Time
In the initial screening, we identified seven BCN samples showing neutralization specificity similar to that of the PGT series of bNAbs (1). However, we could collect second time point sample only from two of these individuals (NAB033 and NAB065). The plasma samples were tested in a neutralization assay with the DU156 WT and DU156 N332A mutant pseudoviruses generally used for this purpose (Figures 6A,C). It was observed that both samples showed significantly stronger neutralization of the DU156 WT pseudovirus than the DU156 N332A mutant; the neutralization activity of NAB033 and NAB065 being 13.6- and 9.2-fold lower with the DU156 N332A mutant as compared to DU156 WT virus (Table 6). Interestingly, both samples showed a significant increase in neutralization property specifically targeting the N332 glycan of the HIV-1 Env in these two samples when compared to the initial time point (p < 0.001) (Figure S2 in Supplementary Material).
Increase in Antibodies Specific to Native HIV-1 Env Trimers
To further characterize the neutralization property of HIV-1 Env-specific antibodies present in the samples, we eluted Env trimer specific (93IN101-gp145 trimer) polyclonal IgG from the plasma and performed cell-surface binding as well as neutralization assays with JR-FL WT and JR-FL E168K mutant Env protein and pseudoviruses, respectively. Analysis of the results of the cell-surface binding assay with eluted IgG showed that one of the samples (NAB033) showed stronger binding to the JR-FL E168K mutant (30.49-fold) than to the JR-FL WT Env (Figure 7; Table 7), similar to what was observed with the positive control mAb PG9, which also exhibited significantly stronger binding (142.43-fold) to JR-FL mutant than to JR-FL WT. We then compared the neutralization potency of the eluted IgG with that of known mAbs using HIV-1 JR-FL WT and single mutant JR-FL E168K pseudoviruses. We observed that the median inhibitory concentration (IC50) of NAB033.2 with JR-FL E168K mutant was 12.4-fold lower (i.e., more potent neutralization) than the IC50 with JR-FL WT, as was also seen with the mAb PG9 (Figure 7; Table 7). Collectively our results suggest strongly that the sample NAB033 contains potent PG9/PG16 like bNAbs.
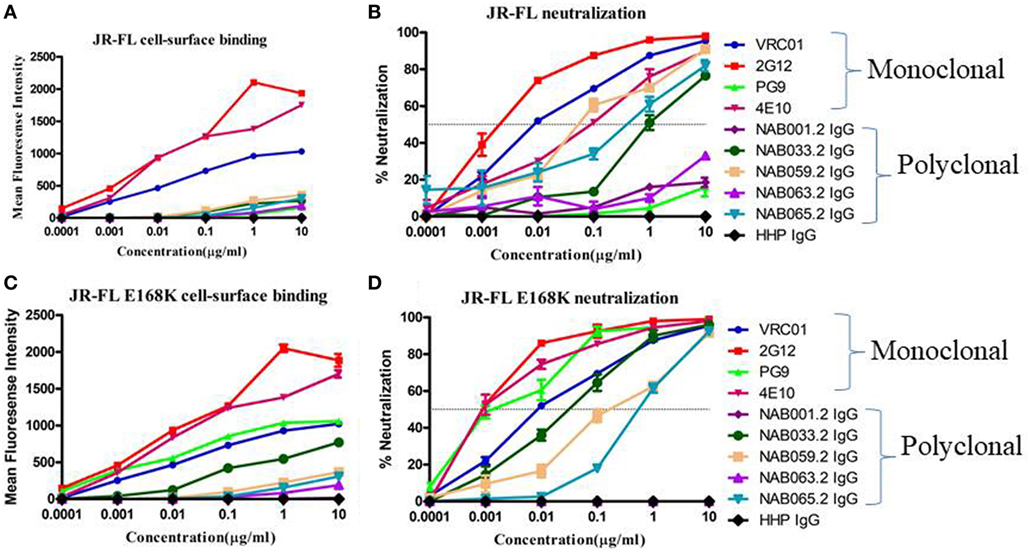
Figure 7. Cell-surface binding and neutralization analysis of IgG eluted from BCN samples. (A,C) Cell-surface binding assay with IgG eluted from BCN samples and control mAbs with JR-FL WT and JR-FL E168K mutant Env trimers expression on the surface of 293 T cells as determined by their mean fluorescence intensity in flow cytometry. (B,D) Neutralization activity of IgG from BCN samples and control mAbs against JR-FL WT and JR-FL E168K mutant pseudoviruses. VRC01, 2G12, PG9, and 4E10 were included for comparison as positive controls and HHP IgG was used as negative control. Each experiment was performed in duplicate on two independent occasions.
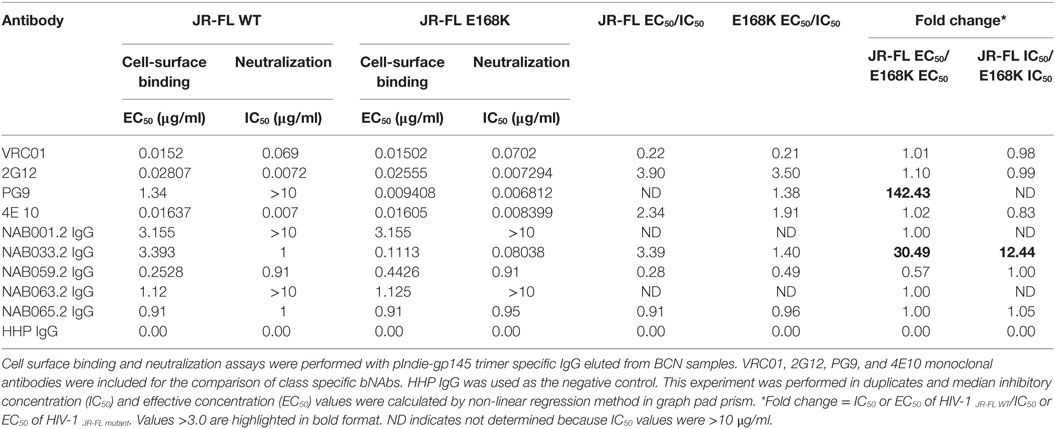
Table 7. Comparison of cell-surface binding (EC50) and neutralization activity (IC50) of IgG eluted from BCN samples against JR-FL WT and JR-FL E168K mutant.
Discussion
We recently published a study in which we identified 12 HIV-1 subtype C infected individuals whose plasma showed broad and potent cross-clade neutralization (BPCN) property (1). Subsequently we obtained a follow-up sample from a subset of these individuals after a four-year interval, to evaluate the evolution of the host NAb response over time. Few studies suggest that evolution of viral variants as well as bNAbs takes place simultaneously, and that acquisition of neutralization breadth and potency continue during ongoing HIV-1 infection (48, 49). However, the evolution of high level of neutralization breadth and potency may take several years in order to attain special features not found in most Ab responses, i.e., neutralization of most of the globally circulating HIV-1 strains at very minimal concentrations (in other words, the Ab concentration needed to achieve 50% neutralization should be less than 1 µg/ml) (17). Liao et al. in 2013 had reported increase in neutralization potency in an African HIV-infected individual (CH505), in a longitudinal study that compared baseline and follow-up samples of those with broad clade neutralization ability and suggested a maturation of the broadly NAb response (48). More recently, Patil et al. (33) reported similar observations in a HIV-infected individual from India. In the present study, we made similar observations of increase in neutralization potency of BCN samples during follow-up as demonstrated by an increase in ID50 values. Interestingly, we observed that one sample (NAB059) which exhibited strong neutralization potency at the time of initial screening (1), showed a tremendous increase in neutralization potency during follow-up with GMT > 7100 (6.75-fold), prompting us to categorize this individual as an elite neutralizer. The remaining four samples (NAB001, NAB033, NAB063, and NAB065) also showed a significant increase in the neutralization potency of the follow-up samples with GMT ranging between 1,100 and 1,800 as compared to 100 and 400 at the initial time point.
We went on to characterize the neutralization specificity in the study subjects in a systematic manner. We first performed a PepScan analysis to determine if any of the samples had acquired newer antibody responses. We found that NAB059 and NAB033 had evolved additional antibody responses with binding reactivity spread across the C2, V4, C4, C5, HR2, and CT of the envelope. Interestingly the plasma (NAB059) retained binding reactivity to the Kennedy epitope (KE; PRGPDRPEGIEEEGGERDRDRS) present in the transmembrane region of gp41 (amino acids 731 to 752as on HXB2) (50, 51), as seen during the initial testing (1). Since most bNAbs are known to bind strongly to conformational epitopes on the HIV-1 envelope than to linear epitopes, and bring about virus neutralization (38, 52), we went on to analyze the antibody response with well-characterized conformational and native forms of the Env protein. We found a significant increase in the binding reactivity of one of the follow-up samples (NAB059) to 93IN101 gp145 trimer, dimer, HXB2 gp120 monomer, and NL4-3 gp41 monomer (p < 0.05), indicating the evolution of a more robust antibody response to the native forms of the envelope glycoprotein over time. Similarly the follow-up sample of NAB033 also showed a significant increase in binding reactivity to the NL4-3 gp41 monomer (p < 0.001).
Further, we characterized the samples for CD4BS neutralization specificity. Three of the five BCN samples (NAB033, NAB059, and NAB063) were found to possess CD4BS-specific antibody response at the initial time point. In the present analysis, the binding reactivity of two of the three samples (NAB033 and NAB059) was found to have increased significantly with the RSC3 protein (p < 0.001). RSC3 WT and RSC3Δ371I/P363N recombinant proteins have been employed widely for the identification of CD4BS NAbs from the HIV-1-infected individuals (1, 7, 53). It was of particular interest to note that these two subjects also maintained a good CD4 + T cell count > 350 cells/mm3 and continued to be naïve to ART, pointing to good correlation between positive evolution of the bNAb response to the CD4BS with clinical outcome. This makes these samples worthy of further investigation for isolation of potent bNAbs that can be used as therapeutic tools. Previous studies have reported that Indian HIV-1 subtype C strains show considerable molecular differences in their envelope as compared to the other HIV-1 subtypes around the globe (53), and that RSC3 reactive antibodies mediating broad neutralization alone do not include all CD4BS antibodies (54). This has necessitated the use of additional recombinant gp120 (rgp120) core molecules for identification of CD4BS antibodies (20). Saha et al. (43) and Bhattacharyya et al. (44) designed a protein called CD4OD, with alterations in the CD4OD of the HIV-1 subtype B Env protein, such that it also binds CD4BS antibodies present in HIV-1 subtype C infected individuals from India and other parts of the globe. When screened with the CD4OD recombinant protein, we observed that a third sample (NAB063), also exhibited stronger binding reactivity to the CD4OD protein as compared to the initial time point (Figure 5B). Bhattacharyya et al. (45) have reported that CD4BS neutralizing antibodies and not n-NAbs show specific binding reactivity to the CD4OD protein, indicating an increase in CD4BS specific antibody response over time in individuals having this class of bNAbs.
The Env trimer is known to be covered in a dense array of glycans constituting about half of its mass (2, 3, 10, 17). The glycans play a critical role in viral escape from the host immune response. Two major classes of bNAbs that target glycan-dependent epitopes have been identified; these include an epitope in the V1/V2 loop containing a glycan at N160 (PG9/PG16-like), and an epitope in the V3 loop containing a glycan at N332 (PGT 128-series). We had identified in our previous study 4 BCN plasma samples (NAB016, NAB033, NAB062 and NAB069) with PG9/PG16-like NAb specificity (1). However, only one of these 4 samples (NAB033) could be obtained for follow-up analysis. When this sample was tested for neutralization against HIV-1 subtype C DU156WT and DU156N160K pseudoviruses, it was observed that there was a fourfold increase in the neutralization efficiency of DU156 WT and 6.7-fold decrease in neutralization efficiency of DU156N160K mutant virus as compared to the first time sample. It was also observed that there was a >fivefold increased dependency on glycan at position 160 in the follow-up sample (6.7-fold) as compared to the first time sample (1.1-fold). Since the PG9/PG16 class of bNAbs are known to strongly neutralize the mutant JR-FL E168K strain but not JR-FL WT (3, 55), we also performed a neutralization assay with the JR-FL WT and JR-FL E168K mutant pseudoviruses to confirm the presence of PG9/PG16-like neutralization property, and found a >10-fold neutralization of the mutant JR-FL E168K than the JR-FL WT. Comparison of the obtained results with that of the first time point sample (1), showed a >onefold increase in neutralization of the JR-FL E168K mutant pseudovirus, indicating that this sample had evolved a stronger bNAb response to the epitope comprising N160 glycan of the HIV-1 Env.
Two of the seven BCN samples (NAB033 and NAB065), which exhibited PGT series-like neutralization property in our earlier study (1), were available for follow-up analysis. These samples were tested with DU156WT and DU156 N332A mutant pseudoviruses that are widely employed for the screening of this class of bNAbs. The two plasma samples neutralized the WT virus with 13.6- and 9.2-fold greater potency than the mutant DU156N332A pseudovirus. When compared to the results of the initial sample, the follow-up samples of NAB033 and NAB065 showed a >threefold and >onefold increase in bNAb specificity toward the N332 glycan, respectively.
Previous studies have reported a strong correlation between antibody responses to the cell-surface expressed native Env trimers and neutralization potency in other systems (56). In order to correlate the bNAbs binding response with neutralizing potency in the present study, we isolated polyclonal IgG from all the five BCN samples using pIndie gp145 trimeric protein coated My One Dynabeads. IgG was eluted and tested for neutralization potency and cell-surface binding affinity to HIV-1 JR-FL WT and its single mutant JR-FL E168K. We observed that median IC50 values were lower than EC50 values, indicating the highly potent nature of the neutralizing antibodies in these samples. Overall, our results suggest that NAB033 has strong neutralization property similar that of the PG9 class of bNAbs, prompting us to further characterize the antibodies in this sample.
To summarize, we observed a positive evolution of neutralization potency in most of the second time samples as seen by increase in ID50 values over time. Interestingly, the sample NAB059 that had the greatest increase in potency of neutralization also showed an increase in CD4BS specific reactivity during follow-up and this was associated with a stable CD4+ T cells count >350 cells/mm3, lower plasma viral load, and good clinical correlation making this a very interesting case to pursue for the isolation of potent bNAbs that can be used as potential therapeutic tools. Overall, our findings are quite encouraging as they suggest that the potency of the BCN response, once initiated, increases and matures with time.
Ethics Statement
The study was approved by the Institutional Ethics Committee of the National Institute for Research in Tuberculosis (NIRT IEC No: 2011001) and all experiments were performed in accordance with relevant guidelines and regulations. Sample collection was done after obtaining written informed consent from the study participants.
Author Contributions
NC, SS, ST, RV, KS, RD, and LH participated in the study design. NC, MS, PS, KM, and NK collected and processed samples, NC, HB, and BA performed the experiments and data analysis. NC and LH drafted the paper. All authors have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank K Lucia Precilla (Dept. of HIV, NIRT-ICMR, Chennai, India) for her help with pIndie gp145 specific IgG elution, cell-surface binding assay and manuscript preparation. We acknowledge Dr. Subash Babu and his team (Dr. Anuradha, Mr. Kadar Moideen and Dr. Pavankumar) for their kind gift of PE-conjugated anti-human IgG secondary antibody and also support in the neutralization assay and FACs analysis for cell-surface binding assay. We acknowledge Mr. Sivasankarn MP for providing Britelite substrate. We acknowledge Dr. Lynn Morris (NICD, Johannesburg) and Dr. Jayantha Battacharya (THSTI, India) for providing DU156 N160 and DU156 N332A pseudovirus plasmids and Muzafar Jan and Dr. Sunil Arora (PGI-Chandigarh, India) for providing JR-FL wild type plasmid. Pseudo viruses belonging to clades A, B, C, A/E, and A/G, RSC3 WT and RSC3 Δ371I/P363N recombinant proteins were obtained from the NIH AIDS Reagent Program.
Funding
NC received fellowship from University Grants Commission (UGC) India for the period of 2010 to 2015, and Senior Research Fellowship from the Indian Council of Medical Research (ICMR) from 2016 onward. The work was carried out with intramural research funds from the National Institute for Research in Tuberculosis, ICMR. This work was supported in part by National Institutes of Health Grant R01A1118366-01to RV.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.00618/full#supplementary-material.
References
1. Cheedarla N, Precilla KL, Babu H, Vijayan KKV, Ashokkumar M, Chandrasekaran P, et al. Broad and potent cross clade neutralizing antibodies with multiple specificities in the plasma of HIV-1 subtype C infected individuals. Sci Rep (2017) 7:46557. doi:10.1038/srep46557
2. Shcherbakov DN, Bakulina AY, Karpenko LI, Ilyichev AA. Broadly neutralizing antibodies against HIV-1 as a novel aspect of the immune response. Acta Naturae (2015) 7:11–21.
3. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science (2009) 326:285–9. doi:10.1126/science.1178746
4. Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature (2011) 477:466–70. doi:10.1038/nature10373
5. Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science (2012) 337:183–6. doi:10.1126/science.1225416
6. Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity (2012) 37:412–25. doi:10.1016/j.immuni.2012.08.012
7. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (2010) 329:856–61. doi:10.1126/science.1187659
8. Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science (2011) 333:1593–602. doi:10.1126/science.1207532
9. Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science (2010) 329:811–7. doi:10.1126/science.1192819
10. Sattentau QJ, McMichael AJ. New templates for HIV-1 antibody-based vaccine design. F1000 Biol Rep (2010) 2:60. doi:10.3410/B2-60
11. Stamatatos L. HIV vaccine design: the neutralizing antibody conundrum. Curr Opin Immunol (2012) 24:316–23. doi:10.1016/j.coi.2012.04.006
12. Shaw GM, Hunter E. HIV transmission. Cold Spring Harb Perspect Med (2012) 2. doi:10.1101/cshperspect.a006965
13. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A (2008) 105:7552–7. doi:10.1073/pnas.0802203105
14. Moody MA, Gao F, Gurley TC, Amos JD, Kumar A, Hora B, et al. Strain-specific V3 and CD4 binding site autologous HIV-1 neutralizing antibodies select neutralization-resistant viruses. Cell Host Microbe (2015) 18:354–62. doi:10.1016/j.chom.2015.08.006
15. Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol (2008) 82:12449–63. doi:10.1128/JVI.01708-08
16. Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One (2010) 5:e8805. doi:10.1371/journal.pone.0008805
17. Simonich CA, Williams KL, Verkerke HP, Williams JA, Nduati R, Lee KK, et al. HIV-1 neutralizing antibodies with limited hypermutation from an infant. Cell (2016) 166:77–87. doi:10.1016/j.cell.2016.05.055
18. Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS (2014) 28:163–9. doi:10.1097/QAD.0000000000000106
19. Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol (2015) 16:571–6. doi:10.1038/ni.3158
20. Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol (2011) 85:4828–40. doi:10.1128/JVI.00198-11
21. Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med (2012) 209:1469–79. doi:10.1084/jem.20120423
22. Lynch RM, Tran L, Louder MK, Schmidt SD, Cohen M, Dersimonian R, et al. The development of CD4 binding site antibodies during HIV-1 infection. J Virol (2012) 86:7588–95. doi:10.1128/JVI.00734-12
23. Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, et al. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat Med (2017) 23:185–91. doi:10.1038/nm.4268
24. Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell (2014) 158(6):1243–53. doi:10.1016/j.cell.2014.08.023
25. Nimmerjahn F, Ravetch JV. Translating basic mechanisms of IgG effector activity into next generation cancer therapies. Cancer Immun (2012) 12:13.
26. Bruel T, Guivel-Benhassine F, Amraoui S, Malbec M, Richard L, Bourdic K, et al. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat Commun (2016) 7:10844. doi:10.1038/ncomms10844
27. Tudor D, Bomsel M. The broadly neutralizing HIV-1 IgG 2F5 elicits gp41 specific antibody-dependent cell cytotoxicity in a FcγRI-dependent manner. AIDS (2011) 25(6):751–9. doi:10.1097/QAD.0b013e32834507bd
28. Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, et al. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol (2007) 81:6187–96. doi:10.1128/JVI.00239-07
29. Gray ES, Moody MA, Wibmer CK, Chen X, Marshall D, Amos J, et al. Isolation of a monoclonal antibody that targets the alpha-2 helix of gp120 and represents the initial autologous neutralizing-antibody response in an HIV-1 subtype C-infected individual. J Virol (2011) 85:7719–29. doi:10.1128/JVI.00563-11
30. Morris L, Chen X, Alam M, Tomaras G, Zhang R, Marshall DJ, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS One (2011) 6:e23532. doi:10.1371/journal.pone.0023532
31. Bhiman JN, Anthony C, Doria-Rose NA, Karimanzira O, Schramm CA, Khoza T, et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat Med (2015) 21:1332–6. doi:10.1038/nm.3963
32. Rusert P, Kouyos RD, Kadelka C, Ebner H, Schanz M, Huber M, et al. Determinants of HIV-1 broadly neutralizing antibody induction. Nat Med (2016) 22:1260–7. doi:10.1038/nm.4187
33. Patil S, Kumar R, Deshpande S, Samal S, Shrivastava T, Boliar S, et al. Conformational epitope-specific broadly neutralizing plasma antibodies obtained from an HIV-1 clade C-infected elite neutralizer mediate autologous virus escape through mutations in the V1 loop. J Virol (2016) 90:3446–57. doi:10.1128/JVI.03090-15
34. Euler Z, van den Kerkhof TLGM, van Gils MJ, Burger JA, Edo-Matas D, Phung P, et al. Longitudinal analysis of early HIV-1-specific neutralizing activity in an elite neutralizer and in five patients who developed cross-reactive neutralizing activity. J Virol (2012) 86(4):2045–55. doi:10.1128/JVI.06091-11
35. de Almeida DV, Macieira KV, Grinsztejn BGJ, Veloso dos Santos VG, Guimarães ML. Cross-neutralizing antibodies in HIV-1 individuals infected by subtypes B, F1, C or the B/Bbr variant in relation to the genetics and biochemical characteristics of the env gene. PLoS One (2016) 11(12):e0167690. doi:10.1371/journal.pone.0167690
36. Almeida DV, Morgado MG, Côrtes FH, Guimarães ML, Mendonça-Lima L, Pilotto JH, et al. Neutralizing antibodies in HIV-1-infected Brazilian individuals. AIDS Res Hum Retroviruses (2013) 29(3):488–92. doi:10.1089/aid.2012.0052
37. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol (1998) 72:2855–64.
38. Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother (2002) 46:1896–905. doi:10.1128/AAC.46.6.1896-1905.2002
39. Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res (1996) 24:596–601. doi:10.1093/nar/24.4.596
40. Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol (2005) Chapter 12:Unit 121. doi:10.1002/0471142735.im1211s64
41. Sneha Priya R, Veena M, Kalisz I, Whitney S, Priyanka D, LaBranche CC, et al. Antigenicity and immunogenicity of a trimeric envelope protein from an Indian clade C HIV-1 isolate. J Biol Chem (2015) 290:9195–208. doi:10.1074/jbc.M114.621185
42. Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology (2005) 332(1):145–56. doi:10.1016/j.virol.2004.10.042
43. Saha P, Bhattacharyya S, Kesavardhana S, Miranda ER, Ali PS, Sharma D, et al. Designed cyclic permutants of HIV-1 gp120: implications for envelope trimer structure and immunogen design. Biochemistry (2012) 51:1836–47. doi:10.1021/bi300003v
44. Bhattacharyya S, Rajan RE, Swarupa Y, Rathore U, Verma A, Udaykumar R, et al. Design of a non-glycosylated outer domain-derived HIV-1 gp120 immunogen that binds to CD4 and induces neutralizing antibodies. J Biol Chem (2010) 285:27100–10. doi:10.1074/jbc.M110.152272
45. Bhattacharyya S, Singh P, Rathore U, Purwar M, Wagner D, Arendt H, et al. Design of an Escherichia coli expressed HIV-1 gp120 fragment immunogen that binds to b12 and induces broad and potent neutralizing antibodies. J Biol Chem (2012) 288:9815–25. doi:10.1074/jbc.M112.425959
46. Shankarappa R, Gupta P, Learn GH Jr, Rodrigo AG, Rinaldo CR Jr, Gorry MC, et al. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology (1998) 241:251–9. doi:10.1006/viro.1997.8996
47. Bachu M, Yalla S, Asokan M, Verma A, Neogi U, Sharma S, et al. Multiple NF-kappaB sites in HIV-1 subtype C long terminal repeat confer superior magnitude of transcription and thereby the enhanced viral predominance. J Biol Chem (2012) 287:44714–35. doi:10.1074/jbc.M112.397158
48. Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature (2013) 496:469–76. doi:10.1038/nature12053
49. Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, et al. Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity (2016) 45:1108–21. doi:10.1016/j.immuni.2016.10.027
50. Kennedy RC, Eichberg JW, Lanford RE, Dreesman GR. Anti-idiotypic antibody vaccine for type B viral hepatitis in chimpanzees. Science (1986) 232:220–3. doi:10.1126/science.3952505
51. Steckbeck JD, Sun C, Sturgeon TJ, Montelaro RC. Topology of the C-terminal tail of HIV-1 gp41: differential exposure of the Kennedy epitope on cell and viral membranes. PLoS One (2010) 5:e15261. doi:10.1371/journal.pone.0015261
52. He L, de Val N, Morris CD, Vora N, Thinnes TC, Kong L, et al. Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat Commun (2016) 7:12041. doi:10.1038/ncomms12041
53. Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med (2008) 358:1590–602. doi:10.1056/NEJMra0706737
54. Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, et al. Broadly neutralizing antibody responses in a large longitudinal sub-Saharan HIV primary infection cohort. PLoS Pathog (2016) 12:e1005369. doi:10.1371/journal.ppat.1005369
55. Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol (2013) 13:487–98. doi:10.1038/nri3478
Keywords: human immunodeficiency virus type 1, broadly neutralizing antibodies, Indian HIV-1 subtype C infection, evolution of neutralization response, CD4-binding site, glycan-dependent neutralization
Citation: Cheedarla N, Hemalatha B, Anangi B, Muthuramalingam K, Selvachithiram M, Sathyamurthi P, Kailasam N, Varadarajan R, Swaminathan S, Tripathy SP, Vaniambadi SK, Vadakkupattu DR and Hanna LE (2018) Evolution of Neutralization Response in HIV-1 Subtype C-Infected Individuals Exhibiting Broad Cross-Clade Neutralization of HIV-1 Strains. Front. Immunol. 9:618. doi: 10.3389/fimmu.2018.00618
Received: 09 October 2017; Accepted: 12 March 2018;
Published: 27 March 2018
Edited by:
Aurelio Cafaro, Istituto Superiore di Sanità, ItalyReviewed by:
Morgane Bomsel, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceLucia Lopalco, San Raffaele Hospital (IRCCS), Italy
Christiane Moog, Institut National de la Santé et de la Recherche Médicale (INSERM), France
Copyright: © 2018 Cheedarla, Hemalatha, Anangi, Muthuramalingam, Selvachithiram, Sathyamurthi, Kailasam, Varadarajan, Swaminathan, Tripathy, Vaniambadi, Vadakkupattu and Hanna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luke Elizabeth Hanna, aGFubmEmI3gwMDA0MDtuaXJ0LnJlcy5pbg==
 Narayanaiah Cheedarla
Narayanaiah Cheedarla Babu Hemalatha
Babu Hemalatha Brahmaiah Anangi2
Brahmaiah Anangi2 Murugesan Selvachithiram
Murugesan Selvachithiram Pattabiraman Sathyamurthi
Pattabiraman Sathyamurthi Raghavan Varadarajan
Raghavan Varadarajan Luke Elizabeth Hanna
Luke Elizabeth Hanna