
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 26 March 2018
Sec. Immunological Memory
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.00515
This article is part of the Research Topic Orchestration of an Immune Response to Respiratory Pathogens View all 15 articles
Tissue-resident memory CD8+ T cells are an important first line of defense from infection in peripheral non-lymphoid tissues, such as the mucosal tissues of the respiratory, digestive, and urogenital tracts. This memory T cell subset is established late during resolution of primary infection of those tissues, has a distinct genetic signature, and is often defined by the cell surface expression of CD69, CD103, CD49a, and CD44 in both mouse and human studies. The stimuli that program or imprint the unique gene expression and cell surface phenotypes on TRM are beginning to be defined, but much work remains to be done. It is not clear, for example, when and where the TRM precursors receive these signals, and there is evidence that supports imprinting in both the lymph node and the peripheral tissue sites. In most studies, expression of CD49a, CD103, and CD69 on T cells in the tissues appears relatively late in the response, suggesting there are precise environmental cues that are not present at the height of the acute response. CD49a and CD103 are not merely biomarkers of TRM, they confer substrate specificities for cell adhesion to collagen and E-cadherin, respectively. Yet, little attention has been paid to how expression affects the positioning of TRM in the peripheral tissues. CD103 and CD49a are not mutually exclusive, and not always co-expressed, although whether they can compensate for one another is unknown. In fact, they may define different subsets of TRM in certain tissues. For instance, while CD49a+CD8+ memory T cells can be found in almost all peripheral tissues, CD103 appears to be more restricted. In this review, we discuss the evidence for how these hallmarks of TRM affect positioning of T cells in peripheral sites, how CD49a and CD103 differ in expression and function, and why they are important for immune protection conferred by TRM in mucosal tissues such as the respiratory tract.
Tissue-resident memory CD8+ T cells (TRM) are a distinct memory population that is generated and persists at the site of infection or vaccination (1–3). Upon exposure to the same or similar diseases, TRM cells provide a first line of adaptive cellular defense and are indispensible in lethal challenge models (2, 4). Since CD8+ T cells mount responses aimed primarily at more conserved internal epitopes of pathogens, eliciting a TRM response may provide increased protection compared with B-cell targeted vaccines, which some pathogens can escape by mutating antigenic sites (5). While research efforts have started to shed light on some of the requisite signals for maintenance of the TRM population, less is understood in regard to the positioning within the tissue that is required for the development of this subset and responses of TRM cells after rechallenge. Here, we will provide an overview of the gross anatomical locations in which TRM cells form, the known interactions that facilitate their development, and the consequences of reactivation, as well as the voids that remain in our understanding of this critical population. Closing these knowledge gaps will allow us to harness the full potential of TRM and elicit improved protective responses to vaccination.
TRM cells are most well known for their roles in barrier sites such as mucosal tissue and skin. While the populations in different tissues display heterogeneity in requirements for migration to the site of infection, the developmental program, maintenance within the tissue, and their surface phenotype, they ultimately perform similar critical functions. In the mucosa, TRM cells have been identified in the intestines, female reproductive tract, salivary glands, tonsils, and lungs (1, 6–10). In addition, TRM or TRM-like cells have been described in a number of other non-mucosal sites, including lymphoid and peripheral tissues such as the thymus, spleen, lymph nodes, liver, kidneys, pancreas, heart, skin, and brain (6, 7, 11–14).
In most mucosal and barrier tissues, the TRM population primarily homes to areas of epithelial surfaces, which represent the common site of infections in these tissues. Influenza, for example, infects the airway epithelium and Herpes simplex viruses, the epithelial cells of the skin or cervix (15–18). In other instances, however, such as with cytomegalovirus, the virus infects epithelial cells within an organ (salivary glands) rather than at the barrier surface (19, 20). As one might anticipate based on the name, salivary glands are home to extensive acinar epithelial-rich glandular structures that provide a location for the infection to persist, but also act as an ideal habitat for the formation and maintenance of TRM cells (8). This opens up the possibility that internal glandular structures within other peripheral tissues could be similar targets for TRM and warrants further investigation.
In the lung, CD8+ TRM cells preferentially localize to regions rich in collagen IV (ColIV), while CD4+ T cells are biased to areas abundant in collagen I (ColI) (21). This coincides with the relative expression of the integrins specific for those collagen types, CD49a and CD49b, respectively (21). In non-mucosal organs, the positioning within the tissue is less well understood. As discussed later in this review, understanding the function of the surface receptors expressed on TRM cells as well as their responses to different chemokine cues may better inform their roles and localization within non-mucosal tissues.
Recent work has focused on identifying the transcriptional regulators of TRM; however, these studies suggest that the specific requirements may vary between mice and humans. In mice, CD8+ TRM cells from a number of tissues express elevated levels of HOBIT (homolog of BLIMP-1 in T cells) compared with peripheral T cells (22). In conjunction with BLIMP-1, HOBIT promotes maintenance of the TRM population through repression of genes associated with tissue egress. In addition, in mouse CD8+ TRM, it has been shown that T-bet and Eomes, two T-box transcription factors, needed to be down regulated for the cell to receive signals from TGF-β and upregulate CD103 (23, 24). However, a low level of T-bet expression is required to maintain expression of the IL-15 receptor β-chain (24, 25). Thus, these two T-box transcription factors control receptiveness to TGF-β and IL-15 signals, which are necessary for proper TRM formation and function (9, 23–25).
In humans, a different set of transcription factors appear to be critical for TRM development and maintenance. Similar to mice, Eomes and T-bet were not expressed in the TRM subset (26). However, the link between HOBIT expression and TRM cells is less evident. HOBIT is expressed in both circulating and resident CD8+ T cell populations in humans, and when identified, associates more with cells lacking markers of residence. Reinforcing this notion, a more recent study evaluated gene profiles of CD69+CD8+ T cells derived from various human tissues and found low to absent levels of HOBIT (27). Instead, these cells expressed NOTCH-1 and HIF-1α, where NOTCH-1 regulated TRM metabolism, which was suggested as its major function (26). In other cases, organ-specific transcriptional regulators were identified. In the lung, RUNX3, BATF, AHR, AP-1, RBPJ, and NF-κB were detected in the TRM subset (26). Many of these regulate T cell effector functions and homing receptors. In the small intestine and vaginal mucosa, a requirement for mTOR was found through inhibition with rapamycin treatment (28). This defect was attributed to an inability to migrate to the site and respond to antigen, and less with a failure to be maintained within the tissue.
Despite some insight into the transcriptional control of TRM cells, few of these transcription factors identified appear to be “master regulators” of TRM differentiation as they are expressed in other CD8+ effector or memory subsets. This suggests that TRM differentiation and maintenance is likely controlled by complex combinations of several transcription factors, and the requirements may differ between mice and humans.
It has become clear that one of the only ways to concretely define a cell as “resident” is through the use of parabiotic mice, which allow for equilibration of all circulating cells, but not cells of residence (29, 30). However, the majority of studies in both mice and humans have used expression of a panel of TRM identifiers: CD103 (integrins αE paired with β7), sphingosine-1-phosphate receptor 1 (S1P1) antagonist CD69, collagen-binding CD49a (integrin α1 paired with β1), and hyaluronic acid (HUA) binding CD44 as surrogates. It is worth noting that not all populations of TRM cells display all of these markers, suggesting there may be some nuance in TRM subsets. The functional ramifications of expressing any of these molecules likely fit with the requirements for the appropriate positioning of the cells and long-term survival in the tissue.
The lectin CD69 is an antagonist of S1P1 and limits egress by blocking responsiveness to sphingosine-1-phosphate gradients (31–33). It complexes with S1P1 on the cell surface, which leads to its internalization and degradation (34). CD69 is initially upregulated on recently activated effector cells that have seen their cognate antigen, perhaps to limit egress from the lymph node, but constitutive expression is only observed on resident cells (35, 36). Downregulation of Krüppel-like factor 2 on these cells ultimately allows for the expression of CD69 within peripheral tissues (37). Interestingly, CD69 expression is not limited to CD8+ T cells, and its presence on other immune subsets including natural killer cells and certain peripheral dendritic cells (DCs) plays a similar functional role of maintaining them within the organ (38, 39). While CD69 is expressed on the majority of TRM subsets, absence of this lectin on CD8+ T cells only limits the size of the population and does not result in complete ablation (12). This suggests that CD69 is not an absolute requirement, and while its expression may be advantageous, it is not mandatory. T cells, including TRM, are dynamic in their movement in different tissue settings, and in peripheral tissues, multiple retention factors are important for maintaining the resident population. As previously insinuated, populations of TRM cells exist within the salivary glands and female reproductive tract that are CD69 negative. This is interesting given the requirement for downregulation of the S1P1 to establish the TRM population in the salivary glands; however, it further suggests that other mechanisms of organ retention are at play (37, 40). So far, little is understood in regard to whether CD69 plays any role in how the cells are positioned within the tissue. In fact, it is quite possible that its primary and only role is limiting their exit from the organ to return to the blood and lymphatics.
Integrin αE (CD103) pairs with integrin β7 and is upregulated upon exposure to the active form of TGF-β (41, 42). The most well-known function of CD103 is as a receptor for E-cadherin, an adherans junctional protein interlocking epithelial cells (41). CD103:E-cadherin interactions can act as a tether, which may aid in positioning, retention, and the shape of cells within the epithelium (43, 44). Skin TRM cells lacking CD103 are fewer in number and exhibit increased motility compared with their wild-type counterparts, corroborating this role in vivo (12, 45). Similarly, CD103 deficiency results in lower numbers of CD8+ TRM cells in the lung after influenza infection (46) and a decrease in intestinal CD8+ T cells responding to oral Listeria infection due to a defect in initial accumulation (47). Since epithelial cells are the targets for a number of mucosal viral infections, adherence and localization of TRM cells to the epithelium positions them to act as the first line of defense in subsequent exposures. In this regard, CD103 also facilitates the generation of a TRM population at tumor sites such as in the case of melanoma (48). In fact, TRM production by mucosal vaccination leads to inhibition of tumor growth in a preclinical model of head and neck cancer, which was substantiated through parabiotic experiments in mice (49).
While physical retention through ligand binding is the most obvious role for CD103, engagement of CD103 may have a number of other functional ramifications outside of adhesion. While the effects of CD103 binding have been primarily studied in tumor models, the identified features of this integrin are likely widespread throughout various disease states. CD103+ tumor-infiltrating CD8+ T cells are more capable of killing tumor cells (50). This is likely attributed to the fact that CD103+ T cells form more stable synapses with target cells than their CD103-negative counterparts (51). Engagement of CD103 also positions cytolytic granules to organize in a polarized fashion, and the addition of signaling through the TCR results in lytic granule exocytosis (52, 53). Although these functions of CD103 are redundant in the presence of CD11a (LFA-1), TRM cells, especially in the airways of the lungs, display low levels of LFA-1 (54). In fact, LFA-1 levels have been used to determine the age of the TRM cells in the airway, functioning as a clock and decreasing over time (3). One hypothesis is that airway TRM cells are not cytolytic because the synapse stability is affected by this defect. However, CD103 expression on TRM may compensate for low LFA-1 levels and promote effective cytolytic responses to secondary infections.
Moreover, engagement of CD103 may also function to directly position the cells within a given tissue. As an example, it has been shown in the tumor microenvironment that binding of CD103 results in the upregulation of the chemokine receptor CCR5 (55). This suggests that the integrin/chemokine axis could greatly affect the downstream consequences of migratory cues received by a cell and looking at each pathway discretely may limit the overall understanding of the response. In the lung, CCR5 is critical for CD8+ T cells to reach the airways (56). Therefore, it would not be unreasonable to hypothesize that CD103 deficiency may alter the localization of the CD8+ T cells and delay clearance of the infection. On the flip side, binding of CCL25 through chemokine receptor CCR9 contributes to expression of CD103 on CD8+ T cells in the intestine (57). While it is relevant that chemokine signals other than TGF-β may contribute to the upregulation of this integrin on the surface, due to limited expression of CCR9 on CD8+ T cells in other organs, this may be a gut-specific mechanism.
In addition to E-cadherin expression on epithelial cells, flow cytometric analysis has demonstrated that the protein is expressed on the surface of specific immune populations such as DCs and, in some instances, TRM cells (58–60). In the salivary glands and gut, E-cadherin is detected on virus-specific CD8+ T cells, a phenomenon specific to mucosal compartments, as their lymphoid counterparts in the spleen do not display this phenotype (60). While it is unclear whether the CD8+ T cells are producing E-cadherin naturally or acquiring the surface phenotype through a process such as trogocytosis, there are likely to be functional consequences of non-epithelial cells adorned with E-cadherin (61, 62). This may allow for the formation of more stable synapses, in this case between T cells and their APC, or potential cell:cell communication via engagement of ligand on other T cells. Surface E-cadherin on the TRM cell could alternatively lead to homotypic interactions (63) in the absence of CD103 expression; however, the downstream functional consequences of this interaction are unclear.
The brain and other epithelium-sparse tissues pose a conundrum in regard to understanding the role that CD103 plays on TRM cells. In the brain, TRM cells are localized to borders of different anatomical regions and in some cases in proximity to vasculature (64). Despite this, expression of CD103 does not appear to corroborate with positioning. However, as just posed, one possibility is that CD103 solely interacts with E-cadherin expressing immune cells in regions devoid of epithelial surfaces and potentially localized similarly to the profile of the TRM cells. Another possibility is that CD103 has other unexplored ligands at play. Although only identified in an in vitro system, it has been suggested that CD103 can bind to microvascular endothelial cells derived from the intestine (65). While this did not hold true with endothelial cells derived from the dermis, it has yet to be investigated in the brain and other non-mucosal organs and could better identify a functional role for CD103 in these locations.
Overall, one of the caveats when studying the roles of CD103 in vivo is that deletion of the gene results in a lack of DCs expressing this marker. CD103+ DCs are critical for the development of TRM cells in the lung during influenza infection and are one of the APC population that drains to the mediastinal lymph node early during infection to present antigen (30, 53). One of the approaches to circumvent this issue is to transfer CD103 knockout transgenic cells into WT mice. This is the approach employed to establish its requirement in the skin, lungs, and gut as previously described (12, 46). However, none of these studies addressed whether CD103 signaling may be required during early stages of development and could ultimately alter the population prior to becoming established at a peripheral site. To fully examine the role of CD103 in the development and persistence of CD8+ T cells, it would be necessary to develop an inducible knockout specific to CD8+ T cells, so that the integrin could be eliminated at specific time points postinfection.
The role of CD49a on memory T cells in peripheral tissues was discovered in 2004 (2). At that time, the term TRM had not yet been coined, although it has since been associated with this subset of memory T cells (2, 9, 12, 66, 67). CD49a, or integrin α1, pairs with CD29 (integrin β1) to form the heterodimer VLA-1. VLA-1 is a collagen-binding integrin, with preference for the non-fibrillar form, ColIV (68, 69), although it can also bind to ColI, the fibrillar form present in the interstitium of almost all tissues (21). Early studies showed that VLA-1 was not only critical for adherence to ColIV but also migration of the cells along the collagen (68–70). The idea that two of the predominant integrins expressed on TRM cells have opposing functions when interacting with the tissue is quite remarkable, and yet both CD49a and CD103 appear to contribute to the development and/or survival of this population.
While CD49a does not have a direct role in attaching to epithelial cells, its ligand ColIV is located in the lamina densa layer of the basement membrane of mucosal epithelium and is the surface to which the epithelial cells are attached (71). The motility features of this interaction could allow for migration along the basement membrane to access additional regions of the epithelium or may be essential for traversing the collagen to reach infected cells. In either scenario, this interaction is believed to be critical for persistence of the resident population as demonstrated in both the lungs and the intestines (2, 72). While some have assumed that CD49a acts to retain T cells at the epithelium, this has not been unequivocally demonstrated experimentally. As mentioned, it could be necessary to migrate within those sites. CD49a regulation of retention and motility is not mutually exclusive as the ability to stay in the tissue and perform surveillance are the functional hallmarks of TRM.
In addition to fostering close proximity to the target cells, engagement of CD49a has a pro-survival role. In conjunction with signaling through the TNF receptor II, binding to CD49a works in a synergistic fashion to protect the cells from undergoing apoptosis (73). Blocking CD49a with antibodies results in a diminution of TRM in mucosal sites (2). Similarly, genetic deletion of CD49a results in the resident population becoming limited (2, 72). Protection requires the presence of a sufficient number of TRM cells, and mice deficient in CD49a become susceptible to secondary heterosubtyptic infections at an earlier time point after primary infection (2–4).
While the requirement for CD49a expression on TRM cells became clear through mouse focused studies, TRM from human subjects share many attributes. In the lung, TRM expressing CXCR6, CD49a, CD103, and CD69 are known to be the major memory population (27, 74). In the skin of healthy individuals, CD8+ TRM cells expressing CD49a identify a population of cells that produce IFNγ (9). Unexpectedly, in addition to IFNγ, when stimulated with IL-15, they produce large amounts of perforin and granzyme B (9), perhaps offering a potential mechanism for their reactivation as effectors in secondary encounters. On the other hand, CD49a-negative CD8+ T cells in the skin instead produced IL-17 and were associated with psoriatic lesions, indicating that CD49a expression defines different functional subsets. Whether CD49a was required to establish or maintain these cells was not investigated. In mice, CD49a+ CD4+ memory T cells in the lung provide rapid effector and innate like functions, probably related to more efficient recruitment of other effector cells. This effector function bias based on CD49a expression has not yet been reported for TRM in other tissues.
A fourth marker of TRM cells is CD44. While CD44 alone does not distinguish TRM cells from other CD8+ T cell populations, its continued expression suggests that similar to other surface receptors, CD44 may have a functional role in TRM biology (75). CD44 is a C-lectin containing glycoprotein, which is expressed on various cell types, including leukocytes and epithelial cells (76). The most well-studied function of CD44 is as a receptor for HUA, a component of the extracellular matrix, and a substance made by vascular endothelial cells and an array of immune cells (77, 78). HUA expression in peripheral tissues is upregulated during inflammation and increases hydration of the tissue (79). On CD8+ T cells, CD44 is a classical marker of previous activation, expressed on newly generated effector cells as well as resting memory cells (80). Unlike other T cell markers, which are either on or off, CD44 is expressed in a gradient depending on whether a cell is naive, an effector, or a memory cell (81, 82). Of note, CD44 exists in alternatively spliced isoforms, with various posttranslational modifications, which result in differential affinity for ligand (83, 84). However, in mice, only the invariant form of CD44 has been identified on T cells, with only suggestions that alternative forms are transiently expressed during periods of immunological challenge (85).
In regard to accessing peripheral tissues during an immune challenge, CD44 can bind HUA expressed on vascular endothelial cells and facilitate transmigration (86). In addition, CD44 can interact with CD49d through their intracellular domains, which may enhance this process. However, complete deletion of CD44 does not limit the accumulation of CD8+ T cells at the site of initial infection, suggesting that this role is redundant when the cells can utilize other selectins expressed on the cell surface. Once within the tissue, CD44 expressed on DCs improves TCR synapse stability, possibly through binding to CD8+ T cell production of HUA (87, 88). Expression of HUA on other immune cell subsets could also aid in cell:cell communication.
Most relevant to migration of CD8+ T cells is that CD44 has been shown to play a critical role in maintaining cell structure through adherence to ECM (89). In addition to binding HUA, CD44 can interact with other matrix proteins such as fibronectin, laminin, and collagen. Loss of CD44 during in vitro experiments demonstrated that cells no longer display stable polarization with extension of a uropod (89). This defect resulted in a decreased migratory capacity within peripheral tissue, which affects initial access of the tissue. CD44 ligand binding led to recruitment of a number of signaling molecules, which transmitted signals to the cytoskeleton to stabilize the cellular morphology (89).
In the TRM population, CD44 is maintained at high levels, suggesting that it may be important for the TRM population; however, the specific function has yet to be elucidated (75). One possibility is that CD44 could be acting as a general receptor for ECM, maintaining cell shape and polarity as the cells interact with collagen and epithelial cells (89). However, this has yet to be directly tested. An essential role for CD44 in survival of Th1 CD4+ T cells has been demonstrated; however, this function was not identified on CD8+ T cells (90). In fact, so far the majority of functional requirements for CD44 are enhanced on Th1 CD4+ T cells and do not translate to the CD8+ T cell compartment (90). However, its role within the TRM subset has not been comprehensively interrogated. Understanding the biology behind the contribution of CD44 to the localization and/or maintenance of the CD8+ TRM population could shed light on a new pathway of regulation and warrants further investigation.
The roles of chemokines in proper positioning within lymphoid organs have been extensively studied (91). However, their contribution within peripheral tissues is less well understood. In the lung, absence of CCR5 on T cells prevents CD8+ T cells from effectively reaching the airway and clearing influenza infection (56). Alternatively, CXCL12 packaged in neutrophil trails facilitates efficient migration of CXCR4+CD8+ T cells to the site of influenza infection (92). However, the chemokine cues that are required for the acute response may or may not translate to the TRM population and long-term protection. Depletion of neutrophils delayed CD8+ T cell infiltration into the infected lung but did not affect the development or protective capacity of TRM (93). By contrast, it is clear in the intestine that expression of CCR9 and consequent binding to CCL25 is essential for migrating T cell subsets to localize to the epithelium and for the development of memory (57).
In the skin, treatment with pertussis toxin decreases the velocity of CD8+ TRM cells and alters their morphology, resulting in a rounded phenotype (45). This suggests that chemokines likely play a role in the formation of dendritic spines on the T cells and interactions with the tissue. Keeping with this, the development of the TRM population in the skin greatly benefits from the presence of CXCR6, which is one of the “core” markers of bona fide TRM from many tissues (27, 45). The chemokine that this receptor binds is CXCL16, which has been shown to position innate lymphoid cells and natural killer T cells in tissues and is released by DCs during viral infection (94, 95). Although a diminished population still develops in the absence of CXCR6, the great reduction suggests that it increases either entry into tissue or proper positioning for survival. Alternatively, in the tonsils constitutive expression of IL-15 in the T cell zones and the subepithelium retained virus-specific CD8+ T cells within these two locations (96). Interestingly, in this organ, CD103 was only expressed on the T cells in close proximity to the epithelial cells and not in the extra-follicular region.
In many of these cases, signalizing through a cytokine or chemokine receptor may have other implications that indicate that the molecule itself is not directly necessary, but rather the downstream effect such as integrin activation or expression. For example, as previously addressed, interplay between TGFβ and CCL25 are associated with an upregulation of CD103 (57). Engagement of CD103 in turn leads to upregulation of CCR5 (55). CCR5 and CX3CR1 on CD8+ T cells in the lung are important for positioning of the cells in proximity to highly inflamed sites and the presence of antigens (56). These consequences suggest that there are complicated signaling pathways and loops at play that have yet to be fully elucidated.
In addition, the presence of various cytokines may provide sufficient inflammatory signals to allow cells access to locations not previously available. Inflammation of the vascular endothelium and changes to the barrier epithelium can occur either through cytokines or early immune cytolytic responses to infection (97, 98). Both of these events could dynamically alter the tissue landscape and allow TRM cells to gain access to previously obscured regions and persist. To fully understand these changes, it is likely that in vivo imaging systems will need to be developed to examine alterations in the ECM over time.
While this was touched on briefly earlier in the review, the ultimate role that TRM cells play in protecting a host is not fully understood. One possibility is that the cells produce antiviral cytokines and control downstream immune responses to eliminate infected cells. Alternatively, TRM cells could directly target the cells. In mice, TRM cells in the lungs make large quantities of IFN-γ in response to antigen-specific rechallenge, and blocking of this antiviral cytokine is detrimental to survival (99). In this instance, cells do not produce granzymes or other markers of cytotoxicity. TRM in the female reproductive tract display a decrease in motility after reexposure suggesting that the cells are interacting with antigen-bearing DCs; however, the direct antiviral outcome was not examined in this context (100). CD4+CD49a+ T cells in the lung activate within hours of secondary challenge with influenza and secrete an array of chemokines that attract other immune effectors (101). In line with this, CD4+-resident memory cells in the lungs are also sufficient for promoting airway hyperresponsiveness in a house dust mite model of asthma (102); however, this response can be partially attributed to TRM activation of local DC subsets (103).
Reactivation of human TRM cells alternatively not only leads to IFNγ production but also degranulation of cytolytic granules, including granzyme B and perforins (9, 74). It is conceivable that the limited responses observed in the mouse models are due to the methods employed to evaluate the cellular responses. It is possible that TRM cells are cytolytic in vivo with the proper stimulus. Other signals may be necessary for a similar response in vitro. For example, TRM cells may require signals from CD103 as well as through the TCR for degranulation of lytic granules. To fully understand the responses, it is necessary to examine these cells in vivo. To achieve this, granzyme B reporter mice and other protein reporter mice are the ideal tools (104, 105). As an alternative approach, transcriptional reporters for cytokine production or calcium reporters to indicate productive interactions with APCs may suffice (106–108). Utilizing these tools in conjunction with reporter pathogens, all the cues and responses necessary for protection of the host can be elucidated (109).
With the current methods employed for examining TRM cells, researchers are likely underrepresenting the populations as demonstrated by Steinert et al. (40). Using histology and/or other imaging methods will better reveal the totality of the TRM cell presence and ultimately their response. Although TRM cells are identified through expression of the discussed surface receptors, it is still not clear how the combination of markers alters the response of a given cell. Not all cells that express these markers at the resolution of an infection persist in the organ, suggesting that it is a combination of expression profiles, localization within a tissue, and perhaps proximity to APCs, which may present persistent antigen.
While CD49a, CD103, CD69, and combinations of chemokine receptors are used to define TRM, the precise positioning of the T cells in mucosal and glandular tissues remains to be directly examined, including demonstration that CD103 actually binds to E-cadherin at epithelial sites. Besides positioning, how these markers regulate cell motility versus retention in different tissues is not well defined. Intravital microscopy may be one approach to answer some of these questions.
It is also still not fully understood what population of cells lead to the TRM subset. In the lung, evidence suggests that TEM may be maintained within the parenchyma and refeed the transitory airway subset (30). Other TRM populations may also be self-renewing, similar to other populations of resident immune cells which seed organs earlier during development (11, 110). In fact, recent evidence in the female reproductive tract suggests that TRM cells divide during antigen-specific challenge and contribute to the secondary TRM population more than circulating memory cells (100).
Given the common features of TRM subsets in different tissues begs the question of whether there is a common developmental program, as no one specific pathway has been identified. How that program is triggered remains unclear, although TGF-β is required for CD49a and CD103 expression (23). Identifying key features, such as route of delivery, will be critical for optimally generating TRM. Previous studies indicate that memory generated through direct infection or exposure of the tissue is distinct from cells that result from systemic priming (47). Ideally, production of TRM could be an effective approach to problems like a universal flu vaccine; however, the field still struggles with eliciting an effective response in a vaccination setting.
TRM cells are important for protection from secondary encounters with various pathogens. They can protect and preserve the integrity of barrier surfaces such as skin, gut, and respiratory tissues. Transcriptional regulation is different in mouse and human systems, although a set of “core” markers have been identified, each having a distinct role in establishing and maintaining TRM. It is clear from these studies that TRM cells are highly regulated, poised to respond, yet suppressed by molecules including PD-1 and CD101 to prevent aberrant activation (27). Although the specific cues that result in loss of that suppression have not been identified, it is likely a combination of signals that may include type 1 interferons, which are one of the earliest innate factors made during infections and TCR engagement. In terms of positioning, there is evidence that CD49a interacts with ColIV, and CD103 adheres to E-cadherin, both retaining the cells near the epithelial surface (Figure 1). CD69 and CD44 do not seem to have the same level of necessity, and they are not included in the list of core markers identified in humans; however, they both likely contribute to persistence through limiting egress and maintenance of cytoskeletal structure. To resolve many of these questions future work will need to be done to elicit a full understanding of how TRM cells function to provide pathogen surveillance and protection.
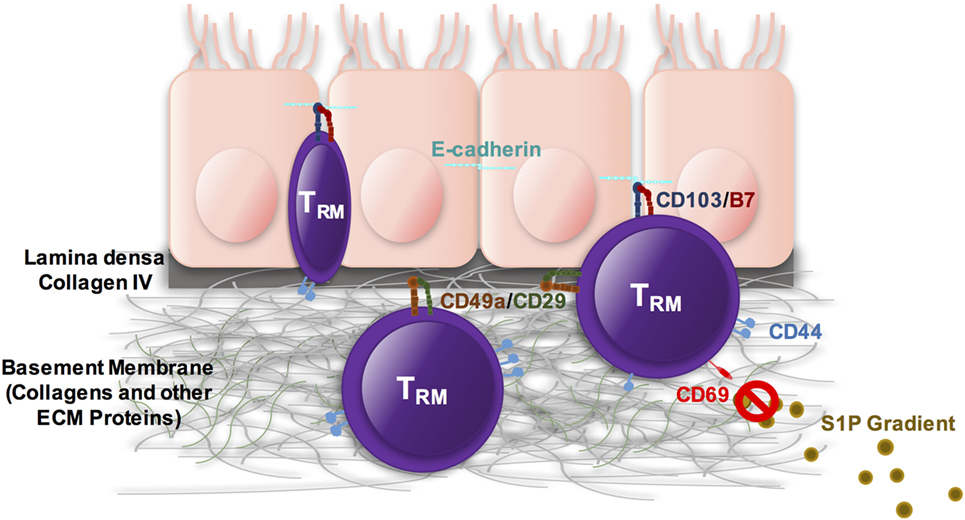
Figure 1. TRM cells are heterogenous both within tissues and also between sites. They express some combination of CD49a, CD103, CD44, and CD69, which cooperate to position the cells and maintain them within the site of initial infection. CD103 can interact with E-cadherin within epithelial surfaces. CD49a interacts with collagens with a preference for collagen IV in the lamina densa underlying the epithelium. CD44 maintains cell shape and integrity and can interact with a number of different tissue components including hyaluronic acid as well as fibronectin and other ECM proteins. CD69 antagonizes S1P1, essentially blocking any response to S1P gradients.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was funded by HHS/NIH/NIAID/AI102851-04.
1. Masopust D, Jiang J, Shen H, Lefrancois L. Direct analysis of the dynamics of the intestinal mucosa CD8 T cell response to systemic virus infection. J Immunol (2001) 166:2348–56. doi:10.4049/jimmunol.166.4.2348
2. Ray SJ, Franki SN, Pierce RH, Dimitrova S, Koteliansky V, Sprague AG, et al. The collagen binding alpha1beta1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity (2004) 20:167–79. doi:10.1016/S1074-7613(04)00021-4
3. Ely KH, Cookenham T, Roberts AD, Woodland DL. Memory T cell populations in the lung airways are maintained by continual recruitment. J Immunol (2006) 176:537–43. doi:10.4049/jimmunol.176.1.537
4. Wu T, Hu Y, Lee YT, Bouchard KR, Benechet A, Khanna K, et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J Leukoc Biol (2014) 95:215–24. doi:10.1189/jlb.0313180
5. Shin H, Iwasaki A. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature (2012) 491:463–7. doi:10.1038/nature11522
6. Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci U S A (2010) 107:17872–9. doi:10.1073/pnas.1010201107
7. Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol (2012) 188:4866–75. doi:10.4049/jimmunol.1200402
8. Thom JT, Weber TC, Walton SM, Torti N, Oxenius A. The salivary gland acts as a sink for tissue-resident memory CD8(+) T cells, facilitating protection from local cytomegalovirus infection. Cell Rep (2015) 13:1125–36. doi:10.1016/j.celrep.2015.09.082
9. Cheuk S, Schlums H, Gallais Serezal I, Martini E, Chiang SC, Marquardt N, et al. CD49a expression defines tissue-resident CD8+ T cells poised for cytotoxic function in human skin. Immunity (2017) 46:287–300. doi:10.1016/j.immuni.2017.01.009
10. Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature (2017) 543:252–6. doi:10.1038/nature21379
11. Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature (2012) 483:227–31. doi:10.1038/nature10851
12. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol (2013) 14:1294–301. doi:10.1038/ni.2744
13. Wakim LM, Woodward-Davis A, Liu R, Hu Y, Villadangos J, Smyth G, et al. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J Immunol (2012) 189:3462–71. doi:10.4049/jimmunol.1201305
14. Landrith TA, Sureshchandra S, Rivera A, Jang JC, Rais M, Nair MG, et al. CD103+ CD8 T cells in the toxoplasma-infected brain exhibit a tissue-resident memory transcriptional profile. Front Immunol (2017) 8:335. doi:10.3389/fimmu.2017.00335
15. Vesterinen E, Leinikki P, Saksela E. Induction of latent herpes simplex virus type 2 infection in human cervical epithelial cells in vitro. Acta Pathol Microbiol Scand B (1977) 85B:289–95.
16. Reiss TF, Gruenert DC, Nadel JA, Jacoby DB. Infection of cultured human airway epithelial cells by influenza A virus. Life Sci (1991) 49:1173–81. doi:10.1016/0024-3205(91)90565-S
17. Cunningham AL, Diefenbach RJ, Miranda-Saksena M, Bosnjak L, Kim M, Jones C, et al. The cycle of human herpes simplex virus infection: virus transport and immune control. J Infect Dis (2006) 194(Suppl 1):S11–8. doi:10.1086/505359
18. Wu NH, Yang W, Beineke A, Dijkman R, Matrosovich M, Baumgartner W, et al. The differentiated airway epithelium infected by influenza viruses maintains the barrier function despite a dramatic loss of ciliated cells. Sci Rep (2016) 6:39668. doi:10.1038/srep39668
19. Henson D, Strano AJ. Mouse cytomegalovirus. Necrosis of infected and morphologically normal submaxillary gland acinar cells during termination of chronic infection. Am J Pathol (1972) 68:183–202.
20. Jonjic S, Mutter W, Weiland F, Reddehase MJ, Koszinowski UH. Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J Exp Med (1989) 169:1199–212. doi:10.1084/jem.169.4.1199
21. Richter M, Ray SJ, Chapman TJ, Austin SJ, Rebhahn J, Mosmann TR, et al. Collagen distribution and expression of collagen-binding alpha1beta1 (VLA-1) and alpha2beta1 (VLA-2) integrins on CD4 and CD8 T cells during influenza infection. J Immunol (2007) 178:4506–16. doi:10.4049/jimmunol.178.7.4506
22. Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science (2016) 352:459–63. doi:10.1126/science.aad2035
23. Zhang N, Bevan MJ. Transforming growth factor-beta signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity (2013) 39:687–96. doi:10.1016/j.immuni.2013.08.019
24. Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, et al. T-box transcription factors combine with the cytokines TGF-beta and IL-15 to control tissue-resident memory T cell fate. Immunity (2015) 43:1101–11. doi:10.1016/j.immuni.2015.11.008
25. Schenkel JM, Fraser KA, Casey KA, Beura LK, Pauken KE, Vezys V, et al. IL-15-independent maintenance of tissue-resident and boosted effector memory CD8 T cells. J Immunol (2016) 196:3920–6. doi:10.4049/jimmunol.1502337
26. Hombrink P, Helbig C, Backer RA, Piet B, Oja AE, Stark R, et al. Programs for the persistence, vigilance and control of human CD8(+) lung-resident memory T cells. Nat Immunol (2016) 17:1467–78. doi:10.1038/ni.3589
27. Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, et al. Human Tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep (2017) 20:2921–34. doi:10.1016/j.celrep.2017.08.078
28. Sowell RT, Rogozinska M, Nelson CE, Vezys V, Marzo AL. Cutting edge: generation of effector cells that localize to mucosal tissues and form resident memory CD8 T cells is controlled by mTOR. J Immunol (2014) 193:2067–71. doi:10.4049/jimmunol.1400074
29. Klonowski KD, Williams KJ, Marzo AL, Blair DA, Lingenheld EG, Lefrancois L. Dynamics of blood-borne CD8 memory T cell migration in vivo. Immunity (2004) 20:551–62. doi:10.1016/S1074-7613(04)00103-7
30. Takamura S, Yagi H, Hakata Y, Motozono C, Mcmaster SR, Masumoto T, et al. Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J Exp Med (2016) 213:3057–73. doi:10.1084/jem.20160938
31. Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature (2006) 440:540–4. doi:10.1038/nature04606
32. Pham TH, Okada T, Matloubian M, Lo CG, Cyster JG. S1P1 receptor signaling overrides retention mediated by G alpha i-coupled receptors to promote T cell egress. Immunity (2008) 28:122–33. doi:10.1016/j.immuni.2007.11.017
33. Mackay LK, Braun A, Macleod BL, Collins N, Tebartz C, Bedoui S, et al. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J Immunol (2015) 194:2059–63. doi:10.4049/jimmunol.1402256
34. Bankovich AJ, Shiow LR, Cyster JG. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J Biol Chem (2010) 285:22328–37. doi:10.1074/jbc.M110.123299
35. Simms PE, Ellis TM. Utility of flow cytometric detection of CD69 expression as a rapid method for determining poly- and oligoclonal lymphocyte activation. Clin Diagn Lab Immunol (1996) 3:301–4.
36. Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity (2013) 38:187–97. doi:10.1016/j.immuni.2012.09.020
37. Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol (2013) 14:1285–93. doi:10.1038/ni.2745
38. Bieber T, Rieger A, Stingl G, Sander E, Wanek P, Strobel I. CD69, an early activation antigen on lymphocytes, is constitutively expressed by human epidermal Langerhans cells. J Invest Dermatol (1992) 98:771–6. doi:10.1111/1523-1747.ep12499948
39. Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife (2014) 3:e01659. doi:10.7554/eLife.01659
40. Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyarto BZ, et al. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell (2015) 161:737–49. doi:10.1016/j.cell.2015.03.031
41. Hadley GA, Bartlett ST, Via CS, Rostapshova EA, Moainie S. The epithelial cell-specific integrin, CD103 (alpha E integrin), defines a novel subset of alloreactive CD8+ CTL. J Immunol (1997) 159:3748–56.
42. El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, et al. TGF-{beta}-dependent CD103 expression by CD8(+) T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J Exp Med (2005) 201:1647–57. doi:10.1084/jem.20041044
43. Cepek KL, Parker CM, Madara JL, Brenner MB. Integrin alpha E beta 7 mediates adhesion of T lymphocytes to epithelial cells. J Immunol (1993) 150:3459–70.
44. Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, Rimm DL, et al. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature (1994) 372:190–3. doi:10.1038/372190a0
45. Zaid A, Hor JL, Christo SN, Groom JR, Heath WR, Mackay LK, et al. Chemokine receptor-dependent control of skin tissue-resident memory T cell formation. J Immunol (2017) 199:2451–9. doi:10.4049/jimmunol.1700571
46. Lee YT, Suarez-Ramirez JE, Wu T, Redman JM, Bouchard K, Hadley GA, et al. Environmental and antigen receptor-derived signals support sustained surveillance of the lungs by pathogen-specific cytotoxic T lymphocytes. J Virol (2011) 85:4085–94. doi:10.1128/JVI.02493-10
47. Sheridan BS, Pham QM, Lee YT, Cauley LS, Puddington L, Lefrancois L. Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity (2014) 40:747–57. doi:10.1016/j.immuni.2014.03.007
48. Malik BT, Byrne KT, Vella JL, Zhang P, Shabaneh TB, Steinberg SM, et al. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci Immunol (2017) 2(10):eaam6346. doi:10.1126/sciimmunol.aam6346
49. Nizard M, Roussel H, Diniz MO, Karaki S, Tran T, Voron T, et al. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nat Commun (2017) 8:15221. doi:10.1038/ncomms15221
50. Djenidi F, Adam J, Goubar A, Durgeau A, Meurice G, De Montpreville V, et al. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J Immunol (2015) 194:3475–86. doi:10.4049/jimmunol.1402711
51. Franciszkiewicz K, Le Floc’h A, Boutet M, Vergnon I, Schmitt A, Mami-Chouaib F. CD103 or LFA-1 engagement at the immune synapse between cytotoxic T cells and tumor cells promotes maturation and regulates T-cell effector functions. Cancer Res (2013) 73:617–28. doi:10.1158/0008-5472.CAN-12-2569
52. Le Floc’h A, Jalil A, Vergnon I, Le Maux Chansac B, Lazar V, Bismuth G, et al. Alpha E beta 7 integrin interaction with E-cadherin promotes antitumor CTL activity by triggering lytic granule polarization and exocytosis. J Exp Med (2007) 204:559–70. doi:10.1084/jem.20061524
53. Le Floc’h A, Jalil A, Franciszkiewicz K, Validire P, Vergnon I, Mami-Chouaib F. Minimal engagement of CD103 on cytotoxic T lymphocytes with an E-cadherin-Fc molecule triggers lytic granule polarization via a phospholipase Cgamma-dependent pathway. Cancer Res (2011) 71:328–38. doi:10.1158/0008-5472.CAN-10-2457
54. Woodland DL, Scott I. T cell memory in the lung airways. Proc Am Thorac Soc (2005) 2:126–31. doi:10.1513/pats.200501-003AW
55. Franciszkiewicz K, Le Floc’h A, Jalil A, Vigant F, Robert T, Vergnon I, et al. Intratumoral induction of CD103 triggers tumor-specific CTL function and CCR5-dependent T-cell retention. Cancer Res (2009) 69:6249–55. doi:10.1158/0008-5472.CAN-08-3571
56. Kohlmeier JE, Miller SC, Smith J, Lu B, Gerard C, Cookenham T, et al. The chemokine receptor CCR5 plays a key role in the early memory CD8+ T cell response to respiratory virus infections. Immunity (2008) 29:101–13. doi:10.1016/j.immuni.2008.05.011
57. Ericsson A, Svensson M, Arya A, Agace WW. CCL25/CCR9 promotes the induction and function of CD103 on intestinal intraepithelial lymphocytes. Eur J Immunol (2004) 34:2720–9. doi:10.1002/eji.200425125
58. Borkowski TA, Van Dyke BJ, Schwarzenberger K, Mcfarland VW, Farr AG, Udey MC. Expression of E-cadherin by murine dendritic cells: E-cadherin as a dendritic cell differentiation antigen characteristic of epidermal Langerhans cells and related cells. Eur J Immunol (1994) 24:2767–74. doi:10.1002/eji.1830241129
59. Siddiqui KR, Laffont S, Powrie F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity (2010) 32:557–67. doi:10.1016/j.immuni.2010.03.017
60. Hofmann M, Pircher H. E-cadherin promotes accumulation of a unique memory CD8 T-cell population in murine salivary glands. Proc Natl Acad Sci U S A (2011) 108:16741–6. doi:10.1073/pnas.1107200108
61. Lis R, Capdet J, Mirshahi P, Lacroix-Triki M, Dagonnet F, Klein C, et al. Oncologic trogocytosis with hospicells induces the expression of N-cadherin by breast cancer cells. Int J Oncol (2010) 37:1453–61. doi:10.3892/ijo_00000797
62. Dopfer EP, Minguet S, Schamel WW. A new vampire saga: the molecular mechanism of T cell trogocytosis. Immunity (2011) 35:151–3. doi:10.1016/j.immuni.2011.08.004
63. Perez TD, Nelson WJ. Cadherin adhesion: mechanisms and molecular interactions. Handb Exp Pharmacol (2004) 165:3–21. doi:10.1007/978-3-540-68170-0_1
64. Steinbach K, Vincenti I, Kreutzfeldt M, Page N, Muschaweckh A, Wagner I, et al. Brain-resident memory T cells represent an autonomous cytotoxic barrier to viral infection. J Exp Med (2016) 213:1571–87. doi:10.1084/jem.20151916
65. Strauch UG, Mueller RC, Li XY, Cernadas M, Higgins JM, Binion DG, et al. Integrin alpha E(CD103)beta 7 mediates adhesion to intestinal microvascular endothelial cell lines via an E-cadherin-independent interaction. J Immunol (2001) 166:3506–14. doi:10.4049/jimmunol.166.5.3506
66. Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, Kupper TS. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One (2011) 6:e16245. doi:10.1371/journal.pone.0016245
67. Haddadi S, Thanthrige-Don N, Afkhami S, Khera A, Jeyanathan M, Xing Z. Expression and role of VLA-1 in resident memory CD8 T cell responses to respiratory mucosal viral-vectored immunization against tuberculosis. Sci Rep (2017) 7:9525. doi:10.1038/s41598-017-09909-4
68. Bank I, Book M, Ware R. Functional role of VLA-1 (CD49A) in adhesion, cation-dependent spreading, and activation of cultured human T lymphocytes. Cell Immunol (1994) 156:424–37. doi:10.1006/cimm.1994.1187
69. Roberts AI, Brolin RE, Ebert EC. Integrin alpha1beta1 (VLA-1) mediates adhesion of activated intraepithelial lymphocytes to collagen. Immunology (1999) 97:679–85. doi:10.1046/j.1365-2567.1999.00812.x
70. Ben-Horin S, Bank I. The role of very late antigen-1 in immune-mediated inflammation. Clin Immunol (2004) 113:119–29. doi:10.1016/j.clim.2004.06.007
71. Kuhn K. Basement membrane (type IV) collagen. Matrix Biol (1995) 14:439–45. doi:10.1016/0945-053X(95)90001-2
72. Meharra EJ, Schon M, Hassett D, Parker C, Havran W, Gardner H. Reduced gut intraepithelial lymphocytes in VLA1 null mice. Cell Immunol (2000) 201:1–5. doi:10.1006/cimm.2000.1630
73. Richter MV, Topham DJ. The alpha1beta1 integrin and TNF receptor II protect airway CD8+ effector T cells from apoptosis during influenza infection. J Immunol (2007) 179:5054–63. doi:10.4049/jimmunol.179.8.5054
74. Jozwik A, Habibi MS, Paras A, Zhu J, Guvenel A, Dhariwal J, et al. RSV-specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat Commun (2015) 6:10224. doi:10.1038/ncomms10224
75. Hogan RJ, Usherwood EJ, Zhong W, Roberts AA, Dutton RW, Harmsen AG, et al. Activated antigen-specific CD8+ T cells persist in the lungs following recovery from respiratory virus infections. J Immunol (2001) 166:1813–22. doi:10.4049/jimmunol.166.3.1813
76. Kennel SJ, Lankford TK, Foote LJ, Shinpock SG, Stringer C. CD44 expression on murine tissues. J Cell Sci (1993) 104(Pt 2):373–82.
77. Lesley J, Howes N, Perschl A, Hyman R. Hyaluronan binding function of CD44 is transiently activated on T cells during an in vivo immune response. J Exp Med (1994) 180:383–7. doi:10.1084/jem.180.1.383
78. Lesley J, Hascall VC, Tammi M, Hyman R. Hyaluronan binding by cell surface CD44. J Biol Chem (2000) 275:26967–75. doi:10.1074/jbc.M002527200
79. Mikecz K, Brennan FR, Kim JH, Glant TT. Anti-CD44 treatment abrogates tissue oedema and leukocyte infiltration in murine arthritis. Nat Med (1995) 1:558–63. doi:10.1038/nm0695-558
80. Budd RC, Cerottini JC, Macdonald HR. Phenotypic identification of memory cytolytic T lymphocytes in a subset of Lyt-2+ cells. J Immunol (1987) 138:1009–13.
81. Walzer T, Arpin C, Beloeil L, Marvel J. Differential in vivo persistence of two subsets of memory phenotype CD8 T cells defined by CD44 and CD122 expression levels. J Immunol (2002) 168:2704–11. doi:10.4049/jimmunol.168.6.2704
82. Baaten BJ, Li CR, Bradley LM. Multifaceted regulation of T cells by CD44. Commun Integr Biol (2010) 3:508–12. doi:10.4161/cib.3.6.13495
83. Skelton TP, Zeng C, Nocks A, Stamenkovic I. Glycosylation provides both stimulatory and inhibitory effects on cell surface and soluble CD44 binding to hyaluronan. J Cell Biol (1998) 140:431–46. doi:10.1083/jcb.140.2.431
84. Katoh S, Miyagi T, Taniguchi H, Matsubara Y, Kadota J, Tominaga A, et al. Cutting edge: an inducible sialidase regulates the hyaluronic acid binding ability of CD44-bearing human monocytes. J Immunol (1999) 162:5058–61.
85. Zoller M, Mcelwee KJ, Engel P, Hoffmann R. Transient CD44 variant isoform expression and reduction in CD4(+)/CD25(+) regulatory T cells in C3H/HeJ mice with alopecia areata. J Invest Dermatol (2002) 118:983–92. doi:10.1046/j.1523-1747.2002.01745.x
86. Nandi A, Estess P, Siegelman M. Bimolecular complex between rolling and firm adhesion receptors required for cell arrest; CD44 association with VLA-4 in T cell extravasation. Immunity (2004) 20:455–65. doi:10.1016/S1074-7613(04)00077-9
87. Mummert ME, Mummert D, Edelbaum D, Hui F, Matsue H, Takashima A. Synthesis and surface expression of hyaluronan by dendritic cells and its potential role in antigen presentation. J Immunol (2002) 169:4322–31. doi:10.4049/jimmunol.169.8.4322
88. Hegde VL, Singh NP, Nagarkatti PS, Nagarkatti M. CD44 mobilization in allogeneic dendritic cell-T cell immunological synapse plays a key role in T cell activation. J Leukoc Biol (2008) 84:134–42. doi:10.1189/jlb.1107752
89. Mrass P, Kinjyo I, Ng LG, Reiner SL, Pure E, Weninger W. CD44 mediates successful interstitial navigation by killer T cells and enables efficient antitumor immunity. Immunity (2008) 29:971–85. doi:10.1016/j.immuni.2008.10.015
90. Baaten BJ, Li CR, Deiro MF, Lin MM, Linton PJ, Bradley LM. CD44 regulates survival and memory development in Th1 cells. Immunity (2010) 32:104–15. doi:10.1016/j.immuni.2009.10.011
91. Stein JV, Nombela-Arrieta C. Chemokine control of lymphocyte trafficking: a general overview. Immunology (2005) 116:1–12. doi:10.1111/j.1365-2567.2005.02183.x
92. Lim K, Hyun YM, Lambert-Emo K, Capece T, Bae S, Miller R, et al. Neutrophil trails guide influenza-specific CD8(+) T cells in the airways. Science (2015) 349:aaa4352. doi:10.1126/science.aaa4352
93. Reilly EC, Lambert-Emo K, Topham DJ. The effects of acute neutrophil depletion on resolution of acute influenza infection, establishment of tissue resident memory (TRM), and heterosubtypic immunity. PLoS One (2016) 11:e0164247. doi:10.1371/journal.pone.0164247
94. Piqueras B, Connolly J, Freitas H, Palucka AK, Banchereau J. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood (2006) 107:2613–8. doi:10.1182/blood-2005-07-2965
95. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol (2014) 32:659–702. doi:10.1146/annurev-immunol-032713-120145
96. Woon HG, Braun A, Li J, Smith C, Edwards J, Sierro F, et al. Compartmentalization of total and virus-specific tissue-resident memory CD8+ T cells in human lymphoid organs. PLoS Pathog (2016) 12:e1005799. doi:10.1371/journal.ppat.1005799
97. Hers JF. Disturbances of the ciliated epithelium due to influenza virus. Am Rev Respir Dis (1966) 93(Suppl):162–77.
98. Sedgwick JB, Menon I, Gern JE, Busse WW. Effects of inflammatory cytokines on the permeability of human lung microvascular endothelial cell monolayers and differential eosinophil transmigration. J Allergy Clin Immunol (2002) 110:752–6. doi:10.1067/mai.2002.128581
99. Mcmaster SR, Wilson JJ, Wang H, Kohlmeier JE. Airway-resident memory CD8 T cells provide antigen-specific protection against respiratory virus challenge through rapid IFN-gamma production. J Immunol (2015) 195:203–9. doi:10.4049/jimmunol.1402975
100. Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, et al. Intravital mucosal imaging of CD8(+) resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat Immunol (2018) 19:173–82. doi:10.1038/s41590-017-0029-3
101. Chapman TJ, Topham DJ. Identification of a unique population of tissue-memory CD4+ T cells in the airways after influenza infection that is dependent on the integrin VLA-1. J Immunol (2010) 184:3841–9. doi:10.4049/jimmunol.0902281
102. Hondowicz BD, An D, Schenkel JM, Kim KS, Steach HR, Krishnamurty AT, et al. Interleukin-2-dependent allergen-specific tissue-resident memory cells drive asthma. Immunity (2016) 44:155–66. doi:10.1016/j.immuni.2015.11.004
103. Turner DL, Goldklang M, Cvetkovski F, Paik D, Trischler J, Barahona J, et al. Biased generation and in situ activation of lung tissue-resident memory CD4 T cells in the pathogenesis of allergic asthma. J Immunol (2018) 200(5):1561–9. doi:10.4049/jimmunol.1700257
104. Mouchacca P, Schmitt-Verhulst AM, Boyer C. Visualization of cytolytic T cell differentiation and granule exocytosis with T cells from mice expressing active fluorescent granzyme B. PLoS One (2013) 8:e67239. doi:10.1371/journal.pone.0067239
105. Mouchacca P, Chasson L, Frick M, Foray C, Schmitt-Verhulst AM, Boyer C. Visualization of granzyme B-expressing CD8 T cells during primary and secondary immune responses to Listeria monocytogenes. Immunology (2015) 145:24–33. doi:10.1111/imm.12420
106. Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol (2009) 10:385–93. doi:10.1038/ni.1715
107. Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity (2001) 15:303–11. doi:10.1016/S1074-7613(01)00186-8
108. Thestrup T, Litzlbauer J, Bartholomaus I, Mues M, Russo L, Dana H, et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat Methods (2014) 11:175–82. doi:10.1038/nmeth.2773
109. Dipiazza A, Nogales A, Poulton N, Wilson PC, Martinez-Sobrido L, Sant AJ. Pandemic 2009 H1N1 influenza venus reporter virus reveals broad diversity of MHC class II-positive antigen-bearing cells following infection in vivo. Sci Rep (2017) 7:10857. doi:10.1038/s41598-017-11313-x
Keywords: T cells, memory, tissue distribution, integrins, infection, immunity, cellular
Citation: Topham DJ and Reilly EC (2018) Tissue-Resident Memory CD8+ T Cells: From Phenotype to Function. Front. Immunol. 9:515. doi: 10.3389/fimmu.2018.00515
Received: 15 December 2017; Accepted: 27 February 2018;
Published: 26 March 2018
Edited by:
Steven Varga, University of Iowa, United StatesReviewed by:
Jeffrey C. Nolz, Oregon Health & Science University, United StatesCopyright: © 2018 Topham and Reilly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David J. Topham, ZGF2aWRfdG9waGFtQHVybWMucm9jaGVzdGVyLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.