
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Immunol., 05 February 2018
Sec. Molecular Innate Immunity
Volume 9 - 2018 | https://doi.org/10.3389/fimmu.2018.00162
This article is part of the Research TopicC1q: A Molecular Bridge to Innate and Adaptive ImmunityView all 10 articles
The immunopathological functions associated with human C1q are still growing in terms of novelty, diversity, and pathologic relevance. It is, therefore, not surprising that C1q is being recognized as an important molecular bridge between innate and adaptive immunity. The secret of this functional diversity, in turn, resides in the elegant but complex structure of the C1q molecule, which is assembled from three distinct gene products: A, B, and C, each of which has evolved from a separate and unique ancestral gene template. The C1q molecule is made up of 6A, 6B, and 6C polypeptide chains, which are held together through strong covalent and non-covalent bonds to form the 18-chain, bouquet-of-flower-like protein that we know today. The assembled C1q protein displays at least two distinct structural and functional regions: the collagen-like region (cC1q) and the globular head region (gC1q), each being capable of driving a diverse range of ligand- or receptor-mediated biological functions. What is most intriguing, however, is the observation that most of the functions appear to be predominantly driven by the A-chain of the molecule, which begs the question: what are the evolutionary modifications or rearrangements that singularly shaped the primordial A-chain gene to become a pluripotent and versatile component of the intact C1q molecule? Here, we revisit and discuss some of the known unique structural and functional features of the A-chain, which may have contributed to its versatility.
C1q is the first subcomponent of the complement classical pathway. In addition to its complement activation mediated immune functions, it has a broad range of developmental homeostatic functions that are not dependent on its ability to activate the classical pathway [reviewed in Ref. (1)]. The functional versatility of C1q depends on several unique structural and functional properties (1–3). It is made up of three chains, A, B, and C, which are the product of three distinct genes, found highly clustered and aligned 5′⇑3′, in the same orientation, in the order A–C–B on a 24 kb stretch of DNA on chromosome 1p at position 36.12 (4, 5). Each chain contains an N-terminal collagen-like region and a C-terminal globular head region. There are 18 chains in the intact C1q molecule: 6A, 6B, and 6C, which are arranged first as a single heterotrimeric strand comprising of A, B, and C, in which the A chain and B chain within the strand are covalently linked to each other, whereas the C-chain of one strand which is non-covalently associated with the AB dimer, nonetheless forms a covalent link with the C chain of a neighboring ABC strand to form an ABC-CBA doublet. Three such doublets are then held together with non-covalent bonds to give rise to the well-recognized hexameric structure of C1q. The globular “heads” of each ABC strand are linked via six collagen-like “stalks” to a fibril-like central region resulting in two unique structural and functional domains: the collagen-like region (cC1q) and the globular “heads” or domains (gC1q) (6, 7). Each of the gC1q domains is a heterotrimeric structure made up of each of the individual chains (ghA, ghB, and ghC). What has become apparently clear is the fact that each of the gh domains is capable of recognizing a gh-specific ligand independent of the other gh domains (3, 8, 9). Therefore, assuming that each of the gh domains recognizes a single target or ligand, the C1q molecule can recognize and bind simultaneously six individual molecular patterns, making it one of the most efficient, and versatile pattern recognition molecules.
The crystal structure of the heterotrimeric gC1q domain revealed a compact jellyroll β-sandwich fold similar to that of the multifunctional tumor necrosis factor (TNF) family of proteins (10, 11). This suggested that C1q not only diverged from a primordial ancestral gene template of the innate immune system that gave birth to the TNF-α and other C1q-like proteins, but also retained some of its ancestral “cytokine-like” functions (2, 10). Therefore, C1q could be considered as a prototype “complekine,” i.e., complement protein with cytokine-like activity, which is capable of mimicking some, if not all, functions of the TNF family of proteins, including the induction of cytokines (IL-6 and IL-8) and chemokines (e.g., MCP-1) that orchestrate a myriad of a rapidly expanding list of pathophysiological processes (12, 13).
There is also an abundance of clinical evidence, which shows that genetic deficiency in C1q is associated with a wide range of clinical syndromes closely related to SLE, with rashes, glomerulonephritis, and CNS disease as well as other autoimmune diseases (14). In addition, C1q also can have a major role in tumor growth and progression (15–19). The role of C1q, being a part of tumor microenvironment, has appeared to be complex so far. In some reports, it has been shown to be protumorigenic (15–17), whereas there are recent reports of antitumor activities of C1q in the case of prostrate (18) and ovarian cancers (19).
Although individuals with congenital C1q deficiency constitute only a small cohort of patients, this strong association nonetheless implicates an important role for complement in general, and C1q in particular, in the development of SLE and other autoimmune diseases (20–24). What is perplexing, however, is the fact that among the C1q deficiencies, the A-chain of C1q should take center stage in significance as homozygous deficiency or mutation in the A-chain is almost invariably associated with various diseases (20–24). The mutation in the A-chain is due to a homogeneous mutation in which the C to T transition in codon 186 of exon 2 results in Gln-to Stop (Q186X) substitution. The question is: what are the structural signatures that make the C1q-A chain such a powerful susceptibility biomarker of these diseases? It is worth noting that although the most prevalent mutation is the C1qA, Gln208X mutation, there are other mutations in B and C chains too (25).
The genes encoding the three chains of C1q are highly conserved from zebrafish to human. Phylogenetic analysis also intimates that the C1qA, C1qB, and C1qC may have originally been generated by gene duplications from a single copy of an ancestral C1qB gene, since the latter is found in the same branch as amphioxus C1q, which is an earlier lower vertebrate than zebrafish (26). Furthermore, the IgG binding properties between fish and mammalian C1q show no difference since substitution of human C1q by fish C1q has the same activity, suggesting that the IgG or IgM recognizing properties have remained conserved throughout the evolutionary history (26, 27). However, more recent studies have shown that there is a preferential binding of the gC1q modules when it comes to IgG binding. Whereas the gC1qA (or ghA) module binds aggregated IgG and IgM in a similar manner, gC1qB (ghB) binds aggregated IgG in preference to IgM (28). The functional preferences of the gC1q domains, therefore, may reflect an evolutionary structural adaptation that resulted in recent history.
In an elegant and in depth review, Trinder et al. (29) analyzed the structural and functional correlates that distinguish the A-chain from the B- and C-chains. First, while the B and C chains are highly conserved, the A-chain is not. This fact alone should support the notion that the A-chain developed to be functionally adaptable throughout evolution. Second, various types of cells including macrophages and dendritic cells among a long list of others, synthesize the C1q molecule. The cell-associated molecule in turn, is anchored in the membrane via a 22 amino acid long leader peptide, which is found only in the A-chain (29). Third, the A-chain contains several antigen recognition sites (Figure 1), but in particular, possesses one major (aa 14–26) and one minor (aa 76–92) promiscuous region (29), which serve as a binding site for a wide range of non-immunoglobulin antigens including lipopolysaccharide, C-reactive protein (CRP), DNA, heparin, fibronectin, monosodium, urate crystals, amyloid P component, von Willebrand factor (30) as well as bacterial and mitochondrial membranes (29–42). Importantly, this A chain region has also been shown to bind specifically by SLE patients’ sera compared to serum derived from healthy control (43). Although recent studies have suggested that the interaction site for CRP is located in the gC1q rather than the cC1q (44–46), it is plausible to assume that certain molecules could actually bind to multiple regions of the A-chain. Regardless, these non-immunoglobulin antigens have been shown to activate the classical pathway by binding to the cC1q region of the A-chain rather than to the globular heads (29–42). In addition, the A-chain contains a collagen-type II-like sequence comprising of residues 26–34, which has been shown to suppress collagen type-II-induced arthritis in a mouse model (47). Interestingly, this same region is also predicted to be a potential MHC class II binding site (48). However, little is known about the significance of this finding but may have potential implications in autoimmunity and tolerance (48), especially since C1q has been shown to keep monocytes in a predendritic or immature phenotype, thus ensuring that unwarranted DC-driven immune response does not occur, a fact that is relevant to the development of SLE (49). Finally, although it is found only in the mouse, and not in the human C1q A-chain, the presence of an RGD sequence may also explain why human C1q still retains its ability to support adhesion of normal endothelial cells and fibroblasts (50–53) in a manner that is inhibited by an RGD peptide but not an RGE (51). Very recently, Agostinis et al. have shown that C1q can act as a bridge between hyaluronic acid (HA), an abundant matrix component of the tumor microenvironment, and the HA receptor on tumor cells, i.e., CD44, thus inducing considerable proliferation of primary tumor cells derived from malignant pleural mesothelioma (MPM) (17). Curiously, the A-chain of the globular region of C1q bound specifically and differentially to a range of LPS-free HA, leaving C-chain to liaise with MPM cells.
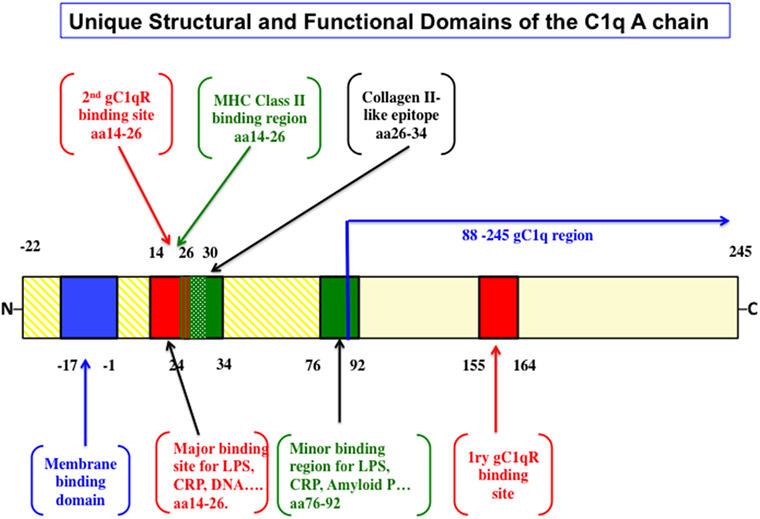
Figure 1. The structural and functional correlates of the C1q A-chain. The intact C1q molecule is anchored to the cell membrane by a leader peptide in the A chain. The major and minor ligand bind sites as well as the putative MHC class II binding domain are highlighted. Although the major gC1qR-binding domain spans residues 155–164, unexpected sites in the promiscuous collagen domain spanning residues 14–26 and another at 76–92 have also been shown very recently [the figure is adapted from Ref. (29)].
Thus, although it may be overly simplistic to suggest that the A-chain is the functional anchor of the C1q molecule, it appears to be clear that of the three chains, the A-chain has singularly undergone systematic and adaptable molecular evolution. Whether the selection of the A-chain to evolve as a master orchestrator of C1q functions was by design or serendipity, or whether the B- and C-chains are also undergoing similar evolution albeit at a much slower rate, are questions still for the future.
BG and EK wrote the first draft; UK and EP revised and edited the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This article is dedicated to Prof. Dr. Michael Loos, who is no longer with us, but whose wisdom and vision still drives us to discover unexpected functions of the molecule he loved, i.e., “C1q.”
The work included in this article was supported in part by grants from the National Institutes of Allergy and Infectious Diseases R01 AI 060866 and R01 AI-084178 (to BG) and the NIH/NCI cancer support grant P30 CA008748 [to Memorial Sloan-Kettering Cancer Center (MSKCC)].
ghA, ghB, and ghC, globular heads of the A, B, and C chains of C1q, respectively; gC1q, the globular heads of C1q; cC1q, the collagen domain of C1q; gC1qR, receptor for gC1q; cC1qR, receptor for cC1q; CRT, calreticulin (another name for cC1qR).
1. Kouser L, Madhukaran SP, Shastri A, Saraon A, Ferluga J, Al-Mozaini M, et al. Emerging and novel functions of complement protein C1q. Front Immunol (2015) 29(6):317. doi:10.3389/fimmu.2015.00317
2. Kishore U, Gaboriaud C, Waters O, Shrive AK, Greenhough TJ, Reid KB, et al. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol (2004) 25:551–61. doi:10.1016/j.it.2004.08.006
3. Kishore U, Gupta SK, Perdikoulis MV, Kojouharova MJ, Urban BC, Reid KB. Modular organization of the carboxy-terminal globular head region of human C1q A, B, and C chains. J Immunol (2003) 171:812–20.
4. Sellar GC, Blake DJ, Reid KBM. Characterization and organization of the genes encoding the A-, B, and C- chains of human complement subcomponent C1q. Biochem J (1991) 274:481–91. doi:10.1042/bj2740481
5. Sellar GC, Cockburn D, Reid KB. Localization of the gene cluster encoding the A, B, and C chains of human C1q to 1p34.1-1p36.3. Immunogenetics (1992) 35:214–6. doi:10.1007/BF00185116
6. Reid KBM. Molecular cloning and characterization of the complementary DNA and gene coding for the B chain subcomponent C1q of the human complement system. Biochem J (1985) 231:729–35. doi:10.1042/bj2310729
8. Kishore U, Leigh LE, Eggleton P, Strong P, Perdikoulis MV, Willis AC, et al. Functional characterization of a recombinant form of the C-terminal, globular head region of the B-chain of human serum complement protein, C1q. Biochem J (1998) 333(Pt 1):27–32.
9. Kojouharova MS, Gadjeva MG, Tsacheva IG, Zlatarova A, Roumenina LT, Tchorbadjieva MI, et al. Mutational analyses of the recombinant globular regions of human C1q A, B, and C chains suggest an essential role for arginine and histidine residues in the C1q-IgG interaction. J Immunol (2004) 172(7):4351–8. doi:10.4049/jimmunol.172.7.4351
10. Shapiro L, Scherer PE. The crystal structure of a complement-1q family of protein suggests an evolutionary link to tumor necrosis factor. Curr Biol (1998) 8:335–8. doi:10.1016/S0960-9822(98)70133-2
11. Gaboraiud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, et al. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem (2003) 278:46974–82. doi:10.1074/jbc.M307764200
12. Ghebrehiwet B, Hosszu KK, Valentino A, Peerschke EIB. The C1q family of proteins: insights into the emerging non-traditional functions. Front Immunol (2012) 3:52.
13. van den Berg RH, Faber-Krol MC, Sim RB, Daha MR. The first subcomponent of complement C1q triggers the production of IL-6, IL-8, and monocyte chemoattractant protein-1 by human umbilical vein endothelial cells. J Immunol Lett (1998) 161:6924–30.
14. Pickering MC, Botto M, Taylor PR, Lachmann PJ, Walport MJ. Systemic lupus erythematosus, complement deficiency, and apoptosis. Adv Immunol (2000) 76:227–34. doi:10.1016/S0065-2776(01)76021-X
15. Bulla R, Tripodo C, Rami D, Ling GS, Agostinis C, Guarnotta C, et al. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat Commun (2016) 7:10346. doi:10.1038/ncomms10346
16. Racilla E, Racilla DM, Ritchie JM, Taylor C, Dahle C, And Weiner GJ. The pattern of clinical breast cancer metastasis correlates with a single nucleotide polymorphism in the C1qA component of complement. Immunogentics (2006) 58:1–8. doi:10.1007/s00251-005-0077-y
17. Agostinis C, Vidergar R, Belmonte B, Mangogna A, Amadio L, Geri P, et al. Complement protein C1q binds to hyaluronic acid in the malignant pleural mesothelioma microenvironment and promotes tumor growth. Front Immunol (2017) 8:1559. doi:10.3389/fimmu.2017.01559
18. Hong Q, Sze CI, Lin SR, Lee MH, He RY, Schultz L, et al. Complement C1q activates tumor suppressor WWOX to induce apoptosis in prostate cancer cells. PLoS One (2009) 4(6):e5755. doi:10.1371/journal.pone.0005755
19. Kaur A, Sultan SH, Murugaiah V, Pathan AA, Alhamlan FS, Karteris E, et al. Human C1q induces apoptosis in an ovarian cancer cell line via tumor necrosis factor pathway. Front Immunol (2016) 7:599. doi:10.3389/fimmu.2016.00599
20. Komatsu A, Komazawa M, Murakami M, Nagaki Y. A case of selective C1q-deficiency with SLE-like symptoms. J Jpn Pediatr Soc (1982) 86:23.
21. Sun-Tan C, Ozqür TT, Kilinç G, Topaloglu R, Gököz O, Ersoy-Evans S, et al. Hereditary C1q deficiency: a new family with C1qA deficiency. Turk J Pediatr (2010) 52:184–6.
22. Petry F, Izzet Berkel A, Loos M. Multiple identification of a particular type of hereditary C1q deficiency in the Turkish population: review of the cases and additional genetic and functional analysis. Hum Genet (1997) 100:51–6. doi:10.1007/s004390050464
23. Topaloglu R, Bakkaloglu A, Slingsby JH, Mihatsch MJ, Pascual M, Norsworthy P, et al. Molecular basis of hereditary C1q deficiency associated with SLE and IgA nephropathy in a Turkish family. Kidney Int (1996) 50:635–42. doi:10.1038/ki.1996.359
24. Trouw LA, Daha N, Kurreeman FA, Bohringer S, Goulielmos GN, Westra HJ, et al. Genetic variants in the region of C1q genes are associated with rheumatoid arthritis. Clin Exp Immunol (2013) 173:76–83. doi:10.1111/cei.12097
25. Schejbel L, Skattum L, Hagelberg S, Åhlin A, Schiller B, Berg S, et al. Molecular basis of hereditary C1q deficiency-revisited: identification of several novel disease-causing mutations. Genes Immun (2011) 12:626–34. doi:10.1038/gene.2011.39
26. Hu Y-L, Pan X-M, Xiang L-X, Xiao J-Z. Characterization of C1q in Teleosts; insight into the molecular and functional evolution of C1q family and classical pathway. J Biol Chem (2010) 285:28777–86. doi:10.1074/jbc.M110.131318
27. Ghai R, Waters P, Roumenina LT, Gadjeva M, Kojouharova MS, Reid KB, et al. C1q and its growing family. Immunobiology (2007) 212(4–5):253–66. doi:10.1016/j.imbio.2006.11.001
28. Zlatarova AS, Rouseva M, Roumenina LT, Gadjeva M, Kolev M, Dobrev I, et al. Existence of different but overlapping IgG-and IgM-binding sites on the globular domain of human C1q. Biochemistry (2006) 45:9979–88. doi:10.1021/bi060539v
29. Trinder PK, Maeurer MJ, Kaul M, Petry E, Loos M. Functional domains of the human C1q A-chain. Behring Inst Mitt (1993) 93:180–8.
30. Kölm R, Schaller M, Roumenina LT, Niemiec I, Kremer Hovinga JA, Khanicheh E, et al. Von Willebrand factor interacts with surface-bound C1q and induces platelet rolling. J Immunol (2016) 197(9):3669–79. doi:10.4049/jimmunol.1501876
31. Kaul M, Loos M. C1q, the collagen-like subcomponent of the first component of complement C1, is a membrane protein of guinea pig macrophages. Eur J Immunol (1993) 23:2166–74. doi:10.1002/eji.1830230918
32. Gewurz H, Mold C, Siegel J, Fiedel B. C-reactive protein and the acute phase response. Adv Intern Med (1982) 27:345.
33. Ying S-C, Gewurz AT, Jiang H, Gewurz H. Human serum amyloid P component oligomers bind and activate the classical complement pathway via residues 14–26 and 76–92 of the A chain collagen-like region of C1q. J Immunol (1993) 150:169–76.
34. Volanakis JE. Complement activation by C-reactive protein complexes. Ann N Y Acad Sci (1982) 389:235–49. doi:10.1111/j.1749-6632.1982.tb22140.x
35. Jiang H, Robey FA, Gewurz H. Localization of sites through which C-reactive protein binds and activates complement to residues 14–26 and 76–92 of the human C1q A chain. J Exp Med (1992) 175:1373–9. doi:10.1084/jem.175.5.1373
36. Jiang H, Siegel JN, Gewurz H. Binding and complement activation by C-reactive protein via the collagen-like region of C1q and inhibition of these reactions by monoclonal antibodies to C-reactive protein and C1q. J Immunol (1991) 146:2324–30.
37. Jiang H, Burdick D, Glabe CG, Cotman CW, Tenner AJ. β-Amyloid activates complement by binding to a specific region of the collagen-like domain of the C1q A chain. J Immunol (1994) 152:5050–9.
38. Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest (2003) 111:1805–12. doi:10.1172/JCI200318921
39. Travis J, Salvesen GS. Human plasma proteinase inhibitors. Annu Rev Biochem (1983) 52:655–709. doi:10.1146/annurev.bi.52.070183.003255
40. Sorvillo J, Gigli I, Pearlstein E. Fibronectin binding to complement subcomponent C1q. Localization of their respective binding sites. Biochem J (1985) 226:207–15. doi:10.1042/bj2260207
41. Jiang H, Ying S-C, Kim YB, Gewurz H. Endotoxin activates the classical complement pathway via residues14-26 of the C1q A-chain and peptide 14-16 inhibits this activation. In: Loos M, editor. International Workshop on C1-The first component of complement. Mainz (1994). p. 47.
42. Cooper NR, Morrison DC. Binding and activation of the first component of human complement by the lipid A region of lipopolysaccharides. J Immunol (1978) 120:1862–8.
43. Vanhecke D, Roumenina LT, Wan H, Osthoff M, Schaller M, Trendelenburg M. Identification of a major linear C1q epitope allows detection of systemic lupus erythematosus anti-C1q antibodies by a specific peptide-based enzyme-linked immunosorbent assay. Arthritis Rheum (2012) 64(11):3706–14. doi:10.1002/art.34605
44. McGrath FDG, Brouwer MC, Arlaud GJ, Daha MR, Haxk CE, Roos A. Evidence that complement protein C1q interacts with C-reactive protein through its globular head region. J Immunol (2006) 176:2950–7. doi:10.4049/jimmunol.176.5.2950
45. Roumenina LT, Ruseva MM, Zlatarova A, Ghai R, Kolev M, Olova N, et al. Interaction of C1q with IgG1, C-reactive protein and pentraxin 3: mutational studies using recombinant globular head modules of human C1q A, B, and C chains. Biochemistry (2006) 45(13):4093–104. doi:10.1021/bi052646f
46. Kishore U, Ghai R, Greenhough TJ, Shrive AK, Bonifati DM, Gadjeva MG, et al. Structural and functional anatomy of the globular domain of complement protein C1q. Immunol Lett (2004) 95(2):113–28. doi:10.1016/j.imlet.2004.06.015
47. Meurer MJ, Trinder PKE, Storkel S, Loos M. Modulation of type II collagen-induced arthritis in DBA/1 mice by intravenous application of a peptide from the C1q A-chain. Immunobiology (1992) 185:103–10. doi:10.1016/S0171-2985(11)80321-9
48. Myers LK, Sturat JM, Seyer JM, Kang KH. Identification of an immunosuppressive epitope of type II collagen that confers protection against collagen-induced arthritis. J Exp Med (1989) 170:199–210. doi:10.1084/jem.170.6.1999
49. Myers LK, Seyer JM, Stuart JM, Terato K, David CS, Kang AH. T cell epitopes of type II collagen that regulate murine collagen-induced arthritis. J Immunol (1993) 151:500–5.
50. Hosszu KK, Santiago-Schwarz F, Peerschke EIB, Ghebrehiwet B. Evidence that a C1q/C1qR system regulates monocyte-derived cell differentiation at the interface of acquired and innate immunity. Innate Immun (2010) 16:115–27. doi:10.1177/1753425909339815
51. Feng X, Tonnesen MG, Peerschke EIB, Ghebrehiwet B. Cooperation of C1q receptors and integrins in C1q-mediated endothelial cell adhesion and spreading. J Immunol (2002) 168:2441–8. doi:10.4049/jimmunol.168.5.2441
52. Bordin S, Ghebrehiwet B, Page RC. Participation of C1q and its receptor in adherence of human diploid fibroblast. J Immunol (1990) 145:2520–6.
Keywords: Complement, classical pathway, C1q, A chain, charge pattern recognition, C1q receptor
Citation: Ghebrehiwet B, Kandov E, Kishore U and Peerschke EIB (2018) Is the A-Chain the Engine That Drives the Diversity of C1q Functions? Revisiting Its Unique Structure. Front. Immunol. 9:162. doi: 10.3389/fimmu.2018.00162
Received: 13 December 2017; Accepted: 18 January 2018;
Published: 05 February 2018
Edited by:
Zvi Fishelson, Tel Aviv University, IsraelReviewed by:
Marten Trendelenburg, University Hospital of Basel, SwitzerlandCopyright: © 2018 Ghebrehiwet, Kandov, Kishore and Peerschke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Berhane Ghebrehiwet, YmVyaGFuZS5naGVicmVoaXdldEBzdG9ueWJyb29rLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.