- 1Department of Pediatrics, Division of Blood and Marrow Transplantation, University of Minnesota Cancer Center, Minneapolis, MN, United States
- 2The Antigen Presentation and Immunoregulation Laboratory and Bone Marrow Transplantation Laboratory, QIMR Berghofer Medical Research Institute, University of Minnesota Cancer Center, Brisbane, QLD, Australia
Regulatory T cells (Tregs) are key mediators of the immune system. MicroRNAs (miRNAs) are a family of ~22 nucleotide non-coding RNAs that are processed from longer precursors by the RNases Drosha and Dicer. miRNA regulates protein expression posttranscriptionally through mRNA destabilization or translational silencing. A critical role for miRNA in Treg function was initially discovered when both Dicer and Drosha knockout (KO) mice were found to develop a fatal autoimmune disease phenotypically similar to Foxp3 KO mice.
Introduction
Regulatory T cells (Tregs), CD4+ 25+ Foxp3+ cells, are key mediators of the immune system, which function to suppress self-reactive lymphocytes (1) and limit immune responses to chronic pathogens and commensal bacteria (2). Treg development and suppressive function are tightly regulated. Genetic mutations that prevent Treg lineage specification lead to severe autoimmune diseases, whereas expanded Treg numbers lead to global immunosuppression and inhibit the clearance of tumors and opportunistic infections (3, 4). Allogeneic hematopoietic stem cell transplantation (HSCT) is a curative option for many hematological malignancies. Graft-versus-host disease (GVHD) (5) occurs in 40–70% of recipients, with skin, liver, and gut representing major GVHD target organs (6). Adoptive Treg transfer is effective at preventing autoimmunity, organ rejection, and GVHD in preclinical models (1, 5), and Treg therapy reduces disease in human clinical trials in GVHD and stabilized C-peptide levels for >2 years in several individuals with diabetes (7–10).
MicroRNAs (miRNAs) are a family of ~22 nucleotide non-coding RNAs that contain a short seed region complementary to mRNAs (typically the 3′ UTR) sequences. miRNA regulates gene expression posttranscriptionally through repression of protein production by mRNA destabilization or translational silencing. miRNAs are produced as longer primary transcripts (pri-miRNAs) by RNA polymerase II or III. pri-miRNAs are processed into mature miRNAs by the RNases Drosha and Dicer and are incorporated into the RNA-induced silencing complex. miRNAs instruct the differentiation, suppressive function, and stability of thymically derived Treg (tTreg) and Treg induced in the periphery (pTreg) or in vitro (iTreg) (11–14). Tregs exhibit a distinct miRNA profile compared with conventional T cells (15), and individual miRNA or miRNA clusters contribute to Treg biology through distinct mechanisms. The expression of the Treg defining transcription factor Foxp3 itself shapes the Treg miRNA profile. Moreover, miRNAs promote Foxp3 expression during iTreg generation. Finally, it is now widely recognized that expression of Foxp3 does not endow a terminal state of differentiation, and Tregs have a degree of plasticity, and miRNAs are required to integrate the external signals that drive this phenomena (16). This review focuses on how our knowledge of Treg pathways controlled by miRNA can be applied to improve the efficacy of Treg cellular therapy, with an emphasis on GVHD.
miRNA Effects on Thymic Treg (tTreg) Differentiation
In mouse models (17–20), the deletion of miRNAs by lineage-specific ablation of Dicer or Drosha in T cells or Treg specifically reduces the number of tTreg and pTreg precipitating fatal multiorgan inflammatory disease. Cell autonomous miRNAs are required for tTreg development in the thymus and Foxp3 induction by TGF-β during iTreg generation (19). These results have been confirmed (21) using Tie2Cre- and CD4Cre-mediated Dicer deletion mouse models where miRNA depletion in either hematopoietic/endothelial cells or thymocytes leads to a twofold to threefold decrease in Foxp3+ Treg frequencies.
miR-155, controlled by Dicer, is regulated by Foxp3 that binds the host gene bic promoter region (19, 22–24). Murine bic/miR-155 deficiency resulted in reduced thymic and splenic Treg as a consequence of impaired tTreg development but did not alter Treg function or in vivo homeostatic proliferation (Table 1) (25). However, as discussed below, miR-155 knockout (KO) Tregs have inferior Foxp3 expression and stability as well as fitness compared with wild type Tregs (24) but without defective in vitro TGF-β-mediated Foxp3 induction in CD4+ T cells (25). miR-155 KO mice have a reduced proportion and absolute number of Foxp3 cells, accompanied by diminished STAT5 signaling, downstream of the IL-2R complex, and higher suppressor of cytokine signaling 1 (SOCS1), a negative regulator of STAT5 signaling (24). Indeed, miR-155 effects are partially mediated by targeting SOCS1, a negative regulator of the IL-2 signaling pathway with a crucial role in Treg development (26). In a positive feedback loop, it has been suggested that during tTreg differentiation, induction and upregulation of Foxp3 expression reciprocally drive high expression of miRNA-155 (24). Similarly, the C-type lectin receptor, CD69, was found to control tTreg development, peripheral Treg homeostasis, and iTreg generation via STAT5 signaling effects; miR-155 induced CD69 (27). Similarly, in human, miR-155 (and miR-124a) was shown to repress the histone deacetylase, sirtuin-1, resulting in higher Foxp3 expression and iTreg generation (28). Strategies to augment miR-155 and miR-124a or conversely to downregulate SOCS1 should increase STAT5 signaling and Treg responses to IL-2 (Figure 1A).
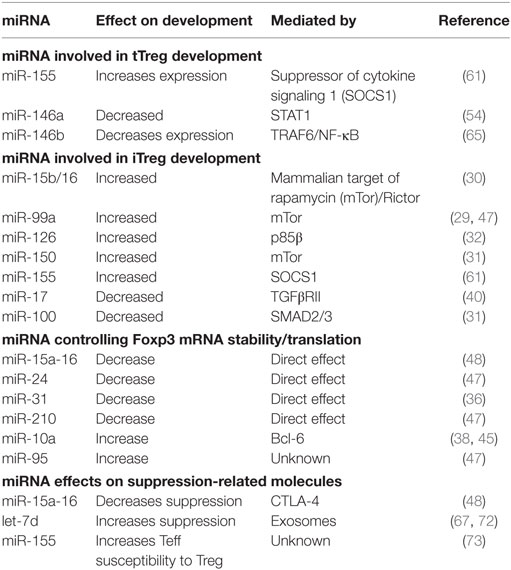
Table 1. Role of microRNA (miRNA) in regulatory T cell (Treg) induction, Foxp3 expression, and suppressive function.
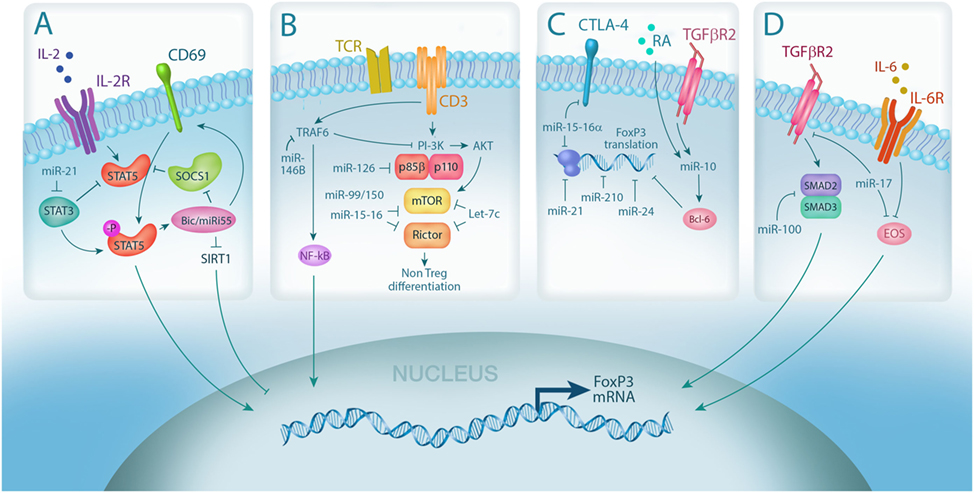
Figure 1. MicroRNA (miRNA) integrates multiple pathways in regulatory T cell (Treg). (A) IL-2 signaling is required for Treg differentiation and is enhanced by miRNA-mediated downregulation of the IL-2R signaling inhibitors, suppressor of cytokine signaling 1 (SOCS1) (miR-155) and STAT3 (miR-21); (B) miRNA expression controls Treg induction by negatively regulating TCR signal strength via downmodulation of PI3Kβ (miR-126) and mammalian target of rapamycin (mTor)/Rictor (miR-99/150, miR-15-16, and Let-7); (C) Foxp3 mRNA stability and translation can also be directly negatively regulated by miRNA via (miR-15-16a, miR-24, and miR-210) or positively regulated through indirect mechanisms (miR-10, via Bcl-6); (D) miRNA also controls Treg plasticity by regulating the expression of transcription factors required for Foxp3 transcription, including SMADs (miR-100) and EOS (miR-17).
miRNAs Effect on the Differentiation of In Vitro Induced Treg (iTreg)
Dicer also regulates iTreg generation and differentiation (Figure 1B) (19). In vitro TGF-β induces Foxp3 less efficiently in naïve Dicer-deficient CD4+ T-cells compared with wild type counterpart with similar results observed in mice harboring CD4-restricted Drosha or Dicer deficiencies (17). With Drosha or Dicer deficiency, the observed decreased Foxp3 expression after TGF-β stimulation could not be overcome by retinoic acid (RA) addition. These studies provided a rationale for an extensive miR screen, which lead to the identification of positive regulators of iTreg generation (29). miR-10b, miR-99a, miR-130a, miR-146b, miR-150, and miR-320 were amongst those found to drive Treg differentiation. With the multitude of miRNAs revealed by the screening, it was proposed that an miRNA network, rather than individual miRNAs, controls iTreg differentiation. For example, miR-99a cooperates with miR-150 to repress the expression of the mammalian target of rapamycin (mTor), a known inhibitor of iTreg differentiation (29). miR-150 antagomir exposure led to a reduced iTreg differentiation. Whereas miR-99a expression was upregulated by RA exposure and repressed mTor by binding to the 3′ UTR (29), miR-150 only repressed mTor in the presence of miR-99a (29). Similarly, miR-15b-16 (30) and miR-15a-16 function to reduce mTor signaling. miRNA-dependent mTor pathway downregulation supports human iTreg generation (31) as evidenced by inhibition of iTreg differentiation in CD4+ T-cells overexpressing miR-100. The specific miR-100 editing (C-to-U transversion) changes the miR-100 target from mTor to SMAD2, augmenting TGF-β signaling and iTreg differentiation. mTor and the PI3K–AKT pathways were identified as miR-126 targets in both mouse and human Treg (32). miR-126 inhibits PI3K p85β, responsible for AKT upregulation, and in so doing, augments Foxp3 and iTreg differentiation. Conversely, miRNA-126 silencing reduced iTreg generation and Foxp3 expression via enhanced p85β, pAKT, and mTOR, consistent with a critical effect of PI3K/Akt pathway on Treg Foxp3 expression (33, 34). As in tTreg generation, miR-155 facilitates iTreg generation (35) by downregulating SOCS1 and hence promoting JAK/STAT signaling (26).
Not all miRNAs have a positive effect on iTreg differentiation. miR-31 and the miR-17–miR-92 cluster function as negative regulators of iTreg differentiation (29). miR-31 represses human Treg Foxp3 expression (36) while the miR-17–miR-92 cluster represses iTreg formation (37–40). miR-17 directly targets TGF-β receptor II and the cAMP-responsive element binding protein 1, both implicated in bolstering Treg differentiation. It has been proposed that strong CD28 signals inhibit Foxp3 induction, which may be influenced by costimulatory signaling pathways that induce miR-17–miR-92 (41). Similarly, miR-23–miR-27–miR-24 cluster overexpression impairs TGF-β-mediated Treg induction (42). Together, these data identify miRNA agonist targets (miR-99a, miR-150, iR-15b-16, miR-100, miR-126, and miR-155) that can be exploited to increase iTreg generation. Moreover, miRNA (miR-31, miR-17–miR-92, and miR-23–miR-27–miR-24) antagomir treatment of T cells in vitro may be exploited to support iTreg generation, while in vivo treatment may foster pTreg generation.
miRNA Effects on Foxp3 Expression
miR-10a, which is induced in Treg following RA and TGF-β exposure, is one of few miRNAs exclusively expressed in tTreg (38, 43, 44). Although not required for Foxp3 expression, miR-10a contributes to Treg stability by targeting the transcriptional repressor Bcl-6 resulting in high and sustained Foxp3 (Figure 1C) (38, 43, 45). In this regard, miR-10a antagomir-treated Tregs exhibit decreased Foxp3 protein expression (46). In addition, miR-95 is also highly expressed in human Treg and, through an unknown mechanism, enhances Foxp3 mRNA and protein expression (47).
By contrast, miRNA can also function as negative regulators of Foxp3 expression (47, 48). Foxp3 is a direct target of miR-15a-16, which is expressed at low levels in human Treg (40). Forced overexpression of this miRNA in Treg markedly reduced Foxp3 and CTLA-4 expression, concordant with reduced suppressor function, whereas miR-15a-16 deletion in conventional T-cells upregulated Foxp3 and CTLA-4 expression (40). Similarly, forced expression of either miR-24 or miR-210 in Treg resulted in a twofold decrease in Foxp3 expression demonstrating Foxp3 as a direct target of these miRNAs (47). Thus, to increase Foxp3 expression and promote Treg stability and suppressor function, approaches can be undertaken to increase miR-10a or miR-95 and/or decrease miR-15a-17, miR-24, or miR-210.
miRNA Regulation of Treg Stability
As briefly discussed earlier, Tregs exhibit some degree of plasticity, especially with the Th17 subset of CD4+ T cells whose differentiation, like inducible Treg, is driven by TGF-β (49). miRNAs also have been shown to be critical in the maintenance of Treg stability that is required for the preservation of suppressor function during inflammation that can occur as a result of GVHD and autoimmune disease. Thus, the following miRNAs present potential new targets for the regulation of Treg stability in vivo.
miR-21, a key regulator of Treg stability (15, 36, 50), is more highly expressed in Treg compared with conventional T-cells and regulates Foxp3 expression and Treg proliferation (36, 50). A significant reduction in miR-21 and Foxp3 mRNA was noted in tTreg from rheumatoid arthritis patients compared with healthy controls (50) and was accompanied by increased STAT3, decreased STAT5 protein expression, and skewing toward Th17 cells. These data led to the hypothesis that miR-21 is part of a negative feedback loop dysregulated during disease that contributes to Th17 and Treg imbalance in rheumatoid arthritis patients (50). Evidence supports that miR-21 regulates Foxp3 expression and Treg homeostasis by modulating STAT3 and STAT5 (36, 50). Peptide nucleic acid (PNA) inhibition was used to block miR-21 expression to delineate its effects on human Treg functions (15). Although PNA-inhibited Treg failed to proliferate under anti-CD3 mAb stimulation conditions (15), suppression was unchanged, supporting the contention that miR-21 expression promotes Treg stability and homeostasis.
Foxp3 regulates gene expression by recruiting corepressors and coactivators, and miRNA directly regulates Treg function by controlling the expression of these cofactors. For example, IL-6, which is an established mediator of acute GVHD in mice and patients (51) and required (with TGF-β) to induce Th17 cells, induces the expression of miR-17. miR-17 targets Eos, a transcription factor that cooperates with Foxp3 to mediate suppressor gene expression (Figure 1D) (52). Decreased Eos expression causes derepression of effector cytokine genes and exuberant cytokine responsiveness. In line with this, transgenic miR-17 overexpression exacerbates pathology in a murine colitis model (52). Conversely, miR-146a targets STAT1 expression and minimizes IFNγ/STAT1-mediated loss of Treg-suppressive function. This plasticity is mediated by T-bet, the Th1-specifying transcription factor, which promotes Treg CXCR3 expression leading to their accumulation in type 1 inflammatory sites (53, 54). Strategically generating Tregs that have low miR-17, miR-16a/16, or miR-142-3p or high miR-146a or let-7d expression may represent an approach to increase Treg suppressor function.
Four other miRNA, miR-7, miR-18a, miR-34a, and miR-155, have been shown to contribute to the stabilization of Treg suppressor function (47, 55, 56). It is suggested that the activity of these miRNA is mediated by Treg expression of SATB1, a genome organizer that regulates chromatin structure and gene expression (57). In mature Tregs, Foxp3 directly suppresses the Satb1 locus. SATB1 is required for Treg suppressive function and to prevent the acquisition of helper T cell characteristics. Under non-activating conditions, Treg SATB1 is expressed at low levels; its forced expression results in complete loss of suppression to a level comparable with Foxp3 deficiency (55). In addition, T cell-specific deficiency of Satb1 impaired Treg-super-enhancer activation and Treg signature gene expression in thymic Treg precursors, resulting in severe autoimmunity due to Treg deficiency (58). Foxp3 repression of SATB1 is achieved by inducing miRNA (miR-155, miR-21, miR-7, miR-34a, and miR-18a) binding to the Satb1 3′ untranslated region, indirectly suppressing SATB1. Thus, increased expression of these miRs may be particularly useful in inflammatory diseases such as GVHD by promoting Foxp3 expression and function.
Effect of miRNA on Treg Fitness
MicroRNAs are instrumental in maintaining Treg fitness and survival. As mentioned earlier, miR-155 has proven functionally relevant to Treg fitness (a measure of survival/death and expansion) (59) with miR-155 KO mice having markedly reduced Treg number and impaired Treg proliferation (24, 60) associated with impaired STAT5 activation (24, 61) and increased SOCS1 (61). Conversely, miR-155 upregulation increases IL-2 sensitivity, promoting Treg fitness and proliferation (24, 59). miR-17–miR-92 also assists in maintaining Treg fitness (62). Treg-specific miR-17–miR-92 deletion increased Treg apoptosis and reduced proliferation, causing loss of Foxp3 expressing Treg in aged mice (63). By contrast, elevated miR-17–miR-92 in murine lymphocytes increased proliferation and reduced cell death (64), resulting in favored Treg accumulation in lymph nodes and non-lymphoid target tissues (63). By contrast, miR-146b impedes human Treg homeostasis (65), offering a potential therapeutic target that could be exploited to augment Treg fitness and survival. miR-146b antagomir treatment of Treg enhanced TRAF6 and the TRAF6–NF-κB–Foxp3 axis, resulting in improved Treg survival and proliferation (65). Thus, superior GVHD control may be acquired by increasing Treg fitness by upregulating miR-155 or reducing miR-17 or miR-146b expression.
miRNA Effects on Genes That Contribute to Treg Suppressive Function
MicroRNA also regulates genes directly involved in Treg-mediated suppression. miR-15a/16 inhibits CTLA-4 expression (Figure 1C), precluding optimal Treg-mediated inhibition of dendritic cell (DC) maturation (48, 66). miR-142-3p inhibits expression of adenylyl cyclase 9, responsible for generating the inhibitory second messenger, cAMP (48, 66). Treg inhibition of Th1 proliferation and cytokine secretion in vivo has been demonstrated after transfer of exosomes containing suppressive miRNA, including let-7d, shown to be required for Treg-mediated amelioration of murine colitis (67).
Concluding Remarks
The potential of miRNA therapeutics in human Treg cellular therapy was demonstrated by ex vivo inhibition of miR-146b that enhanced in vitro tTreg function and, upon adoptive tTreg transfer, superior in vivo xenogeneic GVHD lethality compared with scrambled miR control treatment (65). Clinical GVHD trials registered in http://clinicaltrials.gov employ in vitro generated iTreg and IL-10, TGFβ-producing CD4+ Foxp3− Treg type 1 (Tr1) cells (NCT01634217: MacMillan; NCT03198234: Roncarolo). Stability of Foxp3 expression and suppressive function are of paramount concern for Treg therapies in the intense GVHD inflammatory environment and would be amenable to miR manipulation. Similar Treg therapies are being used to treat diabetes (7) and could be used to restore normal expression of miR-342, miR-191, and miR-510 in Treg from diabetes patients (68). In addition, while outside the purview of this review, it should be noted that manipulating miRNA expression in other cell types (e.g., DCs) can have a significant impact on Treg induction and stability.
Regulatory T cell therapy, especially for GVHD, may benefit from miRNA therapeutics. First, because Tregs can be substantially expanded in vitro, permitting a higher Treg:Teffector ratio needed for optimal GVHD suppression and miRNA increases or knockdown can be tightly controlled. By treating just the cell of interest, the miR reagents needed are minimized. In contrast to autoimmunity and organ transplantation, acute GVHD typically is high risk in 1–3 months post-allo-HSCT until central and peripheral tolerance mechanisms become operative. Although clinical development of miRNA-based therapeutics has been slow, progress is being made. Locked nucleic acid-based miRNA and antagomirs prolong half-life, miRNA encapsulated in anionic nanoparticles greatly increases uptake, and antibodies can be incorporated into the nanoparticles to aid targeting, providing a new avenue for maximizing the efficacy and safety of Treg infusional therapies for GVHD (69–71).
Author Contributions
KH, ML, and JN drafted the manuscript. KM prepared illustration. All the authors critically revised the manuscript for intellectual content and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by grants from the Children’s Cancer Research Fund and National Institutes of Health, National Heart, Lung and Blood Institute grant R01 HL114512-01 (KH), National Cancer Institute grants P01 CA142106, P01 CA047741, and P01 CA065493, National Institutes of Health, National Institute of Allergy and Infectious Diseases grants P01 AI056299, R01 AI11879, and R37 AI344495, Leukemia & Lymphoma Society Translational Research grant 6458-15 and 6462-15 (BB), and the Australian National Health and Medical Research Council (NH&MRC) APP1031728 (KM).
References
1. Bluestone JA, Tang Q, Sedwick CE. T regulatory cells in autoimmune diabetes: past challenges, future prospects. J Clin Immunol (2008) 28(6):677–84. doi:10.1007/s10875-008-9242-z
2. Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity (2009) 30(5):626–35. doi:10.1016/j.immuni.2009.05.002
3. Richer MJ, Straka N, Fang D, Shanina I, Horwitz MS. Regulatory T-cells protect from type 1 diabetes after induction by coxsackievirus infection in the context of transforming growth factor-beta. Diabetes (2008) 57(5):1302–11. doi:10.2337/db07-1460
4. Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol (2006) 6(4):295–307. doi:10.1038/nri1806
5. Riley JL, June CH, Blazar BR. Human T regulatory cell therapy: take a billion or so and call me in the morning. Immunity (2009) 30(5):656–65. doi:10.1016/j.immuni.2009.04.006
6. Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol (2012) 12(6):443–58. doi:10.1038/nri3212
7. Bluestone JA, Buckner JH, Fitch M, Gitelman SE, Gupta S, Hellerstein MK, et al. Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci Transl Med (2015) 7(315):315ra189. doi:10.1126/scitranslmed.aad4134
8. Brunstein CG, Miller JS, Cao Q, McKenna DH, Hippen KL, Curtsinger J, et al. Infusion of ex vivo expanded T regulatory cells in adults transplanted with umbilical cord blood: safety profile and detection kinetics. Blood (2011) 117(3):1061–70. doi:10.1182/blood-2010-07-293795
9. Brunstein CG, Miller JS, McKenna DH, Hippen KL, DeFor TE, Sumstad D, et al. Umbilical cord blood-derived T regulatory cells to prevent GVHD: kinetics, toxicity profile, and clinical effect. Blood (2016) 127(8):1044–51. doi:10.1182/blood-2015-06-653667
10. Di Ianni M, Falzetti F, Carotti A, Terenzi A, Castellino F, Bonifacio E, et al. Tregs prevent GVHD and promote immune reconstitution in HLA-haploidentical transplantation. Blood (2011) 117(14):3921–8. doi:10.1182/blood-2010-10-311894
11. Buoli Comani G, Panceri R, Dinelli M, Biondi A, Mancuso C, Meneveri R, et al. miRNA-regulated gene expression differs in celiac disease patients according to the age of presentation. Genes Nutr (2015) 10(5):482. doi:10.1007/s12263-015-0482-2
12. Jasinski-Bergner S, Stoehr C, Bukur J, Massa C, Braun J, Huttelmaier S, et al. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology (2015) 4(6):e1008805. doi:10.1080/2162402X.2015.1008805
13. Tang X, Tang R, Xu Y, Wang Q, Hou Y, Shen S, et al. MicroRNA networks in regulatory T cells. J Physiol Biochem (2014) 70(3):869–75. doi:10.1007/s13105-014-0348-x
14. de Candia P, Torri A, Pagani M, Abrignani S. Serum microRNAs as biomarkers of human lymphocyte activation in health and disease. Front Immunol (2014) 5:43. doi:10.3389/fimmu.2014.00043
15. Bhairavabhotla R, Kim YC, Glass DD, Escobar TM, Patel MC, Zahr R, et al. Transcriptome profiling of human FoxP3+ regulatory T cells. Hum Immunol (2016) 77(2):201–13. doi:10.1016/j.humimm.2015.12.004
16. Lu L, Barbi J, Pan F. The regulation of immune tolerance by FOXP3. Nat Rev Immunol (2017) 17(11):703–17. doi:10.1038/nri.2017.75
17. Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med (2008) 205(9):2005–17. doi:10.1084/jem.20081219
18. Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med (2008) 205(9):1993–2004. doi:10.1084/jem.20081062
19. Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, et al. A role for Dicer in immune regulation. J Exp Med (2006) 203(11):2519–27. doi:10.1084/jem.20061692
20. Zhou L, Seo KH, Wong HK, Mi QS. MicroRNAs and immune regulatory T cells. Int Immunopharmacol (2009) 9(5):524–7. doi:10.1016/j.intimp.2009.01.017
21. Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG, et al. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci U S A (2009) 106(25):10266–71. doi:10.1073/pnas.0811119106
22. Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature (2007) 445(7130):931–5. doi:10.1038/nature05478
23. Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature (2007) 445(7130):936–40. doi:10.1038/nature05563
24. Lu L-F, Thai T-H, Calado DP, Chaudhry A, Kubo M, Tanaka K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity (2009) 30(1):80–91. doi:10.1016/j.immuni.2008.11.010
25. Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol (2009) 182(5):2578–82. doi:10.4049/jimmunol.0803162
26. Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X, et al. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One (2012) 7(10):e46082. doi:10.1371/journal.pone.0046082
27. Sanchez-Diaz R, Blanco-Dominguez R, Lasarte S, Tsilingiri K, Martin-Gayo E, Linillos-Pradillo B, et al. Thymus-derived regulatory T cell development is regulated by C-type lectin-mediated BIC/microRNA 155 expression. Mol Cell Biol (2017) 37(9):1–18. doi:10.1128/MCB.00341-16
28. Heyn J, Luchting B, Hinske LC, Hübner M, Azad SC, Kreth S. miR-124a and miR-155 enhance differentiation of regulatory T cells in patients with neuropathic pain. J Neuroinflammation (2016) 13(1):248. doi:10.1186/s12974-016-0712-6
29. Warth SC, Hoefig KP, Hiekel A, Schallenberg S, Jovanovic K, Klein L, et al. Induced miR-99a expression represses Mtor cooperatively with miR-150 to promote regulatory T-cell differentiation. EMBO J (2015) 34(9):1195–213. doi:10.15252/embj.201489589
30. Singh Y, Garden OA, Lang F, Cobb BS. MicroRNA-15b/16 enhances the induction of regulatory T cells by regulating the expression of Rictor and mTOR. J Immunol (2015) 195(12):5667–77. doi:10.4049/jimmunol.1401875
31. Negi V, Paul D, Das S, Bajpai P, Singh S, Mukhopadhyay A, et al. Altered expression and editing of miRNA-100 regulates iTreg differentiation. Nucleic Acids Res (2015) 43(16):8057–65. doi:10.1093/nar/gkv752
32. Qin A, Wen Z, Zhou Y, Li Y, Li Y, Luo J, et al. MicroRNA-126 regulates the induction and function of CD4(+) Foxp3(+) regulatory T cells through PI3K/AKT pathway. J Cell Mol Med (2013) 17(2):252–64. doi:10.1111/jcmm.12003
33. Merkenschlager M, von Boehmer H. PI3 kinase signalling blocks Foxp3 expression by sequestering Foxo factors. J Exp Med (2010) 207(7):1347–50. doi:10.1084/jem.20101156
34. Crellin NK, Garcia RV, Levings MK. Altered activation of AKT is required for the suppressive function of human CD4+CD25+ T regulatory cells. Blood (2007) 109(5):2014–22. doi:10.1182/blood-2006-07-035279
35. Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol (2011) 187(5):2213–21. doi:10.4049/jimmunol.1003952
36. Rouas R, Fayyad-Kazan H, El Zein N, Lewalle P, Rothe F, Simion A, et al. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol (2009) 39(6):1608–18. doi:10.1002/eji.200838509
37. Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood (2011) 118(20):5487–97. doi:10.1182/blood-2011-05-355644
38. Baumjohann D, Ansel KM. MicroRNA regulation of T helper cell differentiation and plasticity. Nat Rev Immunol (2013) 13(9):666. doi:10.1038/nri3494
39. Kang SG, Liu WH, Lu P, Jin HY, Lim HW, Shepherd J, et al. MicroRNAs of the miR-17 approximately 92 family are critical regulators of T(FH) differentiation. Nat Immunol (2013) 14(8):849–57. doi:10.1038/ni.2648
40. Liu SQ, Jiang S, Li C, Zhang B, Li QJ. miR-17-92 cluster targets phosphatase and tensin homology and Ikaros family zinc finger 4 to promote TH17-mediated inflammation. J Biol Chem (2014) 289(18):12446–56. doi:10.1074/jbc.M114.550723
41. Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med (2007) 204(8):1765–74. doi:10.1084/jem.20070719
42. Cho S, Wu CJ, Yasuda T, Cruz LO, Khan AA, Lin LL, et al. miR-23 approximately 27 approximately 24 clusters control effector T cell differentiation and function. J Exp Med (2016) 213(2):235–49. doi:10.1084/jem.20150990
43. Gao Y, Lin F, Su J, Gao Z, Li Y, Yang J, et al. Molecular mechanisms underlying the regulation and functional plasticity of FOXP3+ regulatory T cells. Genes Immun (2012) 13(1):1. doi:10.1038/gene.2011.77
44. Monticelli S. MicroRNAs in T helper cell differentiation and plasticity. Semin Immunol (2013) 25(4):291–8. doi:10.1016/j.smim.2013.10.015
45. Kelada S, Sethupathy P, Okoye IS, Kistasis E, Czieso S, White SD, et al. miR-182 and miR-10a are key regulators of Treg specialisation and stability during schistosome and Leishmania-associated inflammation. PLoS Pathog (2013) 9(6):e1003451. doi:10.1371/journal.ppat.1003451
46. Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G, et al. MicroRNA 10a marks regulatory T cells. PLoS One (2012) 7(5):e36684. doi:10.1371/journal.pone.0036684
47. Fayyad-Kazan H, Rouas R, Fayyad-Kazan M, Badran R, El Zein N, Lewalle P, et al. MicroRNA profile of circulating CD4-positive regulatory T cells in human adults and impact of differentially expressed microRNAs on expression of two genes essential to their function. J Biol Chem (2012) 287(13):9910–22. doi:10.1074/jbc.M111.337154
48. Liu X, Robinson S, Setoyama T, Tung S, D’abundo L, Shah M, et al. FOXP3 is a direct target of miR15a/16 in umbilical cord blood regulatory T cells. Bone Marrow Transplant (2014) 49(6):793. doi:10.1038/bmt.2014.57
49. Muranski P, Restifo NP. Essentials of Th17 cell commitment and plasticity. Blood (2013) 121(13):2402–14. doi:10.1182/blood-2012-09-378653
50. Dong L, Wang X, Tan J, Li H, Qian W, Chen J, et al. Decreased expression of microRNA-21 correlates with the imbalance of Th17 and Treg cells in patients with rheumatoid arthritis. J Cell Mol Med (2014) 18(11):2213–24. doi:10.1111/jcmm.12353
51. Kennedy GA, Varelias A, Vuckovic S, Le Texier L, Gartlan KH, Zhang P, et al. Addition of interleukin-6 inhibition with tocilizumab to standard graft-versus-host disease prophylaxis after allogeneic stem-cell transplantation: a phase 1/2 trial. Lancet Oncol (2014) 15(13):1451–9. doi:10.1016/S1470-2045(14)71017-4
52. Yang H-Y, Barbi J, Wu C-Y, Zheng Y, Vignali PD, Wu X, et al. MicroRNA-17 modulates regulatory T cell function by targeting Co-regulators of the Foxp3 transcription factor. Immunity (2016) 45(1):83–93. doi:10.1016/j.immuni.2016.06.022
53. Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol (2009) 10(6):595–602. doi:10.1038/ni.1731
54. Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell (2010) 142(6):914–29. doi:10.1016/j.cell.2010.08.012
55. Dooley J, Linterman MA, Liston A. MicroRNA regulation of T-cell development. Immunol Rev (2013) 253(1):53–64. doi:10.1111/imr.12049
56. Lu L-F, Gasteiger G, Yu I-S, Chaudhry A, Hsin J-P, Lu Y, et al. A single miRNA-mRNA interaction affects the immune response in a context-and cell-type-specific manner. Immunity (2015) 43(1):52–64. doi:10.1016/j.immuni.2015.04.022
57. Beyer M, Thabet Y, Muller RU, Sadlon T, Classen S, Lahl K, et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol (2011) 12(9):898–907. doi:10.1038/ni.2084
58. Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol (2017) 18(2):173–83. doi:10.1038/ni.3646
59. Belver L, Papavasiliou FN, Ramiro AR. MicroRNA control of lymphocyte differentiation and function. Curr Opin Immunol (2011) 23(3):368–73. doi:10.1016/j.coi.2011.02.001
60. Shan J, Feng L, Luo L, Wu W, Li C, Li S, et al. MicroRNAs: potential biomarker in organ transplantation. Transpl Immunol (2011) 24(4):210–5. doi:10.1016/j.trim.2011.03.004
61. Zhang M, Zhang Q, Liu F, Yin L, Yu B, Wu J. MicroRNA-155 may affect allograft survival by regulating the expression of suppressor of cytokine signaling 1. Med Hypotheses (2011) 77(4):682–4. doi:10.1016/j.mehy.2011.07.016
62. Skinner JP, Keown AA, Chong MM. The miR-17~92a cluster of microRNAs is required for the fitness of Foxp3+ regulatory T cells. PLoS One (2014) 9(2):e88997. doi:10.1371/journal.pone.0088997
63. de Kouchkovsky D, Esensten JH, Rosenthal WL, Morar MM, Bluestone JA, Jeker LT. MicroRNA-17–92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. J Immunol (2013) 191(4):1594–605. doi:10.4049/jimmunol.1203567
64. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with elevated miR-17−92 expression in lymphocytes. Nat Immunol (2008) 9(4):405. doi:10.1038/ni1575
65. Lu Y, Hippen KL, Lemire AL, Gu J, Wang W, Ni X, et al. miR-146b antagomir-treated human Tregs acquire increased GVHD inhibitory potency. Blood (2016) 128(10):1424–35. doi:10.1182/blood-2016-05-714535
66. Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX, et al. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep (2009) 10(2):180–5. doi:10.1038/embor.2008.224
67. Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T, et al. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity (2014) 41(1):89–103. doi:10.1016/j.immuni.2014.05.019
68. Hezova R, Slaby O, Faltejskova P, Mikulkova Z, Buresova I, Raja KR, et al. MicroRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol (2010) 260(2):70–4. doi:10.1016/j.cellimm.2009.10.012
69. Huang X, Schwind S, Yu B, Santhanam R, Wang H, Hoellerbauer P, et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clin Cancer Res (2013) 19(9):2355–67. doi:10.1158/1078-0432.CCR-12-3191
70. Luck ME, Muljo SA, Collins CB. Prospects for therapeutic targeting of microRNAs in human immunological diseases. J Immunol (2015) 194(11):5047–52. doi:10.4049/jimmunol.1403146
71. Ranganathan P, Heaphy CE, Costinean S, Stauffer N, Na C, Hamadani M, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood (2012) 119(20):4786–97. doi:10.1182/blood-2011-10-387522
72. Guan H, Fan D, Mrelashvili D, Hao H, Singh NP, Singh UP, et al. MicroRNA let-7e is associated with the pathogenesis of experimental autoimmune encephalomyelitis. Eur J Immunol (2013) 43(1):104–14. doi:10.1002/eji.201242702
Keywords: regulatory T cell, tTreg, iTreg, microRNA, graft-versus-host disease
Citation: Hippen KL, Loschi M, Nicholls J, MacDonald KPA and Blazar BR (2018) Effects of MicroRNA on Regulatory T Cells and Implications for Adoptive Cellular Therapy to Ameliorate Graft-versus-Host Disease. Front. Immunol. 9:57. doi: 10.3389/fimmu.2018.00057
Received: 31 October 2017; Accepted: 09 January 2018;
Published: 31 January 2018
Edited by:
Pavan Reddy, University of Michigan, United StatesReviewed by:
Sergio Querol, Banc de Sang i Teixits, SpainPhilippe Saas, INSERM UMR1098 Interactions Hôte-Greffon-Tumeur & Ingénierie Cellulaire et Génique, France
Copyright: © 2018 Hippen, Loschi, Nicholls, MacDonald and Blazar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keli L. Hippen, aGlwcGUwMDJAdW1uLmVkdQ==;
Bruce R. Blazar, YmxhemEwMDFAdW1uLmVkdQ==
 Keli L. Hippen
Keli L. Hippen Michael Loschi
Michael Loschi Jemma Nicholls
Jemma Nicholls Kelli P. A. MacDonald
Kelli P. A. MacDonald Bruce R. Blazar
Bruce R. Blazar