
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 22 December 2017
Sec. Cancer Immunity and Immunotherapy
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.01858
This article is part of the Research Topic NKT Cells in Cancer Immunotherapy View all 18 articles
 Amrendra Kumar1,2
Amrendra Kumar1,2 Naveenchandra Suryadevara2
Naveenchandra Suryadevara2 Timothy M. Hill2,3
Timothy M. Hill2,3 Jelena S. Bezbradica4
Jelena S. Bezbradica4 Luc Van Kaer2
Luc Van Kaer2 Sebastian Joyce1,2*
Sebastian Joyce1,2*
Type I natural killer T (NKT) cells are innate-like T lymphocytes that recognize glycolipid antigens presented by the MHC class I-like protein CD1d. Agonistic activation of NKT cells leads to rapid pro-inflammatory and immune modulatory cytokine and chemokine responses. This property of NKT cells, in conjunction with their interactions with antigen-presenting cells, controls downstream innate and adaptive immune responses against cancers and infectious diseases, as well as in several inflammatory disorders. NKT cell properties are acquired during development in the thymus and by interactions with the host microbial consortium in the gut, the nature of which can be influenced by NKT cells. This latter property, together with the role of the host microbiota in cancer therapy, necessitates a new perspective. Hence, this review provides an initial approach to understanding NKT cells from an ecological evolutionary developmental biology (eco-evo-devo) perspective.
The evolutionary appearance of the vertebrate immune system equipped complex organisms with the ability to resist invasion by pathogenic microbes and to sense and respond to a loss of tissue integrity due to infection, aberrant cell growth, or mechanical injury. As organisms became increasingly more complex and lived beyond their fecund years, a finer ability to discriminate self from non-self was required (1, 2). Thus, the maintenance of homeostasis in such organisms requires the concerted action of multiple cell types that stand poised to respond to a hostile world filled with a seemingly endless array of infectious agents, toxic chemicals, and biologics. The first responders in this elaborate defensive network have historically been classified as members of the more archaic, multi-modular innate immune system. Should the innate defenses prove insufficient, the evolutionarily younger, adaptive immune system—consisting of B and T lymphocytes—is recruited to restore the homeostatic state. The quick-acting cells of the innate immune system senses an altered homeostatic state with pattern recognition receptors to detect conserved molecular structures shared by many pathogens alike (3, 4). By contrast, the slow-responding, adaptive immune system senses alterations in homeostasis by using diverse, clonally distributed B cell receptors (BCR and their secreted counterparts, antibodies), and T cell receptors (TCRs), respectively.
Bridging the gap between innate and adaptive immune responses are the innate-like B and T lymphocytes. These are a group of cells that express a relatively restricted repertoire of receptors generated through somatic recombination, yet unlike conventional T and B cells, exhibit innate-like recognition principles and functional responses (5). Innate-like lymphocytes include both T cells (γδ T cells, natural killer T cells, mucosal-associated invariant T lymphocytes, and CD8αα-expressing intestinal intraepithelial lymphocytes) and B cells (B-1 B cells and marginal zone B cells). The evolutionary appearance of this group of immune cells, including natural killer T cells (NKT cells) endowed upon vertebrates the capacity to initiate and amplify both the innate and adaptive immune responses. By virtue of their immunoregualtory functions, innate-like lymphocytes can fine-tune the nature and magnitude of these immune responses (6). Although each immune module plays a specific role, it is the controlled integration of multiple modules that results in an effective inflammatory response that is essential in maintaining a stable milieu intérieur (7).
NKT cells—originally defined as cells that co-express key natural killer (NK) cell surface markers and a conserved αβ TCR repertoire—are thymus-derived, innate-like T lymphocytes. The functions of NKT cells are controlled by self and non-self-lipid agonists presented by CD1d molecules (8). The majority of NKT cells (type I, invariant NKT) express an invariant TCR α-chain (Vα14Jα18 in mice; Vα24Jα18 in humans). The invariant α-chain pairs predominantly with Vβ8.2, Vβ7, or Vβ2 in mouse NKT cells, or Vβ11 almost exclusively in human NKT cells. A small NKT cell population—referred to as type II NKT cells—expresses a more diverse TCR repertoire and recognizes a distinct group of lipid antigens; these, however, are the focus of other reviews (9–14). The recognition of lipid agonists rapidly activates NKT cells, which respond just as quickly by secreting a variety of cytokines and chemokines, and upregulate costimulatory molecules. By acting promptly, NKT cells alert and regulate the effector functions of myeloid and lymphoid cells. In so doing, NKT cells play a critical role in controlling microbial and tumor immunity as well as autoimmune and inflammatory diseases (6, 15–17).
The functions of NKT cells are controlled by CD1d molecules. CD1d molecules bind to and present a variety of lipid ligands to reactive T cells (18). Numerous in vitro and in vivo studies using the synthetic lipid α-galactosylceramide (αGalCer, KRN7000) and its analogs (Table 1 and references therein) has led to our current understanding of NKT cell biology. αGalCer is a natural product isolated from the marine sponge, A. mauritianus. The gut bacterium, Bacteroides fragilis, and the fungus, Aspergillus fumigatus, also biosynthesise αGalCers and/or related compounds (Table 1 and references therein). Hence, αGalCer and related compounds may be more prevalent in nature than previously thought and the NKT cell biology so gleaned may be highly relevant.
αGalCer is a potent NKT cell agonist, which when presented by CD1d molecules directly activates NKT cells in a TCR-dependent manner without need for additional signals. This activation mechanism is considered TCR-dominated mode of NKT cell activation (Figure 1).
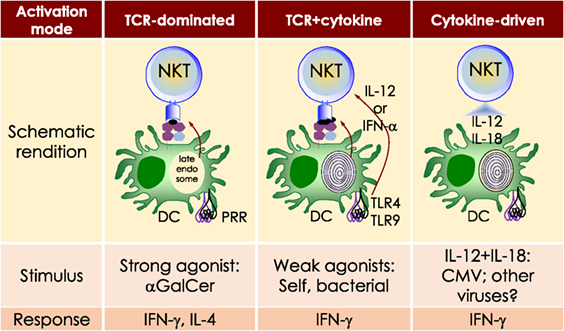
Figure 1. Three distinct strategies activate mouse NKT cells. Potent NKT cell agonists—such as αGalCer—directly activate NKT cells without the need for a second signal, in a T cell receptor (TCR)-signaling dominated fashion (left panel). Alternatively, microbes containing toll-like receptor (TLR) ligands such as LPS activate NKT cells by inducing IL-12 production by DCs, which amplifies weak responses elicited upon the recognition of CD1d bound with self-glycolipids by the NKTCR. Several endogenous lipid agonists have been identified and characterized (see Table 1). Some microbes such as Sphingomonas capsulata, which are α-Proteobacteria, synthesize α-anomeric glycolipids for their cell walls. These glycolipids, when presented by CD1d, weakly activate NKT cells directly. In the presence of a second signal—generally a pro-inflammatory cytokine such as IL-12—such weak agonists strongly activate NKT cells (middle panel). Intriguingly, NKT cells can be activated solely by cytokines—mainly IL-12— in a TCR-independent manner (right panel). This diagram rendering the different strategies to NKT cells is an adaptation of past reviews (8, 44) and is based on works cited in the text.
Sphingomonas spp. biosynthesises an αGalCer-related compound, α-galacturonosylceramide (αGalACer). Other weak NKT cell agonists include microbial glycosphingolipid [GSL; e.g., αGalCer-related asparamide B (A fumigatus)], diacylglycerolipids [e.g., α-galactosyl- (Borrelia burgdorferi—the agent of Lyme disease) and α-glucosyl-diacylglycerol (Streptococcus pneumoniae)] and cholesteryl-α-glycoside [e.g., cholesteryl-6-O-acyl α-glucoside (Helicobacter pylori)] (Table 1 and references therein). Being a weak agonist, NKT cell activation by these microbial glycolipids requires a second activation signal from inflammatory cytokines. Such inflammatory cytokines result from dendritic cells (DCs) that are activated through their pattern recognition receptors (45–47). This activation mechanism is considered TCR- and cytokine-mediated mode of NKT cell activation (Figure 1)—a feature that is important for NKT cell activation by weak microbial and self-lipid agonists.
NKT cells react to CD1d molecules presenting self-lipids on host APCs in the presence of a second signal (6, 48). The inability to activate NKT cell hybridomas by using artificial APCs lacking βGlcCer synthase (49) and impaired NKT cell development in mice lacking βGlcCer synthase in their thymocytes (50), suggested that a cellular, βGlcCer-derived GSL is an endogenous mouse NKT cell agonist (49, 50). Several microbes—bacteria (e.g., Staphylococcus aureus, Salmonella typhimurium, Listeria monocytogenes, etc.), fungi (e.g., A. fumigatus) and viruses—activate NKT cells but do not biosynthesise NKT cell agonists. Such microbes induce the biosynthesis and/or presentation of self-lipids, which are thought to be mammalian αGalCer and perhaps iGb3 (19, 28, 35). As self-lipids are weak NKT cell agonists, NKT cell activation is bolstered by IL-12 secreted by DCs activated through dectin-1 DCs (31, 47) or toll-like receptor (TLR)-4 (45, 46). This activation mechanism is a variation on the TCR- and cytokine-mediated mode of NKT cell activation and a feature that is important for NKT cell activation by microbes that do not themselves biosynthesise an NKT cell agonist.
Type I interferon (IFN)—produced by DCs activated by the TLR9 ligand CpG—can serve as a second signal for NKT cell activation in conjunction with the presentation of sialylated cellular glycolipids by CD1d molecules (51). This finding is significant because almost all viral infections induce type I IFN response. Even though viruses do not biosynthesise NKT cell agonists, or any lipid for that matter, viral infections also activate NKT cells (52–62). Perchance, in such a circumstance, NKT cell activation occurs via the recognition of a self-lipid(s) presented by CD1d in the presence of inflammatory signals relayed by type I IFNs.
NKT cells are activated by the combined actions of IL-12 and IL-18. Under such conditions, NKT cell activation does not require the recognition of a CD1d-restricted agonist (63–65). This latter mechanism is considered cytokine-driven NKT cell activation (Figure 1). This mechanism is important for immunity to cytomegalovirus (65). Summarily, these multiple modes of activation suggest that NKT cells have evolved many different mechanisms to sense an altered homeostatic state caused by microbial infections. How activated NKT cells steer downstream innate and adaptive immune responses is described below.
NKT cells form immune synapses upon recognition of lipid agonists presented by CD1d molecules displayed on APCs or planar membranes. The kinetics NKTCR/ligand interactions determine the functional outcome (66). Positive cooperative engagement of CD1d-lipid agonistic complexes by the NKTCR allows NKT cells to recognize subtle changes in cellular lipid content and to actuate a response (67). Upon activation, NKT cells rapidly polarize IFN-γ and lytic granules to the immune synapse to transmit an effector response (66, 68, 69). The synaptic transmission of effector molecules controls downstream innate and adaptive immune responses as described below.
Akin to the cells of the innate immune system (e.g., neutrophils, Mϕ, DCs, and NK cells), NKT cells respond within the first several hours of agonist recognition and secrete copious amounts of effector cytokines and chemokines (Figure 2). The nature of the activating NKT cell agonist controls the nature of the cytokine response (see Table 1). For example, the synthetic agonist αGalCer, within 30–90 min, elicits a wide variety of cytokines (Figure 2). Nonetheless, αGalCer variants containing different lipid chain length or unsaturation typically induce an IL-4 cytokine response (24, 25). By contrast, other αGalCer variants that have an altered glycosidic linkage, a chemically modified acyl-chain, or a modified sphingoid base, potently induce an IFN-γ response (Table 1 and references therein). Thus, it is possible to steer desirable immune responses against cancers by harnessing lipid agonists that induce therapeutic cytokine responses. This feature of αGalCer variants is further accentuated by the ability of activated NKT cell responses to transactivate cells of the innate and adaptive immune systems as narrated briefly below (see Figure 2).
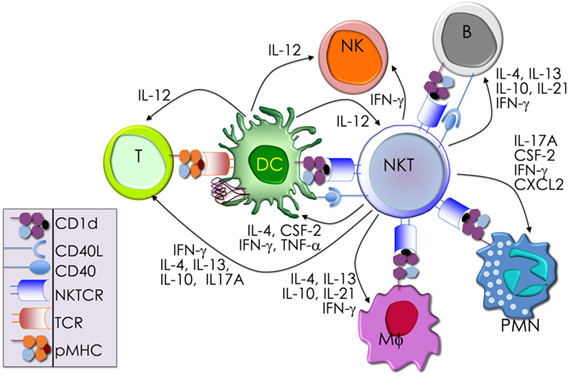
Figure 2. The immunological effector functions of mouse NKT cells. The interactions between the invariant natural killer T (NKT) cell receptor and its cognate antigen, as well as interactions between costimulatory molecules CD28 and CD40 and their cognate ligands CD80/86 (B7.1/7.2) and CD40L, respectively, activate NKT cells. Activated NKT cells participate in crosstalk with members of the innate and the adaptive immune systems by deploying cytokine and chemokine messengers. Upon activation in vivo, NKT cells rapidly secrete a variety of cytokines and chemokines, which influence the polarization of CD4+ T cells toward T helper (Th)1 or Th2 cells as well as the differentiation of precursor CD8+ T cells to effector lymphocytes, and B cells to antibody-secreting plasma cells. Some of these mediators facilitate the recruitment, activation and differentiation of macrophages and dendritic cells (DCs), which results in the production of interleukin (IL)-12 and possibly other factors. Interleukin (IL)-12, in turn, stimulates NK cells to secrete interferon (IFN)-γ. Thus, activated NKT cells have the potential to enhance as well as temper the immune response. This schematic rendition of NKT cell effector functions is an adaptation of past reviews (6, 8, 44, 70) and is based on works cited in the text.
Dendritic cells, especially CD8α+ DCs, which are a major producer of IL-12 (71), play a critical role in glycolipid agonist presentation and NKT cell activation (72–78). Activated NKT cells reciprocate by activating the interacting DCs. DCs so activated rapidly mature. Hence, they upregulate costimulatory molecules CD40, CD80, and CD86; several molecules critical for protein antigen capture and peptide presentation, such as DEC205 and MHC class II molecules (79); and induce the production of IFN-γ, tumor necrosis factor (TNF)-α, and IL-12 (80–83). IFN-γ produced by activated NKT cells coupled with CD154 (CD40 ligand on NKT cells) and CD40 (on DCs) mediate the NKT-DC crosstalk (81, 84). This crosstalk steers multiple downstream immune responses: (1) the number and phenotype of DCs after tumor induction (85). (2) IL-12 and IL-18 resulting from NKT-DC crosstalk transactivates NK cells to produce IFN-γ (82). (3) NKT-DC crosstalk can result in IL-4, IL-6, IL-13, and IL-21, which together can enhance B cells responses to protein antigens by B cells (86–93). (4) NKT-DC cross talk licenses DCs for antigen cross-presentation to CD8+ T cells (94–96), and the activation and differentiation of CD4 and CD8 T cells (79, 95–97). Through these bidirectional interactions, NKT cells and DCs cooperate to amplify and direct both innate and adaptive immune responses. Hence, NKT cells are an attractive target for cancer immunotherapies (98–102).
NKT cells have long represented an attractive target for tumor immunotherapy (103, 104). Numerous studies in both humans and mice have demonstrated their ability to directly target CD1d-expressing tumor cells (105–108), recruit and activate anti-tumor effector cells of the innate and adaptive immune systems (100, 109–114), and control the activity of immunosuppressive cells in the tumor microenvironment. After in vivo administration of αGalCer, NKT-DC cross-talk-mediated NK cell activation results in IFN-γ response (82) and, potentially, the anti-tumor effect of αGalCer (85, 115).
The potent anti-metastatic activity of αGalCer in mice (20, 116), which is NKT cell mediated (22), prompted investigations in the role of NKT cells in natural immunity against tumors. Such investigations include chemically induced tumors, transplanted tumors, and tumors arising in genetically engineered animals (115). The outcomes of these studies have been promising because NKT cells exhibit natural immunity against different cancer models. Independent studies have sometimes reported conflicting results as to the importance of NKT cells in the anti-tumor response, particularly with carcinomas induced by the topical carcinogen methylcholanthrene (117, 118). Such conflicting results were likely due to unknown environmental and/or genetic factors present in the mice used as controls in similar experiments by different groups (117). Studies in mice revealed that αGalCer variants that induce type I inflammatory response (see Table 1) were protective against tumor metastases. The mechanistic basis of this anti-metastatic effect remains elusive. Nonetheless, the ability of NKT cells activated by αGalCer variants to steer desirable downstream effector functions, such as NK cells, cytotoxic T cells, Th1 and Th17 cells, γδ T cells, IFN-γ, and direct lysis of myeloid lineage cells may underlie the outcome (100, 115). The anti-tumor activities of NKT cell agonists have already been exploited in a variety of clinical trials. The outcomes of these trials have also been promising (103, 104, 119–121).
NKT cells development and maturation occurs in the thymus (122, 123). Thus, genetically altered mice in which thymocytes do not develop beyond the double-negative (DN)2/DN3 stage also fail to develop NK1.1+ T cells (124). [Note: historically, prior to the development of CD1d-lipid tetramers (125, 126), NKT cells were identified by co-expression of the NK1.1 marker and a TCR. Hence, in pre-tetramer literature, they were referred to as NK1.1+ T cells (127).] Thymic NK1.1− NKT cells were later recognized as a CD1d tetramer+ NK1.1− subset that precedes NK1.1+ NKT cells in development (128, 129). Current literature refers to the IFN-γ-producing, mature, stage 3 (st3) NKT cells as NK1.1+ NKT cells (Figure 3). Furthermore, NKT cells do not develop in mice harboring mutations in genes (e.g., Myb, that encodes the transcription factor c-Myb, Rorc, which encodes RORγt, and Tcf12 that codes for HEB) that impair survival of immature double-positive (DP) thymocytes—cells that co-express both CD4 and CD8 co-receptors— (130–133). Moreover, Vα14 and Jα18 rearrangement occurs at a late DP stage (130, 132). Consistent with this finding, NKT cells develop in NKT cell-deficient Jα18-deficient (Ja18−/−) mice that receive highly purified tetramer-negative, DP-high thymocytes (134). These findings together support the notion that commitment to the NKT cell lineage occurs at the DP stage much alike conventional T cells (135). That notwithstanding, compelling new data indicate that Vα14 and Jα18 rearrangement can occur within CD4- and CD8-negative (DN) thymocytes. Additional data indicate that a fraction (~15%) of NKT cells that differentiate into NKT1 cells emerge from DN thymocytes (136). Hence, an alternative precursor can give rise to functional NKT cells.
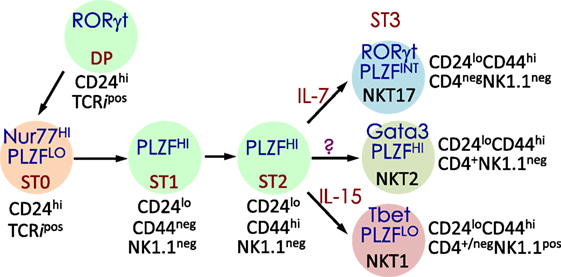
Figure 3. Schematic rendition of mouse NKT cell developmental stages: precursor ST0, immature ST1 and ST2 and mature ST3 and NKT1, 2, and 17 are functional subsets. Early developmental steps are common to both NKT cell and conventional T cell lineages as commitment to the NKT lineage occurs at the CD4 and CD8 double-positive (DP) stage. NKT cell ontogeny begins with rearrangement of the Vα14 to Jα18 T cell receptor (TCR) α-chain gene segments and after its interaction with the positively selecting CD1d-self-lipid complex. Stage-specific NKT cell markers—e.g., CD24, CD44, and NK1.1—and subset-specific differentiation signals and transcription factors are indicated. Interleukin (IL)-7 and IL-15 are cytokines that mediate intercellular communication. NKTCR signaling turns-on the master transcription factor promyelocytic leukemia zinc finger (PLZF), which controls multiple molecular events that distinguish NKT cells from all of the other thymus-derived T lymphocytes. Additional molecular cues include Fyn and Lck, which are Src (cellular protein homologous to the Rous sarcoma virus oncogene) kinases (protein phosphorylation enzymes) essential for transmitting NKTCR signals from the plasma membrane to inside of the cell. Fyn also transmits signals relayed from SLAM (signaling lymphocyte activation molecule) through the adapter protein SAP (SLAM-associated protein). Protein kinase C (PKC)-θ processes NKTCR signaling and activates the transcription factor nuclear factor-κB (NF-κB). Other transcription factors, such as Egr-2, Ets-1, GATA3, Id2, Id3, MEF, Nur77, RORγt, and T-bet, some of which are also essential for functional differentiation of NKT cell subsets (refer to Figures 4 and 6) and act at distinct stages of NKT cell development. This diagrammatic rendition of NKT cell development is an adaptation of a past review (8) and is based on works cited in the text.
Positive selection of NK1.1+ T cells depends on DP thymocytes (122). Developing NKT cell-DP thymocyte interactions involve both self-lipid-bound CD1d/NKTCR (22, 116, 137–139) and signaling lymphocytic activation molecule (SLAM)–SLAM interactions (140–142). These interactions are critical to NKT cell maturation, which involves protein kinase Cθ-NF-κB (143) and NFAT-Egr2 (144–146) activation downstream of the NKTCR, and SLAM-associated protein-Fyn activation downstream of SLAM (140, 141, 147, 148). Signals so transmitted from the cell surface are relayed through multiple signaling nodes in the cytoplasm and integrated in the nucleus into a unique transcriptional program (Figure 3). A key nuclear event involves the activation of the zinc finger BTB domain-containing-16 (Zbtb16) gene that codes for promyelocytic leukemia zinc finger (PLZF). The PLZF-mediated genomic control distinguishes the unique NKT cell functions from those of the other T lymphocytes (149, 150). NK1.1− NKT cells undergo several rounds of cell division, retaining some in the thymus with the remaining emigrating and populating the peripheral lymphatic organs. Thence, NK1.1− NKT cells mature to become NK1.1+ NKT cells, both in the thymus and the periphery (Figure 3). A key feature of this maturation process is the acquisition of cytokine secretion function in a less well-understood mechanism (148) and the differentiation into three functional subsets: NKT1, NKT2, and NKT17 (discussed below). These NKT cell subsets marked by the same subset-specific transcription factors and cell surface markers expressed by the corresponding T helper cell subsets (151–156).
Gene regulatory networks (GRNs) are composed of trans-regulatory factors—generally made up of transcription factors and regulatory RNA such as microRNAs and long non-coding RNA—and cis-regulatory regions generally found upstream of genes whereupon transcription factors bind to control lineage-specific gene expression. GRNs unveil the origins and evolution of cell lineages (157). Many transcription factors have been studied in relation to NKT cell development and function. Among these, PLZF works as a master transcription factor controlling the development of innate-like functions within NKT cells (Figure 4) (149, 150, 158). Mice harboring a loss-of-function PLZF mutation or lacking PLZF demonstrated poor NKT cell development, and those NKT cells that developed were NK1.1− and homed to lymph nodes but not to tissues such as thymus and liver where they are found abundantly in wild type (wt) mice (149, 150). Additional studies indicated that PLZF binds to cis elements of effector cytokine and homing receptor genes to direct their expression within NKT cells (Figure 4) (158). Furthermore, forced expression of a Zbtb16 transgene in all T cells during thymic development resulted in the acquisition of an innate-like phenotype and function in conventional T cells (158). These findings heralded PLZF as a lineage-specific master regulator of transcription (149, 150, 158), and has led to the unveiling of a GRN that controls effector differentiation in developing NKT cells (Figure 4).
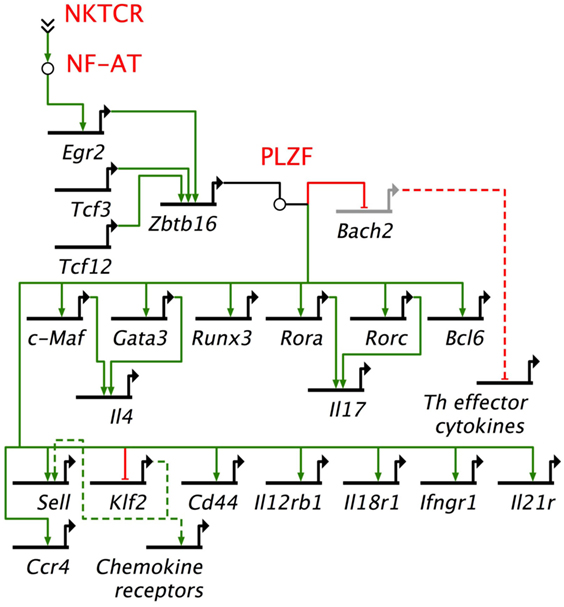
Figure 4. Promyelocytic leukemia zinc finger (PLZF)-driven gene regulatory network directs innate effector function in mouse NKT cells. NKTCR stimulation by self-glycolipid antigen activates downstream nuclear factor of activated T cells (NF-AT), which activates Egr2 gene. Egr2 is essential for Zbtb16 gene expression downstream of NKTCR stimulation. PLZF encoded by Zbtb16 activates T helper (Th) lineage-specific transcription factors, except Tbx21, which codes for the Th1 master regulator Tbet. Promyelocytic leukemia zinc finger (PLZF) also binds to multiple cis-regulatory elements to repress Bach2, which is a repressor of Th cytokine genes. In addition, PLZF binds to cis-regulatory elements of a variety of cytokine and chemokine receptor genes. NF-AT, Tcf3, Tcf12, Egr2, Zbtb16, Bach2, c-Maf, Gata 3, Runx3, Rora, Rorc, Bcl6, and Klf2 encode transcription factors. Other genes under PLZF control encode effector proteins, mostly cytokines (e.g., Il4), cytokine receptor (Il12Rb1, Ifngr1, Il21r), chemockine (Ccr4) or cell adhesins [Cd44, Sell (L-selectin)]. Bach2 represses the induction of T helper (Th) effector cytokine genes. Thick black lines, cis-regulatory elements of genes; solid green lines, enhancer; solid red lines, repressor; dashed green lines, indirect evidence for enhancement; dashed red lines, indirect evidence for repression. Based on Ref. (145, 149, 150, 158).
The induction of Zbtb16 is controlled in part by acetylated Egr2 (159), which is induced downstream of NKTCR signaling (144). A recent study demonstrated that the gene encoding the histone acetylase GCN (general control non-derepressible) 5 acetylates a critical lysine residue in Egr2. DP thymocyte-specific depletion of GCN5 blocked the progression of NKT cell development from stage 0 to stage 1 in a cell intrinsic manner. This stage 0 to stage 1 developmental block was due to transcriptional downregulation of the lineage driving gene Zbtb16 and other genes such as Runx1, Tbx21, and Il2rb that are essential for proper NKT cell development (159). GCN5 itself is an acetylated protein. Whether its function during NKT cell development depends on acetylation is currently unknown. In some models, the function of GCN5 depends on its deactylation (160). Should GCN5 function in NKT cells depend on deacetylation, whether and which sirtuins [silent mating type information regulation 2 homologs 1–7 (160)] play this role in NKT cells remains to be established.
Even though the mouse invariant Vα14i TCR α-chain has the potential to pair with virtually all available TCR β-chains, the peripheral NKT cell repertoire consists of Vα14i paired with a restricted set of β-chains, viz., Vβ8, Vβ7, and Vβ2 (161). There are two views to the events that sculpt this semi-invariant NKTCR repertoire: the predominant view is that such a semi-invariant NKTCR repertoire is built exclusively by positive selection (162). The competing hypothesis—that both positive and negative selections sculpt the semi-invariant NKTCR repertoire—is supported by indirect evidence (163–166).
Two lines of evidence support the notion that positive selection sculpts the NKT cell repertoire. CD1d molecules have a recycling motif in their cytoplasmic tail, which is essential for the endo/lysosomal exchange of CD1d-bound lipids and their subsequent presentation to NKT cells. Transgenic mice expressing a mutant CD1d molecule that has lost the ability to recycle do not develop NKT cells, suggesting that positive selection requires a recycling CD1d molecule (167). Another line of support comes from the study of CD1d-null mice, which contain a small number of CD1d-tetramer+ thymocytes. These pre-selection thymocytes also express only the Vβ8, Vβ7, and Vβ2 β-chains expressed by the post-selection NKT cells. Such pre-selection thymocytes expand the same NKTCR repertoire when stimulated with a putative self-glycolipid called isogloboside-3 in vitro (35, 161). These lines of evidence support positive selection as the sole model for sculpting the NKT cell repertoire.
Deletion of the gene coding for NKAP (NF-κB activating protein) in DP thymocytes specifically blocks the development of NKT cells but not conventional T cells (168). NKAP colludes with HDAC3 (histone deacetylase 3) to function as a transcriptional repressor (169). Accordingly, deletion of the Hdac3 gene in DP thymocytes completely blocks NKT cell development, while conventional T cell development proceeds normally (168). Hence, the repression of target genes at the DP thymocytes stage by the combined action of NKAP and HDAC3 is essential for positive selection of the NKT cell lineage.
Three lines of evidence support a potential role for negative selection in pruning self-reactive NKT cells for sculpting a functional repertoire: first, all available TCR β-chains can pair with the Vα14i TCR α-chain and react with CD1d tetramer, yet only Vβ8, Vβ7, and Vβ2 β-chains are expressed by the post-selection NKT cells (161). This finding can be explained only by negative selection of the majority of the β-chains and not by the failure to survive owing to the inability to interact with CD1d or to failed positive selection (38, 161). Second, transgenic over expression of either mouse or human CD1d in DP thymocytes and thymic myeloid cells results in fewer NKT cells and, those that remain, display altered Vβ usage (163, 170). Furthermore, only wt 16.5-day post-coitus mouse fetal thymic organ cultures (FTOCs), but not FTOCs from CD1d-overexpressing transgenic animals, fostered NKT cell development (163). Finally, exogenous addition of αGalCer, to wt mouse FTOCs resulted in NKT cell depletion (163, 164). Likewise, in vivo αGalCer injections into neonatal mice also resulted in the intra-thymic depletion of NKT cells (164). Together, these findings provide compelling evidence, albeit indirect, supporting a role for negative selection in sculpting a functional NKT cell repertoire.
Agonistic ligand(s)—those that positively select in the thymus being similar or identical to ligands that activate in the periphery (19, 27, 171)—selects NKT cells, which strikingly contrast antagonist ligand-mediated selection of conventional T cells. Further, SLAM–SLAM interactions, which activate PKC-θ via the SAP-FynT signaling module, mediate persistent interactions between developing NKT cells and the selecting DP cells (140, 141, 147, 172–175). NF-κB provides a survival signal to escape death that could result from these high affinity interactions (166, 176–182). Current evidence suggests that signals relayed through the TCR–PKCθ–CARMA1 axis are integrated by NF-κB to prevent death of developing NKT cells (143, 166, 183). But the signals relayed by the TCR-PKCθ-CARMA1 axis only partially accounts for such death signals. Consistent with this conclusion is the finding that TNF-α ligation of TNF receptor superfamily member 1a (TNFR1) relays caspase 8 and caspase 9 activation signals to mediate NKT cell death. This death signal is also obviated by NF-κB activation (183). Additional signals also mediate NKT cell survival during development (181, 184–192). Hence, escaping cell death from multiple signals may be a key feature of thymic NKT cell development. Whether this cell death is the basis for negative selection of NKT cells currently remains unknown.
NKT cells must tightly regulate NF-κB activation as mice that lack RelA or cannot activate NF-κB poorly develop NKT cells (143, 176, 177). On the other hand, mice that express overactive NF-κB or lack the negative regulator of NF-κB signaling CYLD, develop NKT cells but fail to mature and populate the lymphoid organs and peripheral tissues (181). Hence, NF-κB may function as a rheostat to set the threshold for peripheral NKT cell activation. Such a threshold may be critical as their selection and function are controlled by agonistic ligand(s) so as to prevent autoreactivity. How NF-κB functions as a rheostat in developing NKT cells needs elucidation.
Recent findings on NKT cell developmental properties may be best understood from an ecological perspective. These properties include, (a) functional NKT cell subsets and the division of labor; (b) NKT cell frequency variation; (c) tissue environment-dependent NKT cell subset frequency variation; and (d) gut microbiota-dependent peripheral NKT cell maturation and reciprocal NKT cell control over gut microbiota.
NKT cell activation results in rapid secretion of pro-inflammatory and regulatory cytokines and chemokines. This property in conjunction with the capacity to transactivate a variety of innate and adaptive immune cells—see subsection on Transactivation—allows NKT cells to steer downstream immune responses. NKT cells are heterogeneous, consisting of at least four distinct subsets—NKT1, NKT2, NKT10, and NKT17. In addition, at least one induced subset, NKTfh, is also recognized. As with conventional CD4+ T cell subsets, NKT cell subsets are characterized by prototypic cytokine responses and subset-specific transcription factors (Figure 5). Each subset is represented at different proportions in various mouse strains (151–155).
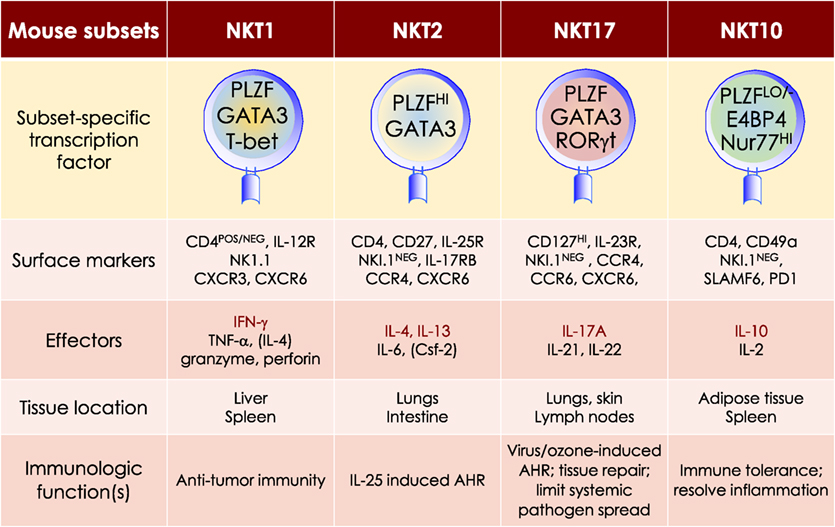
Figure 5. At least four mouse NKT cell subsets divide-up the labor. The four NKT cell subsets reflect T helper (Th)1, Th2, Th17, and Treg subsets. The transcription factors and the prototypic effector molecules as well as the locations and functions, which define the four NKT cell subsets, are represented. The text contains detailed description of each subset. This diagrammatic rendition of NKT cell subsets is an adaptation of a past review (8) and is based on works cited in the text.
MOUSE NKT1 CELLS are marked by either the expression of CD4 or the absence of CD4/CD8 co-receptors. NKT1 cell activation results in a Th1-like cytokine response. The majority of mouse splenic and hepatic NKT cells are NKT1 subset, especially in the C57Bl/6 strain. NKT1 cell differentiation depends on T-bet (Tbx21) and IL-15 but less on GATA3 (151, 152, 187, 189–191). Unlike HDAC3 depletion in DP thymocytes, NKT cell lineage-specific deletion of Hdac3 (derived with the use if Zbtb-Cre) results in selective impairment in NKT1 cell development. The selective absence of HDAC3 in NKT cells resulted from reduced autophagy (193–195)—a cytoplasm recycling process essential to T and NKT cell development —and decreased GLUT1, CD71, and CD98 nutrient receptor expression (196). Moreover, the anti-tumor effect of αGalCer (109) is potentially mediated by IFNγ- and TNFα-producing NKT1 cells.
MOUSE NKT2 CELLS express the CD4 co-receptor. NKT2 cell actiation results in a Th2-like cytokine and chemokine response. This subset is enriched in mouse lungs and the intestine (152). IL-13 and IL-4 as well as CCL17, CCL22, CCL10/CCL6, and eosinophil chemotactic factor-L secreted by activated NKT2 cells may mediate airway hyperresponsiveness (151, 197–200). This Th2-type response recruits Mϕs, eosinophils, neutrophils, and lymphocytes into the lungs to incite tissue damage (197). Coincidently, in BALB/c mouse that is sensitive to airway hyperresponsiveness, NKT2 cells predominate (152).
NKT cells constitutively express Il4 and Ifng transcripts. This constitutively expressed cytokine genes may explain the rapid NKT cell response to agonistic stimulation in vivo (201). Epigenetic changes in the two cytokine genes control their transcription. For example, the conserved non-coding sequence (CNS) 2 located downstream of the mouse Il4 locus is constitutively active in NKT cells, which thereby constitutively transcribe the Il4 gene. CNS 2 activity depends on NOTCH and Rbp-j (recombination signal binding protein for immunoglobulin kappa J region)—a transcriptional regulator of NOTCH signaling. Hence, DP thymocyte-specific deletion of Rbp-j abolished CNS 2 activity and the ability to transcribe Il4 (202).
A similar epigenetic control of the human Ifng locus using CNS-30 and CNS +18–20 transcribes the Ifng locus in NKT cells (203, 204). Consistent with this finding, NKT cells showed acetylated histone 4 marks upstream and downstream of the Ifng coding region only when activated by weak (self agonists) or strong signals (phorbolmyristate acetate + ionomycin) but not in resting NKT cells. Furthermore, NKT cells rested after stimulation returned the Ifng locus to an unmarked state (205). H4 acetylation occurs at CNS +18–20, a site essential for human Ifng transcription in NKT cells and conserved within the mouse Ifng locus (203, 205). These findings notwithstanding, it is unclear whether human NKT cells constitutively transcribe the Ifng locus and how mouse NKT cells constitutively transcribe its Ifng locus.
MOUSE NKT17 CELLS do not express CD4 or CD8 co-receptors. They are enriched in the lungs, skin, and peripheral lymph nodes, and are poorly represented in the spleen and liver (206–208). These cells require IL-7, not IL-15, for survival (151, 209). The development of NKT17 cells also requires mTORC2 signaling and the transcription factors Runx1 and NKAP (168, 210–213). Thus, NKT cell-specific Runx1 deletion results in decreased IL-7Rα, BATF, and c-Maf expression against the backdrop of increased Lef and Bcl11b expression (211). On the other hand, how NKAP controls NKT17 cell development is not understood, but appears not to require mTOR, IL-7, and TGF-β signaling (210).
Akin to Th17 cells, NKT17 cells constitutively express RORγt (206), rapidly produce IL-17A in response to certain bacterial infections, and induce airway neutrophilia when challenged with synthetic glycolipid or LPS (37, 206, 214). NKT17 cells may contribute to ozone-induced airway hypersensitivity (215), the development of experimental autoimmune encephalomyelitis (214), and the pathogenesis of acute hepatitis in mice (216).
MOUSE NKT10 CELLS, the PLZF-independent subset (154), are found in low frequency in unchallenged mice and in human peripheral blood mononuclear cells (PBMCs). Upon re-activation, NKT10 cells that previously responded to αGalCer in vivo, secrete IL-10 (155). IL-10 produced by activated NKT10 is thought to maintain immune-privilege sites. This NKT cell subset may also control Treg cell functions in adipose tissues (154).
Mouse NKT cells can provide cognate (lipid antigens) or non-cognate (protein antigens) help to B cells and regulate antibody responses (89, 90, 92, 217, 218). Upon immunization with αGalCer a subset of NKT cells acquire a phenotype similar to T follicular helper T cells (Tfh) referred to as NKT follicular helper (NKTfh) cells (218–220). NKTfh are characterized by the expression of CXCR5, ICOS, PD1, Bcl6, and BTLA. Their development is dependent on same factors that drive Tfh development (219). NKTfh cells induce rapid production of germinal centers through IL-21 production that yields detectable levels of antigen-specific IgG (91, 219, 220). Nonetheless, NKTfh cell-induced antibody responses are short-lived and inferior to Tfh cell-induced responses (91, 219, 220). NKTfh cells may play a role in antibody responses against human pathogens such as Borrelia hermsii, Streptococcus pneumoniae, and Plasmodium falciparum (91, 219, 220). NKTfh and Tfh cells can act synergistically to induce robust antigen-specific antibody responses underscoring the use of αGalCer as a vaccine adjuvant (218).
Human NKT cell responses are as diverse as those of mouse (221), yet NKT cell subsets have not been formalized in humans. Functional dichotomy has been reported in human CD4+ and DN NKT cell subsets: activated human CD4+ NKT cells secrete IL-4. A pathological role has been attributed to human CD4+ NKT cells, which accumulate in the lungs of chronic asthmatic patients and produce IL-4 and IL-13 (222). Hence, human CD4+ NKT cell resembles the mouse NKT2 cell subset. On the other hand, the activated DN NKT cells secrete IFN-γ and TNF-α. Furthermore, both CD4 and DN human NKT cell subsets upregulate perforin in the presence of inflammatory signals. The DN NKT cells also upregulate NKG2D expression, which together with perforin may mediate cytotoxicity against infected cells and cancer cells (223, 224). These functions of human NKT cells resemble those of mouse NKT1 cells. Activated human NKT cells can also secrete IL-17 (221), suggesting the presence of an NKT17-like subset.
In summary, mouse NKT cells divide labor into four subsets. Global and single cell transcriptome analyzes demonstrated that the thymic NKT1, NKT2, and NKT17 cells were distinct subsets (156, 225). Even though not formalized, human NKT cells also have the potential to mirror mouse NKT cell subsets, but this requires further investigation. That the tissue environment plays a role in the differentiation of NKT cell subsets is supported by the finding that NKT17 differentiation required mammalian target of rapamycin complex-2 (213) or is suppressed by Tet enzymes that modify 5-methylcytosine in DNA by controlling the expression of Tbet and ThPOK transcription factors (226). Another study using somatic cell nuclear transfer to generate mice with monoclonal NKT cell populations demonstrated that tissue homing pattern, and, to a lesser extent, TCR avidity governed NKT cell subset differentiation (208). That NKT1, NKT2, and NKT17 cells differentiated within peripheral tissues of each of the three monoclonal mouse lines, derived from somatic cell nuclear transfer, suggests that the subsets are perhaps NKT cell “reaktionsnorm [German for reaction norm or norm of reaction; Woltereck 1909 cited in Ref. (227)]” induced by the tissue-specific environment, potentially by local cytokine/chemokine milieu in conjunction with the host microbiota.
An intriguing property of NKT cells is their frequency variation observed in lymphoid tissues of different inbred strains of similar age: low in 129 and NOD, intermediate in C57Bl/6, and high in BALB/c, CBA, and DBA/2 mice (152, 153, 228–230). Likewise, NKT cells show striking frequency variation that can range from as little as 0.001% to 5–10% within human PBMCs (221, 231, 232).
Mice show inter-strain variation in thymic NKT cell subset numbers (152). C57Bl/6 mice have high proportion of NKT1 cells and low frequency of NKT2 cells, whereas BALB/c have high frequency of NKT2 and NKT17 suggesting an inverse correlation between frequency of NKT1 cells versus NKT2 cells and mouse strains. Curiously, mouse strains that have a high frequency of NKT2 cells (BALB/c, CBA, and DBA/2) showed high numbers of eomesodermin-expressing memory-like CD8+ thymocytes (152) which was attributed to the steady-state production of IL-4 by the expanded NKT2 population in these mice. In an effort to understand whether genetic polymorphisms between mouse strains controlled NKT cell frequency, recombinant inbred and co-isogenic strains begotten from NOD (low NKT cell frequency) X C57Bl/10 (intermediate NKT cell frequency) crosses were analyzed. The outcomes of several such studies indicated that NKT cell frequency segregated with the genetic background of the mouse (153, 229, 230). Whereas this outcome suggests that NKT cell frequency is under genetic control, whether this control is direct or indirect remains to be ascertained.
NKT cells surveil barrier mucosae such as that of the small and large intestine (233, 234). The number, phenotype, and functional maturation of NKT cells in the gut epithelium and lamina propria are controlled by neonatal colonization of the gut by bacterial symbionts. Thus, germ-free (GF) mice have high numbers of NKT cells in the gut epithelium and lamina propria that are immature and, hence, hypo-responsive to αGalCer (233). Curiously, reconstitution of young, but not adult mouse gut by bacteria that biosynthesise αGalCer or related compounds reverses the hypo-responsiveness of NKT cells found in GF intestinal mucosae (234). Similarly, GF mice also harbor high hepatic and pulmonary, but not thymic and splenic NKT cell frequencies (234). Additional evidence implicates the CXCR6 ligand CXCL16, whose expression is under the control of gut microbiota, in regulating gut NKT cell frequency and maturation (234, 235). Furthermore, αGalCer compounds (see Table 1) synthesized by the bacterial symbiont Bacteriodes fragilis, exert either an inhibitory effect preventing proliferation, or are stimulatory on developing NKT cells (26, 27). As the gut microbiota varies between individuals of different genetic, ethnic, and geographic backgrounds (236), the above findings in mice suggest the intriguing possibility that the human symbionts may impart an epistatic control over human NKT cell frequency and maturation as well. Because the frequency and functional status are environmentally controlled even though the genotype of the differentiating NKT cells remains the same, NKT cell frequency and proper maturation are potentially polyphenic (227, 237) properties.
Early-life microbial ecology has implications for health. Thus, GF mice are prone to severe airway hypersensitivity and dextran sodium sulfate-induced colitis (233–235). The latter phenotype is obviated by the interaction of NKT cells with B. fragilis-derived glycosphingolipid(s) during early life (26). Not surprisingly, NKT cells can, in turn, control gut microbial ecology and gut physiology (238). Whether similar reciprocal interactions between NKT cells and the gut microbiota occur in humans currently remains unknown.
Microbial ecology has emerged as an important deterministic factor in the outcome of chemotherapy, radiation therapy, and immunotherapy against cancers (239). NKT cells have been targeted in the clinic for immunotherapy (see Implications for Cancer Immunotherapy), but how each of these therapies impact NKT cells is not known. It is noteworthy that a fraction of NKT cells are radiation resistant (130). This feature can be exploited for NKT cell-targeted immunotheraphy against lymphomas and leukemias. Clinical trials have shown that the outcome of NKT cell-targeted immunotherapy varied between recipients (103, 104). Hence, what roles the gut microbiota played in the outcome is worthy of investigation. So also, considering that NKT cells can impact microbial ecology (238), what roles NKT cells play in tumorigenesis and metastasis are also worthy of investigation. Insights into how the microbial community assembles and forms the host–symbiont ecosystem will facilitate an essential understanding of the molecular underpinnings that govern reciprocal interactions between the host and its internal ecosystem. These new insights can, in turn, impact the way by which new cancer therapies are designed, developed, and refined.
… the struggle against diseases, and especially infectious diseases, has been a very important evolutionary agent and that some of its results have been unlike those of the struggle for life … [(240) within a collection of papers in genetics by Haldane (241)].
Comparative vertebrate genomics, enabled by recent advances in whole-genome sequencing, have revealed molecular signatures of selection upon genes that control many biologic functions, including immune responses. Hence, pathobionts can apply immense selection pressure and play significant roles in the evolution of immune response genes and cells. As early-life symbionts can impact health, microbial ecology may also play roles in the evolution of the immune response genes and cells.
The NKTCR engages its ligand, CD1d-lipid co-complex, with conserved germline-encoded residues in four-to-five of the six complementarity-determining regions of the combined TCR α- and β-chains (242). Hence, phylogenetic studies of genes that encode CD1 molecules and the invariant NKTCR α-chain can reveal the origin and evolution of NKT cells. A recent phylogenomic analysis revealed that the Cd1 gene is an amniote innovation that evolved in the Mesozoic reptiles and was retained in the extant anapsid (green anole lizard Anolis carolinensis) and synapsid (Siamese crocodile Crocodylus siamensis and Chinese alligator Alligator sinensis) reptilians (243). Cd1 genes diversified in mammals, wherein evolved the Cd1d gene that encodes the lipid agonist presenting molecule that controls the functions of NKT cells in eutherians (of placental mammals; Figure 6) (244). Curiously however, the reptilian Cd1 gene has no orthology with avian or mammalian Cd1 genes (243), suggesting that Cd1 genes may have emerged multiple times during amniote evolution. Or alternatively, Cd1 genes may have evolved rapidly and diverged substantially from the reptilian form within extinct synapsid and mammal-like reptiles prior to stabilization within eutherian species. The latter view is supported by the finding that egg-laying monotremes such as platypuses and echidnas do not have Cd1 genes while a CD1d-like gene exists in a few metatherian (of marsupial mammals) species such as the opossum.
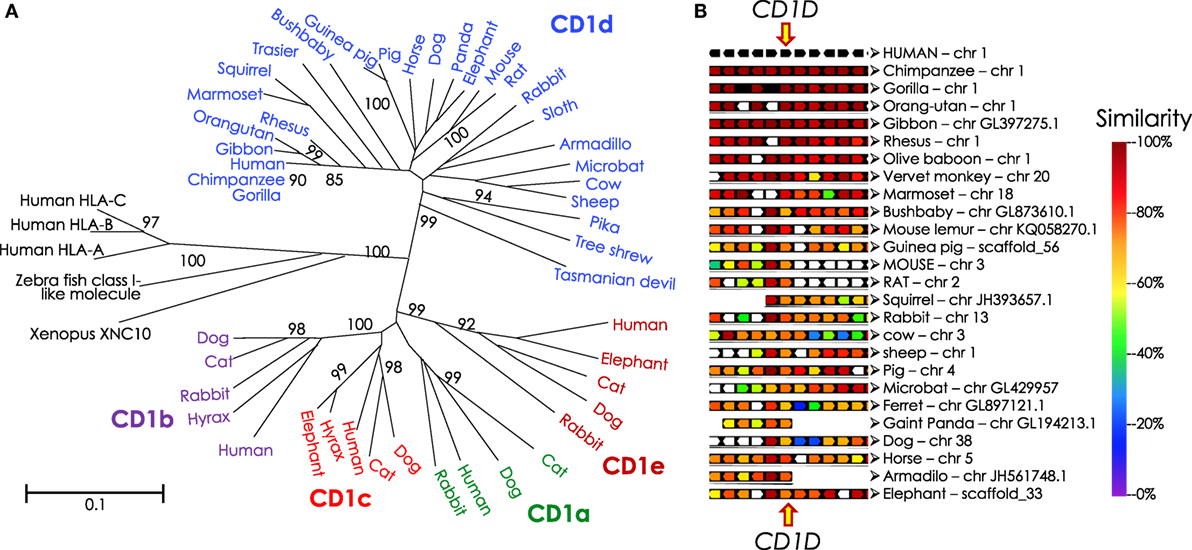
Figure 6. Phylogenetic tree of CD1 and syntenic relationship of eutherian Cd1d. (A) Protein sequences of α1-α2 domains of mammalian and reptilian CD1, human and zebra fish MHC class I and Xenopus XNC10 were retrieved from NCBI using PSI-BLAST. Sequences with E-value of ≤0.05 were considered mostly similar to the human CD1D query sequence; significant bootstrap values are indicated for critical nodes. Sequences were aligned using ClustalW. Phylogenetic analysis (pairwise deletion, bootstrap: n = 1,000) was performed to construct the optimal tree with the sum of branch length = 9.42589507. Neighbor-Joining method (245) was used to infer evolutionary history. An optimal tree with the sum of branch length = 9.42589507 is shown. p-distance method was used to compute evolutionary distances (246, 247). The tree is drawn to scale, with branch length units the same as those of the evolutionary distances used to infer the phylogenetic tree. A total of 51 sequences and 210 positions were included in the final dataset. Evolutionary analyses were performed using MEGA6 (248). Mammalian species names according to Ref. (249), and the Order/Family to which they belong according to Ref. (250, 251) as well as homology to human CD1D are as follows: northern brown bandicoot (Isoodon macrourus), Peramelemorphia/Peramelidae, 50%; Tasmanian devil (Sarcophilus harrisii), Marsupialia/Dasyuridae, 42%; Hoffmann’s two-toed sloth (Choloepus hoffmanni), Pilosa (Xenarthra)/Megalonychidae, missing various parts of the protein; nine-banded armadillo (Dasypus novemcinctus), Cingulata (Xenarthra)/Dasypodidae, 62%; tailless tenrec (Echinops telfairi), Afrosoricida (Insectivora)/Tenrecidae, missing N- and C-terminal ends of the protein; western European hedgehog (Erinaceus europaeus), Erinaceomorpha (Insectivora)/Erinaceidae; Eurasian shrew (Sorex araneus), Soricomorpha (Insectivora)/Soricidae, has only 181 aa, missing N- and C-terminal ends of the protein; northern tree Shrew (Tupaia belangeri), Scandentia/Tupiidae, 68%, missing leader sequence; large flying fox (Pteropus vampyrus): Chiroptera/Petropodidae; little brown bat (Myotis lucifugus), Chiroptera/Vespertilionidae 62%; dog (Canis lupus familiaris), Carnivora/Canidae, 57%, missing initiator methionine; giant panda (Ailuropoda melanoleuca), Carnivora/Ursidae, 60%, missing initiator methionine; ferret (Mustela putorius furo), Carnivora/Mustelidae, 60%, missing leader sequence; cat (Felis catus), Carnivora/Filidae; bottlenose dolphin (Tursiops truncatus), Cetacea/Delphinidae; African bush elephant (Loxodonta africana) Probosidea/Eliphantidae 65%; horse (Equus caballus), Perissodactyla/Equidae, 73%; rock hyrax (Procavia capensis), Hyracoidea/Procavidae, 64%; wild boar (Sus scrofa), Artiodactyla/Suidae, 65%; alpaca—Andean paca (Vicugna pacos), Artiodactyla/Camelidae; cow (Bos taurus), Artiodactyla/Bovidae, 65%, valine substitution of initiator methionine; squirrel (Ictidomys tridecemlineatus), Rodentia/Sciuridae, 64%, missing last 12 aa including the recycling motif; Ord’s kangaroo rat (Dipodomys ordii): Rodentia/Heteromyidae; brown rat (Rattus norvegicus), Rodentia/Muridae; 64%; mouse (Mus musculus domesticus), Rodentia/Muridae, 61%; guinea pig (Cavia porcellus), Rodentia/Cavidae, 65%; American pika (Ochotona princeps), Logomorpha/Ochotonidae, 64%, missing last 17 aa including the recycling motif; European rabbit (Oryctolagus cuniculus), Logomorpha/Leporidae, 67%; gray mouse lemur (Microcebus murinus), Primates/Cheirogaleidae, 78%; small-eared galago—a bushbaby (Otolemur garnettii), Primates/Galagidae (Loridae), 71%, missing last 31 aa including the recycling motif; Philippine tarsier (Tarsius syrichta), Primates/Tarsiidae, 71%; white-tufted-ear marmoset (Callithrix jacchus), Primates/Callitrichidae, 83%; rhesus monkey/macaque (Macaca mulatta), Primates/Cercopithecidae, 88%; northern white-cheeked gibbon (Nomascus leucogenys), Primates/Hylobatidae, 95%; western gorilla (Gorilla gorilla gorilla), Primates/Hominidae, 99%; Sumatran orang utan (Pongo abelii), Primates/Hominidae, 96%; chimpanzee (Pan troglodytes), Primates/Hominidae, 98%; human (Homo sapiens), Primates/Hominidae. (B) Syntenic map was drawn using Genomicus v89.01 Phyloview (252). Human CD1D gene was used as reference gene and species, respectively, Eutheria as the root species. Upstream of CD1D gene are KIRREL, CD5L, FCRL1, FCRL2, and FCRL3 genes, and downstream from CD1D are CD1A, CD1C, CD1B, CD1E, and ORIOT2 genes in the syntenic map.
A phylogenetic analysis of TRAV10 (encoding the human Vα24 gene segment) or TRAV11 (encoding the mouse Vα14 gene segment) and TRAJ18 (encoding the Jα18 gene segment) revealed that gene elements related to TRAV10/11 and TRAJ18 were found only in placental mammals (244). This finding suggests that NKT cells are a eutherian innovation. As the host–gut microbiota controls NKT cell terminal functional differentiation and NKT cells impact gut microbial ecology, it is postulated that placental development, sudden perinatal exposure to maternal and environmental microbiota, and lactation may have contributed to the evolution of CD1d-restricted type I NKT cells.
The foregoing discusses recent advances in developmental biology of NKT cells and the environmental context in which it develops, matures and differentiates. A final section discusses their evolutionary path and how developmental biology and ecology may have contributed to this unique developmental plan. In addition, how the eco-evo-devo perspective on NKT cells may contribute to cancer immunotherapy is touched upon. Finally, areas that will benefit from further investigation are also pin pointed in their respective sections. Summarily, such areas include, (a) what early events specify NKT cell lineage commitment and turn on the unique lineage-specific GRN?; (b) what signals do symbionts relay to developing NKT cells to specify physiologic functions?; (c) in turn, what signals do NKT cells relay to the microbial community in the gut, and potentially to the microbionts in skin and lungs, to ensure physiologic community assembly, structure, and organization in early, young, and adult life?; (d) what tissue environmental signals underlie NKT cell subset differentiation?; (e) can radiation resistance of NKT cells be used in cancer immunotherapy?; and (f) what NKT cell intrinsic and environmental signals have retained NKT cells in certain mammalian species but not in others?
As a final note to the devo-eco-evo synthesis, we observed that the unique behavior of a group of innate-like T lymphocytes and innate lymphoid cells (ILCs) are under the control of PLZF (253–255). These include γδ T cells, NKT cells, MAIT cells, and certain ILCs. In addition, the development (MAIT cells, and potentially γδ T cells) and functional differentiation (NKT cells, MAIT cells, and ILCs) of these cells are determined by gut and potentially other barrier (skin and lungs) symbionts. As these immune cells, all of lymphoid origin, function at the edge (limbus in Latin) of the innate and adaptive immune systems, a proposal to group them into the “limbic immune system” is made here. Curiously, γδ T, NK, and NKT cells localize to the inter-follicular region of the lymph nodes, straddling the cells of the innate and adaptive immune systems (256). By virtue of their physiologic functions, other tissue-restricted innate-like lymphocytes, such as CD8αα innate-type lymphocytes (257) as well as B1 cells and NK cells (258), can be included in the “limbic immune system” even though their development and function may not be controlled by PLZF or the microbiota. In other words, the “limbic immune system” is anglicized Latin for the “inbetweeners” (259) and, hence, synonymous with it.
All authors wrote and edited the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by VA Merit Award (BX001444) to SJ and by research (AI042284, AI061721, AI070305, HL089667, AI068149, AI074754, and AI064639) grants.
1. Ottaviani E, Valensin S, Franceschi C. The neuro-immunological interface in an evolutionary perspective: the dynamic relationship between effector and recognition systems. Front Biosci (1998) 3:d431–5. doi:10.2741/A289
2. Boehm T. Evolution of vertebrate immunity. Curr Biol (2012) 22(17):R722–32. doi:10.1016/j.cub.2012.07.003
3. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol (2015) 33:257–90. doi:10.1146/annurev-immunol-032414-112240
4. Medzhitov R, Janeway CA Jr. Innate immunity: the virtues of a nonclonal system of recognition. Cell (1997) 91(3):295–8. doi:10.1016/S0092-8674(00)80412-2
5. Lanier LL. Shades of grey – the blurring view of innate and adaptive immunity. Nat Rev Immunol (2013) 13(2):73–4. doi:10.1038/nri3389
6. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol (2013) 13(2):101–17. doi:10.1038/nri3369
7. Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell (2015) 160(5):816–27. doi:10.1016/j.cell.2015.02.010
8. Hill TM, Bezbradica JS, Van Kaer L, Joyce S. CD1d-restricted natural killer T cells. In: eLS. Chichester: John Wiley & Sons, Ltd (2016). doi:10.1002/9780470015902.a0020180.pub2
9. Marrero I, Ware R, Kumar V. Type II NKT cells in inflammation, autoimmunity, microbial immunity, and cancer. Front Immunol (2015) 6:316. doi:10.3389/fimmu.2015.00316
10. Rhost S, Sedimbi S, Kadri N, Cardell SL. Immunomodulatory type II natural killer T lymphocytes in health and disease. Scand J Immunol (2012) 76(3):246–55. doi:10.1111/j.1365-3083.2012.02750.x
11. Terabe M, Berzofsky JA. The immunoregulatory role of type I and type II NKT cells in cancer and other diseases. Cancer Immunol Immunother (2014) 63(3):199–213. doi:10.1007/s00262-013-1509-4
12. Macho-Fernandez E, Brigl M. The extended family of CD1d-restricted NKT cells: sifting through a mixed Bag of TCRs, antigens, and functions. Front Immunol (2015) 6:362. doi:10.3389/fimmu.2015.00362
13. Dasgupta S, Kumar V. Type II NKT cells: a distinct CD1d-restricted immune regulatory NKT cell subset. Immunogenetics (2016) 68(8):665–76. doi:10.1007/s00251-016-0930-1
14. Dhodapkar MV, Kumar V. Type II NKT cells and their emerging role in health and disease. J Immunol (2017) 198(3):1015–21. doi:10.4049/jimmunol.1601399
15. Cohen NR, Garg S, Brenner MB. Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv Immunol (2009) 102:1–94. doi:10.1016/S0065-2776(09)01201-2
16. Kohlgruber AC, Donado CA, LaMarche NM, Brenner MB, Brennan PJ. Activation strategies for invariant natural killer T cells. Immunogenetics (2016) 68(8):649–63. doi:10.1007/s00251-016-0944-8
17. Kim EY, Lynch L, Brennan PJ, Cohen NR, Brenner MB. The transcriptional programs of iNKT cells. Semin Immunol (2015) 27(1):26–32. doi:10.1016/j.smim.2015.02.005
18. Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol (2004) 22:817–90. doi:10.1146/annurev.immunol.22.012703.104608
19. Kain L, Webb B, Anderson BL, Deng S, Holt M, Costanzo A, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian alpha-linked glycosylceramides. Immunity (2014) 41(4):543–54. doi:10.1016/j.immuni.2014.08.017
20. Natori T, Koezuka Y, Higa T. Agelasphins, novel a-galactosylceramides from the marine sponge Agelas mauritianus. Tetrahedron Lett (1993) 34:5591–2. doi:10.1016/S0040-4039(00)73889-5
21. Morita M, Motoki K, Akimoto K, Natori T, Sakai T, Sawa E, et al. Structure-activity relationship of alpha-galactosylceramides against B16-bearing mice. J Med Chem (1995) 38(12):2176–87. doi:10.1021/jm00012a018
22. Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science (1997) 278(5343):1626–9. doi:10.1126/science.278.5343.1626
23. Schmieg J, Yang G, Franck RW, Tsuji M. Superior protection against malaria and melanoma metastases by a C-glycoside analogue of the natural killer T cell ligand alpha-galactosylceramide. J Exp Med (2003) 198(11):1631–41. doi:10.1084/jem.20031192
24. Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature (2001) 413(6855):531–4. doi:10.1038/35097097
25. Yu KO, Im JS, Molano A, Dutronc Y, Illarionov PA, Forestier C, et al. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of alpha-galactosylceramides. Proc Natl Acad Sci U S A (2005) 102(9):3383–8. doi:10.1073/pnas.0407488102
26. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell (2014) 156(1–2):123–33. doi:10.1016/j.cell.2013.11.042
27. Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, et al. Production of α-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol (2013) 11(7):e1001610. doi:10.1371/journal.pbio.1001610
28. Mattner J, Debord KL, Ismail N, Goff RD, Cantu C III, Zhou D, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature (2005) 434(7032):525–9. doi:10.1038/nature03408
29. Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature (2005) 434(7032):520–5. doi:10.1038/nature03407
30. Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol (2005) 35(6):1692–701. doi:10.1002/eji.200526157
31. Albacker LA, Chaudhary V, Chang YJ, Kim HY, Chuang YT, Pichavant M, et al. Invariant natural killer T cells recognize a fungal glycosphingolipid that can induce airway hyperreactivity. Nat Med (2013) 19(10):1297–304. doi:10.1038/nm.3321
32. Chang YJ, Kim HY, Albacker LA, Lee HH, Baumgarth N, Akira S, et al. Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest (2011) 121(1):57–69. doi:10.1172/JCI44845
33. Ortaldo JR, Young HA, Winkler-Pickett RT, Bere EW Jr, Murphy WJ, Wiltrout RH. Dissociation of NKT stimulation, cytokine induction, and NK activation in vivo by the use of distinct TCR-binding ceramides. J Immunol (2004) 172(2):943–53. doi:10.4049/jimmunol.172.2.943
34. Parekh VV, Singh AK, Wilson MT, Olivares-Villagómez D, Bezbradica JS, Inazawa H, et al. Quantitative and qualitative differences in the in vivo response of NKT cells to distinct alpha- and beta-anomeric glycolipids. J Immunol (2004) 173(6):3693–706. doi:10.4049/jimmunol.173.6.3693
35. Zhou D, Mattner J, Cantu C III, Schrantz N, Yin N, Gao Y, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science (2004) 306(5702):1786–9. doi:10.1126/science.1103440
36. Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol (2006) 7(9):978–86. doi:10.1038/ni1380
37. Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol (2011) 12(10):966–74. doi:10.1038/ni.2096
38. Mallevaey T, Clarke AJ, Scott-Browne JP, Young MH, Roisman LC, Pellicci DG, et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity (2011) 34(3):315–26. doi:10.1016/j.immuni.2011.01.013
39. Gumperz JE, Roy C, Makowska A, Lum D, Sugita M, Podrebarac T, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity (2000) 12(2):211–21. doi:10.1016/S1074-7613(00)80174-0
40. Facciotti F, Ramanjaneyulu GS, Lepore M, Sansano S, Cavallari M, Kistowska M, et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol (2012) 13(5):474–80. doi:10.1038/ni.2245
41. Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, et al. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol (2009) 7(10):e1000228. doi:10.1371/journal.pbio.1000228
42. Kumar A, Bezbradica JS, Stanic AK, Joyce S. Characterization and functional analysis of mouse semi-invariant natural T cells. Curr Protoc Immunol (2017) 117:1–14. doi:10.1002/cpim.22
43. Joyce S, Girardi E, Zajonc DM. NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J Immunol (2011) 187(3):1081–9. doi:10.4049/jimmunol.1001910
44. Florence WC, Bhat RK, Joyce S. CD1d-restricted glycolipid antigens: presentation principles, recognition logic and functional consequences. Expert Rev Mol Med (2008) 10:e20. doi:10.1017/S1462399408000732
45. Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol (2003) 4(12):1230–7. doi:10.1038/ni1002
46. Brigl M, Tatituri RV, Watts GF, Bhowruth V, Leadbetter EA, Barton N, et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med (2011) 208(6):1163–77. doi:10.1084/jem.20102555
47. Cohen NR, Tatituri RV, Rivera A, Watts GF, Kim EY, Chiba A, et al. Innate recognition of cell wall beta-glucans drives invariant natural killer T cell responses against fungi. Cell Host Microbe (2011) 10(5):437–50. doi:10.1016/j.chom.2011.09.011
48. Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol (2007) 25:297–336. doi:10.1146/annurev.immunol.25.022106.141711
49. Stanic AK, De Silva AD, Park JJ, Sriram V, Ichikawa S, Hirabyashi Y, et al. Defective presentation of the CD1d1-restricted natural Va14Ja18 NKT lymphocyte antigen caused by β-D-glucosylceramide synthase deficiency. Proc Natl Acad Sci U S A (2003) 100(4):1849–54. doi:10.1073/pnas.0430327100
50. Popovic ZV, Rabionet M, Jennemann R, Krunic D, Sandhoff R, Gröne HJ, et al. Glucosylceramide synthase is involved in development of invariant natural killer T cells. Front Immunol (2017) 8:848. doi:10.3389/fimmu.2017.00848
51. Paget C, Mallevaey T, Speak AO, Torres D, Fontaine J, Sheehan KC, et al. Activation of invariant NKT cells by toll-like receptor 9-stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity (2007) 27(4):597–609. doi:10.1016/j.immuni.2007.08.017
52. Grubor-Bauk B, Simmons A, Mayrhofer G, Speck PG. Impaired clearance of herpes simplex virus type 1 from mice lacking CD1d or NKT cells expressing the semivariant V alpha 14-J alpha 281 TCR. J Immunol (2003) 170(3):1430–4. doi:10.4049/jimmunol.170.3.1430
53. Cornish AL, Keating R, Kyparissoudis K, Smyth MJ, Carbone FR, Godfrey DI. NKT cells are not critical for HSV-1 disease resolution. Immunol Cell Biol (2006) 84(1):13–9. doi:10.1111/j.1440-1711.2005.01396.x
54. Ashkar AA, Rosenthal KL. Interleukin-15 and natural killer and NKT cells play a critical role in innate protection against genital herpes simplex virus type 2 infection. J Virol (2003) 77(18):10168–71. doi:10.1128/JVI.77.18.10168-10171.2003
55. Kim EY, Battaile JT, Patel AC, You Y, Agapov E, Grayson MH, et al. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nat Med (2008) 14(6):633–40. doi:10.1038/nm1770
56. Johnson TR, Hong S, Van Kaer L, Koezuka Y, Graham BS. NK T cells contribute to expansion of CD8(+) T cells and amplification of antiviral immune responses to respiratory syncytial virus. J Virol (2002) 76(9):4294–303. doi:10.1128/JVI.76.9.4294-4303.2002
57. Paget C, Ivanov S, Fontaine J, Renneson J, Blanc F, Pichavant M, et al. Interleukin-22 is produced by invariant natural killer T lymphocytes during influenza A virus infection: potential role in protection against lung epithelial damages. J Biol Chem (2012) 287(12):8816–29. doi:10.1074/jbc.M111.304758
58. Paget C, Ivanov S, Fontaine J, Blanc F, Pichavant M, Renneson J, et al. Potential role of invariant NKT cells in the control of pulmonary inflammation and CD8+ T cell response during acute influenza A virus H3N2 pneumonia. J Immunol (2011) 186(10):5590–602. doi:10.4049/jimmunol.1002348
59. De Santo C, Salio M, Masri SH, Lee LY, Dong T, Speak AO, et al. Invariant NKT cells reduce the immunosuppressive activity of influenza A virus-induced myeloid-derived suppressor cells in mice and humans. J Clin Invest (2008) 118(12):4036–48. doi:10.1172/JCI36264
60. Ho LP, Denney L, Luhn K, Teoh D, Clelland C, McMichael AJ. Activation of invariant NKT cells enhances the innate immune response and improves the disease course in influenza A virus infection. Eur J Immunol (2008) 38(7):1913–22. doi:10.1002/eji.200738017
61. Kok WL, Denney L, Benam K, Cole S, Clelland C, McMichael AJ, et al. Pivotal advance: invariant NKT cells reduce accumulation of inflammatory monocytes in the lungs and decrease immune-pathology during severe influenza A virus infection. J Leukoc Biol (2012) 91(3):357–68. doi:10.1189/jlb.0411184
62. Zeissig S, Murata K, Sweet L, Publicover J, Hu Z, Kaser A, et al. Hepatitis B virus-induced lipid alterations contribute to natural killer T cell-dependent protective immunity. Nat Med (2012) 18(7):1060–8. doi:10.1038/nm.2811
63. Nagarajan NA, Kronenberg M. Invariant NKT cells amplify the innate immune response to lipopolysaccharide. J Immunol (2007) 178(5):2706–13. doi:10.4049/jimmunol.178.5.2706
64. Tyznik AJ, Tupin E, Nagarajan NA, Her MJ, Benedict CA, Kronenberg M. The mechanism of invariant NKT cell responses to viral danger signals. J Immunol (2008) 181(7):4452–6. doi:10.4049/jimmunol.181.7.4452
65. Wesley JD, Tessmer MS, Chaukos D, Brossay L. NK cell-like behavior of Valpha14i NK T cells during MCMV infection. PLoS Pathog (2008) 4(7):e1000106. doi:10.1371/journal.ppat.1000106
66. McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, et al. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med (2007) 204(5):1131–44. doi:10.1084/jem.20062342
67. Stanic AK, Shashidharamurthy R, Bezbradica JS, Matsuki N, Yoshimura Y, Miyake S, et al. Another view of T cell antigen recognition: cooperative engagement of glycolipid antigens by Va14Ja18 natural T(iNKT) cell receptor [corrected]. J Immunol (2003) 171(9):4539–51. doi:10.4049/jimmunol.171.9.4539
68. Bezbradica JS, Gordy LE, Stanic AK, Dragovic S, Hill T, Hawiger J, et al. Granulocyte-macrophage colony-stimulating factor regulates effector differentiation of invariant natural killer T cells during thymic ontogeny. Immunity (2006) 25(3):487–97. doi:10.1016/j.immuni.2006.06.017
69. Das R, Bassiri H, Guan P, Wiener S, Banerjee PP, Zhong MC, et al. The adaptor molecule SAP plays essential roles during invariant NKT cell cytotoxicity and lytic synapse formation. Blood (2013) 121(17):3386–95. doi:10.1182/blood-2012-11-468868
70. Mashayekhi M, Sandau MM, Dunay IR, Frickel EM, Khan A, Goldszmid RS, et al. α-Galactosylceramide therapy for autoimmune diseases: prospects and obstacles. Nat Rev Immunol (2005) 5(1):31–42. doi:10.1038/nri1531
71. Mashayekhi M, et al. CD8alpha(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity (2011) 35(2):249–59. doi:10.1016/j.immuni.2011.08.008
72. Bezbradica JS, Stanic AK, Matsuki N, Bour-Jordan H, Bluestone JA, Thomas JW, et al. Distinct roles of dendritic cells and B cells in Va14Ja18 natural T cell activation in vivo. J Immunol (2005) 174(8):4696–705. doi:10.4049/jimmunol.174.8.4696
73. Bai L, Constantinides MG, Thomas SY, Reboulet R, Meng F, Koentgen F, et al. Distinct APCs explain the cytokine bias of alpha-galactosylceramide variants in vivo. J Immunol (2012) 188(7):3053–61. doi:10.4049/jimmunol.1102414
74. Fujii S, Shimizu K, Kronenberg M, Steinman RM. Prolonged IFN-gamma-producing NKT response induced with alpha-galactosylceramide-loaded DCs. Nat Immunol (2002) 3(9):867–74. doi:10.1038/ni827
75. Arora P, Baena A, Yu KO, Saini NK, Kharkwal SS, Goldberg MF, et al. A single subset of dendritic cells controls the cytokine bias of natural killer T cell responses to diverse glycolipid antigens. Immunity (2014) 40(1):105–16. doi:10.1016/j.immuni.2013.12.004
76. Bialecki E, Macho Fernandez E, Ivanov S, Paget C, Fontaine J, Rodriguez F, et al. Spleen-resident CD4+ and CD4- CD8alpha- dendritic cell subsets differ in their ability to prime invariant natural killer T lymphocytes. PLoS One (2011) 6(10):e26919. doi:10.1371/journal.pone.0026919
77. Macho-Fernandez E, Cruz LJ, Ghinnagow R, Fontaine J, Bialecki E, Frisch B, et al. Targeted delivery of alpha-galactosylceramide to CD8alpha+ dendritic cells optimizes type I NKT cell-based antitumor responses. J Immunol (2014) 193(2):961–9. doi:10.4049/jimmunol.1303029
78. Zajonc DM, Cantu C III, Mattner J, Zhou D, Savage PB, Bendelac A, et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol (2005) 6(8):810–8. doi:10.1038/ni1224
79. Fujii S, Shimizu K, Smith C, Bonifaz L, Steinman RM. Activation of natural killer T cells by alpha-galactosylceramide rapidly induces the full maturation of dendritic cells in vivo and thereby acts as an adjuvant for combined CD4 and CD8 T cell immunity to a coadministered protein. J Exp Med (2003) 198(2):267–79. doi:10.1084/jem.20030324
80. Fujii S, Liu K, Smith C, Bonito AJ, Steinman RM. The linkage of innate to adaptive immunity via maturing dendritic cells in vivo requires CD40 ligation in addition to antigen presentation and CD80/86 costimulation. J Exp Med (2004) 199(12):1607–18. doi:10.1084/jem.20040317
81. Vincent MS, Leslie DS, Gumperz JE, Xiong X, Grant EP, Brenner MB. CD1-dependent dendritic cell instruction. Nat Immunol (2002) 3(12):1163–8. doi:10.1038/ni851
82. Carnaud C, Lee D, Donnars O, Park SH, Beavis A, Koezuka Y, et al. Cutting edge: cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J Immunol (1999) 163(9):4647–50.
83. Singh N, Hong S, Scherer DC, Serizawa I, Burdin N, Kronenberg M, et al. Cutting edge: activation of NK T cells by CD1d and α-galactosylceramide directs conventional T cells to the acquisition of a Th2 phenotype. J Immunol (1999) 163(5):2373–7.
84. Kitamura H, Iwakabe K, Yahata T, Nishimura S, Ohta A, Ohmi Y, et al. The natural killer T (NKT) cell ligand alpha-galactosylceramide demonstrates its immunopotentiating effect by inducing interleukin (IL)-12 production by dendritic cells and IL-12 receptor expression on NKT cells. J Exp Med (1999) 189(7):1121–8. doi:10.1084/jem.189.7.1121
85. Shimizu K, Asakura M, Shinga J, Sato Y, Kitahara S, Hoshino K, et al. Invariant NKT cells induce plasmacytoid dendritic cell (DC) cross-talk with conventional DCs for efficient memory CD8+ T cell induction. J Immunol (2013) 190(11):5609–19. doi:10.4049/jimmunol.1300033
86. Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc Natl Acad Sci U S A (2008) 105(32):11287–92. doi:10.1073/pnas.0801631105
87. Chaudhry MS, Karadimitris A. Role and regulation of CD1d in normal and pathological B cells. J Immunol (2014) 193(10):4761–8. doi:10.4049/jimmunol.1401805
88. Lang GA, Exley MA, Lang ML. The CD1d-binding glycolipid alpha-galactosylceramide enhances humoral immunity to T-dependent and T-independent antigen in a CD1d-dependent manner. Immunology (2006) 119(1):116–25. doi:10.1111/j.1365-2567.2006.02413.x
89. Tonti E, Galli G, Malzone C, Abrignani S, Casorati G, Dellabona P. NKT-cell help to B lymphocytes can occur independently of cognate interaction. Blood (2009) 113(2):370–6. doi:10.1182/blood-2008-06-166249
90. Leadbetter EA, Brigl M, Illarionov P, Cohen N, Luteran MC, Pillai S, et al. NK T cells provide lipid antigen-specific cognate help for B cells. Proc Natl Acad Sci U S A (2008) 105(24):8339–44. doi:10.1073/pnas.0801375105
91. King IL, Fortier A, Tighe M, Dibble J, Watts GF, Veerapen N, et al. Invariant natural killer T cells direct B cell responses to cognate lipid antigen in an IL-21-dependent manner. Nat Immunol (2012) 13(1):44–50. doi:10.1038/ni.2172
92. Barral P, Eckl-Dorna J, Harwood NE, De Santo C, Salio M, Illarionov P, et al. B cell receptor-mediated uptake of CD1d-restricted antigen augments antibody responses by recruiting invariant NKT cell help in vivo. Proc Natl Acad Sci U S A (2008) 105(24):8345–50. doi:10.1073/pnas.0802968105
93. Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol (2012) 13(1):35–43. doi:10.1038/ni.2166
94. Semmling V, Lukacs-Kornek V, Thaiss CA, Quast T, Hochheiser K, Panzer U, et al. Alternative cross-priming through CCL17-CCR4-mediated attraction of CTLs toward NKT cell-licensed DCs. Nat Immunol (2010) 11(4):313–20. doi:10.1038/ni.1848
95. Gilchuk P, Hill TM, Guy C, McMaster SR, Boyd KL, Rabacal WA, et al. A distinct lung-interstitium-resident memory CD8(+) T cell subset confers enhanced protection to lower respiratory tract infection. Cell Rep (2016) 16(7):1800–9. doi:10.1016/j.celrep.2016.07.037
96. Gilchuk P, Spencer CT, Conant SB, Hill T, Gray JJ, Niu X, et al. Discovering naturally processed antigenic determinants that confer protective T cell immunity. J Clin Invest (2013) 123(5):1976–87. doi:10.1172/JCI67388
97. Hermans IF, Silk JD, Gileadi U, Salio M, Mathew B, Ritter G, et al. NKT cells enhance CD4+ and CD8+ T cell responses to soluble antigen in vivo through direct interaction with dendritic cells. J Immunol (2003) 171(10):5140–7. doi:10.4049/jimmunol.171.10.5140
98. Yu KO, Porcelli SA. The diverse functions of CD1d-restricted NKT cells and their potential for immunotherapy. Immunol Lett (2005) 100(1):42–55. doi:10.1016/j.imlet.2005.06.010
99. Wu L, Gabriel CL, Parekh VV, Van Kaer L. Invariant natural killer T cells: innate-like T cells with potent immunomodulatory activities. Tissue Antigens (2009) 73(6):535–45. doi:10.1111/j.1399-0039.2009.01256.x
100. Van Kaer L, Parekh VV, Wu L. Invariant NK T cells: potential for immunotherapeutic targeting with glycolipid antigens. Immunotherapy (2011) 3(1):59–75. doi:10.2217/imt.10.85
101. Singh AK, Gaur P, Das SN. Natural killer T cell anergy, co-stimulatory molecules and immunotherapeutic interventions. Hum Immunol (2014) 75(3):250–60. doi:10.1016/j.humimm.2013.12.004
102. Carreno LJ, Kharkwal SS, Porcelli SA. Optimizing NKT cell ligands as vaccine adjuvants. Immunotherapy (2014) 6(3):309–20. doi:10.2217/imt.13.175
103. Nair S, Dhodapkar MV. Natural killer T cells in cancer immunotherapy. Front Immunol (2017) 8:1178. doi:10.3389/fimmu.2017.01178
104. Waldowska M, Bojarska-Junak A, Roliński J. A brief review of clinical trials involving manipulation of invariant NKT cells as a promising approach in future cancer therapies. Cent Eur J Immunol (2017) 42(2):181–95. doi:10.5114/ceji.2017.69361
105. Bassiri H, Das R, Guan P, Barrett DM, Brennan PJ, Banerjee PP, et al. iNKT cell cytotoxic responses control T-lymphoma growth in vitro and in vivo. Cancer Immunol Res (2014) 2(1):59–69. doi:10.1158/2326-6066.CIR-13-0104
106. Wingender G, Krebs P, Beutler B, Kronenberg M. Antigen-specific cytotoxicity by invariant NKT cells in vivo is CD95/CD178 dependent and is correlated with antigenic potency. J Immunol (2010) 185(5):2721–9. doi:10.4049/jimmunol.1001018
107. Hagihara M, Gansuvd B, Ueda Y, Tsuchiya T, Masui A, Tazume K, et al. Killing activity of human umbilical cord blood-derived TCRValpha24+ NKT cells against normal and malignant hematological cells in vitro: a comparative study with NK cells or OKT3 activated T lymphocytes or with adult peripheral blood NKT cells. Cancer Immunol Immunother (2002) 51(1):1–8. doi:10.1007/s00262-001-0246-2
108. Haraguchi K, Takahashi T, Nakahara F, Matsumoto A, Kurokawa M, Ogawa S, et al. CD1d expression level in tumor cells is an important determinant for anti-tumor immunity by natural killer T cells. Leuk Lymphoma (2006) 47(10):2218–23. doi:10.1080/10428190600682688
109. Crowe NY, Coquet JM, Berzins SP, Kyparissoudis K, Keating R, Pellicci DG, et al. Differential antitumor immunity mediated by NKT cell subsets in vivo. J Exp Med (2005) 202(9):1279–88. doi:10.1084/jem.20050953
110. Keller CW, Freigang S, Lünemann JD. Reciprocal crosstalk between dendritic cells and natural killer T cells: mechanisms and therapeutic potential. Front Immunol (2017) 8:570. doi:10.3389/fimmu.2017.00570
111. Gottschalk C, Mettke E, Kurts C. The role of invariant natural killer T cells in dendritic cell licensing, cross-priming, and memory cd8(+) t cell generation. Front Immunol (2015) 6:379. doi:10.3389/fimmu.2015.00379
112. Chung Y, Kim BS, Kim YJ, Ko HJ, Ko SY, Kim DH, et al. CD1d-restricted T cells license B cells to generate long-lasting cytotoxic antitumor immunity in vivo. Cancer Res (2006) 66(13):6843–50. doi:10.1158/0008-5472.CAN-06-0889
113. Kim Y-J, Ko HJ, Kim YS, Kim DH, Kang S, Kim JM, et al. α-Galactosylceramide-loaded, antigen-expressing B cells prime a wide spectrum of antitumor immunity. Int J Cancer (2008) 122(12):2774–83. doi:10.1002/ijc.23444
114. Iyoda T, Yamasaki S, Hidaka M, Kawano F, Abe Y, Suzuki K, et al. Amelioration of NK cell function driven by Vα24+ invariant NKT cell activation in multiple myeloma. Clin Immunol (2017). doi:10.1016/j.clim.2017.10.007
115. Fujii S, Shimizu K, Okamoto Y, Kunii N, Nakayama T, Motohashi S, et al. NKT cells as an ideal anti-tumor immunotherapeutic. Front Immunol (2013) 4:409. doi:10.3389/fimmu.2013.00409
116. Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, et al. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science (1997) 278(5343):1623–6. doi:10.1126/science.278.5343.1623
117. Kammertoens T, Qin Z, Briesemeister D, Bendelac A, Blankenstein T. B-cells and IL-4 promote methylcholanthrene-induced carcinogenesis but there is no evidence for a role of T/NKT-cells and their effector molecules (Fas-ligand, TNF-alpha, perforin). Int J Cancer (2012) 131(7):1499–508. doi:10.1002/ijc.27411
118. Smyth MJ, Thia KY, Street SE, Cretney E, Trapani JA, Taniguchi M, et al. Differential tumor surveillance by natural killer (NK) and NKT cells. J Exp Med (2000) 191(4):661–8. doi:10.1084/jem.191.4.661
119. Motohashi S, Nagato K, Kunii N, Yamamoto H, Yamasaki K, Okita K, et al. A phase I-II study of alpha-galactosylceramide-pulsed IL-2/GM-CSF-cultured peripheral blood mononuclear cells in patients with advanced and recurrent non-small cell lung cancer. J Immunol (2009) 182(4):2492–501. doi:10.4049/jimmunol.0800126
120. Kunii N, Horiguchi S, Motohashi S, Yamamoto H, Ueno N, Yamamoto S, et al. Combination therapy of in vitro-expanded natural killer T cells and alpha-galactosylceramide-pulsed antigen-presenting cells in patients with recurrent head and neck carcinoma. Cancer Sci (2009) 100(6):1092–8. doi:10.1111/j.1349-7006.2009.01135.x
121. Lam PY, Nissen MD, Mattarollo SR. Invariant natural killer T cells in immune regulation of blood cancers: harnessing their potential in immunotherapies. Front Immunol (2017) 8:1355. doi:10.3389/fimmu.2017.01355
122. Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med (1995) 182(6):2091–6. doi:10.1084/jem.182.6.2091
123. Coles MC, Raulet DH. NK1.1+ T cells in the liver arise in the thymus and are selected by interactions with class I molecules on CD4+CD8+ cells. J Immunol (2000) 164(5):2412–8. doi:10.4049/jimmunol.164.5.2412
124. Boesteanu A, Silva AD, Nakajima H, Leonard WJ, Peschon JJ, Joyce S. Distinct roles for signals relayed through the common cytokine receptor gamma chain and interleukin 7 receptor alpha chain in natural T cell development. J Exp Med (1997) 186(2):331–6. doi:10.1084/jem.186.2.331
125. Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med (2000) 191(11):1895–903. doi:10.1084/jem.191.11.1895
126. Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med (2000) 192(5):741–54. doi:10.1084/jem.192.5.741
127. Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nat Rev Immunol (2004) 4(3):231–7. doi:10.1038/nri1309
128. Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science (2002) 296(5567):553–5. doi:10.1126/science.1069017
129. Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J Exp Med (2002) 195(7):835–44. doi:10.1084/jem.20011544
130. Bezbradica JS, Hill T, Stanic AK, Van Kaer L, Joyce S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc Natl Acad Sci U S A (2005) 102(14):5114–9. doi:10.1073/pnas.0408449102
131. D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol (2010) 11(3):240–9. doi:10.1038/ni.1845
132. Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity (2005) 22(6):705–16. doi:10.1016/j.immuni.2005.03.011
133. Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol (2010) 11(5):435–41. doi:10.1038/ni.1865
134. Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol (2001) 2(10):971–8. doi:10.1038/ni710
135. Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol (2007) 7(7):505–18. doi:10.1038/nri2116
136. Dashtsoodol N, Shigeura T, Aihara M, Ozawa R, Kojo S, Harada M, et al. Alternative pathway for the development of Valpha14(+) NKT cells directly from CD4(-)CD8(-) thymocytes that bypasses the CD4(+)CD8(+) stage. Nat Immunol (2017) 18(3):274–82. doi:10.1038/ni.3668
137. Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity (1997) 6(4):459–67. doi:10.1016/S1074-7613(00)80289-7
138. Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity (1997) 6(4):469–77. doi:10.1016/S1074-7613(00)80290-3
139. Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4-secreting CD1-dependent cells. Science (1997) 275(5302):977–9. doi:10.1126/science.275.5302.977
140. Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity (2007) 27(5):751–62. doi:10.1016/j.immuni.2007.08.020
141. Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, et al. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med (2004) 199(9):1255–64. doi:10.1084/jem.20031835
142. Jordan MA, Fletcher JM, Jose R, Chowdhury S, Gerlach N, Allison J, et al. Role of SLAM in NKT cell development revealed by transgenic complementation in NOD mice. J Immunol (2011) 186(7):3953–65. doi:10.4049/jimmunol.1003305
143. Stanic AK, Bezbradica JS, Park JJ, Van Kaer L, Boothby MR, Joyce S. Cutting edge: the ontogeny and function of Va14Ja18 natural T lymphocytes require signal processing by protein kinase C theta and NF-kappa B. J Immunol (2004) 172(8):4667–71. doi:10.4049/jimmunol.172.8.4667
144. Lazarevic V, Zullo AJ, Schweitzer MN, Staton TL, Gallo EM, Crabtree GR, et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol (2009) 10(3):306–13. doi:10.1038/ni.1696
145. Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol (2012) 13(3):264–71. doi:10.1038/ni.2230
146. Qi Q, Kannan AK, August A. Tec family kinases: Itk signaling and the development of NKT alphabeta and gammadelta T cells. FEBS J (2011) 278(12):1970–9. doi:10.1111/j.1742-4658.2011.08074.x
147. Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med (2005) 11(3):340–5. doi:10.1038/nm1189
148. Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol (2010) 11(3):197–206. doi:10.1038/ni.1841
149. Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity (2008) 29(3):391–403. doi:10.1016/j.immuni.2008.07.011
150. Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol (2008) 9(9):1055–64. doi:10.1038/ni.1641
151. Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, et al. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol (2012) 10(2):e1001255. doi:10.1371/journal.pbio.1001255
152. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol (2013) 14(11):1146–54. doi:10.1038/ni.2731
153. Hammond KJ, Pellicci DG, Poulton LD, Naidenko OV, Scalzo AA, Baxter AG, et al. CD1d-restricted NKT cells: an interstrain comparison. J Immunol (2001) 167(3):1164–73. doi:10.4049/jimmunol.167.3.1164
154. Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of Treg cells and macrophages in adipose tissue. Nat Immunol (2015) 16(1):85–95. doi:10.1038/ni.3047
155. Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest (2014) 124(9):3725–40. doi:10.1172/JCI72308
156. Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, et al. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol (2016) 17(6):728–39. doi:10.1038/ni.3437
157. Peter I, Davidson E. Genomic Control Process: Development and Evolution. London: Academic Press (2015).
158. Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, et al. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc Natl Acad Sci U S A (2016) 113(27):7602–7. doi:10.1073/pnas.1601504113
159. Wang Y, Yun C, Gao B, Xu Y, Zhang Y, Wang Y, et al. The lysine acetyltransferase GCN5 is required for iNKT cell development through EGR2 acetylation. Cell Rep (2017) 20(3):600–12. doi:10.1016/j.celrep.2017.06.065
160. Dominy JE Jr, Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell (2012) 48(6):900–13. doi:10.1016/j.molcel.2012.09.030
161. Mallevaey T, Scott-Browne JP, Matsuda JL, Young MH, Pellicci DG, Patel O, et al. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity (2009) 31(1):60–71. doi:10.1016/j.immuni.2009.05.010
162. Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. Mechanisms imposing the Vbeta bias of Valpha14 natural killer T cells and consequences for microbial glycolipid recognition. J Exp Med (2006) 203(5):1197–207. doi:10.1084/jem.20060418
163. Chun T, et al. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J Exp Med (2003) 197(7):907–18. doi:10.1084/jem.20021366
164. Pellicci DG, Uldrich AP, Kyparissoudis K, Crowe NY, Brooks AG, Hammond KJ, et al. Intrathymic NKT cell development is blocked by the presence of alpha-galactosylceramide. Eur J Immunol (2003) 33(7):1816–23. doi:10.1002/eji.200323894
165. Schümann J, Pittoni P, Tonti E, Macdonald HR, Dellabona P, Casorati G. Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Valpha14i NKT cells. J Immunol (2005) 175(11):7303–10. doi:10.4049/jimmunol.175.11.7303
166. Stanic AK, Bezbradica JS, Park JJ, Matsuki N, Mora AL, Van Kaer L, et al. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J Immunol (2004) 172(4):2265–73. doi:10.4049/jimmunol.172.4.2265
167. Chiu YH, Park SH, Benlagha K, Forestier C, Jayawardena-Wolf J, Savage PB, et al. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat Immunol (2002) 3(1):55–60. doi:10.1038/ni740
168. Thapa P, Das J, McWilliams D, Shapiro M, Sundsbak R, Nelson-Holte M, et al. The transcriptional repressor NKAP is required for the development of iNKT cells. Nat Commun (2013) 4:1582. doi:10.1038/ncomms2580
169. Pajerowski AG, Nguyen C, Aghajanian H, Shapiro MJ, Shapiro VS. NKAP is a transcriptional repressor of notch signaling and is required for T cell development. Immunity (2009) 30(5):696–707. doi:10.1016/j.immuni.2009.02.011
170. Napolitano A, Pittoni P, Beaudoin L, Lehuen A, Voehringer D, MacDonald HR, et al. Functional education of invariant NKT cells by dendritic cell tuning of SHP-1. J Immunol (2013) 190(7):3299–308. doi:10.4049/jimmunol.1203466
171. Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A, et al. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med (2005) 202(2):239–48. doi:10.1084/jem.20050413
172. Cannons JL, Yu LJ, Hill B, Mijares LA, Dombroski D, Nichols KE, et al. SAP regulates TH2 differentiation and PKC-θ-mediated activation of NF-κB1. Immunity (2004) 21(5):693–706. doi:10.1016/j.immuni.2004.09.012
173. Huang B, Gomez-Rodriguez J, Preite S, Garrett LJ, Harper UL, Schwartzberg PL. CRISPR-mediated triple knockout of SLAMF1, SLAMF5 and SLAMF6 supports positive signaling roles in NKT cell development. PLoS One (2016) 11(6):e0156072. doi:10.1371/journal.pone.0156072
174. Chen S, Cai C, Li Z, Liu G, Wang Y, Blonska M, et al. Dissection of SAP-dependent and SAP-independent SLAM family signaling in NKT cell development and humoral immunity. J Exp Med (2017) 214(2):475–89. doi:10.1084/jem.20161312
175. Michel ML, Lenoir C, Massot B, Diem S, Pasquier B, Sawa S, et al. SLAM-associated protein favors the development of iNKT2 over iNKT17 cells. Eur J Immunol (2016) 46(9):2162–74. doi:10.1002/eji.201646313
176. Vallabhapurapu S, Hammond KJL, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor κB family members in natural killer T cell development. J Exp Med (2003) 197(12):1613–21. doi:10.1084/jem.20022234
177. Vallabhapurapu S, Powolny-Budnicka I, Riemann M, Schmid RM, Paxian S, Pfeffer K, et al. Rel/NF-kappaB family member RelA regulates NK1.1- to NK1.1+ transition as well as IL-15-induced expansion of NKT cells. Eur J Immunol (2008) 38(12):3508–19. doi:10.1002/eji.200737830
178. Elewaut D, Shaikh RB, Hammond KJ, De Winter H, Leishman AJ, Sidobre S, et al. NIK-dependent RelB activation defines a unique signaling pathway for the development of V alpha 14i NKT cells. J Exp Med (2003) 197(12):1623–33. doi:10.1084/jem.20030141
179. Schmidt-Supprian M, Tian J, Grant EP, Pasparakis M, Maehr R, Ovaa H, et al. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc Natl Acad Sci U S A (2004) 101(13):4566–71. doi:10.1073/pnas.0400885101
180. Stankovic S, Gugasyan R, Kyparissoudis K, Grumont R, Banerjee A, Tsichlis P, et al. Distinct roles in NKT cell maturation and function for the different transcription factors in the classical NF-kappaB pathway. Immunol Cell Biol (2011) 89(2):294–303. doi:10.1038/icb.2010.93
181. Lee AJ, Zhou X, Chang M, Hunzeker J, Bonneau RH, Zhou D, et al. Regulation of natural killer T-cell development by deubiquitinase CYLD. EMBO J (2010) 29(9):1600–12. doi:10.1038/emboj.2010.31
182. Qi Q, Huang W, Bai Y, Balmus G, Weiss RS, August A. A unique role for ITK in survival of invariant NKT cells associated with the p53-dependent pathway in mice. J Immunol (2012) 188(8):3611–9. doi:10.4049/jimmunol.1102475
183. Kumar A, Gordy LE, Bezbradica JS, Stanic AK, Hill TM, Boothby MR, et al. NF-kappaB protects NKT cells from tumor necrosis factor receptor 1-induced death. Sci Rep (2017) 7(1):15594. doi:10.1038/s41598-017-15461-y
184. Crawford G, Enders A, Gileadi U, Stankovic S, Zhang Q, Lambe T, et al. DOCK8 is critical for the survival and function of NKT cells. Blood (2013) 122(12):2052–61. doi:10.1182/blood-2013-02-482331
185. Okamura K, Kitamura A, Sasaki Y, Chung DH, Kagami S, Iwai K, et al. Survival of mature T cells depends on signaling through HOIP. Sci Rep (2016) 6:36135. doi:10.1038/srep36135
186. Lan P, Fan Y, Zhao Y, Lou X, Monsour HP, Zhang X, et al. TNF superfamily receptor OX40 triggers invariant NKT cell pyroptosis and liver injury. J Clin Invest (2017) 127(6):2222–34. doi:10.1172/JCI91075
187. Gordy LE, Bezbradica JS, Flyak AI, Spencer CT, Dunkle A, Sun J, et al. IL-15 regulates homeostasis and terminal maturation of NKT cells. J Immunol (2011) 187(12):6335–45. doi:10.4049/jimmunol.1003965
188. Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, et al. Homeostasis of V[alpha]14i NKT cells. Nat Immunol (2002) 3(10):966–74. doi:10.1038/ni837
189. Castillo EF, Acero LF, Stonier SW, Zhou D, Schluns KS. Thymic and peripheral microenvironments differentially mediate development and maturation of iNKT cells by IL-15 transpresentation. Blood (2010) 116(14):2494–503. doi:10.1182/blood-2010-03-277103
190. Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, et al. Reversible defects in natural killer and memory Cd8 T cell lineages in interleukin 15–deficient mice. J Exp Med (2000) 191(5):771–80. doi:10.1084/jem.191.5.771
191. Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, et al. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity (1998) 9(5):669–76. doi:10.1016/S1074-7613(00)80664-0
192. Anderson CK, Salter AI, Toussaint LE, Reilly EC, Fugère C, Srivastava N, et al. Role of SHIP1 in invariant NKT cell development and functions. J Immunol (2015) 195(5):2149–56. doi:10.4049/jimmunol.1500567
193. Parekh VV, Wu L, Boyd KL, Williams JA, Gaddy JA, Olivares-Villagómez D, et al. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J Immunol (2013) 190(10):5086–101. doi:10.4049/jimmunol.1202071
194. Pei B, Zhao M, Miller BC, Véla JL, Bruinsma MW, Virgin HW, et al. Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation. J Immunol (2015) 194(12):5872–84. doi:10.4049/jimmunol.1402154
195. Salio M, Puleston DJ, Mathan TS, Shepherd D, Stranks AJ, Adamopoulou E, et al. Essential role for autophagy during invariant NKT cell development. Proc Natl Acad Sci U S A (2014) 111(52):E5678–87. doi:10.1073/pnas.1413935112
196. Thapa P, Romero Arocha S, Chung JY, Sant’Angelo DB, Shapiro VS. Histone deacetylase 3 is required for iNKT cell development. Sci Rep (2017) 7(1):5784. doi:10.1038/s41598-017-06102-5
197. Terashima A, Watarai H, Inoue S, Sekine E, Nakagawa R, Hase K, et al. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. J Exp Med (2008) 205(12):2727–33. doi:10.1084/jem.20080698
198. Matangkasombut P, Marigowda G, Ervine A, Idris L, Pichavant M, Kim HY, et al. Natural killer T cells in the lungs of patients with asthma. J Allergy Clin Immunol (2009) 123(5):1181–5. doi:10.1016/j.jaci.2009.02.013
199. Kim HY, Pichavant M, Matangkasombut P, Koh YI, Savage PB, DeKruyff RH, et al. The development of airway hyperreactivity in T-bet-deficient mice requires CD1d-restricted NKT cells. J Immunol (2009) 182(5):3252–61. doi:10.4049/jimmunol.0803339
200. Akbari O, Stock P, Meyer E, Kronenberg M, Sidobre S, Nakayama T, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med (2003) 9:582. doi:10.1038/nm851
201. Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, et al. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med (2003) 198(7):1069–76. doi:10.1084/jem.20030630
202. Tanaka S, Tsukada J, Suzuki W, Hayashi K, Tanigaki K, Tsuji M, et al. The interleukin-4 enhancer CNS-2 is regulated by Notch signals and controls initial expression in NKT cells and memory-type CD4 T cells. Immunity (2006) 24(6):689–701. doi:10.1016/j.immuni.2006.04.009
203. Collins PL, Chang S, Henderson M, Soutto M, Davis GM, McLoed AG, et al. Distal regions of the human IFNG locus direct cell type-specific expression. J Immunol (2010) 185(3):1492–501. doi:10.4049/jimmunol.1000124
204. Collins PL, Henderson MA, Aune TM. Lineage-specific adjacent IFNG and IL26 genes share a common distal enhancer element. Genes Immun (2012) 13(6):481–8. doi:10.1038/gene.2012.22
205. Wang X, Bishop KA, Hegde S, Rodenkirch LA, Pike JW, Gumperz JE. Human invariant natural killer T cells acquire transient innate responsiveness via histone H4 acetylation induced by weak TCR stimulation. J Exp Med (2012) 209(5):987–1000. doi:10.1084/jem.20111024
206. Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, et al. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med (2007) 204(5):995–1001. doi:10.1084/jem.20061551
207. Doisne JM, Becourt C, Amniai L, Duarte N, Le Luduec JB, Eberl G, et al. Skin and peripheral lymph node invariant NKT cells are mainly retinoic acid receptor-related orphan receptor (gamma)t+ and respond preferentially under inflammatory conditions. J Immunol (2009) 183(3):2142–9. doi:10.4049/jimmunol.0901059
208. Clancy-Thompson E, Chen GZ, Tyler PM, Servos MM, Barisa M, Brennan PJ, et al. Monoclonal invariant NKT (iNKT) cell mice reveal a role for both tissue of origin and the TCR in development of iNKT functional subsets. J Immunol (2017) 199(1):159–71. doi:10.4049/jimmunol.1700214
209. Webster KE, Kim HO, Kyparissoudis K, Corpuz TM, Pinget GV, Uldrich AP, et al. IL-17-producing NKT cells depend exclusively on IL-7 for homeostasis and survival. Mucosal Immunol (2014) 7(5):1058–67. doi:10.1038/mi.2013.122
210. Thapa P, Chen MW, McWilliams DC, Belmonte P, Constans M, Sant’Angelo DB, et al. NKAP regulates invariant NKT cell proliferation and differentiation into ROR-gammat-expressing NKT17 cells. J Immunol (2016) 196(12):4987–98. doi:10.4049/jimmunol.1501653
211. Thapa P, Manso B, Chung JY, Romera Arocha S, Xue HH, Angelo DBS, et al. The differentiation of ROR-gammat expressing iNKT17 cells is orchestrated by Runx1. Sci Rep (2017) 7(1):7018. doi:10.1038/s41598-017-07365-8
212. Sklarz T, Guan P, Gohil M, Cotton RM, Ge MQ, Haczku A, et al. mTORC2 regulates multiple aspects of NKT-cell development and function. Eur J Immunol (2017) 47(3):516–26. doi:10.1002/eji.201646343
213. Wei J, Yang K, Chi H. Cutting edge: discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. J Immunol (2014) 193(9):4297–301. doi:10.4049/jimmunol.1402042
214. Price AE, Reinhardt RL, Liang HE, Locksley RM. Marking and quantifying IL-17A-producing cells in vivo. PLoS One (2012) 7(6):e39750. doi:10.1371/journal.pone.0039750
215. Pichavant M, Goya S, Meyer EH, Johnston RA, Kim HY, Matangkasombut P, et al. Ozone exposure in a mouse model induces airway hyperreactivity that requires the presence of natural killer T cells and IL-17. J Exp Med (2008) 205(2):385–93. doi:10.1084/jem.20071507
216. Milosavljevic N, Gazdic M, Simovic Markovic B, Arsenijevic A, Nurkovic J, Dolicanin Z, et al. Mesenchymal stem cells attenuate acute liver injury by altering ratio between interleukin 17 producing and regulatory natural killer T cells. Liver Transpl (2017) 23(8):1040–50. doi:10.1002/lt.24784
217. Galli G, Pittoni P, Tonti E, Malzone C, Uematsu Y, Tortoli M, et al. Invariant NKT cells sustain specific B cell responses and memory. Proc Natl Acad Sci U S A (2007) 104(10):3984–9. doi:10.1073/pnas.0700191104
218. Tangye SG, Ma CS, Brink R, Deenick EK. The good, the bad and the ugly – TFH cells in human health and disease. Nat Rev Immunol (2013) 13(6):412–26. doi:10.1038/nri3447
219. Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol (2011) 13(1):35–43. doi:10.1038/ni.2166
220. Tonti E, Fedeli M, Napolitano A, Iannacone M, von Andrian UH, Guidotti LG, et al. Follicular helper NKT cells induce limited B cell responses and germinal center formation in the absence of CD4(+) T cell help. J Immunol (2012) 188(7):3217–22. doi:10.4049/jimmunol.1103501
221. Chan AC, Leeansyah E, Cochrane A, d’Udekem d’Acoz Y, Mittag D, Harrison LC, et al. Ex-vivo analysis of human natural killer T cells demonstrates heterogeneity between tissues and within established CD4(+) and CD4(-) subsets. Clin Exp Immunol (2013) 172(1):129–37. doi:10.1111/cei.12045
222. Akbari O, Faul JL, Hoyte EG, Berry GJ, Wahlström J, Kronenberg M, et al. CD4+ invariant T-cell-receptor+ natural killer T cells in bronchial asthma. N Engl J Med (2006) 354(11):1117–29. doi:10.1056/NEJMoa053614
223. Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med (2002) 195(5):625–36. doi:10.1084/jem.20011786
224. Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human V(alpha)24 natural killer T cells. J Exp Med (2002) 195(5):637–41. doi:10.1084/jem.20011908
225. Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, Kang J, et al. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol (2013) 14(1):90–9. doi:10.1038/ni.2490
226. Tsagaratou A, González-Avalos E, Rautio S, Scott-Browne JP, Togher S, Pastor WA, et al. TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat Immunol (2017) 18(1):45–53. doi:10.1038/ni.3630
228. Lee YJ, Wang H, Starrett GJ, Phuong V, Jameson SC, Hogquist KA. Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity (2015) 43(3):566–78. doi:10.1016/j.immuni.2015.06.025
229. Matsuki N, Stanic AK, Embers ME, Van Kaer L, Morel L, Joyce S. Genetic dissection of V alpha 14J alpha 18 natural T cell number and function in autoimmune-prone mice. J Immunol (2003) 170(11):5429–37. doi:10.4049/jimmunol.170.11.5429
230. Poulton LD, Smyth MJ, Hawke CG, Silveira P, Shepherd D, Naidenko OV, et al. Cytometric and functional analyses of NK and NKT cell deficiencies in NOD mice. Int Immunol (2001) 13(7):887–96. doi:10.1093/intimm/13.7.887
231. Lee PT, Putnam A, Benlagha K, Teyton L, Gottlieb PA, Bendelac A. Testing the NKT cell hypothesis of human IDDM pathogenesis. J Clin Invest (2002) 110(6):793–800. doi:10.1172/JCI0215832
232. Motsinger A, Haas DW, Stanic AK, Van Kaer L, Joyce S, Unutmaz D. CD1d-restricted human natural killer T cells are highly susceptible to human immunodeficiency virus 1 infection. J Exp Med (2002) 195(7):869–79. doi:10.1084/jem.20011712
233. Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology (2012) 143(2):418–28. doi:10.1053/j.gastro.2012.04.017
234. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science (2012) 336(6080):489–93. doi:10.1126/science.1219328
235. Zeissig S, Blumberg RS. Commensal microbiota and NKT cells in the control of inflammatory diseases at mucosal surfaces. Curr Opin Immunol (2013) 25(6):690–6. doi:10.1016/j.coi.2013.09.012
236. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486(7402):207–14. doi:10.1038/nature11234
237. Sultan SE. Organism & Environment: Ecological Development, Niche Construction, and Adaptation. Oxford, UK: Oxford University Press (2015).
238. Selvanantham T, Lin Q, Guo CX, Surendra A, Fieve S, Escalante NK, et al. NKT cell-deficient mice harbor an altered microbiota that fuels intestinal inflammation during chemically induced colitis. J Immunol (2016) 197(11):4464–72. doi:10.4049/jimmunol.1601410
239. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer (2017) 17(5):271–85. doi:10.1038/nrc.2017.13
241. Dronamraju KR, editor. Selected Genetic Papers of J.B.S. Haldane. New York/London: Garland Publishing (1990). 542 p.
242. Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol (2015) 33:169–200. doi:10.1146/annurev-immunol-032414-112334
243. Yang Z, Wang C, Wang T, Bai J, Zhao Y, Liu X, et al. Analysis of the reptile CD1 genes: evolutionary implications. Immunogenetics (2015) 67(5–6):337–46. doi:10.1007/s00251-015-0837-2
244. Boudinot P, Mondot S, Jouneau L, Teyton L, Lefranc MP, Lantz O. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc Natl Acad Sci U S A (2016) 113(21):E2983–92. doi:10.1073/pnas.1600674113
245. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol (1987) 4(4):406–25.
246. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution (1985) 39(4):783–91. doi:10.1111/j.1558-5646.1985.tb00420.x
247. Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press (2000).
248. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol (2013) 30(12):2725–9. doi:10.1093/molbev/mst197
249. Wund M, Myers P. The Animal Diversity Web. (2017). Available from: http://animaldiversity.org/accounts/Mammalia/
250. Corbert G, Hill J. A World List of Mammalian Species. New York/London: Facts on File Publications/British Museum of Natural History (1991).
252. Louis A, Nguyen NT, Muffato M, Roest Crollius H. Genomicus update 2015: KaryoView and MatrixView provide a genome-wide perspective to multispecies comparative genomics. Nucleic Acids Res (2015) 43(Database issue):D682–9. doi:10.1093/nar/gku1112
253. Alonzo ES, Sant’Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol (2011) 23(2):220–7. doi:10.1016/j.coi.2010.12.016
254. Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol (2013) 25(2):161–7. doi:10.1016/j.coi.2013.01.003
255. Ishizuka IE, Constantinides MG, Gudjonson H, Bendelac A. The innate lymphoid cell precursor. Annu Rev Immunol (2016) 34:299–316. doi:10.1146/annurev-immunol-041015-055549
256. Kastenmuller W, Torabi-Parizi P, Subramanian N, Lammermann T, Germain RN. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell (2012) 150(6):1235–48. doi:10.1016/j.cell.2012.07.021
257. Van Kaer L, Algood HMS, Singh K, Parekh VV, Greer MJ, Piazuelo MB, et al. CD8alphaalpha(+) innate-type lymphocytes in the intestinal epithelium mediate mucosal immunity. Immunity (2014) 41(3):451–64. doi:10.1016/j.immuni.2014.08.010
258. Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: innate B and T lymphocytes. Nat Rev Immunol (2001) 1(3):177–86. doi:10.1038/35105052
Keywords: NKT cells, cancer immunotherapy, microbiota, infectious diseases, evolution
Citation: Kumar A, Suryadevara N, Hill TM, Bezbradica JS, Van Kaer L and Joyce S (2017) Natural Killer T Cells: An Ecological Evolutionary Developmental Biology Perspective. Front. Immunol. 8:1858. doi: 10.3389/fimmu.2017.01858
Received: 16 October 2017; Accepted: 07 December 2017;
Published: 22 December 2017
Edited by:
Paolo Dellabona, Scientific Institute San Raffaele (IRCCS), ItalyReviewed by:
Graham Robert Leggatt, The University of Queensland, AustraliaCopyright: © 2017 Kumar, Suryadevara, Hill, Bezbradica, Van Kaer and Joyce. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian Joyce, c2ViYXN0aWFuLmpveWNlQHZhbmRlcmJpbHQuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.