
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 14 November 2017
Sec. NK and Innate Lymphoid Cell Biology
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.01571
This article is part of the Research TopicDifferentiation and Mechanisms of Activation of Innate Lymphoid CellsView all 11 articles
Innate lymphoid cells (ILCs) are regarded as the innate counterpart of effector CD4 T helper (Th) cells. Just as Th cells, ILCs are classified into distinct subsets based on their functions that are delivered mainly through effector cytokine production. Both ILCs and Th cells play critical roles in various protective immune responses and inflammatory diseases. Similar to Th cell differentiation, the development of ILC subsets depends on several master transcription factors, among which GATA3 is critical for the development and maintenance of type 2 ILCs (ILC2s). However, GATA3 is expressed by all ILC subsets and ILC progenitors, albeit at different levels. In a striking parallel with GATA3 function in T cell development and differentiation, GATA3 also has multiple functions in different ILCs at various stages. In this review, I will discuss how quantitative and dynamic expression of GATA3 regulates the development and functions of ILC subsets.
Innate lymphoid cells (ILCs) are innate counterparts of CD4 T helper (Th) cells, which are considered professional cytokine-producing cells during adaptive immune responses. Based on their cytokine production and functions, just as Th cells, ILCs are divided into at least three major groups—group 1 ILC (ILC1), group 2 ILC (ILC2), and group 3 ILC (ILC3) subsets (1–4). ILC1s mainly produce IFN-γ; ILC2s produce IL-5 and IL-13; whereas ILC3s primarily produce IL-22. T-bet, GATA3, and RORγt are the master transcription factors for the development and functions of Th1, Th2, and Th17 (IL-17-producing Th) cells, respectively (5). Similarly, these master regulators are also critical for the development of ILC1s, ILC2s, and ILC3s, respectively (Figure 1). Natural killer (NK) cells are considered as the innate counterpart of CD8 T cells since both have cytolytic activity and depend on the transcription factor Eomes for their development.
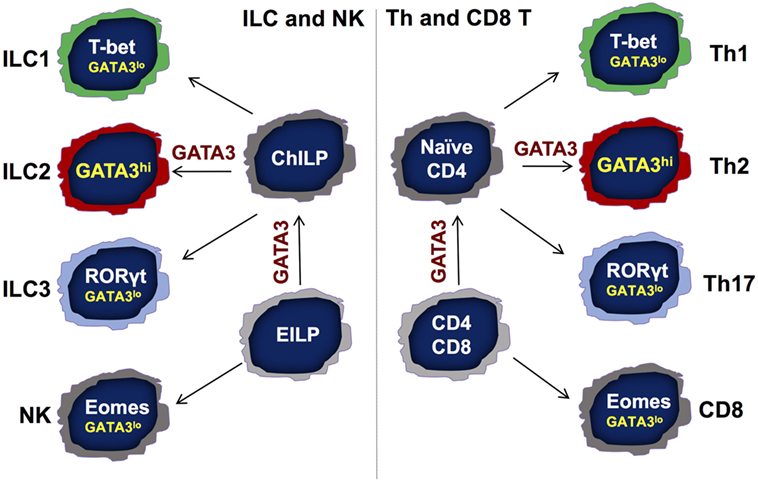
Figure 1. Symmetry between the development of adaptive and innate lymphocytes. There are following three major T helper (Th) subsets: Th1, Th2, and Th17 cells. Transcription factors T-bet, GATA3, and RORγt are the master regulators for the differentiation of Th1, Th2, and Th17 cells, respectively, from naïve CD4 T cells. Th2 cells express high levels of GATA3 that is critical for the development and functional maintenance of Th2 cells. The importance of low GATA3 expression in Th1 and Th17 cells is not clear. During T cell development, GATA3 is critical for the development of CD4 but not CD8 T cells in the thymus. Similarly, GATA3 is indispensable for the development of all IL-7Rα-expressing helper-like innate lymphoid cells (ILCs) but not natural killer (NK) cells. While GATA3 is critical for the functional maintenance of ILC2s, it is also required for the homeostasis, function, and further maturation of other ILCs, including ILC1s and ILC3s. Although GATA3 is not necessary for the development of NK and CD8 T cells, GATA3 expression at low levels contributes to the homeostasis and maturation of these cells. EILP, early ILC progenitor that expresses TCF7; ChILP, common helper-like ILC progenitor that expresses Id2.
Genome-wide analyses indicate that, in addition to the similarity in their transcriptomes, the epigenome of a given ILC subset is very similar to that of the Th counterpart especially at the cytokine loci (6, 7). Because of their similar capacity in producing effector cytokines (6–8), same class of ILC and Th subset may participate in specific type of immune responses (1, 6–11). Like Th1 cells, ILC1s may be involved in protective immunity against intracellular pathogens (12). Similar to Th2 cells, ILC2s are involved in type 2 immune responses to helminth infections (13–16). ILC3s and Th17 cells are important in dealing with infections of extracellular bacteria and fungi (17, 18). While ILC2s contribute to allergic inflammation (19–23), activation and expansion of ILC1s and ILC3s may induce certain types of autoimmunity (24). Interestingly, ILC activation may be sufficient to induce inflammation or to mount an effective immune response to infections in the absence of T cells under certain conditions (16, 25). Among the ILC3s, there is a distinct subset that expresses chemokine receptor CCR6 in mice (26). These mouse CCR6+ ILC3s represent lymphoid tissue inducer (LTi) or LTi-like cells that play a critical role in the development of lymphoid tissues including lymph nodes and Peyer’s patches (27, 28).
T helper and ILC subsets also differ from each other in many aspects. Because ILCs lack antigen receptors, they mainly respond to inflammatory cytokines to produce their own effector cytokines; IL-1 and IL-12 family cytokines including IL-1, IL-18, IL-33, and IL-23, as well as IL-25, are the major players in activating ILC subsets (1, 29, 30). Although Th cells mainly respond to antigens through activation of their T cell receptors, they can also respond to inflammatory cytokines in an antigen-independent manner, and these cytokines may play an important role in the maturation of Th cells (31, 32). Since antigen-specific Th cells need to be activated and expanded during an immune response while ILCs preexist in the tissue (33, 34), ILCs provide first line of host defense.
Another striking difference between Th cells and ILCs in their development is their dependence on cytokine signaling. While Th cell differentiation critically depends on cytokine-mediated activation of signal transducer and activator of transcription (STAT) family proteins (35), many STATs including STAT3, STAT4, and STAT6 are not required for ILC development (36–38). However, the response of mature ILCs to cytokines may still require the activation of STAT proteins (36–38). Nevertheless, STAT proteins that are important for cell proliferation and survival, especially STAT5 proteins (STAT5A and STAT5B), are important for the homeostasis of both Th cells and ILCs (39). In fact, ILCs are largely absent in common gamma chain (γc)-deficient animals and STAT5 is a major downstream molecule activated by cytokines that utilize γc (40).
Id2 plays an important role in ILC development but has a minimal effect on T cell development (12, 26, 41, 42). Some Id2-expressing progenitors in the bone marrow represent common helper-like ILC progenitors (12). On the other hand, the essential factor for T cell development Bcl11b only controls the development of ILC2s but not of other ILCs (26, 41, 43–47). Nevertheless, the development of ILC and Th cell subsets depends on many shared transcription factors such as TCF7, Tox, and GATA3 (8, 41, 48–50). Some TCF7-expressing cells in the bone marrow represent early ILC progenitor with a potential to become either NK cells or helper-like ILCs (48). Possibly because LEF1 (a TCF7 family member) (51) and Tox2 (52–54) are also expressed by ILCs and T cells, the blockage of ILC and T cell development in TCF7- or Tox-deficient animals is incomplete. On the other hand, since GATA3 is the only GATA protein expressed in T cells and ILCs, GATA3 is absolutely required for the development of T cells and ILCs (8, 55). In mature ILCs, while ILC2s express high levels of GATA3, all other ILCs express low levels of GATA3. In this review, I will discuss the functions of quantitative GATA3 expression in different ILC subsets and ILC progenitors in parallel with its important functions in the development and functions of T cell subsets.
ILC2s are enriched in the gut, lung, skin, and adipose tissues with few of them found in lymphoid tissues and in blood, and all the ILC2s express high levels of GATA3 (1, 13, 20). Like Th2 cells, ILC2s depend on GATA3 for their development. Even after ILC2s are fully mature, GATA3 remains important for their maintenance and functions, just as the essential role of GATA3 in differentiated Th2 cells (8, 56–64). In fact, GATA3 regulates a common set of important genes in both cell types, including Il5, Il13, Areg, Il1rl1, and Ccr8, which are well-known genes important for type 2 immune responses (8). This may explain identical functions of Th2 cells and ILC2s. Interestingly, the transcriptomes of ILC2s and Th2 cells harvested during helminth infection are remarkably similar (6).
Genome-wide analysis of GATA3 binding through ChIP-Seq shows that GATA3 binds to the Il4/Il13 gene locus at multiple sites in both Th2 cells and ILC2s (65, 66). GATA3 also binds to the Il5 and Il13 promoters to induce their transcription (67, 68). Although the function of GATA3 in regulating epigenetic modifications at cytokine gene loci in ILC2s is not clear, GATA3 plays an important role in chromatin remodeling at the Il4/Il13 locus in Th2 cells (65). GATA3 also directly binds to many genes that are involved in type 2 immune responses including Il1rl1, which encodes the IL-33 receptor subunit T1/ST2 (65, 66) and Il17rb, which encodes IL-25R (8). By regulating IL-33R and IL-25R expression, GATA3 is necessary for mature ILC2s to respond to inflammatory cytokines IL-33 and IL-25. ILC2s die quickly after GATA3 removal (8). It is possible that GATA3-deficient ILC2s fail to respond to cytokines. Alternatively, GATA3 may directly regulate cell proliferation and/or survival related genes in ILC2s (8).
The functions of GATA3 in lymphocyte development and functional regulation are far beyond its critical role in Th2 cells and ILC2s, because all T cells and ILCs express different levels of GATA3 (66, 69, 70). We have previously reported that the development of IL-7Rα-expressing ILCs (or Th-like ILCs) but not of NK cells depends on GATA3 (8). Another report also indicates that ILC3 development requires GATA3 expression (50). This is consistent with the critical function of GATA3 during CD4 but not CD8 cell development (69, 71–73). It also supports the notion that ILC1s are developmentally distinct from conventional NK cells. Therefore, the development of innate (ILC) and adaptive (T cells) lymphocytes is highly symmetrical (Figure 1).
GATA3 promotes IL-7Rα expression in all T cells and ILCs, indicating that this may be a common mechanism through which GATA3 regulates lymphocyte homeostasis (66, 74). GATA3 expressed at low levels is sufficient to bind to the Il7r locus and regulates IL-7Rα expression in all ILCs and T lymphocytes (66, 74); the fact that the GATA3 binding pattern to the Il7r gene in ILC3s is identical to that in ILC2s and Th2 cells suggesting the existence of a high-affinity GATA3 binding site at the Il7r gene (66). However, GATA3-mediated IL-7Rα expression does not explain its critical role in the development of IL-7Rα-expressing ILCs because we have found that IL-7Rα transgene fails to rescue the ILC developmental defect in the absence of GATA3.
It has been reported that the ILC1s, ILC2s, and non-LTi ILC3s are derived from ILC progenitors that express both PLZF (75) and PD-1 (76). These PLZF-expressing progenitors are known as common precursors to ILCs (ILCPs). However, CCR6+ LTi or LTi-like cells do not have a history of PLZF expression according to PLZF-fate-mapping experiments (75). We have previously reported that ILC numbers are dramatically reduced but not absent in the Gata3fl/fl-Vav-Cre conditional knockout mice (Vav-Cre is active at the hematopoietic stem cell stage) (8). Our recent results further demonstrate that LTi or LTi-like cells are the only ILCs that still remain in the GATA3 conditional knockout mice. By contrast, ILC1s, ILC2s, and CCR6− ILC3s are undetectable in these mice. Interestingly, the PLZF-expressing ILCPs express high levels of GATA3 expression whereas the LTi or LTi-like progenitors express low levels of GATA3 in the bone marrow. Strikingly, the PLZF-expressing progenitors are completely lost in the bone marrow of Gata3fl/fl-Vav-Cre mice. The critical function of GATA3 for the generation of PLZF-expressing ILC progenitors is independent of cytokine signaling and/or IL-7Rα regulation since we have observed that these progenitors are present in normal numbers in γc-deficient animals. Thus, GATA3 is a critical regulator for the generation of PLZF-expressing ILC progenitors (Figure 2).
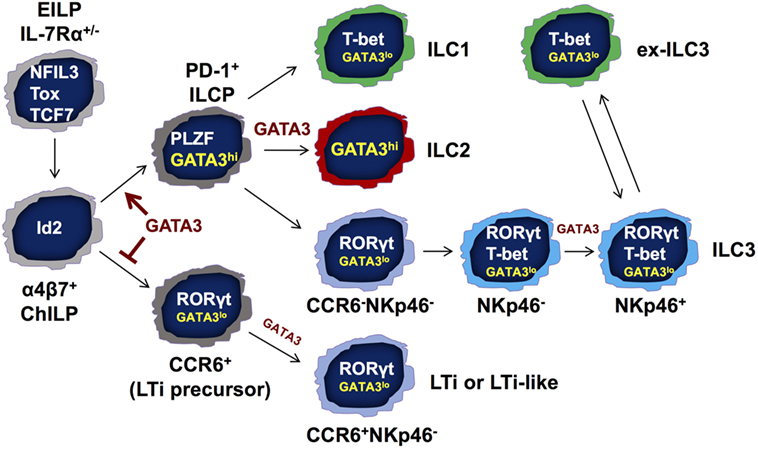
Figure 2. GATA3 regulates innate lymphoid cell (ILC) development and functions at multiple stages. GATA3 is expressed by all ILCs including lymphoid tissue inducer (LTi) cells and natural killer (NK) cells, albeit at different levels. GATA3 is required for the development of IL-7Rα-expressing ILCs but not NK cells as shown in Figure 1. At a possible developmental branch point for the bifurcation of PD-1+ PLZF-expressing ILC progenitors (ILCPs) and CCR6+ LTi progenitors, GATA3 is highly expressed in and absolutely required for the generation of “non-LTi” PLZF-expressing ILCPs. High level of GATA3 expression may suppress LTi developmental program. Nevertheless, GATA3 expression at low levels during the early stage of LTi cell development regulates LTi function. In mature ILCs, GATA3 is required for the functions and maintenance of ILC2s. GATA3 is also important for the homeostasis of ILC1s. Furthermore, the development of NKp46+ ILC3s that co-express T-bet and RORγt depends on low levels of GATA3 expression. Finally, GATA3 promotes IL-22 production in RORγt-expressing ILCs and regulates IL-7Rα expression in all ILC subsets. EILP, early ILC progenitor that expresses TCF7, NFIL3, and Tox; ChILP, common helper-like ILC progenitor that expresses Id2; ILCP, common ILC progenitor that expresses PLZF and GATA3.
ILC3s are the major ILC population highly enriched in the gut (8, 18). RORγt, the master regulator of Th17 cells (77), is critical for the development and functions of ILC3s including both NKp46-expressing ILC3s and CCR6-expressing LTi or LTi-like cells (27). Although NKp46+ ILC3s may have some specialized functions in inflammation and gut homeostasis, by producing IL-22, NKp46+ ILC3s and CCR6+ ILC3s are functionally redundant in host defense (54, 78). NKp46-expressing ILC3s co-express T-bet and RORγt, whereas CCR6− ILC3s in mice only express RORγt (66, 79, 80). Interestingly, both types of ILC3s express low levels of GATA3 (66).
T-bet and RORγt are critical for the development of NKp46+ ILC3s. During Th cell differentiation, transcription factors induced in one lineage can repress the expression and/or functions of transcription factors that are expressed by other lineages. For example, T-bet suppresses GATA3 expression as well as its function (81–83). In addition, T-bet and GATA3 bind to Th1- or Th2-specific genes at the same regions (83–85). T-bet also inhibits RORγt during T cell differentiation (86). NKp46+ ILC3s express slightly lower levels of RORγt than CCR6+ ILC3s, suggesting that T-bet may inhibit RORγt expression in these cells (80). Such inhibition may explain how “ILC1-like” “ex-ILC3s” can be generated through turning off RORγt expression (80). Interestingly, these “ex-ILC3s” may re-express RORγt and become ILC3s by IL-1/IL-23 stimulation (87).
Since the master transcription factors cross-regulate each other and they can be co-expressed at the single cell level, quantitative expression of different master regulators may alter the balance among these factors resulting in phenotypical changes. Indeed, haplo-insufficiency of the master regulators, including T-bet, GATA3, and RORγt has been reported (58, 66, 88). Interestingly, all three transcription factors, T-bet, GATA3, and RORγt, are expressed by NKp46+ ILC3s. More strikingly, low levels of GATA3 are also required for the development of NKp46+ ILC3s, and GATA3 regulates the balance between RORγt and T-bet (66). GATA3-deficient ILC3s express ~2-fold higher RORγt indicating that GATA3 inhibits RORγt expression in ILC3s. Correcting RORγt expression levels by breeding GATA3-deficient mice onto the Rorc heterozygous background restores the development of NKp46+ ILC3s, indicating that GATA3 regulates the balance between RORγt and T-bet during NKp46+ ILC3 development (Figure 2).
As mentioned earlier, GATA3 is not required for the development of LTi or LTi-like cells. However, these LTi cells are nonfunctional, since Gata3fl/fl-VavCre mice do not have lymph node structures (8). Single cell analysis of gene expression may address whether GATA3-deficient LTi cells fail to express some LTi-specific genes that determine LTi function. On the other hand, high levels of GATA3 may be inhibitory for the development of LTi progenitors since we have found that the expression levels of GATA3 are negatively associated with many LTi-specific genes at the single cell level. Furthermore, deletion of Gata3 in NKp46+ ILC3s results in upregulation of CCR6+ ILC3-specific genes (66). Therefore, high levels of GATA3 expression at the PLZF-expressing progenitor stage are important for suppressing LTi lineage fate, and low expression of GATA3 in NKp46+ ILC3s is continuously required to maintain NKp46+ ILC3 cell identity by repressing the expression of LTi lineage-related genes.
GATA3 is also important for the optimal expression of Il22 (66). Interestingly, GATA3 binds to the Il22 promoter only in ILC3s but not ILC2s. Since GATA3 promotes IL-22 expression in both CCR6+ ILC3s and NKp46+ ILC3s, mice with ILC3-specific Gata3 deletion mediated by RORγt-Cre are susceptible to Citrobacter rodentium infection. However, these mice develop normal lymph node structures. These results suggest that while GATA3 regulates LTi function at an early stage of their development, maintenance of LTi functions does not need continuous expression of GATA3 in LTi cells (66).
ILC1s including tissue-resident NK cells are enriched in the liver and T-bet is the master regulator for the development of ILC1s (12, 34). Similar to ILC3s, ILC1s also express low levels of GATA3 (12, 66). It has been reported that GATA3 is important for the maintenance of ILC1s (12). However, it is not known whether such GATA3 function is related to its effect on IL-7Rα expression in ILC1s. As discussed earlier, GATA3 is not necessary for the development of conventional NK cells (8, 89, 90). However, GATA3 is also expressed by NK cells, and they need GATA3 for their maturation and cytokine production (89).
Since GATA3 plays important roles in different ILC subsets and progenitors, and its function is associated with its dynamic and quantitative expression, it is critical to understand signals that regulate GATA3 expression. During Th2 differentiation, IL-4-mediated STAT6 activation is the major driving force responsible for the upregulation of GATA3 expression. TCR-mediated signaling especially triggered by low dose of antigens can also upregulate GATA3 expression (91). However, ILCs do not express antigen receptors, and ILC2 development seems to be IL-4-STAT6 independent (37).
Notch signaling induces whereas TGFβ downregulates GATA3 expression (92, 93). These signaling pathways may be important in regulating GATA3 expression in different ILC subsets at different stages. Indeed, it has been reported that TCF7, which can be induced by Notch signaling, positively regulates GATA3 expression during early stages of ILC development (48, 59). Besides transcriptional regulation of Gata3 gene expression, posttranscriptional and posttranslational regulations of GATA3 expression and functions should be also considered. For example, GATA3 activity is regulated by tyrosine kinase p38 in both T cells and ILCs (60, 94). Furthermore, GATA3 protein quickly disappears in ILC2s upon Gata3 deletion indicating that there is an active mechanism for GATA3 protein degradation in these cells.
As another critical transcription factor involved in ILC development, Id2 may regulate GATA3 expression or vice versa. In the future, it is essential to understand the relationship between Id2 and GATA3. Because gene regulation mediated by GATA3 is highly cell type and stage specific, its induction may also depend on cell context. Identification of the cofactors for GATA3 and the gene regulatory networks in different cell types at various developmental stages is essential to understand the biology of GATA3 in ILCs.
T-bet, GATA3, and RORγt are the lineage-defining factors that are critical for the development and functions of ILC1s, ILC2s, and ILC3s, respectively. Besides its critical role in the development and maintenance of ILC2s, GATA3 is also indispensable for the generation of PLZF-expressing ILC progenitors that give rise to non-LTi ILCs. Furthermore, GATA3 regulates the acquisition of LTi cell function, ILC3 effector function, and ILC homeostasis (Figure 2).
T-bet, GATA3, and RORγt can be co-expressed at the single cell level resulting in heterogeneity and possible plasticity of ILC subsets. In particular, NKp46+ ILC3s express all three transcription factors. GATA3 expression in NKp46+ ILC3s cells, albeit at low levels, is important for regulating the balance between T-bet and RORγt. Because quantitative changes in GATA3 expression may result in qualitative developmental effects, a model with titratable GATA3 expression may be needed to study its dose effects on the development and functions of ILC subsets.
In addition to studying the dose effect of GATA3 in ILC development and functions, investigating cell type- and stage-specific GATA3 regulation and function will greatly enhance our knowledge on ILC development. It is important to identify cell type- or stage-specific enhancers within the 1Mb Gata3 gene locus through epigenetic analyses. Future studies should also identify the similarities and differences between the actions of GATA3 in different cell types at various developmental stages, including both T cell and ILC lineages. Revealing cell type-specific gene regulation and epigenetic modifications mediated by GATA3 as well as cell type-specific binding patterns of GATA3 at a genome level will help us understand detail mechanisms. Finding GATA3 partners in different cell types, either by using co-immunoprecipitation followed by mass spectrometry or by genome-wide screening through bimolecular fluorescence complementation methods, may explain cell type-specific gene regulation mediated by GATA3.
Besides GATA3, many other transcription factors such as Id2, TCF7, NFIL3, Tox, and Runx proteins are also involved in ILC development (8, 41, 42, 48, 49, 95, 96). Further investigation on the relationship between GATA3 and other important transcription factors that are involved in ILC development and functions, including studies at a single cell level, may yield a deeper understanding of the ILC biology.
The author confirms being the sole contributor of this work and approved it for publication.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
JZ is supported by the Division of Intramural Research (DIR), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), USA.
1. Artis D, Spits H. The biology of innate lymphoid cells. Nature (2015) 517:293–301. doi:10.1038/nature14189
2. Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity (2014) 41:354–65. doi:10.1016/j.immuni.2014.09.005
3. McKenzie AN, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity (2014) 41:366–74. doi:10.1016/j.immuni.2014.09.006
4. Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol (2016) 17:765–74. doi:10.1038/ni.3489
5. Fang D, Zhu J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J Exp Med (2017) 214:1861–76. doi:10.1084/jem.20170494
6. Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR, et al. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell (2016) 165:1120–33. doi:10.1016/j.cell.2016.04.029
7. Koues OI, Collins PL, Cella M, Robinette ML, Porter SI, Pyfrom SC, et al. Distinct gene regulatory pathways for human innate versus adaptive lymphoid cells. Cell (2016) 165:1134–46. doi:10.1016/j.cell.2016.04.014
8. Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity (2014) 40:378–88. doi:10.1016/j.immuni.2014.01.012
9. Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood (2008) 112:1557–69. doi:10.1182/blood-2008-05-078154
10. Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol (2010) 28:445–89. doi:10.1146/annurev-immunol-030409-101212
11. Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol (2002) 2:933–44. doi:10.1038/nri954
12. Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell (2014) 157:340–56. doi:10.1016/j.cell.2014.03.030
13. Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature (2010) 463:540–4. doi:10.1038/nature08636
14. Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature (2010) 464:1367–70. doi:10.1038/nature08900
15. Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci U S A (2010) 107:11489–94. doi:10.1073/pnas.1003988107
16. Fallon PG, Ballantyne SJ, Mangan NE, Barlow JL, Dasvarma A, Hewett DR, et al. Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. J Exp Med (2006) 203:1105–16. doi:10.1084/jem.20051615
17. Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell (2015) 163:381–93. doi:10.1016/j.cell.2015.08.061
18. Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity (2013) 39:386–99. doi:10.1016/j.immuni.2013.08.002
19. Chang YJ, Kim HY, Albacker LA, Baumgarth N, McKenzie AN, Smith DE, et al. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol (2011) 12:631–8. doi:10.1038/ni.2045
20. Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity (2012) 36:451–63. doi:10.1016/j.immuni.2011.12.020
21. Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol (2011) 12:1045–54. doi:10.1031/ni.2131
22. Kim BS, Wang K, Siracusa MC, Saenz SA, Brestoff JR, Monticelli LA, et al. Basophils promote innate lymphoid cell responses in inflamed skin. J Immunol (2014) 193:3717–25. doi:10.4049/jimmunol.1401307
23. Roediger B, Kyle R, Yip KH, Sumaria N, Guy TV, Kim BS, et al. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol (2013) 14:564–73. doi:10.1038/ni.2584
24. Shikhagaie MM, Germar K, Bal SM, Ros XR, Spits H. Innate lymphoid cells in autoimmunity: emerging regulators in rheumatic diseases. Nat Rev Rheumatol (2017) 13:164–73. doi:10.1038/nrrheum.2016.218
25. Bando JK, Colonna M. Innate lymphoid cell function in the context of adaptive immunity. Nat Immunol (2016) 17:783–9. doi:10.1038/ni.3484
26. Zook EC, Kee BL. Development of innate lymphoid cells. Nat Immunol (2016) 17:775–82. doi:10.1038/ni.3481
27. Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi YW, Littman DR. An essential function for the nuclear receptor ROR gamma t in the generation of fetal lymphoid tissue inducer cells. Nat Immunol (2004) 5:64–73. doi:10.1038/ni1022
28. Mebius RE. Organogenesis of lymphoid tissues. Nat Rev Immunol (2003) 3:292–303. doi:10.1038/nri1054
29. Guo L, Wei G, Zhu J, Liao W, Leonard WJ, Zhao K, et al. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc Natl Acad Sci U S A (2009) 106(32):13463–8. doi:10.1073/pnas.0906988106
30. Guo L, Junttila IS, Paul WE. Cytokine-induced cytokine production by conventional and innate lymphoid cells. Trends Immunol (2012) 33(12):598–606. doi:10.1016/j.it.2012.07.006
31. Guo L, Huang Y, Chen X, Hu-Li J, Urban JF Jr, Paul WE. Innate immunological function of TH2 cells in vivo. Nat Immunol (2015) 16:1051–9. doi:10.1038/ni.3244
32. Van Dyken SJ, Nussbaum JC, Lee J, Molofsky AB, Liang HE, Pollack JL, et al. A tissue checkpoint regulates type 2 immunity. Nat Immunol (2016) 17:1381–7. doi:10.1038/ni.3582
33. Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science (2015) 350:981–5. doi:10.1126/science.aac9593
34. Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife (2014) 3:e01659. doi:10.7554/eLife.01659
35. Shih HY, Sciume G, Poholek AC, Vahedi G, Hirahara K, Villarino AV, et al. Transcriptional and epigenetic networks of helper T and innate lymphoid cells. Immunol Rev (2014) 261:23–49. doi:10.1111/imr.12208
36. Guo X, Qiu J, Tu T, Yang X, Deng L, Anders RA, et al. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity (2014) 40:25–39. doi:10.1016/j.immuni.2013.10.021
37. Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature (2013) 502:245–8. doi:10.1038/nature12526
38. O’Sullivan TE, Rapp M, Fan X, Weizman OE, Bhardwaj P, Adams NM, et al. Adipose-resident group 1 innate lymphoid cells promote obesity-associated insulin resistance. Immunity (2016) 45:428–41. doi:10.1016/j.immuni.2016.06.016
39. Villarino AV, Sciume G, Davis FP, Iwata S, Zitti B, Robinson GW, et al. Subset- and tissue-defined STAT5 thresholds control homeostasis and function of innate lymphoid cells. J Exp Med (2017) 214:2999–3014. doi:10.1084/jem.20150907
40. Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol (2009) 9:480–90. doi:10.1038/nri2580
41. Zhong C, Zhu J. Transcriptional regulators dictate innate lymphoid cell fates. Protein Cell (2017) 8(4):242–54. doi:10.1007/s13238-017-0369-7
42. Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature (1999) 397:702–6. doi:10.1038/17812
43. Zhong C, Zhu J. Bcl11b drives the birth of ILC2 innate lymphocytes. J Exp Med (2015) 212:828. doi:10.1084/jem.2126insight1
44. Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science (2010) 329:89–93. doi:10.1126/science.1188989
45. Walker JA, Oliphant CJ, Englezakis A, Yu Y, Clare S, Rodewald HR, et al. Bcl11b is essential for group 2 innate lymphoid cell development. J Exp Med (2015) 212:875–82. doi:10.1084/jem.20142224
46. Yu Y, Wang C, Clare S, Wang J, Lee SC, Brandt C, et al. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J Exp Med (2015) 212:865–74. doi:10.1084/jem.20142318
47. Califano D, Cho JJ, Uddin MN, Lorentsen KJ, Yang Q, Bhandoola A, et al. Transcription factor Bcl11b controls identity and function of mature type 2 innate lymphoid cells. Immunity (2015) 43:354–68. doi:10.1016/j.immuni.2015.07.005
48. Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol (2015) 16:1044–50. doi:10.1038/ni.3248
49. Seehus CR, Aliahmad P, de la Torre B, Iliev ID, Spurka L, Funari VA, et al. The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nat Immunol (2015) 16:599–608. doi:10.1038/ni.3168
50. Serafini N, Klein Wolterink RG, Satoh-Takayama N, Xu W, Vosshenrich CA, Hendriks RW, et al. Gata3 drives development of RORgammat+ group 3 innate lymphoid cells. J Exp Med (2014) 211:199–208. doi:10.1084/jem.20131038
51. Steinke FC, Xue HH. From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunol Res (2014) 59:45–55. doi:10.1007/s12026-014-8545-9
52. Vong QP, Leung WH, Houston J, Li Y, Rooney B, Holladay M, et al. TOX2 regulates human natural killer cell development by controlling T-BET expression. Blood (2014) 124:3905–13. doi:10.1182/blood-2014-06-582965
53. Bjorklund AK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol (2016) 17:451–60. doi:10.1038/ni.3368
54. Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A, et al. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat Immunol (2016) 17:179–86. doi:10.1038/ni.3332
55. Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature (1996) 384:474–8. doi:10.1038/384474a0
56. Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity (2012) 37:634–48. doi:10.1016/j.immuni.2012.06.020
57. Mjosberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, Blom B, et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity (2012) 37:649–59. doi:10.1016/j.immuni.2012.08.015
58. Klein Wolterink RG, Serafini N, van Nimwegen M, Vosshenrich CA, de Bruijn MJ, Fonseca Pereira D, et al. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc Natl Acad Sci U S A (2013) 110:10240–5. doi:10.1073/pnas.1217158110
59. Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, Tang J, et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity (2013) 38:694–704. doi:10.1016/j.immuni.2012.12.003
60. Furusawa J, Moro K, Motomura Y, Okamoto K, Zhu J, Takayanagi H, et al. Critical role of p38 and GATA3 in natural helper cell function. J Immunol (2013) 191:1818–26. doi:10.4049/jimmunol.1300379
61. Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol (2004) 5:1157–65. doi:10.1038/ni1128
62. Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci U S A (2004) 101:1993–8. doi:10.1073/pnas.0308697100
63. Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem (1997) 272:21597–603. doi:10.1074/jbc.272.34.21597
64. Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell (1997) 89:587–96. doi:10.1016/S0092-8674(00)80240-8
65. Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity (2011) 35:299–311. doi:10.1016/j.immuni.2011.08.007
66. Zhong C, Cui K, Wilhelm C, Hu G, Mao K, Belkaid Y, et al. Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat Immunol (2016) 17:169–78. doi:10.1038/ni.3318
67. Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, et al. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem (2002) 277:42399–408. doi:10.1074/jbc.M205876200
68. Tanaka S, Motomura Y, Suzuki Y, Yagi R, Inoue H, Miyatake S, et al. The enhancer HS2 critically regulates GATA-3-mediated Il4 transcription in T(H)2 cells. Nat Immunol (2011) 12:77–85. doi:10.1038/ni.1966
69. Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol (2009) 9:125–35. doi:10.1038/nri2476
70. Tindemans I, Serafini N, Di Santo JP, Hendriks RW. GATA-3 function in innate and adaptive immunity. Immunity (2014) 41:191–206. doi:10.1016/j.immuni.2014.06.006
71. Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity (2003) 19:863–75. doi:10.1016/S1074-7613(03)00328-5
72. Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol (2008) 9:1122–30. doi:10.1038/ni.1647
73. Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity (2003) 19:83–94. doi:10.1016/S1074-7613(03)00176-6
74. Wang Y, Misumi I, Gu AD, Curtis TA, Su L, Whitmire JK, et al. GATA-3 controls the maintenance and proliferation of T cells downstream of TCR and cytokine signaling. Nat Immunol (2013) 14:714–22. doi:10.1038/ni.2623
75. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature (2014) 508:397–401. doi:10.1038/nature13047
76. Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, et al. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature (2016) 539:102–6. doi:10.1038/nature20105
77. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell (2006) 126:1121–33. doi:10.1016/j.cell.2006.07.035
78. Song C, Lee JS, Gilfillan S, Robinette ML, Newberry RD, Stappenbeck TS, et al. Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J Exp Med (2015) 212:1869–82. doi:10.1084/jem.20151403
79. Sciume G, Hirahara K, Takahashi H, Laurence A, Villarino AV, Singleton KL, et al. Distinct requirements for T-bet in gut innate lymphoid cells. J Exp Med (2012) 209:2331–8. doi:10.1084/jem.20122097
80. Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature (2013) 494:261–5. doi:10.1038/nature11813
81. Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O’Shea JJ, et al. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription (vol 23, pg 755, 2006). J Exp Med (2006) 203:754–754. doi:10.1084/jem.20052165
82. Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science (2005) 307:430–3. doi:10.1126/science.1103336
83. Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity (2012) 37:660–73. doi:10.1016/j.immuni.2012.09.007
84. Kanhere A, Hertweck A, Bhatia U, Gokmen MR, Perucha E, Jackson I, et al. T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat Commun (2012) 3:1268. doi:10.1038/ncomms2260
85. Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, et al. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Natl Acad Sci U S A (2009) 106:17876–81. doi:10.1073/pnas.0909357106
86. Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, et al. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol (2011) 12:96–104. doi:10.1038/ni.1969
87. Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, et al. Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity (2015) 43:146–60. doi:10.1016/j.immuni.2015.06.019
88. Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science (2002) 295:338–42. doi:10.1126/science.1065543
89. Samson SI, Richard O, Tavian M, Ranson T, Vosshenrich CA, Colucci F, et al. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity (2003) 19:701–11. doi:10.1016/S1074-7613(03)00294-2
90. Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, Richard-Le Goff O, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol (2006) 7:1217–24. doi:10.1038/ni1395
91. Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med (2005) 202:793–804. doi:10.1084/jem.20051304
92. Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity (2007) 27:89–99. doi:10.1016/j.immuni.2007.05.021
93. Gorelik L, Fields PE, Flavell RA. Cutting edge: TGF-beta inhibits Th type 2 development through inhibition of GATA-3 expression. J Immunol (2000) 165:4773–7. doi:10.4049/jimmunol.165.9.4773
94. Chen CH, Zhang DH, LaPorte JM, Ray A. Cyclic AMP activates p38 mitogen-activated protein kinase in Th2 cells: phosphorylation of GATA-3 and stimulation of Th2 cytokine gene expression. J Immunol (2000) 165:5597–605. doi:10.4049/jimmunol.165.10.5597
95. Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med (2014) 211:1723–31. doi:10.1084/jem.20140212
Keywords: GATA3 transcription factor, innate lymphoid cells, development, cytokines, transcriptional activation
Citation: Zhu J (2017) GATA3 Regulates the Development and Functions of Innate Lymphoid Cell Subsets at Multiple Stages. Front. Immunol. 8:1571. doi: 10.3389/fimmu.2017.01571
Received: 22 September 2017; Accepted: 01 November 2017;
Published: 14 November 2017
Edited by:
Chiara Romagnani, Deutsches Rheuma-Forschungszentrum (DRFZ), GermanyReviewed by:
Barbara L. Kee, University of Chicago, United StatesCopyright: © 2017 Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinfang Zhu, amZ6aHVAbmlhaWQubmloLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.