
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Immunol. , 09 November 2017
Sec. Molecular Innate Immunity
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.01501
This article is part of the Research Topic Host and Microbe Adaptations in the Evolution of Immunity View all 21 articles
 Juris A. Grasis1,2*
Juris A. Grasis1,2*
Animals live in symbiosis with the microorganisms surrounding them. This symbiosis is necessary for animal health, as a symbiotic breakdown can lead to a disease state. The functional symbiosis between the host, and associated prokaryotes, eukaryotes, and viruses in the context of an environment is the holobiont. Deciphering these holobiont associations has proven to be both difficult and controversial. In particular, holobiont association with viruses has been of debate even though these interactions have been occurring since cellular life began. The controversy stems from the idea that all viruses are parasitic, yet their associations can also be beneficial. To determine viral involvement within the holobiont, it is necessary to identify and elucidate the function of viral populations in symbiosis with the host. Viral metagenome analyses identify the communities of eukaryotic and prokaryotic viruses that functionally associate within a holobiont. Similarly, analyses of the host in response to viral presence determine how these interactions are maintained. Combined analyses reveal how viruses interact within the holobiont and how viral symbiotic cooperation occurs. To understand how the holobiont serves as a functional unit, one must consider viruses as an integral part of disease, development, and evolution.
All animals interact with a consortium of microbes at all times and have done so since the dawn of animal life (1). Animal life has evolved from and in intimate association with microorganisms, while these same microorganisms have evolved in part to the resources provided by their animal surroundings. This symbiosis allows for a sharing of resources, including metabolic products and genes. These interactions have been of intense research and speculation; however, an important player in these symbiotic interactions is often overlooked, the effects of viruses. None of these interactions occur in the absence of viruses, so to inquire about symbioses requires discussion of viruses.
Viruses are seemingly universal in the biosphere (2). Their numbers are so staggering that when speaking of large numbers, one should use the term “viral” rather than “astronomical.” There are an estimated 1031 viruses on the planet, which may be an underestimation due to our inability to properly enumerate RNA viruses and viral elements that persist in cells and genomes (3). Further, viral genomes are worldwide reservoirs of genetic diversity (4). Considering viral abundances, diversity, and ubiquitous presence (5), understanding symbioses is lacking without taking into account the effects of viruses on host and associated microbe metabolism, and genetic flow between organisms.
Viruses infect all animals, from Poriferans to Cnidarians to Bilaterans to Chordates. There is ever-increasing evidence that viral infections have occurred during all of cellular life, as the presence of viral elements are often found in genomes throughout evolution (6). Host–viral infections or associations are not adequately quantified, but in most host-associated systems it seems that the number of viruses is equivalent to or slightly less than the number of bacteria associating with a eukaryotic host (2, 7). In most cases, the enumerable viral populations are the free DNA prokaryotic viruses, which are likely involved with the regulation of the host-associated bacteria. In host-associated systems, it seems that Lotka–Volterra “kill-the-winner” predator–prey dynamics of the prokaryotic virus and bacteria are atypical. Many prokaryotic viruses found in these systems display temperate lifestyles in which the virus becomes latent and integrates into a host chromosome or exists as an episomal element, as indicated by the large abundance of integrase genes in viral genomes (8, 9). Additionally, the presence of latent viruses may allow for bacterial dominance of a niche in the presence of related strains (10). Experimental evidence in non-host-associated systems supports this idea, as increasing concentrations of bacteria favor prokaryotic virus temperate lifestyles (11). While most viral research focuses on lytic/virulent infections, it is useful to explore both the temperate dynamics of prokaryotic viruses and latent eukaryotic viral infection, and their role in symbiosis.
The functional association between a host, prokaryotic, eukaryotic, and viral entities within a particular environment is the holobiont. This functional association helps to define the phenotypic unit. Casual associations may not define the phenotype, so functional associations (and the genes used) help define the phenotype. This functional symbiosis is involved in animal development (12), nervous system regulation (13, 14), immune system development and regulation (15, 16), and many other biological processes (17). When this functional association breaks down, a dysbiotic state occurs, leading to grave effects on animal health, ranging from coral bleaching (18), to stunted immune system development (19), to nervous and immunological disorders (20), to effects on human health (21). Further, the holobiont is not static; it is in a constant state of genetic flux. Viruses predominately affect this genetic flow and the acquisition of evolutionary traits (22, 23). Therefore, understanding the holobiont requires investigation of the effects viruses have on gene flow occurring within it. This is evaluated through viral metagenomics (viromics), where culture-independent viral isolations from host systems are sequenced and the viral genomes are analyzed. Not only can host-associated viral populations be identified, but how these populations change under dysbiotic conditions (24, 25), the identification of new viruses (26, 27), and the effects these viruses have on cellular systems (28) can all be learned through viral metagenome (virome) analyses.
Viruses act as parasites; they infect and either replicate within the host cell or integrate within the host genome. Viruses propagate by one of two different lifestyles, either lytic/virulent or temperate/latent. The lytic/virulent lifestyle involves the infection, replication, and lysis of the cell, leading to the death of the cell and release of viral progeny. The temperate/latent lifestyle involves the integration of the virus into the genome in a proviral form, which can be activated at a later time to become a lytic/virulent replicative virus. Either one of these scenarios affects the host; replication leads to cellular damage, while integration leads to genomic damage. The host defense against parasitism limits cellular or genomic damage (29). These viral parasitic lifestyles cause a molecular arms race, the virus seeking a new host to continue propagation, while the host immune system recognizes the virus to minimize damage (30).
There are many direct causes of pathogenesis by parasitic viruses, but there are many indirect causes as well. Proviral endogenous retroelements can have negative effects on the genome by inserting, deleting, or rearranging portions of the genome (31). The large number of freely associating viruses found interacting with host systems also presents a conundrum, that the presence of large amounts of viral material, be it nucleic acid or protein, makes it unlikely that they would not cause an immune response. Microbial-associated molecular patterns (MAMPs) on prokaryotic and eukaryotic viruses can cause immune system recognition that can lead to immune related pathogenesis. Further, lysis of cells, be it of a bacterial cell or of a eukaryotic cell, or apoptosis of a virally infected cell can cause activation of the immune system leading to pathogenesis (32). Cellular lysis is often considered in the aftermath of eukaryotic viral infection, but prokaryotic lysis of bacteria is commonly overlooked. Release of bacterial antigens, such as LPS, peptidoglycans, lipopeptides, lipoteichoic acid, flagellin, and bacterial DNA, can easily activate the immune system, and in extreme cases lead to sepsis (33). There are many direct and indirect causes of viral pathogenesis, but given the sheer numbers of viruses within a holobiont, and the limited pathogenesis that actually occurs, it seems more likely that viral pathogenesis is not as common as viral commensalism and mutualism.
Most consider viruses to be parasites, where infection benefits the virus, but decreases the fitness of the host. Now consider other scenarios, such as commensals and mutualists. A virus can be commensal, the virus benefits while host fitness is unaffected. A virus can be mutualistic, in which both organisms benefit and fitness increases. Such viral associations may provide advantages that promote evolution and biodiversity (34, 35). Also consider that one virulent virus among a sea of non-virulent viruses does not equate to pathogenesis. Unless transmission and recovery rates are high, pathogenicity may be an evolutionarily poor strategy for viral survival. More likely, pathogenesis is the exception and not the rule, with more instances being discovered of viruses having cooperative roles with the host (34, 36).
There are many instances where an organism cannot exist without beneficial viruses. Polydnavirus integration into parasitoid wasp genomes counters the effects of the caterpillar host immune system where the wasp has laid its eggs (37). Without this polydnavirus presence, the caterpillar immune system would eliminate the wasp eggs, but when the polydnavirus endogenous viral element becomes active upon egg deposition, the host immune response to the eggs is negated. Similarly, endogenous retrovirus syncytin expression in the placenta of mammals allows for the development of the placental syncytium (38). This syncytial fusion creates a barrier for the placenta, which in part keeps the fetus from being rejected by the mother’s immune system. Viruses can also modulate the immune system and restore dysbiotic conditions. Kernbauer et al. have shown that an enteric murine norovirus can restore normal mucosal immunity and intestinal morphology in germ-free mice, essentially replacing the immune stimulatory effects of gut microbiota (39). Viruses can also protect against or impede further infection or pathogenesis, such as Hepatitis G virus slowing the progress of HIV infection (40), and latent herpesviruses protecting against bacterial infections (41). It is becoming evident that viruses have the potential to be something more than parasites in a holobiont, which revises conceptions of how viruses impact host interactions.
Viruses can also integrate into cellular genomes and act as genetic elements associating with genomes. The amount of DNA of viral origin within the human genome is similar to that of human coding domains (42). One major discovery in viromes is the persistence of viral genetic elements, either latently integrated into host genomes or surviving as chromosomal episomes. Host-associated viral populations seem to be dominated by temperate prokaryotic viruses or latent eukaryotic viruses. This is attributed to a large abundance of integrase sequences in prokaryotic viromes (8) and a large abundance of transposase sequences in eukaryotic viromes (43).
Integrated viral DNA in the host genome are endogenous viral elements (EVEs), which have the potential to drive evolutionary processes, such as speciation, resulting in the emergence of new traits (44–46). In addition to these evolutionary transitions, EVE integration can affect gene expression through their long terminal repeats (LTRs). These LTRs are repetitive viral DNA sequences that flank integrated EVEs, serving as promoters to both viral and host genes. These LTRs can affect stem cells (47), development (48), and immunity (49, 50). There are many individual genes affected by EVEs, though their major impact on evolutionary traits may be on gene regulatory networks, or the cellular regulators that impact RNA and protein expression (51, 52). The effects of EVEs and transposable elements in all these biological processes are being recognized as vitally important (53).
Genomically integrated viral elements are reminders that viruses affect everything in biology, but what about free viruses that associate with hosts? Viromics allow researchers to analyze the viral populations and effects these viruses have on the holobiont. These studies have been conducted in many host systems, from the base of animal life in the Cnidarian phylum (54) to mammals (55). Often, the viruses found freely associating are prokaryotic viruses, which regulate the number and strains of bacteria in a holobiont (56). These viruses are likely selected by the host to maintain bacterial populations (26). Further, viromics show the sphere of viral involvement in gene flow and gene shuffling in an ever-changing environment, often from within bacterial cells and sometimes from within eukaryotic cells.
Many viruses can persist in host cells and influence the host without symptoms of disease. Chronic systemic viruses continuously stimulate the immune system (57), driving the emergence of many viral recognition systems over evolutionary time (58). These recognition systems give a host integrity to coexist with viruses while minimizing pathogenesis and protecting genomic information. Antisense RNA encoded by genomic transposable elements allows for specific regulation of viral amplification products (59). This evolved into use of antisense RNAs with Argonaute nucleases. Piwi-interacting RNAs utilize transposon derived small RNAs to defend against integration events by binding to complementary RNAs and cleaving the complex with a bound Argonaute nuclease. This system seems to be restricted to the germ-line and protects genomic integrity. Similarly, the RNAi system processes RNAs by binding to small RNA fragments and cleaving these complexes with an RNase III nuclease, Dicer (60). While controversial, it appears that chordates may not have retained RNAi antiviral function. However, there are many immune functions additionally used in both chordates and non-chordates to regulate viral presence (Figure 1). These systems rely on host pattern-recognition receptors (PRRs) evolved to recognize MAMPs. These PRRs include the Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), cGAS-STING pathway, NOD-like receptors (NLRs), C-type lectin receptors (CLRs), and absent-in-melanoma-like receptors (ALRs). TLRs recognize viruses endosomally once viral nucleic acids are released (61), cytoplasmic RLRs recognize viral genomic RNA or double-stranded RNA intermediates (62), cGAS-STING senses retroviral and double-stranded DNA (63), NLRs recognize viral DNA genomes (64), ALRs can also recognize viral genomic DNA (65), while CLRs recognize carbohydrates (66). In the biological arms race that caused the development of the adaptive immune system capable of tracking evolutionary changes in pathogens, antiviral cytokines such as interferons (IFNs) became prominent signals alerting the host of viral infection and inhibit viral propagation (67). With IFNs came recombination events to generate antibodies and major histocompatability complexes in vertebrates to increase the recognition possibilities that came with increased pathogen complexity. Although viral recognition research is often focused on the adaptive immune system in mammals, the overwhelming majority of animals has multiple pathways to recognize, regulate, and maintain viral associations and may not necessarily use canonical adaptive systems to structure the holobiont. Continuing research will involve the 95% of Metazoans that do not possess such an adaptive immune system to recognize viruses, yet are able to adapt to ever-changing viral populations through mechanisms, such as trained innate immunity (68).
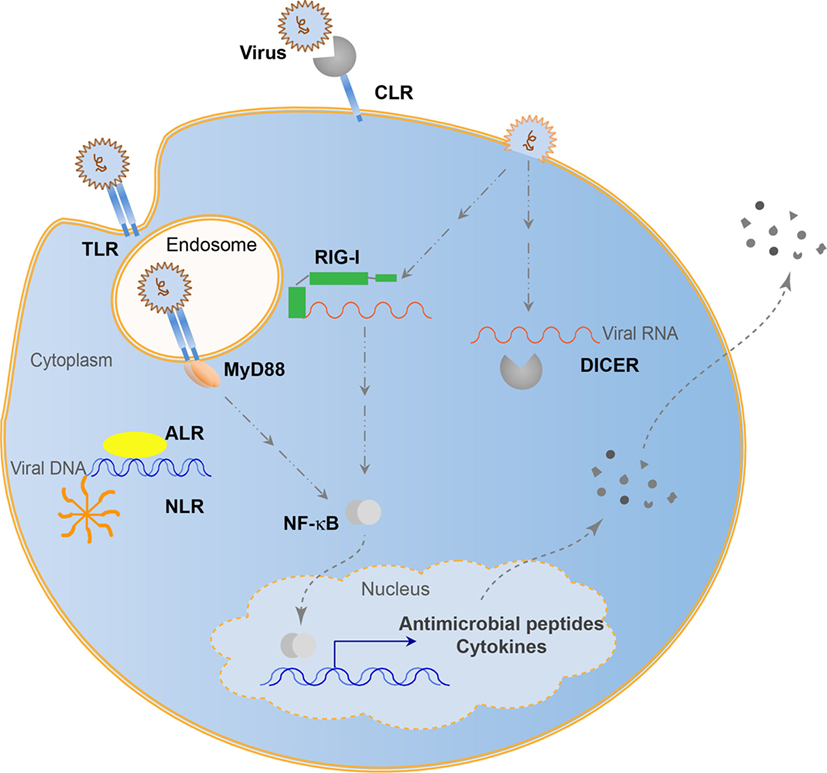
Figure 1. Evolutionarily conserved antiviral innate immune systems. Toll-like receptors (TLRs) recognize endosomal viral nucleic acids, NOD-like receptors (NLRs) form an inflammasome and recognize viral DNA, absent-in-melanoma-like receptors (ALRs) recognize viral DNA, retinoic acid-inducible gene I (RIG-I) and RNAi (Dicer) pathways recognize viral RNA, while C-type lectins (CLRs) recognize viral carbohydrates. Some pathways can lead to the direct elimination of viral entities, while others lead to transcriptional activation resulting in cytokine and antimicrobial peptide secretion.
Holobiont studies are complex. If one considers the sheer number of associated prokaryotes, eukaryotes, viruses, and all of their respective genomes, the number of potential interactions is overwhelming. Therefore, if one can use a model system with a limited number of microbial partners to deconstruct the holobiont and if this can be studied in an ancient animal phylum for conserved holobiont interactions, it could simplify these studies while retaining informative and predictive capabilities. The use of a basal metazoan allows research on mechanisms of holobiont assembly, holobiont effects on microbiota and host health, and metabolic interactions between the host and microbiota. This helps to elucidate symbiosis in healthy states and dysbiosis in disease states.
There are many useful systems that meet the above criteria to investigate the holobiont, including ascidians (69), anemones (70), and sponges (71). The basal model organism Hydra is another useful system. Hydra are freshwater Cnidarians practical for developmental, neural, aging, and stem cell studies (72). Importantly, the findings made using Hydra translate well into host–microbe interaction studies due to its diploblastic morphology (73), conserved mucosal immunity (74), and limited number of microbial partners (75). Additionally, Hydra are clonal, have a well-annotated genome (76), can be made transgenic (77), germ-free (78), and due to its limited number of microbial interactions, Hydra can be used in symbiosis studies (79). Hydra display distinct microbial colonization patterns dependent on host factors (78), which are primarily driven by antimicrobial peptide selection at the epithelium (80). Hydra have many evolutionarily conserved receptor pathways to regulate microbial interactions, including a TLR pathway (81) and a large repertoire of NLRs (82). Further, Hydra utilize many uniquely identified classes of antimicrobial peptides to regulate its microbial interactions (81, 83, 84). Finally, 57% of the Hydra genome are transposable elements, one of the largest percentages found in an animal genome (76). These factors make Hydra a useful system to deconstruct and reconstruct an organismal holobiont (Figure 2).
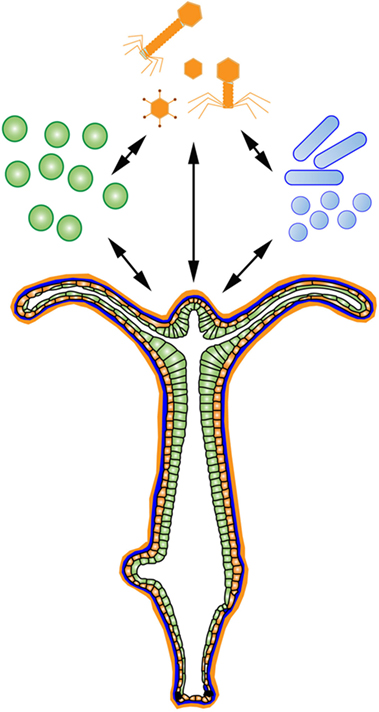
Figure 2. The Hydra Holobiont. Hydra are an ideal system to deconstruct and reconstruct an organismal holobiont consisting of associated eukaryotes (green), prokaryotes (blue), and viruses (orange) at an exposed epithelium.
Understanding the complete Hydra-associated virome has commenced. The Hydra DNA virome consists primarily of prokaryotic viruses in the Caudovirales order, the majority of the eukaryotic viruses are of the Herpesviridae family, the diversity of the viruses increases upon environmental heat stress, and each species of Hydra associates with a specific community of viruses (25). Further, these Hydra-associated viruses affect Hydra-microbiome metabolism (25, 85). Studies on the RNA virome, germ-free eukaryotic virome, and prokaryotic virome of Hydra-associated bacteria are ongoing to create a comprehensive Hydra virome [J. Grasis, in preparation; (86)]. Combining the virome with in vivo viral infection transcriptomes and the ability to induce inflammatory conditions makes Hydra a useful system to structure viral–holobiont interactions related to animal health conditions. The Hydra model system may shed light on novel aspects of holobiont formation, maintenance, and dysbiosis, while integrating viral involvement within the holobiont.
There has been much discussion about the holobiont recently, particularly as it relates to selective units of animal host and microbiome (87–93). Much of the focus has been placed on host–bacterial associations, but what of the viruses? They are intrinsically part of the genome and part of the holobiont, and yet, extrinsically exist beyond the genome and the holobiont. This duality exists because both the host and the microbiome are under their own selective pressures, each are selecting for the environment that benefits them, establishing or propagating a phenotype, and allowing for co-existence to continue. It is neither eukaryo-centric, prokaryo-centric, nor viro-centric, each member has a role to play within the holobiont. Therefore, the holobiont is a coordination of integrated functions by all members to suit adaptation to an environment.
Viruses are genetic parasites constantly sampling their environments. Functional aspects of their genomes can be selected by their prokaryotic and eukaryotic hosts, and in this way, viruses are symbionts to these hosts. Viruses can also transfer DNA in the form of lateral gene transfer, which can be important for adaptations to new environments (94). For example, prophages can promote genetic transfer between prokaryotic viruses and eukaryotes. Wolbachia prophage WO in arthropods contains a eukaryotic association module, which among other genes, contains a spider toxin gene that can form pores in both prokaryotic and eukaryotic membranes to facilitate viral escape (95). There are many more instances of viral drivers of adaptation (96), which makes viral dynamics in the holobiont fluid. Free viruses can be acquired from the environment through horizontal transmission, while viral elements can be vertically transmitted through genomically integrated viral elements and episomes. Such horizontal and vertical transmissions allow for a fully functional range of symbioses, from obligate (both need each other to survive) to facultative (both benefit from the association, but it is not absolute).
Viruses are remarkable symbionts. Viral elements exist intra-genomically, intra-cellularly, extra-cellularly, and environmentally. They persist in all of these realms, and yet, are vital to the holobiont. As mentioned earlier, viromics teaches us that viruses are involved in gene flow and shuffling in a changing environment, and that the elements in the holobiont are in a constant ecological flux. In all cases, viruses provide balance to the holobiont, keeping the host and associating prokaryotes and eukaryotes functioning together as a unit.
JG wrote, did the artwork, and is responsible for the content of this manuscript.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
JG would like to thank Dr. Benjamin Knowles for critical reading of the manuscript and to the reviewers of the manuscript who provided fantastic feedback. Support for JG is through NIH F32AI098418. JG would also like to acknowledge the many related articles not cited here due to word restrictions, particularly in plant research areas, which have been vital in our burgeoning understanding of viral symbioses.
Support for JG is through NIH F32AI098418.
2. Cobián-Güemes AG, Youle M, Cantú VA, Felts B, Nulton J, Rohwer F. Viruses are winners in the game of life. Annu Rev Virol (2016) 3:197–214. doi:10.1146/annurev-virology-100114-054952
3. Suttle CA. Marine viruses – major players in the global ecosystem. Nat Rev Microbiol (2007) 5:801–12. doi:10.1038/nrmicro1750
4. Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, et al. Uncovering earth’s virome. Nature (2016) 536:425–30. doi:10.1038/nature19094
5. Wasik BR, Turner PE. On the biological success of viruses. Annu Rev Microbiol (2013) 67:519–41. doi:10.1146/annurev-micro-090110-102833
6. Katzourakis A. Paleovirology: inferring viral evolution from host genome sequence data. Philos Trans R Soc Lond B Biol Sci (2013) 368:20120493. doi:10.1098/rstb.2012.0493
7. Kim M-S, Park E-J, Roh SW, Bae J-W. Diversity and abundance of single-stranded DNA viruses in human feces. Appl Environ Microbiol (2011) 77:8062–70. doi:10.1128/AEM.06331-11
8. Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature (2010) 466:334–40. doi:10.138/nature09199
9. Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res (2011) 21:1616–25. doi:10.1101/gr.122705.111
10. Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci U S A (2012) 109:17621–6. doi:10.1073/pnas.1206136109
11. Knowles B, Silveira CB, Bailey BA, Barott K, Cantu VA, Cobián-Güemes AG, et al. Lytic to temperate switching of viral communities. Nature (2016) 531:466–70. doi:10.1038/nature17193
12. Gilbert SF, Bosch TCG, Ledon-Rettig C. Eco-evo-devo: developmental symbiosis and developmental plasticity as evolutionary agents. Nat Rev Genet (2015) 16:611–22. doi:10.1038/nrg3982
13. Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci (2012) 13:701–12. doi:10.1038/nrn3346
14. Eisthen HL, Theis KR. Animal-microbe interaction and the evolution of nervous systems. Philos Trans R Soc Lond B Biol Sci (2016) 371:20150052. doi:10.1098/rstb.2015.0052
15. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol (2016) 16:341–52. doi:10.1038/nri.2016.42
16. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature (2016) 535:65–74. doi:10.1038/nature18847
17. Dheilly NM. Holobiont-holobiont interactions: redefining host-parasite interactions. PLoS Pathog (2014) 10:e1004093. doi:10.1371/journal.ppat.1004093
18. Egan S, Gardiner M. Microbial dysbiosis: rethinking disease in marine ecosystems. Front Microbiol (2016) 7:991. doi:10.3389/fmicb.2016.00991
19. Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science (2010) 330:1768–73. doi:10.1126/science.1195568
20. Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci (2017) 20:145–55. doi:10.1038/nn.4476
21. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol (2017) 17:219–32. doi:10.1038/nri.2017.7
22. Poole AM. Horizontal gene transfer and the earliest stages of the evolution of life. Res Microbiol (2009) 160:473–80. doi:10.1016/j.resmic.2009.07.009
23. Boto L. Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc Biol Sci (2014) 281:20132450. doi:10.1098/rspb.2013.2450
24. Vega Thurber RL, Barrott KL, Hall D, Liu H, Rodiguez-Mueller B, Desnues C, et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc Natl Acad Sci U S A (2008) 105:18413–8. doi:10.1073/pnas.0808985105
25. Grasis JA, Lachnit T, Anton-Erxleben F, Lim YM, Schmieder R, Fraune S, et al. Species-specific viromes in the ancestral holobiont Hydra. PLoS One (2014) 9:e109952. doi:10.1371/journal.pone.0109952
26. Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol (2012) 2:63–77. doi:10.1016/j.coviro.2011.12.004
27. Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, Boling L, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun (2014) 5:4498. doi:10.1038/ncomms5498
28. Quistad SD, Lim YW, Silva GG, Nelson CE, Haas AF, Kelly LW, et al. Using viromes to predict novel immune proteins in non-model organisms. Proc Biol Sci (2016) 283:20161200. doi:10.1098/rspb.2016.1200
29. Aiewsakun P, Katzourakis A. Endogenous viruses: connecting recent and ancient viral evolution. Virology (2015) 479-480:26–37. doi:10.1016/j.virol.2015.02.011
30. Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet (2012) 46:677–700. doi:10.1146/annurev-genet-110711-155522
31. Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol (2014) 15:415–22. doi:10.1038/ni.2872
32. Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol (2013) 14:654–9. doi:10.1038/ni.2614
33. Wiersinga WJ, Leopold SJ, Cranendonk DR, van der Poll T. Host innate immune responses to sepsis. Virulence (2014) 5:36–44. doi:10.4161/viru.25436
34. Roossinck MJ. Move over bacteria! Viruses make their mark as mutualistic microbial symbionts. J Virol (2015) 89:6532–5. doi:10.1128/JVI/02974-14
35. Pradeu T. Mutualistic viruses and the heteronomy of life. Stud Hist Philos Biol Biomed Sci (2016) 59:80–8. doi:10.1016/j.shpsc.2016.02.007
36. Roossinck MJ. The good viruses: viral mutualistic symbioses. Nat Rev Microbiol (2011) 9:99–108. doi:10.1038/nrmicro2491
37. Herniou EA, Huguet E, Thézé J, Bézier A, Periquet G, Drezen JM. When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Philos Trans R Soc Lond B Biol Sci (2013) 368:20130051. doi:10.1098/rstb.2013.0051
38. Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, et al. Syncytin-A knockout mice demonstrate the critical role in plancentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A (2009) 106:12127–32. doi:10.1073/pnas.0902925106
39. Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature (2014) 516:94–8. doi:10.1038/nature13960
40. Tillmann HL, Heiken H, Knapik-Botor A, Heringlake S, Ockenga J, Wilber JC, et al. Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med (2001) 345:715–24. doi:10.1056/NEJMoa010398
41. Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature (2007) 447:326–9. doi:10.1038/nature05762
42. Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet (2012) 13:283–96. doi:10.1038/nrg3199
43. Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res (2010) 38:4207–17. doi:10.1093/nar/gkq140
44. Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet (2008) 42:709–32. doi:10.1146/annurev.genet.42.110807.091501
45. Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet (2010) 6:e1001191. doi:10.1371/journal.pgen.1001191
46. Huang CRL, Burns KH, Boeke JD. Active transposons in genomes. Annu Rev Genet (2012) 46:651–75. doi:10.1146/annurev-genet-110711-155616
47. Schlesinger S, Goff SP. Retroviral transcriptional regulation and embryonic stem cells: war and peace. Mol Cell Biol (2015) 35:770–7. doi:10.1128/MCB.01293-14
48. Emera D, Wagner GP. Transposable element recruitments in the mammalian placenta: impacts and mechanisms. Brief Funct Genomics (2012) 11:267–76. doi:10.1093/bfgp/els013
49. Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science (2016) 351:1083–7. doi:10.1126/science.aad5497
50. Kassiotis G, Stoye JP. Immune response to endogenous retroelements: taking the bad with the good. Nat Rev Immunol (2016) 16:207–19. doi:10.1038/nri.2016.27
51. Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature (2009) 457:818–23. doi:10.1038/nature0791
52. Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet (2012) 46:21–42. doi:10.1146/annure-genet-110711-155621
53. Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet (2017) 18:71–86. doi:10.1038/nrg.2016.139
54. Vega Thurber R, Payet JP, Thurber AR, Correa AMS. Virus-host interactions and their roles in coral reef health and disease. Nat Rev Microbiol (2017) 15:205–16. doi:10.1038/nrmicro.2016.176
55. Virgin HW. The virome in mammalian physiology and disease. Cell (2014) 157:142–50. doi:10.1016/j.cell.2014.02.032
56. Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasić L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol (2009) 7:828–36. doi:10.1038/nrmicro2235
57. Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell (2009) 238:30–50. doi:10.1016/j.cell.2009.06.036
58. tenOever BR. The evolution of antiviral defense systems. Cell Host Microbe (2016) 19:142–9. doi:10.1016/j.chom.2016.01.006
59. Zhou R, Rana TM. RNA-based mechanisms regulating host-virus interactions. Immunol Rev (2013) 253:97–111. doi:10.1111/imr.12053
61. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity (2011) 34:637–50. doi:10.1016/j.immuni.2011.05.006
62. Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol (2014) 32:461–88. doi:10.1146/annurev-immunol-032713-120156
63. Margolis SR, Wilson SC, Vance RE. Evolutionary origins of cGAS-STING signaling. Trends Immunol (2017) 38:733–43. doi:10.1016/j.it.2017.03.004
64. Motta V, Soares F, Sun T, Philpott DJ. NOD-like receptors: versatile cytosolic sentinels. Physiol Rev (2015) 95:149–78. doi:10.1152/physrev.00009.2014
65. McFadden MJ, Gokhale NS, Horner SM. Protect this house: cytosolic sensing of viruses. Curr Opin Virol (2017) 22:36–43. doi:10.1016/j.coviro.2016.11.012
66. Geijtenbeek TBH, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol (2009) 9:465–79. doi:10.1038/nri2569
67. Fensterl V, Chattopadhyay S, Sen GC. No love lost between viruses and interferons. Annu Rev Virol (2015) 2:549–72. doi:10.1146/annurev-virology-100114-055249
68. Quintin J, Cheng SC, van der Meer JW, Netea MG. Innate immune memory: towards a better understanding of host defense mechanisms. Curr Opin Immunol (2014) 29:1–7. doi:10.1016/j.coi.2014.02.006
69. Dishaw LJ, Flores-Torres J, Lax S, Gemayel K, Leigh B, Melillo D, et al. The gut of geographically disparate Ciona intestinalis harbors a core microbiota. PLoS One (2014) 9:e93386. doi:10.1371/journal.pone.0093386
70. Mortzfeld BM, Urbanski S, Reitzel AM, Künzel S, Technau U, Fraune S. Response of bacterial colonization in Nematostella vectensis to development, environment and biogeography. Environ Microbiol (2015) 18:1764–81. doi:10.1111/1462-2920.12926
71. Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C, et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun (2016) 7:11870. doi:10.1038/ncomms11870
72. Galliot B. Hydra, a fruitful model system for 270 years. Int J Dev Biol (2012) 56:411–23. doi:10.1387/ijdb.120094bg
73. Bosch TCG. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu Rev Microbiol (2013) 67:499–518. doi:10.1146/annurev-micro-092412-155626
74. Schröder K, Bosch TCG. The origin of mucosal immunity: lessons from the holobiont Hydra. MBio (2016) 7:e1184–1116. doi:10.1128/mBio.01184-16
75. Fraune S, Bosch TCG. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci U S A (2007) 104:13146–51. doi:10.1073/pnas.0703375104
76. Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, et al. The dynamic genome of Hydra. Nature (2010) 464:592–6. doi:10.1038/nature08830
77. Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TCG. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci U S A (2006) 103:6208–11. doi:10.1073/pnas.0510163103
78. Franzenburg S, Fraune S, Künzel S, Baines JF, Domazet-Lozo T, Bosch TCG. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci U S A (2012) 109:19374–9. doi:10.1073/pnas.1213110109
79. Kovacevic G. Value of the Hydra model system for studying symbiosis. Int J Dev Biol (2012) 56:627–35. doi:10.1387/ijdb.123510gk
80. Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TC, et al. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci U S A (2013) 110:E3730–8. doi:10.1073/pnas.1304960110
81. Bosch TCG, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol (2009) 33:559–69. doi:10.1016/j.dci.2008.10.004
82. Lange C, Hemmrich G, Klostermeier UC, Lopez-Quintero JA, Miller DJ, Rahn T, et al. Defining the origins of the NOD-like receptor system at the base of animal evolution. Mol Biol Evol (2011) 28:1687–702. doi:10.1093/molbev/msq349
83. Augustin R, Anton-Erxleben F, Jungnickel S, Hemmrich G, Spudy B, Podschun R, et al. Activity of the novel peptide arminin against multiresistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob Agents Chemother (2009) 53:5245–50. doi:10.1128/AAC.00826-09
84. Fraune S, Augustin R, Anton-Erxleben F, Wittleib J, Gelhaus C, Klimovich VB, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci U S A (2010) 107:18067–72. doi:10.1073/pnas.1008573107
85. Bosch TCG, Grasis JA, Lahnit T. Microbial ecology in Hydra: why viruses matter. J Microbiol (2015) 53:193–200. doi:10.1007/s12275-015-4695-2
86. Deines P, Bosch TCG. Transitioning from microbiome composition to microbial community interactions: the potential of the metaorganism Hydra as an experimental model. Front Microbiol (2016) 7:1610. doi:10.3389/fmicb.2016.01610
87. Bordenstein SR, Theis KR. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol (2015) 13:e1002226. doi:10.1371/journal.pbio.1002226
88. Douglas AE, Werren JH. Holes in the hologenome: why host-microbe symbioses are not holobionts. MBio (2016) 7:e2099. doi:10.1128/mBio.02099-15
89. Hester ER, Barott KL, Nulton J, Vermeij MJ, Rohwer FL. Stable and sporadic symbiotic communities of coral and algal holobionts. ISME J (2016) 10:1157–69. doi:10.1038/ismej.2015.190
90. Moran NA, Sloan DB. The hologenome concept: helpful or hollow? PLoS Biol (2015) 13:e1002311. doi:10.1371/journal.pbio.1002311
91. Rosenberg E, Zilber-Rosenberg I. Microbes drive evolution of animals and plants: the hologenome concept. MBio (2016) 7:e01395. doi:10.1128/mBio.01395-15
92. Shropshire JD, Bordenstein SR. Speciation by symbiosis: the microbiome and behavior. MBio (2016) 7:e01785. doi:10.1128/mBio.01785-15
93. Theis KR, Dheilly NM, Klassen JL, Brucker RM, Baines JF, Bosch TCG, et al. Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiome. mSystems (2016) 1:e28–16. doi:10.1128/mSystems.00028-16
94. Gilbert C, Cordaux R. Viruses as vectors of horizontal transfer of genetic material in eukaryotes. Curr Opin Virol (2017) 25:16–22. doi:10.1016/j.coviro.2017.06.005
95. Bordenstein SR, Bordenstein SR. Eukaryotic association module in phage WO genomes from Wolbachia. Nat Commun (2016) 7:13155. doi:10.1038/ncomms13155
Keywords: holobiont, virome, symbiosis, viral metagenomics, host–microbe interactions, innate immunity, antiviral immunity, bacteriophage
Citation: Grasis JA (2017) The Intra-Dependence of Viruses and the Holobiont. Front. Immunol. 8:1501. doi: 10.3389/fimmu.2017.01501
Received: 29 April 2017; Accepted: 24 October 2017;
Published: 09 November 2017
Edited by:
Larry J. Dishaw, University of South Florida St. Petersburg, United StatesReviewed by:
Jonathan L. Klassen, University of Connecticut, United StatesCopyright: © 2017 Grasis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juris A. Grasis, amFncmFzaXNAdWNtZXJjZWQuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.