- 1Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 2Nursing School, Jiangsu Vocational College of Medicine, Yancheng, China
Evidence has shown that dysbiosis of the urinary microbiota existed in female type 2 diabetes mellitus (T2DM) patients. Perturbations of intestinal microbiota are linked to proinflammatory chemokine interleukin-8 (IL-8); however, the correlations between urinary microbiota and IL-8 are not well studied. Here, we investigated the associations between the altered urinary microbiota and urinary IL-8 in female T2DM patients. A modified four-tube midstream urine technique was used to collect urine specimens from 70 female T2DM patients and 70 matched healthy controls (HCs). Bacterial genomic DNA from urine specimens was isolated using magnetic beads and the urinary microbiota was assessed using Illumina MiSeq platform targeting on the 16S rRNA gene V3–V4 region. Urinary IL-8 was determined by enzyme linked immunosorbent assay. Subsequently, the T2DM patients were separated into urine IL-8 detectable (WIL8) and undetectable (NIL8) groups, and the composition of urinary microbiota between the two groups was compared. Meanwhile, the levels of IL-8 between the “≥HCs” group (those specific bacterial genera were more than or equal to the HCs) and the “<HCs” group (those specific bacterial genera were less than the HCs) was also compared. Of 70 urine samples from T2DM patients without urinary tract infections, 46 patients had detectable IL-8 in their urine (64.31 ± 70.43 pg/mL), while 24 patients had undetectable IL-8. Compared to the NIL8 group, 11 bacterial genera increased in the WIL8 group, including Corynebacterium, Akkermansia, Enterococcus, etc., whereas 10 genera, such as Faecalibacterium, Bacteroides, and Pseudomonas decreased. One species of Lactobacillus, Lactobacillus iners, increased obviously in the WIL8 group. The “≥HCs” group showed 17 genera increased and 16 genera decreased. In addition, 18 genera contributed to the presence of urinary IL-8 in T2DM patients, which explained 95.60% of the total variance of urinary microbiota. Our study demonstrated that dysbiosis of the urinary microbiota with several key bacteria was associated with urinary IL-8 in female T2DM patients, which might be useful to explore the interactions between urinary microbiota and inflammatory responses and shed light on novel diagnosis and therapy for urinary microbiota associated with infections in T2DM patients.
Introduction
Diabetes mellitus (DM) is a common, serious, and costly disease, which is a major public health issue (1). In recent decades, the global prevalence of DM has increased from 4.7% in 1980 to 8.5% in 2014, while the number of people with diabetes has risen from 108 million in 1980 to 422 million in 2014 (1). In 2012, 1.5 million deaths were directly caused by DM, which has been projected as the seventh leading cause of death in 2030 (1). A previous study has reported that type 2 diabetes mellitus (T2DM) comprises the majority of the people with DM around the world, approximately 90% of DM patients are T2DM patients (2).
Patients with DM are prone to various infections, with the urinary tract as the most common infection site (3, 4). Urinary tract infection (UTI) in hospitalized DM patients was nearly two times higher than that caused by other factors (5). The probability of UTIs in T2DM patients was 60% higher than that in non-DM individuals. A survey showed that the incidence of UTIs in T2DM patients was 46.9 per 1,000 person-years, compared with 29.9 for non-T2DM subjects (6).
Cytokines are small, soluble proteins produced by various cells in response to infections and inflammation (7). Interleukin (IL)-8, a potent proinflammatory chemokine and activator of neutrophils, can be stimulated by lipopolysaccharide, IL-1, and tumor necrosis factor alpha (TNF-α). Previous studies have found that interleukin-8 (IL-8) causes migration of neutrophils to the place of inflammation, leading to pyuria in patients with UTIs (8, 9). Elevated urine levels of IL-8 were detected in febrile children with UTIs compared to children with asymptomatic bacteriuria (7). It is reported that urinary IL-8 can be identified as a potential novel biomarker for the diagnosing of UTIs with 93% sensitivity and 90% specificity (10, 11). Ko et al. found that confirmation of the presence of bioactive IL-8 in urine suggests the participation of IL-8 in UTI, providing additional evidence of the role of IL-8 in inflammation (12). Consequently, the measurement of urinary IL-8 is thought to be a potential bioindicator of the localization and severity of inflammation within the urinary tract (9). Epithelial cell lines secrete interleukin in response to stimulation with bacteria (13). Therefore, it is possible that urinary microbiota modulate the presence and levels of IL-8 in the urinary tract.
Previous studies show that the production of cytokines is related to human microbiota. For instance, Schueller et al. found that Veillonellaceae and Neisseriaceae are the most abundant indicators for high IL-8 status, whereas Erysipelotrichaceae is the most abundant bacteria in low IL-8 subjects in an oral microbiota study (14). One study that explores the cytokine secretion of vaginal epithelial cells induced by commensal bacteria shows that colonization of parallel multilayer cultures with Staphylococcus epidermidis resulted in a significant increase in IL-8 relative to non-colonized cultures (15). Cervicovagina those dominated by Gardnerella and Prevotella induced higher levels of IL-8, whereas Lactobacillus iners induced moderate IL-8 secretion and Lactobacillus crispatus did not elicit IL-8 secretion (16). On the other hand, IL-8 in combination with other cytokines plays crucial roles in regulating the microbiota. Previous studies have shown that IL-6 and IL-10 play essential roles for the maintenance of intestinal homeostasis and the prevention of colitis (17, 18), while proinflammatory cytokines, such as IL-17 and IL-15 promote intestinal dysbiosis associated with increased susceptibility to colitis (19, 20). In addition, levels of IL-α, IL-β, and IL-6 are able to stimulate the expression of antimicrobial peptides, which help to shape the skin microbiota and have a protective role against potential pathogen attack (21).
Seventy female T2DM patients were recruited in our previous study, since female patients are known to have higher prevalence of UTI than males (22). We explored whether dysbiosis of urinary microbiota was related to T2DM females. We found that both diversity and richness of urinary microbiota declined in T2DM patients. Actinobacteria, Flavobacteriales, and Flavobacteria were recognized as potential distinguishing biomarkers for T2DM patients. Fourteen bacterial genera, including Lactobacillus, Prevotella, and Streptococcus, were enriched in the T2DM patients, while 19 bacterial genera, such as Pseudomonas, Klebsiella, and Akkermansia, decreased. Meanwhile, the relative abundance of Actinobacteria, Lactobacillus, and Akkermansia muciniphila correlated to the levels of fasting blood and urine glucose (23). The correlations between cytokines and microbiota from intestine, oral cavity, vagina, and skin have been extensively investigated, while the relationship between IL-8 and urinary microbiota remains poorly studied. Here, we investigated whether dysbiosis of urinary microbiota was associated with the presence of urinary IL-8, which might be useful to explore the interactions between the urinary microbiota and immune system and shed light on potential novel diagnosis and therapy for UTIs in T2DM patients.
Materials and Methods
Recruitment of Subjects
We used an individually matched case–control design in our previous study, with one control for each T2DM patient. The matching attributes were age in years (based on decade) and marital and menstrual status. In this study, 70 female T2DM patients and 70 healthy controls (HCs) were recruited from the First Affiliated Hospital, School of Medicine, Zhejiang University from June 28, 2015 to January 2, 2016. Both groups ranged from 26 to 85 years old. The body mass index (BMI) in the HCs was 23.10 ± 4.49 kg/m2 and in the T2DM group was 23.87 ± 3.65 kg/m2. Subjects with the following attributes were excluded: UTIs in the previous month; use of antibiotics, probiotics, prebiotics, or synbiotics in the previous 3 months; unable to complete the questionnaire; menstruation; urinary incontinence; known anatomic urinary tract abnormalities (e.g., cystoceles, hydronephrosis, renal atrophy, or neurogenic bladder); urinary catheter (23). The Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University approved the study (reference number: 295). Written informed consent based on the Declaration of Helsinki was obtained from each patient before enrollment, and all patients were given informed consent.
Collection of Urinary Specimens and DNA Sequencing
A modified four-tube midstream urine collection technique was used for the first urine of the day, which guaranteed that the real midstream urine was collected. A urine sample was discarded when it was confirmed to be contaminated. Samples were given anonymous identification codes and were transferred immediately to the laboratory and stored at −80°C until DNA extraction. For urinary microbiota analysis, total DNA was extracted from the pellet of urine from tubes 2 and 3, and 40 mL of urine was aspirated from each tube, separated into three sections, and injected into three 15 mL sterile centrifuge tubes. Each tube was pelleted by centrifugation at 4,000 × g for 15 min at 4°C. 10 mL of the supernatant was decanted, and the pellet was obtained by centrifugation for 15 min at 4,000 × g at 4°C. The pellet was transferred into a 2 mL sterile centrifugation tube which contained 500 µL of lysis buffer. Magnetic bead isolation of genomic DNA from bacteria was applied according to the manufacturer’s protocol with minor modifications (Supplementary Material: Protocol of DNA Isolation). The 16S rRNA gene V3–V4 regions were amplified from microbial genomic DNA (forward primer, 5'–ACTCCTACGGGAGGCAGCAG–3'; reverse primer, 5'–GGACTACHVGGGTWTCTAAT–3') (23). Urinary IL-8 was determined by enzyme-linked immunosorbent assay kits (RayBiotech, Inc., Norcross, GA, USA), following the manufacturer’s instructions. All measurements were performed in duplicate wells. The lower limit of detection for each assay was 1 pg/mL. Standard curves were generated for every plate and the average 0 standard optical densities were subtracted from the rest of the standards, controls and samples to obtain a corrected concentration.
Bioinformatic Analysis
Sequencing reads were processed using QIIME (version 1.9.0), and included additional quality trimming, demultiplexing, and taxonomic assignments. KW rank sum test and pairwise Wilcoxon test were used for the identification of the different markers, and LDA was used to score each feature in the LEfSe analysis. An index of alpha diversity was calculated with QIIME based on sequence similarity at 97%. Beta diversity was measured by unweighted UniFrac distance, which was also calculated by QIIME. Diversity and richness of bacteria in the urine specimens were calculated using several estimates. These consisted of the level of operational taxonomic units (which provides a measure of bacterial richness), Chao1 (which is also an estimate of bacteria richness) and the Shannon and Simpson indices (which are measures of bacterial diversity). The output file was further analyzed using Statistical Analysis of Metagenomic Profiles software package (version 2.1.3) (24). Sequence data from this study are deposited in the GenBank Sequence Read Archive with accession number SRP 087709.
Grouping
From the concentration of urinary IL-8, the T2DM patients were separated into “with IL-8” (WIL8) group, indicating there was detectable IL-8 in their urine; and “no IL-8” (NIL8) group, indicating there was no detectable IL-8 in their urine. Our previous study showed that the relative abundance of 33 bacterial genera in urine were significantly different between the HCs and the T2DM patients (Figure S1 in Supplementary Material) (23). After obtaining the results of differences in relative abundance of bacterial genera in urine between the HCs and T2DM patients (p < 0.05) (23), the patients were divided into two groups: “≥HCs” group, indicating relative abundance of bacterial genera were not less than the HCs; and “<HCs” group, indicating the relative abundance of bacterial genera were less than the HCs. Thereafter, the levels of IL-8 in the “≥HCs” group and “<HCs” group were compared.
Statistical Analysis
Statistical analysis was performed using the SPSS data analysis program (version 21.0) and Statistical Analysis of Metagenomic Profiles software. For continuous variables, independent t-test, Welch’s t-test, and White’s non-parametric t-test were applied. For categorical variables between groups, either the Pearson chi-square or Fisher’s exact test was used depending on assumption validity. For taxon among subgroups, ANOVA test was applied (Tukey–Kramer was used in Post hoc test, effect size was Eta-squared) with Benjamini–Hochberg FDP false discovery rate correction (25, 26). All tests of significance were two-sided, and p < 0.05, or corrected p < 0.05, was considered statistically significant. In addition, stepwise multiple linear regression analysis was used to determine the factors that significantly affected urine microbiota in T2DM patients. All potential variables (p < 0.05) were entered into the analysis.
Results
Characteristics of T2DM Patients in NIL8 and WIL8 Groups
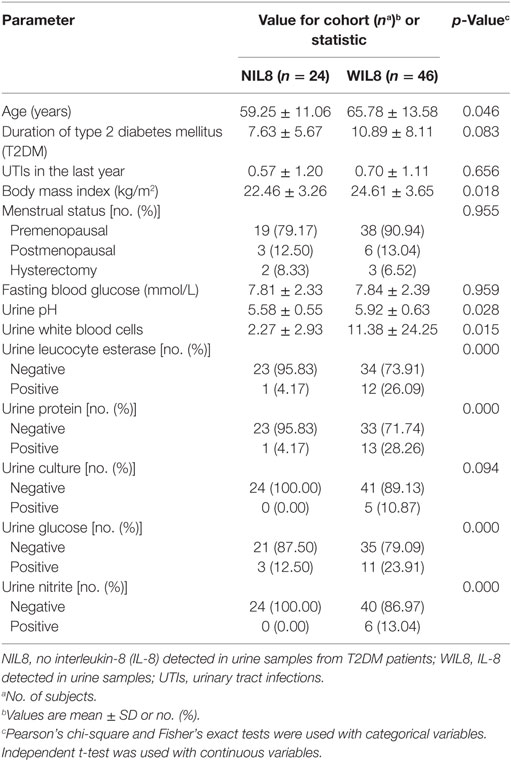
There were significant differences in age, BMI, urine pH, urine white blood cells, urine leukocyte esterase, urine protein, urine glucose, and urine nitrite between T2DM patients with detectable IL-8 in their urine (WIL8) and those without detectable urine IL-8 (NIL8) (p < 0.05, Table 1).
Sequencing Data and Concentrations of Urinary IL-8
Briefly, we obtained 3,981,519 reads for microbiota analysis, which accounted for 76.93% of the valid reads. The mean read length was 438 bp (range 423–486 bp). The Good’s coverage estimator was 98% (23).
The average urinary IL-8 concentration in the 70 patients was 42.26 ± 64.66 pg/mL. Urinary IL-8 was found in 46 samples and concentration was 64.31 ± 70.43 pg/mL. No significant difference was found in Shannon and Simpson indices between the NIL8 and WIL8 group (p > 0.05, Table S1 in Supplementary Material). However, principal coordinate analysis (PCoA) indicated that most of the samples from NIL8 and WIL8 groups could be clustered together (Figure 1).
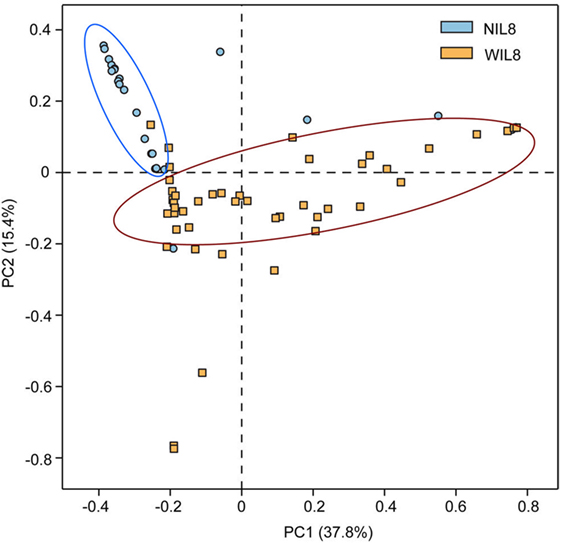
Figure 1. Principal coordinate analysis (PCoA) plot. PCoA plot of the urinary microbiota based on the unweighted UniFrac metric. Blue and yellow dots represent NIL8 and WIL8 specimens, respectively.
Urinary IL-8 Associated Biomarkers
To identify the specific bacterial taxa associated with urinary IL-8, the urinary microbiota in the WIL8 and NIL8 groups were compared using LEfSe. A cladogram representative of the structure of the urinary microbiota and their predominant bacteria is shown in Figure 2. The greatest differences in taxa between two groups are displayed. Bifidobacteriaceae, Shuttleworthia, Thermus, Thermales, and Thermaceae could be used as potential distinguishing biomarkers between the WIL8 and NIL8 groups.
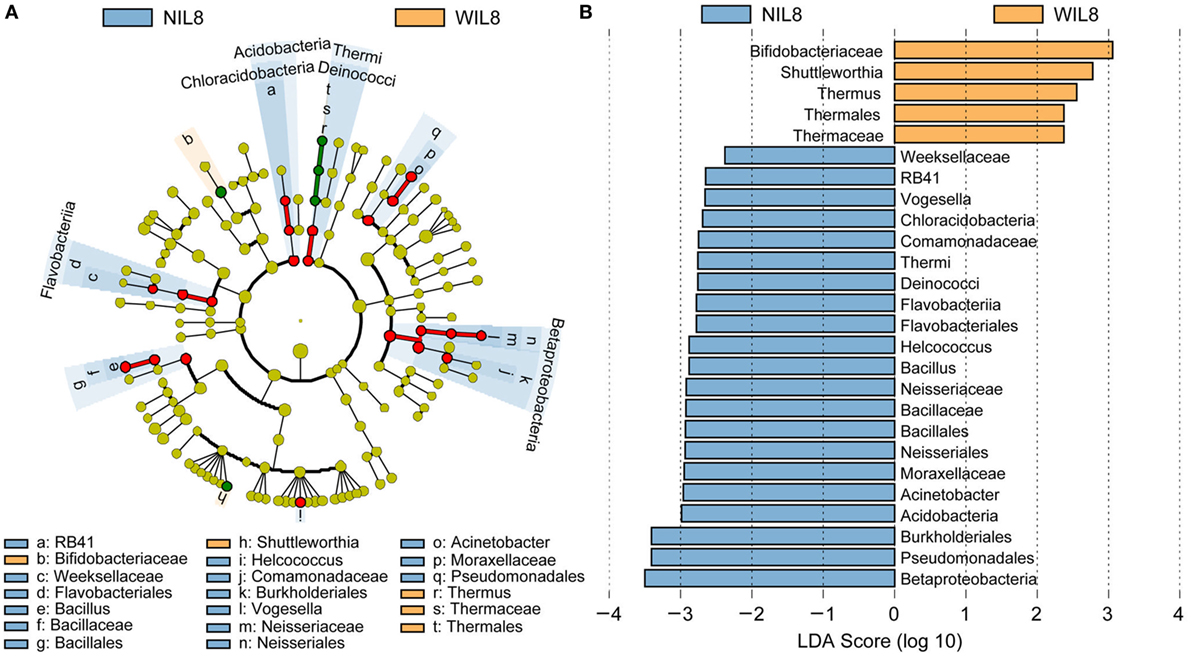
Figure 2. Cladogram showing differentially abundant taxa of urinary microbiota in type 2 diabetes mellitus patients. (A) LEfSe cladogram showed the most differentially abundant taxa between WIL8 and NIL8 groups. Taxonomic cladogram obtained from LEfSe analysis of 16S rDNA sequences. Taxa enriched for WIL8 in yellow; NIL8 enriched taxa in blue. The size of each dot is proportional to its effect size. (B) Only taxa meeting an LDA threshold > 2.0 are shown.
Associations between Urinary Microbiota and IL-8
At the phylum level, Proteobacteria was significantly higher in the WIL8 group than the NIL8 group, while Bacteroidetes was dramatically decreased in the WIL8 group (p < 0.05, Figure 3). At the genus level, 11 genera were enriched in the WIL8 group compared to the NIL8 group, including Shuttleworthia, Mobiluncus, Peptoniphilus, Corynebacterium, Thermus, Gemella, Enterococcus, Acinetobacter, Akkermansia, Aquaspirillum, and Geobacillus, while 10 genera were decreased, including Faecalibacterium, Megamonas, Comamonas, Bacteroides, Coprococcus, Sutterella, Pseudomonas, Phascolarctobacterium, Prevotella, and Parabacteroides (p < 0.05, Figure 4). Five bacterial species, Streptococcus anginosus, Acinetobacter rhizosphaerae, Acinetobacter schindleri, L. iners, and A. muciniphila, showed a significant increase in the WIL8 group than the NIL8 group, while another five species showed a significant decrease, including Prevotella copri, Faecalibacterium prausnitzii, Prevotella stercorea, and Bacteroides uniformis, and Coprococcus eutactus (p < 0.05, Figure S2 in Supplementary Material). L. iners dramatically increased in the WIL8 group. In addition, the Lactobacillus species including Lactobacillus mucosae and Lactobacillus reuteri showed a trend increase in the WIL8 group, whereas Lactobacillus ruminis showed a trend decrease, but these species did not reach statistical differences (Figure S3 in Supplementary Material).
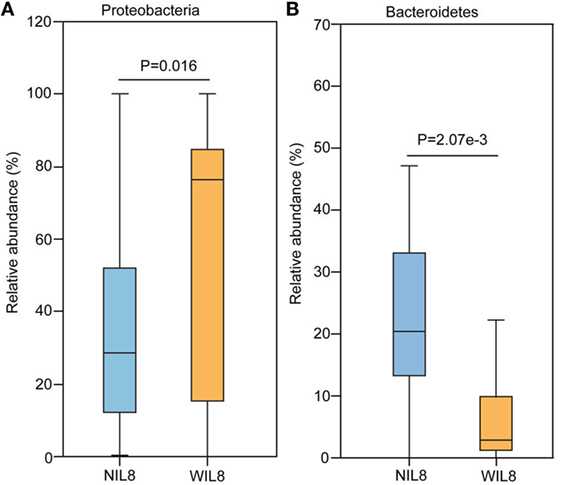
Figure 3. Phylum-level operational taxonomic units different between NIL8 and WIL8 groups. STAMP software was used to calculate the bacterial phylum proportions in the two groups: Proteobacteria (A) and Bacteroidetes (B). Welch’s t-test was used to compare abundance at the bacterial phylum level for NIL8 urine samples and WIL8 samples. The different phyla were assigned only to those presenting a minimum variation at a significant level [p (corrected) < 0.05].
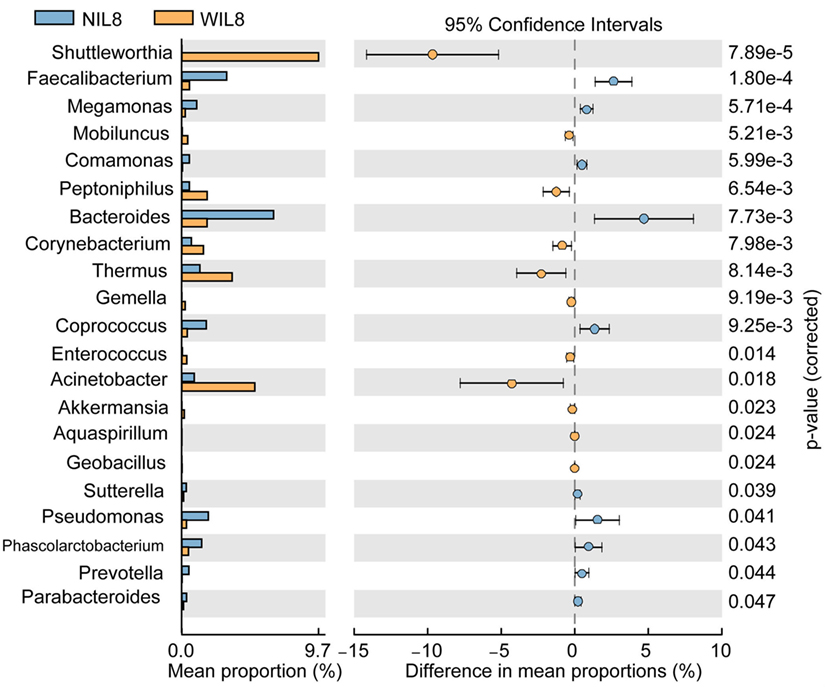
Figure 4. Genus-level operational taxonomic units different between NIL8 and WIL8 groups. STAMP software was used to calculate the genus proportions in the two groups. Welch’s t-test was used to compare abundance at the genus level for NIL8 and WIL8 specimens. The different genera were assigned only to those presenting a minimum variation at a significant level [p (corrected) < 0.05].
Interestingly, 17 bacterial genera were enriched in the “≥HCs” group, while 16 genera were enriched in the “<HCs” group. Specifically, those patients with Bacteroides “≥HCs” group, Klebsiella “≥HCs” group, Pseudomonas “≥HCs” group and Akkermansia “≥HCs” group had higher concentrations of urinary IL-8, while those patients with Lactobacillus “<HCs” group, Megamonas “<HCs” group and Microbacterium “<HCs” group had higher levels of IL-8 (Figure 5).
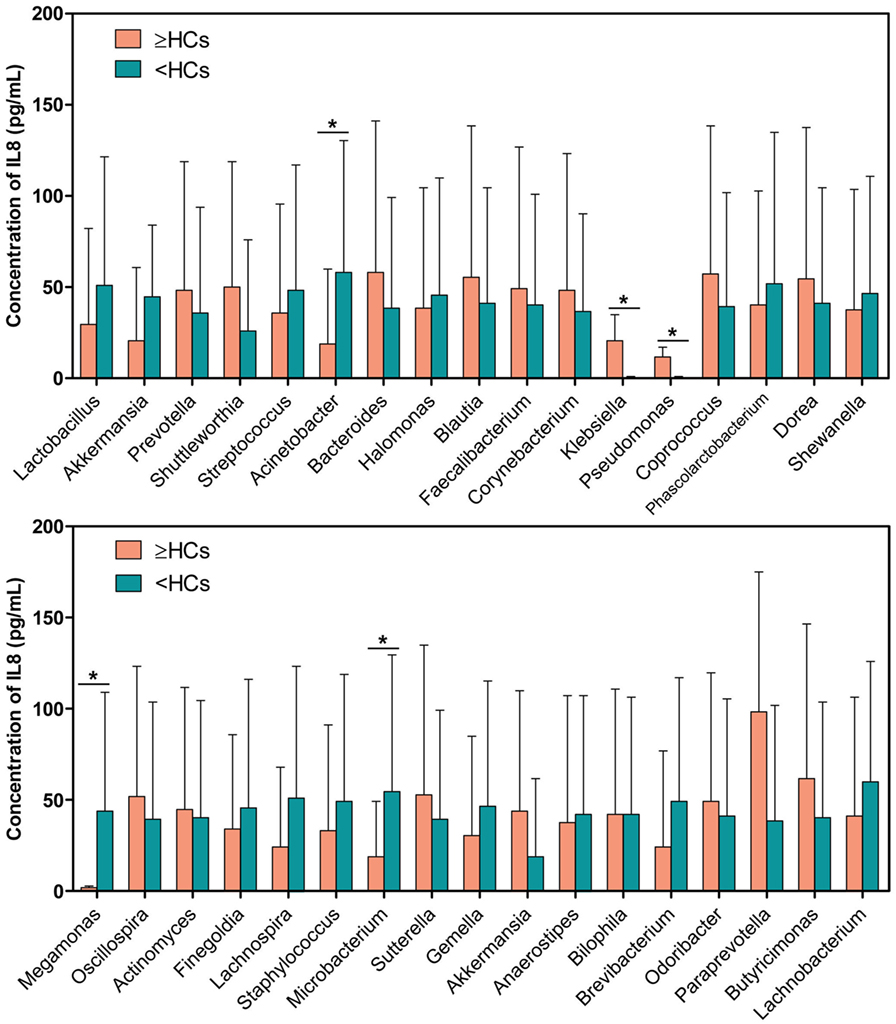
Figure 5. Comparison of interleukin-8 (IL-8) levels between genus “≥HCs” group and “<HCs” group. SPSS software was used to calculate the difference in IL-8 concentrations between bacterial genus “≥HCs” group and “<HCs” group. “≥HCs” group indicates the relative abundance of a bacterial genus in this group was not less than the healthy controls (HCs), and “<HCs” group indicates relative abundance of a bacterial genus was less than the HCs. Asterisks indicate significant differences between the two groups (p < 0.05).
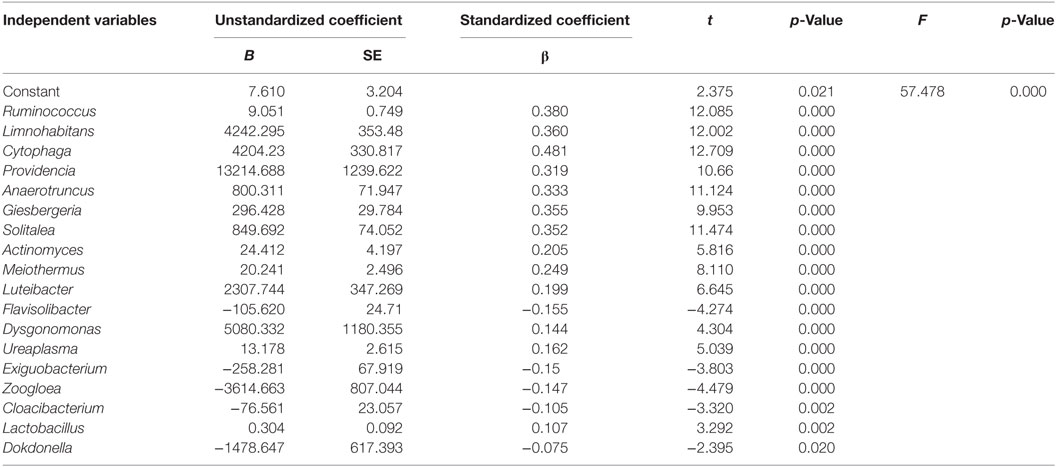
Multiple regression analysis showed that the genera Ruminococcus, Limnohabitans, Cytophaga, Providencia, Anaerotruncus, Giesbergeria, Solitalea, Actinomyces, Meiothermus, Luteibacter, Flavisolibacter, Dysgonomonas, Ureaplasma, Exiguobacterium, Zoogloea, Cloacibacterium, Lactobacillus, and Dokdonella significantly affected urinary microbiota in T2DM patients and explained 95.60% of the total variance of urinary microbiota in this population (Table 2). In addition, the relative abundance of Ruminococcus was significantly positively associated with the levels of urinary IL-8 (Figure S4 in Supplementary Material).
Discussion
Type 2 diabetes mellitus patients are prone to a higher occurrence of certain infections compared with the healthy population (27). The hospitalization rate for UTIs caused by diabetes is over twice those caused by other factors (5). Defects in maintaining the integrity of mucosal barriers can result in systemic endotoxemia that contributes to chronic low-grade inflammation (28). Recent advances in understanding the pathophysiology of T2DM have established the involvement of low-grade inflammation due to an increased production of proinflammatory cytokines (29, 30). This study focused on the correlations between urinary microbiota and the proinflammatory chemokine IL-8 for the first time, with the aim of providing new insights on host–urinary microbiota interactions in T2DM patients.
Urine specimens from 46 T2DM patients had detectable levels of IL-8 (WIL8 group), while IL-8 was not detected in 24 T2DM patients (NIL8 group). The alpha diversity indices, such as Shannon and Simpson, did not show significant differences between the NIL8 and WIL8 groups, indicating that bacterial diversity was not affected by the presence of IL-8 in urine. The beta diversity index such as PCoA analysis indicated that most of the patients with urinary IL-8 formed their own cluster, which reflected a contribution from IL-8 was prominent in differentiating urinary microbiota in groups.
Interestingly, the distinguishing biomarker, Bifidobacteriaceae, which is associated with diabetic patients with higher BMIs (31), had a higher abundance in WIL8 subjects, and those patients also had higher BMIs than NIL8 participants in our study. Elfeky et al. reported that exosomes increased IL-8 release from endothelial cells, and the effect was even higher when exosomes were isolated from obese women compared to lean subjects (32). Thus, BMI might play a role in regulating IL-8 levels, and subsequently IL-8 modulates the abundance of urinary Bifidobacteriaceae.
A higher abundance of Proteobacteria was found in the WIL8 group in this study. Fricke et al. and Rani et al. have demonstrated that Proteobacteria in urine decreased in patients that had renal transplantation and were exposed to high dose immunosuppressant (33, 34), during which cytokine production might be inhibited by the suppressing T cells induced by administrating the immunosuppressant. In addition, a recent study on intestinal microbiota reported that Proteobacteria was detected in feces of rats at the peak of experimental autoimmune encephalomyelitis (35). Demmer et al. demonstrated that inflammation explained 30–98% of the observed associations between levels of microbiota in a subgingival microbiome study, and the percentages of the overall phyla associations with inflammation were 27% for Proteobacteria (36). The above findings illustrated that Proteobacteria might be correlated to the onset or development of the inflammatory process. It is demonstrated that a rise in species belonging to the phylum Proteobacteria may have a larger impact on host autoimmunity which may make a protein molecule non-functional and thereby may be involved in the onset of inflammatory disorders, including diabetes (37).
The relative abundance of Bacteroidetes was lowered in the WIL8 group. A similar alteration was found in a study that showed that patients with urgency urinary incontinence had decreased abundance of Bacteroidetes compared to HCs (38). The patients suffering from renal transplantation and successive immunosuppressing therapy also had lowered levels of Bacteroidetes (34). Bacteroidetes can suppress enteric inflammation, suggesting that members of the phylum play a similar role in regulating the level of IL-8 in urinary tract system. Interestingly, there was a noticeable decrease in the phylum Bacteroidetes in newly diagnosed diabetics (39), and in our present study, we observed that patients with detectable IL-8 had T2DM for a longer duration than the NIL8 patients, suggesting that in the early state of diabetes, Bacteroidetes play a minor role in regulating the inflammatory process in diabetes.
We found that the genus of Corynebacterium in the urinary microbiota was enriched in WIL8 patients. A recent study reported that the prevalence of Corynebacterium correlated with concentrations of IL-6 and C-reaction protein in cancer patients (40). In our present study, the WIL8 group had a higher level of urine white blood cells, more leukocyte esterase and urine nitrite-positive cases, which indicated an inflammatory reaction was present in the urinary tract system, although no UTI was diagnosed currently. This might suggest that an inflammatory reaction and its correlation with urinary microbiota is established before a patient is diagnosed with UTI and the presence of obvious clinical manifestation, thus early detection of the interaction of the inflammatory response with urinary microbiota is valuable for clinicians to diagnose and treat UTIs in the early stages.
A higher abundance of A. muciniphila was correlated with a higher concentration of IL-8. Moreover, Akkermansia in the “≥HCs” group had a higher level of IL-8 than the “<HC” group. An in vitro model demonstrated that A. muciniphila could induce IL-8 production by enterocytes at cell concentrations 100-fold higher than those for Escherichia coli (41). Another study demonstrated that mice with low levels of inflammation were enriched for Akkermansia (42). Surprisingly, the patients from the WIL8 group had higher levels of urine leukocyte esterase and nitrite than the NIL8 patients, suggesting Akkermansia might play important roles in protecting patients away from UTIs, since patients in either WIL8 or NIL8 groups were not currently diagnosed with UTIs while they were recruited to the present study.
Lin et al. reported that the overgrowth of Enterococcus in diabetic mice was accompanied with increased IL-1β and TNF-α expression from Kupffer cells in intestine (43). Interestingly, our data indicated that Enterococcus was dramatically increased in the WIL8 group. Therefore, the abundance of Enterococcus might be regulating the level of cytokines in diabetic populations. Nienhouse et al. reported that Pseudomonas was enriched in positive urine culture specimens comparing to negative specimens. Also, Pseudomonas might cause the most severe inflammation which was accompanied by an increase in the number of inflammatory cells and IL-6 (44). This trend was similar to our study, in which patients with Pseudomonas in the “≥HCs” group had a higher level of IL-8. Interestingly, Pseudomonas aeruginosa were the predominant isolates of non-healing ulcers in diabetic foot patients (45), and diabetic foot infection has demonstrated higher concentrations of IL-6 and IL-1β than controls (46). Thus, Pseudomonas might be correlated to the inflammatory process in diabetic patients and might regulate the levels of cytokines. Klebsiella, being a recognized uropathogen (47), was enriched in female participants who developed UTIs after pelvic floor surgery (48). Interestingly, patients with Klebsiella in the “≥HCs” group had a higher concentration of IL-8. Huang et al. reported that diabetic patients was associated with relapse of recurrent bacteremia caused by Klebsiella pneumonia (49), suggesting that this bacteria is responsible for the inflammation process in diabetes. S. anginosus was also increased in the WIL8 group compared to the NIL8 group. A similar result was reported in the study conducted by Price et al., in which S. anginosus increased in subjects with UTIs compared to the non-UTI participants (50).
Members of the genus Lactobacillus exhibit probiotic effects in epithelial attachment, pathogen inhibition, and intestinal immunomodulation (51–53). However, their relative abundance was increased in the WIL8 group compared to the NIL8 group. Furthermore, Lactobacillus was one of the predictors of the presence of IL-8 in urine in our study. It was reported that Lactobacillus showed a rise in interstitial cystitis patients than HCs (54). However, a recent study reported that there were no associations between the presence of Lactobacillus and urinary cytokine levels in patients with interstitial cystitis (55). Therefore, Lactobacillus might play distinctive roles in regulating cytokine production in urine in different health statuses. L. iners, which may induce moderate IL-8 secretion and has moderate proinflammatory activity in the cervicovaginal bacterial community (16), was also increased in the WIL8 group. L. mucosae which has been demonstrated to possess IL-6 induction ability in macrophages (56), was found enriched in the WIL8 group. L. reuteri can suppress intestinal inflammation in a trinitrobenzene sulfonic acid-induced mouse colitis model via downregulation of gene expression of mucosal cytokine IL-6 and IL-1β in the colon (57). In another study, L. reuteri could produce molecules that had potential anti-TNF-α activity in vitro and antimicrobial compounds in diabetes (58, 59). Furthermore, intake of L. reuteri can increase insulin secretion, which might be due to augmented incretin release (60). Low-grade chronic inflammation is accepted as an internal metabolic adaptation pathway in T2DM, so the increased levels of members of the genus Lactobacillus might be believed to be players in controlling the development of inflammation in diabetes. Moreover, patients with the Lactobacillus “≥HCs” group had lower levels of IL-8 than the “<HCs” group, which also demonstrated that Lactobacillus contributed to inhibit the inflammatory process in the bladder.
In total, 18 bacterial genera contributed to the presence of IL-8 in the urine of T2DM patients. Most of these genera have not been reported by human urinary microbiota studies, except for Actinomyces (61) and Lactobacillus (47, 50, 61–65). Furthermore, very few of these genera have been reported in terms of their correlations with inflammatory cytokines. Interestingly, the abundance of Ruminococcus was positively correlated with the concentration of urinary IL-8, and can be considered as an important contributor to the presence of IL-8 as well. Ruminococcus spp. could induce dominant effector ex vivo mesenteric lymph node T-helper 17 responses (66), which is correlated to inducing the expression of IL-8 (67). In addition, Ruminococcus was more abundant in diabetic mice, and correlated negatively with delayed diabetes onset age (68). Thus, Ruminococcus might play a role in diabetes development by regulating the level of cytokines. Actinomyces spp. has been demonstrated to induce inflammatory cytokines by researchers (69), which were linked to the presence of urinary IL-8 in the present study suggesting therapy for inflammatory development in diabetes involving urinary microbiota should take a considerable to the 18 bacteria included in multiple analysis model.
There were several limitations in our study. First, the sample size in the WIL8 and NIL8 groups was not equal, which might affect the reliability of the results. Second, although all participants were not currently diagnosed with UTIs, we could not completely rule out the influence caused by previous occurrence of UTIs since it takes 6 months for diabetic patients to revert to normal glomerular filtration rate trends after an infection is cured (70); this might affect the growth environment of urinary microbiota and urine IL-8.
Conclusion
To our knowledge, this is the first study focused on the associations between urinary microbiota and concentrations of urine IL-8 in T2DM patients. The bacterial community showed differences in the WIL8 and the NIL8 groups, and the concentrations of IL-8 were different in the “≥HCs” group and “<HCs” group. Findings from this study indicated that urine IL-8 is interplayed with urinary microbiota among T2DM patients. Future studies should focus on how the urinary microbiota affects the inflammatory cytokine excretion in the urinary tract, which might be conducive to explore novel therapies to regulate inflammation in T2DM patients.
Ethics Statement
Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University approved the study (Reference Number: 295).
Author Contributions
LL and FL conceived and designed the study. ZL generated the sequencing data. FL and SL collected the samples. FL and YC conducted urine cultures and the urinalysis. FL extracted the bacterial DNA. ZL and FL analyzed the data, carried out the computational analysis, interpreted the data, and drafted the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge the volunteers who participated in our study.
Funding
This present work was funded by National Natural Science Foundation of China under Grant No. 81400586, and the Opening Foundation of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases under Grant No. 2015KF05.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.01032/full#supplementary-material.
Materials and Methods
Protocol of DNA Isolation
The tube was placed in liquid nitrogen for 1 min, and transferred into a water bath at 65°C for 5 min, with vigorous mixing. This last process was repeated three times with a final bath for 30 min. 50 µL Agencourt AMPure XP (Beckman Coulter, USA) was added to 100 µL of the urine pellet, vortexed for 30 s, and incubated for 5 min at room temperature. The tube was placed into a magnetic separator for 5 min, and DNA was bound to magnetic beads which were drawn to the wall of the microcentrifuge tube. The supernatant was carefully removed without disrupting the magnetic beads. The sample was washed twice with 200 µL 80% ethanol for 30 s, being placed on a magnet separator between each washing. The purified DNA was eluted with 50 µL ddH2O for 1 min. The beads, now released from the DNA, were collected with the magnet. The DNA-containing supernatant was transferred to a clean tube.
Figure S1. Genus-level operational taxonomic units different between healthy controls (HCs) and type 2 diabetes mellitus (T2DM) patients. Welch’s t-test was used to compare the abundance at the bacterial genus level between HCs and T2DM patients. The different genera were assigned only to those presenting a minimum variation at a significant level [p (corrected) < 0.05]. H and Pt represent HCs and T2DM patients, respectively.
Figure S2. Species-level operational taxonomic units different between NIL8 and WIL8 groups. STAMP software was used to calculate the species proportions in the two groups. Welch’s t-test was used to compare abundance at the species level for NIL8 and WIL8 specimens. The different species were assigned only to those presenting a minimum variation at a significant level [p (corrected) < 0.05].
Figure S3. Differences of Lactobacillus spp. between NIL8 and WIL8 groups. STAMP software was used to calculate the proportions of Lactobacillus spp. in NIL8 and WIL8 groups. Welch’s t-test was used to compare abundance of Lactobacillus spp. level for NIL8 and WIL8 specimens. The different levels were assigned only to those presenting a minimum variation at a significant level [p (corrected) < 0.05].
Figure S4. Correlation between the relative abundance of Ruminococcus and the concentration of urinary IL-8. A correlation analysis was carried out and a significance level of p < 0.05 was used.
References
2. Rubino F. Is type 2 diabetes an operable intestinal disease? A provocative yet reasonable hypothesis. Diabetes Care (2008) 31:S290–6. doi:10.2337/dc08-s271
3. Shah BR, Hux JE. Quantifying the risk of infectious diseases for people with diabetes. Diabetes Care (2003) 26(2):510–3. doi:10.2337/diacare.26.2.510
4. Boyko EJ, Fihn SD, Scholes D, Abraham L, Monsey B. Risk of urinary tract infection and asymptomatic bacteriuria among diabetic and nondiabetic postmenopausal women. Am J Epidemiol (2005) 161(6):557–64. doi:10.1093/aje/kwi078
5. Korbel L, Spencer JD. Diabetes mellitus and infection: an evaluation of hospital utilization and management costs in the United States. J Diabetes Complications (2015) 29(2):192–5. doi:10.1016/j.jdiacomp.2014.11.005
6. Hirji I, Guo ZC, Andersson SW, Hammar N, Gomez-Caminero A. Incidence of urinary tract infection among patients with type 2 diabetes in the UK general practice research database (GPRD). J Diabetes Complications (2012) 26(6):513–6. doi:10.1016/j.jdiacomp.2012.06.008
7. Krzemien G, Szmigielska A, Turczyn A, Panczyk-Tomaszewska M. Urine interleukin-6, interleukin-8 and transforming growth factor beta1 in infants with urinary tract infection and asymptomatic bacteriuria. Cent Eur J Immunol (2016) 41(3):260–7. doi:10.5114/ceji.2016.63125
8. Gurgoze MK, Akarsu S, Yilmaz E, Godekmerdan A, Akca Z, Ciftci I, et al. Proinflammatory cytokines and procalcitonin in children with acute pyelonephritis. Pediatr Nephrol (2005) 20(10):1445–8. doi:10.1007/s00467-005-1941-6
9. Gokce I, Alpay H, Biyikli N, Unluguzel G, Dede F, Topuzoglu A. Urinary levels of interleukin-6 and interleukin-8 in patients with vesicoureteral reflux and renal parenchymal scar. Pediatr Nephrol (2010) 25(5):905–12. doi:10.1007/s00467-009-1396-2
10. Rao WH, Evans GS, Finn A. The significance of interleukin 8 in urine. Arch Dis Child (2001) 85(3):256–62. doi:10.1136/adc.85.3.256
11. Nanda N, Juthani-Mehta M. Novel biomarkers for the diagnosis of urinary tract infection-a systematic review. Biomark Insights (2009) 4:111–21.
12. Ko YC, Mukaida N, Ishiyama S, Tokue A, Kawai T, Matsushima K, et al. Elevated interleukin-8 levels in the urine of patients with urinary-tract infections. Infect Immun (1993) 61(4):1307–14.
13. Hedges S, Svensson M, Svanborg C. Interleukin-6 response of epithelial cell lines to bacterial stimulation in vitro. Infect Immun (1992) 60(4):1295–301.
14. Schueller K, Riva A, Pfeiffer S, Berry D, Somoza V. Members of the oral microbiota are associated with IL-8 release by gingival epithelial cells in healthy individuals. Front Microbiol (2017) 8:416. doi:10.3389/Fmicb.2017.00416
15. Rose WA, Mcgowin CL, Spagnuolo RA, Eaves-Pyles TD, Popov VL, Pyles RB. Commensal bacteria modulate innate immune responses of vaginal epithelial cell multilayer cultures. PLoS One (2012) 7(3):e32728. doi:10.1371/journal.pone.0032728
16. Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity (2015) 42(5):965–76. doi:10.1016/j.immuni.2015.04.019
17. Morrison PJ, Ballantyne SJ, Kullberg MC. Interleukin-23 and T helper 17-type responses in intestinal inflammation: from cytokines to T-cell plasticity. Immunology (2011) 133(4):397–408. doi:10.1111/j.1365-2567.2011.03454.x
18. Kole A, Maloy KJ. Control of intestinal inflammation by interleukin-10. Curr Top Microbiol Immunol (2014) 380:19–38. doi:10.1007/978-3-662-43492-5_2
19. Puel A, Cypowyj S, Bustamante J, Wright JF, Liu LY, Lim HK, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science (2011) 332(6025):65–8. doi:10.1126/science.1200439
20. Meisel M, Mayassi T, Fehlner-Peach H, Koval JC, O’brien SL, Hinterleitner R, et al. Interleukin-15 promotes intestinal dysbiosis with butyrate deficiency associated with increased susceptibility to colitis. ISME J (2017) 11(1):15–30. doi:10.1038/ismej.2016.114
21. Percoco G, Merle C, Jaouen T, Ramdani Y, Benard M, Hillion M, et al. Antimicrobial peptides and pro-inflammatory cytokines are differentially regulated across epidermal layers following bacterial stimuli. Exp Dermatol (2013) 22(12):800–6. doi:10.1111/exd.12259
22. Sewify M, Nair S, Warsame S, Murad M, Alhubail A, Behbehani K, et al. Prevalence of urinary tract infection and antimicrobial susceptibility among diabetic patients with controlled and uncontrolled glycemia in Kuwait. J Diabetes Res (2016) 2016:Article ID 65732157. doi:10.1155/2016/6573215
23. Liu FP, Ling ZX, Xiao YH, Lv LX, Yang Q, Wang BH, et al. Dysbiosis of urinary microbiota is positively correlated with type 2 diabetes mellitus. Oncotarget (2017) 8(3):3798–810. doi:10.18632/oncotarget.14028
24. Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics (2014) 30(21):3123–4. doi:10.1093/bioinformatics/btu494
25. White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol (2009) 5(4):e1000352. doi:10.1371/journal.pcbi.1000352
26. Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics (2010) 26(6):715–21. doi:10.1093/bioinformatics/btq041
27. Wilke T, Boettger B, Berg B, Groth A, Mueller S, Botteman M, et al. Epidemiology of urinary tract infections in type 2 diabetes mellitus patients: an analysis based on a large sample of 456,586 German T2DM patients. J Diabetes Complications (2015) 29(8):1015–23. doi:10.1016/j.jdiacomp.2015.08.021
28. Wang XT, Ota N, Manzanillo P, Kates L, Zavala-Solorio J, Eidenschenk C, et al. Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature (2014) 514(7521):237–41. doi:10.1038/nature13564
29. Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol (2011) 11(2):98–107. doi:10.1038/nri2925
30. Nunemaker CS. Considerations for defining cytokine dose, duration, and milieu that are appropriate for modeling chronic low-grade inflammation in type 2 diabetes. J Diabetes Res (2016) 2016:2846570. doi:10.1155/2016/2846570
31. Long J, Cai Q, Steinwandel M, Hargreaves MK, Bordenstein SR, Blot WJ, et al. Association of oral microbiome with type 2 diabetes risk. J Periodontal Res (2017) 52(3):636–43. doi:10.1111/jre.12432
32. Elfeky O, Longo S, Lai A, Rice GE, Salomon C. Influence of maternal BMI on the exosomal profile during gestation and their role on maternal systemic inflammation. Placenta (2017) 50:60–9. doi:10.1016/j.placenta.2016.12.020
33. Fricke WF, Maddox C, Song Y, Bromberg JS. Human microbiota characterization in the course of renal transplantation. Am J Transplant (2014) 14(2):416–27. doi:10.1111/ajt.12588
34. Rani A, Ranjan R, Mcgee HS, Andropolis KE, Panchal DV, Hajjiri Z, et al. Urinary microbiome of kidney transplant patients reveals dysbiosis with potential for antibiotic resistance. Transl Res (2017) 181:59–70. doi:10.1016/j.trsl.2016.08.008
35. Stanisavljevic S, Lukic J, Sokovic S, Mihajlovic S, Mostarica Stojkovic M, Miljkovic D, et al. Correlation of gut microbiota composition with resistance to experimental autoimmune encephalomyelitis in rats. Front Microbiol (2016) 7:363–73. doi:10.3389/fmicb.2016.02005
36. Demmer RT, Breskin A, Rosenbaum M, Zuk A, Leduc C, Leibel R, et al. The subgingival microbiome, systemic inflammation and insulin resistance: the oral infections, glucose intolerance and insulin resistance study. J Clin Periodontol (2017) 44(3):255–65. doi:10.1111/jcpe.12664
37. Negi S, Singh H, Mukhopadhyay A. Gut bacterial peptides with autoimmunity potential as environmental trigger for late onset complex diseases: in-silico study. PLoS One (2017) 12(7):e0180518. doi:10.1371/journal.pone.0180518
38. Karstens L, Asquith M, Davin S, Stauffer P, Fair D, Gregory WT, et al. Does the urinary microbiome play a role in urgency urinary incontinence and its severity? Front Cell Infect Microbiol (2016) 6:78. doi:10.3389/Fcimb.2016.00078
39. Bhute SS, Suryavanshi MV, Joshi SM, Yajnik CS, Shouche YS, Ghaskadbi SS. Gut microbial diversity assessment of Indian type-2-diabetics reveals alterations in eubacteria, archaea, and eukaryotes. Front Microbiol (2017) 8:214. doi:10.3389/fmicb.2017.00214
40. Chua LL, Rajasuriar R, Azanan MS, Abdullah NK, Tang MS, Lee SC, et al. Reduced microbial diversity in adult survivors of childhood acute lymphoblastic leukemia and microbial associations with increased immune activation. Microbiome (2017) 5:35. doi:10.1186/s40168-017-0250-1
41. Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, et al. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol (2015) 81(11):3655–62. doi:10.1128/Aem.04050-14
42. Borton MA, Sabag-Daigle A, Wu J, Solden LM, O’banion BS, Daly RA, et al. Chemical and pathogen-induced inflammation disrupt the murine intestinal microbiome. Microbiome (2017) 5(1):47. doi:10.1186/s40168-017-0264-8
43. Lin SH, Chung PH, Wu YY, Fung CP, Hsu CM, Chen LW. Inhibition of nitric oxide production reverses diabetes-induced Kupffer cell activation and Klebsiella pneumonia liver translocation. PLoS One (2017) 12(5):e0177269. doi:10.1371/journal.pone.0177269
44. Song C, Li H, Zhang Y, Yu J. Effects of Pseudomonas aeruginosa and Streptococcus mitis mixed infection on TLR4-mediated immune response in acute pneumonia mouse model. BMC Microbiol (2017) 17(1):82. doi:10.1186/s12866-017-0999-1
45. Noor S, Ahmad J, Maazozair IP, Ozair M. Culture-based screening of aerobic microbiome in diabetic foot subjects and developing non-healing ulcers. Front Microbiol (2016) 7:1792. doi:10.3389/Fmicb.2016.01792
46. Tiwari S, Pratyush DD, Gupta SK, Singh SK. Vitamin D deficiency is associated with inflammatory cytokine concentrations in patients with diabetic foot infection. Br J Nutr (2014) 112(12):1938–43. doi:10.1017/S0007114514003018
47. Wolfe AJ, Toh E, Shibata N, Rong RC, Kenton K, Fitzgerald M, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol (2012) 50(4):1376–83. doi:10.1128/Jcm.05852-11
48. Ottman N, Reunanen J, Meijerink M, Pietila TE, Kainulainen V, Klievink J, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One (2017) 12(3):e0173004. doi:10.1371/journal.pone.0173004
49. Huang YT, Liao CH, Teng LJ, Hsu HS, Hsueh PR. Reinfection and relapse of recurrent bacteremia caused by Klebsiella pneumoniae in a medical center in Taiwan. Future Microbiol (2016) 11(9):1157–65. doi:10.2217/fmb-2016-0036
50. Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, Brincat C, et al. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol (2016) 54(5):1216–22. doi:10.1128/Jcm.00044-16
51. Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet AC, et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc Natl Acad Sci U S A (2004) 101(8):2512–7. doi:10.1073/pnas.0307327101
52. Watanabe M, Kinoshita H, Nitta M, Yukishita R, Kawai Y, Kimura K, et al. Identification of a new adhesin-like protein from Lactobacillus mucosae ME-340 with specific affinity to the human blood group A and B antigens. J Appl Microbiol (2010) 109(3):927–35. doi:10.1111/j.1365-2672.2010.04719.x
53. Lee JH, Valeriano VD, Shin YR, Chae JP, Kim GB, Ham JS, et al. Genome sequence of Lactobacillus mucosae LM1, isolated from piglet feces. J Bacteriol (2012) 194(17):4766. doi:10.1128/JB.01011-12
54. Siddiqui H, Lagesen K, Nederbragt AJ, Jeansson SL, Jakobsen KS. Alterations of microbiota in urine from women with interstitial cystitis. BMC Microbiol (2012) 12:205. doi:10.1186/1471-2180-12-205
55. Abernethy MG, Rosenfeld A, White JR, Mueller MG, Lewicky-Gaupp C, Kenton K. Urinary microbiome and cytokine levels in women with interstitial cystitis. Obstet Gynecol (2017) 129(3):500–6. doi:10.1097/AOG.0000000000001892
56. Chiang ML, Chen HC, Chen KN, Lin YC, Lin YT, Chen MJ. Optimizing production of two potential probiotic lactobacilli strains isolated from piglet feces as feed additives for weaned piglets. Asian-Australas J Anim Sci (2015) 28(8):1163–70. doi:10.5713/ajas.14.0780
57. Gao CX, Major A, Rendon D, Lugo M, Jackson V, Shi ZC, et al. Histamine H2 receptor-mediated suppression of intestinal inflammation by probiotic Lactobacillus reuteri. MBio (2015) 6(6):e1358–1315. doi:10.1128/mBio.01358-15
58. Schaefer L, Auchtung TA, Hermans KE, Whitehead D, Borhan B, Britton RA. The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology (2010) 156:1589–99. doi:10.1099/mic.0.035642-0
59. Thomas CM, Hong T, Van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, et al. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS One (2012) 7(2):e31951. doi:10.1371/journal.pone.0031951
60. Simon MC, Strassburger K, Nowotny B, Kolb H, Nowotny P, Burkart V, et al. Intake of Lactobacillus reuteri improves incretin and insulin secretion in glucose-tolerant humans: a proof of concept. Diabetes Care (2015) 38(10):1827–34. doi:10.2337/dc14-2690
61. Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, Fok C, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio (2014) 5(4):e1283–1214. doi:10.1128/mBio.01283-14
62. Lewis DA, Brown R, Williams J, White P, Jacobson SK, Marchesi JR, et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol (2013) 3:41. doi:10.3389/fcimb.2013.00041
63. Nienhouse V, Gao X, Dong QF, Nelson DE, Toh E, Mckinley K, et al. Interplay between bladder microbiota and urinary antimicrobial peptides: mechanisms for human urinary tract infection risk and symptom severity. PLoS One (2014) 9(12):e114185. doi:10.1371/journal.pone.0114185
64. Siddiqui H, Lagesen K, Nederbragt AJ, Eri LM, Jeansson SL, Jakobsen KS. Pathogens in urine from a female patient with overactive bladder syndrome detected by culture-independent high throughput sequencing: a case report. Open Microbiol J (2014) 8:148–53. doi:10.2174/1874285801408010148
65. Pearce MM, Zilliox MJ, Rosenfeld AB, Thomas-White KJ, Richter HE, Nager CW, et al. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol (2015) 213(3):347.e1–11. doi:10.1016/j.ajog.2015.07.009
66. Eun CS, Mishima Y, Wohlgemuth S, Liu B, Bower M, Carroll IM, et al. Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10(-/-) mice. Infect Immun (2014) 82(6):2239–46. doi:10.1128/Iai.01513-13
67. Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, Albesiano E, et al. Cutting edge: an in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol (2007) 179(7):4313–7. doi:10.4049/jimmunol.179.7.4313
68. Krych L, Nielsen DS, Hansen AK, Hansen CH. Gut microbial markers are associated with diabetes onset, regulatory imbalance, and IFN-gamma level in NOD mice. Gut Microbes (2015) 6(2):101–9. doi:10.1080/19490976.2015.1011876
69. Sato T, Watanabe K, Kumada H, Toyama T, Tani-Ishii N, Hamada N. Peptidoglycan of Actinomyces naeslundii induces inflammatory cytokine production and stimulates osteoclastogenesis in alveolar bone resorption. Arch Oral Biol (2012) 57(11):1522–8. doi:10.1016/j.archoralbio.2012.07.012
Keywords: Akkermansia, interleukin-8, Lactobacillus, type 2 diabetes mellitus, urinary microbiota
Citation: Ling Z, Liu F, Shao L, Cheng Y and Li L (2017) Dysbiosis of the Urinary Microbiota Associated With Urine Levels of Proinflammatory Chemokine Interleukin-8 in Female Type 2 Diabetic Patients. Front. Immunol. 8:1032. doi: 10.3389/fimmu.2017.01032
Received: 27 May 2017; Accepted: 10 August 2017;
Published: 25 August 2017
Edited by:
Haruki Kitazawa, Tohoku University, JapanReviewed by:
Maryam Dadar, Razi Vaccine and Serum Research Institute, IranAshok Munjal, Barkatullah University, India
Copyright: © 2017 Ling, Liu, Shao, Cheng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengping Liu, MTE0MTgyMjlAemp1LmVkdS5jbg==;
Lanjuan Li, bGpsaUB6anUuZWR1LmNu
†These authors have contributed equally to this work.
 Zongxin Ling
Zongxin Ling Fengping Liu
Fengping Liu Li Shao1
Li Shao1