
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 14 August 2017
Sec. Inflammation
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.00942
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder of the gastrointestinal tract. Although the etiology and pathogenesis of IBD remain unclear, both genetic susceptibility and environmental factors are implicated in the initiation and progression of IBD. Recent studies with experimental animal models and clinical patients indicated that the intestinal microbiota is one of the critical environmental factors that influence nutrient metabolism, immune responses, and the health of the host in various intestinal diseases, including ulcerative colitis and Crohn’s disease. The objective of this review is to highlight the crosstalk between gut microbiota and host immune response and the contribution of this interaction to the pathogenesis of IBD. In addition, potential therapeutic strategies targeting the intestinal micro-ecosystem in IBD are discussed.
Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), is a chronic and relapsing inflammatory condition of the gastrointestinal (GI) tract characterized by abdominal pain, diarrhea, and bloody stools (1–3). IBD affects approximately 3.7 million people in North America and Europe (4–6). Furthermore, an increasing incidence of IBD has been observed in Asian countries, such as China (7), India, South Korea (4, 8), and Saudi Arabia (9), over the past two decades. Although CD and UC share partially overlapping pathological and clinical symptoms, there are distinct clinical characteristics; for example, CD can affect one or several segments of the digestive tract, whereas UC is mainly restricted to the mucosal layer of the colon or rectum without affecting other areas of the GI tract (2, 10). The clinical manifestations of IBD have been well-defined many years ago. Nevertheless, the etiologies and pathogenesis of this disorder remain largely elusive (11). The identification of genetic loci associated with susceptibility to IBD has indicated the role of genetic background in the pathophysiology of IBD (12). However, genetic polymorphism alone does not predict the development of IBD, thus highlighting the involvement of other environmental factors in pathogenesis (13).
The human gut is the habitat for 100 trillion of different microbial organisms, including bacteria, viruses, fungi, and protozoans, which are 10 times more than all of the cells in the human body (14, 15). It is generally believed that the gut of human is colonized by commensal microbes from the vaginal canal, skin, feces, or breast milk during delivery or after birth (16, 17). The phylogenetic diversity of the intestinal microbiota increases with growth and development and ultimately leads to a complex and relatively stable community of microorganisms at the age of roughly 2–3 years (18, 19). Based on culture-independent molecular profiling methods (20, 21), it is estimated that more than 1,000 species of bacteria are represented in the GI tract, and the majority of them are obligate anaerobic organisms, including Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (22). Among them, Firmicutes (Gram-positive) and Bacteroidetes (Gram-negative) are the predominant phyla across all mammalian species, accounting for over 90% of all intestinal bacteria (12, 23–25). The total number and composition of bacteria vary in different segments of the GI tract, with a relatively low number and few species of bacteria residing in the stomach and upper part of the small intestine. The number of resident microbes increases from the jejunum to each subsequent part of the gut, reaching up to 1012/g in the feces (18, 26).
Changes in the composition of intestinal bacteria could influence intestinal homeostasis through various signaling pathways, thus affecting the interactions between bacteria and the host (27, 28). Intestinal bacteria are responsible for the degradation of indigestible carbohydrates to produce short-chain fatty acids (SCFAs), synthesis of vitamins (vitamin K, vitamin B12, and folic acid), synthesis of amino acids, and regulation of fat metabolism, which are required for the integrity of intestinal barrier function (29–31). Accordingly, studies have observed that the small intestinal crypts of germ-free mice are atrophic and have a decreased proliferation rate and impaired angiogenesis (32, 33). In addition, the bacteria in the gut protect the intestinal epithelium from the harmful effects of pathogens by inhibiting the colonization of pathogenic bacteria and producing antimicrobial compounds, thus contributing to homeostasis (34). Furthermore, the intestinal microbiota actively stimulates and promotes the development and maturation of the immune system, which is required for the body’s defense against pathogens and peripheral immune tolerance against potential antigens in the lumen (35–37). The resident intestinal microbiota is vital for the physiology and health of the host. However, this does not mean that a particular species is entirely beneficial (38). Bacteroides fragilis, a commensal bacterium that is well controlled under normal conditions, has been reported to invade intestinal tissues and pose a serious threat in immunocompromised individuals (39).
The diversity and composition of the intestinal flora can be altered by diet, environmental factors, stress, lifestyle, exogenous probiotics, and antibiotic use (40, 41), which in turn would affect intestinal homeostasis. The mutual relationship and interaction between the intestinal flora and host have increasingly attracted the attention of both research scientists and clinical practitioners, and this has led to a greater understanding of the human microbiome in recent years (42). Consistently, the dysfunction of the interaction between the intestinal microbiota and host can lead to an excessive inflammatory response and contribute to the initiation and/or progression of IBD (28), intestinal bowel syndrome, and functional dyspepsia in humans (43), thus highlighting the critical role of the intestinal microbiota in health and IBD (44). Given these observations, IBD is regarded as an excessive activation of the host immune response to the intestinal microbiota in genetically susceptible patients (7). Herein, we summarize the current literature that describes the association between gut microbiota and host immune response in the pathogenesis of IBD as well as the potential therapeutic strategies involving the modulation of the intestinal micro-ecosystem.
A large number of studies have shown that the disturbance of normal microbial populations in the GI tract is linked to both acute infections, such as Clostridium difficile infection (CDI) and to chronic diseases, including IBD, irritable bowel syndrome, metabolic diseases, and autoimmune disorders (45, 46). The term “dysbiosis” was first coined by Metchnikoff in the early twentieth century to describe changes in intestinal bacteria, which has been further defined and extended by others (47). The concept of dysbiosis implies that an imbalance in the microbial ecosystem disrupts immune homeostasis and leads to intestinal disorders, including both CD and UC (48). It emphasizes the critical role of the interactions between the intestinal flora and host immune system in the pathogenesis of intestinal diseases (49). In an initial study conducted in 2002, Swidsinski et al. demonstrated that the abundance of mucosal microbiota in patients affected with IBD is positively correlated with disease severity (50). Although they did not observe substantial changes in the composition of the intestinal flora in IBD patients because of technological limitations (24, 25), this is the first study showing a possible relationship between bacteria and IBD. In the last decade, the application of culture-independent molecular approaches in the study of intestinal microbiota diversity has improved our understanding of the intestinal microbiota and immune response in intestinal disorders (51). Metagenomic studies demonstrated that microbial diversity and intestinal microbiota stability decrease in IBD patients compared with individuals without IBD (52). Consistently, 25% fewer genes were detected in the fecal samples of IBD patients than in those of control patients (26). Further studies have shown that IBD patients have fewer bacteria with anti-inflammatory properties (bacteria in phyla Firmicutes) and/or more bacteria with pro-inflammatory properties (24, 53–55). Joossens et al. demonstrated that the fecal microbiota in CD patients has a reduced abundance of anti-inflammatory F. prausnitzii, B. adolescentis, and D. invisus and an increased abundance of potentially pro-inflammatory R. gnavus (56). This change in the intestinal flora might contribute to chronic inflammation as observed in the GI tract of IBD patients.
Another bacterium with pro-inflammatory properties is E. coli AIEC (adherent/invasive E. coli), a mucosa-associated E. coli with strong adhesive–invasive properties, which was originally isolated from adult CD patients (57). In comparison with normal controls and patients with colonic CD, about 38% of patients with active ileal CD have been found to have an increased concentration of AIEC (58, 59). Campylobacter concisus, another invasive proteobacterium, has also been reported to preferentially colonize CD patients (60–62). The increase in pathogenic bacteria that adhere to intestinal epithelial cells affects intestinal permeability, alters the diversity and composition of gut microbiota, and induces inflammatory responses by regulating pro-inflammatory gene expression, ultimately resulting in colitis (63). In addition, the disruption of intestinal bacteria affects the pathogenesis of IBD by their metabolites. For example, the SCFAs produced by commensal bacteria exert anti-inflammatory activity and serve as a major energy source for the colonic epithelium. The production of SCFAs has been reported to decrease in IBD-affected patients because of decreased F. prausnitzii, a butyrate-producing bacterium in the gut (64–67). By contrast, the concentration of sulfate-reducing bacteria is higher in IBD patients, which can result in the metabolic production of hydrogen sulfide that is toxic to intestinal epithelial cells and induce mucosal inflammation, thus leading to UC (68). Taken together, the overall data have indicated a strong correlation between intestinal microbiota alteration and the pathogenesis of IBD (69–71). However, it should be kept in mind that the diversity and composition of bacteria among experimental animal studies or IBD patients are not consistent, and the results are contradictory under some conditions (72). Moreover, it remains unclear whether the observed changes in phylogenetic composition are causative for the development of IBD or simply a consequence of an altered intestinal environment during the progression of IBD (18, 72, 73). Additional studies and an in-depth analysis of the gut flora are needed to address these issues before any conclusions can be drawn.
Intestinal epithelial cells are in direct contact with a variety of xenobiotic factors, including pathogenic microorganisms, dietary antigens, or toxic components, which can trigger and activate the immune system of the host (74, 75). Inflammation is a protective response of the immune system to infection or tissue injury. However, prolonged or chronic inflammation is detrimental and associated with the development of IBD (76). Therefore, an appropriate immune response to intestinal pathogens without eliciting an inflammatory response to commensal bacteria is critical for the maintenance of intestinal homeostasis (77). Host immune cells have evolved various mechanisms to ensure intestinal homeostasis, including the mucosal and epithelial barrier, pro-inflammatory signaling pathways, and intestinal innate and adaptive components (78) (Figure 1). These mechanisms allow a relative stable bacterial population to form and limit the colonization of pathogenic bacteria and microbiota-driven inflammation (76, 79). First, the intestinal barrier formed by epithelial cells provides a physical barrier separating the luminal contents from the underlying immune compartments, thereby blocking the entry of microflora into the lamina propria (76). Second, the specialized secretory cells in the GI tract, such as plasma cells, goblet cells, and Paneth cells, produce and secret IgA, mucus, and antimicrobial proteins, respectively; these make up the main components of the intestinal mucosa, which function as a defense line to reduce the microbial burden of the epithelium (38). The expression of antimicrobial proteins in the gut is regulated by distinct mechanisms (Figure 1). For example, most α-defensins are constitutively expressed, whereas the expression of β-defensins, C-type lectin regenerating islet-derived protein 3γ, and a subset of α-defensins is regulated by bacteria-activated toll-like receptor or nucleotide-binding oligomerization domain-containing protein 2 (NOD2) signaling (80). Third, microbiome-derived metabolites or small molecules with microbicidal or microbiostatic properties promote resistance to colonization by pathogenic species. For instance, polysaccharide A, a microbial molecule synthesized by B. fragilis, has been reported to prevent colitis induced by Helicobacter hepaticus through suppression of interleukin (IL)-17 production and enhancement of IL-10-producing CD4+ T cells (81). Fourth, when commensal bacteria penetrate the intestinal epithelial cells, the innate and adaptive immunity systems can be activated to eliminate the microbiota (38). A combination of the above-mentioned actions in epithelial cells, secretory cells, and immune cells ensures a state of limited mucosal response to commensal bacteria (38, 76). Importantly, immune responses induced by commensal flora can regulate the composition of the intestinal microbiota, thus maintaining the dynamic balance between commensal bacteria and the host immune system and ensuring gut homeostasis and health (82).
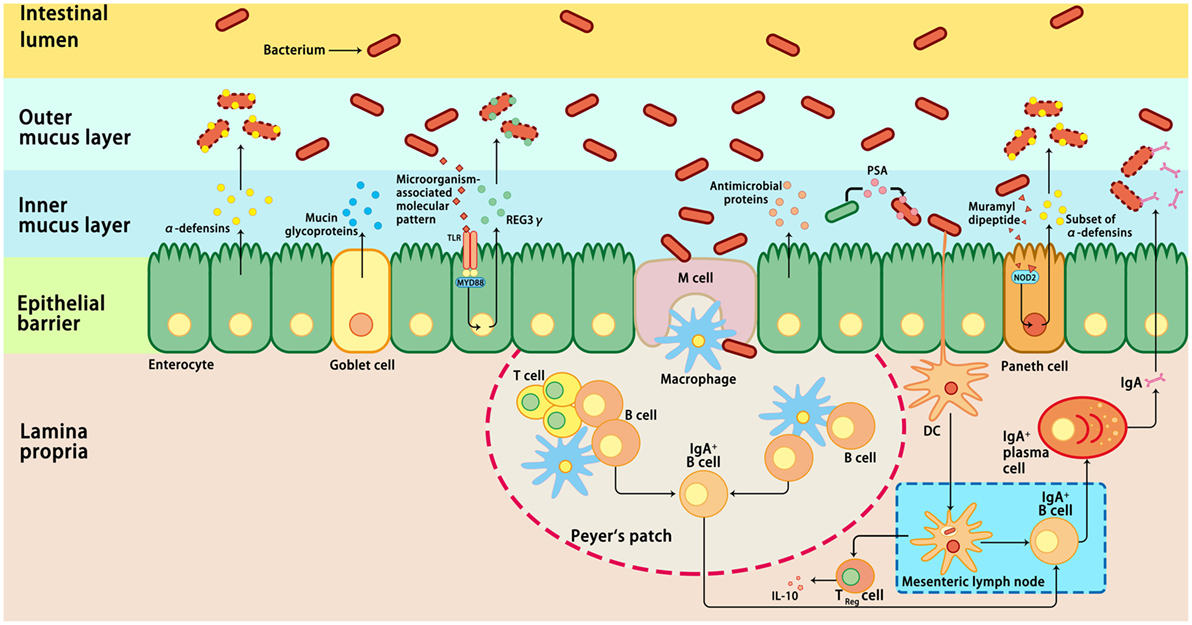
Figure 1. Host immune responses to intestinal microbiota. Several immune mechanisms work in concert to the intestinal microbiota and contribute to intestinal homeostasis. Goblet cells secret mucin glycoproteins, plasma cells secret IgA, and epithelial cells secrete antimicrobial proteins through toll-like receptors (TLRs), or nucleotide-binding oligomerization domain-containing protein 2 (NOD2)-dependent mechanisms. Dendritic cells (DCs) take up bacteria migrate to Peyer’s patches and mesenteric lymph nodes where B cells are differentiated into IgA-secreting plasma cells. In addition, sampling of polysaccharide A (PSA) from Bacteroides fragilis by intestinal DCs leads to induction of regulatory T (Treg), which is responsible for the production of IL-10. In addition, the antimicrobial proteins secreted by the host cells can modulate the composition of the microbiota. IL-10, interleukin 10; M cell, microfold cell; MYD88, myeloid differentiation primary-response protein 88; REG3γ, C-type lectin regenerating islet-derived protein 3γ.
Despite the mucus, antimicrobial proteins and secretory IgA are critical components for maintaining intestinal homeostasis, several GI pathogens can penetrate the epithelial barrier and contribute to the pathogenesis of IBD if they are not eliminated through host immune reactions (38). By using non-aqueous Carnoy fixative, Swidsinski et al. demonstrated increased bacterial adherence to the mucosal surface of IBD patients, indicating a decreased ability to limit direct contact between the epithelium and intestinal microbiota and the potential over-activation of the host immune response (83) (Figure 2). Based on this observation, IBD is regarded as a chronic inflammatory disorder caused by an excessive immune response to intestinal flora. Many susceptibility genes and environmental factors have been identified to interfere with the interactions between the microbiota and host immune system through various signaling pathways in IBD patients (27, 28, 82, 84, 85).
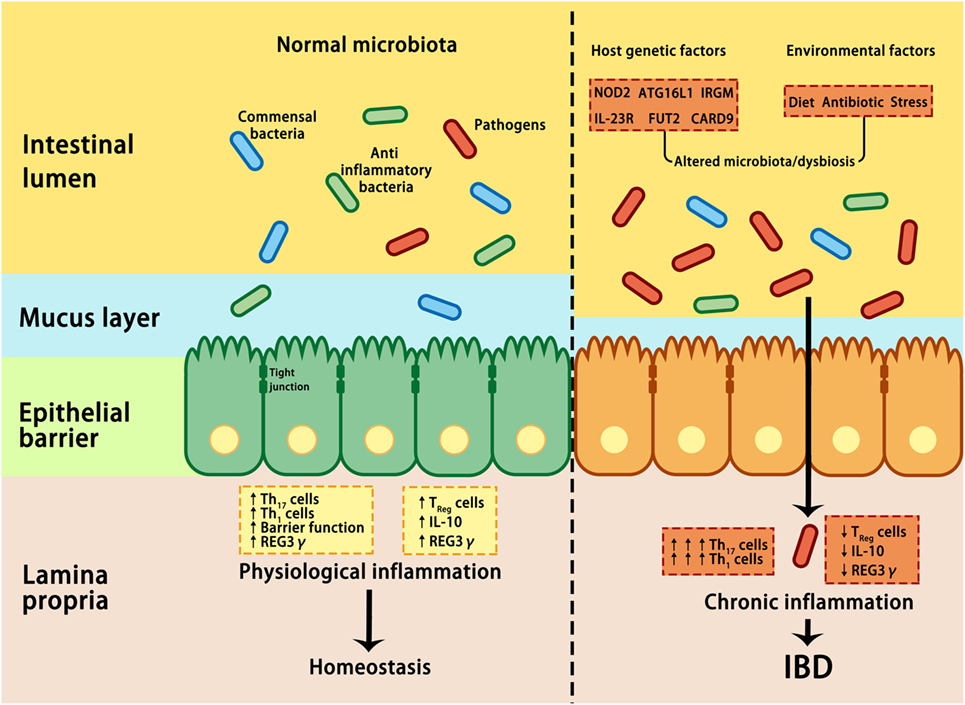
Figure 2. Interactions between microbiota and host genetic and environmental factors contribute to the pathogenesis of IBD. Under healthy conditions (left panel), pathogens are suppressed by beneficial commensal bacteria through the induction of antimicrobial proteins, such as IL-10 and REG3γ, thus maintaining homeostasis. In IBD (right panel), a combination of genetic factors and environmental factors (such as stress, diet, and antibiotic) lead to dysbiosis, which in turn affects barrier integrity, innate, and adaptive immunity, resulting in uncontrolled chronic inflammation and hyper-activation of T helper 1 (Th1) and Th17 cells, increase in tight junction permeability, reduction in regulatory T (Treg) cells, and decrease in REG3γ and IL-10. ATG16L, autophagy-related 16-like; CARD9, caspase recruitment domain family member 9; FUT2, fucosyltransferase 2; IBD, inflammatory bowel disease; IL-10, interleukin 10; IRGM, immunity-related GTPase M; NOD2, nucleotide-binding oligomerization domain-containing protein 2; REG3γ, C-type lectin regenerating islet-derived protein 3γ.
Nucleotide-binding oligomerization domain-containing protein 2 plays an important role in intestinal homeostasis by activating nuclear factor-κB or the mitogen-activated protein kinase (MAPK)-mediated immune response upon sensing specific bacterial peptidoglycan motifs in the cytosol (86). The activation of NOD2 signaling also enhances the production of antimicrobial peptides and mucin, two critical components of the intestinal mucosa that prevent the bacteria from colonizing epithelial cells (86). As a critical regulator of inflammation, mutations of NOD2 are associated with decreased Clostridium group XIVa and IV (SCFA-producing bacterial strains) and increased Actinobacteria and Proteobacteria, resulting in an intestinal flora shift in patients with CD and UC and thus leading to a higher susceptibility to inflammation (87–91). Consistently, NOD2-deficient mice have compromised epithelial barrier, reduced intraepithelial lymphocytes, decreased α-defensin production, and impaired immune responses to pathogenic bacteria, which are indicative of experimental colitis (92–94). In addition to NOD2, other IBD susceptibility genes, such as ATG16L1, XBP1, IRGM, CARD9, and FUT2, have been reported to affect innate and/or adaptive immune functions as well as the composition and diversity of the intestinal flora (95, 96). The ATG16L1 or XBP1 gene variants are associated with disturbances in Paneth and/or goblet cell function (97–99) and increased IL-1β production in response to bacterial peptidoglycan muramyl dipeptide (95), thus contributing to the ileal disease phenotype in CD. Recently, fucosyltransferase 2 (FUT2), a gene that regulates the expression of H antigens (precursors of blood group A and B antigens) in the GI mucosa, has been reported to be involved in regulating the structure and composition of the intestinal microbiota, thereby significantly contributing to CD susceptibility (100). Genome-wide association studies have identified over 163 single nucleotide polymorphisms (SNPs) associated with different susceptibilities to CD or UC, and more loci will no doubt be reported in the future (101–103). Most of the identified genes (110 of 163 gene loci) are shared by both CD and UC, whereas only 23 and 30 loci are specifically correlated with susceptibility to UC and CD, respectively (103, 104). These susceptibility genes primarily regulate host immune response signaling (101–103). Recent studies have shown that dysregulation of the IL-23/Th17 axis is associated with multiple genetic susceptibility SNPs in patients with CD and UC because of impairment in the innate and adaptive immune response (105, 106). Notably, intestinal dysbiosis is associated with the elevated generation of reactive oxygen species (107), which in turn can lead to changes in the composition and diversity of the intestinal microbiota (108), increased mucosal permeability (73), and increased immune stimulation, thus forming a vicious cycle. Nevertheless, more studies are required to elucidate the mechanism of the bidirectional regulation between the microbiota and host immune response under specific conditions (109, 110). An example of how specific microbes induce intestinal inflammation and influence the pathogenesis of IBD was reported by Bloom et al. (111). In their study, commensal Bacteroides species were isolated in IL-10r2- and Tgfbr2-deficient mice (111). Despite the colonization of the isolated bacteria in both IBD-susceptible and non-susceptible mice, IBD induction was exclusively observed in susceptible animals, thus providing an important insight into the role of intestinal dysbiosis in IBD induction (112). Considering that commensal Bacteroides species are present in abundance in the mammalian intestine and have beneficial effects on the host through the breakdown of complex dietary carbohydrates, modulation of mucosal glucosylation, and immune maturation (46), classically beneficial bacteria might have detrimental effects under specific conditions and could contribute to IBD (112). These data also support the critical role of genetic factors in the development of IBD via regulation of intestinal bacterial composition and diversity as well as immune responses in the GI tract. It should be kept in mind that the susceptibility alleles are not sufficient on their own to trigger IBD. The evidence thus far indicates that the development of IBD is a combined effect of both genetic and non-genetic factors, which act together and lead to changes in the structure and function of the human intestinal microbiota (113).
The composition of the intestinal flora is affected by various environmental factors, such as diet, stress, age, and antibiotic treatment (114). In this section, we will mainly focus on diet and stress as well as their effects on the interactions between the intestinal microbiota and host immune response.
It is well known that dietary components affect the structure and activity of intestinal bacteria (115, 116). Western food intake (high sugar/fat and low dietary fibers, fruits, and vegetables) is associated with the altered structure and function of commensal flora in the gut (113, 117), which might favor an increased incidence of IBD (6, 8, 118). In a recent study, David et al. reported that the short-term (5 days) consumption of animal-based foods is associated with an increased abundance of bile-tolerant microorganisms (Alistipes, Bilophila, and Bacteroides) and reduced Firmicutes that metabolize dietary plant polysaccharides, demonstrating the rapid modulatory effect of dietary nutrients on intestinal bacteria (116). Considering that the production of H2S by Bilophila wadsworthia, a sulfite-reducing bacterium, might trigger inflammatory responses in the intestine (119), these data provide a plausible explanation for the higher prevalence of IBD in Western populations. Similar results were observed when switching from a plant-based diet to a typical Western diet for 1 day (115), thus highlighting the possibility of restoring the gut microbiota in IBD by nutritional interventions.
Dietary fibers can be converted to SCFAs by anaerobic bacteria (Bacteroidetes and Firmicutes) fermentation in the intestine, which in turn could regulate the expression of genes involved in the proliferation, differentiation, and apoptosis of intestinal epithelial cells and affect the composition of the gut microbiota and host inflammatory response (65). Various studies have reported decreased SCFA production or decreased SCFA-producing bacteria (Roseburia hominis and Faecalibacterium prausnitzii) in IBD-affected patients (64, 65, 120, 121), suggesting a role for SCFAs in inflammatory responses in IBD. The beneficial therapeutic effects of SCFAs have also been observed following the administration of SCFAs or prebiotics in animal models or UC-affected patients (122–126). The effect of SCFAs on IBD is predominantly mediated by regulating both the innate immune response and adaptive immune response (127). In addition, several metabolites produced by intestinal bacteria have been identified to have the ability to affect host metabolism and immunity in experimental colitis or IBD patients (128). The aryl hydrocarbon receptor (AhR) is a transcription factor that resides in the cytoplasm of the intestinal epithelium, macrophages, B cells, T cells, and dendritic cells (129). Kynurenine, a metabolic product derived from the essential amino acid tryptophan, has been identified as an endogenous AhR ligand (130) that regulates the expansion of intraepithelial lymphocytes, innate lymphoid cells, and immune and inflammatory reactions, and maintains normal mucosal function in the gut (131). In the GI tract, diet-derived AhR ligands promote local IL-22 production, which in turn stimulates the production of antimicrobial peptides and mucin (132), thus conferring pathogen resistance and mucosal protection (132). CARD9 null mice have been reported to have impaired immune responses to Citrobacter rodentium as shown by the decreased production of colonic IL-6 and IL-17A as well as fewer IL-22-producing innate lymphoid cells in the colon lamina propria (133). In a recent study, Lamas et al. showed that microbiota dysfunction and susceptibility to IBD in CARD9 knockout mice are mainly attributed to their inability to metabolize tryptophan into metabolites that act as AhR ligands. By contrast, the addition of tryptophan-metabolizing Lactobacillus strains can attenuate intestinal inflammation in CARD9 null mice (96). These data suggest that tryptophan metabolites are bioactive mediators that regulate the crosstalk between the host immune response and intestinal microbiota ecosystem (134).
Stress is thought to be another risk factor in the development of IBD (135). Data from preclinical and clinical studies have revealed that stress reduction is associated with decreased relapse in patients with UC or CD (136, 137). This effect of stress on IBD is mainly mediated by corticotropin-releasing factor (CRF) signaling (138). In response to stress, CRF is synthesized and released from multiple brain regions including the paraventricular nucleus and hypothalamus. The released CRF stimulates the production of adrenocorticotropic hormone from the pituitary gland, which is transported to the adrenal cortex to induce the synthesis and secretion of cortisol in response to stress (139). CRF signaling can act in peripheral tissues, including the stomach, pancreas, small intestine, and lymphocytes (139–142). Our study and others have shown that CRF signaling can be activated by stress in the intestines of rodents and pigs (143, 144). Importantly, the activation of corticotropin-releasing hormone has been reported to be associated with inflammation of the colonic mucosa and increased intestinal permeability in patients with CD or UC (145–147), thus suggesting the involvement of stress in the pathophysiology of IBD through several mechanisms. First, CRF activation can lead to TNF-α release and protease secretion (hallmarks of IBD) from mast cells, which in turn act on epithelial cells and result in bacterial translocation and over-activation of the immune response due to increased permeability (11, 41, 148). Second, stress-induced HPA activation, alterations in neurotransmitters, and immune function activation can modulate the intestinal microbiota composition and metabolism as well as permeability (149–151). Considering the effect of stress-induced microbiota dysfunction in the pathophysiology of IBD, CRF antagonistic or anti-TNF-α agents have been found to result in favorable outcomes in IBD patients (152). In our recent study, we demonstrated that glutamine supplementation attenuates the stress-induced downregulation of tight junction proteins, increase in permeability, and release of CRF in intestinal tissues (144, 153). Considering the essential role of tight junction proteins in the maintenance of the intestinal barrier and prevention of bacterial translocation, glutamine might be a promising adjuvant in IBD therapeutics. Taken together, environmental factors are emerging as critical contributors to the development of IBD through complex interactions between intestinal bacteria and the host. In addition to dietary factors, the intake of drugs and antibiotics, age, and other environmental factors can influence the intestinal microbiota by interacting with commensal microorganisms (154–157).
It is increasingly evident that a delicate balance between the gut microbiota and intestinal immune system is required to protect against pathogenic bacteria and contribute to intestinal homeostasis and functions (84). Recent studies in both animal models and clinical patients have highlighted the critical role of the intestinal microbiota in initiating, maintaining, and determining the severity of IBD (72). Restoration of the diversity and composition of the commensal microbiota is emerging as a novel therapeutic intervention for microbial imbalance involved GI diseases, including IBD and IBS (72). Manipulation of the gut microbiota can be achieved by antibiotics, fecal microbiota transplantation (FMT), or probiotics.
The rationale for antibiotic therapy in IBD is based on evidence showing that intestinal microbes, including luminal bacteria, have an important role in the pathogenesis of IBD (27, 73, 158). Antibiotic treatment can decrease the abundance of pathogenic bacteria to favor the growth of beneficial bacteria (159, 160). Although antibiotics are commonly used in clinical practice to improve the life quality of patients, their benefits have not been well-established in carefully designed clinical trials with patients affected with IBD (159, 161). Moreover, antibiotic treatment has also been reported to have adverse outcomes due to currently unknown reasons or lead to various side effects in fetuses and children affected by IBD (162). A meta-analysis has demonstrated that long-term exposure to antibiotics is associated with a higher incidence of CD because of their interfering effect on the intestinal flora (163). Moreover, antibiotic intervention early in life might influence the development and maturation of the intestinal immune system of the host (164). Another concern regarding antibiotic treatment for IBD is the development and spread of bacterial resistance to antibiotics as well as the rebound of intestinal bacteria after the cessation of therapy (162). Although antibiotics are potent in regulating microbiome ecological diversity, these issues should be carefully addressed for IBD treatment in the future.
Fecal microbiota transplantation, also known as “fecal bacteriotherapy” or “fecal infusion,” is the transfer of intestinal bacteria from a healthy donor to restore the intestinal microbiota of a diseased individual (165, 166). FMT has been adapted in clinical practice for CDI, which cannot be eliminated with antibiotics alone, and has been proven to be more effective than antibiotic treatment (166, 167). Thereafter, FMT has been evaluated in several microbiota-driven diseases, including IBD (168, 169) and metabolic syndrome (170), and has gained interest as a novel therapy option. In contrast to the impressive results of treatment for CDI (92%) (171), FMT has been reported to reduce symptoms in about 20% of IBD patients (172). This huge difference might be attributed to several reasons. First, the severity and duration of symptoms in clinical trials may influence the outcome; thus, positive outcomes are observed only in some patients enrolled in the clinical trials (173). Second, IBD is a chronic disease associated with various factors, such as gut microbiota impairment and genetic and/or environmental factors; thus, the development of IBD is a synergetic effect of multiple factors instead of a single one (4, 169). Third, the immune system of IBD patients is upregulated due to chronic inflammation (174), which might affect the therapeutic effect of FMT. Fourth, the composition of the gut microbiota from donors is different (175) and might affect the clinical outcome. Fifth, the FMT protocol used in each trial (including the criterion for donor selection, patient preparation, the number and composition of bacteria infused, and route of administration) is different for each patient (172), which will result in different therapeutic effects. All these factors could contribute to a lower clinical outcome compared with that of CDI. Nevertheless, it is too early to draw conclusions on the potential of FMT therapy. Additional clinical studies using a standard protocol are required to evaluate its efficacy and safety before prescribing it for IBD patients (173).
Probiotics are living microorganisms that exert beneficial effects on the host by modulating the intestinal microflora (176, 177). Lactobacillus and Bifidobacterium species, two of the most common probiotic bacteria, have been found not only to improve immune system responses but also to exert a positive effect on the preexisting microflora stability, to inhibit pathogen colonization, and to enhance mucosal trophic effects by stimulating intestinal epithelial cell barrier responses (178, 179). Tao et al. showed that the probiotic Lactobacillus GG releases soluble factors, which in turn stimulate the synthesis of heat-shock proteins through the p38 MAPK pathway, thus exerting a cytoprotective effect on intestinal epithelial cells (180). In a randomized study, 39 UC patients were treated with L. rhamnosus GG daily, while the remaining patients (N = 78) were not treated; the progression of UC was inhibited by probiotic administration, demonstrating the significant clinical benefit of probiotics (181). In another study, VSL#3, a high-potency probiotic medical food containing eight different strains, could induce remission and prevent the relapse of inflammatory disease in patients with mild to moderately active UC (182–184). Other probiotics, such as Bifidobacterium bifidum, L. acidophilus (185), and L. reuteri ATCC 55730 (186), have also been reported to be associated with beneficial effects in IBD patients.
The effects of probiotics are mediated by modulating the mucosal inflammation system, enhancing competitive exclusion to pathogenic bacteria, regulating the secretion of cytokines involved in the development of IBD, or modulating intestinal permeability (109, 178) (Figure 3). It is well known that probiotics express pathogen-associated molecular patterns; they can mimic the function of commensal bacteria by engaging and/or activating the pattern recognition receptors on epithelial mucosal surfaces, thus regulating the expression of genes involved in the host immune response (178). In addition, the beneficial effects of probiotics have been attributed to the restoration of the number and function of goblet cells and the stimulation of the mucosal immune system to secrete protective immunoglobulins, such as secretory IgA, protective defensins, and bacteriocidins in the intestinal tract (187). Although the practical application of probiotics has been encouraged by positive results in clinical trials involving UC patients, clinical trials involving CD patients have not inspired much enthusiasm (178, 188). Further studies on the efficacy, safety, and underlying mechanisms of action are required before probiotics may be recommended for the treatment of IBD patients (188).
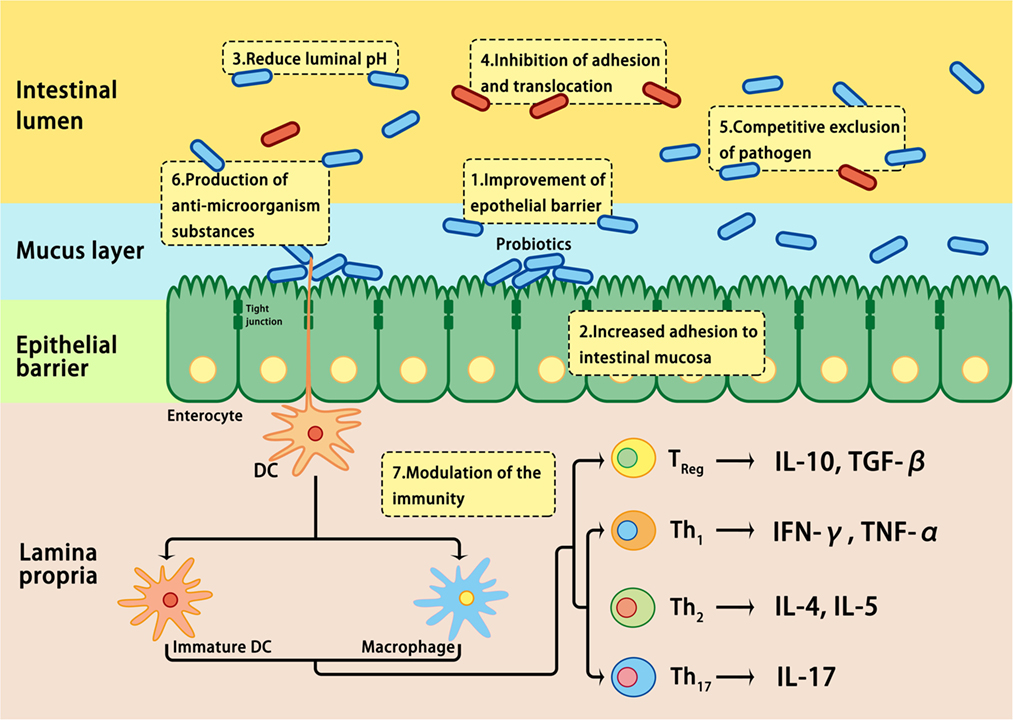
Figure 3. Mechanisms involved in probiotic-induced protection against intestinal dysbiosis. Probiotics suppress pathogens through various actions, including lowering luminal pH, production of antimicrobial proteins, inhibition of adhesion and translocation of flora, competitive exclusion of pathogens, improvement of epithelial barrier, enhancement of adhesion of commensal bacteria to the intestinal mucosa, and modulation of gastrointestinal mucosal immune system.
The role of the interaction between the intestinal microbiota and host immune response in the pathogenesis of IBD has attracted much attention because of the application of next-generation sequencing technology and the availability of genetically engineered animal models. Intestinal dysbiosis is emerging as a risk factor because of its functional role in the maintenance of intestinal homeostasis, activation of the immune response of the intestinal epithelium, and its crosstalk with other factors through genetic or epigenetic mechanisms. Based on advances in our understanding of the microbiota in IBD pathogenesis, several therapeutic interventions have been investigated for restoring the commensal microecology, and some of them have shown impressive results in clinical trials or experimental animal models. However, various questions need to be addressed. The intestinal microbiota is subjected to changes in both host and exogenous factors, and it is largely unknown how dysbiosis is triggered and leads to chronic inflammation. Furthermore, an imbalance in bacterial populations is associated with various diseases; the pathogenic implications of specific microbes in CD or UC, as well as underlying mechanisms, remain to be determined. Despite the occurrence of dysbiosis in patients with CD and UC, it is unclear whether alterations in the intestinal flora contribute to the development of IBD or are instead a consequence of this disorder. The treatment of dysbiosis through FMT or prebiotic administration has produced favorable results in clinical trials involving IBD patients; however, both their safety and efficacy have to be determined before they can be considered as a therapeutic strategy. Additional studies on the interplay between the microbiota and intestinal epithelium are of great importance to advance our understanding of the role of the microbiota in the pathogenesis of IBD and to identify potential therapeutic strategies by manipulating the intestinal microbiota.
YY and ZW designed the study; MZ and YW contributed to the literature search; KS, YW, and PT generated the figures and discussed on the revision; ZW revised and finalized the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Basic Research Program of China (No. 2013CB127302), the National Natural Science Foundation of China (No. 31572423, 31572410, 31625025, 31372327), the Chinese Universities Scientific Fund (2015DK001), the Program for New Century Excellent Talents in University (NCET-12-0522), and the “111” Project (B16044). We are also grateful to NIH for their generous support of the Cincinnati Mouse Metabolic Phenotype Center DK059630.
1. Wilson JC, Furlano RI, Jick SS, Meier CR. Inflammatory bowel disease and the risk of autoimmune diseases. J Crohns Colitis (2016) 10:186–93. doi:10.1093/ecco-jcc/jjv193
2. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol (2010) 28:573–621. doi:10.1146/annurev-immunol-030409-101225
3. Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology (2011) 140:1785–94. doi:10.1053/j.gastro.2011.01.055
4. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol (2015) 12:205–17. doi:10.1038/nrgastro.2015.34
5. Nee J, Feuerstein JD. Optimizing the care and health of women with inflammatory bowel disease. Gastroenterol Res Pract (2015) 2015:435820. doi:10.1155/2015/435820
6. Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology (2012) 142:46–54.e42; quiz e30. doi:10.1053/j.gastro.2011.10.001
7. Wong SH, Ng SC. What can we learn from inflammatory bowel disease in developing countries? Curr Gastroenterol Rep (2013) 15:313. doi:10.1007/s11894-013-0313-9
8. Hou JK, Abraham B, El-Serag H. Dietary intake and risk of developing inflammatory bowel disease: a systematic review of the literature. Am J Gastroenterol (2011) 106:563–73. doi:10.1038/ajg.2011.44
9. Al-Mofarreh MA, Al-Mofleh IA. Emerging inflammatory bowel disease in Saudi outpatients: a report of 693 cases. Saudi J Gastroenterol (2013) 19:16–22. doi:10.4103/1319-3767.105915
10. Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol (2003) 3:521–33. doi:10.1038/nri1132
11. Clevers H. Inflammatory bowel disease, stress, and the endoplasmic reticulum. N Engl J Med (2009) 360:726–7. doi:10.1056/NEJMcibr0809591
12. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature (2011) 474:307–17. doi:10.1038/nature10209
13. Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology (2011) 140:1704–12. doi:10.1053/j.gastro.2011.02.046
14. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science (2008) 320:1647–51. doi:10.1126/science.1155725
15. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science (2005) 308:1635–8. doi:10.1126/science.1110591
16. Floch MH. Intestinal microecology in health and wellness. J Clin Gastroenterol (2011) 45:S108–10. doi:10.1097/MCG.0b013e3182309276
17. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature (2012) 489:220–30. doi:10.1038/nature11550
18. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell (2006) 124:837–48. doi:10.1016/j.cell.2006.02.017
19. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature (2012) 486:222–7. doi:10.1038/nature11053
20. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods (2010) 7:335–6. doi:10.1038/nmeth.f.303
21. Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol (2008) 26:1135–45. doi:10.1038/nbt1486
22. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol (2016) 14:e1002533. doi:10.1371/journal.pbio.1002533
23. Eckburg PB, Relman DA. The role of microbes in Crohn’s disease. Clin Infect Dis (2007) 44:256–62. doi:10.1086/510385
24. Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A (2007) 104:13780–5. doi:10.1073/pnas.0706625104
25. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature (2011) 473:174–80. doi:10.1038/nature09944
26. Qin JJ, Li RQ, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464:59–65. doi:10.1038/nature08821
27. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology (2014) 146:1489–99. doi:10.1053/j.gastro.2014.02.009
28. Koboziev I, Reinoso Webb C, Furr KL, Grisham MB. Role of the enteric microbiota in intestinal homeostasis and inflammation. Free Radic Biol Med (2014) 68:122–33. doi:10.1016/j.freeradbiomed.2013.11.008
29. Sansonetti PJ, Medzhitov R. Learning tolerance while fighting ignorance. Cell (2009) 138:416–20. doi:10.1016/j.cell.2009.07.024
30. Hamer HM, Jonkers DMAE, Bast A, Vanhoutvin SALW, Fischer MAJG, Kodde A, et al. Butyrate modulates oxidative stress in the colonic mucosa of healthy humans. Clin Nutr (2009) 28:88–93. doi:10.1016/j.clnu.2008.11.002
31. Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature (2016) 535:56–64. doi:10.1038/nature18846
32. Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A (2005) 102:99–104. doi:10.1073/pnas.0405979102
33. Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A (2002) 99:15451–5. doi:10.1073/pnas.202604299
34. Frick JS, Autenrieth IB. The gut microflora and its variety of roles in health and disease. Curr Top Microbiol Immunol (2013) 358:273–89. doi:10.1007/82_2012_217
35. Zaneveld J, Turnbaugh PJ, Lozupone C, Ley RE, Hamady M, Gordon JI, et al. Host-bacterial coevolution and the search for new drug targets. Curr Opin Chem Biol (2008) 12:109–14. doi:10.1016/j.cbpa.2008.01.015
36. Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol (2004) 12:129–34. doi:10.1016/j.tim.2004.01.001
37. Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science (2001) 292:1115–8. doi:10.1126/science.1058709
38. Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol (2010) 10:159–69. doi:10.1038/nri2710
39. Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, et al. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci U S A (2004) 101:14919–24. doi:10.1073/pnas.0404172101
40. Veldhoen M, Ferreira C. Influence of nutrient-derived metabolites on lymphocyte immunity. Nat Med (2015) 21:709–18. doi:10.1038/nm.3894
41. Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol (2015) 50:495–507. doi:10.1007/s00535-015-1064-1
42. Huttenhower C, Knight R, Brown CT, Caporaso JG, Clemente JC, Gevers D, et al. Advancing the microbiome research community. Cell (2014) 159:227–30. doi:10.1016/j.cell.2014.09.022
43. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev (2010) 90:859–904. doi:10.1152/physrev.00045.2009
44. Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut (2013) 62:1505–10. doi:10.1136/gutjnl-2012-303954
45. Ursell LK, Clemente JC, Rideout JR, Gevers D, Caporaso JG, Knight R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol (2012) 129:1204–8. doi:10.1016/j.jaci.2012.03.010
46. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell (2012) 148:1258–70. doi:10.1016/j.cell.2012.01.035
47. Hawrelak JA, Myers SP. The causes of intestinal dysbiosis: a review. Altern Med Rev (2004) 9:180–97.
48. Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology (2011) 140:1720–8. doi:10.1053/j.gastro.2011.01.054
49. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A (2007) 104:13780–5. doi:10.1073/pnas.0706625104
50. Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology (2002) 122:44–54. doi:10.1053/gast.2002.30294
51. Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature (2011) 478:250–4. doi:10.1038/nature10434
52. Hansen R, Thomson JM, El-Omar EM, Hold GL. The role of infection in the aetiology of inflammatory bowel disease. J Gastroenterol (2010) 45:266–76. doi:10.1007/s00535-009-0191-y
53. Sokol H, Lepage P, Seksik P, Dore J, Marteau P. Temperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J Clin Microbiol (2006) 44:3172–7. doi:10.1128/JCM.02600-05
54. Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe (2008) 3:417–27. doi:10.1016/j.chom.2008.05.001
55. Mondot S, Kang S, Furet JP, de Carcer DA, McSweeney C, Morrison M, et al. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm Bowel Dis (2011) 17:185–92. doi:10.1002/ibd.21436
56. Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut (2011) 60:631–7. doi:10.1136/gut.2010.223263
57. Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infect Immun (1999) 67:4499–509.
58. Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J (2007) 1:403–18. doi:10.1038/ismej.2007.52
59. Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology (2004) 127:412–21. doi:10.1053/j.gastro.2004.04.061
60. Man SM, Zhang L, Day AS, Leach ST, Lemberg DA, Mitchell H. Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn’s disease. Inflamm Bowel Dis (2010) 16:1008–16. doi:10.1002/ibd.21157
61. Zhang L, Man SM, Day AS, Leach ST, Lemberg DA, Dutt S, et al. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn’s disease. J Clin Microbiol (2009) 47:453–5. doi:10.1128/JCM.01949-08
62. Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol (2012) 9:219–30. doi:10.1038/nrgastro.2012.14
63. Ahmed I, Roy BC, Khan SA, Septer S, Umar S. Microbiome, metabolome and inflammatory bowel disease. Microorganisms (2016) 4:20. doi:10.3390/microorganisms4020020
64. Thibault R, Blachier F, Darcy-Vrillon B, de Coppet P, Bourreille A, Segain JP. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflamm Bowel Dis (2010) 16:684–95. doi:10.1002/ibd.21108
65. Rios-Covian D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilan CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol (2016) 7:185. doi:10.3389/fmicb.2016.00185
66. Agus A, Denizot J, Thevenot J, Massier S, Denis S, Bernalier-Donadille A, et al. Western diet reduces short-chain fatty acids production and G-protein-coupled receptor GPR43 expression promoting intestinal inflammation and host susceptibilty to E. Coli infection. Gastroenterology (2015) 148:S706–7. doi:10.1016/S0016-5085(15)32398-2
67. Takaishi H, Matsuki T, Nakazawa A, Takada T, Kado S, Asahara T, et al. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int J Med Microbiol (2008) 298:463–72. doi:10.1016/j.ijmm.2007.07.016
68. Pitcher MC, Beatty ER, Cummings JH. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut (2000) 46:64–72. doi:10.1136/gut.46.1.64
69. Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut (2003) 52:237–42. doi:10.1136/gut.52.2.237
70. Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe (2007) 2:119–29. doi:10.1016/j.chom.2007.06.010
71. Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJO. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol (2006) 44:4136–41. doi:10.1128/JCM.01004-06
72. Becker C, Neurath MF, Wirtz S. The intestinal microbiota in inflammatory bowel disease. ILAR J (2015) 56:192–204. doi:10.1093/ilar/ilv030
73. Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology (2008) 134:577–94. doi:10.1053/j.gastro.2007.11.059
74. Sansonetti PJ. Host-bacteria homeostasis in the healthy and inflamed gut. Curr Opin Gastroenterol (2008) 24:435–9. doi:10.1097/MOG.0b013e32830007f7
75. Cario E. Innate immune signalling at intestinal mucosal surfaces: a fine line between host protection and destruction. Curr Opin Gastroenterol (2008) 24:725–32. doi:10.1097/MOG.0b013e32830c4341
76. Feng T, Elson CO. Adaptive immunity in the host-microbiota dialog. Mucosal Immunol (2011) 4:15–21. doi:10.1038/mi.2010.60
77. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science (2016) 352:539–44. doi:10.1126/science.aad9378
78. Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med (2008) 5:e54. doi:10.1371/journal.pmed.0050054
79. Konrad A, Cong Y, Duck W, Borlaza R, Elson CO. Tight mucosal compartmentation of the murine immune response to antigens of the enteric microbiota. Gastroenterology (2006) 130:2050–9. doi:10.1053/j.gastro.2006.02.055
80. Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity (2015) 42:28–39. doi:10.1016/j.immuni.2014.12.028
81. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature (2008) 453:620–5. doi:10.1038/nature07008
82. Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol (2013) 13:321–35. doi:10.1038/nri3430
83. Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol (2005) 43:3380–9. doi:10.1128/JCM.43.7.3380-3389.2005
84. Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol (2011) 12:5–9. doi:10.1038/ni0111-5
85. Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe (2011) 10:311–23. doi:10.1016/j.chom.2011.10.004
86. Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol (2014) 14:9–23. doi:10.1038/nri3565
87. Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, Schlee M, et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut (2004) 53:1658–64. doi:10.1136/gut.2003.032805
88. Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature (2001) 411:603–6. doi:10.1038/35079114
89. Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature (2001) 411:599–603. doi:10.1038/35079107
90. Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis (2011) 17:179–84. doi:10.1002/ibd.21339
91. Li E, Hamm CM, Gulati AS, Sartor RB, Chen H, Wu X, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One (2012) 7:e26284. doi:10.1371/journal.pone.0026284
92. Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, et al. NOD2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A (2009) 106:15813–8. doi:10.1073/pnas.0907722106
93. Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest (2013) 123:700–11. doi:10.1172/JCI62236
94. Jiang W, Wang X, Zeng B, Liu L, Tardivel A, Wei H, et al. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J Exp Med (2013) 210:2465–76. doi:10.1084/jem.20122490
95. Palomino-Morales RJ, Oliver J, Gomez-Garcia M, Lopez-Nevot MA, Rodrigo L, Nieto A, et al. Association of ATG16L1 and IRGM genes polymorphisms with inflammatory bowel disease: a meta-analysis approach. Genes Immun (2009) 10:356–64. doi:10.1038/gene.2009.25
96. Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med (2016) 22:598–605. doi:10.1038/nm.4102
97. Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology (2007) 132:1665–71. doi:10.1053/j.gastro.2007.03.034
98. Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell (2010) 141:1135–45. doi:10.1016/j.cell.2010.05.009
99. Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell (2008) 134:743–56. doi:10.1016/j.cell.2008.07.021
100. Rausch P, Rehman A, Kunzel S, Hasler R, Ott SJ, Schreiber S, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A (2011) 108:19030–5. doi:10.1073/pnas.1106408108
101. Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet (2011) 43:246–52. doi:10.1038/ng.764
102. Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet (2010) 42:1118–25. doi:10.1038/ng.717
103. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature (2012) 491:119–24. doi:10.1038/nature11582
104. Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology (2013) 145:293–308. doi:10.1053/j.gastro.2013.05.050
105. Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med (2006) 203:2473–83. doi:10.1084/jem.20061099
106. Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest (2006) 116:1310–6. doi:10.1172/JCI21404
107. Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature (2016) 535:85–93. doi:10.1038/nature18849
108. Molloy MJ, Grainger JR, Bouladoux N, Hand TW, Koo LY, Naik S, et al. Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host Microbe (2013) 14:318–28. doi:10.1016/j.chom.2013.08.003
109. Serban DE. Microbiota in inflammatory bowel disease pathogenesis and therapy: is it all about diet? Nutr Clin Pract (2015) 30:760–79. doi:10.1177/0884533615606898
110. Yadav V, Varum F, Bravo R, Furrer E, Bojic D, Basit AW. Inflammatory bowel disease: exploring gut pathophysiology for novel therapeutic targets. Transl Res (2016) 176:38–68. doi:10.1016/j.trsl.2016.04.009
111. Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe (2011) 9:390–403. doi:10.1016/j.chom.2011.04.009
112. Ferreira RB, Willing BP, Finlay BB. Bringing Koch’s postulates to the table in IBD. Cell Host Microbe (2011) 9:353–4. doi:10.1016/j.chom.2011.05.002
113. Albenberg LG, Lewis JD, Wu GD. Food and the gut microbiota in inflammatory bowel diseases: a critical connection. Curr Opin Gastroenterol (2012) 28:314–20. doi:10.1097/MOG.0b013e328354586f
114. Kappelman MD. Environmental factors and inflammatory bowel disease: elusive or nonexistent? Inflamm Bowel Dis (2013) 19:548–9. doi:10.1097/MIB.0b013e318281ce99
115. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med (2009) 1:6ra14. doi:10.1126/scitranslmed.3000322
116. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature (2014) 505:559–63. doi:10.1038/nature12820
117. Cordain L, Eaton SB, Sebastian A, Mann N, Lindeberg S, Watkins BA, et al. Origins and evolution of the Western diet: health implications for the 21st century. Am J Clin Nutr (2005) 81:341–54.
118. Lee D, Albenberg L, Compher C, Baldassano R, Piccoli D, Lewis JD, et al. Diet in the pathogenesis and treatment of inflammatory bowel diseases. Gastroenterology (2015) 148:1087–106. doi:10.1053/j.gastro.2015.01.007
119. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in IL10−/− mice. Nature (2012) 487:104–8. doi:10.1038/nature11225
120. Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients (2011) 3:858–76. doi:10.3390/nu3100858
121. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut (2014) 63:1275–83. doi:10.1136/gutjnl-2013-304833
122. Breuer RI, Soergel KH, Lashner BA, Christ ML, Hanauer SB, Vanagunas A, et al. Short chain fatty acid rectal irrigation for left-sided ulcerative colitis: a randomised, placebo controlled trial. Gut (1997) 40:485–91. doi:10.1136/gut.40.4.485
123. Scheppach W. Treatment of distal ulcerative colitis with short-chain fatty acid enemas. A placebo-controlled trial. German-Austrian SCFA Study Group. Dig Dis Sci (1996) 41:2254–9. doi:10.1007/BF02071409
124. Vernia P, Marcheggiano A, Caprilli R, Frieri G, Corrao G, Valpiani D, et al. Short-chain fatty acid topical treatment in distal ulcerative colitis. Aliment Pharmacol Ther (1995) 9:309–13. doi:10.1111/j.1365-2036.1995.tb00386.x
125. Vernia P, Annese V, Bresci G, d’Albasio G, D’Inca R, Giaccari S, et al. Topical butyrate improves efficacy of 5-ASA in refractory distal ulcerative colitis: results of a multicentre trial. Eur J Clin Invest (2003) 33:244–8. doi:10.1046/j.1365-2362.2003.01130.x
126. Scheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, Christl S, et al. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology (1992) 103:51–6. doi:10.1016/0016-5085(92)91094-K
127. Galvez J, Rodriguez-Cabezas ME, Zarzuelo A. Effects of dietary fiber on inflammatory bowel disease. Mol Nutr Food Res (2005) 49:601–8. doi:10.1002/mnfr.200500013
128. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A (2009) 106:3698–703. doi:10.1073/pnas.0812874106
129. Monteleone I, Pallone F, Monteleone G. Aryl hydrocarbon receptor and colitis. Semin Immunopathol (2013) 35:671–5. doi:10.1007/s00281-013-0396-2
130. Hubbard TD, Murray IA, Perdew GH. Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug Metab Dispos (2015) 43:1522–35. doi:10.1124/dmd.115.064246
131. Quintana FJ, Sherr DH. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacol Rev (2013) 65:1148–61. doi:10.1124/pr.113.007823
132. Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of notch. Nat Immunol (2011) 13:144–51. doi:10.1038/ni.2187
133. Sokol H, Conway KL, Zhang M, Choi M, Morin B, Cao Z, et al. CARD9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology (2013) 145:591–601.e3. doi:10.1053/j.gastro.2013.05.047
134. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity (2013) 39:372–85. doi:10.1016/j.immuni.2013.08.003
135. Abegunde AT, Muhammad BH, Bhatti O, Ali T. Environmental risk factors for inflammatory bowel diseases: evidence based literature review. World J Gastroenterol (2016) 22:6296–317. doi:10.3748/wjg.v22.i27.6296
136. Gerbarg PL, Jacob VE, Stevens L, Bosworth BP, Chabouni F, DeFilippis EM, et al. The effect of breathing, movement, and meditation on psychological and physical symptoms and inflammatory biomarkers in inflammatory bowel disease: a randomized controlled trial. Inflamm Bowel Dis (2015) 21:2886–96. doi:10.1097/MIB.0000000000000568
137. Bitton A, Dobkin PL, Edwardes MD, Sewitch MJ, Meddings JB, Rawal S, et al. Predicting relapse in Crohn’s disease: a biopsychosocial model. Gut (2008) 57:1386–92. doi:10.1136/gut.2007.134817
138. Fink G. Selye’s general adaptation syndrome: stress-induced gastro-duodenal ulceration and inflammatory bowel disease. J Endocrinol (2017) 232:F1–5. doi:10.1530/JOE-16-0547
139. Tache Y, Brunnhuber S. From Hans Selye’s discovery of biological stress to the identification of corticotropin-releasing factor signaling pathways: implication in stress-related functional bowel diseases. Ann N Y Acad Sci (2008) 1148:29–41. doi:10.1196/annals.1410.007
140. Bunnett NW. The stressed gut: contributions of intestinal stress peptides to inflammation and motility. Proc Natl Acad Sci U S A (2005) 102:7409–10. doi:10.1073/pnas.0503092102
141. Lukewich MK, Rogers RC, Lomax AE. Divergent neuroendocrine responses to localized and systemic inflammation. Semin Immunol (2014) 26:402–8. doi:10.1016/j.smim.2014.01.004
142. Tache Y, Larauche M, Yuan PQ, Million M. Brain and gut CRF signaling: biological actions and role in the gastrointestinal tract. Curr Mol Pharmacol (2017). doi:10.2174/1874467210666170224095741
143. Smith F, Clark JE, Overman BL, Tozel CC, Huang JH, Rivier JE, et al. Early weaning stress impairs development of mucosal barrier function in the porcine intestine. Am J Physiol Gastrointest Liver Physiol (2010) 298:G352–63. doi:10.1152/ajpgi.00081.2009
144. Wang H, Zhang C, Wu G, Sun Y, Wang B, He B, et al. Glutamine enhances tight junction protein expression and modulates corticotropin-releasing factor signaling in the jejunum of weanling piglets. J Nutr (2015) 145:25–31. doi:10.3945/jn.114.202515
145. Moss AC, Anton P, Savidge T, Newman P, Cheifetz AS, Gay J, et al. Urocortin II mediates pro-inflammatory effects in human colonocytes via corticotropin-releasing hormone receptor 2alpha. Gut (2007) 56:1210–7. doi:10.1136/gut.2006.110668
146. Kawahito Y, Sano H, Mukai S, Asai K, Kimura S, Yamamura Y, et al. Corticotropin releasing hormone in colonic mucosa in patients with ulcerative colitis. Gut (1995) 37:544–51. doi:10.1136/gut.37.4.544
147. Saruta M, Takahashi K, Suzuki T, Torii A, Kawakami M, Sasano H. Urocortin 1 in colonic mucosa in patients with ulcerative colitis. J Clin Endocrinol Metab (2004) 89:5352–61. doi:10.1210/jc.2004-0195
148. Brzozowski B, Mazur-Bialy A, Pajdo R, Kwiecien S, Bilski J, Zwolinska-Wcislo M, et al. Mechanisms by which stress affects the experimental and clinical inflammatory bowel disease (IBD): role of brain-gut axis. Curr Neuropharmacol (2016) 14:892–900. doi:10.2174/1570159X14666160404124127
149. Karl JP, Margolis LM, Madslien EH, Murphy NE, Castellani JW, Gundersen Y, et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am J Physiol Gastrointest Liver Physiol (2017) 312:G559–71. doi:10.1152/ajpgi.00066.2017
150. Allen-Blevins CR, You X, Hinde K, Sela DA. Handling stress may confound murine gut microbiota studies. PeerJ (2017) 5:e2876. doi:10.7717/peerj.2876
151. Rea K, Dinan TG, Cryan JF. The microbiome: a key regulator of stress and neuroinflammation. Neurobiol Stress (2016) 4:23–33. doi:10.1016/j.ynstr.2016.03.001
152. Cohen BL, Sachar DB. Update on anti-tumor necrosis factor agents and other new drugs for inflammatory bowel disease. BMJ (2017) 357:j2505. doi:10.1136/bmj.j2505
153. Wang B, Wu Z, Ji Y, Sun K, Dai Z, Wu G. l-glutamine enhances tight junction integrity by activating CaMK kinase 2-AMP-activated protein kinase signaling in intestinal porcine epithelial cells. J Nutr (2016) 146:501–8. doi:10.3945/jn.115.224857
154. Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, et al. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun (2009) 77:4668–78. doi:10.1128/IAI.00493-09
155. Morton ER, Lynch J, Froment A, Lafosse S, Heyer E, Przeworski M, et al. Variation in rural African gut microbiota is strongly correlated with colonization by Entamoeba and subsistence. PLoS Genet (2015) 11:e1005658. doi:10.1371/journal.pgen.1005658
156. Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell (2013) 152:39–50. doi:10.1016/j.cell.2012.10.052
157. Vangay P, Ward T, Gerber JS, Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe (2015) 17:553–64. doi:10.1016/j.chom.2015.04.006
158. Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol (2012) 9:599–608. doi:10.1038/nrgastro.2012.152
159. Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology (2004) 126:1620–33. doi:10.1053/j.gastro.2004.03.024
160. Scott KP, Antoine JM, Midtvedt T, van Hemert S. Manipulating the gut microbiota to maintain health and treat disease. Microb Ecol Health Dis (2015) 26:25877. doi:10.3402/mehd.v26.25877
161. Khan KJ, Ullman TA, Ford AC, Abreu MT, Abadir A, Marshall JK, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol (2011) 106:661–73. doi:10.1038/ajg.2011.72
162. Nitzan O, Elias M, Peretz A, Saliba W. Role of antibiotics for treatment of inflammatory bowel disease. World J Gastroenterol (2016) 22:1078–87. doi:10.3748/wjg.v22.i3.1078
163. Ungaro R, Bernstein CN, Gearry R, Hviid A, Kolho KL, Kronman MP, et al. Antibiotics associated with increased risk of new-onset Crohn’s disease but not ulcerative colitis: a meta-analysis. Am J Gastroenterol (2014) 109:1728–38. doi:10.1038/ajg.2014.246
164. Hviid A, Svanstrom H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut (2011) 60:49–54. doi:10.1136/gut.2010.219683
165. Khoruts A, Sadowsky MJ. Therapeutic transplantation of the distal gut microbiota. Mucosal Immunol (2011) 4:4–7. doi:10.1038/mi.2010.79
166. Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat Rev Gastroenterol Hepatol (2016) 13:508–16. doi:10.1038/nrgastro.2016.98
167. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med (2013) 368:407–15. doi:10.1056/NEJMoa1205037
168. Anderson JL, Edney RJ, Whelan K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment Pharmacol Ther (2012) 36:503–16. doi:10.1111/j.1365-2036.2012.05220.x
169. Lopez J, Grinspan A. Fecal microbiota transplantation for inflammatory bowel disease. Gastroenterol Hepatol (N Y) (2016) 12:374–9.
170. Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology (2012) 143:913–6.e7. doi:10.1053/j.gastro.2012.06.031
171. Cammarota G, Ianiro G, Gasbarrini A. Fecal microbiota transplantation for the treatment of Clostridium difficile infection: a systematic review. J Clin Gastroenterol (2014) 48:693–702. doi:10.1097/MCG.0000000000000046
172. Ianiro G, Bibbo S, Scaldaferri F, Gasbarrini A, Cammarota G. Fecal microbiota transplantation in inflammatory bowel disease: beyond the excitement. Medicine (Baltimore) (2014) 93:e97. doi:10.1097/MD.0000000000000097
173. Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis (2014) 8:1569–81. doi:10.1016/j.crohns.2014.08.006
174. Gerich ME, McGovern DP. Towards personalized care in IBD. Nat Rev Gastroenterol Hepatol (2014) 11:287–99. doi:10.1038/nrgastro.2013.242
175. Angelberger S, Reinisch W, Makristathis A, Lichtenberger C, Dejaco C, Papay P, et al. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol (2013) 108:1620–30. doi:10.1038/ajg.2013.257
176. Schrezenmeir J, de Vrese M. Probiotics, prebiotics, and synbiotics—approaching a definition. Am J Clin Nutr (2001) 73:361s–4s.
177. Tamboli CP, Caucheteux C, Cortot A, Colombel JF, Desreumaux P. Probiotics in inflammatory bowel disease: a critical review. Best Pract Res Clin Gastroenterol (2003) 17:805–20. doi:10.1016/S1521-6918(03)00076-3
178. Shanahan F. Probiotics in perspective. Gastroenterology (2010) 139:1808–12. doi:10.1053/j.gastro.2010.10.025
179. Sheil B, Shanahan F, O’Mahony L. Probiotic effects on inflammatory bowel disease. J Nutr (2007) 137:819S–24S.
180. Tao Y, Drabik KA, Waypa TS, Musch MW, Alverdy JC, Schneewind O, et al. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am J Physiol Cell Physiol (2006) 290:C1018–30. doi:10.1152/ajpcell.00131.2005
181. Gosselink MP, Schouten WR, van Lieshout LM, Hop WC, Laman JD, Ruseler-van Embden JG. Delay of the first onset of pouchitis by oral intake of the probiotic strain Lactobacillus rhamnosus GG. Dis Colon Rectum (2004) 47:876–84. doi:10.1007/s10350-004-0525-z
182. Sood A, Midha V, Makharia GK, Ahuja V, Singal D, Goswami P, et al. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol (2009) 7:1202–9, 1209.e1. doi:10.1016/j.cgh.2009.07.016
183. Tursi A, Brandimarte G, Papa A, Giglio A, Elisei W, Giorgetti GM, et al. Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: a double-blind, randomized, placebo-controlled study. Am J Gastroenterol (2010) 105:2218–27. doi:10.1038/ajg.2010.218
184. Shen J, Zuo ZX, Mao AP. Effect of probiotics on inducing remission and maintaining therapy in ulcerative colitis, Crohn’s disease, and pouchitis: meta-analysis of randomized controlled trials. Inflamm Bowel Dis (2014) 20:21–35. doi:10.1097/01.MIB.0000437495.30052.be
185. Kato K, Mizuno S, Umesaki Y, Ishii Y, Sugitani M, Imaoka A, et al. Randomized placebo-controlled trial assessing the effect of bifidobacteria-fermented milk on active ulcerative colitis. Aliment Pharmacol Ther (2004) 20:1133–41. doi:10.1111/j.1365-2036.2004.02268.x
186. Oliva S, Di Nardo G, Ferrari F, Mallardo S, Rossi P, Patrizi G, et al. Randomised clinical trial: the effectiveness of Lactobacillus reuteri ATCC 55730 rectal enema in children with active distal ulcerative colitis. Aliment Pharmacol Ther (2012) 35:327–34. doi:10.1111/j.1365-2036.2011.04939.x
187. Nami Y, Abdullah N, Haghshenas B, Radiah D, Rosli R, Khosroushahi AY. Assessment of probiotic potential and anticancer activity of newly isolated vaginal bacterium Lactobacillus plantarum 5BL. Microbiol Immunol (2014) 58:492–502. doi:10.1111/1348-0421.12175
Keywords: intestinal microbiota, host immune response, inflammatory bowel disease, intestinal barrier function, epithelial cells
Citation: Zhang M, Sun K, Wu Y, Yang Y, Tso P and Wu Z (2017) Interactions between Intestinal Microbiota and Host Immune Response in Inflammatory Bowel Disease. Front. Immunol. 8:942. doi: 10.3389/fimmu.2017.00942
Received: 09 May 2017; Accepted: 24 July 2017;
Published: 14 August 2017
Edited by:
Jixin Zhong, Case Western Reserve University, United StatesReviewed by:
Bo-Zong Shao, Second Military Medical University, ChinaCopyright: © 2017 Zhang, Sun, Wu, Yang, Tso and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Yang, Y2F1dmV0QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.