
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 03 May 2017
Sec. Inflammation
Volume 8 - 2017 | https://doi.org/10.3389/fimmu.2017.00501
 Li Hanyang1,2,3
Li Hanyang1,2,3 Liu Xuanzhe1,2,3
Liu Xuanzhe1,2,3 Chen Xuyang1,2,3
Chen Xuyang1,2,3 Qiu Yujia1,2,3
Qiu Yujia1,2,3 Fu Jiarong1,2,3
Fu Jiarong1,2,3 Shen Jun1,2,3*
Shen Jun1,2,3* Ran Zhihua1,2,3
Ran Zhihua1,2,3
Inflammatory bowel disease (IBD) is a chronic, recurrent, and remitting inflammatory disease with unclear etiology. As a clinically frequent disease, it can affect individuals throughout their lives, with multiple complications. Unfortunately, traditional murine models are not efficient for the further study of IBD. Thus, effective and convenient animal models are needed. Zebrafish have been used as model organisms to investigate IBD because of their suggested highly genetic similarity to humans and their superiority as laboratory models. The zebrafish model has been used to study the composition of intestinal microbiota, novel genes, and therapeutic approaches. The pathogenesis of IBD is still unclear and many risk factors remain unidentified. In this review, we compare traditional murine models and zebrafish models in terms of advantages, pathogenesis, and drug discovery screening for IBD. We also review the progress and deficiencies of the zebrafish model for scientific applications.
Inflammatory bowel disease (IBD) is a general description of gastrointestinal tract diseases that show a chronic or recurring immune response and inflammatory symptoms. The two most common types of IBD are ulcerative colitis (UC) and Crohn’s disease (CD). In CD, the inflammation affects the whole digestive tract, whereas in UC, only the colon is involved. Both these diseases are characterized by an abnormal response by the immune system, compromising diarrhea or rectal urgency, bleeding, abdominal pain, constipation, and loss of appetite (1). In addition, IBD can cause a number of non-digestive tract complications. Parenteral complications severely interfere with patients’ quality of life. As a far-ranging disease, IBD can occur at any age and affects both sexes. About 600,000 new cases of IBD are diagnosed in America every year (2). However, the pathogenesis and etiology of IBD remain incompletely understood. Controversy concerning the etiology of IBD exists because of its multifactorial pathogenesis. It is believed that susceptible genes and environmental factors are responsible for the pathogenesis of IBD (2). Both of these factors lead to an abnormal chronic or recurring immune response, resulting in tissue injury with ulceration and bowel inflammation (2). Animal models can help investigators to identify disease mechanisms and progression, and to develop drugs and targeted therapy (3). So far, the most well-established model in IBD is the murine model (3). As a valuable animal model, it has contributed a lot to the understanding of IBD, in terms of the aspects including gut microbiota, immunoreaction, infection, and inheritance. However, as IBD studies have progressed, limitations of the traditional murine model have emerged. For example, the pathological changes in the murine model cannot be determined by continuous observation. The traditional murine model breeds a generation in about 2 months (4). To some extent, this is too slow for active IBD research (5). For genetic studies, the genetic manipulation of the murine models is difficult. As the drug screening research continues, the traditional murine model has become less useful in terms of modeling speed and modeling quantity (6). These defects of the murine model have prompted investigators to find better models to study the pathogenesis and therapeutic approaches for IBD.
Zebrafish (Danio rerio) larvae have emerged as a useful tool to study IBD and gastrointestinal diseases (7). Compared with humans, the zebrafish has a highly similar gastrointestinal system, with a liver, gall bladder, pancreas, and intestinal tract with comparable absorptive and secretory functions (Figure 1) (8, 9).
In addition, a genome alignment showed that zebrafish orthologs exist for over 70% of human genes (10). This high similarity, and a clearer understanding of the critical genomic features, could promote the identification of susceptibility genes of IBD (11). For example, NOD1 and NOD2 encode two types of nucleotide-binding oligomerization domain-containing proteins, both of which recognize bacterial molecules and stimulate an immune reaction (11). Thus, zebrafish have the potential to act as a model in studies of the genetic and environmental aspects of IBD pathogenesis.
Zebrafish have already attracted the attention of researchers because of their susceptibility to mutagenesis and the availability of transgenic technology (12, 13). Commonly, the strains used in zebrafish models for IBD studies have included three major types: wild-type strains (WT strains), mutant strains, and transgenic strains. The most common type for scientific study is the AB WT strain, which is usually exposed to a sequence of colitogenic agents, containing live bacteria, bacterial products, and chemicals (12). Notably, n-ethyl-n-nitrosourea (ENU), an advanced genetic mutagenesis agent, can generate zebrafish mutants and has been used to develop spontaneous intestinal neoplasia models (13). These heterozygous mutations were confirmed to have a relationship with truncated forms of adenomatous polyposis coli (APC). The accumulated effects included an increase of nuclear β-protein and excessive expression of downstream genes such as c-MYC and AXIN2 (13). When the carcinogen 7,12-dimethylbenzanthracene was used in the zebrafish model, the frequency of gastrointestinal disease development increased, especially those involving the hepatic system, bile duct, and intestinal tract (13). APC mutations cause the spontaneous development of intestinal polyps (14–16). Therefore, the APC zebrafish model could be applied to genetic, drug screening, and toxicology studies for other intestinal diseases, including IBD.
In addition to ENU, intestinal damage models are often induced by dextran sodium sulfate (DSS) or trinitrobenzene sulfonic acid (TNBS) (17). Chemical models of enterocolitis are induced conveniently by oxazolone, lipopolysaccharide (LPS), and glafenine. Agents and chemical models cause non-specific intestinal defects. Chemical genetic methods can also manipulate the host response to injury, which is convenient for use in IBD models (17). The effect of certain chemicals can be analyzed in a specific biological system, and small molecules are then used to disturb the biological processes directly. Compared with the pure chemical model, chemical genetic methods have a more widespread application in drug screening.
Transgenic technology is the preferred method for epigenetic research and to develop more accurate disease models. Forward and reverse genetics have been used in zebrafish models, exploiting the fully sequenced genome and high-throughput sequencing (18). These approaches have provided a better view of the pathogenesis of IBD, which will be discussed in detail in the following section. The zebrafish model as drug screener is also a hot research topic. The key point is that providing specific exogenous substances to the transgenic zebrafish would lead to specific tissue changes associated with different quantities or qualities of a drug. For example, the TNBS-induced zebrafish model has been used to screen mesalazine (5-ASA) and prednisolone combined with immunofluorescence imaging (19).
Thus, zebrafish models show promise in IBD research in terms of pathogenesis and treatment. The important goals of using zebrafish models of IBD are determining and exploiting the pathogenesis of IBD, and reversing the growth and development of IBD in patients via drug screening or by immunosuppression.
Investigators have used IBD animal models for over 100 years, from drug-induced methods to genetic engineering models (20). Although several models are similar to humans, none of them perfectly mimic the pathogenesis of IBD. In nature, enteritis can develop spontaneously in some animals, and these are termed spontaneous IBD models. Pathological changes analogous to UC found in the cotton-top tamarin (CTT) and the high morbidity rate of colon adenocarcinoma in adult CTTs make them an ideal model for UC-related colon carcinoma (21). However, their scarcity has hindered their use.
Murine models have gained popularity among IBD investigators. For instance, C3H/HeJ Bir mice were derived from C3H/HeJ mice with occasional enteritis (22). Pathological changes analogous to human CD are observed in the SAMP1/Yit mouse model, especially in the distal ileum (23). Although murine models have revealed the role of epithelial barrier function, the adaptive immune response and microbiota in the pathogenesis and susceptibility of IBD, and certain limitations, such as high cost, imaging limitations, and the difficulty of genetic manipulation techniques, have impeded their use in IBD research (24). In murine models, the pathological changes cannot be determined until the mice are killed, because the pathological development of IBD cannot be tracked in a direct and dynamic way (22).
Zebrafish are easier to cultivate than mice. Zebrafish are highly fecund: a female lays hundreds of eggs each time with a generation time of approximately 3–4 months and the eggs can be fertilized in vitro. It means zebrafish model can be established more efficient than traditional murine model. It is more suitable to the researches, in which large quantities of sample data are required. Besides, the optical transparency of the embryos and their rapid development facilitate observation, manipulation, and drug screening (25).
The zebrafish intestine is a long tube composed of intestinal bulb, mid-intestine, and caudal intestine, which folds twice in the abdominal cavity (7). Three types of differentiated cells have been identified in the intestinal epithelium: absorptive enterocytes, goblet cells, and endocrine cells. The lamina propria beneath the gut epithelium contains macrophages and neutrophils (26). No Paneth cells, crypts, or submucosal glands have been detected in zebrafish (27, 28).
The immune system is the basis of the interaction between the host and microbes. The zebrafish immune system is highly analogous to that of humans. Adaptive immunity is not functional until 4 weeks after fertilization. Thus, during these 4 weeks, attention can be focused on the effects of innate immunity without the interference of adaptive immunity (24). According to current evidence, zebrafish have orthologs of mammalian toll-like receptors (TLRs), which are the innate immune receptors that recognize specific microbial molecules (29), and have functionally conserved orthologs of human IBD susceptibility genes NOD1 and NOD2 (11). Genetic techniques are easier to perform in zebrafish compared with mice. The improvement in genetic manipulation techniques for forward genetics has permitted the identification of over 160 IBD susceptibility genes in the human genome, most of which are associated with autophagy, host responses to bacteria, and the immune response: many of these have orthologs in the zebrafish genome (7, 24).
Owing to its aqueous living environment, it is easier performing experiments where fish are exposed to chemicals or microbes in real time. In addition, aseptic techniques provide a convenient control of the microbial system in zebrafish (30). The rapid development of a series of bioscience techniques, including mutation techniques (insertional mutagenesis and ENU chemical mutagenesis), transgenic technology, and fluorescence labeling technique have accelerated the use of zebrafish as a new model system (31). Mutant strains of zebrafish are obtained to build models mimicking human disease or models for drug screening. One large-scale zebrafish genetic mutation research center, Cardiovascular Research Center, Massachusetts General Hospital, which preserves thousands of zebrafish mutants with embryonic or organ defects, is located in Massachusetts. This mutant collection will enable studies of zebrafish tissue or organ structure, morphology, function, metabolism, development, ethology, and pathological mechanism (25).
Recently, IBD studies based on zebrafish models have focused mainly on five aspects: genes, development, interactions between gut microbiota and host immunity, and endoplasmic reticulum (ER) stress.
To date, 163 IBD susceptibility regions that were marked by single-nucleotide polymorphisms (SNPs) have been detected by genome-wide association studies (GWASs) in humans (32). Proteins and other functional molecules expressed in these regions have suggested the mechanisms of intestinal epithelial damage and defense, including epigenetic changes, the presence of endothelium stress, and the interaction between microbes and host immunity (33). The conservation of several core gut genes, such as NOD1 and NOD2, in zebrafish and mammals makes zebrafish an interesting model to study the pathogenesis of intestinal inflammation and injury (11, 34). Although several studies have been performed based on the GWASs, their results may not be reliable. Heritability might be missing because the GWASs were poorly designed to reveal epistasis, which would underestimate the effects of non-coding regulatory regions (35). Thus, it is believed that the variants in protein-coding genes and non-coding DNA regulatory regions rich in SNPs might play a role in the pathogenesis (36).
Marjoram et al. developed a novel inflammation-responsive transgenic zebrafish line TgBAC (TNF-α: GFP), analysis of which confirmed the notion that IBD results from the loss of epigenetic repression and TNF overproduction in the intestinal epithelial cells. They used a forward genetic screen to identify mutants characterized by deficiencies in intestinal epithelial integrity (37). One of the mutants, named aa51.3pd1092, had a significantly disrupted intestinal epithelium. A mutation in ubiquitin-like protein including PHD and RING finger domains 1 (UHRF1), a highly conserved gene in methylation, was identified as the affected locus. The dysfunctional UHRF1 leads to hypomethylation of TNF-α promoter, which releases the transcription inhibition of the promoter, thereby leading to increased TNF-α in the intestinal epithelia. Increased TNF-α expression leads to the appearance of hallmarks of microbiota-dependent chronic inflammation, such as shedding and apoptosis of epithelial cells, immune cell recruitment, and barrier dysfunction (37). Therefore, this study identified a possible susceptibility gene for IBD. The unbiased study conducted by Marjoram and his colleagues encouraged other investigators to use GWASs or high-throughput sequencing to identify potential disease-associated genes. These studies could reveal novel genetic risk factors in non-coding regions and define their distinct functions in IBD pathogenesis.
Another genetic screen identified a mutant, cdipthi559, which lacks phosphatidylinositol (PI) synthesis (38). Disturbance of PI signaling has been identified in gastrointestinal disease and inflammation (39). The cdipthi559 mutants showed abnormal villous architecture and some features of IBD, including apoptosis of goblet cells, reduced mucus secretion, bacterial overgrowth, and leukocyte infiltration. Moreover, acute phase genes were upregulated and ER stress markers, such as hspa5 and xbp1, were activated strongly (40). This study identified a previously unknown link between intracellular PI signaling and ER stress-mediated gastrointestinal inflammation.
In addition, GWASs have revealed that SNPs close to MST1 [encoding the macrophage-stimulating protein (MSP)] and MST1R [encoding the MSP receptor Ron (recepteur d’origine nantais)] were susceptibility factors of CD (41, 42). The identification of susceptibility factors in the MSP–RON signaling pathway implicates host responses to tissue lesions in CD. Current murine models with MSP and RON deficiency are incapable of uncovering the function of MSP–RON signaling pathway in intestinal inflammation. Nevertheless, positional cloning has led to the creation of a novel zebrafish MSP mutant (mspt34230) with a premature stop mutation (43). These zebrafish demonstrate intestinal eosinophilia, upregulated expression of the inflammatory marker MMP9 and goblet cell alterations. Additionally, ethanol-induced epithelial damage appears to be more severe in the MSP mutant. Intrarectal ethanol administration (20%) resulted in reduced survival, upregulated infiltration and proliferation of immune cells, and prolonged inflammatory cytokine responses in some MSP-deficient zebrafish (44). The innate cell recruitment and barrier dysfunction in the MSP and Ron mutants require further investigation to understand how disturbance of this pathway leads to chronic inflammation.
While NOD2 was the first human gene to be associated strongly with CD (45, 46), mutations in NOD1 also show a disease-specific association with UC. NOD1/2 is conserved in the human and zebrafish genomes and encodes intracellular bacterial sensor proteins that initiate the innate immunity (Figure 2) (11).
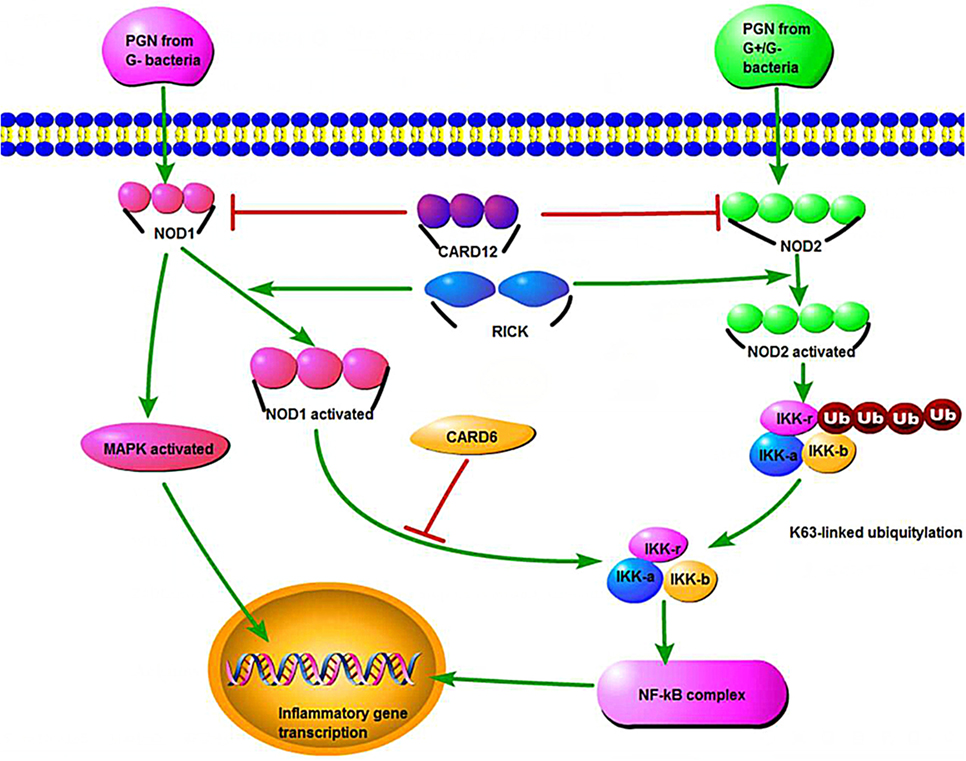
Figure 2. The NOD1 and NOD2 signaling pathways, host defense, and inflammatory defense. PGN represents peptidoglycan, and CARD12 is a member of the CED4/Apaf-1 family that can induce apoptosis. MAPK refers to mitogen-activated protein kinase, which stimulates the inflammatory process of cells. IKK refers to inhibitor of nuclear factor kappa-B (NF-κB) kinase, which participates in the cellular immune response caused by cytokines. NF-κB is a nuclear transcription factor that regulates the genes that are believed to be necessary in promoting inflammation.
NOD signaling genes are expressed in intestinal epithelial cells and neutrophils. Morpholino oligomer (MO) knockdown is a useful tool to evaluate the zebrafish NOD1/2 functions. In NOD1/2 MO knockdown models, susceptibility to bacterial infections (like Salmonella enterica) increased and the expression of a dual oxidase that produces bactericidal reactive oxygen species was impaired (11).
The use of zebrafish to study IBD takes advantage of the combination of the growing list of immune cell reporter fish and the optical transparency of the larvae early in life (<2 weeks). Live imaging in zebrafish aids the study of intestinal immune cell function, the production of pro-inflammatory or anti-inflammatory cytokines, the interaction between the microbe and host immunity, and the integrity of the intestinal barrier (7). Hence, immune responses can be traced in vivo in a complete organism.
Several zebrafish models have demonstrated intestinal immune disorders in recent years. Chemically induced and genetic models have been established to determine the possible pathogenesis of IBD. Chemical zebrafish models have been induced by oxazolone, TNBS, and DSS (19, 20, 47–51). These models are characterized by the upregulated expression of cytokines, leukocytosis, and different degrees of morphological changes (19, 20, 48, 50).
Inflammatory bowel disease pathogenesis was studied initially in an oxazolone-induced enterocolitis zebrafish model. Brugman et al. administered the hapten oxazolone intrarectally to adult wild-type and myeloperoxidase-reporter transgenic zebrafish (20). They established an enterocolitis model successfully using histological and molecular evidence. Severe bowel-wall thickening, disappearance of the intestinal-fold architecture, depletion of goblet cells, and severe infiltration of eosinophils and neutrophils were observed. Oxazolone upregulated the expression of a series of genes encoding colitis-related cytokines, including pro-inflammatory (i.e., IL-1β, TNF-α) and anti-inflammatory cytokines (i.e., IL-10) in the intestine.
Another useful chemically induced zebrafish model was established by immersing the zebrafish in defined media mixed with TNBS. TNBS can induce intestinal inflammation and impairs gut functions in zebrafish larvae (50). Fleming reported morphological changes and increased numbers of goblet cells under exposure to 75 µg/mL TNBS (48). Oehlers documented that the same dose of TNBS exposure shortened the mid-intestine (segment II) and recruited neutrophils significantly in the intestine, but they did not observe significant intestinal morphological changes (19). Despite these differences, inflammation was confirmed by induction of the pro-inflammatory cytokines IL-1β and TNF-α, the degradative enzyme MMP9, and leukocytosis (19, 48). In addition to inflammation, loss of peristalsis and alteration of lipid metabolism have been documented (19, 48).
Dextran sodium sulfate has been used widely on mice and rats in studies of IBD pathogenesis (52). In zebrafish, DSS immersion is also useful to induce an enterocolitis model. Oheler reported the intestinal infiltration of neutrophils, global cell proliferation reduction, and upregulation of pro-inflammatory genes (i.e., TNF-α, IL-1β, IL-8, CCL20, and MMP9) after DSS exposure. Unlike the TNBS models, acidic mucins accumulated strongly in the intestinal bulb; however, the goblet cell number was relatively unchanged (50). The redundancy of mucins has a protective role in the gut epithelium during intestinal inflammation. The distinct mucin phenotype of the DSS zebrafish model permitted studies to uncover the regulation of mucins by various agents. For example, retinoic acid can suppress both basal and intestinal mucus production; thus exacerbating experimental enterocolitis in the zebrafish model (50).
Furthermore, using MO knockdown techniques, researchers can study the pathogenesis of IBD pathogenesis at the level of cytokines. Pro-inflammatory and anti-inflammatory cytokines maintain a dynamic balance of the immune response in the health body, and this balance is dysregulated in IBD patients.
Genome-wide association studies have proved that IL-23R is a susceptibility allele in IBD patients (26). IL-23 participates in the development of Th17 cells and the innate immune response (53). Using a synteny (conservation of gene order) approach, the IL-23 p19 subunit was identified in zebrafish. However, the exact function of IL-23 has not yet been determined. IL-23 is expressed constitutively in the zebrafish intestine and is induced by LPSs administration or Mycobacterium marinum infection (54). Hence, such a model could be used to study adaptive immunity in IBD pathogenesis.
IL-10 is an anti-inflammatory cytokine in the intestine. Lack of IL-10 might lead to IBD in humans (55). As a classic study object, IL-10 has been discussed a lot in the researches with murine models before. In recent years, with the warming up of zebrafish models, the researches of IL-10 can also been taken in the usage of zebrafish. IL-10 is conserved in the zebrafish intestine, and its expression was upregulated after LPS stimulation (56, 57). Using the zebrafish IL-10 mutant model, further study of IL-10 functions might increase our understanding of the host–microbe interactions and intestinal inflammation in IBD (58).
IL-22 is an immuomodulatory and multifunctional molecule in the intestine that is upregulated in CD patients (59). Studies in mice showed that IL-22 stimulates epithelial cells to produce antibacterial proteins, enhances mucus secretion, and strengthens epithelial regeneration (60). Studies using loss-of-function and gain-of-function approaches indicated that IL-22 could promote mucosal healing in acute intestinal injury. Recently, IL-22’s functions in intestinal inflammation in innate immunity were revealed (61). Silencing of IL-22 increased the expressions of pro-inflammatory cytokines (IL-1β and TNF-α) in bacteria-stimulated fish. Further studies should focus on how this cytokine maintains the intestinal homeostasis in zebrafish.
In addition to the well-known pro-inflammatory and anti-inflammatory cytokines, inflammation-related factors and signaling pathway might also be associated with the pathogenesis of IBD. Zebrafish models could help investigators to study CXCL8, CCL13, and CCL14, which are difficult to study in murine models (62). For example, human CXCL8 regulates the chemotaxis of neutrophils. Unfortunately, murine models of CXCL8 deficiency are difficult to investigate. Surprisingly, the expression of CXCL8 is significantly enhanced in the larval intestine of TNBS-induced enterocolitis zebrafish models (49).
The heat shock proteins (HSPs) also have anti-inflammatory effects and can regulate intestinal immune function (63). Studies in mice showed that HSF1 (an HSP) and HSP70 play a protective role in IBD (64). Interestingly, the expressions of the HSP70 and HSP110 family genes HSPa4 and HSP4b were increased in TNBS-induced enterocolitis zebrafish models (47).
Host responses to inflammation are dependent on the highly conserved neuropeptides of the brain–gut axis (64). Orthologs of corticotrophin-releasing hormone and urocortins (65), Substance P (66), neuropeptide Y (67), ghrelin (68), α-MSH (69), and gastrointestinal peptide (68) have been documented in zebrafish. The involvement of these neuropeptides in the host reaction to intestinal inflammation has been demonstrated in both murine and human studies (24, 70–75). Furthermore, an adult zebrafish model created by treatment with intrarectal injection of TNBS demonstrated that Melanin-concentrating hormone (MCH) expression is highly conserved among fish, mice, and humans (70). MCH is a conserved appetite-regulating neuropeptide that is implicated in the pathogenesis of IBD (71). The expression of MCH and the MCH receptor were upregulated in the intestine of model of zebrafish. Immunostaining revealed a reinforcement effect of colitis in zebrafish that was analogous to human IBD and TNBS-induced mouse colitis. However, the signaling pathways involved in zebrafish have not been determined (70).
Myeloid differentiation factor 88 (MYD88) acts as an adaptor protein in interleukin 1 receptor (IL-1R) and TLR signaling in human and mammalian models. Thus, MYD88 is one of the major molecules of the innate immune response. In a zebrafish model for signaling pathway studies, investigators first focused on the MYD88 signaling pathway. MYD88 is highly conserved in zebrafish (72, 73). This property of initiating the innate immune response makes MYD88 a response factor of the host–microbe interaction. Studies have revealed that the expressions of transcription factors (including NF-κB and AP-1) in the core of innate immunity and pro-inflammatory cytokines (including IL-1β, MMP9) depend on MYD88 signaling during bacterial infection. Acute bacterial infection induced by Edwardsiella tarda and Salmonella typhimurium, as well as chronic bacterial infections induced by M. marinum, induces severe intestinal lesions in zebrafish. MYD88-MO and MYD88 mutants had deficiencies of innate immunity and were increasingly susceptible to microbial infection (73). Moreover, knockdown of MYD88 caused zebrafish to develop more severe intestinal lesions under TNBS exposure, suggesting that MYD88 has a protective role in pathogenesis of colitis. Hence, the MYD88 mutant zebrafish will be useful in studies of IL-1R and TLR signaling in the host–microbe interaction and in intestinal inflammation.
Studies performed in gnotobiotic animals, such as mice and zebrafish, have demonstrated that the microbiota have an enormous impact on the host (74). The maturation of gnotobiotic techniques, particularly in vitro fertilization, has helped researchers to manipulate the species and to quantify the host–microbe interaction in zebrafish (24). Therefore, it has been suggested that zebrafish could be a more powerful animal model to study the aspects of the interaction between host and microbes.
To understand the effect of microbial colonization on the zebrafish host, Rawls et al. investigated gene expression changes in response to microbial colonization (75). The gene profiles demonstrated that microbial colonization altered the expressions of 212 genes. Among them, the responses of 59 genes were conserved between mice and zebrafish. These conserved genes were mainly involved in epithelial proliferation, promotion of nutrient metabolism, and innate immune responses. These data suggested that the response to microbes is highly conserved.
Experiments in zebrafish demonstrated that microbes influence fat absorption in the intestine. The presence of a microbiota resulted in increased fat storage in adipose tissue (76). Meanwhile, other studies showed that microbes stimulate fatty acid uptake in the intestinal epithelium and liver (77). These studies implied that diet-induced alterations of microbiota could lead to IBD by disturbing the host energy balance. Microbes can also induce intestinal development. Cheesman et al. documented that the presence of microbiota and Wnt signaling could stimulate epithelial cell proliferation during zebrafish intestinal development (78). Importantly, their findings demonstrated that homeostatic innate immune responses, but not inflammatory signals, contribute to the effects on epithelial proliferation. This could be explained by the fact that epithelial turnover depends on microbes via MYD88, but not via the TNF receptor (78). The absence of microbes hinders the differentiation of the intestinal epithelium (79). However, Bates et al. found several deficiencies in microbe-free intestines. There was a lack of brush border intestinal alkaline phosphate (IAP) activity and glycan expression was immature. The numbers of goblet cells and enteroendocrine cells decreased. Additionally, germ-free (GF) intestines failed to take up protein macromolecules in the distal intestine and presented a faster rate of peristalsis (79). Interestingly, colonization by microbes could completely reverse these GF manifestations. However, exposure to heat-killed microbes and LPS could restore the IAP activity, but not the glycan expression, suggesting that LPS induces IAP. IAP-deficient zebrafish are more sensitive to LPS exposure. Abundant neutrophils are observed under LPS exposure, while neutrophils are insufficient in the GF IAP-deficient phenotype. The phenotype of the IAP-deficient zebrafish was shown to depend on the proteins involved in LPS sensitivity in mammals (i.e., MYD88 and TNFR) (79). The above findings suggested that the presence of zebrafish intestinal endogenous microbiota maintains a normal level of neutrophils in the intestine through the involvement of three molecules: IAP, MYD88, and TNFR. Thus, IAP is involved in the protecting the intestine from microbe-related lesions and is suggested that abnormal microbiota cause IBD by influencing IAP activity.
Microbes intensify the host’s immune responses. The gut microbiota influences the host immune system through mutual interactions (74). In TNBS-induced enterocolitis zebrafish models, administration of ampicillin and kanamycin before TNBS in the fish media increased the survival of the fish and downregulated the expression of pro-inflammatory cytokines such as IL-1β, TNF-α, CCL20, and IL-8 (49). Studies in experimental animals have indicated that the intestinal microbiota play a critical role in the pathogenesis of intestinal inflammation (80). Dysbiosis potentially contributes to the pathogenesis of IBD by augmenting host pro-inflammatory immune responses.
Resident microbiota maintain the homeostasis of the host’s health (81). The composition of microbes in the host depends on the environment. Zebrafish are enriched in Proteobacteria, while mice and human have more Bacteroidetes and Firmicutes (75, 82). A study of intestinal microbiota dysbiosis in zebrafish induced by TNBS demonstrated an increased proportion of Proteobacteria and a decrease in the Firmicutes (83). Further analyses of this dysbiosis indicated a significant relationship with enterocolitis. Further studies confirmed the view that TLR signaling pathways recognize bacteria, resulting in the induced expression of immune molecules, including NF-κB and inflammatory cytokines (30). Investigators compared conventionally reared (CV) zebrafish, GF zebrafish, and TNBS-induced enterocolitis zebrafish models. TNBS exposure of CV zebrafish induced MYD88 and TRIF expression, NF-κB activation, and TNF-α expression. However, TNBS exposure of GF zebrafish and the control showed the absence of a TLR3 signaling pathway response and no detectable expression in MYD88 or TRIF, no NF-κB activation, or no TNF-α expression. Most importantly, TLR signaling pathways, including MYD88-related and MYD88-non-related pathways, appear to be the principal pathogenic pathways of enterocolitis.
In the oxalone-induced zebrafish model, the microbiota can influence the degree of enterocolitis. Brugman et al. established an enterocolitis score to evaluate inflammation. The score takes several factors into consideration, including the depletion of goblet cells, the infiltration of inflammatory cells and upregulation of pro-inflammatory cytokines (20). In vancomycin-treated zebrafish, the population of microbes was dominated by the Fusobacteria. In this model, decreased neutrophil infiltrations correlated with a reduced enterocolitis score. In the colistin sulfate-treated zebrafish model, the population of microbes was dominated by the Proteobacteria. Surprisingly, reduced eosinophil and lymphocyte infiltration was observed, while the enterocolitis scores remained unchanged. These findings indicated that different compositions of microbes determine the severity of enterocolitis and the composition of intestinal infiltration. Collectively, the results suggest that a deeper insight into chemokines and immune homologs in zebrafish is required.
Defects in the ER stress response and autophagy result in intestinal inflammation (84). Components of the ER stress response in the intestinal epithelium have been documented in IBD patients and murine models (85, 86).
The cdipthi559 zebrafish mutant has a deficiency in PI synthesis and shows characteristics of ER stress in the intestinal epithelium (39). The abnormal intestinal morphologies comprise reduction of epithelial proliferation, apoptosis of epithelial and goblet cells, severe inflammation, and disruption of the normal bacteria–intestine balance. Two ER stress factors, HSPα5 and XBP1, are increased in the intestinal epithelium. Temporal electron microscopy analyses demonstrated that PI-deficient intestinal epithelium cells undergo chronic ER stress-mediated cytopathology, including disruption of the ER and Golgi apparatus, mitochondrial damage, autophagy, and apoptosis. Furthermore, the pharmacological induction of ER stress using tunicamycin to inhibit protein glycosylation or PI synthase inhibition in leukocyte-specific reporter lines produced similar phenotypes to those of the cdipthi559 mutants. Antibiotics were administered to confirm the bacteria’s exacerbation of inflammation. In addition, chemical chaperones could reduce inflammation, indicating the therapeutic potential for small molecular agents in IBD (39).
In murine models, phospholipids showed a potential to resist inflammatory lesions (87). PI alleviates pathological ER stress, allowing the ER to perform its anti-inflammatory effect. Goldsmith et al. showed that the μ-opioid receptor agonist DALDA, a dermorphin analog (H-Tyr-d-Arg-Phe-Lys-NH2), alleviated ER stress to protect against intestinal injury in the glafenine-induced zebrafish model by inducing the unfolded protein response (UPR) (87). Investigators first established the zebrafish model by induction using non-steroidal anti-inflammatory drugs to simulate an impaired mucosal barrier function (88). The glafenine-induced phenotype was characterized by increased intestinal epithelial apoptosis and ER stress, but not by a reduction in the epithelial barrier function. Treatment with DALDA had no effect on the early UPR marker BIP, confirming the glafenine-induced effect. Surprisingly, the expressions of downstream proteins ATF6 and s-XBP1 did not increase, explaining the impaired UPR. These two studies clearly illustrated the feasibility of using zebrafish to study the barrier function in IBD.
In developmental biology, fluorescence-activated cell sorting is combined with microarray profiling to identify genes and proteins involved in the intestinal development (89). The phosphatidylinositol 3 kinase pathway is implicated in this process and inhibition of it by LY294002 led to gastrointestinal defects. microRNAs (miR-217 and miR-122), tight junction protein claudin C, FAM136a, and zebrafish tetraspanin, are involved the process of gastrointestinal development. A protein complex containing claudin-7, the tetraspanin CO-029, and others in membrane microdomains has been identified (90). Investigators demonstrated that the claudin family member CLDNC and the tetraspanin LOC565274 are both preferentially expressed in the zebrafish gastrointestinal tract. The claudins participate in epithelial tight junctions and tetraspanins act on proteins in transmembrane signaling (91, 92). Dysregulation of these two proteins might lead to tumor metastasis and disruption of epithelial tight junction, contributing to the pathogenesis of IBD. Genes containing a putative transcription factor binding sequence, GGAANCGGAANY and a nucleolar gene network were identified in novel pathways, such as the PI3 signaling and inositol metabolism pathway, by activating the key mediator AKT2 (89). Multiple factors might tip the balance between intestinal homeostasis and inflammation (93). Thus, the complexity of cytokines and proteins maintains the normal functions of intestinal epithelial cells.
Different therapeutic methods can be developed according to the experimental evidence. Current pharmaceutical research is mainly focused on the microbiota or the host inflammation response. However, drugs to treat these facets of IBD will not achieve mucosal healing. Thus, continuous drug screening is needed to prevent adverse events, to substantiate treatment strategies, and to find innovative treatments (91).
The predominant advantage for TNBS-induced zebrafish models is consistency with high-throughput technology and the capacity to build a poly-phenotype library (93). Although there is no formal report of any subclinical human IBD medication tested to date, 5-ASA and prednisolone have been administrated and investigated for their pharmacodynamic responses. The effectiveness of these two widely used chemical treatments has been proved by evaluating the level of cytokines and the redistribution of leukocytes in intestinal regions of the inflamed larvae. Anti-inflammatory effects were observed for 2 days after administration, which was consistent with their curative effects in humans. This not only demonstrated the fidelity of the chemically induced zebrafish enterocolitis models to IBD but also the feasibility of using zebrafish to identify novel drugs for treating IBD (19).
Despite their advantages, zebrafish have certain limitations. The immune system of the zebrafish has some unique characteristics compared with mammals. Duplicated genes are one of the essential parts of the zebrafish genome, which indicates that homologs of mammal immune factors might not have conserved functions in zebrafish. In addition, TLR4 of the zebrafish does not bind with LPS, which is the TLR4 ligand of mammals (44). In terms of anatomy, zebrafish have no lymph glands and their gut lacks Peyer’s patch. There are also differences between the zebrafish and mammals in the sites of T, B lymphocytes maturation and in subtypes of antibodies. For instance, B lymphocytes of zebrafish are produced in the kidney, while in mammals, they are produced in the bone marrow. Besides, researches of the stem cells located at the base of the crypt of mammalian intestinal epithelium are heating up in recent years (94). And these intestinal stem cells have also been proved to be significant drivers of epithelial homeostasis and regeneration (95). While the zebrafish intestinal epithelium is lack of crypts, which contributes to obvious limitation in the research of these popular field. In addition, the adaptive immune system does not develop until 4 weeks after fertilization; however, this delay benefits the study of innate immunity. For the intestinal microorganisms, those living in mammals do not survive in the zebrafish gut, making it difficult to study the regulation and function of adaptive immunity in host–microbe interactions (37). Moreover, current technology cannot produce sufficient nutritional information to permit the raising of GF zebrafish to adulthood.
Despite some limitations, zebrafish are superior to the existing models of intestinal inflammation. The genetic and immunological similarities between zebrafish and humans mean that zebrafish are replacing mice as model organisms. With the assistance of newly developed strains, and high-throughput genetic and chemical screens, investigators are able to clarify the suppressors and the enhancers of inflammation (32). By contrast, the transgenic lines offer more unique features compared with other experimental platforms. In addition, we can take advantages of the similarities between the immune systems of zebrafish and humans (45). Zebrafish models can be used to determine whether the products of IBD susceptibility genes are pathogenic. Although chemically induced models construct an inflammatory environment, they do not match well with the criteria of IBD. Zebrafish is a low-cost experimental animal with a huge potential; however, the establishment and development of aseptic zebrafish farming require further investment.
Conception and design: SJ and LH. Acquisition of information; analysis and interpretation of information: LH, LX, CX, QY, FJ, and RZ. Writing, review, and/or revision of manuscript: SJ, LH, LX, CX, and QY. Format proofread: QY, LH, and SJ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer, KL, and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
This work was supported by the National Natural Science Foundation of China (grant number 8147082).
1. Hanauer SB. Inflammatory bowel disease: epidemiology, pathogenesis, and therapeutic opportunities. Inflamm Bowel Dis (2006) 12(Suppl 1):S3–9. doi:10.1097/01.MIB.0000195385.19268.68
2. Hutfless S, Almashat S, Berger Z, Wilson LM, Bonson E, Chen Q, et al. Pharmacologic Therapies for the Management of Crohn’s Disease: Comparative Effectiveness. Rockville, MD: Johns Hopkins University Evidence-based Practice Center (2014).
3. Elson CO, Sartor RB, Tennyson GS, Riddell RH. Experimental models of inflammatory bowel disease. Gastroenterology (1995) 109:1344–67. doi:10.1016/0016-5085(95)90599-5
4. Ghia JE, Blennerhassett P, Kumar-Ondiveeran H, Verdu EF, Collins SM. The vagus nerve: a tonic inhibitory influence associated with inflammatory bowel disease in a murine model. Gastroenterology (2006) 131:1122–30. doi:10.1053/j.gastro.2006.08.016
5. Mennigen R, Nolte K, Rijcken E, Utech M, Loeffler B, Senninger N, et al. Probiotic mixture VSL#3 protects the epithelial barrier by maintaining tight junction protein expression and preventing apoptosis in a murine model of colitis. Am J Physiol Gastrointest Liver Physiol (2009) 296:G1140–9. doi:10.1152/ajpgi.90534.2008
6. Brugman S. The zebrafish as a model to study intestinal inflammation. Dev Comp Immunol (2016) 64:82–92. doi:10.1016/j.dci.2016.02.020
7. Marjoram L, Bagnat M. Infection, inflammation and healing in zebrafish: intestinal inflammation. Curr Pathobiol Rep (2015) 3:147–53. doi:10.1007/s40139-015-0079-x
8. Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, et al. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol (2005) 286:114–35. doi:10.1016/j.ydbio.2005.07.013
9. Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev (2005) 122:157–73. doi:10.1016/j.mod.2004.10.009
10. Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature (2013) 496:498–503. doi:10.1038/nature12111
11. Oehlers SH, Flores MV, Hall CJ, Swift S, Crosier KE, Crosier PS. The inflammatory bowel disease (IBD) susceptibility genes NOD1 and NOD2 have conserved anti-bacterial roles in zebrafish. Dis Model Mech (2011) 4:832–41. doi:10.1242/dmm.006122
12. Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet (2007) 8:353–67. doi:10.1038/nrg2091
13. Haramis AP, Hurlstone A, van der Velden Y, Begthel H, van den Born M, Offerhaus GJ, et al. Adenomatous polyposis coli-deficient zebrafish are susceptible to digestive tract neoplasia. EMBO Rep (2006) 7:444–9. doi:10.1038/sj.embor.7400638
14. Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell (1991) 66:589–600. doi:10.1016/0092-8674(81)90021-0
15. Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, et al. Identification of FAP locus genes from chromosome 5q21. Science (1991) 253:661–5. doi:10.1126/science.1651562
16. Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science (1992) 256:668–70. doi:10.1126/science.1350108
17. Oehlers SH, Flores MV, Hall CJ, Okuda KS, Sison JO, Crosier KE, et al. Chemically induced intestinal damage models in zebrafish larvae. Zebrafish (2013) 10:184–93. doi:10.1089/zeb.2012.0824
18. Ryan S, Willer J, Marjoram L, Bagwell J, Mankiewicz J, Leshchiner I, et al. Rapid identification of kidney cyst mutations by whole exome sequencing in zebrafish. Development (2013) 140:4445–51. doi:10.1242/dev.101170
19. Oehlers SH, Flores MV, Okuda KS, Hall CJ, Crosier KE, Crosier PS. A chemical enterocolitis model in zebrafish larvae that is dependent on microbiota and responsive to pharmacological agents. Dev Dyn (2011) 240:288–98. doi:10.1002/dvdy.22519
20. Brugman S, Liu KY, Lindenbergh-Kortleve D, Samsom JN, Furuta GT, Renshaw SA, et al. Oxazolone-induced enterocolitis in zebrafish depends on the composition of the intestinal microbiota. Gastroenterology (2009) 137:1757–67.e1. doi:10.1053/j.gastro.2009.07.069
21. Madara JL, Podolsky DK, King NW, Sehgal PK, Moore R, Winter HS. Characterization of spontaneous colitis in cotton-top tamarins (Saguinus oedipus) and its response to sulfasalazine. Gastroenterology (1985) 88:13–9. doi:10.1016/S0016-5085(85)80126-8
22. Sundberg JP, Elson CO, Bedigian H, Birkenmeier EH. Spontaneous, heritable colitis in a new substrain of C3H/HeJ mice. Gastroenterology (1994) 107:1726–35. doi:10.1016/0016-5085(94)90813-3
23. Yamada Y, Marshall S, Specian RD, Grisham MB. A comparative analysis of two models of colitis in rats. Gastroenterology (1992) 102:1524–34. doi:10.1016/0016-5085(92)91710-L
24. Yang Y, Tomkovich S, Jobin C. Could a swimming creature inform us on intestinal diseases? Lessons from zebrafish. Inflamm Bowel Dis (2014) 20:956–66. doi:10.1097/01.MIB.0000442923.85569.68
26. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature (2012) 491:119–24. doi:10.1038/nature11582
27. Oehlers SH, Flores MV, Chen T, Hall CJ, Crosier KE, Crosier PS. Topographical distribution of antimicrobial genes in the zebrafish intestine. Dev Comp Immunol (2011) 35:385–91. doi:10.1016/j.dci.2010.11.008
28. Wang Z, Du J, Lam SH, Mathavan S, Matsudaira P, Gong Z. Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine. BMC Genomics (2010) 11:392. doi:10.1186/1471-2164-11-392
29. Allen IC. A NOD to zebrafish models of inflammatory bowel disease pathogenesis. Dis Model Mech (2011) 4:711–2. doi:10.1242/dmm.008805
30. He Q, Wang L, Wang F, Li Q. Role of gut microbiota in a zebrafish model with chemically induced enterocolitis involving toll-like receptor signaling pathways. Zebrafish (2014) 11:255–64. doi:10.1089/zeb.2013.0917
31. Clark MD, Hennig S, Herwig R, Clifton SW, Marra MA, Lehrach H, et al. An oligonucleotide fingerprint normalized and expressed sequence tag characterized zebrafish cDNA library. Genome Res (2001) 11:1594–602. doi:10.1101/gr.186901
32. Van Limbergen J, Radford-Smith G, Satsangi J. Advances in IBD genetics. Nat Rev Gastroenterol Hepatol (2014) 11:372–85. doi:10.1038/nrgastro.2014.27
33. Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology (2011) 140:1704–12. doi:10.1053/j.gastro.2011.02.046
34. Cheesman SE, Guillemin K. We know you are in there: conversing with the indigenous gut microbiota. Res Microbiol (2007) 158:2–9. doi:10.1016/j.resmic.2006.10.005
35. Sadee W, Hartmann K, Seweryn M, Pietrzak M, Handelman SK, Rempala GA. Missing heritability of common diseases and treatments outside the protein-coding exome. Hum Genet (2014) 133:1199–215. doi:10.1007/s00439-014-1476-7
36. Mokry M, Middendorp S, Wiegerinck CL, Witte M, Teunissen H, Meddens CA, et al. Many inflammatory bowel disease risk loci include regions that regulate gene expression in immune cells and the intestinal epithelium. Gastroenterology (2014) 146:1040–7. doi:10.1053/j.gastro.2013.12.003
37. Marjoram L, Alvers A, Deerhake ME, Bagwell J, Mankiewicz J, Cocchiaro JL, et al. Epigenetic control of intestinal barrier function and inflammation in zebrafish. Proc Natl Acad Sci U S A (2015) 112:2770–5. doi:10.1073/pnas.1424089112
38. Pack M. IBD. Fishing for missing heritability in IBD. Nat Rev Gastroenterol Hepatol (2015) 12:318–20. doi:10.1038/nrgastro.2015.72
39. Thakur PC, Davison JM, Stuckenholz C, Lu L, Bahary N. Dysregulated phosphatidylinositol signaling promotes endoplasmic-reticulum-stress-mediated intestinal mucosal injury and inflammation in zebrafish. Dis Model Mech (2014) 7:93–106. doi:10.1242/dmm.012864
40. van Dieren JM, Simons-Oosterhuis Y, Raatgeep HC, Lindenbergh-Kortleve DJ, Lambers ME, van der Woude CJ, et al. Anti-inflammatory actions of phosphatidylinositol. Eur J Immunol (2011) 41:1047–57. doi:10.1002/eji.201040899
41. Goyette P, Lefebvre C, Ng A, Brant SR, Cho JH, Duerr RH, et al. Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. Mucosal Immunol (2008) 1:131–8. doi:10.1038/mi.2007.15
42. Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet (2008) 40:955–62. doi:10.1038/ng.175
43. Huitema LF, Renn J, Logister I, Gray JK, Waltz SE, Flik G, et al. Macrophage-stimulating protein and calcium homeostasis in zebrafish. FASEB J (2012) 26:4092–101. doi:10.1096/fj.11-202663
44. Witte M, Huitema LF, Nieuwenhuis EE, Brugman S. Deficiency in macrophage-stimulating protein results in spontaneous intestinal inflammation and increased susceptibility toward epithelial damage in zebrafish. Zebrafish (2014) 11:542–50. doi:10.1089/zeb.2014.1023
45. Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature (2001) 411:599–603. doi:10.1038/35079107
46. Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature (2001) 411:603–6. doi:10.1038/35079114
47. Crawford KC, Vega Flores M, Oehlers SH, Hall CJ, Crosier KE, Crosier PS. Zebrafish heat shock protein a4 genes in the intestinal epithelium are up-regulated during inflammation. Genesis (2011) 49:905–11. doi:10.1002/dvg.20767
48. Fleming A, Jankowski J, Goldsmith P. In vivo analysis of gut function and disease changes in a zebrafish larvae model of inflammatory bowel disease: a feasibility study. Inflamm Bowel Dis (2010) 16:1162–72. doi:10.1002/ibd.21200
49. Oehlers SH, Flores MV, Hall CJ, O’Toole R, Swift S, Crosier KE, et al. Expression of zebrafish cxcl8 (interleukin-8) and its receptors during development and in response to immune stimulation. Dev Comp Immunol (2010) 34:352–9. doi:10.1016/j.dci.2009.11.007
50. Oehlers SH, Flores MV, Hall CJ, Crosier KE, Crosier PS. Retinoic acid suppresses intestinal mucus production and exacerbates experimental enterocolitis. Dis Model Mech (2012) 5:457–67. doi:10.1242/dmm.009365
51. Oehlers SH, Flores MV, Hall CJ, Wang L, Ko DC, Crosier KE, et al. A whole animal chemical screen approach to identify modifiers of intestinal neutrophilic inflammation. FEBS J (2017) 284:402–13. doi:10.1111/febs.13976
52. Mizoguchi A. Animal models of inflammatory bowel disease. Prog Mol Biol Transl Sci (2012) 105:263–320. doi:10.1016/B978-0-12-394596-9.00009-3
53. Hundorfean G, Neurath MF, Mudter J. Functional relevance of T helper 17 (Th17) cells and the IL-17 cytokine family in inflammatory bowel disease. Inflamm Bowel Dis (2012) 18:180–6. doi:10.1002/ibd.21677
54. Holt A, Mitra S, van der Sar AM, Alnabulsi A, Secombes CJ, Bird S. Discovery of zebrafish (Danio rerio) interleukin-23 alpha (IL-23alpha) chain, a subunit important for the formation of IL-23, a cytokine involved in the development of Th17 cells and inflammation. Mol Immunol (2011) 48:981–91. doi:10.1016/j.molimm.2010.12.012
55. Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med (2009) 361:2033–45. doi:10.1056/NEJMoa0907206
56. Grayfer L, Belosevic M. Identification and molecular characterization of the interleukin-10 receptor 1 of the zebrafish (Danio rerio) and the goldfish (Carassius auratus L.). Dev Comp Immunol (2012) 36:408–17. doi:10.1016/j.dci.2011.08.006
57. Zhang DC, Shao YQ, Huang YQ, Jiang SG. Cloning, characterization and expression analysis of interleukin-10 from the zebrafish (Danio rerion). J Biochem Mol Biol (2005) 38:571–6. doi:10.5483/BMBRep.2005.38.5.571
58. Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature (2013) 496:494–7. doi:10.1038/nature11992
59. Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, et al. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol (2006) 290:G827–38. doi:10.1152/ajpgi.00513.2005
60. Mizoguchi A. Healing of intestinal inflammation by IL-22. Inflamm Bowel Dis (2012) 18:1777–84. doi:10.1002/ibd.22929
61. Costa MM, Saraceni PR, Forn-Cuni G, Dios S, Romero A, Figueras A, et al. IL-22 is a key player in the regulation of inflammation in fish and involves innate immune cells and PI3K signaling. Dev Comp Immunol (2013) 41:746–55. doi:10.1016/j.dci.2013.08.021
62. Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol (2006) 7:243. doi:10.1186/gb-2006-7-12-243
63. van Eden W. Diet and the anti-inflammatory effect of heat shock proteins. Endocr Metab Immune Disord Drug Targets (2015) 15:31–6. doi:10.2174/1871530314666140922145333
64. Tanaka K, Mizushima T. Protective role of HSF1 and HSP70 against gastrointestinal diseases. Int J Hyperthermia (2009) 25:668–76. doi:10.3109/02656730903213366
65. Gross KJ, Pothoulakis C. Role of neuropeptides in inflammatory bowel disease. Inflamm Bowel Dis (2007) 13:918–32. doi:10.1002/ibd.20129
66. Brautigam L, Hillmer JM, Soll I, Hauptmann G. Localized expression of urocortin genes in the developing zebrafish brain. J Comp Neurol (2010) 518:2978–95. doi:10.1002/cne.22375
67. Silva AB, Aw D, Palmer DB. Evolutionary conservation of neuropeptide expression in the thymus of different species. Immunology (2006) 118:131–40. doi:10.1111/j.1365-2567.2006.02351.x
68. Amole N, Unniappan S. Fasting induces preproghrelin mRNA expression in the brain and gut of zebrafish, Danio rerio. Gen Comp Endocrinol (2009) 161:133–7. doi:10.1016/j.ygcen.2008.11.002
69. Forlano PM, Cone RD. Conserved neurochemical pathways involved in hypothalamic control of energy homeostasis. J Comp Neurol (2007) 505:235–48. doi:10.1002/cne.21447
70. Geiger BM, Gras-Miralles B, Ziogas DC, Karagiannis AK, Zhen A, Fraenkel P, et al. Intestinal upregulation of melanin-concentrating hormone in TNBS-induced enterocolitis in adult zebrafish. PLoS One (2013) 8:e83194. doi:10.1371/journal.pone.0083194
71. Kokkotou E, Moss AC, Torres D, Karagiannides I, Cheifetz A, Liu S, et al. Melanin-concentrating hormone as a mediator of intestinal inflammation. Proc Natl Acad Sci U S A (2008) 105:10613–8. doi:10.1073/pnas.0804536105
72. van der Sar AM, Stockhammer OW, van der Laan C, Spaink HP, Bitter W, Meijer AH. MyD88 innate immune function in a zebrafish embryo infection model. Infect Immun (2006) 74:2436–41. doi:10.1128/IAI.74.4.2436-2441.2006
73. van der Vaart M, van Soest JJ, Spaink HP, Meijer AH. Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis Model Mech (2013) 6:841–54. doi:10.1242/dmm.010843
74. Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol (2015) 159:122–7. doi:10.1016/j.clim.2015.05.014
75. Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A (2004) 101:4596–601. doi:10.1073/pnas.0400706101
76. Camp JG, Jazwa AL, Trent CM, Rawls JF. Intronic cis-regulatory modules mediate tissue-specific and microbial control of angptl4/fiaf transcription. PLoS Genet (2012) 8:e1002585. doi:10.1371/journal.pgen.1002585
77. Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe (2012) 12:277–88. doi:10.1016/j.chom.2012.08.003
78. Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci U S A (2011) 108(Suppl 1):4570–7. doi:10.1073/pnas.1000072107
79. Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol (2006) 297:374–86. doi:10.1016/j.ydbio.2006.05.006
80. Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol (2013) 13:321–35. doi:10.1038/nri3430
81. Butto LF, Haller D. Dysbiosis in intestinal inflammation: cause or consequence. Int J Med Microbiol (2016) 306:302–9. doi:10.1016/j.ijmm.2016.02.010
82. Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell (2006) 127:423–33. doi:10.1016/j.cell.2006.08.043
83. He Q, Wang L, Wang F, Wang C, Tang C, Li Q, et al. Microbial fingerprinting detects intestinal microbiota dysbiosis in Zebrafish models with chemically-induced enterocolitis. BMC Microbiol (2013) 13:289. doi:10.1186/1471-2180-13-289
84. Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol (2013) 13:722–37. doi:10.1038/nri3532
85. Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, et al. Paneth cells as a site of origin for intestinal inflammation. Nature (2013) 503:272–6. doi:10.1038/nature12599
86. Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell (2008) 134:743–56. doi:10.1016/j.cell.2008.07.021
87. Goldsmith JR, Cocchiaro JL, Rawls JF, Jobin C. Glafenine-induced intestinal injury in zebrafish is ameliorated by mu-opioid signaling via enhancement of Atf6-dependent cellular stress responses. Dis Model Mech (2013) 6:146–59. doi:10.1242/dmm.009852
88. Kuhn S, Koch M, Nubel T, Ladwein M, Antolovic D, Klingbeil P, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res (2007) 5:553–67. doi:10.1158/1541-7786.MCR-06-0384
89. Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol (2006) 68:403–29. doi:10.1146/annurev.physiol.68.040104.131404
90. Zoller M. Gastrointestinal tumors: metastasis and tetraspanins. Z Gastroenterol (2006) 44:573–86. doi:10.1055/s-2006-926795
91. Chandel S, Prakash A, Medhi B. Current scenario in inflammatory bowel disease: drug development prospects. Pharmacol Rep (2015) 67:224–9. doi:10.1016/j.pharep.2014.09.005
92. Atreya R, Neurath MF. IBD pathogenesis in 2014: molecular pathways controlling barrier function in IBD. Nat Rev Gastroenterol Hepatol (2015) 12:67–8. doi:10.1038/nrgastro.2014.201
93. MacRae CA, Peterson RT. Zebrafish as tools for drug discovery. Nat Rev Drug Discov (2015) 14:721–31. doi:10.1038/nrd4627
94. Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell (2013) 154:274–84. doi:10.1016/j.cell.2013.07.004
Keywords: zebrafish, inflammatory bowel disease, murine model, pathogenesis, drug screening
Citation: Hanyang L, Xuanzhe L, Xuyang C, Yujia Q, Jiarong F, Jun S and Zhihua R (2017) Application of Zebrafish Models in Inflammatory Bowel Disease. Front. Immunol. 8:501. doi: 10.3389/fimmu.2017.00501
Received: 10 February 2017; Accepted: 11 April 2017;
Published: 03 May 2017
Edited by:
Kai Fang, University of California at Los Angeles, USAReviewed by:
Anne Cooke, University of Cambridge, UKCopyright: © 2017 Hanyang, Xuanzhe, Xuyang, Yujia, Jiarong, Jun and Zhihua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shen Jun, c2hlbmp1bjc5QHNpbmEuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.