- 1Department of Molecular Medicine, Sapienza University of Rome, Rome, Italy
- 2Italian Institute of Technology, Sapienza University of Rome, Rome, Italy
- 3Istituto Pasteur-Fondazione Cenci Bolognetti, Sapienza University of Rome, Rome, Italy
Natural killer (NK) cells, the prototypic member of innate lymphoid cells, are important effectors of anticancer immune response. These cells can survey and control tumor initiation due to their capability to recognize and kill malignant cells and to regulate the adaptive immune response via cytokines and chemokines release. However, several studies have shown that tumor-infiltrating NK cells associated with advanced disease can have profound functional defects and display protumor activity. This evidence indicates that NK cell behavior undergoes crucial alterations during cancer progression. Moreover, a further level of complexity is due to the extensive heterogeneity and plasticity of these lymphocytes, implying that different NK cell subsets, endowed with specific phenotypic and functional features, may be involved and play distinct roles in the tumor context. Accordingly, many studies reported the enrichment of selective NK cell subsets within tumor tissue, whereas the underlying mechanisms are not fully elucidated. A malignant microenvironment can significantly impact NK cell activity, by recruiting specific subpopulations and/or influencing their developmental programming or the acquisition of a mature phenotype; in particular, neoplastic, stroma and immune cells, or tumor-derived factors take part in these processes. In this review, we will summarize and discuss the recently acquired knowledge on the possible contribution of distinct NK cell subsets in the control and/or progression of solid and hematological malignancies. Moreover, we will address emerging evidence regarding the role of different components of tumor microenvironment on shaping NK cell response.
Introduction
Natural killer (NK) cells are innate lymphoid cells (ILCs) (1) with a crucial role in immunosurveillance. They display cytotoxic activities against transformed or viral infected cells but are also an important source of chemokines and cytokines highly impacting on adaptive immune responses (2, 3).
Natural killer cell activity is dependent on activating and inhibitory signals transmitted by a large repertoire of surface receptors. Inhibitory receptors prevent NK cells from killing healthy cells and include KIRs, CD94/NKG2A, and ILT2/CD85. The activating receptors recognize self-proteins mainly expressed on stressed target cells and include NCRs (NKp46, NKp30, NKp44), NKG2D, and DNAM1, among others (4).
Natural killer cells develop in the bone marrow (BM) from lineage restricted progenitors, although maturation can also occur in the periphery (5–7). Fully mature NK cells circulate in the peripheral blood (PB), where they represent 5–20% of total lymphocytes, but they are also found in several lymphoid and non-lymphoid organs (8, 9).
Phenotypically, NK cells are defined by the expression of CD56 and the lack of CD3–TCR complex. Moreover, based on CD16 and CD56 expression levels, they are classically distinguished in two subsets: CD56highCD16± and CD56lowCD16high. The CD56lowCD16high NK cell subset expresses high levels of KIRs, the maturation marker CD57, and mediates natural and antibody-dependent cellular cytotoxicity, exhibiting high levels of perforin and enhanced killing; CD56highCD16± NK cells are characterized by NKG2A, low levels of perforin, and are primarily specialized for cytokine production. It is still debated whether these subsets are functionally distinct NK cells or different stages of maturation. A linear differentiation relationship between CD56high CD16± NK cells and CD56lowCD16high NK cells has been proposed (10, 11), but it is not supported by observations on human NK cell deficiencies (12, 13); moreover, the possibility that tissue-resident NK cells develop locally is also considered (14).
Besides CD56highCD16± and CD56lowCD16high, additional NK cell subpopulations have been identified under normal and pathological conditions, based on their receptor repertoire (15–17). Thus, human NK cells emerge as a highly heterogeneous and plastic population including subtypes with different and specific functions.
Natural killer cell subsets also differ in tissue distribution that is related to distinct homing properties and/or in situ maturation. Tissue-resident NK cells express a different pattern of chemokine and adhesion receptors and also differ from their blood-circulating counterpart (18, 19). PB CD56highCD16± NK cells express CD62L, CCR7, CXCR4, and CXCR3 that allow their preferential recruitment to secondary lymphoid organs, tumor, and inflamed tissues (8, 20, 21). Conversely, resident CD56high NK cells lack CD62L but express other adhesion molecules, including the α integrin subunit CD49a and CD103 (22). The CD56lowCD16high NK cell subset expresses low level of CD62L and lacks CCR7, but it is characterized by CXCR4, CX3CR1, CXCR2, and CXCR3 chemokine receptors responsible for their migration into the inflamed sites.
Natural killer cells play a major role in tumor immunosurveillance. They can control tumor initiation but are often inefficacious in advanced disease. More recently, strong NK cell infiltration in established cancers also suggested a role in disease progression (23, 24). Tumor-infiltrating NK cells (TINKs) share phenotypic and functional properties with decidual NK cells (dNKs), well known for their regulatory, pro-angiogenic, and low cytotoxic activities (23, 25, 26).
In tumor microenvironment, several cellular and soluble factors affect NK cell phenotype and function and promote tumor cell evasion from NK cell-mediated recognition and killing (27).
Because the capability of distinct NK cell subsets to exert specific functions, it is extremely important to understand which subpopulations mediate the antitumor response and which environmental factors modulate their activity. Here, we review the role of distinct NK cell subsets in human solid and hematological cancers and the impact of tumor microenvironment on their phenotypic and functional features.
NK Cell Subsets in Solid Tumors
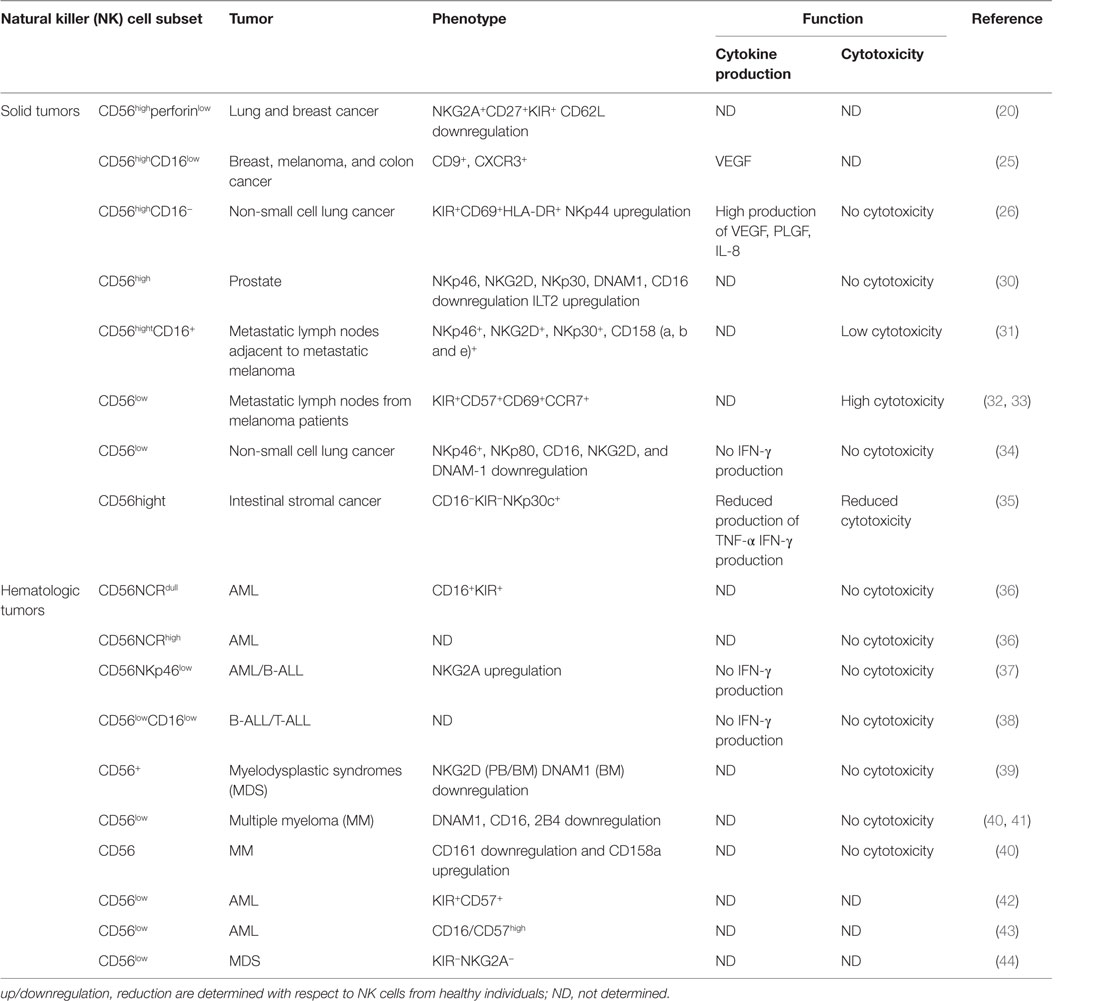
Neoplastic transformation was shown to significantly alter NK cell subset localization (Table 1), though the exact role of the TINKs subsets remains poorly characterized (28, 29).
The study of TINKs in solid tumors is rather complex as phenotypic alterations can occur following isolation and a comparison with the healthy tissue counterpart is difficult to perform.
Like tissue-resident NK cells that are generally CD56highCD16low and more specialized for cytokine production, a prevalence of CD56high NK cells can infiltrate solid malignancies, although they can exhibit features and/or functions other than those of their circulating and/or healthy counterpart tissue. Thus, a significantly higher frequency of CD56highperforinlow NK cells was observed in breast and lung cancers, with respect to normal tissues. CD56high NK cells were poorly cytotoxic, but cytokine producers, and were mainly localized within the stromal compartment. CD56highperforinlow accumulation was not attributed to major tumor microenvironment-driven NK cell developmental alterations, but rather to a peculiar chemokine milieu. Indeed, downregulation of CXCL2 that specifically attracts CD56low NK cells and upregulation of CXCL9 and CXCL10 that specifically support CD56high NK cells homing were observed (20, 45).
In breast cancer patients, five different circulating NK subsets were also identified (46): CD56lowCD16+, CD56lowCD16−, CD56highCD16−, CD56highCD16+, and CD56−CD16+. A higher percentage of CD56lowCD16− and CD56highCD16− subsets were observed both in PB and in advanced invasive mammary tumors. Furthermore, by phenotypic and functional analysis, both subpopulations emerged as more immature (CD117highCD27highCD57low) and less functional (low levels of activating receptors, perforin, and granzyme B and degranulation capability). Collectively, these observations suggest that breast tumor microenvironment blocks or reverses NK cell maturation, favoring the emergence of non-cytotoxic NK cells.
Changes in the expression patterns of activating and inhibitory receptors have been also described in tumor-associated CD56high NK cells and have been implicated in their functional deficits. CD56high NK cells, displaying an immature and activated phenotype associated with low or null degranulation potential, were found in prostate tumor and area selected out of the tumor site (30). However, in prostate cancer, lower expression of some activating receptors (NKp46, NKp30, NKG2D, DNAM-1, CD16) and higher expression of the inhibitory receptor ILT2 were observed, with more pronounced effects in NK cells infiltrating metastatic than localized tumors; these latter data indicate that tumor microenvironment can impair NK cytotoxic functions by altering the balance between NK activating and inhibitory receptors. The analysis of NK cell subsets in the lymph nodes of cancer patients revealed comparable numbers of CD56high NK cells in the regional metastatic lymph nodes from stage III melanoma patients (M-LN) and mediastinal lymph nodes from healthy donors (HD). However, 40–60% of CD56high NK cells in M-LN also expressed CD16. CD56highCD16+ NK cells displayed an activated phenotype, and their ex vivo degranulating capacity inversely correlated with the percentage of malignant cells, suggesting a local tumor-induced suppression of NK cell activation. The prevalence of CD56highCD16+ NK cells in M-LNs was attributed to the maturation and activation of tumor resident CD56highCD16− NK cells and/or to the migration of PB CD56+CD62L+ NK cells to M-LNs, where CD16 expression could be upregulated (31).
A relevant property of CD56highCD16− NK cells within different solid tumors, such as breast, melanoma, colon cancer (25), non-small lung cancer (26) is their pro-angiogenic phenotype possibly responsible for their tumor-promoting role. Indeed, unlike circulating CD56highCD16− but similar to dNK cells, CD56highCD16− TINK cells express high levels of CD9, CXCR3, produce VEGF, and have a lower cytotoxic potential, suggesting that similar maturation/polarization mechanisms occur in the decidua and tumor microenvironment of PB NK cells (47–49).
Although substantial evidence indicates CD56highCD16low NK cells as the major TINK, there are also reports on tumor infiltration by CD56low NK cells (32, 33, 46). Enrichment in the tumor infiltrated lymph nodes (TILN) and concomitant reduction of CD56low NK cells in PB were observed in melanoma patients. These CD56low (CD57+CD69+CCR7+KIR+) NK cells were highly cytotoxic against autologous melanoma cells, and, in accordance with their homing into TILN, they expressed CCR7. The reduced proportion of CD56low NK cells in the PB supports the possibility of a selective recruitment of this subset in TILN. However, in situ maturation of CD56high NK into more cytotoxic CD56low NK cells was also suggested because the different chemokine milieu dominated by CXCL8 and CCL2, which may recruit both CD56high and CD56dim CXCR2+/CCR2+ PB NK cells into the TILN (32).
The emerging concept of tissue-specific functions of NK cells together with the selective enrichment of specific subsets in neoplastic tissues indicate that the outcome of antitumor NK cell effector functions is not always predictable and largely depends on the particular tumor microenvironment.
NK Cell Subsets in Hematological Malignancies
A large body of evidence indicates that NK cells play a preferential role in the control of the onset and progression of hematological tumors. Moreover, unlike solid cancers where monitoring of PB NK cells could not provide correct information on their tumor-infiltrating counterpart, evaluation of circulating NK cell status can be highly relevant in the context of hematological malignancies.
Abnormal NK cell cytolytic function was observed in acute and chronic leukemia (AML-ALL and CLL-CML), myelodysplastic syndromes (MDS), and multiple myeloma (MM). Yet, most of the studies are focused on PB, but not BM, and poorly address NK cell phenotypic and functional heterogeneity.
The main receptors involved in NK cell recognition and killing of leukemic blasts are NCRs, NKG2D, and DNAM1. According to NCR surface density, unlike NK cells from HD that are mainly NCRhigh (50), a NCRlowCD16+KIR+ NK cell subset that failed to recognize and kill autologous and allogeneic blasts was described in AML patients (36). A smaller cohort of AML patients was also characterized by the presence of the NCRhigh NK cell subset that showed impaired cytotoxic activity, probably due to NCR ligand down-modulation on leukemic cells. In addition, significant reduction of NKp46 together with increased NKG2A expression was associated with functionally impaired PB NK cells from AML patients with respect to HD (37). Similar to AML, the frequency of PB NCR+ and in particular NKp46+ NK cells from B-ALL patients was lower. Moreover, they also displayed increased NKG2A expression. These phenotypic abnormalities were associated with impaired NK cell killer ability and IFN-γ production in response to autologous blasts (51). As regards to other activating NK receptors, a lower frequency of NKG2D+ and DNAM-1+ NK cells was observed in the context of MDS, AML, and MM (39, 52); moreover, NK cells from MM patients also displayed reduced levels of CD244, CD16, and CD161 (40, 41, 53, 54).
A different scenario was observed with CLL and CML CD56low NK cells which exhibited the same profile of activating and inhibitory receptors of HD but reduced NK cytotoxic ability (55).
Recently, we reported an increased frequency of a newly identified NK cell subset characterized by low levels of CD56 and CD16 (CD56lowCD16low) and NKG2A+ in both BM and PB of pediatric B-ALL and T-ALL. In HD, this subset was endowed with both higher cytotoxic activity and IFN-γ producing ability, but it resulted functionally impaired in leukemic patients (38). Similarly, a higher frequency of non-cytotoxic CD56lowCD16low NK cells was found in advanced breast cancer (46), suggesting both a preferential homing and functional alterations of this subset in tumor-microenvironment.
Overall, these findings suggest that several mechanisms, including downregulation of activating receptors and/or upregulation of inhibitory receptors on NK cells or modulation of their ligands on cancer cells are responsible for tumor escape from NK cell recognition in hematological malignancies.
In the context of hematological cancers where tumor cells are present in the BM that represents the main site of NK cell differentiation, an important question to address is whether tumor growth also affects NK cell development. Most of the studies, however, addressed this issue examining PB and not BM NK cells. In this regard, Chretien et al. (42) compared the presence of five different stages of NK cell development (CD56high, CD56low/KIR−/CD57−, CD56low/KIR+/CD57−, CD56low/KIR−/CD57+, and CD56low/KIR+/CD57+) in the PB of AML patients and found that one-third of the patients exhibited a significant increase in the proportion of the more mature CD56low/KIR+/CD57+ NK cells at the expenses of more immature CD56high NK cell subset. In addition, a recent study on NK cells from the BM of AML patients showed a reduced frequency of the more mature CD56lowCD16/57high NK cell subset that did not correlate with a good prognosis (43). Collectively, these findings, although suggestive of a possible influence of AML cells on NK cell development, are still incomplete, as BM and PB NK cell subsets from the same patient have not been examined, and the possibility that the observed phenotype is due to a preferential migration of more mature CD56low/KIR+/CD57+ NK cells from BM to PB is still open (56).
Moreover, it is increasingly understood the impact of hematological tumors on BM stromal cells, which are crucial for an optimal NK cell differentiation. In this regard, evidences on altered chemokine and cytokine production by BM stromal cells were provided (44), suggesting that effects on NK cell differentiation can be due to the lack of a proper stromal support for NK cell progenitors and/or an altered NK cell subset trafficking.
NK Cell Subsets and Tumor Microenvironment
Several studies indicate that tumor-induced impairment of NK cell functions correlates with alterations of NK cell subset distribution. On the other hand, different immunosuppressive mechanisms can be also responsible for functional NK cell impairment in solid and hematologic malignancies.
Tumor-related soluble factors may be responsible for phenotypic and functional alterations of NK cells, moreover different tumor-resident immune cells, such as M2-polarized macrophages, MDSC, DC, and Treg, may affect NK cell activity, by releasing soluble factors (e.g., IL-10, IDO, PGE2) or by direct contact-dependent mechanisms (57–59) (Figure 1). Although higher amount of TGF-β1, PGE2, IL-10, and IDO were detected in supernatants of solid and hematological tumors, the impact of these soluble factors on NK cell subset distribution was suggested only based on in vitro observations (30, 51). Differently, Mamessier et al. performed in vivo correlation studies demonstrating that in breast cancer patients decreased expression of activating NK cell receptors (NKp30, NKp46, NKG2D, DNAM-1) or cytotoxic molecules (GZMB) and increased levels of the inhibitory receptor NKG2A on NK cells were associated with high amount of TGF-β1 and PGE2 in tumor supernatants. In particular, TGF-β1 and PGE2 were shown to negatively correlate with molecules related to NK cell cytoxicity and positively correlate with NKG2A receptor expression (60), thus suggesting that these molecules play a role in these regulatory mechanisms.
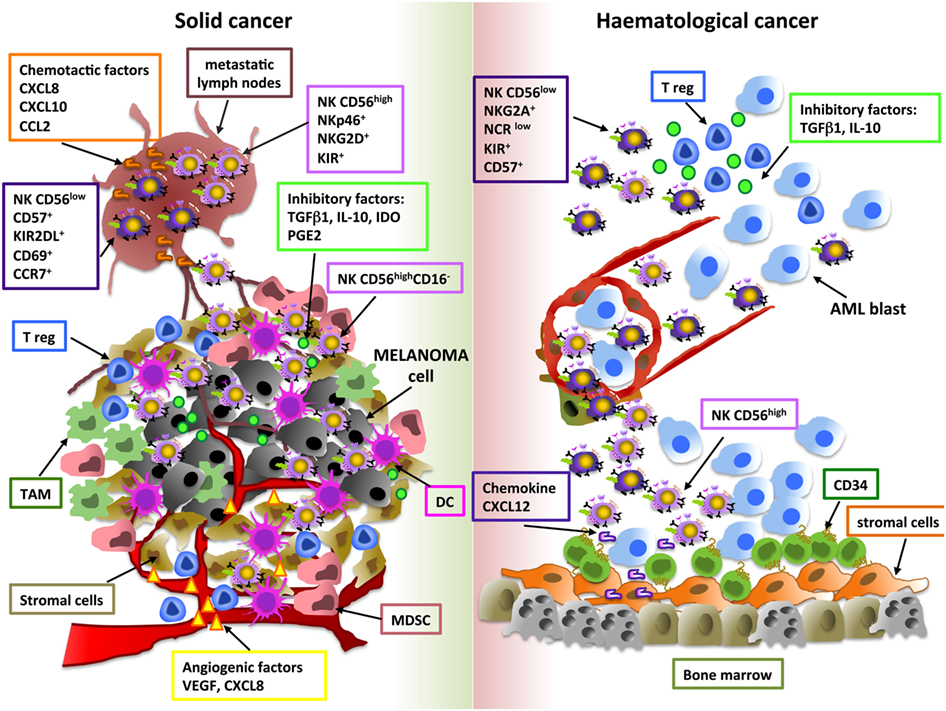
Figure 1. Shaping natural killer (NK) cell subsets in tumor microenvironment. The main cellular and soluble factors affecting NK cell subset distribution in tumor microenvironment are shown. Melanoma and AML are reported as examples of solid and hematological tumor, respectively. For melanoma, tumor and metastatic lymph node (M-LN) infiltrating NK cell subsets are represented. For AML, bone marrow and peripheral blood NK cell subpopulations are indicated.
Among these factors, particular attention has been given to TGF-β1, which has been shown to exert several effects on NK cells, including inhibition of proliferation and in vitro NK cell development and differentiation. In this regard, this cytokine was found to reduce the number of NK cells developing from human CD34+ progenitor cells and to promote the conversion of PB CD56highCD16+ NK cells into a dNK-like CD56highCD16− phenotype (61, 62). Thus, also at tumor site, TGF-β1 may take into account of the pro-angiogenic dNK-like phenotype of tumor-infiltrating NK cells. A number of studies suggest that tumor-derived TGF-β1 also impacts NK development in the context of hematological malignancies. In particular, TGF-β1 overexpression in the BM tumor-microenvironment (MDS, CML, and MM) may be responsible for the suppressive effect of cancer cells on BM stromal cells, thus compromising their supportive role on NK cell maturation (44, 63, 64). Finally, this cytokine may also interfere with intra-tumoral NK cell infiltration via modulation of their chemokine receptors (65). In this regard, a peculiar chemokine milieu has been proposed to be important for the recruitment of specific NK cell subpopulations in a number of solid tumors; moreover, altered chemokine expression patterns may also affect NK cell trafficking in hematological malignancies (32, 44, 45). However, higher concentration of chemokines does not always correlate with the presence of these lymphocytes in tumor microenvironment, thus suggesting that other and more complex mechanisms can affect their recruitment (66).
Tumor Escape from NK Cell-Mediated Recognition and Killing
Elusion of NK cell recognition is a major mechanism of tumor immune evasion. NK cell-activating ligands are expressed on malignant cells, but they can be also released in a soluble form through metalloproteinase-mediated cleavage, exosome secretion, or alternative splicing. Indeed, soluble forms of these ligands are present in the serum or peritoneal fluids of various cancer patients, and their levels positively correlate with tumor stage, metastasis, and poor prognosis (67–69). Reduction of activating ligand expression on cancer cells leads to a less efficient recognition and killing by cytotoxic lymphocytes. Concomitantly, soluble ligands can engage their receptors and cause their internalization in NK cells; accordingly, a negative correlation between soluble ligands and NKG2D expression on NK cells was largely documented in both solid and hematological tumors (70, 71). However, conflicting results have described either inhibition or promotion of NK cell activation following NKG2D endocytosis (72). An additional escape strategy used by cancer cells is based on the dominance of NK cell inhibitory signals. In several cancer cells, expression of MHC class I molecules binding to inhibitory KIR receptors (KIR2DL2/3, KIR3DL1, and KIR2DL1) results in switch off NK cell effector functions (37, 73–75). Moreover, high levels of non-classical antigens HLA-G (ligand of ILT-2 and KIRDL-4) and HLA-E (ligand of NKG2A/CD94) were found in tumor and serum of cancer patients and were considered independent markers of poor prognosis in various malignancies (76–79). Finally, tumor cell overexpression of other ligands triggering inhibitory signals on NK cells, such as PDL-1/2, contributes to inhibit their susceptibility to NK cell-mediated killing (80, 81).
Conclusion
Accumulating evidence indicates that, far from the simple and first distinction in two subsets, NK cells are a very highly heterogeneous population, and different marker combinations can be used to identify distinct subpopulations endowed with specific functional properties. Based on these observations, the role of different NK cell subsets in pathological contexts, including cancer, is increasingly elucidated. Moreover, the emerging evidence about different ILC populations further raise the necessity of a more detailed molecular phenotypic and functional characterization of innate lymphoid subsets in the cancer context. The identification of the role played by the different NK cells both in solid and hematological malignancies would be valuable for the design of novel NK cell targeted therapeutic interventions.
Author Contributions
HS, CF, AG, and AS contributed equally to writing and critically revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by grants from the Italian Association for Cancer Research (AIRC: project #16014 and AIRC 5xmille: project #9962), Istituto Pasteur-Fondazione Cenci Bolognetti and Ministero dell’Istruzione, dell’Università e della Ricerca (Ricerche Universitarie), and the Italian Institute of Technology (A2 project).
References
1. Artis D, Spits H. The biology of innate lymphoid cells. Nature (2015) 517:293–301. doi:10.1038/nature14189
2. Caligiuri MA. Human natural killer cells. Blood (2008) 112:461–9. doi:10.1182/blood-2007-09-077438
3. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science (2011) 331:44–9. doi:10.1126/science.1198687
4. Lanier LL. NK cell recognition. Annu Rev Immunol (2005) 23:225–74. doi:10.1146/annurev.immunol.23.021704.115526
5. Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev (2006) 214:56–72. doi:10.1111/j.1600-065X.2006.00451.x
6. Freud AG, Yu J, Caligiuri MA. Human natural killer cell development in secondary lymphoid tissues. Semin Immunol (2014) 26:132–7. doi:10.1016/j.smim.2014.02.008
7. Di Santo JP, Vosshenrich CA. Bone marrow versus thymic pathways of natural killer cell development. Immunol Rev (2006) 214:35–46. doi:10.1111/j.1600-065X.2006.00461.x
8. Fehniger TA, Cooper MA, Nuovo GJ, Cella M, Facchetti F, Colonna M, et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood (2003) 101:3052–7. doi:10.1182/blood-2002-09-2876
9. Santoni A, Carlino C, Gismondi A. Uterine NK cell development, migration and function. Reprod Biomed Online (2008) 16:202–10. doi:10.1016/S1472-6483(10)60575-5
10. Romagnani C, Juelke K, Falco M, Morandi B, D’Agostino A, Costa R, et al. CD56bright. J Immunol (2007) 178:4947–55. doi:10.4049/jimmunol.178.8.4947
11. Michel T, Poli A, Cuapio A, Briquemont B, Iserentant G, Ollert M, et al. Human CD56bright NK cells: an update. J Immunol (2016) 196:2923–31. doi:10.4049/jimmunol.1502570
12. Gineau L, Cognet C, Kara N, Lach FP, Dunne J, Veturi U, et al. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest (2012) 122:821–32. doi:10.1172/JCI61014
13. Mace EM, Hsu AP, Monaco-Shawver L, Makedonas G, Rosen JB, Dropulic L, et al. Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56(bright) subset. Blood (2013) 121:2669–77. doi:10.1182/blood-2012-09-453969
14. Sojka DK, Tian Z, Yokoyama WM. Tissue-resident natural killer cells and their potential diversity. Semin Immunol (2014) 26:127–31. doi:10.1016/j.smim.2014.01.010
15. Lugthart G, Melsen JE, Vervat C, van Ostaijen-Ten Dam MM, Corver WE, Roelen DL, et al. Human lymphoid tissues harbor a distinct CD69+CXCR6+ NK cell population. J Immunol (2016) 197:78–84. doi:10.4049/jimmunol.1502603
16. Stegmann KA, Robertson F, Hansi N, Gill U, Pallant C, Christophides T, et al. CXCR6 marks a novel subset of T-bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci Rep (2016) 6:26157. doi:10.1038/srep26157
17. Harmon C, Robinson MW, Fahey R, Whelan S, Houlihan DD, Geoghegan J, et al. Tissue-resident Eomes(hi) T-bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol (2016) 46:2111–20. doi:10.1002/eji.201646559
18. Melsen JE, Lugthart G, Lankester AC, Schilham MW. Human circulating and tissue-resident CD56(bright) natural killer cell populations. Front Immunol (2016) 7:262. doi:10.3389/fimmu.2016.00262
19. Bernardini G, Sciume G, Santoni A. Differential chemotactic receptor requirements for NK cell subset trafficking into bone marrow. Front Immunol (2013) 4:12. doi:10.3389/fimmu.2013.00012
20. Carrega P, Bonaccorsi I, Di CE, Morandi B, Paul P, Rizzello V, et al. CD56(bright)perforin(low) noncytotoxic human NK cells are abundant in both healthy and neoplastic solid tissues and recirculate to secondary lymphoid organs via afferent lymph. J Immunol (2014) 192:3805–15. doi:10.4049/jimmunol.1301889
21. Hudspeth K, Donadon M, Cimino M, Pontarini E, Tentorio P, Preti M, et al. Human liver-resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimmun (2016) 66:40–50. doi:10.1016/j.jaut.2015.08.011
22. Cichocki F, Sitnicka E, Bryceson YT. NK cell development and function – plasticity and redundancy unleashed. Semin Immunol (2014) 26:114–26. doi:10.1016/j.smim.2014.02.003
23. Bruno A, Ferlazzo G, Albini A, Noonan DM. A think tank of TINK/TANKs: tumor-infiltrating/tumor-associated natural killer cells in tumor progression and angiogenesis. J Natl Cancer Inst (2014) 106:dju200. doi:10.1093/jnci/dju200
24. Cantoni C, Huergo-Zapico L, Parodi M, Pedrazzi M, Mingari MC, Moretta A, et al. NK cells, tumor cell transition, and tumor progression in solid malignancies: new hints for NK-based immunotherapy? J Immunol Res (2016) 2016:4684268. doi:10.1155/2016/4684268
25. Levi I, Amsalem H, Nissan A, Darash-Yahana M, Peretz T, Mandelboim O, et al. Characterization of tumor infiltrating natural killer cell subset. Oncotarget (2015) 6:13835–43. doi:10.18632/oncotarget.3453
26. Bruno A, Focaccetti C, Pagani A, Imperatori AS, Spagnoletti M, Rotolo N, et al. The proangiogenic phenotype of natural killer cells in patients with non-small cell lung cancer. Neoplasia (2013) 15:133–42. doi:10.1593/neo.121758
27. Hasmim M, Messai Y, Ziani L, Thiery J, Bouhris JH, Noman MZ, et al. Critical role of tumor microenvironment in shaping NK cell functions: implication of hypoxic stress. Front Immunol (2015) 6:482. doi:10.3389/fimmu.2015.00482
28. Carrega P, Ferlazzo G. Natural killer cell distribution and trafficking in human tissues. Front Immunol (2012) 3:347. doi:10.3389/fimmu.2012.00347
29. Tallerico R, Garofalo C, Carbone E. A new biological feature of natural killer cells: the recognition of solid tumor-derived cancer stem cells. Front Immunol (2016) 7:179. doi:10.3389/fimmu.2016.00179
30. Pasero C, Gravis G, Granjeaud S, Guerin M, Thomassin-Piana J, Rocchi P, et al. Highly effective NK cells are associated with good prognosis in patients with metastatic prostate cancer. Oncotarget (2015) 6:14360–73. doi:10.18632/oncotarget.3965
31. Messaoudene M, Fregni G, Fourmentraux-Neves E, Chanal J, Maubec E, Mazouz-Dorval S, et al. Mature cytotoxic CD56(bright)/CD16(+) natural killer cells can infiltrate lymph nodes adjacent to metastatic melanoma. Cancer Res (2014) 74:81–92. doi:10.1158/0008-5472.CAN-13-1303
32. Ali TH, Pisanti S, Ciaglia E, Mortarini R, Anichini A, Garofalo C, et al. Enrichment of CD56(dim)KIR + CD57 + highly cytotoxic NK cells in tumour-infiltrated lymph nodes of melanoma patients. Nat Commun (2014) 5:5639. doi:10.1038/ncomms6639
33. Holtan SG, Creedon DJ, Thompson MA, Nevala WK, Markovic SN. Expansion of CD16-negative natural killer cells in the peripheral blood of patients with metastatic melanoma. Clin Dev Immunol (2011) 2011:316314. doi:10.1155/2011/316314
34. Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, et al. Profound coordinate alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res (2011) 15:5412–22. doi:10.1158/0008-5472.CAN-10-4179
35. Delahaye NF, Rusakiewicz S, Martins I, Ménard C, Roux S, Lyonnet L, et al. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med (2011) 17:700–7. doi:10.1038/nm.2366
36. Costello RT, Sivori S, Marcenaro E, Lafage-Pochitaloff M, Mozziconacci MJ, Reviron D, et al. Defective expression and function of natural killer cell-triggering receptors in patients with acute myeloid leukemia. Blood (2002) 99:3661–7. doi:10.1182/blood.V99.10.3661
37. Stringaris K, Sekine T, Khoder A, Alsuliman A, Razzaghi B, Sargeant R, et al. Leukemia-induced phenotypic and functional defects in natural killer cells predict failure to achieve remission in acute myeloid leukemia. Haematologica (2014) 99:836–47. doi:10.3324/haematol.2013.087536
38. Stabile H, Nisti P, Morrone S, Pagliara D, Bertaina A, Locatelli F, et al. Multifunctional human CD56 low CD16 low natural killer cells are the prominent subset in bone marrow of both healthy pediatric donors and leukemic patients. Haematologica (2015) 100:489–98. doi:10.3324/haematol.2014.116053
39. Epling-Burnette PK, Bai F, Painter JS, Rollison DE, Salih HR, Krusch M, et al. Reduced natural killer (NK) function associated with high-risk myelodysplastic syndrome (MDS) and reduced expression of activating NK receptors. Blood (2007) 109:4816–24. doi:10.1182/blood-2006-07-035519
40. Konjevic G, Vuletic A, Mirjacic MK, Colovic N, Colovic M, Jurisic V. Decreased CD161 activating and increased CD158a inhibitory receptor expression on NK cells underlies impaired NK cell cytotoxicity in patients with multiple myeloma. J Clin Pathol (2016) 69:1009–16. doi:10.1136/jclinpath-2016-203614
41. Costello RT, Boehrer A, Sanchez C, Mercier D, Baier C, Le TT, et al. Differential expression of natural killer cell activating receptors in blood versus bone marrow in patients with monoclonal gammopathy. Immunology (2013) 139:338–41. doi:10.1111/imm.12082
42. Chretien AS, Granjeaud S, Gondois-Rey F, Harbi S, Orlanducci F, Blaise D, et al. Increased NK cell maturation in patients with acute myeloid leukemia. Front Immunol (2015) 6:564. doi:10.3389/fimmu.2015.00564
43. Aggarwal N, Swerdlow SH, TenEyck SP, Boyiadzis M, Felgar RE. Natural killer cell (NK) subsets and NK-like T-cell populations in acute myeloid leukemias and myelodysplastic syndromes. Cytometry B Clin Cytom (2016) 90:349–57. doi:10.1002/cyto.b.21349
44. Geyh S, Oz S, Cadeddu RP, Frobel J, Bruckner B, Kundgen A, et al. Insufficient stromal support in MDS results from molecular and functional deficits of mesenchymal stromal cells. Leukemia (2013) 27:1841–51. doi:10.1038/leu.2013.193
45. Carrega P, Morandi B, Costa R, Frumento G, Forte G, Altavilla G, et al. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(-) cells and display an impaired capability to kill tumor cells. Cancer (2008) 112:863–75. doi:10.1002/cncr.23239
46. Mamessier E, Pradel LC, Thibult ML, Drevet C, Zouine A, Jacquemier J, et al. Peripheral blood NK cells from breast cancer patients are tumor-induced composite subsets. J Immunol (2013) 190:2424–36. doi:10.4049/jimmunol.1200140
47. Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med (2006) 12:1065–74. doi:10.1038/nm1452
48. Carlino C, Stabile H, Morrone S, Bulla R, Soriani A, Agostinis C, et al. Recruitment of circulating NK cells through decidual tissues: a possible mechanism controlling NK cell accumulation in the uterus during early pregnancy. Blood (2008) 111:3108–15. doi:10.1182/blood-2007-08-105965
49. Carlino C, Trotta E, Stabile H, Morrone S, Bulla R, Soriani A, et al. Chemerin regulates NK cell accumulation and endothelial cell morphogenesis in the decidua during early pregnancy. J Clin Endocrinol Metab (2012) 97:3603–12. doi:10.1210/jc.2012-1102
50. Sivori S, Pende D, Bottino C, Marcenaro E, Pessino A, Biassoni R, et al. NKp46 is the major triggering receptor involved in the natural cytotoxicity of fresh or cultured human NK cells. Correlation between surface density of NKp46 and natural cytotoxicity against autologous, allogeneic or xenogeneic target cells. Eur J Immunol (1999) 29:1656–66. doi:10.1002/(SICI)1521-4141(199905)29:05<1656::AID-IMMU1656>3.0.CO;2-1
51. Rouce RH, Shaim H, Sekine T, Weber G, Ballard B, Ku S, et al. The TGF-beta/SMAD pathway is an important mechanism for NK cell immune evasion in childhood B-acute lymphoblastic leukemia. Leukemia (2016) 30:800–11. doi:10.1038/leu.2015.327
52. Sanchez-Correa B, Gayoso I, Bergua JM, Casado JG, Morgado S, Solana R, et al. Decreased expression of DNAM-1 on NK cells from acute myeloid leukemia patients. Immunol Cell Biol (2012) 90:109–15. doi:10.1038/icb.2011.15
53. El-Sherbiny YM, Meade JL, Holmes TD, McGonagle D, Mackie SL, Morgan AW, et al. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res (2007) 67:8444–9. doi:10.1158/0008-5472.CAN-06-4230
54. Fauriat C, Mallet F, Olive D, Costello RT. Impaired activating receptor expression pattern in natural killer cells from patients with multiple myeloma. Leukemia (2006) 20:732–3. doi:10.1038/sj.leu.2404096
55. Chen CI, Koschmieder S, Kerstiens L, Schemionek M, Altvater B, Pscherer S, et al. NK cells are dysfunctional in human chronic myelogenous leukemia before and on imatinib treatment and in BCR-ABL-positive mice. Leukemia (2012) 26:465–74. doi:10.1038/leu.2011.239
56. Hejazi M, Manser AR, Frobel J, Kundgen A, Zhao X, Schonberg K, et al. Impaired cytotoxicity associated with defective natural killer cell differentiation in myelodysplastic syndromes. Haematologica (2015) 100:643–52. doi:10.3324/haematol.2014.118679
57. Szczepanski MJ, Szajnik M, Welsh A, Whiteside TL, Boyiadzis M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica (2011) 96:1302–9. doi:10.3324/haematol.2010.039743
58. Smyth MJ, Teng MW, Swann J, Kyparissoudis K, Godfrey DI, Hayakawa Y. CD4+CD25+ T regulatory cells suppress NK cell-mediated immunotherapy of cancer. J Immunol (2006) 176:1582–7. doi:10.4049/jimmunol.176.3.1582
59. Shenghui Z, Yixiang H, Jianbo W, Kang Y, Laixi B, Yan Z, et al. Elevated frequencies of CD4(+) CD25(+) CD127lo regulatory T cells is associated to poor prognosis in patients with acute myeloid leukemia. Int J Cancer (2011) 129:1373–81. doi:10.1002/ijc.25791
60. Mamessier E, Sylvain A, Thibult ML, Houvenaeghel G, Jacquemier J, Castellano R, et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest (2011) 121:3609–22. doi:10.1172/JCI45816
61. Allan DS, Rybalov B, Awong G, Zuniga-Pflucker JC, Kopcow HD, Carlyle JR, et al. TGF-beta affects development and differentiation of human natural killer cell subsets. Eur J Immunol (2010) 40:2289–95. doi:10.1002/eji.200939910
62. Keskin DB, Allan DS, Rybalov B, Andzelm MM, Stern JN, Kopcow HD, et al. TGFbeta promotes conversion of CD16+ peripheral blood NK cells into. Proc Natl Acad Sci U S A (2007) 104:3378–83. doi:10.1073/pnas.0611098104
63. Diaz-Blanco E, Bruns I, Neumann F, Fischer JC, Graef T, Rosskopf M, et al. Molecular signature of CD34(+) hematopoietic stem and progenitor cells of patients with CML in chronic phase. Leukemia (2007) 21:494–504. doi:10.1038/sj.leu.2404549
64. Bruns I, Cadeddu RP, Brueckmann I, Frobel J, Geyh S, Bust S, et al. Multiple myeloma-related deregulation of bone marrow-derived CD34(+) hematopoietic stem and progenitor cells. Blood (2012) 120:2620–30. doi:10.1182/blood-2011-04-347484
65. Castriconi R, Dondero A, Bellora F, Moretta L, Castellano A, Locatelli F, et al. Neuroblastoma-derived TGF-beta1 modulates the chemokine receptor repertoire of human resting NK cells. J Immunol (2013) 190:5321–8. doi:10.4049/jimmunol.1202693
66. Halama N, Michel S, Kloor M, Zoernig I, Benner A, Spille A, et al. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res (2011) 71:5670–7. doi:10.1158/0008-5472.CAN-11-0268
67. Baragano RA, Suarez-Alvarez B, Lopez-Larrea C. Secretory pathways generating immunosuppressive NKG2D ligands: new targets for therapeutic intervention. Oncoimmunology (2014) 3:e28497. doi:10.4161/onci.28497
68. Zingoni A, Cecere F, Vulpis E, Fionda C, Molfetta R, Soriani A, et al. Genotoxic stress induces senescence-associated ADAM10-dependent release of NKG2D MIC ligands in multiple myeloma cells. J Immunol (2015) 195:736–48. doi:10.4049/jimmunol.1402643
69. Fionda C, Soriani A, Zingoni A, Santoni A, Cippitelli M. NKG2D and DNAM-1 ligands: molecular targets for NK cell-mediated immunotherapeutic intervention in multiple myeloma. Biomed Res Int (2015) 2015:178698. doi:10.1155/2015/178698
70. Hilpert J, Grosse-Hovest L, Grunebach F, Buechele C, Nuebling T, Raum T, et al. Comprehensive analysis of NKG2D ligand expression and release in leukemia: implications for NKG2D-mediated NK cell responses. J Immunol (2012) 189:1360–71. doi:10.4049/jimmunol.1200796
71. Paschen A, Baingo J, Schadendorf D. Expression of stress ligands of the immunoreceptor NKG2D in melanoma: regulation and clinical significance. Eur J Cell Biol (2014) 93:49–54. doi:10.1016/j.ejcb.2014.01.009
72. Molfetta R, Quatrini L, Zitti B, Capuano C, Galandrini R, Santoni A, et al. Regulation of NKG2D expression and signaling by endocytosis. Trends Immunol (2016) 37:790–802. doi:10.1016/j.it.2016.08.015
73. Varbanova V, Naumova E, Mihaylova A. Killer-cell immunoglobulin-like receptor genes and ligands and their role in hematologic malignancies. Cancer Immunol Immunother (2016) 65:427–40. doi:10.1007/s00262-016-1806-9
74. Sanchez-Correa B, Morgado S, Gayoso I, Bergua JM, Casado JG, Arcos MJ, et al. Human NK cells in acute myeloid leukaemia patients: analysis of NK cell-activating receptors and their ligands. Cancer Immunol Immunother (2011) 60:1195–205. doi:10.1007/s00262-011-1050-2
75. Verheyden S, Demanet C. Susceptibility to myeloid and lymphoid leukemia is mediated by distinct inhibitory KIR-HLA ligand interactions. Leukemia (2006) 20:1437–8. doi:10.1038/sj.leu.2404279
76. Schutt P, Schutt B, Switala M, Bauer S, Stamatis G, Opalka B, et al. Prognostic relevance of soluble human leukocyte antigen-G and total human leukocyte antigen class I molecules in lung cancer patients. Hum Immunol (2010) 71:489–95. doi:10.1016/j.humimm.2010.02.015
77. Yie SM, Hu Z. Human leukocyte antigen-G (HLA-G) as a marker for diagnosis, prognosis and tumor immune escape in human malignancies. Histol Histopathol (2011) 26:409–20. doi:10.14670/HH-26.409
78. Maki G, Hayes GM, Naji A, Tyler T, Carosella ED, Rouas-Freiss N, et al. NK resistance of tumor cells from multiple myeloma and chronic lymphocytic leukemia patients: implication of HLA-G. Leukemia (2008) 22:998–1006. doi:10.1038/leu.2008.15
79. McWilliams EM, Mele JM, Cheney C, Timmerman EA, Fiazuddin F, Strattan EJ, et al. Therapeutic CD94/NKG2A blockade improves natural killer cell dysfunction in chronic lymphocytic leukemia. Oncoimmunology (2016) 5:e1226720. doi:10.1080/2162402X.2016.1226720
80. Kearl TJ, Jing W, Gershan JA, Johnson BD. Programmed death receptor-1/programmed death receptor ligand-1 blockade after transient lymphodepletion to treat myeloma. J Immunol (2013) 190:5620–8. doi:10.4049/jimmunol.1202005
Keywords: natural killer cell subset, tumor microenvironment, natural killer cells, hematological malignancies, solid tumors
Citation: Stabile H, Fionda C, Gismondi A and Santoni A (2017) Role of Distinct Natural Killer Cell Subsets in Anticancer Response. Front. Immunol. 8:293. doi: 10.3389/fimmu.2017.00293
Received: 12 December 2016; Accepted: 28 February 2017;
Published: 16 March 2017
Edited by:
Sandra Laurence Lopez-Verges, Instituto Conmemorativo Gorgas de Estudios de la Salud, PanamaReviewed by:
Christine Susanne Falk, Hannover Medical School, GermanyKerry S. Campbell, Fox Chase Cancer Center, USA
Copyright: © 2017 Stabile, Fionda, Gismondi and Santoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela Gismondi, YW5nZWxhLmdpc21vbmRpQHVuaXJvbWExLml0;
Angela Santoni, YW5nZWxhLnNhbnRvbmlAdW5pcm9tYTEuaXQ=
†These authors have contributed equally to this work.
 Helena Stabile
Helena Stabile Cinzia Fionda
Cinzia Fionda Angela Gismondi
Angela Gismondi Angela Santoni
Angela Santoni