- 1The Walter and Eliza Hall Institute of Medical Research, Parkville, VIC, Australia
- 2Department of Medical Biology, University of Melbourne, Melbourne, VIC, Australia
Natural killer (NK) cells are effector lymphocytes of the innate immune system that are known for their ability to kill transformed and virus-infected cells. NK cells originate from hematopoietic stem cells in the bone marrow, and studies on mouse models have revealed that NK cell development is a complex, yet tightly regulated process, which is dependent on both intrinsic and extrinsic factors. The development of NK cells can be broadly categorized into two phases: lineage commitment and maturation. Efforts to better define the developmental framework of NK cells have led to the identification of several murine NK progenitor populations and mature NK cell subsets, each defined by a varied set of cell surface markers. Nevertheless, the relationship between some of these NK cell subsets remains to be determined. The classical approach to studying both NK cell development and function is to identify the transcription factors involved and elucidate the mechanistic action of each transcription factor. In this regard, recent studies have provided further insight into the mechanisms by which transcription factors, such as ID2, FOXO1, Kruppel-like factor 2, and GATA-binding protein 3 regulate various aspects of NK cell biology. It is also becoming evident that the biology of NK cells is not only transcriptionally regulated but also determined by epigenetic alterations and posttranscriptional regulation of gene expression by microRNAs. This review summarizes recent progress made in NK development, focusing primarily on transcriptional regulators and their mechanistic actions.
Introduction to Natural Killer (NK) Cells
Natural killer cells in mice were first described in 1975 (1–3), following further investigation into splenocytes that were able to kill tumor and virus-infected cells without prior sensitization (4–6). NK cells exert their cytotoxic effect on target cells by inducing apoptosis. Upon formation of an immunological synapse with the target cell, NK cells become activated and release cytolytic granules containing perforin and granzymes (7–9). Perforin forms pores in the membrane of target cells, thereby allowing granzymes to enter the cell, activate caspases, and initiate apoptosis (8). In a similar process known as antibody-dependent cell cytotoxicity, NK cells are able to release cytolytic granules and initiate apoptosis in opsonized cells, following recognition of the opsonized cells via low-affinity Fc receptors (CD16) expressed on the surface of NK cells (10). NK cells can also initiate apoptosis in target cells through the respective engagement of Fas ligands and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) on their cell surface with Fas and TRAIL receptors on the target cells (11, 12). In addition to inducing apoptosis, NK cells can indirectly mediate the clearance of target cells by producing pro-inflammatory cytokines [e.g., interferon-gamma (IFN-γ)], which boost the innate response and recruit adaptive immune responses (13–15).
The surface markers that are commonly used to identify murine NK cells by flow cytometry vary depending on the mouse strain. C57B/6 and SJL mice express the surface markers NK1.1, NKp46, and CD49b, but not CD3, which is a surface marker of T cells. CD3 is used to exclude contaminating T cell subsets, such as natural killer T cells and NK-like T cells, that, respectively, express NK1.1 and NKp46 (16). As for other mouse strains, such as BALB/c, NK cells are identified with only CD49b and NKp46 as these strains possess allelic variants of NK1.1 that cannot be detected with the widely used PK136 antibody (16, 17).
Murine NK Cell Development
Murine NK cells can be found in all lymphoid organs and many non-lymphoid tissues, such as salivary glands, liver, and kidney. The more recent discovery of related innate lymphoid cells (ILCs) places NK cells within this family, specifically in the IL-15 dependent, IFN-γ producing group 1 ILCs. ILCs are lymphoid cells that lack rearranged antigen receptors and are dependent on the transcription factors inhibitor of DNA-binding 2 (ID2) and nuclear factor, interleukin 3 regulated (NFIL3) for their development. While NK cells are phenotypically heterogeneous and previously categorized based on their tissue of origin or location (bone marrow, thymus, fetal liver, adult liver), we appreciate that some of this heterogeneity stems from NK cells (Eomes+) and other ILC1s (Eomes−) being viewed as the same cell type. As much of our current understanding of murine NK cell development is built upon studies on bone marrow-derived NK cells [referred to here as conventional NK (cNK) cells], which represent the majority of NK cells within the body, this review will focus primarily on progress made in our understanding of cNK development.
cNK Development in the Bone Marrow—Lineage Commitment
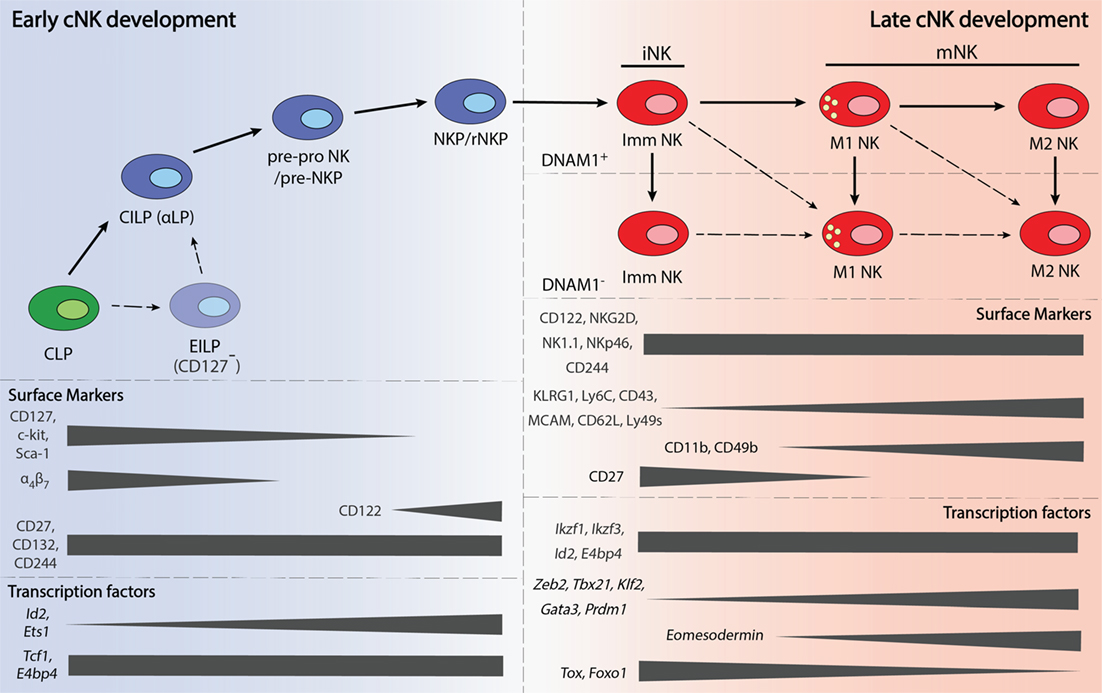
Conventional NK cells develop from HSCs in the bone marrow, through a sequential order of intermediate progenitors. The first progenitor to arise from HSCs is the lymphoid-primed multipotent progenitor, which then gives rise to the common lymphoid progenitor (CLP) (18). The earliest NK lineage committed progenitor to arise from CLPs is known as pre-pro NK (19), which was subdivided into pre-pro A and pre-pro B (19, 20). Differing only in c-kit (CD117) expression, the relationship between pre-pro A and B remains unclear and requires further investigation. Pre-pro NK cells then differentiate into the NK progenitor (NKP) (19, 21). NKPs give rise to immature NK (iNK) cells that either undergo further development within the bone marrow (22) or enter the periphery and develop into mature NK cells (23, 24).
As the early stages of murine NK development are still poorly defined, the developmental pathway outlined above is by no means the definitive model. Heterogeneity within existing progenitor populations, along with the discovery of new distinguishing cell markers, have led to the identification of new sub-populations and, therefore, refinements to the developmental pathway of NK cells. For instance, the common innate lymphoid progenitor (CILP) was found to possess the capacity to give rise to all lineages within the ILC family, of which NK cells are the founding member, but not B and T cells, thus making it an earlier progenitor than the pre-pro NK (25). As the CILP is expresses α4β7, it is also alternatively referred to as the α-lymphoid precursor (αLP) (25). However, it has been postulated that there could be an even earlier progenitor population, as there were only about 50 CILPs per mice, and only 2.5% of CILPs efficiently developed into all ILC lineages (25).
The early innate lymphoid progenitor (EILP) is proposed to be the earliest known progenitor for ILCs and was identified using a TCF1 (T-cell factor 1, encoded by Tcf7) transgenic mouse strain that expresses a green fluorescent protein reporter (26). Like CILPs, EILPs give rise to all ILC lineages both in vivo and in vitro, albeit more efficiently (26). Nonetheless, EILPs have not been shown to differentiate into CILPs and hence, the relationship between these two progenitors remains unresolved. As most of the surface markers used to identify the EILP and CILP were different, a detailed comparison of the surface marker phenotype between the two progenitors may also provide further insight.
The discovery of pre–pro NKPs was the outcome of efforts to better understand why only 8–40% of NKPs had solely NK cell potential (19). Similar studies resulted in the identification of a pre-NKP population that preceded a “streamlined” population of NKPs known as refined-NKP (rNKP) (27). Even though many parallels have been drawn between the pre-NKP and pre-pro NK, likewise with NKP and rNKP, it remains to be determined if these populations are exactly identical (20). A summary of the surface markers that are expressed on the various progenitors are provided in Table 1.
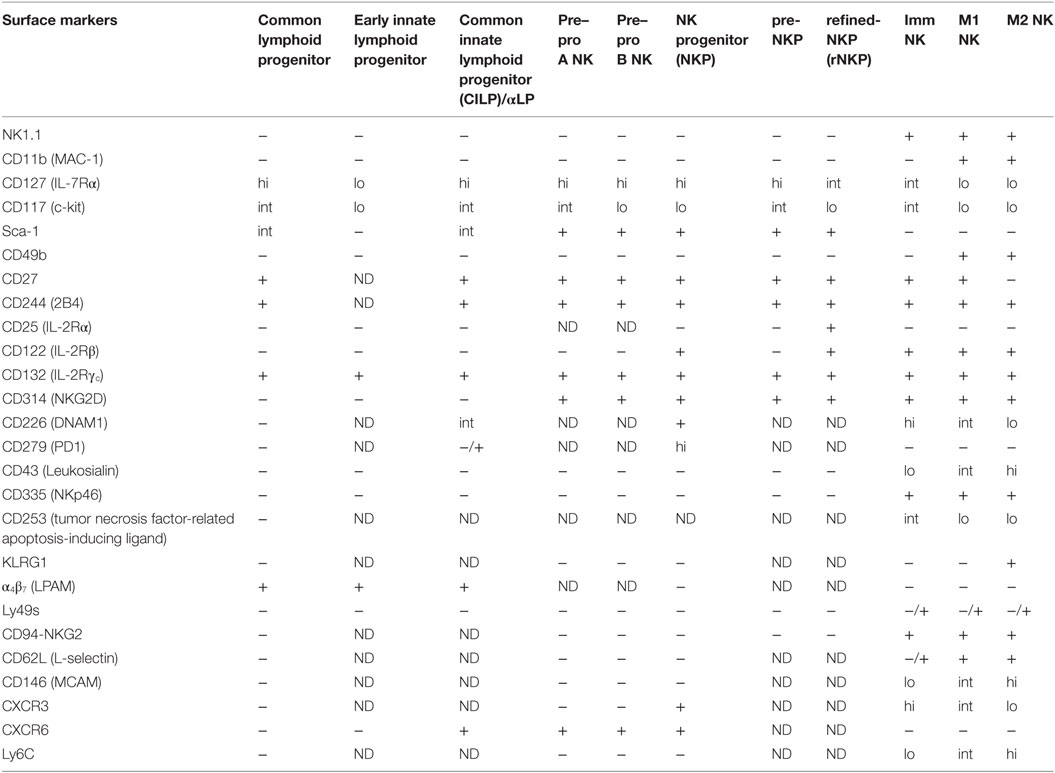
Table 1. Surface markers expressed by different natural killer (NK) cell populations reported in the literature.
cNK Development in the Periphery—NK Cell Maturation
Natural killer cell maturation is a process by which lineage committed NK cells acquire their full effector functions. This process is also accompanied by the expression of different cell surface markers, which have helped in the identification of different NK cell maturation subsets. At present, most studies use CD11b and CD27 to divide cNK cells into three maturation subsets: immature (Imm), mature 1 (M1), and mature 2 (M2). Low (lo) and high (hi) CD11b expression divides cNK cells into immature and mature subsets, respectively (22). Heterogeneity in CD27 expression further delineates the mature NK compartment into CD27hi and CD27lo subsets, which has also been referred to as M1 and M2 NK subsets (24, 28). CD27 and CD11b expressing M1 NK cells have also been termed double-positive NK cells (24). The three subsets differ in proliferative and cytotoxic capacity. In general, NK cells lose proliferative potential and produce less cytokine, but become more cytotoxic against target cells as they mature (22, 24, 28).
Apart from CD27 and CD11b, markers such as KLRG1 (23, 29), CD62L (30), MCAM (31), CD49b (21), CD43 (32, 33), Ly6C (34), DNAM1 (35), and CD160 (36) have further dissected maturing NK cells into various phenotypic subsets (37, 38). NK cells that express CD160 exhibit enhanced IFN-γ production (36), while the opposite is true for mature NK cells that express higher levels of Ly6C (34), and KLRG1 (39). Like Ly6C, MCAM is also more highly expressed on mature NK cells, although MCAM+ and MCAM− NK cells differ in their ability to kill target cells rather than cytokine production (31). Interestingly, the expression of DNAM1 appears to be independent of NK cell maturation that is defined by CD27 and CD11b, as DNAM-1+ and DNAM-1− NK cells were observed in both the immature and mature NK compartments (35). As the correlation between these markers have not been studied in detail, the relationships between these phenotypic subsets remain unclear and warrants further investigation. A summary of the surface markers that are expressed on the various mature NK cell subsets are provided in Table 1.
Transcriptional Regulation of Murine cNK Cell Development
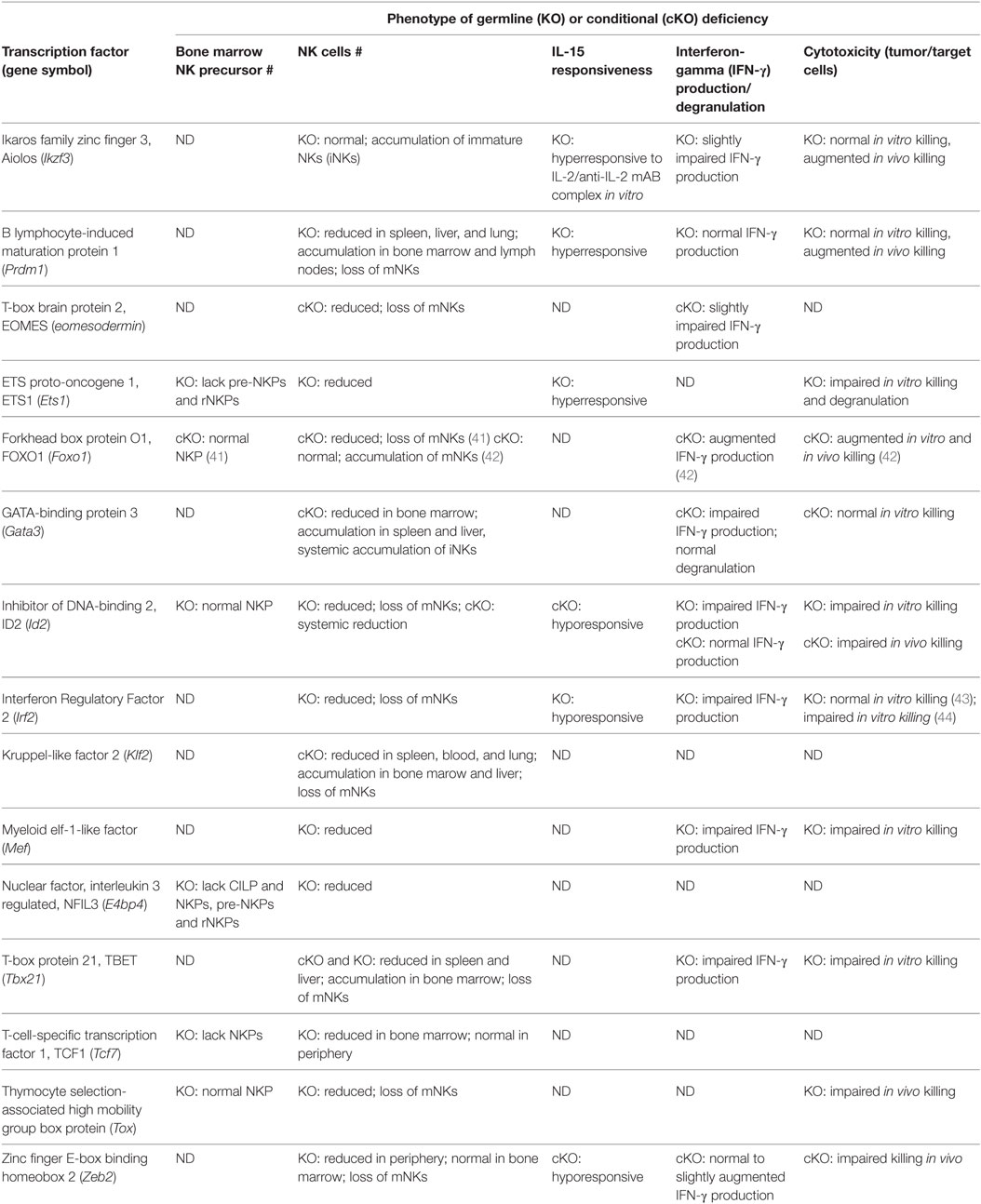
Transcription factors control gene expression by either activating or repressing gene transcription. This is achieved by first binding to specific DNA sequences in the enhancer or promoter regions that lay upstream of target genes, then promoting or blocking the recruitment of RNA polymerases that transcribe those genes (40). In terms of murine NK cell development, several TFs have been shown to play crucial roles in regulating NK cell lineage specification, NK cell maturation, or even both. Conventional NK cell development occurs mostly in the bone marrow, under the coordinated control of the TFs and cytokines. TFs like ID2, NFIL3, T-box brain protein 2 (EOMES), and T-box protein 21 (TBET) to name a few, fall into the category of intrinsic factors that regulate NK cell development. A summary of the transcription factors that are implicated in NK cell development is provided in Table 2.
Transcription Factors Regulating NK Cell Lineage Specification
The TFs that are involved in regulating NK cell lineage specification include ETS proto-oncogene 1 (ETS1), NFIL3, and TCF1. ETS1 is a key regulator of early NK cell development as ETS1-deficient mice have normal CLP numbers but lack NK cells (45). Further investigation into the impact of ETS1 deficiency on NK lineage specification revealed a reduction in pre-pro NK, pre-NKP and rNKP, and mature NK cell numbers, thereby supporting the role of ETS1 in NK cell lineage specification (46). ETS1 is believed to impact early NK cell development by regulating the expression of ID2 and TBET, which are also important TFs for NK cell development (46).
TCF1 and NFIL3 are also key regulators of NK cell lineage specification, as marked reductions in pre-pro NK, pre-NKP, and rNKP numbers were observed within the bone marrow of NFIL3- and TCF1-deficient mice, although only the former mouse strain exhibited an additional reduction in CILP numbers (25, 26, 47, 48). NFIL3 appears to be dispensable for mature NK cell as their numbers remained unchanged after its deletion in mature NKp46+ NK cells (49). Despite reduced progenitor and mature NK cell numbers in the bone marrow of TCF1-deficient mice, mature NK cells have been found in the periphery at frequencies comparable to littermate controls (26, 50). As most NK cells are derived from bone marrow precursors at steady state, an investigation into the source of NK cells in TCF1-deficient mice might perhaps shed some light on alternative pathways of NK development. The mechanism by which TCF1 mediates NK lineage specification remains poorly understood. However, TCF1 has been shown to regulate T-lineage specification by promoting the expression of genes, such as Gata3, Bcl11b, Il2ra, and Cd3e, that are critical for T cell development (51). Similarly, the mechanism by which NFIL3 specifically mediates lineage specification in NK cells remains unclear, although NFIL3 was found to promote the expression of ID2 and EOMES (47), transcription factors known to be involved in the later stages of NK cell development (52, 53). The role of NFIL3 in ID2 expression remains to be clarified given a contradicting report that ID2 expression is normal in NK cells lacking NFIL3 (48).
The role of ID2 in the lineage specification of NK cells is unclear, due to contradicting reports following deletion of the encoding gene Id2. While Yokota et al. (54). reported poor reconstitution of NK cells following bone marrow transplantation, implying a defect early on during NK cell development, Boos et al. (55) did not observe any reduction in NKP and iNK cell numbers. A recent demonstration of low ID2 levels in CLPs but consistently high levels in pre-pro NK, NKP, and immature and mature NK cells lends further support for the hypothesis that ID2 could indeed be important for NK lineage specification/maintenance (53). Interestingly, ID2 has been found to suppress T and B cell development through heterodimer formation with the E-box protein E2A (55–57), although how the interaction specifically promotes commitment to the NK cell lineage remains unknown.
Transcription Factors Regulating NK Cell Maturation
A greater number of TFs have been shown to play a role in NK cell maturation. These factors include ID2, TBET, EOMES, Zinc finger E-box-binding homeobox 2 (ZEB2), Thymocyte selection-associated high mobility group box (TOX), IKAROS family zinc finger 3 (AIOLOS), Interferon regulatory factor 2 (IRF2), B lymphocyte-induced maturation protein 1 (BLIMP1), Forkhead box O1 (FOXO1), Kruppel-like factor 2 (KLF2), and GATA-binding protein 3 (GATA3). An overview of the expression these TFs during NK cell maturation is presented in Figure 1.
The role of ID2 in NK cell maturation is better understood than its role in early NK cell development. A recent study provided additional insight into the underlying mechanism of ID2 by demonstrating that it modulates the expression of E2A target genes (i.e., Socs3, Tcf7, and Cxcr5) by titrating E-protein activity, thereby controlling the responsiveness of NK cells to IL-15 that is crucial for survival (53, 58, 59).
TBET and EOMES are members of the T-box family of transcription factors that appear to regulate distinct checkpoints in NK cell maturation. TBET- and EOMES-deficient mice exhibited a similar phenotype where NK cell numbers were reduced in the all lymphoid tissues (52, 60, 61), except in the bone marrow of the former where there was an increase in NK cell numbers (60, 61). Detailed analyses of the bone marrow from TBET-deficient mice revealed that the increase in NK cell numbers was due to an accumulation of iNK cells, which the authors attributed to a developmental block (60). However, the possibility of a defect in NK cell trafficking remains unaddressed, given that TBET plays a role in the expression of sphigosine-1-phosphate receptor 5 (S1P5) that is required for NK cell egression from the bone marrow (62). Nonetheless, both TFs are crucial for maturation, as mice that are deficient for both have a systemic lack of NK cells despite normal progenitor numbers (52, 61). As TBET and EOMES are, respectively, required by immature and mature NK cells, the two TFs are believed to function in a sequential manner for NK cell maturation (52, 60). Quite fittingly, both TBET and EOMES were shown to be essential for IL-15 responsiveness by enforcing high CD122 expression, with Il2rb (gene encoding CD122) being a direct target of EOMES (63).
Similar to that of TBET-deficient mice, NK cell numbers in the bone marrow of mice deficient for ZEB2 or BLIMP1 were higher than littermate controls (64, 65). The unusual accumulation of NK cells within the bone marrow of ZEB2-deficient mice was due to reduced S1P5 expression (64), while enhanced proliferation was found to be the underlying cause in BLIMP1-deficient mice (65). Profound losses in terminally mature NK cells were observed on closer examination of the bone marrow NK cells from both strains (64, 65). The lack of mature NK cells in ZEB2-deficient was further attributed to poor responsiveness to IL-15, which resulted in poor survival (64).
The lack of mature NK cells was reported in mice that were deficient for TOX (66), AIOLOS (67), IRF2 (43, 44), KLF2 (68), or GATA3 (69, 70). A similar lack of mature NK cells in FOXO1-deficient mice was reported most recently (41), although this remains to be clarified against an earlier study, which instead found an accumulation of mature NK cells (42). Interestingly, the iNK cells in AIOLOS-deficient mice retained their expression of KLRG1, which is typically expressed on terminally mature NK cells (67). NK cells from IRF2-deficient mice have been shown to undergo accelerated apoptosis, indicating a role for IRF2 in regulating NK cell survival as well as maturation (43). Unlike the foregoing TFs, the temporal requirement for myeloid elf-1-like factor (MEF) by maturing NK cells has not been determined, as only an overall reduction in NK cell numbers was reported (71).
FOXO1 was recently found to be directly involved in the initiation of autophagy in iNK cells, most likely via cytosolic interactions between FOXO1 and the autophagy protein ATG7 (41). KLF2 appears to regulate NK cell maturation via a different mechanism, influencing the expression of homing receptors such as CD62L (i.e., L-selectin) on maturing NK cells, thereby dictating their access to IL-15 signaling that is essential for survival (68). Current knowledge of how GATA3 regulates NK cell maturation is limited to disturbances in expression of the TFs ID2, TBET, and NFIL3 (70). Nevertheless, GATA3 has also been shown to regulate NK cell egression from the bone marrow in a CXCR4-dependent manner, and also NK cell proliferation in response to IL-15 via CD25 expression (70).
Transcription Factors Regulating NK Cell Effector Function
Natural killer cell effector function is also regulated by many of the TFs outlined above. FOXO1 has also been proposed to negatively regulate NK cell effector function, as its absence was correlated with augmented IFN-γ production in response to murine cytomegalovirus (MCMV) infections and anti-metastatic activity against the B16F10 mouse melanoma cell line (42). Augmented anti-metastatic activity against the same melanoma cell line also was reported in mice deficient for either BLIMP1 or AIOLOS, despite the lack of any significant impact on cytokine production (65, 67). On the other hand, MEF is required for normal cytokine production and cytotoxicity, as it positively regulates IFN-γ and perforin expression, which corresponds to poorer cytotoxicity against tumor cell targets in MEF-deficient mice (71). Normal cytotoxicity but reduced IFN-γ production has been observed in IRF2-deficient mice and mice specifically lacking GATA3 in NK cells (43, 70). NK cell function is also regulated by TBET and EOMES, as TBET has been shown to bind to the regulatory regions of genes encoding granzyme B and perforin, while the expression of EOMES as NK cells mature is associated with increased transcription of mRNA (52, 60).
Posttranscriptional Regulation of NK Cell Development by microRNAs (miRs)
microRNAs are short non-coding RNAs (19–26 nt) that modulate gene expression at a posttranscriptional level. Recent studies have shown that miRs are also important for NK cell development and function. Using a Dicer1-deficient mouse model that abrogates miR biogenesis in NK cells, Degouve et al. (72). showed that a 10-fold global reduction in miR expression resulted in reduced NK cell numbers, aberrant NK cell maturation, along with attenuated IFN-γ production and cytotoxicity against target cells. Given that IL-15 signaling via the STAT5 and mTOR pathways was significantly affected, it was proposed that miRs regulate NK cell survival by modulating IL-15 sensitivity (72). Although it remains unclear as to whether NK cell survival is dependent on specific miRs, miR-155 and miR-15/16 are unlikely candidates since mice that are deficient for either miR have normal NK cell numbers (73, 74).
Rather than NK cell survival, mIR-155 and miR15/16 appear to be essential for normal NK cell maturation, as NK cells lacking miR-15/16 are unable to fully mature into M2 NK cells (74), much like Dicer1-deficient mice, while miR-155-deficient NK cells undergo accelerated maturation (73). In contrast to the accumulation of M2 NK cells in miR-155 deficient mice, an accumulation of Imm NK cells was observed in mice that over-expressed miR-155, thereby providing additional evidence for the role of miR-155 in NK cell maturation (75). More importantly, the opposing effect that miR-15/16 and miR-155 have on NK cell maturation highlights the pleiotropic effects of miRs and suggests that there is still much to learn about the role of miRs in NK cell biology, particularly about redundancies between miRs. Nevertheless, it has been shown that miR-15/16 controls NK maturation by directly regulating levels of the transcription factor MYB, since the overexpression of miR-15/16 or MYB deficiency in miR-15/16-deficient NK cells rescues the maturation defect (74).
The mechanistic action of miR-155 can be appreciated in the context of NK cell proliferation and homeostasis, as NK cells deficient for miR-155 were unable to proliferate in response to MCMV infections and were also outcompeted by wild-type NK cells when cotransferred into homeostatic or lymphopenic environments (73). This dependency on miR-155 for proliferation under both homeostatic and infectious conditions appears to be mediated through the direct suppression of its target genes suppressor of cytokine signaling 1 (Socs1) and pro-apoptotic molecule phorbol-12-myristate-13-acetate-induced protein 1 (Pmaip1; encoding NOXA) (73). Interestingly, an accumulation of NK cells was observed in transgenic mice that over-expressed miR-155, lending further support for the regulatory role of miR-155 in NK cell proliferation (75).
Epigenetic Regulation of NK Cell Development
Histone modifications have previously been shown to be essential events in B and T cell development (76, 77). Recent studies have demonstrated that defects in histone modification also impacts NK cell development with respect to lineage commitment (78) and maturation (79). Mice deficient for enhancer of zeste homolog 2 (EZH2), a H3K27 methyltransferase, were observed to have higher numbers NKPs and NK cells. Microarray analysis revealed that the difference was associated with the upregulation of genes essential for NK cell development and function, thereby resulting in earlier lineage commitment and enhanced survival of NKPs (78). This increase in NK cell production was also observed when hematopoietic progenitors from human and wild-type mice were treated in vitro with EZH2 inhibitors (78).
A different type of histone modification, deubiquitination, also appears to be involved in the epigenetic regulation of NK cell maturation. The histone deubiquitinase, MYSM1 (Myb-like, SWIRM, and MPN domains-containing protein 1), was found to regulate NK cell maturation as MYSM1-deficient mice possessed fewer NK cells that were mostly immature (79). Given that no defects were observed in the NKP compartment, MYSM1 was proposed to be uniquely required during NK cell maturation. Mechanistically, MYSM1 regulates NK cell maturation by binding directly to the Id2 gene locus, as revealed by chromatin immunoprecipitation, thereby maintaining expression of the TF (79). However, the mechanism by which MYSM1 is selectively directed to the Id2 gene locus remains unclear and thus requires further investigation.
Conclusion
The discovery of new members within the ILC family has rekindled efforts to better understand the development of NK cells, the founding member of the ILC family. Many of the recent breakthroughs made in the transcriptional regulation of NK cell development have been aided by key tools and techniques such as single cell RNA-seq, in vitro differentiation conditions, transcription factor reporter mice and conditional alleles and lineage specific Cre-expressing mouse strains. As these techniques and tools become commonplace in the field of NK cell biology, our understanding of the temporal–spatial transcriptional regulation of NK cell development and the key target genes that govern NK cell fate, homeostasis, and function becomes increasingly more complete. Recent studies have advanced our understanding of how individual TFs may be regulating NK cell commitment and NK cell lineage maintenance. However, how these various TFs form a transcriptional network and act in concert to ensure NK cell homeostasis remains unclear.
On a translational front, defining the extrinsic cues and TFs that regulate NK cell maturation, proliferation, cytokine responsiveness, and priming of effector functions represents an area of therapeutic interest. Proteins that negatively regulate NK cell maturation and fitness are tangible drug targets in cancer immunotherapy as recently evidenced by our group. Building on these potential targets will increase the likelihood of developing specific inhibitors for clinical translation.
Author Contributions
WG wrote the review. NH conceived and edited the review.
Conflict of Interest Statement
NH is a cofounder of oNKo-Innate. WG declares no conflict of interest.
Funding
This work is supported by project grants from the National Health and Medical Research Council (NHMRC) of Australia (1049407, 1066770, 1057852, 1027472), as well as an NHMRC Independent Research Institute Infrastructure Support scheme grant and a Victorian State Government Operational Infrastructure Scheme grant. NH is a recipient of a Melanoma Research Grant from the Harry J Lloyd Charitable Trust and a CLIP grant from Cancer Research Institute.
References
1. Kiessling R, Klein E, Pross H, Wigzell H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol (1975) 5:117–21. doi: 10.1002/eji.1830050209
2. Herberman RB, Nunn ME, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. I. Distribution of reactivity and specificity. Int J Cancer (1975) 16:216–29. doi:10.1002/ijc.2910160204
3. Kiessling R, Klein E, Wigzell H. Natural. Eur J Immunol (1975) 5:112–7. doi:10.1002/eji.1830050208
4. Hellström I, Hellström KE, Pierce GE, Yang JP. Cellular and humoral immunity to different types of human neoplasms. Nature (1968) 220:1352–4. doi:10.1038/2201352a0
5. Herberman RB, Nunn ME, Lavrin DH, Asofsky R. Effect of antibody to theta antigen on cell-mediated immunity induced in syngeneic mice by murine sarcoma virus. J Natl Cancer Inst (1973) 51:1509–12. doi:10.1093/jnci/51.5.1509
6. Kiessling R, Bataillon G, Lamon EW, Klein E. The lymphocyte response to primary Moloney sarcoma virus tumors: definition of a non-specific component of the in vitro cellular hyporeactivity of tumor-bearing hosts. Int J Cancer (1974) 14:642–8. doi:10.1002/ijc.2910140511
7. Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity (2007) 26:798–811. doi:10.1016/j.immuni.2007.04.010
8. Thiery J, Keefe D, Boulant S, Boucrot E, Walch M, Martinvalet D, et al. Perforin pores in the endosomal membrane trigger the release of endocytosed granzyme B into the cytosol of target cells. Nat Immunol (2011) 12:770–7. doi:10.1038/ni.2050
9. Henkart MP, Henkart PA. Lymphocyte mediated cytolysis as a secretory phenomenon. In: Clark WR, Golstein P, editors. Mechanisms of Cell-Mediated Cytotoxicity Advances in Experimental Medicine and Biology. Boston, MA: Springer (1982). p. 227–47.
10. Ojo E, Wigzell H. Natural killer cells may be the only cells in normal mouse lymphoid cell populations endowed with cytolytic ability for antibody-coated tumour target cells. Scand J Immunol (1978) 7:297–306. doi:10.1111/j.1365-3083.1978.tb00457.x
11. Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, et al. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood (2005) 105:2082–9. doi:10.1182/blood-2004-08-3262
12. Arase H, Arase N, Saito T. Fas-mediated cytotoxicity by freshly isolated natural killer cells. J Exp Med (1995) 181:1235–8. doi:10.1084/jem.181.3.1235
13. Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang Z-E, Gapin L, et al. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med (2003) 198:1069–76. doi:10.1084/jem.20030630
14. Martín-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-|[gamma]| for TH1 priming. Nat Immunol (2004) 5:1260–5. doi:10.1038/ni1138
15. Moretta L, Ferlazzo G, Bottino C, Vitale M, Pende D, Mingari MC, et al. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol Rev (2006) 214:219–28. doi:10.1111/j.1600-065X.2006.00450.x
16. Walzer T, Bléry M, Chaix J, Fuseri N, Chasson L, Robbins SH, et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci U S A (2007) 104:3384–9. doi:10.1073/pnas.0609692104
17. Walzer T, Jaeger S, Chaix J, Vivier E. Natural killer cells: from CD3− NKp46+ to post-genomics meta-analyses. Curr Opin Immunol (2007) 19:365–72. doi:10.1016/j.coi.2007.04.004
18. Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell (1997) 91:661–72. doi:10.1016/S0092-8674(00)80291-3
19. Carotta S, Pang SHM, Nutt SL, Belz GT. Identification of the earliest NK-cell precursor in the mouse BM. Blood (2011) 117:5449–52. doi:10.1182/blood-2010-11-318956
20. Huntington ND, Nutt SL, Carotta S. Regulation of murine natural killer cell commitment. Front Immunol (2013) 4:14. doi:10.3389/fimmu.2013.00014
21. Rosmaraki EE, Douagi I, Roth C, Colucci F, Cumano A, Di Santo JP. Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol (2001) 31:1900–9. doi:10.1002/1521-4141(200106)31:6<1900::AID-IMMU1900>3.0.CO;2-M
22. Kim S, Iizuka K, Kang H-SP, Dokun A, French AR, Greco S, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol (2002) 3:523–8. doi:10.1038/ni796
23. Huntington ND, Tabarias H, Fairfax K, Brady J, Hayakawa Y, Degli-Esposti MA, et al. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J Immunol (2007) 178:4764–70. doi:10.4049/jimmunol.178.8.4764
24. Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood (2009) 113:5488–96. doi:10.1182/blood-2008-10-187179
25. Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, et al. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife (2014) 3:945. doi:10.7554/eLife.04406
26. Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol (2015) 16:1044–50. doi:10.1038/ni.3248
27. Fathman JW, Bhattacharya D, Inlay MA, Seita J, Karsunky H, Weissman IL. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood (2011) 118:5439–47. doi:10.1182/blood-2011-04-348912
28. Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol (2006) 176:1517–24. doi:10.4049/jimmunol.176.3.1517
29. Robbins SH, Tessmer MS, Mikayama T, Brossay L. Expansion and contraction of the NK cell compartment in response to murine cytomegalovirus infection. J Immunol (2004) 173:259–66. doi:10.4049/jimmunol.173.1.259
30. Peng H, Sun R, Tang L, Wei H, Tian Z. CD62L is critical for maturation and accumulation of murine hepatic NK cells in response to viral infection. J Immunol (2013) 190:4255–62. doi:10.4049/jimmunol.1202395
31. Despoix N, Walzer T, Jouve N, Blot-Chabaud M, Bardin N, Paul P, et al. Mouse CD146/MCAM is a marker of natural killer cell maturation. Eur J Immunol (2008) 38:2855–64. doi:10.1002/eji.200838469
32. Zafirova B, Mandarić S, Antulov R, Krmpotić A, Jonsson H, Yokoyama WM, et al. Altered NK cell development and enhanced NK cell-mediated resistance to mouse cytomegalovirus in NKG2D-deficient mice. Immunity (2009) 31:270–82. doi:10.1016/j.immuni.2009.06.017
33. Shehata HM, Hoebe K, Chougnet CA. The aged nonhematopoietic environment impairs natural killer cell maturation and function. Aging Cell (2015) 14:191–9. doi:10.1111/acel.12303
34. Omi A, Enomoto Y, Kiniwa T, Miyata N, Miyajima A. Mature resting Ly6Chigh natural killer cells can be reactivated by IL-15. Eur J Immunol (2014) 44:2638–47. doi:10.1002/eji.201444570
35. Martinet L, De Andrade LF, Guillerey C, Lee JS, Liu J, Souza-Fonseca-Guimaraes F, et al. DNAM-1 expression marks an alternative program of NK cell maturation. Cell Rep (2015) 11:85–97. doi:10.1016/j.celrep.2015.03.006
36. Tu TC, Brown NK, Kim T-J, Wroblewska J, Yang X, Guo X, et al. CD160 is essential for NK-mediated IFN-γ production. J Exp Med (2015) 212:415–29. doi:10.1084/jem.20131601
37. Seillet C, Belz GT, Huntington ND. Development, homeostasis, and heterogeneity of NK cells and ILC1. Curr Top Microbiol Immunol (2015) 395:37–61. doi:10.1007/82_2015_474
38. Huntington ND, Martinet L, Smyth MJ. DNAM-1: would the real natural killer cell please stand up! Oncotarget (2015) 6:28537–8. doi:10.18632/oncotarget.5952
39. Robbins SH, Nguyen KB, Takahashi N, Mikayama T, Biron CA, Brossay L. Cutting edge: inhibitory functions of the killer cell lectin-like receptor G1 molecule during the activation of mouse NK cells. J Immunol (2002) 168:2585–9. doi:10.4049/jimmunol.168.6.2585
40. Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev (2000) 14:2551–69. doi:10.1101/gad.831000
41. Wang S, Xia P, Huang G, Zhu P, Liu J, Ye B, et al. FoxO1-mediated autophagy is required for NK cell development and innate immunity. Nat Commun (2016) 7:11023. doi:10.1038/ncomms11023
42. Deng Y, Kerdiles Y, Chu J, Yuan S, Wang Y, Chen X, et al. Transcription factor Foxo1 is a negative regulator of natural killer cell maturation and function. Immunity (2015) 42:457–70. doi:10.1016/j.immuni.2015.02.006
43. Taki S, Nakajima S, Ichikawa E, Saito T, Hida S. IFN regulatory factor-2 deficiency revealed a novel checkpoint critical for the generation of peripheral NK cells. J Immunol (2005) 174:6005–12. doi:10.4049/jimmunol.174.10.6005
44. Lohoff M, Duncan GS, Ferrick D, Mittrücker HW, Bischof S, Prechtl S, et al. Deficiency in the transcription factor interferon regulatory factor (IRF)-2 leads to severely compromised development of natural killer and T helper type 1 cells. J Exp Med (2000) 192:325–36. doi:10.1084/jem.192.3.325
45. Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, et al. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity (1998) 9:555–63. doi:10.1016/S1074-7613(00)80638-X
46. Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, et al. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity (2012) 36:921–32. doi:10.1016/j.immuni.2012.04.006
47. Male V, Nisoli I, Kostrzewski T, Allan DS, Carlyle JR, Lord GM, et al. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med (2014) 211:635–42. doi:10.1084/jem.20132398
48. Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJ, et al. Differential requirement for Nfil3 during NK cell development. J Immunol (2014) 192:2667–76. doi:10.4049/jimmunol.1302605
49. Firth MA, Madera S, Beaulieu AM, Gasteiger G, Castillo EF, Schluns KS, et al. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J Exp Med (2013) 210:2981–90. doi:10.1084/jem.20130417
50. Held W, Kunz B, Lowin-Kropf B, van de Wetering M, Clevers H. Clonal acquisition of the Ly49A NK cell receptor is dependent on the trans-acting factor TCF-1. Immunity (1999) 11:433–42. doi:10.1016/S1074-7613(00)80118-1
51. Weber BN, Chi AW-S, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature (2011) 476:63–8. doi:10.1038/nature10279
52. Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity (2012) 36:55–67. doi:10.1016/j.immuni.2011.11.016
53. Delconte RB, Shi W, Sathe P, Ushiki T, Seillet C, Minnich M, et al. The helix-loop-helix protein ID2 governs NK cell fate by tuning their sensitivity to interleukin-15. Immunity (2016) 44:103–15. doi:10.1016/j.immuni.2015.12.007
54. Sathe P, Delconte RB, Souza-Fonseca-Guimaraes F, Seillet C, Chopin M, Vandenberg CJ, et al. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nat Commun (2014) 5:4539. doi:10.1038/ncomms5539
55. Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med (2007) 204:1119–30. doi:10.1084/jem.20061959
56. Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell (1989) 58:537–44. doi:10.1016/0092-8674(89)90434-0
57. Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Annu Rev Immunol (2003) 20:301–22. doi:10.1146/annurev.immunol.20.092501.162048
58. Huntington ND, Puthalakath H, Gunn P, Naik E, Michalak EM, Smyth MJ, et al. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nat Immunol (2007) 8:856–63. doi:10.1038/ni1487
59. Sathe P, Delconte RB, Souza-Fonseca-Guimaraes F, Seillet C, Chopin M, Vandenberg CJ, et al. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nat Commun (2014) 5:1–10. doi:10.1038/ncomms5539
60. Townsend MJ, Weinmann AS, Matsuda JL, Salomon R. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity (2004) 20(4):477–94. doi:10.1016/S1074-7613(04)00076-7
61. Pikovskaya O, Chaix J, Rothman NJ, Collins A, Chen Y-H, Scipioni AM, et al. Cutting edge: eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J Immunol (2016) 196:1449–54. doi:10.4049/jimmunol.1502396
62. Jenne CN, Enders A, Rivera R, Watson SR, Bankovich AJ, Pereira JP, et al. T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J Exp Med (2009) 206:2469–81. doi:10.1084/jem.20090525
63. Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol (2005) 6:1236–44. doi:10.1038/ni1268
64. van Helden MJ, Goossens S, Daussy C, Mathieu A-L, Faure F, Marçais A, et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med (2015) 212:2015–25. doi:10.1084/jem.20150809
65. Kallies A, Carotta S, Huntington ND, Bernard NJ, Tarlinton DM, Smyth MJ, et al. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood (2011) 117:1869–79. doi:10.1182/blood-2010-08-303123
66. Aliahmad P, la Torre de B, Kaye J. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue-inducer cell and NK cell lineages. Nat Immunol (2010) 11:945–52. doi:10.1038/ni.1930
67. Holmes ML, Huntington ND, Thong RP, Brady J, Hayakawa Y, Andoniou CE, et al. Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J (2014) 33:2721–34. doi:10.15252/embj.201487900
68. Rabacal W, Pabbisetty SK, Hoek KL, Cendron D, Guo Y, Maseda D, et al. Transcription factor KLF2 regulates homeostatic NK cell proliferation and survival. Proc Natl Acad Sci U S A (2016) 113:5370–5. doi:10.1073/pnas.1521491113
69. Samson SI, Richard O, Tavian M, Ranson T, Vosshenrich CAJ, Colucci F, et al. GATA-3 promotes maturation, IFN-γ production, and liver-specific homing of NK cells. Immunity (2003) 19:701–11. doi:10.1016/S1074-7613(03)00294-2
70. Ali AK, Oh JS, Vivier E, Busslinger M, Lee SH. NK cell-specific Gata3 ablation identifies the maturation program required for bone marrow exit and control of proliferation. J Immunol (2016) 196:1753–67. doi:10.4049/jimmunol.1501593
71. Lacorazza HD, Miyazaki Y, Di Cristofano A, Deblasio A, Hedvat C, Zhang J, et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity (2002) 17:437–49. doi:10.1016/S1074-7613(02)00422-3
72. Degouve S, Tavares A, Viel S, Walzer T, Marçais A. NKp46-mediated Dicer1 inactivation results in defective NK-cell differentiation and effector functions in mice. Eur J Immunol (2016) 46:1902–11. doi:10.1002/eji.201546163
73. Zawislak CL, Beaulieu AM, Loeb GB, Karo J, Canner D, Bezman NA, et al. Stage-specific regulation of natural killer cell homeostasis and response against viral infection by microRNA-155. Proc Natl Acad Sci U S A (2013) 110:6967–72. doi:10.1073/pnas.1304410110
74. Sullivan RP, Leong JW, Schneider SE, Ireland AR, Berrien-Elliott MM, Singh A, et al. microRNA-15/16 antagonizes Myb to control NK cell maturation. J Immunol (2015) 195:2806–17. doi:10.4049/jimmunol.1500949
75. Trotta R, Chen L, Costinean S, Josyula S, Mundy-Bosse BL, Ciarlariello D, et al. Overexpression of miR-155 causes expansion, arrest in terminal differentiation and functional activation of mouse natural killer cells. Blood (2013) 121:3126–34. doi:10.1182/blood-2012-12-467597
76. Su I-H, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol (2003) 4:124–31. doi:10.1038/ni876
77. Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4+ T helper type 1 and type 2 cells. Immunity (2013) 39:819–32. doi:10.1016/j.immuni.2013.09.012
78. Yin J, Leavenworth JW, Li Y, Luo Q, Xie H, Liu X, et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc Natl Acad Sci U S A (2015) 112:15988–93. doi:10.1073/pnas.1521740112
Keywords: NK cell, transcription factors, ontogeny, maturation, homeostasis, IL-15
Citation: Goh W and Huntington ND (2017) Regulation of Murine Natural Killer Cell Development. Front. Immunol. 8:130. doi: 10.3389/fimmu.2017.00130
Received: 18 November 2016; Accepted: 25 January 2017;
Published: 15 February 2017
Edited by:
Antoine Toubert, Paris Diderot University, FranceReviewed by:
Evelyn Ullrich, Goethe University Frankfurt, GermanyReem Al-Daccak, Institut national de la santé et de la recherche médicale, France
Jean Villard, University of Geneva, Switzerland
Copyright: © 2017 Goh and Huntington. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicholas D. Huntington, aHVudGluZ3RvbkB3ZWhpLmVkdS5hdQ==
 Wilford Goh1,2
Wilford Goh1,2 Nicholas D. Huntington
Nicholas D. Huntington