
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 18 October 2016
Sec. Microbial Immunology
Volume 7 - 2016 | https://doi.org/10.3389/fimmu.2016.00431
This article is part of the Research TopicFacultative Intracellular Bacteria: Exception or Rule?View all 5 articles
Pathogenic bacteria possess intricate regulatory networks that temporally control the production of virulence factors and enable the bacteria to survive and proliferate within host cell. Small non-coding RNAs (sRNAs) have been identified as important regulators of gene expression in diverse biological contexts. Recent research has shown bacterial sRNAs involved in growth and development, cell proliferation, differentiation, metabolism, cell signaling, and immune response through regulating protein–protein interactions or via their ability to base pair with RNA and DNA. In this review, we provide a brief overview of mechanism of action employed by immune-related sRNAs, their known functions in immunity, and how they can be integrated into regulatory circuits that govern virulence, which will facilitate our understanding of pathogenesis and the development of novel, more effective therapeutic approaches to treat infections caused by intracellular bacterial pathogens.
Precise control of gene expression is an essential feature of the immune system. The immune system depends on a sophisticated gene expression program equipped with an arsenal of strategies to fight against infections, mainly controlled by well-described transcriptional and posttranscriptional mechanisms (1). Cells of the immune system are able to undergo dramatic changes in transcription mechanisms to efficiently organize expression of genes critical to defense. Innate and adaptive immune cell differentiation and activation depends largely on these transcription events (2). Neutrophils, macrophages, and dendritic cells (DCs) exhibit both common and unique sets of toll-like receptors (TLRs), chemokines, and cytokines in innate immune responses. microRNAs (miRNA) and RNA-binding proteins determine the specific region of the gene accessible to transcription factors that ultimately regulate transcription (1).
Small non-coding RNAs (sRNAs) play critical roles in bacterial gene expression and are recognized as key regulators in bacteria. Typically, these RNA regulators range from 50 to 200 nt in length and act on independently expressed targets, often encoded in the intergenic region (3, 4). sRNA controls bacterial gene expression by employing multiple molecular strategies to regulate the expression of gene targets, including binding directly to complementary sequences present in target mRNA molecules (5, 6). sRNAs interact with their specific target to exert both positive and negative effects on gene expression. In positive regulation, sRNAs bind with target mRNA at the 5′-untranslated region (UTR) and alter the secondary structure of the mRNA, to access a ribosome-binding site (RBS) that allows translation (7, 8). In addition, sRNA can bind to the 3′-UTR of target mRNA, ultimately increasing gene expression and stabilizing the transcript (9). Alternatively, sRNAs can also exert inhibitory effects by binding with mRNA 5′-UTR, resulting in decreased stability, degradation of the mRNA, occlusion of the RBS, and inhibition of translation (10). sRNAs are widely identified and their number constantly growing due to their involvement in diverse biological contexts including cell proliferation, development, differentiation, apoptosis, metabolism, stress response and signal transduction (11, 12).
Pathogenic bacteria have to face hostile and changing environments characterized by high concentrations of reactive oxygen and nitrogen species, low pH, and limited nutrient availability that hinder in their replication and infection to succeed (13). Pathogens have evolved a variety of strategies to survive and replicate within eukaryotic cells, establishing mechanisms to manipulate the host-cell machinery for their own benefit (14). After internalization in host cells, pathogenic bacteria modulate their trafficking to avoid lysosomal fusion by occupying a specialized membrane-bound vesicle. Intracellular bacteria are divided in two classes: vacuolar intracellular bacteria, such as Salmonella, Mycobacterium, Legionella, Brucella, and Coxiella, that survive and replicate either by avoiding vacuole–lysosome fusion or by altering the phagolysosome environment; and cytosolic intracellular bacteria, including Francisella, Shigella, Listeria, Burkholderia, and Rickettsia, that usually escape to proliferate within the cytosol of host cell (15, 16).
Recent developments in biocomputation have revealed a large number of regulatory sRNAs and have highlighted their potential links to bacterial pathogenesis (17, 18). Bacterial adaptation to intracellular environment niches is efficiently regulated in both time and space. These newly identified sRNAs play an integral part in virulence expression and bacterial stress responses that are ultimately advantageous for pathogens in adaptation and modification of the host-immune response (19). Understanding the mechanisms adopted by sRNAs to control immune cell function and how immune-related cells maintain cell viability and competitiveness in varying environmental niches is critical. However, roles of sRNA regulators in pathogenesis and immune response mechanisms have only begun to be investigated. In this review, we summarize the mechanisms employed by bacterial sRNAs in gene regulation and sRNA-based strategies to counter host immune response mechanisms as well as their implications in the pathogenesis of intracellular bacteria.
Bacterial regulatory sRNAs operate at all layers of gene regulation to modulate translation, transcription, DNA maintenance or silencing, and mRNA stability. They use different mechanisms to achieve these outstanding regulatory functions (5). The major mechanisms employed by bacterial sRNAs for gene regulation are as follows:
(i) Trans-encoded sRNA are usually encoded on the genome in trans location distinct from their targets and share partial complementarity with their target mRNAs. They potentially establish base pairing to the Shine-Dalgarno (SD) sequence of target mRNAs usually 10–25 nt in length to sequester the RBS (4). Additionally, trans-acting sRNAs are firmly coupled with the RNases activity to exploit their regulatory functions resulting in RNA turnover through RNA cleavage (20). In many cases, it is thought that trans-encoded RNA molecules engage RNA chaperone Hfq to facilitate sRNA-mediated regulation due to limited complementarity between sRNAs and their mRNA targets (21).
(ii) Cis-encoded sRNAs are another class of intracellular bacterial sRNAs complementary to their target encoded in the same region of DNA. They have functional ability to interact autonomously as they are transcribed from DNA strand opposite to genes they regulate (22). Cis-encoded sRNAs vary greatly in size and usually located in the UTRs of the corresponding gene to establish firm RNA duplex formation which in turn affects ribosome-binding/translation and rearranges the secondary structures to affect mRNA stability or termination events as shown in Figure 1 (23).
(iii) In addition to RNA–RNA regulation characterized by base pairing, sRNA can also interact with regulatory proteins to directly modify their activities by mimicking and, thus, efficiently compete with DNA or RNA targets. The best suited example for such interaction is CsrA/RsmA family regulators (global carbon storage regulator). All Csr/Rsm-regulatory networks represent a common feature in pathogenic bacteria, the two-component system (TCS) regulate the transcription of the small RNAs to sequester CsrA. The sRNAs directly bind with CsrA/RsmA to sequester it from interacting with mRNA targets usually in close immediate vicinity of the RBS. This phenomena result in the enhancing the translation of the previously blocked transcripts (24).
(iv) Recent studies indicates that a class of sRNAs participates in an adaptive microbial immune system known as clustered regularly interspaced short palindromic repeats (CRISPR), which provide the bacteria with RNA-based acquired immunity against invading DNA elements, such as from bacteriophages, plasmids, and mobile genetic elements (25). This system is composed of an array of conserved short DNA repeat sequences originating from foreign DNA and interspaced by variable spacer regions. Furthermore, together with the conserved Cas proteins, the crRNA can recognize the complementary DNA target to mediate its degradation (26).
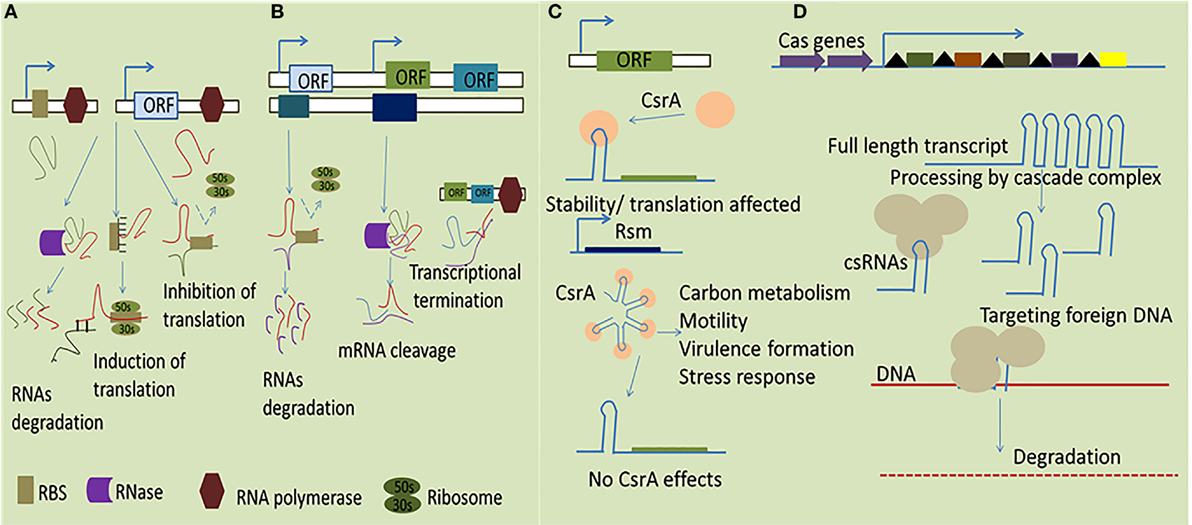
Figure 1. Simplified representation of mechanisms by which sRNAs function in bacteria. (A) The trans-encoded sRNAs interact with their specific target through imperfect base pairing which ultimately results in both positive and negative effect in altering gene expression to promote RNase degradation of the double-stranded RNA molecules. (B) The cis-encoded sRNAs share extensive complementarity by binding with target mRNA resulting in transcriptional termination, degradation of the sRNA–target RNA complex, and affecting translation through a putative loop formation and, consequently, the cessation of the RNA polymerase activity. (C) The dimeric RNA-binding protein CsrA interacts with target mRNA, typically represented in a hairpin loop structure, transcription termination, leading to an alteration of the accessibility of the translation machinery, and/or the stability of the RNA. (D) Mechanism of action of CRISPR arrays for transcription full length RNA which directly target the foreign DNA via CAS proteins resulting in subsequent degradation of the exogenous DNA.
After internalization in the host cell, intracellular bacterial pathogens are challenged by diverse changing environmental conditions. Host cells have established mechanisms to counteract the intracellular bacteria, including degradation of pathogens within the lysosomal compartments. Conversely, as successful pathogens, intracellular bacteria have acquired strategies to avoid lysosomal degradation, such as the arrest or delay of vacuolar maturation in Salmonella (late endosome) and Mycobacterium (early endosome), control of intracellular trafficking in Brucella and Legionella, and resistance to lysosome action in Coxiella (27, 28). These pathogens have developed strategies to evade host protective mechanisms for adaptation, survival, replication, and persistence within host cells to establish chronic infection that is mainly dictated by the presence of certain structural components and virulence factors (29, 30).
The Csr-type system is the most common posttranscriptional regulatory network in intracellular bacteria and is well-characterized in Legionella and Salmonella. Legionella uses effector proteins to modulate host-cell function and establish a replicative niche by forming a membrane-bound vacuole designated the Legionella-containing vacuole (LCV) (31). These effector proteins are under the control of CsrA, which participates in the type IVB secretion system (T4SS) to modulate endoplasmic reticulum (ER)–Golgi vesicular trafficking, with involvement of sRNA-binding protein in survival and replication of intracellular pathogens (32). The VipA, RalF, and YlfA effector proteins have been linked directly to vesicular trafficking to alter the host-cell activity (33). The CsrA controls expression of virulence regulatory genes near the Shine-Dalgarno (SD) sequence of target mRNA located on Salmonella pathogenicity islands (SPIs). Also the transition from a sessile to a motile life form of Salmonella enterica serovar Typhimurium is strongly affected by CsrA (34). In S. enterica serovar Typhimurium, CsrA seems to act positively on swarming motility, and it is required for accurate flagella expression by stabilizing the flhDC and the fliA mRNAs, which are the regulators of the flagella operon. This stabilizing effect leads to an increased production of flagellar proteins (34). In Legionella, CsrA also affects the flagella sigma factor FliA, but in contrast to S. enterica serovar Typhimurium, overexpression of CsrA in Legionella pneumophila resulted in lower fliA transcription and subsequently to reduced levels of FlaA, the major structural flagellar protein controlled by FliA (35). Although CsrA is apparently a common regulator for flagella expression in different bacteria, the regulatory function differs significantly between them, as it can be either positive or negative (36, 37). sRNAs, such as RsmY and RsmZ, regulate the expression of effector protein RsmA to affect the replication of Legionella in macrophages (38), and directly target T4SS regulatory genes to facilitate intracellular survival of pathogen (39, 40).
The RfrA and RfrB are two RybB homologous sRNAs in Salmonella enterica that play an essential role in the intracellular replication in macrophages. Additionally, Fur, a well-known repressor of RybB sRNA is also required for phagocytosis and intracellular survival of Salmonella in human macrophages (Figure 2) (41). These conserved sRNAs regulate expression of virulence genes involved in the oxidative stress, iron homeostasis, and acid resistance within host cells. Furthermore, they have been induced in THP-1 macrophages, fibroblasts, and murine macrophages, suggesting a complementary role of sRNAs in intracellular replication (42). Caswell et al. (43) reported two homologous sRNA in Brucella, named AbcR1 and AbcR2, that play significant role in pathogenicity and establishment of chronic infection. Deletion of AbcR1 or AbcR2 alone in Brucella did not affect intracellular growth in macrophages, but deletion of both resulted in its significant attenuation in macrophages, as well as in mouse model of chronic Brucella infection (43).
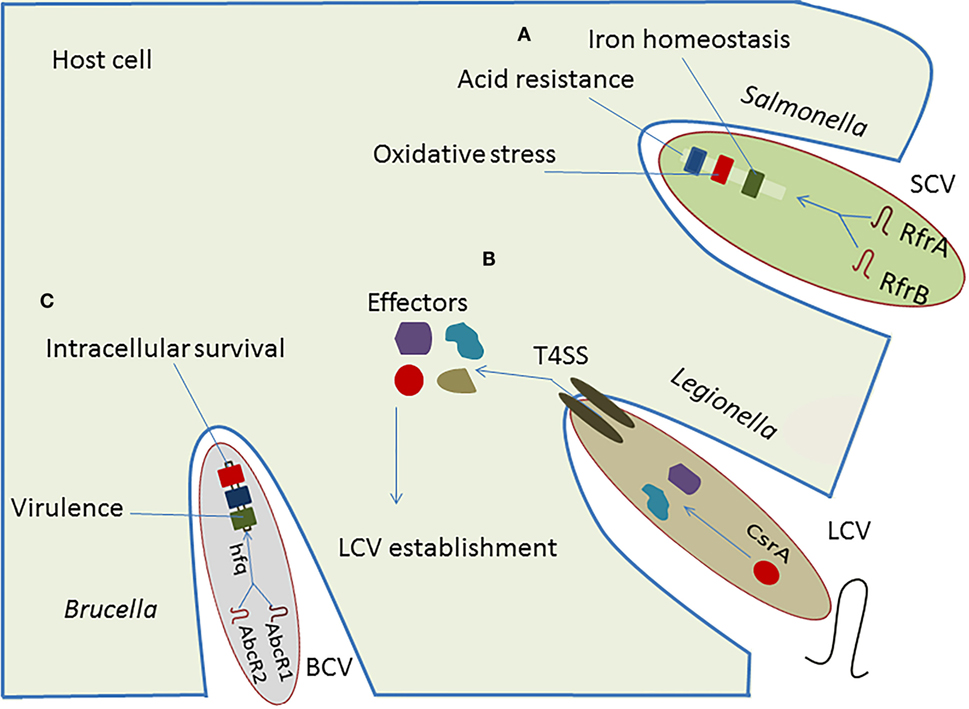
Figure 2. Implication of RNA-mediated regulation in pathogenesis of intracellular bacteria. (A) The regulatory cascade of Salmonella is composed of sRNAs, RfrA and RfrB, plays vital role in the establishment of Salmonella-containing vacuole (SCV), which facilitates adoption of pathogen within host cell resulting in regulation of iron homeostasis, oxidative stress, and acid resistance. (B) The regulatory sRNAs, RsmY and RsmZ, control the action of effectors secreted in cytosol of host which helps in establishing the Legionella-containing vacuole (LCV). (C) The regulatory sRNAs, AbcR1 and AbcR2, interact with hfq to modulate the virulence and intracellular survival of Brucella.
Transcriptional regulators are the subject of considerable study at the molecular level, and the number of newly discovered sRNAs is increasing, with more than 100 identified in Salmonella (19). In many cases, the ubiquitous RNA-binding protein, Hfq, establishes dynamic interactions with RNA molecules to function in virulence of intracellular pathogens. Due to limited complementarity between sRNA and target mRNA, Hfq is essential to facilitate RNA–RNA interactions. Indeed, deletion of hfq has dramatic impact on virulence and intracellular survival in Brucella abortus (44), L. pneumophila (45), and Salmonella typhimurium (46). In addition, Hfq-bound sRNAs are directly involved in regulating metabolic systems and gene expression such as that required for two-component regulatory systems, lipopolysaccharide biosynthesis, host-cell invasion, fatty acid metabolism, central carbon metabolism, and in motility of bacteria (47, 48).
The sRNA GcvBs regulate the ABC transport system through direct binding with extended C/A-rich regions at the mRNA level resulting in lower expression and inhibition of the ABC transport system in S. typhimurium (49). Furthermore, IsrJ sRNA-dependent temporal regulation has been reported in SPIs affecting pathogen invasion of intestinal epithelial cells (50).
IsrM is the SPI-encoding sRNA involved in direct regulation of HilE and SopA virulence genes, which are key regulators of Salmonella virulence and essential for bacteria to evade the host immune system. Specifically, Salmonella hinders production of HilE and SopA for invasion of epithelial cells, which facilitates its survival within the host macrophage (51).
AmgR, a cis-encoded 1.2 kb long antisense RNA in S. typhimurium, which is complementary to mgtCBR mRNA, specifies the MgtC protein. The AmgR sRNA plays an essential role in survival of Salmonella within macrophages and in its virulence in mice and is required for replication in a low Mg2+ environment (52). Surprisingly, transcription of mgtCBR mRNA and AmgR sRNA is controlled by the two-component regulatory system PhoP/PhoQ. In detail, when PhoQ senses decrease level of Mg2+ in cytoplasm, it phosphorylates PhoP initiating transcription of mgtCBR mRNA by direct binding with mgtC and amgR promoters. As MgtB and MgtC protein levels are decreased by long RNA regulatory elements, FtsH protease promotes degradation of the MgtR binding to the MgtC protiens. Consequently, AmgR acts as a timing device for sense-encoded MgtC protein, which was shown to diminish virulence in mice (52). Recently, regulatory sRNAs have been identified in Mycobacterium that act in pathogenesis by regulating the target gene Rv0485 that participates in mediation of virulence in mice (53). These studies suggest that intracellular pathogens utilize sRNA-based strategies to establish productive intracellular infection within host cells.
Intracellular pathogens have developed well-organized strategies to cope and interfere with host innate immune mechanisms that ultimately facilitate establishment of an environment favorable for an effective, long-lasting, adaptive immune response (54). The mammalian innate immune response mechanism provides first line of defense against invading bacterial pathogens. The recognition of molecules typical of a microbe pathogen-associated molecular pattern is obtained via pattern-recognition receptors (PRR) through TLRs that are expressed at high levels on DC and macrophages. The TLRs transmit signals via MyD88 to activate the NF-κB, MAPKs, and IRF signaling pathways to initiate microbial clearance (54). Study of a class of sRNAs designated miRNAs, typically 20–22 nt in length, has greatly expanded our understanding of mechanisms involved in gene expression through posttranscription and translation regulation of protein coding genes. miRNAs play pivotal roles in modulation of innate as well as in adaptive immune response mechanisms (55).
At the initial stages of infection, innate immune response mechanisms effectively control the replication and survival of Salmonella. In vitro studies indicate that Salmonella modulates miRNAs in both epithelial cells and macrophages. Key host miRNAs, such as miR-155 and miR-146, are upregulated in immune cells in response to intracellular bacterial pathogens, apparently co-induced during physiological processes stimulated by lipopolysaccharides to repress TLR-mediated recognition of bacterial molecules and NF-κB activity (56). Modulation of miR-146a/b, miR-155, and miR-21 were first reported in Salmonella infection, with NF-κB-dependent miRNAs significantly induced upon infection in mouse macrophages (57, 58). miRNAs are usually triggered in response to sense extracellular stimulus. This phenomenon was observed in Salmonella mutant strains defective in cell invasion (ΔSPI-1) and replication (ΔSPI-2), as well as in human monocytes (58, 59). It was subsequently found that vaccination of miR-155-null mice with attenuated Salmonella vaccine did not confer protection, and mice showed severely defective T-cell cytokine production, indicating the complex role of miR-155 in innate immune responses to Salmonella (59, 60).
Schulte et al. (56) reported the importance of miR-155 activation by the sensing bacterial peptidoglycan via cytoplasmic NOD2 receptor, suggesting its potential role in innate immune response (56). miR-146 function in zebrafish embryos infected with S. typhimurium was found to be disrupted by knockdown of the TRAF6-MyD88 pathway that mediates transduction of TLR signals and cytokine activation (61). miR-146 function characterization has revealed that it acts as a negative regulator of IRAK1 and TRAF6 expression, which, in turn, affects NF-κB signaling pathway (62). miRNA let-7 appears to be a factor in the acute innate immune response that participates in the TLR signaling pathway via lipopolysaccharide action. In Salmonella infection, let-7 was downregulated in a cell-type dependent manner in HeLa and murine macrophage cells, suggesting that repression of this miRNA family constitutes a common signature of the infection of phagocytic and non-phagocytic cells by Salmonella (57). Zhang and colleagues reported miR-128 upregulated expression upon Salmonella infection that led to reduced secretion of macrophage colony-stimulating factor-mediated macrophage recruitment, in turn suppressing the host immune response mechanisms (63).
Although innate immunity effectively controls replication and survival of Salmonella at the initial stages of infection, a well-organized adaptive immune response is also required at later stages of chronic infection (54). Salmonella shows the ability to infect phagocytic and non-phagocytic cells, residing inside phagocytic cells in the so-called Salmonella-containing vacuole (SCV). Salmonella secretes several virulent SPI-2-encoded proteins into the host cytoplasm to facilitate its intracellular replication and invasion into host cells through two distinct type-III secretion systems (T3SS) (64). Mature DCs may have unique tolerogenic properties for initiation and control of adaptive immune responses. Salmonella has developed mechanisms to counteract the function of DCs, such as subversion of cellular trafficking by preventing fusion of the SCV with lysosomes, which ultimately facilitates pathogen entry into the host (65). Additionally, MHC-I and MHC-II molecules expressed on the surface of DCs show elevated numbers of bacterial-derived antigens that facilitate activation of T cells, resulting in enhanced host adaptive immune response (66). The B cell intrinsic requirement of miR-155 is essential for IgG1 antibody production in response to thymus-dependent and -independent antigens, following vaccination with attenuated Salmonella (67).
Rodriguez et al. reported bic/miR-155-deficient mice show diminished adaptive immune response against S. typhimurium after intravenous immunization that failed to establish strong adaptive immunity, likely due to defective antigen presentation as well as impaired B and T cell functioning by DCs (60). In addition to miR-146 and miR-155, miR-21, miR-23b, miR-27a, miR-24, miR-222, and miR-29 showed upregulation upon Salmonella infection in human monocytes. The upregulated miRNA shows that monocytes differentiation is involved in modulation of the TGF-β signaling pathway to counteract host defense mechanisms (58).
The miRNA let-7 family members directly target major immunomodulatory cytokines IL-6 and IL-10, and their downregulation results in increased expression of both cytokines in response to Salmonella infection (68). miR-21, miR-146a, and miR-155 show strong induction of NF-κB, leading to decreased regulation of B cell and T cell proliferation in murine macrophages upon Salmonella infection (69). These findings suggest that Salmonella uses miRNA as a strategy to modulate TLR-NF-κB signaling pathways as well as to counteract the function of DCs to subvert its cellular trafficking, and miRNAs play a significant role in the interaction of innate and adaptive immune mechanisms.
Recently, several studies have highlighted the role of miRNAs in mycobacterial infection (70). Mycobacterium also modulates miRNAs associated with signaling pathways, which enhances its survival in the host. miR-155 was found to be upregulated in Mycobacterium infection, resulting in increased apoptosis of infected cells by the involvement of the TLR2 and NF-κB signaling pathways (71). Downregulation of miR-155 result in decreased TNF-α production in response to lipomannan, a component of the bacterial cell wall, was reported in human macrophages, affecting the TLR-MAPK/Akt signaling pathway (72). In addition, MiR-155 is involved in regulation of autophagy-mediated mycobacterial elimination by the repression of the negative regulator Rheb (73). miR-142-3p participates in an effective strategy in mycobacterial infection to control early events of phagolysosome biogenesis via targeting the N-Wasp and actin-binding protein (74). On the other hand, down regulation of miR-142-3p negatively regulates the production of NF-κB, TNF-α, and IL-6 in macrophages upon Mycobacterium bovis infection, resulting in the activation of the NF-κB pathway via the de-repression of the target IRAK1 (75). miR-124 has been found to serve as a potent modulator of the immune response in an M. bovis BCG-infected macrophage cell line (Raw 264.7) by targeting components of the TLR signaling pathway including MyD88, TLR6, TRAF6, and TNF-α (76).
miR-146a modulates the inflammatory response upon Mycobacterium infection in Raw 264.7 cells by targeting IRAK1 and TRAF6, resulting in remarkably reduced translation of IL-6, IL-1β, and TNF-α (77). Kumar and colleagues reported that Mycobacterium tuberculosis down regulates the expression of miRNA let-7f, which targets mRNA of A20, an inhibitor of NF-κB. Significantly, the downregulation of let-7f is accompanied by concomitant upregulation of A20 in mice infected with M. tuberculosis (78).
miR-21 was found to induce inhibition of IL-12 production in a NF-κB-dependent manner in DCs and T-cells upon Mycobacterium infection, and thus suppress host Th1 responses (79). Interestingly, miR-21 upregulation promotes DCs apoptosis by targeting Bcl-2 in Mycobacterium-infected cells (80). miR-99b upregulation has been observed to stimulate the production of proinflammatory cytokines such as IL-6, IL-12, IL-1β, and TNF-α in macrophages and DCs upon Mycobacterium infection. Inhibition of TNF-α production is a key strategy of Mycobacterium to promote growth within DCs that, in turn, facilitates evasion of host adaptive immune response mechanisms (81). Transfection of T cells with miR-144 precursor has demonstrated that miR-144 possibly regulates antituberculosis immunity by inhibition of IFN-γ and TNF-α production and T cell proliferation (82). Wang et al. (83) reported that miR-223 and miR-424 promotes monocyte differentiation and subsequently downregulates the expression of transcription factor NFI-A78. The downregulation of miRNAs involved in disorder proportions of T cells and B cells in active tuberculosis patients (83). Collectively, these finding indicates that Mycobacterium use miRNA-based strategies for completion of intracellular replication that, in turn, facilitates evasion of the immune response.
We studied the expression of miRNAs expression in Brucella melitensis-infected RAW 264 macrophages cells and found several miRNAs such as miR-let-7b, miR-93, miR-92a, miR-181b, and miR-1981, differentially expressed compared to mock-infected cells, and purposed that these miRNAs are involved in regulation of autophagy, apoptosis, innate and adaptive immune response mechanisms (Figure 3) (84). Liu et al. (85) reported that the downregulation of miR-125b-5p during B. abortus infection enhances the expression of the A20 protein, thereby inhibiting NF-κB activation and facilitating bacterial intracellular survival (85).
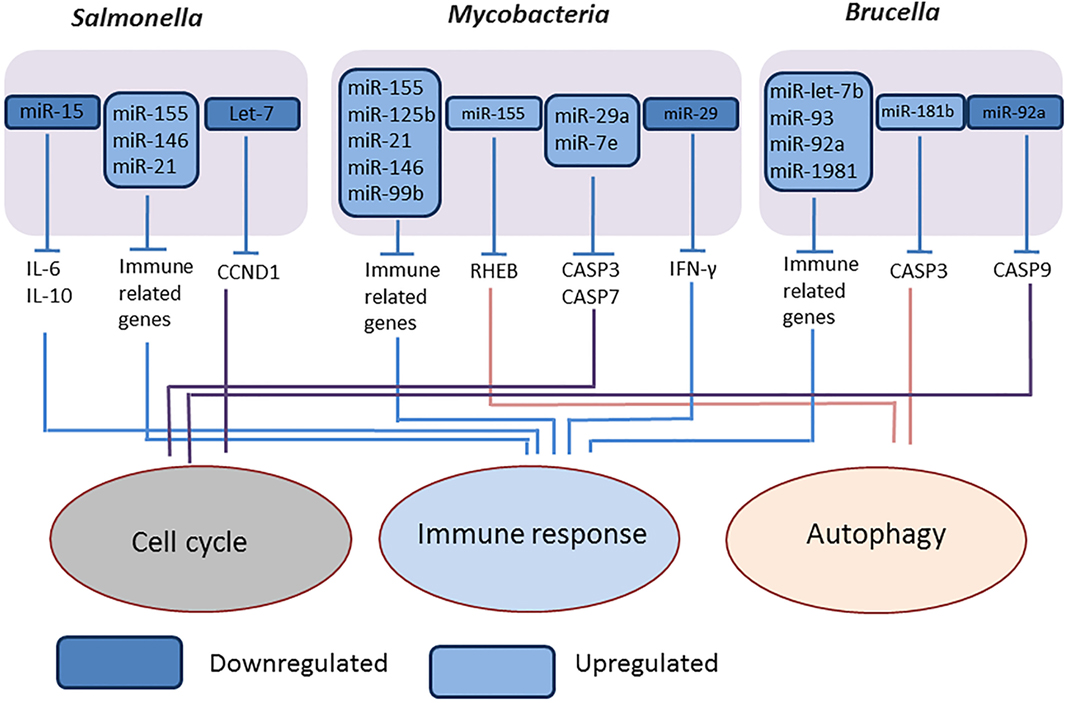
Figure 3. Overview of modulation of host miRNAs by intracellular bacterial pathogens. The representative figure of modulation of host miRNAs by intracellular bacterial pathogens, Salmonella (Gram negative, intracellular), Mycobacteria (intracellular), and Brucella (Gram negative, intracellular).
Now that a number of actively transcribed sRNAs have been identified in intracellular bacteria, further research should focus on the determination of their functions and their specific targets. TargetRNA2 was the first webserver specifically designed for identification of bacterial sRNA target which determine sRNA–target interactions through the straight forward hybridization model. It determines the seed region between two putative sRNA targets which is composed of very small short series of consecutive base (86). IntaRNA is also used to identify sRNA targets which work on the principle of hybridization energy between base pairings of sRNA–target interactions to the hybridization energy of interacting regions being unpaired in intramolecular structures. Further, it conform the seed regions more efficiently as compared all other available webtools (87). Target prediction could also be used in conjunction with experimental genome-wide approaches, such as transcriptional profiling (88). Comparison of the transcription profile of a mutant strain or an over-expresser strain to the wild-type strain can highlight putative targets (9). Comparison of the sRNA deletion mutant, the over-expresser strain and the wild-type strain by SDS-PAGE and Coomassie staining may be sufficient to suggest a putative target (89). The proteomic studies could be undertaken or more direct approaches, such as the streptavidin-binding aptamer tag described above could be used (90). Said et al. (91) have performed a systematic analysis for the use of different aptamers and configurations to identify protein targets of sRNA. Further, we need to establish more accurate identification methods for the bacterial sRNA prediction.
Recent advancements in research have revealed diverse functions, wide distribution, and high variability of sRNAs and described their crucial role in biological processes, such as infectivity and virulence of intracellular bacteria, stress adaptation, and environmental sensing, as well as in modulation of innate and adaptive immune response mechanisms. Intracellular bacteria are divided in two classes: vacuolar intracellular bacteria, such as Salmonella, Mycobacterium, Legionella, Brucella, and Coxiella, which survive and replicate either by avoiding vacuole–lysosome fusion or by altering the phagolysosome environment; and cytosolic intracellular bacteria, including Francisella, Shigella, Listeria, Burkholderia, and Rickettsia, which usually escape to proliferate within the cytosol of host cell. Infection is a multidimensional complex event, with host cells having developed immune mechanisms to counteract invading intracellular pathogens via lysosomal degradation to maintain a balance between host resistance and bacterial virulence. Intracellular pathogens have evolved several sRNA-based strategies to survive and replicate within phagocytic cells and to manipulate the host-cell machinery for their own benefit. Upon internalization in host cells, the pathogenic bacteria are usually surrounded by a membrane-bound vacuole that protects against proteolytic degradation. Bacterial regulatory sRNAs operate at all levels of gene regulation to modulate translation, transcription, DNA maintenance or silencing, and mRNA stability. They use different mechanisms to perform regulatory functions, including changes in RNA conformation, base pairing with other RNAs, protein binding, and interactions with DNA.
The Csr-type system is the most common posttranscriptional regulator network in intracellular bacteria which participates with type IVB secretion system to modulate the ER–Golgi vesicular trafficking highlighting the involvement of sRNA-binding protein in survival and replication of intracellular pathogens. The effector proteins, VipA, RalF, and YlfA, have been linked directly in vesicular trafficking to affect the host-cell activity. Additionally, sRNA, RsmY and RsmZ, in Legionella, RybB sRNA in Salmonella, and sRNA AbcR in Brucella regulate the expression of effector protein for survival and intracellular replication in macrophages.
At the initial stage of infectious pathogenesis, intracellular pathogens modulate the immune response mechanism of host to quickly translocate through the mucosal immune barrier and are endocytosed by mucosal macrophages and DCs. Intracellular pathogens infect the host-cell machinery by targeting IRAK1 and TRAF6 to limit PRRs which, in turn, affects the TLR/NF-κB signaling cascade. Inhibition of antigen presentation to T cells is a further strategy of intracellular bacteria to dampen innate and adaptive immune mechanisms. They have the ability to hinder activation of DCs to subvert the immune response mechanism by averting function of T-cells and secreting IL-12 to establish a strong Th1 immune response.
Additionally, inhibition of TNF-α production and modulation of MHC-I and MHC-II expression is a key strategy of intracellular pathogen to promote growth within DCs that, in turn, favors cytokine regulation to facilitate evasion of host adaptive immune response mechanisms. Although numerous strategies employed by bacterial sRNAs have been reported, there are many mysteries that are still veiled, including how are sRNAs involved in modulation of innate immune signaling? What is the role of sRNAs in regulation of apoptosis and autophagy mechanisms? To date, nothing is known about the role of sRNA interfering with innate and adaptive immune mechanism of Brucella and Legionella and it will be an open question for the next few years. In-depth identification of novel immune evasion strategies employed by bacterial sRNAs will facilitate our understanding of pathogenesis and designing of novel effective therapeutic approaches to combat diseases caused by intracellular pathogens.
Z-FL and WA conceived the research. WA and KZ wrote the manuscript. Z-FL revised the manuscript critically for relevant intellectual content. All authors have read and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AF and handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review.
This work was supported by Natural Science Foundation of China (31270193 and 31470259), Technique Innovation Program of Hubei Province (2016), National Key Research and Development Program (2016YFD0500105), and Fundamental Research Funds for the Central Universities (2016PY052) to Z-FL.
1. Fitzgerald KA, Caffrey DR. Long noncoding RNAs in innate and adaptive immunity. Curr Opin Immunol (2014) 26:140–6. doi: 10.1016/j.coi.2013.12.001
2. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature (2012) 482:339–46. doi:10.1038/nature10887
3. Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol (2004) 58:303–28. doi:10.1146/annurev.micro.58.030603.123841
4. Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell (2011) 43:880–91. doi:10.1016/j.molcel.2011.08.022
5. Waters LS, Storz G. Regulatory RNAs in bacteria. Cell (2009) 136:615–28. doi:10.1016/j.cell.2009.01.043
6. Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol (2011) 3. doi:10.1101/cshperspect.a003798
7. Guillier M, Gottesman S. The 5 end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res (2008) 36:6781–94. doi:10.1093/nar/gkn742
8. Frohlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol (2009) 12:674–82. doi:10.1016/j.mib.2009.09.009
9. Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell (2013) 153:426–37. doi:10.1016/j.cell.2013.03.003
10. Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol (2007) 10:134–9. doi:10.1016/j.mib.2007.03.010
11. Johansson J, Cossart P. RNA-mediated control of virulence gene expression in bacterial pathogens. Trends Microbiol (2003) 11:280–5. doi:10.1016/S0966-842X(03)00118-5
12. Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell (2012) 149:515–24. doi:10.1016/j.cell.2012.04.005
13. Agbor TA, McCormick BA. Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol (2011) 13:1858–69. doi:10.1111/j.1462-5822.2011.01701.x
14. Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science (2004) 304:242–8. doi:10.1126/science.1090124
15. Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol (2010) 26:261–83. doi:10.1146/annurev-cellbio-100109-104034
16. Fredlund J, Enninga J. Cytoplasmic access by intracellular bacterial pathogens. Trends Microbiol (2014) 22:128–37. doi:10.1016/j.tim.2014.01.003
17. Sharma CM, Vogel J. Experimental approaches for the discovery anal characterization of regulatory small RNA. Curr Opin Microbiol (2009) 12:536–46. doi:10.1016/j.mib.2009.07.006
18. Chao YJ, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3’UTRs as a genomic reservoir of regulatory small RNAs. EMBO J (2012) 31:4005–19. doi:10.1038/emboj.2012.229
19. Chao YJ, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol (2010) 13:24–33. doi:10.1016/j.mib.2010.01.001
20. Saramago M, Barria C, dos Santos RF, Silva IJ, Pobre V, Domingues S, et al. The role of RNases in the regulation of small RNAs. Curr Opin Microbiol (2014) 18:105–15. doi:10.1016/j.mib.2014.02.009
21. Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol (2007) 10:125–33. doi:10.1016/j.mib.2007.03.015
22. Caldelari I, Chao YJ, Romby P, Vogel J. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb Perspect Med (2013) 3:a010298. doi:10.1101/cshperspect.a010298
24. Duss O, Michel E, Konte NDD, Schubert M, Allain FHT. Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition. Nucleic Acids Res (2014) 42:5332–46. doi:10.1093/nar/gku141
25. Jinek M, Jiang FG, Taylor DW, Sternberg SH, Kaya E, Ma EB, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science (2014) 343:1247997. doi:10.1126/science.1247997
26. Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet (2010) 26:335–40. doi:10.1016/j.tig.2010.05.008
27. Alix E, Mukherjee S, Roy CR. Host-pathogen interactions subversion of membrane transport pathways by vacuolar pathogens. J Cell Biol (2011) 195:943–52. doi:10.1083/jcb.201105019
28. Ahmed W, Zheng K, Liu ZF. Establishment of chronic infection: Brucella’s stealth strategy. Front Cell Infect Microbiol (2016) 6:30. doi:10.3389/fcimb.2016.00030
29. Faucher SP, Mueller CA, Shuman HA. Legionella pneumophila transcriptome during intracellular multiplication in human macrophages. Front Microbiol (2011) 2:60. doi:10.3389/fmicb.2011.00060
30. Ortega AD, Quereda JJ, Pucciarelli MG, Garcia-del Portillo F. Non-coding RNA regulation in pathogenic bacteria located inside eukaryotic cells. Front Cell Infect Microbiol (2014) 4:162. doi:10.3389/fcimb.2014.00162
31. Faucher SP, Friedlander G, Livny J, Margalit H, Shuman HA. Legionella pneumophila 6S RNA optimizes intracellular multiplication. Proc Natl Acad Sci U S A (2010) 107:7533–8. doi:10.1073/pnas.0911764107
32. Nevo O, Zusman T, Rasis M, Lifshitz Z, Segal G. Identification of Legionella pneumophila effectors regulated by the LetAS-RsmYZ-CsrA regulatory cascade, many of which modulate vesicular trafficking. J Bacteriol (2014) 196:681–92. doi:10.1128/JB.01175-13
33. Heidtman M, Chen EJ, Moy MY, Isberg RR. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol (2009) 11:230–48. doi:10.1111/j.1462-5822.2008.01249.x
34. Jonas K, Edwards AN, Ahmad I, Romeo T, Romling U, Melefors O. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ Microbiol (2010) 12:524–40. doi:10.1111/j.1462-2920.2009.02097.x
35. Forsbach-Birk V, McNealy T, Shi C, Lynch D, Marre R. Reduced expression of the global regulator protein CsrA in Legionella pneumophila affects virulence-associated regulators and growth in Acanthamoeba castellanii. Int J Med Microbiol (2004) 294:15–25. doi:10.1016/j.ijmm.2003.12.003
36. Martinez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, et al. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol (2011) 80:1637–56. doi:10.1111/j.1365-2958.2011.07674.x
37. Kulkarni PR, Jia T, Kuehne SA, Kerkering TM, Morris ER, Searle MS, et al. A sequence-based approach for prediction of CsrA/RsmA targets in bacteria with experimental validation in Pseudomonas aeruginosa. Nucleic Acids Res (2014) 42:6811–25. doi:10.1093/nar/gku309
38. Rasis M, Segal G. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol (2009) 72:995–1010. doi:10.1111/j.1365-2958.2009.06705.x
39. Jayakumar D, Early JV, Steinman HM. Virulence phenotypes of Legionella pneumophila associated with noncoding RNA lpr0035. Infect Immun (2012) 80:4143–53. doi:10.1128/IAI.00598-12
40. Trigui H, Mendis N, Li L, Saad M, Faucher SP. Facets of small RNA-mediated regulation in Legionella pneumophila. Molecular Mechanisms in Legionella Pathogenesis. (Vol. 376) (2014). p. 53–80.
41. Leclerc JM, Dozois CM, Daigle F. Role of the Salmonella enterica serovar Typhi Fur regulator and small RNAs RfrA and RfrB in iron homeostasis and interaction with host cells. Microbiology (2013) 159:591–602. doi:10.1099/mic.0.064329-0
42. Ortega AD, Gonzalo-Asensio J, Garcia-del Portillo F. Dynamics of Salmonella small RNA expression in non-growing bacteria located inside eukaryotic cells. RNA Biol (2012) 9:469–88. doi:10.4161/rna.19317
43. Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, et al. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol (2012) 85:345–60. doi:10.1111/j.1365-2958.2012.08117.x
44. Caswell CC, Gaines JM, Roop RM. The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J Bacteriol (2012) 194:3–14. doi:10.1128/Jb.05623-11
45. McNealy TL, Forsbach-Birk V, Shi CW, Marre R. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J Bacteriol (2005) 187:1527–32. doi:10.1128/JB.187.4.1527-1532.2005
46. Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet (2008) 4:e1000163. doi:10.1371/journal.pgen.1000163
47. Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol (2007) 63:193–217. doi:10.1111/j.1365-2958.2006.05489.x
48. Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, et al. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One (2009) 4(3):e4809. doi:10.1371/journal.pone.0004809
49. Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JCD, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol (2011) 81:1144–65. doi:10.1111/j.1365-2958.2011.07751.x
50. Gong H, Vu GP, Bai Y, Chan E, Wu RB, Yang E, et al. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog (2011) 7:e1002120. doi:10.1371/journal.ppat.1002120
51. Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, et al. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res (2008) 36:1913–27. doi:10.1093/nar/gkn050
52. Lee EJ, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol (2010) 76:1020–33. doi:10.1111/j.1365-2958.2010.07161.x
53. DiChiara JM, Contreras-Martinez LM, Livny J, Smith D, McDonough KA, Belfort M. Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucleic Acids Res (2010) 38:4067–78. doi:10.1093/nar/gkq101
54. Diacovich L, Gorvel JP. Bacterial manipulation of innate immunity to promote infection. Nat Rev Microbiol (2010) 8:117–28. doi:10.1038/nrmicro2295
55. Voinnet O. Micro-balancing innate immunity to Salmonella. EMBO J (2011) 30:1877–9. doi:10.1038/emboj.2011.134
56. Schulte LN, Westermann AJ, Vogel J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res (2013) 41:542–53. doi:10.1093/nar/gks1030
57. Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J (2011) 30:1977–89. doi:10.1038/emboj.2011.94
58. Sharbati S, Sharbati J, Hoeke L, Bohmer M, Einspanier R. Quantification and accurate normalisation of small RNAs through new custom RT-qPCR arrays demonstrates Salmonella-induced microRNAs in human monocytes. BMC Genomics (2012) 13:23. doi:10.1186/1471-2164-13-23
59. Hoeke L, Sharbati J, Pawar K, Keller A, Einspanier R, Sharbati S. Intestinal Salmonella typhimurium infection leads to miR-29a induced caveolin 2 regulation. PLoS One (2013) 8:e67300. doi:10.1371/journal.pone.0067300
60. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science (2007) 316:608–11. doi:10.1126/science.1139253
61. Ordas A, Kanwal Z, Lindenberg V, Rougeot J, Mink M, Spaink HP, et al. microRNA-146 function in the innate immune transcriptome response of zebrafish embryos to Salmonella typhimurium infection. BMC Genomics (2013) 14:696. doi:10.1186/1471-2164-14-696
62. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappa B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A (2006) 103:12481–6. doi:10.1073/pnas.0605298103
63. Zhang TF, Yu JX, Zhang YQ, Li LM, Chen YY, Li DH, et al. Salmonella enterica serovar Enteritidis modulates intestinal epithelial miR-128 levels to decrease macrophage recruitment via macrophage colony-stimulating factor. J Infect Dis (2014) 209:2000–11. doi:10.1093/infdis/jiu006
64. Curtale G, Citarella F. Dynamic nature of noncoding RNA regulation of adaptive immune response. Int J Mol Sci (2013) 14:17347–77. doi:10.3390/ijms140917347
65. Mastroeni P, Grant A, Restif O, Maskell D. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat Rev Microbiol (2009) 7:73–80. doi:10.1038/nrmicro2034
66. Riquelme SA, Bueno SM, Kalergis AM. IgG keeps virulent Salmonella from evading dendritic cell uptake. Immunology (2012) 136:291–305. doi:10.1111/j.1365-2567.2012.03578.x
67. Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity (2007) 27:847–59. doi:10.1016/j.immuni.2007.10.009
68. Maudet C, Mano M, Eulalio A. microRNAs in the interaction between host and bacterial pathogens. FEBS Lett (2014) 588:4140–7. doi:10.1016/j.febslet.2014.08.002
69. Bao H, Kommadath A, Liang GX, Sun X, Arantes AS, Tuggle CK, et al. Genome-wide whole blood microRNAome and transcriptome analyses reveal miRNA-mRNA regulated host response to foodborne pathogen Salmonella infection in swine. Sci Rep (2015) 5:12620. doi:10.1038/srep12620
70. Harapan H, Fitra F, Ichsan I, Mulyadi M, Miotto P, Hasan NA, et al. The roles of microRNAs on tuberculosis infection: meaning or myth? Tuberculosis (Edinb) (2013) 93:596–605. doi:10.1016/j.tube.2013.08.004
71. Ghorpade DS, Leyland R, Kurowska-Stolarska M, Patil SA, Balaji KN. microRNA-155 is required for Mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol Cell Biol (2012) 32:2239–53. doi:10.1128/MCB.06597-11
72. Rajaram MVS, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, et al. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci U S A (2011) 108:17408–13. doi:10.1073/pnas.1112660108
73. Wang JL, Yang K, Zhou L, Minhaowu, Wu YJ, Zhu M, et al. microRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog (2013) 9:e1003697. doi:10.1371/journal.ppat.1003697
74. Bettencourt P, Marion S, Pires D, Santos LF, Lastrucci C, Carmo N, et al. Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142-3p. Front Cell Infect Microbiol (2013) 3:19. doi:10.3389/fcimb.2013.00019
75. Zhang Y, Li YK. microRNAs in the regulation of immune response against infections. J Zhejiang Univ Sci B (2013) 14:1–7. doi:10.1631/jzus.B1200292
76. Ma F, Xu S, Liu XG, Zhang Q, Xu XF, Liu MF, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol (2011) 12:861–U865. doi:10.1038/ni.2073
77. Li S, Yue Y, Xu W, Xiong SD. microRNA-146a represses mycobacteria-induced inflammatory response and facilitates bacterial replication via targeting IRAK-1 and TRAF-6. PLoS One (2013) 8:e81438. doi:10.1371/journal.pone.0081438
78. Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, et al. microRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappa B pathway. Cell Host Microbe (2015) 17:345–56. doi:10.1016/j.chom.2015.01.007
79. Wu ZW, Lu HF, Sheng JF, Li LJ. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett (2012) 586:2459–67. doi:10.1016/j.febslet.2012.06.004
80. Riendeau CJ, Kornfeld H. THP-1 cell apoptosis in response to mycobacterial infection. Infect Immun (2003) 71:254–9. doi:10.1128/IAI.71.1.254-259.2003
81. Singh Y, Kaul V, Mehra A, Chatterjee S, Tousif S, Dwivedi VP, et al. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J Biol Chem (2013) 288:5056–61. doi:10.1074/jbc.C112.439778
82. Liu YH, Wang XJ, Jiang J, Cao ZH, Yang BF, Cheng XX. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol Immunol (2011) 48:1084–90. doi:10.1016/j.molimm.2011.02.001
83. Wang C, Yang SY, Sun G, Tang XY, Lu SH, Neyrolles O, et al. Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PLoS One (2011) 6:e25832. doi:10.1371/journal.pone.0025832
84. Zheng K, Chen DS, Wu YQ, Xu XJ, Zhang H, Chen CF, et al. microRNA expression profile in RAW264.7 cells in response to Brucella melitensis infection. Int J Biol Sci (2012) 8:1013–22. doi:10.7150/ijbs.3836
85. Liu N, Wang L, Sun C, Yang L, Sun W, Peng Q. microRNA-125b-5p suppresses Brucella abortus intracellular survival via control of A20 expression. BMC Microbiol (2016) 16:171. doi:10.1186/s12866-016-0788-2
86. Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, et al. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res (2006) 34:2791–802. doi:10.1093/nar/gkl356
87. Wright PR, Georg J, Mann M, Sorescu DA, Richter AS, Lott S, et al. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res (2014) 42:W119–23. doi:10.1093/nar/gku359
88. Modi SR, Camacho DM, Kohanski MA, Walker GC, Collins JJ. Functional characterization of bacterial sRNAs using a network biology approach. Proc Natl Acad Sci U S A (2011) 108:15522–7. doi:10.1073/pnas.1104318108
89. Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res (2007) 35:1018–37. doi:10.1093/nar/gkl1040
90. Wang J, Rennie W, Liu C, Carmack CS, Prevost K, Caron MP, et al. Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Res (2015) 43:10308–20. doi:10.1093/nar/gkv1158
Keywords: small RNAs, Salmonella, Mycobacterium, Brucella, innate immunity, adaptive immunity
Citation: Ahmed W, Zheng K and Liu Z-F (2016) Small Non-Coding RNAs: New Insights in Modulation of Host Immune Response by Intracellular Bacterial Pathogens. Front. Immunol. 7:431. doi: 10.3389/fimmu.2016.00431
Received: 14 August 2016; Accepted: 03 October 2016;
Published: 18 October 2016
Edited by:
Alexandre Morrot, Federal University of Rio de Janeiro, BrazilReviewed by:
Alessandra D’Almeida Filardy, Federal Univesity of Rio de Janeiro, BrazilCopyright: © 2016 Ahmed, Zheng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng-Fei Liu, bHpmNjc4OUBtYWlsLmh6YXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.