- 1Department of Parasitology, National Institute of Infectious Diseases, Tokyo, Japan
- 2Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Japan
Entamoeba histolytica is a protozoan parasite and the causative agent of amebiasis. It is estimated approximately 1% of humans are infected with E. histolytica, resulting in an estimate of 100,000 deaths annually. Clinical manifestations of amebic infection range widely from asymptomatic to severe symptoms, including dysentery and extra-intestinal abscesses. Like other infectious diseases, it is assumed that only ~20% of infected individuals develop symptoms, and genetic factors of both the parasite and humans as well as the environmental factors, e.g., microbiota, determine outcome of infection. There are multiple essential steps in amebic infection: degradation of and invasion into the mucosal layer, adherence to the intestinal epithelium, invasion into the tissues, and dissemination to other organs. While the mechanisms of invasion and destruction of the host tissues by the amebae during infection have been elucidated at the molecular levels, it remains largely uncharacterized how the parasite survive in the host by evading and attacking host immune system. Recently, the strategies for immune evasion by the parasite have been unraveled, including immunomodulation to suppress IFN-γ production, elimination of immune cells and soluble immune mediators, and metabolic alterations against reactive oxygen and nitrogen species to fend off the attack from immune system. In this review, we summarized the latest knowledge on immune reaction and immune evasion during amebiasis.
Introduction
Entamoeba histolytica is an enteric protozoan parasite that infects humans, and is the etiological agent of amebiasis. Amebiasis remains a worldwide health problem accounting for up to 100,000 deaths annually (1, 2). Transmission occurs via ingestion of food and water contaminated with amebic cysts (1, 3, 4). In endemic areas, exposure can be extremely high: an annual incidence of 40% was estimated among children in an urban slum in Bangladesh (5). In some parts of Asia and Australia, amebiasis is endemic among men who have sex with men (MSM) and can be transmitted sexually (6–9). Majority of infections with E. histolytica remain asymptomatic, while ~20% of the cases develop clinical manifestations, such as dysentery, which is characterized by colonic mucosal invasion and tissue destruction (10). Invasive disease includes dysentery and extra-intestinal amebiasis, most commonly amebic liver abscesses (ALAs), which occur in approximately 1% of symptomatic cases in developing countries and around 17% in Japan (11, 12).
When amebic trophozoites invade the colonic epithelium, they activate immune response in the human host. In order to survive in the host, the repression of host immune systems and the control of the environment of parasitism are crucial. For instance, during extraintestinal dissemination, the amebae must transiently survive in the blood vessels and the spleen, in which a network of immune cells and humoral factors are present, and the amebae are exposed to high concentrations of oxygen (E. histolytica are anaerobic or microaerophilic). To persist in such environment, amebae must subvert detection by antibody and complement, and resist oxidative and nitrosative attack. In this review, we summarize our current knowledge on immune response during amebic infection (Figure 1) and the parasite’s strategies to evade from host immune system (Figure 2).
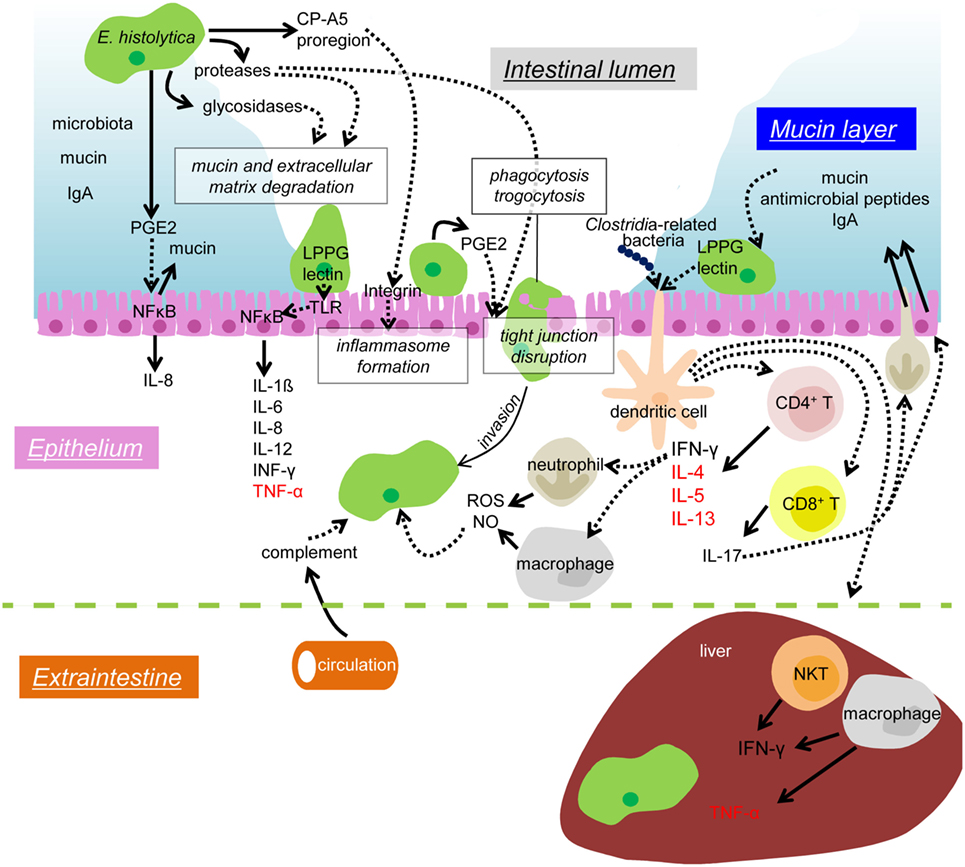
Figure 1. Mechanisms of colonization and invasion by E. histolytica trophozoites and host immune responses to suppress and control amebic infection. In the lumen of the large intestine, the IEC layer is covered by the mucus layer (blue), which contains secreted mucin and IgA from the host and commensal microbiota. Proteases and glycosidases secreted from the amebae are involved in the degradation of mucin and extracellular matrix. The pro-domain of EhCP-A5 binds to and activate integrin and enhances the inflammasome formation of leading to pro-inflammatory responses. PGE2 also secreted from the amebae causes mucin hypersecretion and depletion of mucin from the IECs. PGE2 also elicits signaling in a cascade leading to NFκB activation in the IECs and induces IL-8 secretion. The Gal/GalNAc lectin (lectin) and LPPG on the ameba’s surface binds to TLR2 and leads to NFκB activation and pro-inflammatory cytokine release for IEC. PGE2 also helps to disrupt tight junction function of the epithelium and enhances the amebic infiltration. Phagocytosis and trogocytosis are also involved in removal of host cells and invasion into the host tissue. Infiltrating trophozoites are attacked by complement from the circulation, ROS and NO from neutrophils and macrophages. The Gal/GalNAc lectin and LPPG activate CD4, CD8 T cells, and NKT cells, and, thus, enhances protective cellular immunity. CD4 T cells produce IFN-γ, IL-4, IL-5, and IL-13, and CD8 T cells produce IL-17. IL-17 induces neutrophil infiltration and enhances secretion of mucin, antimicrobial peptides, and IgA into the colonic lumen. When disseminated to the liver, the amebae are attached and removed by the dense mediated by IFN-γ secreted by NKT cells. TNF-α secreted from hepatic macrophages leads to abscess formation. Solid arrows depict secretion of soluble proteins and dotted arrows indicate interaction or signal transduction. Cytokines mainly beneficial for an elimination of the amebae are shown in black, while those involved in disease manifestations are shown in red.
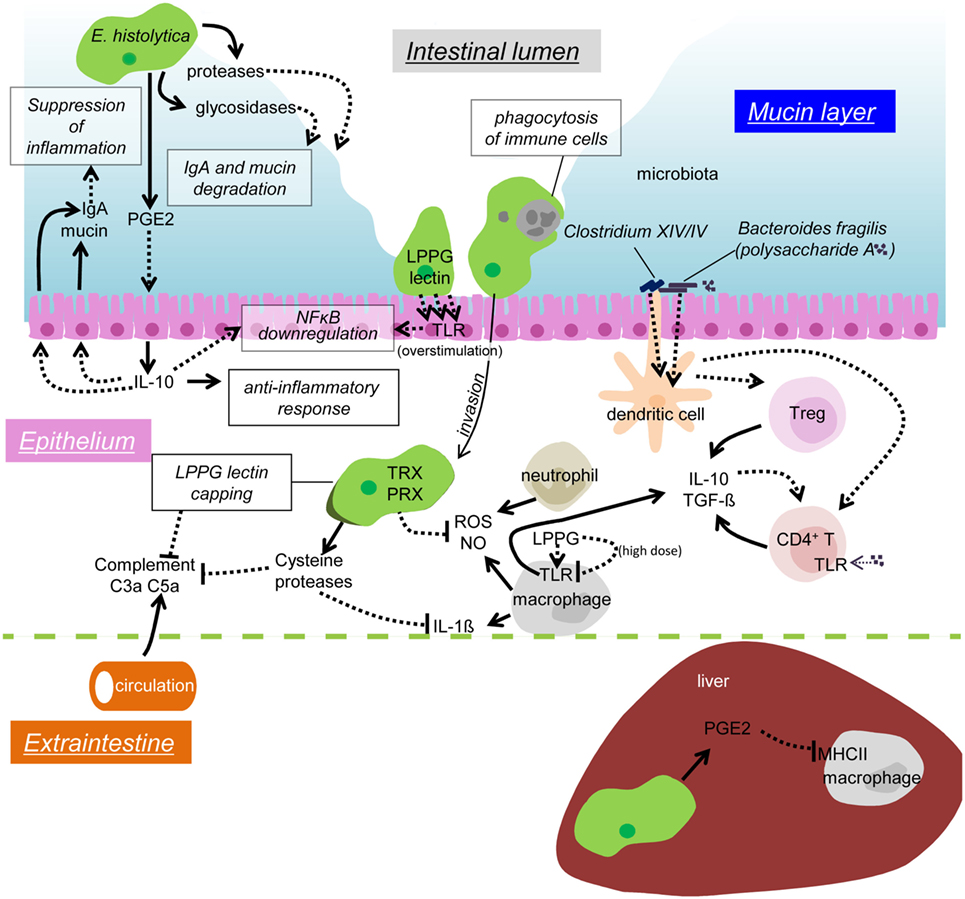
Figure 2. Possible mechanisms of immune evasion during amebiasis. Secreted or surface proteases of the amebae degrade IgA in the mucosal layer. PGE2 from the amebae induces IL-10 secretion from the IECs, and in turn stimulates mucin and IgA secretion, which likely prevents unnecessary inflammation. Overstimulation of TLR causes downregulation of NFκB activation. Removal of infiltrating immune cells by phagocytosis/trogocytosis helps to reduce immune responses. Some commensal microbiota, namely Clostridium XIV and IV groups and Bacteroides fragilis, induce Treg cells to downregulate immune responses. Polysaccharide A from B. fragilis binds to TLR2 on CD4 T cells and induces IL-10 production. The amebae in the tissues and the blood stream evade from complement by surface receptor capping (LPPG, lectin) and degradation of C3a and C5a by cysteine proteases. Cysteine proteases also degrade IL-1β, antioxidative stress defense by the TRX and PRX systems fends off the attack from ROS and NO from activated neutrophils and macrophages. LPPG binds to TLR2 on monocytes and macrophages, which leads to secretion of cytokines, including IL-10 and TGF-β. High doses of LPPG downregulate TLR2 gene expression in monocyte and cause negative feedback of protective immune responses. PGE2 from the amebae and the host causes downregulation of MHC class II expression on macrophages in the liver, which results in anti-inflammation.
Immune Response During Amebic Infection
Course of Amebic Infection
Entamoeba histolytica infection is initiated by parasite adherence to the colonic mucin layer. Trophozoites express a galactose and N-acetyl-d-galactosamine specific lectin (Gal/GalNAc lectin) on the cell surface and attach to host mucin and colonic epithelial cells (13). The colonized parasites are capable of extensive tissue destruction. Beside the pore-forming proteins, amoebapores (14, 15), hydrolytic enzymes, particularly cysteine proteases (CP), are considered to be essential weapons of the parasite to penetrate the epithelium and destroy components of the host’s extracellular matrix (ECM) (16–20). During and after penetration into the submucosal region, amebic trophozoites interact directly and indirectly with host immune and non-immune cells.
Humoral Immunity
While the mucosal layer in the gastrointestinal tract generally serves as a primary physical barrier against intestinal pathogens, the intestinal immune response is the secondary defense to E. histolytica infection. Mucosal immunoglobulins (Ig) are the major component of the human intestinal defense mechanism (21). Among them, secretory IgA is one of the most abundant Ig produced by plasma cells and functions by preventing pathogens from adhering and removing the mucosal barrier (21). Haque and colleagues showed that the presence of Gal/GalNAc lectin-specific IgA antibodies in the stool correlated with reduced re-infection rates with E. histolytica in a study on susceptible children from Bangladesh (5, 22, 23). This implication was also confirmed with patients who had recovered from ALA. Increases in anti-Gal/GalNAc lectin IgA antibodies in post-ALA patients were associated with clearance of subsequent amebic infections, demonstrating that post-ALA patients developed a higher immune responsiveness and maintained immunological memory (24, 25). On the other hand, IgG levels have either protective or non-protective effects on the susceptibility to amebic infections depending upon major IgG subclasses induced by infection (i.e., IgG1 and IgG2 induced by Th2 and Th1, respectively) (26, 27).
Cell-Mediated Immunity
Cell-mediated immune responses are also important for host defense against E. histolytica. During the initial stage of infection, intestinal epithelial cells (IECs) bind to and recognize the carbohydrate recognition domain of the Gal/GalNAc lectin via toll-like receptor (TLR)-2/4, which activates NFκB and leads to the production of inflammatory cytokines, including IL-1β, IL-6, IL-8, IL-12, IFN-γ, and TNF-α (28–30). IECs are the second line of barriers against pathogens after the mucosal layer and the first line of host cells to encounter microbial/parasite antigens, they express an array of pathogen recognition receptors (PRRs), including TLRs (31). IFN-γ is involved in clearance of infection, whereas IL-4 and TNF-α are associated with disease (32–35). In fact, IFN-γ production by peripheral mononuclear cells was shown to be correlated with protection from future E. histolytica infection in children (36) and the serum level of IL-4 was high in patients with invasive amebiasis (27, 37). It has been also shown that IFN-γ-producing CD4+ T cells and IL-17-producing CD8+ T cells are involved protection in vaccinated mice (38, 39). IL-17 plays multiple roles in protection against amebic infection, including induction of secretion of mucin and antimicrobial peptides, increase in IgA transport across the intestinal epithelium, and promotion of neutrophil infiltration (40–43).
IFN-γ-activated neutrophils and macrophages have amebicidal activity in vitro (44, 45). In vivo, neutrophils predominated in amebic lesions where the macrophages were infrequent, suggesting importance of neutrophils for clearance of amebae (46). Production of reactive oxygen species (ROS) and nitric oxide (NO) via NAD(P)H oxidase complex and iNOS, respectively, play critical roles in killing trophozoites (45, 47). In experimental ALA, protection was mediated by IFN-γ from natural killer T cells (NKTs), while TNF-α-producing macrophages increased tissue damage (32, 33). Taken together, both humoral and cell-mediated immune responses play important roles against amebic infection.
Microbiota-Mediated Modifications of Parasite Virulence and Host Immune Response
Microbiota Affects Energy Metabolism and Growth of the Amebae
The adult human intestine has trillions of bacteria composed of more than hundreds of species. Recent studies have suggested that the intestinal bacterial microbiota may influence the outcome of protozoan infections (48, 49). The growth and survival of E. histolytica trophozoites depends on nutrients from the host and the microbiota. The bacterial microbiota produces glycosidases that degrade complex polysaccharides into forms available for the absorption by the amebae and the host (50). Microbial glycosidase activity determines the levels of free colonic carbohydrates (the glycobiome). Thus, microbiota potentially influences central energy metabolism of E. histolytica trophozoites. Since E. histolytica has numerous glycosidases, including amylases, β-hexosaminidases, and lysozymes encoded in its genome (51–55), and can degrade a panel of polysaccharides to yield monocarbohydrates, the activity and regulation of amebic glycosidases also influence available carbohydrate concentrations.
Microbiota Influences the Parasite’s Virulence
The commensal bacteria are generally protective against enteric pathogens. However, E. histolytica infection requires the presence of enteric bacteria. Germ-free animals were resistant to E. histolytica infection, but the introduction of a single bacterial species restored amebic pathogenesis (56, 57).
It has been shown that axenization (i.e., removal of associating bacteria) of xenically cultivated trophozoites decreases virulence, and incubation of axenic trophozoites with live bacteria restored virulence in a contact-dependent manner (58, 59). Incubation of E. histolytica trophozoites with the enteropathogenic Escherichia coli (EHEC) or Shigella dysenteriae increased amebic adherence to and cytotoxicity against MDCK cell monolayer (60). These observations indicate the enteric microbiota influence E. histolytica virulence during human infection (also see Perturbation of the Enteric Microbiota by E. histolytica).
The microbiota-dependent glycobiome has an emerging role in regulating the virulence of enteric pathogenic bacteria, such as EHEC, Clostridium difficile, and Salmonella enterica serovar Typhimurium (S. typhimurium) (61–63). EHEC has a fucose-responsive regulator of virulence genes, while C. difficile growth is promoted with high concentrations of free sialic acid reproduced by sialidase from associating bacteria (Bacteroides thetaiotaomicron). Similarly, the in vivo virulence of S. typhimurium was shown to be dependent on both fucose and sialic acid (63). It has been shown that glucose starvation enhances E. histolytica virulence, motility, and lectin expression via URE-3BP (64, 65). This finding suggests that the ameba has an ability to sense glucose (and possibly other monosugars) to modulate its virulence. The amebae and the bacterial microbiota influence each other by providing energy source and degrading available carbohydrates.
Microbiota Affects Host Immune Response
Gut microbiota plays a number of physiological roles involving digestion, metabolism, extraction of nutrients, synthesis of vitamins, prevention against colonization by pathogens, and immunomodulation (66–68). It has been demonstrated that Bacteroides fragilis and cluster XIV and IV of Clostridium species induce the development of regulatory T (Treg) cells in the colon (69, 70). Treg cells have the ability to suppress inflammatory responses through the production of anti-inflammatory cytokines, including IL-10 and TGF-β, and are considered to be involved in self-tolerance (71, 72). The beneficial effect of B. fragilis depends on the expression of polysaccharide A, which is a unique surface polysaccharide that binds to TLR2 on CD4+ T cells (72). Microbiota-mediated immunomodulation is evident in several diseases, e. g., Rheumatoid arthritis, diabetes, obesity, and cancer (73–78). It seems that E. histolytica requires the intestinal microbiota for pathogenesis, and, conversely, the parasite also needs to disrupt the homeostasis of the microbiota during infection.
Strategies for Immune Evasion of E. histolytica
Disruption of Host Physical Barriers and Soluble Immune Mediators by Hydrolases
Glycosidases
Hydrolases secreted by E. histolytica trophozoites are involved in the elimination of immune cells and degradation and/or activation of soluble immune mediators, as well as disruption of the host gut and liver epithelia (29, 55, 79–84). The mucosal layer between the lumen and the epithelia forms a physical barrier. Degradation of carbohydrates in the barrier is crucial for the initiation of colonization by the amebae. Human intestinal mucus is mainly composed of highly glycosylated mucins (85). Among >20 human mucins, MUC2 is the major gel-forming mucin secreted by goblet cells of the small and large intestines (86, 87). When the amebae colonize the colonic epithelia, they binds to secreted mucin oligosaccharides with the Gal/GalNAc lectin and penetrate through the mucosal layer. In this process, the amebae decompose the mucin barrier to finally reach and subsequently attach on IEC (13).
Secreted proteins by E. histolytica trophozoites displayed glycosidase activities, including β-N-acetyl-d-glucosaminidase, α-d-glucosidase, β-d-galactosidase, β-l-fucosidase, and α-N-acetyl-d-galactosaminidase (88). Among these glycosidases, β-N-acetyl-d-glucosaminidase showed the highest activity (88, 89). Thus, β-N-acetyl-d-glucosaminidase activity likely have a central role in degrading carbohydrates on mucin and exposing its protein backbone (88). It was previously demonstrated that the amount of intracellular and secreted β-N-acetyl-d-glucosaminidase activity increased by complement in the serum (90, 91). Huldt et al. also suggested that hexosaminidase activity plays a role in the amebic virulence (90).
Recently, it has been shown that knock down of a β-amylase gene by siRNA caused reduction in the degradation of the mucosal layer and the invasion into the human colon in an ex vivo experiment (55). Furthermore, the β-amylase gene was found upregulated after contact with colon tissues (55). Mucin degradation by amebic glycosidases may also affect the central metabolism of the amebae per se and also the microbiota equilibrium in the colon since highly glycosylated mucin is a carbon source for the amebae and the colonic microbiota (92, 93).
Cysteine Proteases
The E. histolytica genome has ~50 genes encoding CPs (20), which likely reflects robust biological importance of CPs. Of these, however, only four proteins, EhCP-A1, EhCP-A2, EhCP-A5, and EhCP-A7, are highly expressed under culture conditions and altogether account for more than 90% of the proteolytic activity in trophozoite extracts (94). After mucin was digested by amebic glycosidases, the protein backbone of mucin is degraded by robust CPs. Altogether, these mucin-digesting glycosidases and proteases are the ameba’s first line strategy to overcome the innate defense of the mucus barrier.
As suggested by various studies, among the four major CPs, EhCP-A5 appears to play a pivotal role in virulence, including immunomodulation (80, 95–102). EhCP-A5 has a capacity to bind integrin via the RGD motif in the pro region, and elicits pro-inflammatory response in Caco-2 cells in vitro and the murine colon via NLRP3 inflammasome activation independent of the CP activity (100, 102, 103). CPs are also known to modulate cell-mediated immunity by activating pro-inflammatory cytokines and also modulate humoral immunity (see below).
Involvement of Glycosidases and Proteases for Extraintestinal Propagation
When E. histolytica trophozoites propagate extraintestinally, they take a route similar to that of cancer metastasis (104), which requires both glycosidases and proteases for the disintegration of the basement membrane and entry into circulation (105–107). In case of ALA, amebic glycosidases and proteases are also needed to survive in the blood vessels (see Degradation of Immunoglobulins and Complements), and to destroy Kupffer cells, the epithelial cells, ECM, and hepatocytes in the liver. Thibeaux and colleagues have recently demonstrated that EhCP-A5 secreted from the amebae activates host matrix metalloproteases (MMP), a well-known mediator of ECM degradation (84). Recombinant EhCP-A5 restored the invasiveness of the EhCP-A5 gene-silenced trophozoites, suggesting that proteases from both the ameba and the host contribute to the tissue invasion process. In contrast to proteases, the roles of glycosidases in pathophysiology of amebiasis are not well demonstrated. It is evident in cancer metastasis that the level of serum β-hexosaminidase correlates with the likeliness of liver metastasis in variety of cancers, including colon, breast, stomach, pancreas, small bowel, kidney, testis, melanoma, lymphoma, and myeloma (108). Increased levels of tissue β-hexosaminidase were also reported for breast, kidney, pancreas, thyroid, colon, ovary, brain, salivary gland, stomach, and larynx cancers (109–112). Thus, it is conceivable by analogy that amebic glycosidases are involved in tissue invasion and extraintestinal dissemination.
Degradation of Immunoglobulins and Complements
As described above, the major component responsible for the intestinal immune response against amebic infection is secreted Igs. It was demonstrated that anti-Gal/GalNAc lectin IgA reduces trophozoite colonization in the colon (5, 23, 25, 113–117). Intriguingly, E. histolytica surface-associated CP [most likely EhCP-A5, (118)] cleaves human IgA (16, 119). Amebic CPs are capable of cleaving both isotypes, i.e., IgA1 and IgA2 (119, 120). Furthermore, amebic CPs can also inactivate circulating IgG and, thus, believed to be involved in the survival during tissue invasion and extraintestinal propagation (18). Degradation of IgG in the blood could prevent activation of the classical pathway of the complement system and immune cells that harbor Fc receptors (19).
When the trophozoites are exposed to the intravascular immune system, complements are the major component that mediates trophozoite destruction. E. histolytica trophozoites evade from a complement attack by cleaving and inactivating anaphylatoxins C5a and C3a with CPs (79). C5a and C3a are potent activators of inflammation and enhance the release of histamine from mast cells, lysosomal enzymes from leukocytes, and pro-inflammatory cytokines, including IL-6 and TNF-α, from macrophages (121–123). C5a and C3a also increase vascular permeability and attract immune cells (122, 123). Reduction of these anaphylatoxins detracts from immune detection of the amebae in the blood and reduces inflammation in amebic lesions. It also partially explains the lack of severe inflammation in advanced colitis and ALA region.
Degradation of Cytokines
Cysteine proteases are also known to modulate cell-mediated immunity by activating pro-inflammatory cytokine IL-1β and inactivation of pro- and mature IL-18 (82, 124). It is not concluded, however, if these changes are protective against or deleterious for amebic infection.
Cell Surface Decorations to Evade Host Immunity
Glycosylphosphatidylinositol-Anchored Proteins
Entamoeba histolytica is also capable of evading from complement attach by decorating their surface with glycosylphosphatidylinositol (GPI)-anchored proteins. GPI is a glycolipid required for anchoring many proteins and glycoconjugates to the cell surface in most of eukaryotes (125–127). E. histolytica trophozoites expose on their cell surface a complex GPI-anchored glycoconjugate, designated lipopeptidophosphoglycan (LPPG) (128, 129). LPPG on the cell surface is a component of glycocalyx that is composed of oligosaccharides of glycoproteins and glycolipids and afford trophozoites protection by creating an impervious layer to complement (130, 131). It was demonstrated that complement-susceptible Entamoeba dispar trophozoites possess a much thinner structure of LPPG-containing glycocalyx, which is consistent with the premise that LPPG is important for the evasion from complement (130). It is also known that antibody against human CD59, a cell surface protein that prevents auto-lysis by inhibiting the formation of the membrane attack complex (MAC) antibody cross-reacts with Gal/GalNAc lectin and a 21 kDa surface protein (132, 133). Later, it was shown that the Gal/GalNAc lectin contains a CD59-like region on the cell surfaces that prevents MAC formation (132). These data suggest that the Gal/GalNAc lectin is a cross-reactive CD59 homolog of the ameba and have a similar function as CD59. In agreement with these results, global inhibition of GPI-anchor formation leaves E. histolytica trophozoites susceptible to complement-mediated lysis (131). However, functionality of 21 kDa protein as an inhibitor of MAC formation and its molecular identity has yet to be elucidated.
Surface Receptor Capping
Surface receptor capping is another strategy to hide from the immune system by disposing of the surface molecules that have been recognized by Igs or complements (134, 135). During cell movement, surface-bound immune complexes are translocated toward the uroid, where capped ligands accumulate (136). This polar re-distribution can be induced by concanavalin A (Con A) or anti-amebic polyclonal antibodies (137). It has been reported that serine protease, E. histolytica rhomboid protease (ROM1), is involved in the translocation of the complex to the base of the caps and subsequent release of the materials in the cap (135, 138). It is of note that ROM1 also cleaves the transmembrane domain of the heavy subunit of the Gal/GalNAc lectin (138). As the lectin heavy subunit is highly immunogenic, its release from the plasma membrane by ROM1 may interfere with host immune response directed to amebae.
Killing and Phago/Togocytosis of Immune Cells
Contact-Dependent Cell Killing
Immobilization and killing of immune cells also serves as an ameba’s strategy for evasion from immune surveillance. Amebic trophozoites are able to kill a variety of cells, including neutrophils, T lymphocytes, macrophages, and a variety of tissue culture lines (116, 139–141). Adherence of the ameba triggers multiple intracellular events leading to cytotoxic effects to the mammalian cells. Such events include increased intracellular Ca2+, production of ROS, loss of membrane integrity, DNA fragmentation, phosphatidylserine exposure on the cell surface, and caspase-3 activation (116, 117, 139–144). It was reported that after host cell killing, E. histolytica preferentially ingest the dead cells (117, 140, 143). This observation is consistent with the theory that clearance of dead cells and debris by phagocytosis helps to minimize pro-inflammatory responses (145, 146). A phagocytosis-defective line of E. histolytica apparently showed decreased virulence in vitro and in vivo, suggesting a potential causal link between phagocytosis and virulence (147, 148).
Huston and colleagues demonstrated that E. histolytica preferentially ingests apoptotic Jurkat cells via recognition of phosphatidylserine and collectins (140, 149). Amebic calreticulin was found to be the surface receptor for host C1q, and required for phagocytosis of apoptotic cells, but it did not directly mediate cell killing (150). A few recent studies have started to unveil the detailed molecular mechanisms involved in the ameba phagocytosis (151, 152). However, the molecular events that take place in host immune cells in particular to suppress (or augment) immune response, together with a missing link between the surface receptor to the internalization machinery, remains totally unknown.
Trogocytosis
Ralston and colleagues have recently reported E. histolytica trophozoites ingested pieces of intact living cells via trogocytosis (“trogo” = nibbling) (153). When trophozoites were incubated with a combination of live and pre-killed host cells (Jurkat T cells), the live cells were ingested by trogocytosis, while the pre-killed host cells were ingested as a whole by canonical phagocytosis. Trogocytosis is an active process that resembles phagocytosis in some ways, i.e., it requires physiological temperature, actin rearrangements, Gal/GalNAc lectin, C2 domain-containing protein kinase, and phosphatidylinositol 3-phosphate kinase signaling, and it is accompanied with a rapid rise in intracellular Ca2+ concentrations. Trogocytosed host cells finally were killed. Trogocytosis of murine IEC was also evident in the in vivo animal model, suggesting that both trogocytosis of live host cells and phagocytosis of dead cells are important for pathogenesis and sustained parasitism of E. histolytica. Since amebic contact can potentially results in multiple outcomes: apoptosis and necrosis, followed by phagocytosis, or trogocytosis, it remains to be elucidated what factors and conditions differentiate these distinct manners of killing and ingestion of target host cells.
IFN-γ
Entamoeba histolytica regulates IFN-γ for survival in the host. In CBA mice, which are susceptible to E. histolytica cecal infection, the amebic infection led to upregulation of Th2 (IL-4, IL-5, and IL-13) and Th17 (IL-17) cytokine responses, while Th1 cytokines, IL-12p35 and IFN-γ, were suppressed (154). This indicates that suppression of INF-γ causes susceptibility of amebiasis. From cohort studies in Bangladesh, susceptible children with malnutrition showed lower IFN-γ levels (36, 155). Analysis of asymptomatic carriers of E. histolytica showed that carriers had higher levels of IFN-γ, while patients with invasive amebiasis displayed higher levels of IL-4 (35). The significance of IFN-γ in susceptibility is also implicated for ALA. It is known that more than 80% of all ALA cases occur in adult males (156–158), and the male predominance is attributable to testosterone (159). Lotter and colleagues showed that testosterone inhibits IFN-γ secretion from invariant natural killer T (iNKT) cells stimulated by LPPG, a physiological ligand for CD1d (159). iNKT cells are a subset of NKT cells that recognize lipid antigens in the context of CD1d and produce IFN-γ and IL-4. E. histolytica LPPG is presented on CD1d to invariant TCR and activates iNKT cells in combination with TLR signaling. αGalCer, a CD1d agonist, stimulates production of both IFN-γ and IL-4, whereas LPPG induces IFN-γ but not IL-4 production (33). These data suggest that iNKT cells provide a link between innate and adaptive immunity due to their capacity to produce large amounts of IFN-γ and IL-4 that can bias the immune response into either a Th1 or Th2 direction. Production of IFN-γ helps clearance of E. histolytica infection and controls abscess formation, whereas an adequate level of IFN-γ reduces the trophozoite number and pro-inflammatory response at a low level, and may balance for trophozoites to survive.
IL-10
It is known that anti-inflammatory cytokine, IL-10, plays a critical role to maintain the mucosal barrier. IL-10-deficient mice have compromised and highly permeable mucosal barriers and develop spontaneous intestinal inflammation in response to normal microflora (160). A murine amebic colitis model demonstrated that IL-10 from hematopoietic cells (CD4+ T cells) acting upon the non-hematopoietic compartment (IEC) is required for innate resistance to parasite invasion (161). Furthermore, it has been shown that IL-10 enhances MUC2 production, suppresses activation of antigen-presenting cells, induces B cell class-switching to IgA, has anti-apoptotic effects on IECs, reduces pro-inflammatory NFκB signaling in IECs, and promotes induction of CD4+ Treg cells (162–165). Interestingly, in asymptomatic carriers, no elevation of IL-10 level was observed. On the other hand, the IL-10 level was increased in dysenteric and ALA patients (27, 37). These studies indicate that invasion of the colon and liver by E. histolytica elicits an anti-inflammatory immune response and may successfully suppress immune reaction to the amebae. Altogether, the ameba needs to balance IL-10 and inflammatory cytokine levels to establish infection. It was shown that peritoneal monocytes and macrophages exposed to LPPG secreted TNF-α, IL-6, IL-8, IL-12, and IL-10 via TLR2 (166). It has been also shown that high doses of LPPG down-regulated TLR2 gene expression (166, 167). Thus, LPPG-driven signaling may activate a negative feedback loop that attenuates inflammatory responses. The mechanisms of the suppression of IL-10 production by the ameba remain to be elucidated (see below).
Suppression of NFκB in IECs
Entamoeba histolytica trophozoites secrete materials that induce a protective response in human IECs (168, 169), the first line of host cells to encounter microbial antigens, via PRRs, including TLRs. Upon binding to their ligand, PRRs trigger activation of a transcription factor NFκB. Gut homeostasis requires continuous activation of NFκB by TLR signaling in response to intestinal bacteria (170), commensal microbes can also disrupt NFκB signaling to attenuate pro-inflammatory IEC responses (171). It has been shown that secreted components from E. histolytica trophozoites induce a protective response in human IECs that primed by macrophage secretions through suppression of NFκB via heat shock protein response and increase resistance of IECs to apoptosis (168). Thus, it appears that E. histolytica elicits a stress response to IECs and promotes a hyporesponsive state toward trophozoites. The amebic factors that induce NFκB suppression have not yet determined. The factors that activate TLR2, i.e., LPPG and Gal/GalNAc lectin, are candidates involved in this pathway (172).
Prostaglandin PGE2
Entamoeba histolytica trophozoites produce and secrete prostaglandin 2 (PGE2), which have contact-independent effects on tight junction integrity and ion absorption. Secreted amebic PGE2 binds to prostaglandin E receptor 4 (EP4) on IECs, disrupts tight junctions, and increases luminal Cl− secretion (173, 174). PGE2 secreted from the amebae elicits inflammatory response in IECs by increasing IL-8 production by IECs (173). PGE2 is a potent mucin secretagogue (175) that can overcome luminal barrier function by causing hypersecretion and, thus, depletion of the protective mucus barrier (176). On the contrary, it has been also reported that during invasive amebiasis, local PGE2 has anti-inflammatory effect. In animal model of chronic ALA, hepatic granuloma macrophages do not respond to IFN-γ and LPS and do not produce inflammatory cytokines, show decrease in MHC class II expression, and are unable to kill trophozoites (47, 177, 178). This suppression is local during chronic ALA and is directly caused by the parasite (47, 177). A culture supernatant and an unknown soluble protein component of E. histolytica trophozoites decrease class II major histocompatibility complex (MHC II) immune-associated (Ia) antigen expression through a PGE2-dependent manner (178). Inhibition of macrophage PGE2 synthesis can partially recover MHC II Ia expression and TNF-α expression (177, 178). However, inhibition of PGE2 synthesis does not recover iNOS expression or amebicidal activity in the deactivated macrophage (177). A continuous supply of parasite-derived PGE2 likely prevents iNOS expression and full recovery of MHC II and TNF-α, possibly through a concentration-dependent effect of PGE2. In short, ameba-secreted PGE2 represses inflammation in ALA, which is beneficial for survival, whereas it likely enhances destruction of the colon.
Perturbation of the Enteric Microbiota by E. histolytica
It has been reported that E. histolytica infection alters the microbiota composition. E. histolytica-induced dysbiosis was characterized by fewer Bacteroides, Clostridia, Lactobacillus, Campylobacter, and Eubacterium species, and increased Bifidobacterium species (179). In vitro experiments have shown that E. histolytica preferentially ingest some bacterial species (59, 180). It is known that amoebapores, a family of the major pore-forming peptides, have differential activity against bacteria and eukaryotes (15). Furthermore, E. histolytica infection induces production of colonic antimicrobial peptides, while the trophozoites degrade them (181). A recent study has shown that dendritic cells from the mouse intestine where Clostridia-related bacteria colonized provide IL-17A-dependent protection against amebic colitis (182). Detailed molecular events remain to be elucidated, however, by examining how alternations of the microbiota modulate host immune responses against amebic intestinal infection. Altogether, microbiota can be modulated by amebic infection, and in turn concentrations of carbohydrates (and other compounds) that affect growth and virulence of the amebae can strongly influence outcome of infection. It remains to be elucidated whether and how the amebae modulate the intestinal microbiota for their survival and parasitism.
Strategy for Oxidative Stress Management and Metabolic Control
Lack of Respiration and Antioxidative Stress Management in E. histolytica
Entamoeba histolytica trophozoites are microaerophilic and consume oxygen. They tolerate low levels of oxygen tension. E. histolytica lacks a conventional respiratory electron transport chain that terminates in the reduction of O2 to H2O. However, it does respire and tolerates up to 5% oxygen in the gas phase (183–185). The parasite lacks most of the components of antioxidant defense mechanisms that are widely present in other prokaryotic and eukaryotic organisms, such as catalase, peroxidase, glutathione, and the glutathione-recycling enzymes glutathione peroxidase and glutathione reductase (184, 185). However, during tissue invasion, trophozoites must fend off reactive oxygen and nitrogen species produced by activated immune cells through the respiratory burst. Thus, trophozoites must use antioxidative stress defense to survive immune surveillance.
Anti-Oxidative Stress Response Contributes to Immune Evasion in E. histolytica
Entamoeba histolytica trophozoites contain high levels of cysteine, instead of glutathione, as the major thiol in the cell. They possess several enzymes to defend from oxidative stress, such as peroxiredoxin (Prx), superoxide dismutase, flavoprotein A, ferredoxin, thioredoxin (Trx), and Trx reductase (186, 187). The Trx/Trx reductase system is crucial for buffering sensitive proteins under oxidative stress (188). The amebicidal drugs, metronidazole and auranofin, are known to disrupt Trx (189, 190). Interestingly, the oxidative stress increases E. histolytica virulence. It has been shown that oxidative stress causes upregulation of a stress-induced adhesion factor and a phospholipid transporting P-type ATPase/flippase (187). Both genes are involved in adhesion and phagocytosis. Oxidative stress also alters metabolic flux, including glycerol and chitin biosynthesis, potentially triggering encystation (191). Furthermore, it has been shown that E. histolytica (HM-1:IMSS) responds more strongly to oxidative stress than E. dispar and E. histolytica non-virulent Rahman strain, and surface localization of Prx in HM-1:IMSS is associated with virulence (186). Altogether, antioxidative defense mechanisms in E. histolytica are associated with pathogenesis. For more details on the antioxidative management in E. histolytica, a recent review should be consulted (192).
Conclusion
Our understanding of molecular mechanisms of the parasite’s pathogenesis, such as adherence to host cells, induction of apoptosis, degradation of mucin and ECM, tissue invasion, and phago/trogocytosis of host cells, has greatly advanced in recent years. So have mechanisms of immune evasion, such as induction of IL-10 and suppression of INF-γ, degradation of Igs, complement, and pro-inflammatory cytokines. In addition, defense against ROS and NO and evasion from antibody and complement-dependent killing also plays important roles in survival in the host. Furthermore, mutual signaling among the three domains in the complex network of the parasite, the human, and the microbiota with polymorphic genetic backgrounds affect outcome of amebic infection. Further research is needed to elucidate the molecular basis of the complex interaction in the intestinal ecosystem.
Author Contributions
KN-T and TN have made substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (23117001, 23117005, 26293093, 26111524), a grant for research on emerging and re-emerging infectious diseases from Japan Agency for Medical Research and Development (AMED), a grant for Science and Technology Research Partnership for Sustainable Development (SATREPS) from AMED and Japan International Cooperation Agency (JICA) to TN.
References
1. Ackers J, Clark CG, Diamond LS, Duchêne M, Cantellano ME, Jackson TF, et al. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28-29 January, 1997. Epidemiol Bull (1997) 18:13–4.
2. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet (2012) 380:2095–128. doi: 10.1016/S0140-6736(12)61728-0
3. Ali IK, Clark CG, Petri WA Jr. Molecular epidemiology of amebiasis. Infect Genet Evol (2008) 8:698–707. doi:10.1016/j.meegid.2008.05.004
4. Ximénez C, Morán P, Rojas L, Valadez A, Gómez A. Reassessment of the epidemiology of amebiasis: state of the art. Infect Genet Evol (2009) 9:1023–32. doi:10.1016/j.meegid.2009.06.008
5. Haque R, Ali IM, Sack RB, Farr BM, Ramakrishnan G, Petri WA Jr. Amebiasis and mucosal IgA antibody against the Entamoeba histolytica adherence lectin in Bangladeshi children. J Infect Dis (2001) 183:1787–93. doi:10.1086/320740
6. Hung CC, Chang SY, Ji DD. Entamoeba histolytica infection in men who have sex with men. Lancet Infect Dis (2012) 12:729–36. doi:10.1016/S1473-3099(12)70147-0
7. Watanabe K, Gatanaga H, Escueta-de Cadiz A, Tanuma J, Nozaki T, Oka S. Amebiasis in HIV-1-infected Japanese men: clinical features and response to therapy. PLoS Negl Trop Dis (2011) 5:e1318. doi:10.1371/journal.pntd.0001318
8. Haghighi A, Kobayashi S, Takeuchi T, Masuda G, Nozaki T. Remarkable genetic polymorphism among Entamoeba histolytica isolates from a limited geographic area. J Clin Microbiol (2002) 40:4081–90. doi:10.1128/JCM.40.11.4081-4090.2002
9. Haghighi A, Kobayashi S, Takeuchi T, Thammapalerd N, Nozaki T. Geographic diversity among genotypes of Entamoeba histolytica field isolates. J Clin Microbiol (2003) 41:3748–56. doi:10.1128/JCM.41.8.3748-3756.2003
10. Haque R, Mondal D, Karim A, Molla IH, Rahim A, Faruque AS, et al. Prospective case-control study of the association between common enteric protozoal parasites and diarrhea in Bangladesh. Clin Infect Dis (2009) 48:1191–7. doi:10.1086/597580
11. Haque R, Huston CD, Hughes M, Houpt E, Petri WA Jr. Amebiasis. N Engl J Med (2003) 348:1565–73. doi:10.1056/NEJMra022710
12. Infectious Disease Surveillance Center, National Institute of Infectious Diseases of Japan. HIV/AIDS in Japan in the Year of 2013. IASR. Vol. 35. (2014). p. 223–4. Available from: http://www.nih.go.jp/niid/ja/id/969-disease-based/a/entamoeba-histolytica/idsc/iasr-in/4978-kj4151.html
13. Petri WA Jr, Haque R, Mann BJ. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu Rev Microbiol (2002) 56:39–64. doi:10.1146/annurev.micro.56.012302.160959
15. Bruhn H, Riekens B, Berninghausen O, Leippe M. Amoebapores and NK-lysin, members of a class of structurally distinct antimicrobial and cytolytic peptides from protozoa and mammals: a comparative functional analysis. Biochem J (2003) 375:737–44. doi:10.1042/BJ20030250
16. Kelsall BL, Ravdin JI. Degradation of human IgA by Entamoeba histolytica. J Infect Dis (1993) 168:1319–22. doi:10.1093/infdis/168.5.1319
17. Schulte W, Scholze H. Action of the major protease from Entamoeba histolytica on proteins of the extracellular matrix. J Protozool (1989) 36:538–43. doi:10.1111/j.1550-7408.1989.tb01092.x
18. Tran VQ, Herdman S, Torian BE, Reed SL. The neutral cysteine proteinase of Entamoeba histolytica degrades IgG and prevents its binding. J Infect Dis (1998) 177:508–11. doi:10.1086/517388
19. Que X, Reed SL. Cysteine proteinases and the pathogenesis of amebiasis. Clin Microbiol Rev (2000) 13:196–206. doi:10.1128/CMR.13.2.196-206.2000
20. Tillack M, Biller L, Irmer H, Freitas M, Gomes MA, Tannich E, et al. The Entamoeba histolytica genome: primary structure and expression of proteolytic enzymes. BMC Genomics (2007) 8:170. doi:10.1186/1471-2164-8-170
21. Lamm ME. Current concepts in mucosal immunity. IV. How epithelial transport of IgA antibodies relates to host defense. Am J Physiol (1998) 274:G614–7.
22. Haque R, Duggal P, Ali IM, Hossain MB, Mondal D, Sack RB, et al. Innate and acquired resistance to amebiasis in bangladeshi children. J Infect Dis (2002) 186:547–52. doi:10.1086/341566
23. Haque R, Mondal D, Duggal P, Kabir M, Roy S, Farr BM, et al. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect Immun (2006) 74:904–9. doi:10.1128/IAI.74.2.904-909.2006
24. Abd-Alla MD, Jackson TF, Rogers T, Reddy S, Ravdin JI. Mucosal immunity to asymptomatic Entamoeba histolytica and Entamoeba dispar infection is associated with a peak intestinal antilectin immunoglobulin A antibody response. Infect Immun (2006) 74:3897–903. doi:10.1128/IAI.02018-05
25. Ravdin JI, Abd-Alla MD, Welles SL, Reddy S, Jackson TF. Intestinal antilectin immunoglobulin A antibody response and immunity to Entamoeba dispar infection following cure of amebic liver abscess. Infect Immun (2003) 71:6899–905. doi:10.1128/IAI.71.12.6899-6905.2003
26. Kaur U, Sharma AK, Sharma M, Vohra H. Distribution of Entamoeba histolytica Gal/GalNAc lectin-specific antibody response in an endemic area. Scand J Immunol (2004) 60:524–8. doi:10.1111/j.0300-9475.2004.01512.x
27. Bernin H, Marggraff C, Jacobs T, Brattig N, Le VA, Blessmann J, et al. Immune markers characteristic for asymptomatically infected and diseased Entamoeba histolytica individuals and their relation to sex. BMC Infect Dis (2014) 14:621. doi:10.1186/s12879-014-0621-1
28. Gay NJ, Gangloff M. Structure and function of toll receptors and their ligands. Annu Rev Biochem (2007) 76:141–65. doi:10.1146/annurev.biochem.76.060305.151318
29. Bansal D, Ave P, Kerneis S, Frileux P, Boché O, Baglin AC, et al. An ex-vivo human intestinal model to study Entamoeba histolytica pathogenesis. PLoS Negl Trop Dis (2009) 3:e551. doi:10.1371/journal.pntd.0000551
30. Galván-Moroyoqui JM, Del CarmenDomínguez-Robles M, Meza I. Pathogenic bacteria prime the induction of toll-like receptor signalling in human colonic cells by the Gal/GalNAc lectin carbohydrate recognition domain of Entamoeba histolytica. Int J Parasitol (2011) 41:1101–12. doi:10.1016/j.ijpara.2011.06.003
31. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol (2011) 30:16–34. doi:10.3109/08830185.2010.529976
32. Helk E, Bernin H, Ernst T, Ittrich H, Jacobs T, Heeren J, et al. TNFα-mediated liver destruction by Kupffer cells and Ly6Chi monocytes during Entamoeba histolytica infection. PLoS Pathog (2013) 9:e1003096. doi:10.1371/journal.ppat.1003096
33. Lotter H, González-Roldán N, Lindner B, Winau F, Isibasi A, Moreno-Lafont M, et al. Natural killer T cells activated by a lipopeptidophosphoglycan from Entamoeba histolytica are critically important to control amebic liver abscess. PLoS Pathog (2009) 5(5):e1000434. doi:10.1371/journal.ppat.1000434
34. Peterson KM, Shu J, Duggal P, Haque R, Mondal D, Petri WA Jr. Association between TNF-α and Entamoeba histolytica diarrhea. Am J Trop Med Hyg (2010) 82:620–5. doi:10.4269/ajtmh.2010.09-0493
35. Sánchez-Guillén Mdel C, Pérez-Fuentes R, Salgado-Rosas H, Ruiz-Argüelles A, Ackers J, Shire A, et al. Differentiation of Entamoeba histolytica/Entamoeba dispar by PCR and their correlation with humoral and cellular immunity in individuals with clinical variants of amoebiasis. Am J Trop Med Hyg (2002) 66:731–7.
36. Haque R, Mondal D, Shu J, Roy S, Kabir M, Davis AN, et al. Correlation of interferon-γ production by peripheral blood mononuclear cells with childhood malnutrition and susceptibility to amebiasis. Am J Trop Med Hyg (2007) 76:340–4.
37. Bansal D, Sehgal R, Chawla Y, Malla N, Mahajan RC. Cytokine mRNA expressions in symptomatic vs. asymptomatic amoebiasis patients. Parasite Immunol (2005) 27:37–43. doi:10.1111/j.1365-3024.2005.00739.x
38. Guo X, Barrosom L, Becker SM, Lyerly DM, Vedvick TS, Reed SG, et al. Protection against intestinal amebiasis by a recombinant vaccine is transferable by T cells and mediated by gamma interferon. Infect Immun (2009) 77:3909–18. doi:10.1128/IAI.00487-09
39. Guo X, Barroso L, Lyerly DM, Petri WA Jr, Houpt ER. CD4+ and CD8+ T cell- and IL-17-mediated protection against Entamoeba histolytica induced by a recombinant vaccine. Vaccine (2011) 29:772–7. doi:10.1016/j.vaccine.2010.11.013
40. Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med (2006) 203(10):2271–9. doi:10.1084/jem.20061308
41. Nishida A, Lau CW, Zhang M, Andoh A, Shi HN, Mizoguchi E, et al. The membrane-bound mucin Muc1 regulates T helper 17-cell responses and colitis in mice. Gastroenterology (2012) 142:865–74.e2. doi:10.1053/j.gastro.2011.12.036
42. Cao AT, Yao S, Gong B, Elson CO, Cong Y. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol (2012) 189:4666–73. doi:10.4049/jimmunol.1200955
43. Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol (2010) 30:196–203. doi:10.1007/s10875-010-9368-7
44. Ghadirian E, Denis M. In vivo activation of macrophages by IFN-γ to kill Entamoeba histolytica trophozoites in vitro. Parasite Immunol (1992) 14:397–404. doi:10.1111/j.1365-3024.1992.tb00014.x
45. Lin JY, Chadee K. Macrophage cytotoxicity against Entamoeba histolytica trophozoites is mediated by nitric oxide from L-arginine. J Immunol (1992) 148:3999–4005.
46. Espinosa-Cantellano M, Martínez-Palomo A. Pathogenesis of intestinal amebiasis: from molecules to disease. Clin Microbiol Rev (2000) 13:318–31. doi:10.1128/CMR.13.2.318-331.2000
47. Denis M, Chadee K. Cytokine activation of murine macrophages for in vitro killing of Entamoeba histolytica trophozoites. Infect Immun (1989) 57:1750–6.
48. Berrilli F, Di Cave D, Cavallero S, D’Amelio S. Interactions between parasites and microbial communities in the human gut. Front Cell Infect Microbiol (2012) 2:141. doi:10.3389/fcimb.2012.00141
49. Gilchrist CA, Petri SE, Schneider BN, Reichman DJ, Jiang N, Begum S, et al. Role of the gut microbiota of children in diarrhea due to the protozoan parasite Entamoeba histolytica. J Infect Dis (2015). doi:10.1093/infdis/jiv772
50. Ouwerkerk JP, de Vos WM, Belzer C. Glycobiome: bacteria and mucus at the epithelial interface. Best Pract Res Clin Gastroenterol (2013) 27:25–38. doi:10.1016/j.bpg.2013.03.001
51. Nakada-Tsukui K, Tsuboi K, Furukawa A, Yamada Y, Nozaki T. A novel class of cysteine protease receptors that mediate lysosomal transport. Cell Microbiol (2012) 14:1299–317. doi:10.1111/j.1462-5822.2012.01800.x
52. Furukawa A, Nakada-Tsukui K, Nozaki T. Novel transmembrane receptor involved in phagosome transport of lysozymes and β-hexosaminidase in the enteric protozoan Entamoeba histolytica. PLoS Pathog (2012) 8:e1002539. doi:10.1371/journal.ppat.1002539
53. Furukawa A, Nakada-Tsukui K, Nozaki T. Cysteine protease-binding protein family 6 mediates the trafficking of amylases to phagosomes in the enteric protozoan Entamoeba histolytica. Infect Immun (2013) 81:1820–9. doi:10.1128/IAI.00915-12
54. Marumo K, Nakada-Tsukui K, Tomii K, Nozaki T. Ligand heterogeneity of the cysteine protease binding protein family in the parasitic protist Entamoeba histolytica. Int J Parasitol (2014) 44:625–35. doi:10.1016/j.ijpara.2014.04.008
55. Thibeaux R, Weber C, Hon CC, Dillies MA, Avé P, Coppée JY, et al. Identification of the virulence landscape essential for Entamoeba histolytica invasion of the human colon. PLoS Pathog (2013) 9:e1003824. doi:10.1371/journal.ppat.1003824
56. Phillips BP, Wolfe PA, Rees CW, Gordon HA, Wright WH, Reyniers JA. Studies on the amebabacteria relationship in amebiasis: comparative results of the intracecal inoculation of germfree, monocontaminated, and conventional guinea pigswith Entamoeba histolytica. Am J Trop Med Hyg (1955) 4:675–92.
57. Phillips BP, Gorstein F. Effects of different species of bacteria on the pathology of enteric amebiasis in monocontaminated guinea pigs. Am J Trop Med Hyg (1966) 15:863–8.
58. Wittner M, Rosenbaum RM. Role of bacteria in modifying virulence of Entamoeba histolytica: studies of amebae from axenic cultures. Am J Trop Med Hyg (1970) 19:755–61.
59. Mirelman D, Feingold C, Wexler A, Bracha R. Interactions between Entamoeba histolytica, bacteria and intestinal cells. Ciba Found Symp (1983) 99:2–30.
60. Galván-Moroyoqui JM, Del Carmen Domínguez-Robles M, Franco E, Meza I. The interplay between Entamoeba and enteropathogenic bacteria modulates epithelial cell damage. PLoS Negl Trop Dis (2008) 2:e266. doi:10.1371/journal.pntd.0000266
61. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes (2012) 3:289–306. doi:10.4161/gmic.19897
62. Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, et al. Fucose sensing regulates bacterial intestinal colonization. Nature (2012) 492:113–7. doi:10.1038/nature11623
63. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature (2013) 502:96–9. doi:10.1038/nature12503
64. Tovy A, Hertz R, Siman-Tov R, Syan S, Faust DM, Guillen N, et al. Glucose starvation boosts Entamoeba histolytica virulence. PLoS Negl Trop Dis (2011) 5:e1247. doi:10.1371/journal.pntd.0001247
65. Gilchrist CA, Baba DJ, Zhang Y, Crasta O, Evans C, Caler E, et al. Targets of the Entamoeba histolytica transcription factor URE3-BP. PLoS Negl Trop Dis (2008) 2:e282. doi:10.1371/journal.pntd.0000282
66. Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, et al. Energy-balance studies reveal associations between gut microbes, caloricload, and nutrient absorption in humans. Am J Clin Nutr (2011) 94:58–65. doi:10.3945/ajcn.110.010132
67. Purchiaroni F, Tortora A, Gabrielli M, Bertucci F, Gigante G, Ianiro G, et al. The role of intestinal microbiota and the immune system. Eur Rev Med Pharmacol Sci (2013) 17:323–33.
68. Donia MS, Fischbach MA. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science (2015) 349:1254766. doi:10.1126/science.1254766
69. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature (2008) 453:620–5. doi:10.1038/nature07008
70. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science (2011) 331:337–41. doi:10.1126/science.1198469
71. Sakaguchi S. The origin of FOXP3-expressing CD4+ regulatory T cells: thymus or periphery. J Clin Invest (2003) 112:1310–2. doi:10.1172/JCI200320274
72. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science (2011) 332:974–7. doi:10.1126/science.1206095
73. Hot A, Miossec P. Effects of interleukin (IL)-17A and IL-17F in human rheumatoid arthritis synoviocytes. Ann Rheum Dis (2011) 70:727–32. doi:10.1136/ard.2010.143768
74. Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol (2011) 7:569–78. doi:10.1038/nrrheum.2011.121
75. Romano-Keeler J, Weitkamp JH, Moore DJ. Regulatory properties of the intestinal microbiome effecting the development and treatment of diabetes. Curr Opin Endocrinol Diabetes Obes (2012) 19:73–80. doi:10.1097/MED.0b013e3283514d43
76. Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes (2012) 3:4–14. doi:10.4161/gmic.19320
77. Jin C, Henao-Mejia J, Flavell RA. Innate immune receptors: key regulators of metabolic disease progression. Cell Metab (2013) 17:873–82. doi:10.1016/j.cmet.2013.05.011
78. Erdman SE, Poutahidis T. Gut bacteria and cancer. Biochim Biophys Acta (2015) 1856:86–90. doi:10.1016/j.bbcan.2015.05.007
79. Reed SL, Ember JA, Herdman DS, DiScipio RG, Hugli TE, Gigli I. The extracellular neutral cysteine proteinase of Entamoeba histolytica degrades anaphylatoxins C3a and C5a. J Immunol (1995) 155:266–74.
80. Ankri S, Stolarsky T, Bracha R, Padilla-Vaca F, Mirelman D. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect Immun (1999) 67:421–2.
82. Que X, Kim SH, Sajid M, Eckmann L, Dinarello CA, McKerrow JH, et al. A surface amebic cysteine proteinase inactivates interleukin-18. Infect Immun (2003) 71:1274–80. doi:10.1128/IAI.71.3.1274-1280.2003
83. Kissoon-Singh V, Mortimer L, Chadee K. Entamoeba histolytica cathepsin-like enzymes: interactions with the host gut. Adv Exp Med Biol (2011) 712:62–83. doi:10.1007/978-1-4419-8414-2_5
84. Thibeaux R, Avé P, Bernier M, Morcelet M, Frileux P, Guillén N, et al. The parasite Entamoeba histolytica exploits the activities of human matrix metalloproteinases to invade colonic tissue. Nat Commun (2014) 5:5142. doi:10.1038/ncomms6142
85. Corfield AP. Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim Biophys Acta (2015) 1850:236–52. doi:10.1016/j.bbagen.2014.05.003
86. Carlstedt I, Herrmann A, Karlsson H, Sheehan J, Fransson L, Hansson G. Characterization of two different glycosylated domains from the insoluble mucin complex of rat small intestine. J Biol Chem (1993) 268:18771–81.
87. Herrmann A, Davies JR, Lindell G, Martensson S, Packer NH, Swallow DM, et al. Studies on the “insoluble” glycoprotein complex from human colon. Identification of reduction-insensitive MUC2 oligomers and C-terminal cleavage. J Biol Chem (1999) 274:15828–36. doi:10.1074/jbc.274.22.15828
88. Moncada D, Keller K, Chadee K. Entamoeba histolytica-secreted products degrade colonic mucin oligosaccharides. Infect Immun (2005) 73:3790–3. doi:10.1128/IAI.73.6.3790-3793.2005
89. Connaris S, Greenwell P. Glycosidases in mucin-dwelling protozoans. Glycoconj J (1997) 14:879–82. doi:10.1023/A:1018554408558
90. Huldt G, Davies P, Allison AC, Schorlemmer HU. Interactions between Entamoeba histolytica and complement. Nature (1979) 18:214–6. doi:10.1038/277214a0
91. Arias-Negrete S, Sabanero-López M, Villagómez-Castro JC. Biochemical analysis of Entamoeba histolytica HM1 strain resistant to complement lysis. Arch Med Res (1992) 23:135–7.
92. Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science (2005) 307:1955–9. doi:10.1126/science.1109051
93. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464:59–65. doi:10.1038/nature08821
94. Irmer H, Tillack M, Biller L, Handal G, Leippe M, Roeder T, et al. Major cysteine peptidases of Entamoeba histolytica are required for aggregation and digestion of erythrocytes but are dispensable for phagocytosisand cytopathogenicity. Mol Microbiol (2009) 72:658–67. doi:10.1111/j.1365-2958.2009.06672.x
95. Bruchhaus I, Jacobs T, Leippe M, Tannich E. Entamoeba histolytica and Entamoeba dispar: differences in numbers and expression of cysteine proteinase genes. Mol Microbiol (1996) 22:255–63. doi:10.1046/j.1365-2958.1996.00111.x
96. Willhoeft U, Hamann L, Tannich E. A DNA sequence corresponding to the gene encoding cysteine proteinase 5 in Entamoeba histolytica is present and positionally conserved but highly degenerated in Entamoeba dispar. Infect Immun (1999) 67:5925–9.
97. Hellberg A, Nickel R, Lotter H, Tannich E, Bruchhaus I. Overexpression of cysteine proteinase 2 in Entamoeba histolytica or Entamoeba dispar increases amoeba-induced monolayer destruction in vitro but does not augment amoebic liver abscess formation in gerbils. Cell Microbiol (2001) 3:13–20. doi:10.1046/j.1462-5822.2001.00086.x
98. Tillack M, Nowak N, Lotter H, Bracha R, Mirelman D, Tannich E, et al. Increased expression of the major cysteine proteinases by stable episomal transfection underlines the important role of EhCP5 for the pathogenicity of Entamoeba histolytica. Mol Biochem Parasitol (2006) 149:58–64. doi:10.1016/j.molbiopara.2006.04.009
99. Freitas MA, Fernandes HC, Calixto VC, Martins AS, Silva EF, Pesquero JL, et al. Entamoeba histolytica: cysteine proteinase activity and virulence. Focus on cysteine proteinase 5 expression levels. Exp Parasitol (2009) 122:306–9. doi:10.1016/j.exppara.2009.04.005
100. Hou Y, Mortimer L, Chadee K. Entamoeba histolytica cysteine proteinase 5 binds integrin on colonic cells and stimulates NFkappaB-mediated pro-inflammatory responses. J Biol Chem (2010) 285:35497–504. doi:10.1074/jbc.M109.066035
101. Thibeaux R, Dufour A, Roux P, Bernier M, Baglin AC, Frileux P, et al. Newly visualized fibrillar collagen scaffolds dictate Entamoeba histolytica invasion route in the human colon. Cell Microbiol (2012) 14:609–21. doi:10.1111/j.1462-5822.2012.01752.x
102. Mortimer L, Moreaum F, Cornick S, Chadee K. The NLRP3 Inflammasome is a pathogen sensor for invasive Entamoeba histolytica via activation of α5β1 integrin at the macrophage-amebae intercellular junction. PLoS Pathog (2015) 11:e1004887. doi:10.1371/journal.ppat.1004887
103. Mortimer L, Moreau F, Cornick S, Chadee K. Gal-lectindependent contact activates the inflammasome by invasive Entamoeba histolytica. Mucosal Immunol (2014) 7:829–41. doi:10.1038/mi.2013.100
104. Leroy A, Mareel M, De Bruyne G, Bailey G, Nelis H. Metastasis of Entamoeba histolytica compared to colon cancer: one more step in invasion. Invasion Metastasis (1995) 14:177–91.
105. Bernacki RJ, Niedbala MJ, Korytnyk W. Glycosidases in cancer and invasion. Cancer Metastasis Rev (1985) 4:81–101. doi:10.1007/BF00047738
106. Liotta LA. Tumor invasion and metastases: role of the basement membrane. Am J Pathol (1984) 117:339–48.
107. Coghlin C, Murray GI. Current and emerging concepts in tumour metastasis. J Pathol (2010) 222:1–15. doi:10.1002/path.2727
108. Plucinsky MC, Prorok JJ, Alhadeff JA. Variant serum beta-hexosaminidase as a biochemical marker of malignancy. Cancer (1986) 58:1484–7. doi:10.1002/1097-0142(19861001)58:7<1484:AID-CNCR2820580718>3.0.CO;2-R
109. Bosmann HB, Hall TC. Enzyme activity in invasive tumors of human breast and colon. Proc Natl Acad Sci USA (1974) 71:1833–7. doi:10.1073/pnas.71.5.1833
110. Gil-Martín E, Rodríguez-Berrocal J, Páez de la Cadena M, Fernández-Briera A. Alterations of glycosidases in human colonic adenocarcinoma. Clin Biochem (1997) 30:17–25. doi:10.1016/S0009-9120(96)00123-3
111. Ramessur KT, Greenwell P, Nash R, Dwek MV. Breast cancer invasion is mediated by beta-N-acetylglucosaminidase (beta-NAG) and associated with a dysregulation in the secretory pathway of cancer cells. Br J Biomed Sci (2010) 67:189–96.
112. Choromańska B, Luto M, Szajda SD, Waszkiewicz N, Kępka A, Janica J, et al. Activity of N-acetyl-β-hexosaminidase and its isoenzymes A and B in cancer. Postepy Hig Med Dosw (Online) (2011) 65:752–8. doi:10.5604/17322693.966833
113. Chadee K, Petri WA Jr, Innes DJ, Ravdin JI. Rat and human colonic mucins bind to and inhibit adherence lectin of Entamoeba histolytica. J Clin Invest (1987) 80:1245–54. doi:10.1172/JCI113199
114. Saffer LD, Petri WA Jr. Role of the galactose lectin of Entamoeba histolytica in adherence-dependent killing of mammalian cells. Infect Immun (1991) 59:4681–3.
115. Guerrant RL, Brush J, Ravdin JI, Sullivan JA, Mandell GL. Interaction between Entamoeba histolytica and human polymorphonuclear neutrophils. J Infect Dis (1981) 143:83–93. doi:10.1093/infdis/143.1.83
116. Ravdin JI, Moreau F, Sullivan JA, Petri WA Jr, Mandell GL. Relationship of free intracellular calcium to the cytolytic activity of Entamoeba histolytica. Infect Immun (1988) 56:505–12.
117. Huston CD, Boettner DR, Miller-Sims V, Petri WA Jr. Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect Immun (2003) 71:964–72. doi:10.1128/IAI.71.2.964-972.2003
118. Jacobs T, Bruchhaus I, Dandekar T, Tannich E, Leippe M. Isolation and molecular characterization of a surface-bound proteinase of Entamoeba histolytica. Mol Microbiol (1998) 27:269–76. doi:10.1046/j.1365-2958.1998.00662.x
119. Garcia-Nieto RM, Rico-Mata R, Arias-Negrete S, Avila EE. Degradation of human secretory IgA1 and IgA2 by Entamoeba histolytica surface-associated proteolytic activity. Parasitol Int (2008) 57:417–23. doi:10.1016/j.parint.2008.04.013
120. Macpherson AJ, McCoy KD, Johansen F, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol (2008) 1:11–22. doi:10.1038/mi.2007.6
121. Walport MJ. Complement: second of two parts. N Engl J Med (2001) 344:1140–4. doi:10.1056/NEJM200104123441506
122. Köhl J. Anaphylatoxins and infectious and non-infectious inflammatory diseases. Mol Immunol (2001) 38:175–87. doi:10.1016/S0161-5890(01)00041-4
123. Gasque P. Complement: a unique innate immune sensor for danger signals. Mol Immunol (2004) 41:1089–98. doi:10.1016/j.molimm.2004.06.011
124. Zhang Z, Wang L, Seydel KB, Li E, Ankri S, Mirelman D, et al. Entamoeba histolytica cysteine proteinases with interleukin-1 beta converting enzyme (ICE) activity cause intestinal inflammation and tissue damage in amoebiasis. Mol Microbiol (2000) 37:542–8. doi:10.1046/j.1365-2958.2000.02037.x
125. Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol (2004) 5:110–20. doi:10.1038/nrm1309
126. Fujita M, Kinoshita T. GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim Biophys Acta (2012) 1821:1050–8. doi:10.1016/j.bbalip.2012.01.004
127. Saha S, Anilkumar AA, Mayor S. GPI-anchored protein organization and dynamics at the cell surface. J Lipid Res (2015). doi:10.1194/jlr.R062885
128. Isibasi A, Santa Cruz M, Soto Montano X, Ramirez A, Kumate J. Localization of a lipopeptidophosphoglycan extracted by phenol-water from trophozoites of the HK-9 strain of Entamoeba histolytica. Arch Invest Med (Mex) (1982) 13:57–62.
129. Guha-Niyogi A, Sullivan DR, Turco SJ. Glycoconjugate structures of parasitic protozoa. Glycobiology (2001) 11:45R–59R. doi:10.1093/glycob/11.4.45R
130. Bhattacharya A, Arya R, Clark CG, Ackers JP. Absence of lipophosphoglycan-like glycoconjugates in Entamoeba dispar. Parasitology (2000) 120:31–5. doi:10.1017/S0031182099005259
131. Weber C, Blazquez S, Marion S, Ausseur C, Vats D, Krzeminski M, et al. Bioinformatics and functional analysis of an Entamoeba histolytica mannosyltransferase necessary for parasite complement resistance and hepatical infection. PLoS Negl Trop Dis (2008) 2:e165. doi:10.1371/journal.pntd.0000165
132. Braga L, Ninomiya H, McCoy JJ, Eacker S, Wiedmer T, Pham C, et al. Inhibition of the complement membrane attack complex by the galactose-specific adhesion of Entamoeba histolytica. J Clin Invest (1992) 90:1131–7. doi:10.1172/JCI115931
133. Ventura-Juárez J, Campos-Rodríguez R, Jarillo-Luna RA, Muñoz-Fernández L, Escario-G-Trevijano JA, Pérez-Serrano J, et al. Trophozoites of Entamoeba histolytica express a CD59-like molecule in human colon. Parasitol Res (2009) 104:821–6. doi:10.1007/s00436-008-1262-3
134. Calderón J, de Lourdes Muñoz M, Acosta HM. Surface redistribution and release of antibody-induced caps in entamoebae. J Exp Med (1980) 151:184–93. doi:10.1084/jem.151.1.184
135. Baxt LA, Baker RP, Singh U, Urban S. An Entamoeba histolytica rhomboid protease with atypical specificity cleaves a surface lectin involved in phagocytosis and immune evasion. Genes Dev (2008) 22:1636–46. doi:10.1101/gad.1667708
136. Bailey GB, Day DB, McCoomer NE. Entamoeba motility: dynamics of cytoplasmic streaming, locomotion and translocation of surface bound particles, and organization of the actin cytoskeleton in Entamoeba invadens. J Protozool (1992) 39:267–72. doi:10.1111/j.1550-7408.1992.tb01313.x
137. Espinosa-Cantellano M, Martínez-Palomo A. Entamoeba histolytica: mechanism of surface receptor capping. Exp Parasitol (1994) 79:424–35. doi:10.1006/expr.1994.1104
138. Baker RP, Wijetilaka R, Urban S. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathog (2006) 2:e113. doi:10.1371/journal.ppat.0020113
139. Ragland BD, Ashley LS, Vaux DL, Petri WA Jr. Entamoeba histolytica: target cells killed by trophozoites undergo DNA fragmentation which is not blocked by Bcl-2. Exp Parasitol (1994) 79:460–7. doi:10.1006/expr.1994.1107
140. Huston CD, Houpt ER, Mann BJ, Hahn CS, Petri WA Jr. Caspase 3-dependent killing of host cells by the parasite Entamoeba histolytica. Cell Microbiol (2000) 2:617–25. doi:10.1046/j.1462-5822.2000.00085.x
141. Sim S, Yong TS, Park SJ, Im KI, Kong Y, Ryu JS, et al. NADPH oxidase-derived reactive oxygen species-mediated activation of ERK1/2 is required for apoptosis of human neutrophils induced by Entamoeba histolytica. J Immunol (2005) 174:4279–88. doi:10.4049/jimmunol.174.7.4279
142. Berninghausen O, Leippe M. Necrosis versus apoptosis as the mechanism of target cell death induced by Entamoeba histolytica. Infect Immun (1997) 65:3615–21.
143. Boettner DR, Huston CD, Linford AS, Buss SN, Houpt E, Sherman NE, et al. Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS Pathog (2008) 4:e8. doi:10.1371/journal.ppat.0040008
144. Ralston KS, Petri WA Jr. Tissue destruction and invasion by Entamoeba histolytica. Trends Parasitol (2011) 27:254–63. doi:10.1016/j.pt.2011.02.006
145. Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest (1998) 101:890–8. doi:10.1172/JCI1112
146. Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol (2014) 14:166–80. doi:10.1038/nri3607
147. Orozco E, Guarneros G, Martinez-Palomo A, Sánchez T. Entamoeba histolytica: phagocytosis as a virulence factor. J Exp Med (1983) 158:1511–21. doi:10.1084/jem.158.5.1511
148. Rodríguez MA, Orozco E. Isolation and characterization of phagocytosis- and virulence-deficient mutants of Entamoeba histolytica. J Infect Dis (1986) 154:27–32. doi:10.1093/infdis/154.1.27
149. Teixeira JE, Heron BT, Huston CD. C1q- and collectin-dependent phagocytosis of apoptotic host cells by the intestinal protozoan Entamoeba histolytica. J Infect Dis (2008) 198:1062–70. doi:10.1086/591628
150. Vaithilingam A, Teixeira JE, Miller PJ, Heron BT, Huston CD. Entamoeba histolytica cell surface calreticulin binds human C1q and functions in amebic phagocytosis of host cells. Infect Immun (2012) 80:2008–18. doi:10.1128/IAI.06287-11
151. Somlata, Bhattacharya S, Bhattacharya A. A C2 domain protein kinase initiates phagocytosis in the protozoan parasite Entamoeba histolytica. Nat Commun (2011) 2:230. doi:10.1038/ncomms1199
152. Mansuri MS, Bhattacharya S, Bhattacharya A. A novel alpha kinase EhAK1 phosphorylates actin and regulates phagocytosis in Entamoeba histolytica. PLoS Pathog (2014) 10:e1004411. doi:10.1371/journal.ppat.1004411
153. Ralston KS, Solga MD, Mackey-Lawrence NM, Somlata, Bhattacharya A, Petri WA Jr. Trogocytosis by Entamoeba histolytica contributes to cell killing and tissue invasion. Nature (2014) 508:526–30. doi:10.1038/nature13242
154. Guo X, Stroup SE, Houpt E. Persistence of Entamoeba histolytica infection in CBA mice owes to intestinal IL-4 production and inhibition of protective IFN-γ. Mucosal Immunol (2008) 1:139–46. doi:10.1038/mi.2007.18
155. Petri WA Jr, Mondal D, Peterson KM, Duggal P, Haque R. Association of malnutrition with amebiasis. Nutr Rev (2009) 67:S207–15. doi:10.1111/j.1753-4887.2009.00242.x
156. Acuna-Soto R, Maguire JH, Wirth DF. Gender distribution in asymptomatic and invasive amebiasis. Am J Gastroenterol (2000) 95:1277–83. doi:10.1111/j.1572-0241.2000.01525.x
157. Blessmann J, Van Linh P, Nu PA, Thi HD, Muller-Myhsok B, Buss H, et al. Epidemiology of amebiasis in a region of high incidence of amebic liver abscess in central Vietnam. Am J Trop Med Hyg (2002) 66:578–83.
158. Blessmann J, Ali IK, Nu PA, Dinh BT, Viet TQ, Van AL, et al. Longitudinal study of intestinal Entamoeba histolytica infections in asymptomatic adult carriers. J Clin Microbiol (2003) 41:4745–50. doi:10.1128/JCM.41.10.4745-4750.2003
159. Lotter H, Helk E, Bernin H, Jacobs T, Prehn C, Adamski J, et al. Testosterone increases susceptibility to amebic liver abscess in mice and mediates inhibition of IFNγ secretion in natural killer T cells. PLoS One (2013) 8:e55694. doi:10.1371/journal.pone.0055694
160. Madsen KL, Malfair D, Gray D, Doyle JS, Jewell LD, Fedorak RN. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm Bowel Dis (1999) 5:262–70. doi:10.1097/00054725-199911000-00004
161. Hamano S, Asgharpour A, Stroup SE, Wynn TA, Leiter EH, Houpt E. Resistance of C57BL/6 mice to amoebiasis is mediated by nonhemopoietic cells but requires hemopoietic IL-10 production. J Immunol (2006) 177:1208–13. doi:10.4049/jimmunol.177.2.1208
162. Ruiz PA, Shkoda A, Kim SC, Sartor RB, Haller D. IL-10 gene-deficient mice lack TGF-beta/Smad signaling and fail to inhibit proinflammatory gene expression in intestinal epithelial cells after the colonization with colitogenic Enterococcus faecalis. J Immunol (2005) 174:2990–9. doi:10.4049/jimmunol.174.5.2990
163. Schwerbrock NM, Makkink MK, van der Sluis M, Buller HA, Einerhand AW, Sartor RB, et al. Interleukin 10-deficient mice exhibit defective colonic Muc2 synthesis before and after induction of colitis by commensal bacteria. Inflamm Bowel Dis (2004) 10:811–23. doi:10.1097/00054725-200411000-00016
164. Grazia Roncarolo M, Bacchetta R, Bordignon C, Narula S, Levings MK. Type 1 T regulatory cells. Immunol Rev (2001) 182:68–79. doi:10.1034/j.1600-065X.2001.1820105.x
165. Zhou P, Streutker C, Borojevic R, Wang Y, Croitoru K. IL-10 modulates intestinal damage and epithelial cell apoptosis in T cell-mediated enteropathy. Am J Physiol (2004) 287:G599–604. doi:10.1152/ajpgi.00063.2004
166. Maldonaldo-Bernal C, Kirschning CJ, Rosenstein Y, Rocha LM, Rios-Sarabia N, Espinosa-Cantellano M, et al. The innate immune response to Entamoeba histolytica lipopeptidophosphoglycan is mediated by toll-like receptors 2 and 4. Parasite Immunol (2005) 2:127–37. doi:10.1111/j.1365-3024.2005.00754.x
167. Maldonado C, Trejo W, Ramirez A, Carrera M, Sánchez J, López-Macías C, et al. Lipophosphopeptidoglycan of Entamoeba histolytica induces an anti-inflammatory innate immune response and downregulation of toll-like receptor 2 (TLR-2) gene expression in human monocytes. Arch Med Res (2000) 31:S71–3. doi:10.1016/S0188-4409(00)00199-5
168. Kammanadiminti SJ, Chadeem K. Suppression of NF-kappaB activation by Entamoeba histolytica in intestinal epithelial cells is mediated by heat shock protein 27. J Biol Chem (2006) 281:26112–20. doi:10.1074/jbc.M601988200
169. Cho KN, Becker SM, Houpt ER. The NF-kappaB p50 subunit is protective during intestinal Entamoeba histolytica infection of 129 and C57BL/6 mice. Infect Immun (2010) 78:1475–81. doi:10.1128/IAI.00669-09
170. Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature (2007) 446:557–61. doi:10.1038/nature05698
171. Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol (2008) 8:411–20. doi:10.1038/nri2316
172. Ivory C, Kammanadiminti S, Chadee K. Innate resistance to Entamoeba histolytica in murine models. Trends Parasitol (2007) 23:46–8. doi:10.1016/j.pt.2006.12.006
173. Dey I, Chadee K. Prostaglandin E2 produced by Entamoeba histolytica binds to EP4 receptors and stimulates interleukin-8 production in human colonic cells. Infect Immun (2008) 6:5158–63. doi:10.1128/IAI.00645-08
174. Lejeune M, Moreau F, Chadee K. Prostaglandin E2 produced by Entamoeba histolytica signals via EP4 receptor and alters claudin-4 to increase ion permeability of tight junctions. Am J Pathol (2011) 179:807–18. doi:10.1016/j.ajpath.2011.05.001
175. Belley A, Chadee K. Prostaglandin E(2) stimulates rat and human colonic mucin exocytosis via the EP(4) receptor. Gastroenterology (1999) 117:1352–62. doi:10.1016/S0016-5085(99)70285-4
176. Moncada D, Keller K, Chadee K. Entamoeba histolytica cysteine proteinases disrupt the polymeric structure of colonic mucin and alter its protective function. Infect Immun (2003) 71:838–44. doi:10.1128/IAI.71.2.838-844.2003
177. Wang W, Keller K, Chadee K. Entamoeba histolytica modulates the nitric oxide synthase gene and nitric oxide production by macrophages for cytotoxicity against amoebae and tumour cells. Immunology (1994) 83:601–10.
178. Wang W, Chadee K. Entamoeba histolytica suppresses gamma interferon-induced macrophage class II major histocompatibility complex Ia molecule and I-Aβ mRNA expression by a prostaglandin E2-dependent mechanism. Infect Immun (1995) 63:1089–94.
179. Verma AK, Verma R, Ahuja V, Paul J. Real-time analysis of gut flora in Entamoeba histolytica infected patients of northern India. BMC Microbiol (2012) 12:183. doi:10.1186/1471-2180-12-183
180. Bracha R, Kobiler D, Mirelman D. Attachment and ingestion of bacteria by trophozoites of Entamoeba histolytica. Infect Immun (1982) 36:396–406.
181. Cobo ER, He C, Hirata K, Hwang G, Tran U, Eckmann L, et al. Entamoeba histolytica induces intestinal cathelicidins but is resistant to cathelicidin-mediated killing. Infect Immun (2012) 80:143–9. doi:10.1128/IAI.05029-11
182. Burgess SL, Buonomo E, Carey M, Cowardin C, Naylor C, Noor Z, et al. Bone marrow dendritic cells from mice with an altered microbiota provide interleukin 17A-dependent protection against Entamoeba histolytica colitis. MBio (2014) 5:e01817. doi:10.1128/mBio.01817-14
183. Band RN, Cirrito H. Growth response of axenic Entamoeba histolytica to hydrogen, carbon dioxide, and oxygen. J Protozool (1979) 26:282–6. doi:10.1111/j.1550-7408.1979.tb02781.x
184. Mehlotra RK. Antioxidant defense mechanisms in parasitic protozoa. Crit Rev Microbiol (1996) 22:295–314. doi:10.3109/10408419609105484
185. Weinbach EC, Diamond LS. Entamoeba histolytica. I. Aerobic metabolism. Exp Parasitol (1974) 35:232–43. doi:10.1016/0014-4894(74)90027-7
186. Vicente JB, Ehrenkaufer GM, Saraiva LM, Teixeira M, Singh U. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: implications for amebic pathogenesis. Cell Microbiol (2009) 11:51–69. doi:10.1111/j.1462-5822.2008.01236.x
187. Rastew E, Vicente JB, Singh U. Oxidative stress resistance genes contribute to the pathogenic potential of the anaerobic protozoan parasite, Entamoeba histolytica. Int J Parasitol (2012) 42:1007–15. doi:10.1016/j.ijpara.2012.08.006
188. Schlosser S, Leitschm D, Duchêne M. Entamoeba histolytica: identification of thioredoxin-targeted proteins and analysis of serine acetyltransferase-1 as a prototype example. Biochem J (2013) 451:277–88. doi:10.1042/BJ20121798
189. Debnath A, Parsonage D, Andrade RM, He C, Cobo ER, Hirata K, et al. A high-throughput drug screen for Entamoeba histolytica identifies a new lead and target. Nat Med (2012) 18:956–60. doi:10.1038/nm.2758
190. Leitsch D, Kolarich D, Wilson IB, Altmann F, Duchêne M. Nitroimidazole action in Entamoeba histolytica: a central role for thioredoxin reductase. PLoS Biol (2007) 5:e211. doi:10.1371/journal.pbio.0050211
191. Husain A, Sato D, Jeelani G, Soga T, Nozaki T. Dramatic increase in glycerol biosynthesis upon oxidative stress in the anaerobic protozoan parasite Entamoeba histolytica. PLoS Negl Trop Dis (2012) 6:e1831. doi:10.1371/journal.pntd.0001831
Keywords: Entamoeba histolytica, cysteine protease, glycosidase, mucin, phagocytosis, oxidative stress, metabolism
Citation: Nakada-Tsukui K and Nozaki T (2016) Immune Response of Amebiasis and Immune Evasion by Entamoeba histolytica. Front. Immunol. 7:175. doi: 10.3389/fimmu.2016.00175
Received: 23 March 2016; Accepted: 20 April 2016;
Published: 12 May 2016
Edited by:
Alexandre Morrot, Federal University of Rio de Janeiro, BrazilReviewed by:
Julio Aliberti, Cincinnati Children’s Hospital Medical Center, USAWilliam Petri, The University of Virginia, USA
Copyright: © 2016 Nakada-Tsukui and Nozaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoyoshi Nozaki, nozaki@nih.go.jp
 Kumiko Nakada-Tsukui
Kumiko Nakada-Tsukui