
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 11 September 2015
Sec. Mucosal Immunity
Volume 6 - 2015 | https://doi.org/10.3389/fimmu.2015.00467
This article is part of the Research Topic Understanding Crohn's disease: immunity, genes and microbes View all 13 articles
The incidence of inflammatory bowel diseases (IBD), including Crohn’s disease (CD), is increasing worldwide, especially in young children and adolescents. Although hospitalized patients are usually provided with enteral or parenteral support, continuing care typically requires a trial-and-error approach to suppressing symptoms and maintaining disease remission. Current nutritional advice does not differ from general population guidelines. International collaborative studies have revealed 163 distinct genetic loci affecting susceptibility to IBD, in some of which host–microbe interactions can be seen to play an important role. The nature of these loci enables a rationale for predicting nutritional requirements that may not be evident through standard therapeutic approaches. Certain recognized nutrients, such as vitamin D and long-chain omega-3 polyunsaturated fatty acids, may be required at higher than anticipated levels. Various phytochemicals, not usually considered in the same class as classic nutrients, could play an important role. Prebiotics and probiotics may also be beneficial. Genomic approaches enable proof of principle of nutrient optimization rather than waiting for disease symptoms to appear and/or progress. We suggest a paradigm shift in diagnostic tools and nutritional therapy for CD, involving a systems biology approach for implementation.
Inflammatory bowel diseases (IBD) cover a spectrum of gastrointestinal disorders, including both Crohn’s disease (CD) and ulcerative colitis (UC), differentiated by their location and behavior. While these diseases were seen as rare in the early twentieth century, they have become increasingly common, causing gut inflammation, ulceration, and other symptoms, in up to 1 in 250 people worldwide. For example, they had not been previously described among Bedouin Arabs in Southern Israel, but numbers are increasing now, almost certainly because of their increasingly urbanized lifestyle (1). A recent commentary described IBD in Asia as the “emergence of a Western disease” (2). While observed previously in Australasia, these diseases are becoming more common in young children and adolescents (3).
There are a number of accepted treatments for CD, aimed not only to reduce the inflammation but also to improve the long-term prognosis (4). The most desirable end point, achieved in only a relatively low proportion of cases at present, is long-term remission. One of the two approaches is used for CD treatment. “Step-up” starts with milder drugs first, while “top-down” gives people stronger drugs earlier in the treatment process. Anti-inflammatory drugs are often the first step in the treatment. These include oral 5-aminosalicylates, such as sulfasalazine and mesalamine, or corticosteroids. However, all of these lead to undesirable side effects. Immune system suppressors also reduce inflammation by targeting the immune system. These drugs include azathioprine (Imuran), mercaptopurine (Purinethol), infliximab (Remicade), adalimumab (Humira), and certolizumab pegol (Cimzia). The latter three drugs are inhibitors of the immune system protein tumor necrosis factor-alpha (TNF-α). It is important to realize, however, that the inflammatory response is a necessary part of response to an infection or injury, and long-term suppression of this may be disadvantageous.
Not only is CD itself debilitating, it also carries with it an increased risk of colorectal cancer (CRC) (5). The nature of this cancer is fundamentally different from the classic disease, which arises from an adenomatous polyp (6). The type of CRC that develops from either form of IBD is almost inevitably lethal. However, like sporadic CRC, it arises through genomic instability.
Genomic instability refers to the accumulation of mutations during the life cycle of cells (7–9). These mutations can involve changes at the level of nucleotide base pairs, chromosome rearrangements, or aneuploidy. Genomic instability can initiate cancer, augment cancer progression, and influence the overall prognosis of the affected patient. It arises from many different pathways, including telomere damage, centrosome amplification, epigenetic modifications, and DNA damage from endogenous and exogenous sources. Protection against genomic instability, or at least reduction of the probability, is an important function of certain nutrients and phytochemicals (7).
Although nutrition is an important consideration in hospitalized patients, non-hospitalized patients are not currently being given nutritional guidelines outside those for the general population. We explore the possibility that thinking outside the current square in terms of nutrition might have significant beneficial effects beyond protection from malnutrition. That is, we suggest that nutrition could move from being an adjunct to therapy, to the point where it becomes primary therapy in its own right, integrated with currently accepted approaches (10). There is reason to suggest that this could have significant benefits in terms of delaying IBD progression and reducing the possibility of CRC formation. Such information would ideally be tailored according to individual patient genetics, and validated using systems biology approaches, as detailed in the following sections.
Either prior to hospitalization or following discharge from hospital, dietary selection is determined by something of a random process (11). While the diet may be based on previous recollections of foods which affected symptoms, either in a positive or negative sense, it is often guided by advice from others with the disease. It has been clear for some time that certain foods may have adverse effects in most subjects, exacerbating disease symptoms by increasing inflammation and/or producing flatulence. In some groups, there has been endorsement of a “low FODMAP” diet, as this appears to reduce some of the symptoms in CD and other functional gastrointestinal disorders, especially irritable bowel syndrome (12, 13). FODMAP refers to the combination of saccharides and polyols and is an acronym of “Fermentable Oligo-, Di-, mono and polyols.” Against the recommendation for a low-FODMAP diet, there is a concern that such a structured dietary regime will have adverse effects in reducing dietary diversity. Of particular concern is the recognition that this same group of excluded nutrients plays a major role in modulating the composition of the gut microbiome (14).
It has been recognized that there is a considerable danger of malnutrition exacerbating the already difficult symptoms of CD. Attention has been drawn to the possible need for higher than normal levels of vitamins such as vitamin D or lipids such as long-chain omega-3 polyunsaturated fatty acids (n-3 PUFAs) (15, 16). What has not always been recognized is the role of dietary components that are not currently included in essential lists. For example, prebiotics and probiotics may be important in regulating the gut microbiota, and their importance may be missed in some assessments (17–19). Phytochemicals are sometimes called “non-nutrients.” Although these are not included in lists of essential nutrients, many of them play key roles in maintaining genomic stability and in modulating the composition of the gut microbiome (7, 20, 21). These could be key factors in helping to prevent the development of CRC in IBD.
Irrespective of nutrient composition, it has repeatedly been observed that foods considered to be beneficial for some individuals may actually cause adverse effects in others (11, 22). Individuals with the diseases differ in genetic, epigenetic, and phenotypic characteristics, so it is not surprising that a single approach is unlikely to be beneficial to all. However, studies showing the importance of genotype in the phenotypic characteristics of the diseases encourage the possibility of tailoring diets based on genetic and genomic information (23).
Crohn’s disease was one of the subjects of early reports on genome-wide association studies (GWAS). At that time, it appeared that there could be up to eight loci affecting disease susceptibility (24). Since that time, large international groupings have performed a meta-analysis of CD and UC GWAS scans, as part of activity of the International IBD Genetics Consortium (IIBDGC) (25). This was followed by extensive validation of significant findings, from data on a combined total of more than 75,000 cases and controls. These efforts initially revealed 163 distinct genetic loci affecting susceptibility to IBD overall (26), but this number has more recently been expanded to 201 (27). A number of these genes are involved in primary immune deficiencies, characterized by a dysfunctional immune system and increased susceptibility to serious infections. In particular, there is a considerable overlap between IBD loci and the immune-mediated disorders, ankylosing spondylitis and psoriasis (27). This study found considerable overlap between susceptibility loci for IBD and mycobacterial infection, which relationships were emphasized by further analyses using coexpression network analysis. Many of the identified loci contain genes involved in microbial handling, and host–microbe interactions clearly play an important role in disease susceptibility (26).
In further analyses of these datasets, the IIBDGC detailed fine-mapping project, clarified the nature of these genes and their interactions, and suggested that phenotypically there should be three rather than just two main classes of IBD and that CD should be categorized into two distinct classes depending upon its location (27). Each of these classes of disease has characteristic genetic and phenotypic characteristics. The detailed nature of the genes and networks involved in disease susceptibility enables a rationale for predicting nutritional requirements that may not be evident through standard therapeutic approaches.
There is a considerable scientific input into the setting of recommended daily nutrient intakes. While these may be good for the general population, they may often not reflect the actual requirements for CD patients. Figure 1 illustrates the identification of specific dietary components or items that may be important for different groups of CD patients, according to their genotype and the nature of the gut microbiome. Genetic testing can indicate the primary single-nucleotide polymorphisms (SNPs) of importance at an individual level. If, for example, some of the interleukin (IL) genes are affected, these are primarily involved in inflammation (28, 29), although they may have a downstream effect on the microbiota. Thus, supplementation with long-chain n-3 PUFA or fish oils may be a nutritional approach to prioritize (30–32). Similarly, identification of NOD2 or ATG16L1 gene variants indicate that ability to respond appropriately to colonic bacteria is compromised (33, 34), and prebiotics and/or probiotics may provide an important starting point (14, 35–37). For all these individuals, especially those carrying a number of variant SNPs, it becomes important to protect against genomic instability since this will reflect a propensity for cancer development. In this respect, certain nutrients, especially vitamin D and phytochemicals, may play an important role (7).
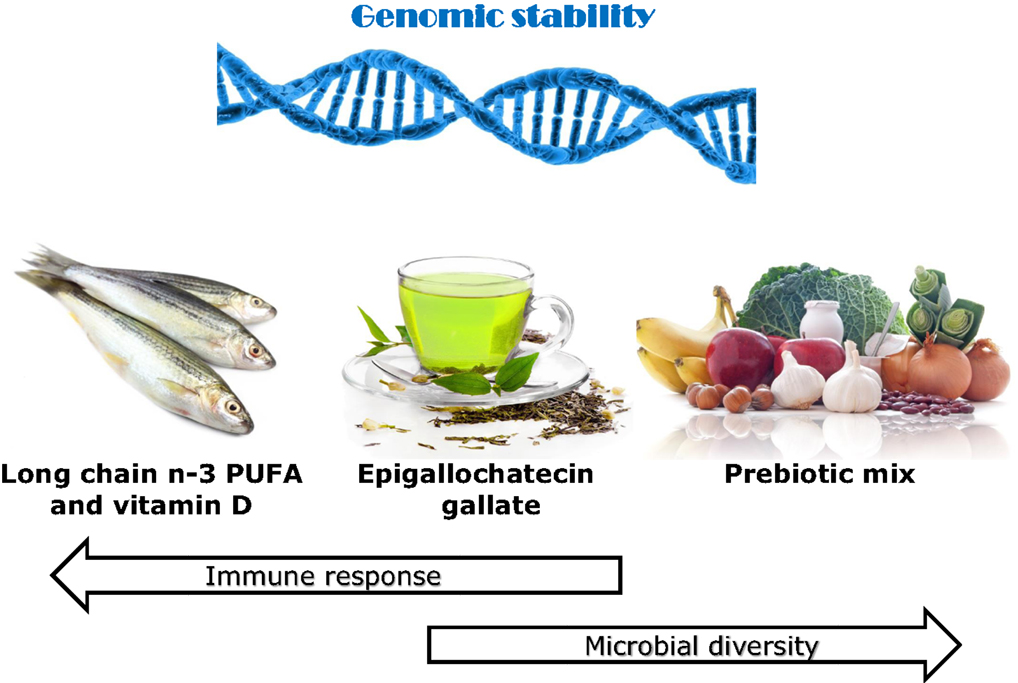
Figure 1. Effects of various identified dietary components on immune response, microbial diversity, and genomic stability, with use justified by genetic testing. The use of the immunochip supplemented with fine mapping enables very detailed understanding of the nature of the genes involved in CD susceptibility of an individual. A number of the affected genes play key roles in inflammatory response. If the majority of the affected genes favor inflammation, then a primary recommendation may be to enhance the intake of long-chain n-3 PUFA, especially EPA and DHA. Both of these play distinctive and major controlling roles in inflammatory response (16). If genes affecting microbial response and signaling pathways predominate, then probiotics may be an especially important dietary component (35). In addition, various prebiotics such as the food combination illustrated here act to stimulate the growth of beneficial gut microbes (14, 37). Both vitamin D and various phytochemicals, including the active component in green tea, EGCG, are recognized as having roles in each of these processes and are likely to be beneficial to CD patients at higher than standard dietary recommendations (15, 38–40). Maintenance of genomic stability will play an important role in slowing progression of CD or the development of CRC (7, 21). Thus, diets containing a good balance of such components, possibly tailored according to genotype, may act to complement or slow the need for recognized therapeutics in CD. Images from Can stock photos.
While it is desirable to have studies in CD patients, in practice much of this work comes from mouse models, developed to reflect our understanding of the genetics of CD (41–44). Two of these models will be described here.
The IL-10 gene-deficient (IL-10−/−) mouse model lacks a functional version of the anti-inflammatory cytokine, IL-10. IL-10−/− mice, inoculated with normal intestinal bacteria, have been used to investigate the role of various dietary components in intestinal inflammation, including mechanistic studies that consider transcriptomic, metabolomic, and proteomic effects (41, 45, 46). The multidrug-resistant (mdr1a−/−) mouse model carries a deletion mutation enabling disease-causing microorganisms (bacteria, viruses, fungi, or parasites) to resist a range of important drugs (antibiotics, antifungal, antiviral, and antiparasitic drugs) targeted at eradicating the organism (47). Mdr1a knockout mice are susceptible to developing a severe spontaneous intestinal inflammation in pathogen-free animal facilities.
Fenech has reviewed the role of various micronutrients in slowing the progress of genomic instability, a key component in the progression of digestive diseases and the initiation of cancer (8, 48, 49). He points to the importance of individualizing dietary components according to genotype and shows increased dietary intake of vitamin E, calcium, folate, retinol, and nicotinic acid being associated with less DNA damage and a need to define the optimal amount being especially important for riboflavin, pantothenic acid, and biotin. These three have been distinguished because of increased DNA damage being especially evident at higher doses. Fenech has described high-throughput nutrient arrays that enable defining, on an individual basis, the optimal combination of nutrients for DNA damage prevention, maintenance of telomere integrity (important in cancer risk), and cancer growth control (48).
We have more generally reviewed vitamin and mineral requirements to maintain genomic stability, especially in the context of the micronutrient genomics project (50). It is noteworthy that certain of these nutrients may be required in higher than usual amounts in CD, since they are utilized in the control of immune response and inflammation. Our own studies have particularly emphasized the importance of getting not only the correct form of selenium but also the appropriate level according to genotype (51, 52).
Vitamin D is an important vitamin that appears to be required at higher than anticipated levels in CD patients (15). This may partly be caused by genetic requirements, and it is of interest that a number of SNPs associated with vitamin D uptake and distribution actually appear on the immunochip, used in the important study of IBD risk genes by Jostins and coworkers (26). Again, there is a specific link with inflammatory processes and with control of microbiota in the identified genes. Higher plasma vitamin D levels have been associated with a reduced risk of Clostridium difficile (53), whereas reduced levels of circulating vitamin D enhance the risk of cancer and other inflammatory diseases (54, 55). We suggest that such an observation may add fuel to an argument that higher than current recommended daily intakes of vitamin D may be particularly appropriate to CD patients.
Many naturally occurring agents directly bind with and activate peroxisome proliferator-activated receptor gamma (PPAR-γ or PPAR gamma), a type II nuclear receptor that in humans is encoded by the PPAR-γ gene. Agents binding this include various PUFAs including arachidonic acid and arachidonic acid metabolites. PPAR-g regulates fatty acid storage and glucose metabolism. The genes activated by PPAR-g stimulate lipid uptake by adipocytes and play an important role in regulating inflammation and cancer cell growth (46).
Peyrin-Biroulet and coworkers (56) demonstrated antimicrobial functions of the PPAR-γ gene products in maintaining epithelial expression of a type of colonic beta-defensin (mDefB10 in mice, DEFB1 in humans). In mutant mice carrying this mutation, these authors showed defective killing of a number of bacteria including Candida albicans, Bacteroides fragilis, Enterococcus faecalis, and Escherichia coli. It appears that colonic involvement in CD is linked to reduced expression of DEFB1, independent of inflammation. Thus, it has been suggested that PPAR-g-targeting by either drugs or nutrients could prevent colonic inflammation by restoring antimicrobial immunity in CD.
There have been variable results across various studies in relation to the association of common PPAR-g variants (C161T and Pro12Ala) with IBD. While Hume and coworkers reported no association in an Australian cohort (57), Shrestha et al. found suggestive relationships in Chinese but not Dutch patients (58). However, the Pro12Ala variants appeared to be protective against the development of CD in European Caucasians (57). Such studies have led support to the suggestion that there are significant ethnic differences in the phenotypic expression of this variant. There is reason to believe that diet, including lipids, may play a key role in this effect.
Conjugated linoleic acid has been shown to modulate immune responses in patients with mild to moderately active CD (59), and there is some support for this being through modulation of PPAR-g. In their Caucasian CD cohort, Ferreira and coworkers (60) found that a high intake of total, saturated, and monounsaturated fats and a higher ratio of n-6/n-3 PUFAs were associated with a more active phenotype. They studied the effects of four genetic polymorphisms, including the two PPAR-g SNPs 161C/T and Pro12Ala SNPs, in a case–control population. Although they reported no significant effects of these SNPs on disease susceptibility per se, they found that the presence of either of these SNPs led to a more detrimental effect of a high intake of total and trans fats.
The effects of omega-3 polyunsaturated fatty acids (n-3 PUFAs) are also modified by genotype, especially the nitric oxide synthase (NOS3) gene (61). These authors reported associations of plasma fatty acid composition and NOS3 SNP genotypes (rs11771443, rs1800783, rs1800779, rs1799983, rs3918227, and rs743507) in 450 individuals with the MetS from the LIPGENE dietary intervention cohort. They found that several markers of inflammation were significantly different between the genotype groups. There was a significant gene–nutrient interaction between the NOS3 rs1799983 SNP and plasma n-3 PUFA status on plasma triacylglycerol (TAG) concentrations. Minor allele carriers (AC + AA) were considerably more responsive to changes in plasma n-3 PUFA than major allele homozygotes. Such individuals are likely to benefit even more from long-chain n-3 PUFA consumption than the general population in order to reduce inflammation in CD (16).
Polyphenols are secondary metabolites of higher plants and one of the largest groups of natural products. Although not usually considered in the same class as essential nutrients, various polyphenols could play an important role in CD susceptibility and progression. A range of polyphenols have been shown to modulate inflammation, especially in genetically deficient mouse models. For example, a green tea extract enriched in polyphenols was able to modulate colonic inflammation in mice lacking the multidrug resistance gene (Mdr1a−/−). Mice were fed control or green tea-enriched diets for 21–24 weeks of age, after which a colonic histological injury score was obtained, colonic gene expression analyzed using microarrays, and colon protein expression also measured (20). The authors reported reduced abundance of transcripts and proteins associated with immune and inflammatory response, which suggested that its anti-inflammatory activity is mediated by multiple molecular pathways.
Cruciferous vegetables, such as cabbage and broccoli, contain two types of sulfur-containing phytochemicals – glucosinolates and S-methyl cysteine sulfoxide. While these chemicals have a range of effects generally considered to be beneficial in the population at large, this group of food plants also polarizes individuals with CD. Laing and coworkers associated SNPs and the beneficial or adverse effects of the 10 most commonly eaten foods in this group (22). One of the SNPs that showed exceptionally beneficial properties to individuals consuming cruciferous vegetables was in the defensin (DEFA6) gene. Conversely, one SNP strongly associated with adverse effects was in the major histocompatibility complex, which characterizes one important group of CD patients (27).
The main polyphenol in green tea is (−)-epigallocatechin gallate (EGCG). Unno and coworkers showed this compound at levels of 0.6% to have strong effects on gut microbiota and biomarkers of colonic fermentation in rats (38). They found a significant reduction of Clostridium spp and an increased gut occupation by Bacteroides. Smaller changes were seen for Bifidobacterium and Prevotella. ECGC also has effects on genomic stability, suggesting that it could be protective against cancer development in IBD (7, 21).
Ellagic acid and ellagitannins are a class of hydrolyzable tannins found in some fruits and nuts. At least in in vitro studies, there is good evidence that these may have the potential to reduce inflammation in genetically modified cells (62). Boysenberries, a hybrid Rubus berry, are among the best food sources of ellagitannins, although chestnuts and pomegranates also have high concentrations. Nasef and coworkers found effects modulated through toll-like receptors 2 and 4 of extracts from blackcurrants and feijoa, which may have been associated with elligitannins (63). The significance of these results awaits confirmation by in vivo testing.
Resveratrol is another polyphenol that has been shown to protect against genomic instability. It acts as an antioxidant protecting against free radical-induced DNA damage and is likely to play a protective role in gut inflammation and progression to CRC (64).
The diversity of microbiota appears as a key characteristic that may predict response to therapy in CD (65). In addition, it has been suggested that a discriminant score of intestinal microbiota may provide an index of disease activity in CD (66). While fecal transplants have received some recognition as therapy for CD generally, these are not sustainable over a long period (67). Additionally, a number of concerns have been raised about their true efficacy (68). Regulating the gut microbiota through diet may provide a more sustainable solution. There is no question but that diet plays a major role in modulating the colonic microbiota (69). Certain nutrients, especially vitamin D, may play an important role in this respect (70).
Probiotics have been defined by FAO/WHO as “live microorganisms that confer a health benefit to the host when administered in adequate amounts.” Especially in combination with prebiotics (plant substrates that enable modulation of colonic microbiota), these may be especially beneficial to CD patients (35, 71). Probiotic bacteria have been shown to produce conjugated linoleic acid in the gut, which in turn plays a role in suppressing disease symptoms through targeting PPAR-γ gamma (42). Prebiotics are digested and fermented to form short-chain fatty acids, which themselves enhance the growth of certain important colonic bacteria (17).
Additionally, an increasing number of plant polyphenols are being identified to have effects on colonic microbiota identification and regulation (64). Innovative prebiotic/probiotic foods are also being developed and may have promise in a clinical context (71).
Not only genetics per se but also certain epigenetic events are associated with susceptibility to CD, disease progression, and CRC development.
DNA methylation has been considered for its role in the development of CD. Whole-blood DNA methylation profiles were compared in treatment-naive children with CD and healthy controls, as measured using the Illumina 450 K platform (72). Sixty-five differentially methylated CpG sites were identified as reaching epigenome-wide significance. The most significantly differentially methylated region in patients with CD involves the transcription start site for microRNA (miR)-21.
microRNAs (miRNAs) are recognized as playing an essential role in the development and control of the innate and adaptive immune system. These are small non-coding RNA molecules that lead to post-translational gene silencing and control gene expression in diverse biological processes, including inflammation. miRNA genes occur within intronic sequences of protein-coding genes, within intronic or exonic regions of non-coding RNAs, are intergenic (72). As well as miR-21, various other miRNAs including miR-192, miR-122, miR-29, and miR-146a may be implicated in CD. Krissansen and coworkers found that increased circulating levels of miR-595 and miR-1246 related to a highly aggressive form of the disease (73). Because miRNAs play a major role in regulating gene expression, they are being looked at as therapeutic targets and also as biomarkers for aggressive disease development.
There are no published reports of individual nutrients affecting gene expression in CD from human studies because the definitive studies would be unethical to perform. However, there are a number of studies in animal models or tissue culture systems, which provide useful information.
The active binding sites of 1,25-dihydroxyvitamin D3 (1,25D) are likely to regulate gene expression at the cellular level (74). This active form of vitamin D has been shown to interact with the epigenome through effects on DNA methylation, histone acetylation and miRNA, as well influencing pre-mRNA splicing. Genomic profiling has identified a set of 1,25D regulated genes that are especially relevant to cancer prevention. For example, Ma and coworkers compared 1,25D-regulated miRNA expression profiles in a human cancer cell line in comparison with a highly tumorigenic and metastatic variant, using miRNA qPCR panels (75). The two lines showed distinctly different miRNA expression profiles that the authors suggest are likely to influence the behavior of different tumor cells and are relevant to both the susceptibility to and subsequent behavior of cancer.
Human studies on vitamin D3 supplementation have enabled dissection of high from low responders in a pre-diabetic study population (76). Only around 60% of these subjects responded to supplementation. While VDR receptor gene polymorphisms played a role in this, other clinical parameters such as the level of parathyroid hormone were also important. The authors suggested that vitamin D3-induced changes in human peripheral blood mononuclear cells can be described by transcriptomics. Added to information from other serum biomarkers, these allow the identification of those subjects who will (or will not) respond to vitamin D supplementation.
These compounds have sometimes been described as master regulators of gene expression (16). Serhan and coworkers have discussed the function of long-chain omega-3 PUFA as novel pro-resolving mediators in the resolution of acute inflammation. They have discovered a new genus of pro-resolving lipid mediators that is temporally produced by resolving exudates that enable a return to homeostasis. These not only have anti-inflammatory actions but also enhance microbial clearance. Such properties make the two long-chain omega-3 PUFAs, EPA and DHA, essential nutrients not only for protection against CD in the first place but also for reducing inflammation and thereby reducing disease progression.
Knoch and coworkers compared the effects of EPA supplementation with that of oleic acid (OA) as control in the IL-10−/− mouse model (77). They found that EPA reversed the decrease in colon fatty acid beta-oxidation gene expression observed in OA-fed mice. The mice fed with the OA diet showed a decrease in the expression of antioxidant enzyme genes, as well as those involved in detoxification, when compared with wild-type (C57Bl) mice on the same diet. In contrast, EPA up-regulated the expression of these same enzymes. The authors suggested that these results imply that EPA might have a potential anti-inflammatory effect on colon tissue. In support of this hypothesis was the observation that EPA also activated expression of the PPAR-α gene, which regulates the expression of proinflammatory and immunomodulatory genes.
Cytochrome P450s (CYPs) play an essential role in the metabolism of endogenous and exogenous molecules (78). They facilitate the biosynthesis of a number of essential molecules, including fatty acids, lipid-soluble vitamins, and steroid hormones. They also metabolize most pharmaceuticals as well as carcinogenic compounds. CYP gene expression is regulated by a number of polyphenols, such as EGCG. This molecule not only has antioxidant properties but can be pro-oxidant at very high levels, although generally beneficial in modulating the risk and progression of inflammatory diseases and cancer (79). The review by Korobkova extensively summarizes the effects of a range of polyphenols on CYP metabolism, with flow-on implications for gene expression (79). Aylissi and coworkers have also summarized the epigenetic effects of natural polyphenols, which have implications for the modulation of gene expression (80).
An extract of green tea, enriched in polyphenols (GrTP), was tested for effects on colonic inflammation in the MDRa−/− mouse model utilizing a transcriptomics approach (20). The histological injury score was significantly lower in the GrTP-fed mice than in the control group. Colon mRNA transcripts were assessed using microarrays. They revealed a reduced abundance of transcripts associated with immune and inflammatory response and an abundance of those associated with xenobiotic metabolism pathways, suggesting an anti-inflammatory effect mediated by diverse mechanisms. PPAR-a and signal activator of transcription 1 (STAT1) appear as two key regulatory molecules in these events.
Curcumin has been studied for effects on gene expression, using the N2A cell line (81). Treatment with this polyphenol leads to suppression of NF-kappaB and its downstream proinflammatory targets including COX-2 and iNOS. Resveratrol has also been shown to alter gene expression patterns in another cancer cell line (82). Microarray analysis showed effects on apoptosis-related genes. The green tea polyphenol, EGCG, has been used as an exemplar of a polyphenol showing effects on gene expression revealing other than antioxidant properties (39). In particular, EGCG regulates signal transduction pathways, transcription factors, DNA methylation, mitochondrial function, and autophagy, several of which effects are relevant to CD patients. ECGC has also been shown to attenuate the activation of STAT3, which again is an important pathway in CD (83). In an overview on the effects of polyphenols on gene expression, Joven and coworkers suggest that these provide an excellent illustration as to how we may be able to eat our way out of chronic diseases (84).
Dietary curcumin and rutin were compared for effects on colonic inflammation and gene expression in the MDR1−/− mouse model (44). Curcumin but not rutin significantly reduced the histological evidence of inflammation in this mouse model. Microarray and pathway analysis implied that the mechanism was likely through up-regulation of xenobiotic metabolizing enzyme expression and a down-regulation of proinflammatory pathways.
Plaza-Diaz and coworkers found evidence that a range of probiotics modulate the expression of immunity and inflammatory genes in the gut, which appear especially relevant to CD. They performed a systematic review to conclude that strains of Bifidobacterium, Lactobacillus, Escherichia coli, Propionibacterium, Bacillus, and Saccharomyces influence the gene expression of mucins, Toll-like receptors, caspases, NF-κB, and ILs, leading to an anti-inflammatory response in cultured enterocytes. Similar results were found in animal models ranging from fish to mice, rats, and piglets (85).
Prebiotics can also modulate host gene expression, with potentially beneficial flow-on effects relevant to CD (86). Sauer and coworkers studied the effects of products formed from an inulin derivative incubated with human gut flora (87). They considered the expression of 96 genes related to biotransformation using cDNA microarrays. The pattern of gene expression suggested various effects likely to protect the cells from carcinogenic compounds. Zenholm et al. used a Caco-2 gastrointestinal cancer cell model to study the effects of an oligosaccharide and a fructo-oligosaccharide in reducing the expression of IL-12p45, IL-8, and TNF-α (88).
Marlow and coworkers reported on gene expression effects of a specific diet containing examples of the nutrient classes under discussion (89). They developed a Mediterranean-inspired diet whose ingredients were delivered to participants each week over a 6-week intervention period along with recipes and tips for maintaining a healthy diet, and participants were also contacted for encouragement on a regular basis. The diet did not contain the amounts of whole grains and diary products common in the Mediterranean but had a seasonal range of fruits and vegetables (various vitamins and polyphenols), two meals of oily fish per week (omega-3 and vitamin D), green tea (EGCG), and probiotic capsules. Marlow and coworkers considered the effects on symptoms, biomarkers of inflammation, and the gut microbiota as well as classic measures of inflammation such as C-reactive protein (CRP), and they also used a transcriptomic end point. They found that adherence to the diet for 6 weeks reduced the established biomarkers of inflammation. However, transcriptomic analysis provided an important biomarker, showing that the expression of certain genes important in the etiology of CD was beneficially modulated. The diet also resulted in a trend of normalizing the microbiota.
In previous sections, we have identified a number of nutrients that are especially important for CD development and progression. Other nutrients will undoubtedly play an additive role. Systems biology links the interactions among genes, gene products, and environmental factors. As Polytarchou et al. have suggested, there is reason to suggest that this is ready for prime time in CD research (90). It is being increasingly used to refine desirable nutrition for an individual and treat complex human diseases, such as CD. Figure 2 illustrates the way in which these technologies have the potential to revolutionize conventional CD diagnosis and treatment by providing a strong scientific basis for nutritional therapy. As indicated in previous sections and Figure 1, genetic screening will inform the desirability of certain foods or nutrients that will become part of the regular diet (26, 27). The benefit of such a combination of foods may be tested, at an individuals or group level, using transcriptomic approaches (89), while the long-term benefits in protection against disease progression may be interrogated using proteomic and/or metabolomic approaches (91, 92). Changes in the gut microbiota will also be important (93).
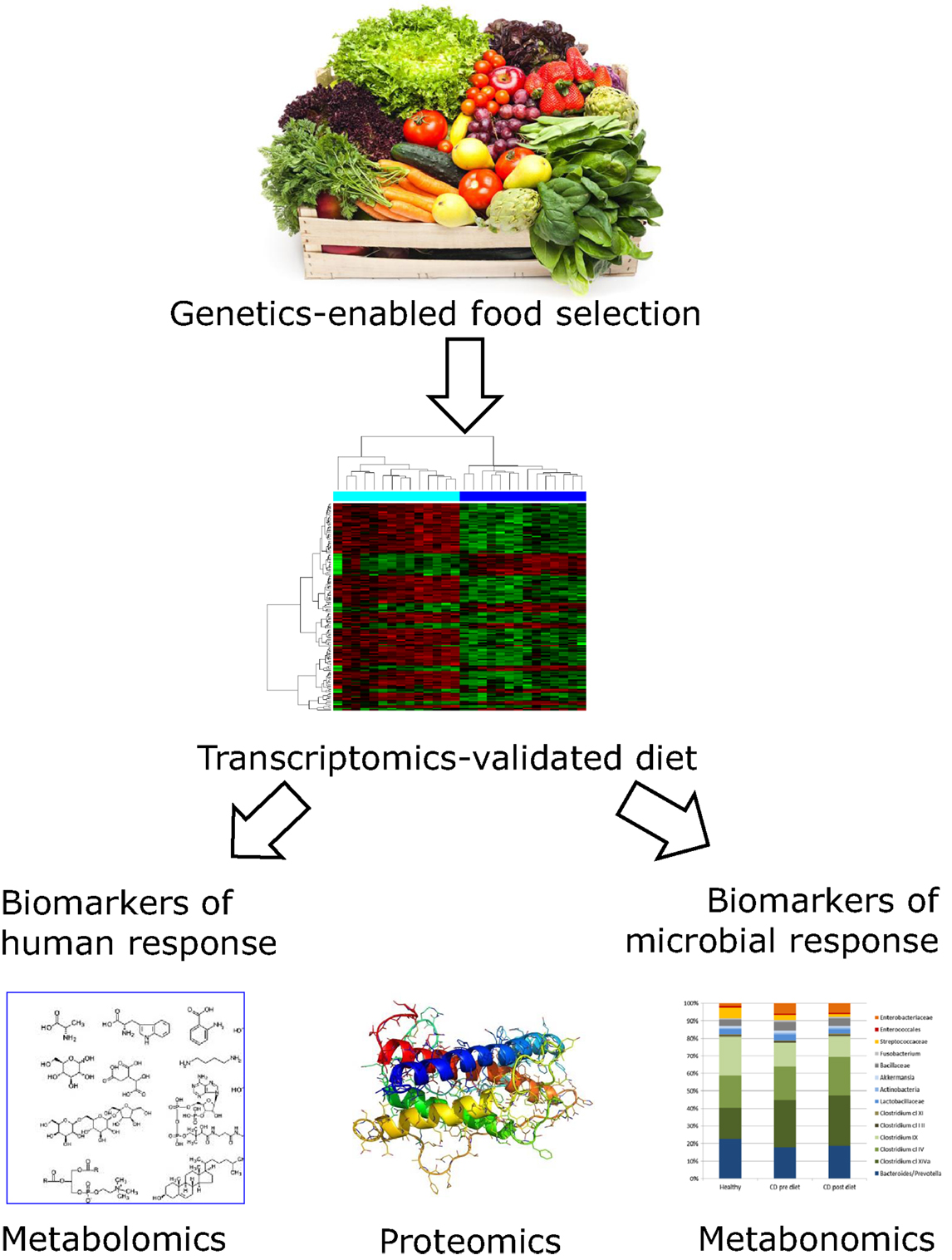
Figure 2. Flowchart showing how genetic and genomic technologies may inform optimal nutritional modulation of CD. Genetic characterization of human DNA and the nature of the associated gut microbiome enable selection of an individualized diet for CD patients. While long-chain omega-3 PUFA containing fish or dietary supplements may be appropriate for most individuals, the exact nature of the most desirable fruits, vegetables, and supplements will vary. Although we are now in a position to make informed guesses, the proof of efficacy of the dietary choice would and probably should be informed by genomic approaches. In particular, there is reason to believe that the inflammation associated with CD will itself consume the various nutrients at higher than average levels. These techniques are now sufficiently sensitive to allow testing the effect of the selected dietary strategy following short-term intervention studies. Blood, urine, and feces samples would be required, before and after the study period. Changes in gene expression as monitored by transcriptomic profiles from peripheral blood mononuclear cells show which CD-related genes have modulated activity (89). Urine or fecal samples can be used for metabolomic and proteomic techniques (91, 94). These may be utilized to monitor dietary compliance and also to provide biomarkers relevant to CD progression. The microbiota profile can be estimated from stool samples, and modulation of microbiota will provide important complementary information on whether the dietary selection has the desired effect on slowing disease progression (67, 89, 95). Images from Adobe stockimages and from Ref. (89).
Fiocchi has identified four components of IBD pathogenesis as environment, genetics, gut microbiota, and mucosal immune response, which can be defined by the terms “expososome,” “genome,” “microbiome,” and “immunome” (94). None of these act independently, but interact (the “interactome”), resulting in the emergence of IBD. Polytarchou and coworkers (90) have stressed the way in which novel computational methodologies can now integrate high-throughput molecular data. They suggest that a systems biology approach could identify the central regulators in the IBD interactome. This work suggests that identification of key nutrients and their interactions with these central networks, possibly at an individual level, might lead to novel therapeutic approaches to CD.
Metabolomic approaches may provide important tools for understanding the differences between individuals in response to dietary components. For example, Lin and coworkers used a mouse model to investigate the effects of feeding kiwifruit-derived extracts from two different species (96). Not only did they find differences in anti-inflammatory activity between the green (Actinidia deliciosa) and gold-fleshed (Actinidia chinensis) kiwifruit extracts, but the efficacy of these extracts was modulated by the variant IL-10 genotype. This same mouse model and experimental approach revealed gradual changes in the metabolome as disease developed, particularly decreased levels of very low-density lipoprotein and increased low-density and very low-density lipoproteins and various PUFAs (97). The metabolome also interrelates with the microbiome, suggesting the possibility of utilizing targeted metabolomics for monitoring the consequences of therapeutically manipulating the microbial community as adjunct therapy in CD (98).
Much of the proof of concept of beneficial effects of certain diets in CD patients has been based on animal or tissue culture studies, rather than humans, since there are obvious ethical constraints in working with people. The classic biomarker of inflammation is considered to be CRP, while fecal calprotectin has gained considerable ground. However, as illustrated in the study by Marlow and coworkers, transcriptomics provides a more sensitive and more informative biomarker of beneficial effects of a selected diet in CD than CRP (89).
Research into gastrointestinal cancer has highlighted the importance of biomarkers, especially miRNAs, usually taken from colonic endoscopies. There is considerable interest in the discovery that miRNAs are present in serum in a cell-free state, highlighting the possibility of their potential use as non-invasive biomarkers (99). At this time, extracellular miRNAs have been identified in most biological fluids, including serum, urine, saliva, and breast milk. They are being considered as novel biomarkers, with considerable potential for predicting disease course and response to therapy.
Metabolites reflect the physiological phenotype, providing a molecular readout of the cell status (95). Such measures, taken from various biological fluids, can lead to the identification of “marker metabolites” that differentiate health from disease. In particular, metabonomics (spectroscopy-based metabolic profiling) of fecal extracts has been used to differentiate active from inactive CD. This method comes across as a powerful non-invasive diagnostic tool able to characterize changes in the metabolic profile associated with malabsorption and dysbiosis (100). Specific metabolites that distinguished groups with active from those with inactive disease included N-acetylated compounds and phenylalanine, low-density lipoproteins, and very low-density lipoproteins (101). More generally, metabolomics is being used in human studies to monitor compliance with required dietary changes and also efficacy of the change in slowing disease progression (102).
Proteomics is defined as the large-scale study of proteins, particularly their structures and functions (103). This technique has been used to validate conclusions from other technologies in a series of mouse studies by Barnett and coworkers (20, 46, 104). In studies on green tea polyphenols, proteomic analysis supported the conclusions from microarrays regarding the mechanism of action of the polyphenol-enriched extract (20). Application of the same technology to studies of n-3 and n-6 PUFAs (arachidonic acid and EPA) showed that the proteomic analysis identified some actions that had not been apparent from transcriptomic studies (46). These authors emphasized the complementary nature of these two approaches and the importance of using the two in concert to better understand the nature of complex diseases such as CD.
The accurate diagnosis of CD has often lagged behind disease presentation. Proteomics is being increasingly used for early and accurate diagnosis of CD as well as for monitoring the course of the disease (92). By providing information on the nature of the disease process, such technology may inform appropriate treatment regimes. Although intestinal tissue will provide the most information, blood, urine, and stool are increasingly providing highly predictive substitutes that are considerably less invasive than the requirement for a colonoscopy.
The toolbox for studying microbiota has become more powerful in recent years, with metagenomics reflecting the study of DNA, metatranscriptomics the total transcribed RNA, and metaproteomics the protein associated with the microbiota. Draft genome sequences have been derived for more than 1,000 human-associated microorganisms derived from the gastrointestinal tract of more than 100 humans (70). Bioinformatics provides an essential tool to tie together these multi-omic analyses of IBD (105).
There is a great deal written about desirable intakes for various classes of food and nutrients. However, the genetic background of CD would appear to make certain nutrients more important than for the general population, albeit differing somewhat according to the genetics of the individual patient. Such nutrients will be especially important during a flare-up of the disease, but will also be crucial in slowing disease progression. Proof of principle of these assertions has been largely dependent on tissue culture or animal models to date. However, improvements in sensitivity of various technologies, especially transcriptomics, proteomics, and metabolomics, enable non-invasive sampling of humans from blood, urine, and/or stool samples, after relatively short-term human intervention studies. We suggest that there is justification for a paradigm shift in diagnostic tools and nutritional therapy for CD, involving a systems biology approach for implementation (106).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Abu Freha N, Schwartz D, Elkrinawi J, Ben Yakov G, Abu Tailakh M, Munteanu D, et al. Inflammatory bowel disease among Bedouin Arabs in southern Israel: urbanization and increasing prevalence rates. Eur J Gastroenterol Hepatol (2015) 27(3):230–4. doi:10.1097/MEG.0000000000000263
2. Ng SC. Emerging leadership lecture: inflammatory bowel disease in Asia: emergence of a “western” disease. J Gastroenterol Hepatol (2015) 30(3):440–5. doi:10.1111/jgh.12859
3. Day AS, Lemberg DA, Gearry RB. Inflammatory bowel disease in Australasian children and adolescents. Gastroenterol Res Pract (2014) 2014:703890. doi:10.1155/2014/703890
4. Quetglas EG, Armuzzi A, Wigge S, Fiorino G, Barnscheid L, Froelich M, et al. Review article: the pharmacokinetics and pharmacodynamics of drugs used in inflammatory bowel disease treatment. Eur J Clin Pharmacol (2015) 71(7):773–99. doi:10.1007/s00228-015-1862-7
5. Lee HS, Park SH, Yang SK, Ye BD, Kim JH, Kim SO, et al. The risk of colorectal cancer in inflammatory bowel disease: a hospital-based cohort study from Korea. Scand J Gastroenterol (2015) 50(2):188–96. doi:10.3109/00365521.2014.989538
6. Itzkowitz S. Colon carcinogenesis in inflammatory bowel disease: applying molecular genetics to clinical practice. J Clin Gastroenterol (2003) 36(5 Suppl):S70–4. doi:10.1097/00004836-200305001-00012
7. Ferguson LR, Chen H, Collins AR, Connell M, Damia G, Dasgupta S, et al. Genomic instability in human cancer: molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Semin Cancer Biol (2015). doi:10.1016/j.semcancer.2015.03.005
8. Fenech MF. Nutriomes and personalised nutrition for DNA damage prevention, telomere integrity maintenance and cancer growth control. Cancer Treat Res (2014) 159:427–41. doi:10.1007/978-3-642-38007-5_24
9. Vasan S, Deem A, Ramakrishnan S, Argueso JL, Malkova A. Cascades of genetic instability resulting from compromised break-induced replication. PLoS Genet (2014) 10(2):e1004119. doi:10.1371/journal.pgen.1004119
10. Ferguson LR, Denny WA. COMMENTARY: pre-emptive nutrition: refining the targets of drugs targeted to colorectal cancer. Curr Cancer Drug Targets (2015) 15(3):173–5. doi:10.2174/156800961503150505120604
11. Triggs CM, Munday K, Hu R, Fraser AG, Gearry RB, Barclay ML, et al. Dietary factors in chronic inflammation: food tolerances and intolerances of a New Zealand Caucasian Crohn’s disease population. Mutat Res (2010) 690(1–2):123–38. doi:10.1016/j.mrfmmm.2010.01.020
12. Muir JG, Gibson PR. The low FODMAP diet for treatment of irritable bowel syndrome and other gastrointestinal disorders. Gastroenterol Hepatol (N Y) (2013) 9(7):450–2.
13. Donnellan CF, Yann LH, Lal S. Nutritional management of Crohn’s disease. Therap Adv Gastroenterol (2013) 6(3):231–42. doi:10.1177/1756283X13477715
14. Rastall RA, Gibson GR. Recent developments in prebiotics to selectively impact beneficial microbes and promote intestinal health. Curr Opin Biotechnol (2015) 32:42–6. doi:10.1016/j.copbio.2014.11.002
15. Carvalho AY, Bishop KS, Han DY, Ellett S, Jesuthasan A, Lam WJ, et al. The role of Vitamin D level and related single nucleotide polymorphisms in Crohn’s disease. Nutrients (2013) 5(10):3898–909. doi:10.3390/nu5103898
16. Serhan CN, Chiang N, Dalli J, Levy BD. Lipid mediators in the resolution of inflammation. Cold Spring Harb Perspect Biol (2015) 7(2):a016311. doi:10.1101/cshperspect.a016311
17. Zhou X, Ruan Z, Huang X, Zhou Y, Liu S, Yin Y. The prebiotic lactosucrose modulates gut metabolites and microbiota in intestinal inflammatory rats. Food Sci Biotechnol (2014) 23(1):157–63. doi:10.1007/s10068-014-0021-8
18. Ferguson LR. Nutrigenetics, nutrigenomics and inflammatory bowel diseases. Expert Rev Clin Immunol (2013) 9(8):717–26. doi:10.1586/1744666X.2013.824245
19. Gentschew L, Ferguson LR. Role of nutrition and microbiota in susceptibility to inflammatory bowel diseases. Mol Nutr Food Res (2012) 56(4):524–35. doi:10.1002/mnfr.201100630
20. Barnett MP, Cooney JM, Dommels YE, Nones K, Brewster DT, Park Z, et al. Modulation of colonic inflammation in Mdr1a (-)/(-) mice by green tea polyphenols and their effects on the colon transcriptome and proteome. J Nutr Biochem (2013) 24(10):1678–90. doi:10.1016/j.jnutbio.2013.02.007
21. Ferguson LR. Role of plant polyphenols in genomic stability. Mutat Res (2001) 475(1–2):89–111. doi:10.1016/S0027-5107(01)00073-2
22. Laing B, Han DY, Ferguson LR. Candidate genes involved in beneficial or adverse responses to commonly eaten brassica vegetables in a New Zealand Crohn’s disease cohort. Nutrients (2013) 5(12):5046–64. doi:10.3390/nu5125046
23. Ferguson LR, Hu R, Lam WJ, Munday K, Triggs CM. Tailoring foods to match people’s genes in New Zealand: opportunities for collaboration. J Nutrigenet Nutrigenomics (2010) 3(4–6):305–11. doi:10.1159/000324369
24. Lewis CM, Whitwell SC, Forbes A, Sanderson J, Mathew CG, Marteau TM. Estimating risks of common complex diseases across genetic and environmental factors: the example of Crohn disease. J Med Genet (2007) 44(11):689–94. doi:10.1136/jmg.2007.051672
25. Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet (2010) 42(12):1118–25. doi:10.1038/ng.717
26. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature (2012) 491(7422):119–24. doi:10.1038/nature11582
27. Goyette P, Boucher G, Mallon D, Ellinghaus E, Jostins L, Huang H, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1 01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet (2015) 47(2):172–9. doi:10.1038/ng.3176
28. Wang J, Yu JC, Kang WM, Ma ZQ. Superiority of a fish oil-enriched emulsion to medium-chain triacylglycerols/long-chain triacylglycerols in gastrointestinal surgery patients: a randomized clinical trial. Nutrition (2012) 28(6):623–9. doi:10.1016/j.nut.2011.08.004
29. Masterson JC, McNamee EN, Fillon SA, Hosford L, Harris R, Fernando SD, et al. Eosinophil-mediated signalling attenuates inflammatory responses in experimental colitis. Gut (2014) 8:1326–47. doi:10.1136/gutjnl-2014-306998
30. Feng Y, Browner P, Teitelbaum DH. Effects on varying intravenous lipid emulsions on the small bowel epithelium in a mouse model of parenteral nutrition. JPEN J Parenter Enteral Nutr (2013) 37(6):775–86. doi:10.1177/0148607113491608
31. Calder PC, Albers R, Antoine JM, Blum S, Bourdet-Sicard R, Ferns GA, et al. Inflammatory disease processes and interactions with nutrition. Br J Nutr (2009) 101(Suppl 1):S1–45. doi:10.1017/S0007114509377867
32. Honda KL, Lamon-Fava S, Matthan NR, Wu D, Lichtenstein AH. EPA and DHA exposure alters the inflammatory response but not the surface expression of toll-like receptor 4 in macrophages. Lipids (2015) 50(2):121–9. doi:10.1007/s11745-014-3971-y
33. Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature (2014) 506(7489):456–62. doi:10.1038/nature13044
34. Folkard DL, Marlow G, Mithen RF, Ferguson LR. Effect of sulforaphane on NOD2 via NF-kappaB: implications for Crohn’s disease. J Inflamm (2015) 12:6. doi:10.1186/s12950-015-0051-x
35. Cammarota G, Ianiro G, Bibbò S, Gasbarrini A. Gut microbiota modulation: probiotics, antibiotics or fecal microbiota transplantation? Intern Emerg Med (2014) 9(4):365–73. doi:10.1007/s11739-014-1069-4
36. Clark MJ, Robien K, Slavin JL. Effect of prebiotics on biomarkers of colorectal cancer in humans: a systematic review. Nutr Rev (2012) 70(8):436–43. doi:10.1111/j.1753-4887.2012.00495.x
37. Johnson LP, Walton GE, Psichas A, Frost GS, Gibson GR, Barraclough TG. Prebiotics modulate the effects of antibiotics on gut microbial diversity and functioning in vitro. Nutrients (2015) 7(6):4480–97. doi:10.3390/nu7064480
38. Unno T, Sakuma M, Mitsuhashi S. Effect of dietary supplementation of (-)-epigallocatechin gallate on gut microbiota and biomarkers of colonic fermentation in rats. J Nutr Sci Vitaminol (Tokyo) (2014) 60(3):213–9. doi:10.3177/jnsv.60.213
39. Kim HS, Quon MJ, Kim JA. New insights into the mechanisms of polyphenols beyond antioxidant properties; lessons from the green tea polyphenol, epigallocatechin 3-gallate. Redox Biol (2014) 2:187–95. doi:10.1016/j.redox.2013.12.022
40. Dougherty KA, Bertolaso C, Schall JI, Smith-Whitley K, Stallings VA. Safety and efficacy of high-dose daily vitamin D3 supplementation in children and young adults with sickle cell disease. J Pediatr Hematol Oncol (2015) 37(5):e308–15. doi:10.1097/MPH.0000000000000355
41. Barnett MP, McNabb WC, Cookson AL, Zhu S, Davy M, Knoch B, et al. Changes in colon gene expression associated with increased colon inflammation in interleukin-10 gene-deficient mice inoculated with Enterococcus species. BMC Immunol (2010) 11:39. doi:10.1186/1471-2172-11-39
42. Bassaganya-Riera J, Viladomiu M, Pedragosa M, De Simone C, Carbo A, Shaykhutdinov R, et al. Probiotic bacteria produce conjugated linoleic acid locally in the gut that targets macrophage PPAR gamma to suppress colitis. PLoS One (2012) 7(2):e31238. doi:10.1371/journal.pone.0031238
43. Baur P, Martin FP, Gruber L, Bosco N, Brahmbhatt V, Collino S, et al. Metabolic phenotyping of the Crohn’s disease-like IBD etiopathology in the TNF(DeltaARE/WT) mouse model. J Proteome Res (2011) 10(12):5523–35. doi:10.1021/pr2007973
44. Nones K, Dommels YE, Martell S, Butts C, McNabb WC, Park ZA, et al. The effects of dietary curcumin and rutin on colonic inflammation and gene expression in multidrug resistance gene-deficient (mdr1a-/-) mice, a model of inflammatory bowel diseases. Br J Nutr (2009) 101(2):169–81. doi:10.1017/S0007114508009847
45. Lin HM, Barnett MP, Roy NC, Joyce NI, Zhu S, Armstrong K, et al. Metabolomic analysis identifies inflammatory and noninflammatory metabolic effects of genetic modification in a mouse model of Crohn’s disease. J Proteome Res (2010) 9(4):1965–75. doi:10.1021/pr901130s
46. Cooney JM, Barnett MP, Brewster D, Knoch B, McNabb WC, Laing WA, et al. Proteomic analysis of colon tissue from interleukin-10 gene-deficient mice fed polyunsaturated fatty acids with comparison to transcriptomic analysis. J Proteome Res (2012) 11(2):1065–77. doi:10.1021/pr200807p
47. Panwala CM, Jones JC, Viney JL. A novel model of inflammatory bowel disease: mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J Immunol (1998) 161(10):5733–44.
48. Fenech MF. Nutriomes and nutrient arrays – the key to personalised nutrition for DNA damage prevention and cancer growth control. Genome Integr (2010) 1(1):11. doi:10.1186/2041-9414-1-11
49. Fenech MF. Dietary reference values of individual micronutrients and nutriomes for genome damage prevention: current status and a road map to the future. Am J Clin Nutr (2010) 91(5):1438S–54S. doi:10.3945/ajcn.2010.28674D
50. van Ommen B, El-Sohemy A, Hesketh J, Kaput J, Fenech M, Evelo CT, et al. The micronutrient genomics project: a community-driven knowledge base for micronutrient research. Genes Nutr (2010) 5(4):285–96. doi:10.1007/s12263-010-0192-8
51. Karunasinghe N, Han DY, Zhu S, Duan H, Ko YJ, Yu JF, et al. Effects of supplementation with selenium, as selenized yeast, in a healthy male population from New Zealand. Nutr Cancer (2013) 65(3):355–66. doi:10.1080/01635581.2013.760743
52. Ferguson LR, Karunasinghe N. Nutrigenetics, nutrigenomics, and selenium. Front Genet (2011) 2:15. doi:10.3389/fgene.2011.00015
53. Ananthakrishnan AN, Cagan A, Gainer VS, Cheng SC, Cai T, Szolovits P, et al. Higher plasma vitamin D is associated with reduced risk of Clostridium difficile infection in patients with inflammatory bowel diseases. Aliment Pharmacol Ther (2014) 39(10):1136–42. doi:10.1111/apt.12706
54. Ananthakrishnan AN, Cheng SC, Cai T, Cagan A, Gainer VS, Szolovits P, et al. Association between reduced plasma 25-hydroxy vitamin D and increased risk of cancer in patients with inflammatory bowel diseases. Clin Gastroenterol Hepatol (2014) 12(5):821–7. doi:10.1016/j.cgh.2013.10.011
55. Meeker S, Seamons A, Paik J, Treuting PM, Brabb T, Grady WM, et al. Increased dietary vitamin D suppresses MAPK signaling, colitis, and colon cancer. Cancer Res (2014) 74(16):4398–408. doi:10.1158/0008-5472.CAN-13-2820
56. Peyrin-Biroulet L, Beisner J, Wang G, Nuding S, Oommen ST, Kelly D, et al. Peroxisome proliferator-activated receptor gamma activation is required for maintenance of innate antimicrobial immunity in the colon. Proc Natl Acad Sci U S A (2010) 107(19):8772–7. doi:10.1073/pnas.0905745107
57. Hume GE, Fowler EV, Griffiths LR, Doecke JD, Radford-Smith GL. Common PPARgamma variants C161T and Pro12Ala are not associated with inflammatory bowel disease in an Australian cohort. J Gastrointestin Liver Dis (2012) 21(4):349–55.
58. Shrestha UK, Karimi O, Crusius JB, Zhou F, Wang Z, Chen Z, et al. Distribution of peroxisome proliferator-activated receptor-gamma polymorphisms in Chinese and Dutch patients with inflammatory bowel disease. Inflamm Bowel Dis (2010) 16(2):312–9. doi:10.1002/ibd.21059
59. Bassaganya-Riera J, Hontecillas R. Dietary conjugated linoleic acid and n-3 polyunsaturated fatty acids in inflammatory bowel disease. Curr Opin Clin Nutr Metab Care (2010) 13(5):569–73. doi:10.1097/MCO.0b013e32833b648e
60. Ferreira P, Cravo M, Guerreiro CS, Tavares L, Santos PM, Brito M. Fat intake interacts with polymorphisms of Caspase9, FasLigand and PPARgamma apoptotic genes in modulating Crohn’s disease activity. Clin Nutr (2010) 29(6):819–23. doi:10.1016/j.clnu.2010.06.008
61. Ferguson JF, Phillips CM, McMonagle J, Pérez-Martínez P, Shaw DI, Lovegrove JA, et al. NOS3 gene polymorphisms are associated with risk markers of cardiovascular disease, and interact with omega-3 polyunsaturated fatty acids. Atherosclerosis (2010) 211(2):539–44. doi:10.1016/j.atherosclerosis.2010.03.027
62. Martin H, Burgess EJ, Smith WA, McGhie TK, Cooney JM, Lunken RC, et al. JAK2 and AMP-kinase inhibition in vitro by food extracts, fractions and purified phytochemicals. Food Funct (2015) 6(1):305–12. doi:10.1039/c4fo00626g
63. Nasef NA, Mehta S, Murray P, Marlow G, Ferguson LR. Anti-inflammatory activity of fruit fractions in vitro, mediated through toll-like receptor 4 and 2 in the context of inflammatory bowel disease. Nutrients (2014) 6(11):5265–79. doi:10.3390/nu6115265
64. Ahmed Nasef N, Mehta S, Ferguson LR. Dietary interactions with the bacterial sensing machinery in the intestine: the plant polyphenol case. Front Genet (2014) 5:64. doi:10.3389/fgene.2014.00064
65. Comito D, Cascio A, Romano C. Microbiota biodiversity in inflammatory bowel disease. Ital J Pediatr (2014) 40:32. doi:10.1186/1824-7288-40-32
66. Fukuda K, Fujita Y. Determination of the discriminant score of intestinal microbiota as a biomarker of disease activity in patients with ulcerative colitis. BMC Gastroenterol (2014) 14:49. doi:10.1186/1471-230X-14-49
67. Rossen NG, MacDonald JK, de Vries EM, D’Haens GR, de Vos WM, Zoetendal EG, et al. Fecal microbiota transplantation as novel therapy in gastroenterology: a systematic review. World J Gastroenterol (2015) 21(17):5359–71. doi:10.3748/wjg.v21.i17.5359
68. Kelly CR, Kahn S, Kashyap P, Laine L, Rubin D, Atreja A, et al. Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology (2015) 149(1):223–37. doi:10.1053/j.gastro.2015.05.008
69. Goldsmith JR, Sartor RB. The role of diet on intestinal microbiota metabolism: downstream impacts on host immune function and health, and therapeutic implications. J Gastroenterol (2014) 49(5):785–98. doi:10.1007/s00535-014-0953-z
70. Ottman N, Smidt H, de Vos WM, Belzer C. The function of our microbiota: who is out there and what do they do? Front Cell Infect Microbiol (2012) 2:104. doi:10.3389/fcimb.2012.00104
71. Marteau P. Therapy: probiotic-enriched artichokes for abdominal discomfort. Nat Rev Gastroenterol Hepatol (2012) 9(5):251–2. doi:10.1038/nrgastro.2012.49
72. Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology (2013) 145(2):293–308. doi:10.1053/j.gastro.2013.05.050
73. Krissansen GW, Yang Y, McQueen FM, Leung E, Peek D, Chan YC, et al. Overexpression of miR-595 and miR-1246 in the sera of patients with active forms of inflammatory bowel disease. Inflamm Bowel Dis (2015) 21(3):520–30. doi:10.1097/MIB.0000000000000285
74. Kalman B, Toldy E. Genomic binding sites and biological effects of the vitamin D – VDR complex in multiple sclerosis [corrected]. Neuromolecular Med (2014) 16(2):265–79. doi:10.1007/s12017-014-8301-2
75. Ma Y, Hu Q, Luo W, Pratt RN, Glenn ST, Liu S, et al. 1alpha,25(OH)2D3 differentially regulates miRNA expression in human bladder cancer cells. J Steroid Biochem Mol Biol (2015) 148:166–71. doi:10.1016/j.jsbmb.2014.09.020
76. Saksa N, Neme A, Ryynänen J, Uusitupa M, de Mello VD, Voutilainen S, et al. Dissecting high from low responders in a vitamin D3 intervention study. J Steroid Biochem Mol Biol (2015) 148:275–82. doi:10.1016/j.jsbmb.2014.11.012
77. Knoch B, Barnett MP, Zhu S, Park ZA, Nones K, Dommels YE, et al. Genome-wide analysis of dietary eicosapentaenoic acid- and oleic acid-induced modulation of colon inflammation in interleukin-10 gene-deficient mice. J Nutrigenet Nutrigenomics (2009) 2(1):9–28. doi:10.1159/000134292
78. Zhang Y, Han L, Qi W, Cheng D, Ma X, Hou L, et al. Eicosapentaenoic acid (EPA) induced apoptosis in HepG2 cells through ROS-Ca(2+)-JNK mitochondrial pathways. Biochem Biophys Res Commun (2015) 456(4):926–32. doi:10.1016/j.bbrc.2014.12.036
79. Korobkova EA. Effect of natural polyphenols on CYP metabolism: implications for diseases. Chem Res Toxicol (2015) 28(7):1359–90. doi:10.1021/acs.chemrestox.5b00121
80. Ayissi VB, Ebrahimi A, Schluesenner H. Epigenetic effects of natural polyphenols: a focus on SIRT1-mediated mechanisms. Mol Nutr Food Res (2014) 58(1):22–32. doi:10.1002/mnfr.201300195
81. Deng Y, Lu X, Wang L, Li T, Ding Y, Cao H, et al. Curcumin inhibits the AKT/NF-kappaB signaling via CpG demethylation of the promoter and restoration of NEP in the N2a cell line. AAPS J (2014) 16(4):649–57. doi:10.1208/s12248-014-9605-8
82. Harati K, Slodnik P, Chromik AM, Goertz O, Hirsch T, Kapalschinski N, et al. Resveratrol induces apoptosis and alters gene expression in human fibrosarcoma cells. Anticancer Res (2015) 35(2):767–74.
83. Lin CH, Chao LK, Hung PH, Chen YJ. EGCG inhibits the growth and tumorigenicity of nasopharyngeal tumor-initiating cells through attenuation of STAT3 activation. Int J Clin Exp Pathol (2014) 7(5):2372–81.
84. Joven J, Micol V, Segura-Carretero A, Alonso-Villaverde C, Menéndez JA, Bioactive Food Components Platform. Polyphenols and the modulation of gene expression pathways: can we eat our way out of the danger of chronic disease? Crit Rev Food Sci Nutr (2014) 54(8):985–1001. doi:10.1080/10408398.2011.621772
85. Plaza-Diaz J, Gomez-Llorente C, Fontana L, Gil A. Modulation of immunity and inflammatory gene expression in the gut, in inflammatory diseases of the gut and in the liver by probiotics. World J Gastroenterol (2014) 20(42):15632–49. doi:10.3748/wjg.v20.i42.15632
86. Druart C, Alligier M, Salazar N, Neyrinck AM, Delzenne NM. Modulation of the gut microbiota by nutrients with prebiotic and probiotic properties. Adv Nutr (2014) 5(5):624S–33S. doi:10.3945/an.114.005835
87. Sauer J, Richter KK, Pool-Zobel BL. Products formed during fermentation of the prebiotic inulin with human gut flora enhance expression of biotransformation genes in human primary colon cells. Br J Nutr (2007) 97(5):928–37. doi:10.1017/S0007114507666422
88. Zenhom M, Hyder A, de Vrese M, Heller KJ, Roeder T, Schrezenmeir J. Prebiotic oligosaccharides reduce proinflammatory cytokines in intestinal Caco-2 cells via activation of PPARgamma and peptidoglycan recognition protein 3. J Nutr (2011) 141(5):971–7. doi:10.3945/jn.110.136176
89. Marlow G, Ellett S, Ferguson IR, Zhu S, Karunasinghe N, Jesuthasan AC, et al. Transcriptomics to study the effect of a Mediterranean-inspired diet on inflammation in Crohn’s disease patients. Hum Genomics (2013) 7:24. doi:10.1186/1479-7364-7-24
90. Polytarchou C, Koukos G, Iliopoulos D. Systems biology in inflammatory bowel diseases: ready for prime time. Curr Opin Gastroenterol (2014) 30(4):339–46. doi:10.1097/MOG.0000000000000081
91. Coughlin SS. Toward a road map for global -omics: a primer on -omic technologies. Am J Epidemiol (2014) 180(12):1188–95. doi:10.1093/aje/kwu262
92. Bennike T, Birkelund S, Stensballe A, Andersen V. Biomarkers in inflammatory bowel diseases: current status and proteomics identification strategies. World J Gastroenterol (2014) 20(12):3231–44. doi:10.3748/wjg.v20.i12.3231
93. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology (2014) 146(6):1489–99. doi:10.1053/j.gastro.2014.02.009
94. Fiocchi C. Integrating omics: the future of IBD? Dig Dis (2014) 32(Suppl 1):96–102. doi:10.1159/000367836
95. De Preter V, Verbeke K. Metabolomics as a diagnostic tool in gastroenterology. World J Gastrointest Pharmacol Ther (2013) 4(4):97–107. doi:10.4292/wjgpt.v4.i4.97
96. Lin HM, Edmunds SJ, Zhu S, Helsby NA, Ferguson LR, Rowan DD. Metabolomic analysis reveals differences in urinary excretion of kiwifruit-derived metabolites in a mouse model of inflammatory bowel disease. Mol Nutr Food Res (2011) 55(12):1900–4. doi:10.1002/mnfr.201100302
97. Martin FP, Rezzi S, Philippe D, Tornier L, Messlik A, Hölzlwimmer G, et al. Metabolic assessment of gradual development of moderate experimental colitis in IL-10 deficient mice. J Proteome Res (2009) 8(5):2376–87. doi:10.1021/pr801006e
98. McHardy IH, Goudarzi M, Tong M, Ruegger PM, Schwager E, Weger JR, et al. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome (2013) 1(1):17. doi:10.1186/2049-2618-1-17
99. Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Therap Adv Gastroenterol (2015) 8(1):4–22. doi:10.1177/1756283X14547360
100. Bjerrum JT, Wang Y, Hao F, Coskun M, Ludwig C, Günther U, et al. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn’s disease and healthy individuals. Metabolomics (2015) 11:122–33. doi:10.1007/s11306-014-0677-3
101. Dawiskiba T, Deja S, Mulak A, Zabek A, Jawień E, Pawełka D, et al. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J Gastroenterol (2014) 20(1):163–74. doi:10.3748/wjg.v20.i1.163
102. Lin HM, Helsby NA, Rowan DD, Ferguson LR. Using metabolomic analysis to understand inflammatory bowel diseases. Inflamm Bowel Dis (2011) 17(4):1021–9. doi:10.1002/ibd.21426
103. Anderson NL, Anderson NG. Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis (1998) 19(11):1853–61. doi:10.1002/elps.1150191103
104. Knoch B, Barnett MP, Cooney J, McNabb WC, Barraclough D, Laing W, et al. Dietary oleic acid as a control fatty acid for polyunsaturated fatty acid intervention studies: a transcriptomics and proteomics investigation using interleukin-10 gene-deficient mice. Biotechnol J (2010) 5(11):1226–40. doi:10.1002/biot.201000066
105. Huang H, Vangay P, McKinlay CE, Knights D. Multi-omics analysis of inflammatory bowel disease. Immunol Lett (2014) 162(2):62–8. doi:10.1016/j.imlet.2014.07.014
Keywords: vitamin D, phytochemicals, probiotics, genetics, genomics, microbiota
Citation: Ferguson LR (2015) Nutritional modulation of gene expression: might this be of benefit to individuals with Crohn’s disease? Front. Immunol. 6:467. doi: 10.3389/fimmu.2015.00467
Received: 17 June 2015; Accepted: 27 August 2015;
Published: 11 September 2015
Edited by:
Amélia M. Sarmento, Universidade Fernando Pessoa, PortugalReviewed by:
Diane Bimczok, Montana State University, USACopyright: © 2015 Ferguson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lynnette R. Ferguson, Discipline of Nutrition and Dietetics, Faculty of Medical and Health Sciences, The University of Auckland, Private Bag 92019, Auckland 1184, New Zealand,bC5mZXJndXNvbkBhdWNrbGFuZC5hYy5ueg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.