
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 17 August 2015
Sec. Antigen Presenting Cell Biology
Volume 6 - 2015 | https://doi.org/10.3389/fimmu.2015.00423
This article is part of the Research TopicDendritic Cell and Macrophage nomenclature and classificationView all 18 articles
In contrast to the past reliance on morphology, the identification and enumeration of blood monocytes are nowadays done with monoclonal antibodies and flow cytometry and this allows for subdivision into classical, intermediate, and non-classical monocytes. Using specific cell surface markers, dendritic cells in blood can be segregated from these monocytes. While in the past, changes in monocyte numbers as determined in standard hematology counters have not had any relevant clinical impact, the subset analysis now has uncovered informative changes that may be used in management of disease.
The term monocyte is used for blood cells of a lineage called monocytes/macrophages or mononuclear phagocytes. These blood monocytes are bone marrow-derived leukocytes that are functionally characterized by the ability to phagocytose, to produce cytokines, and to present antigen. In early studies, they had been identified based on glass adherence and morphology (1). Also, cytochemistry for specific enzymes like monocyte-specific esterase (2, 3) has been employed, while the standard approach in clinical hematology relies on physical properties of these cells including light scatter.
In bone marrow, the monocytes derive from myelo-monocytic stem cells, which give rise to more direct precursors like monoblasts and pro-monocytes. These cells earlier were identified based on morphology (4) such that the monoblast was an ill-defined cell type. More recently in the mouse model, a Ly6C+ CD115+ CD117+ monoblast-type cell, termed common monocyte progenitor (cMoP), was identified in bone marrow and spleen and this cell is able to proliferate and give rise to the different monocyte subsets (5). A cMoP monoblast type of cell remains to be identified for man and other species.
The number of circulating blood monocytes in man can strongly increase within minutes by stress or exercise followed by a rapid return to baseline levels. These recruited cells are thought to come from what is called the marginal pool (6). This compartment describes areas of reduced blood velocity close to the endothelium of venules and here cells can loosely adhere and can be mobilized in a catecholamine-dependent fashion (7). These marginal pool monocytes can have an adhesion molecule pattern distinct from monocytes found in blood at rest.
In addition, CD11bhigh (CD90, B220, CD49b, NK1.1, Ly-6G, F4/80, I-Ab, CD11c)low cells are mobilized from the spleen after severe injury (8). These cells have monocyte morphology and their transcriptome matches with that of blood monocytes. Furthermore, CD11b+ Ly6Chi monocytes can be mobilized from bone marrow to blood in infectious disease models (9), and adoptively transferred monocytes were shown to return to the bone marrow (10) in the mouse. What remains to be determined is whether the spleen and bone marrow compartments also contribute to the pool of monocytes that can be mobilized by stress and exercise.
When under homeostatic or inflammatory conditions, the monocytes have migrated into tissue; then by definition, these cells are called macrophages. Cells newly emigrated into the lung have been termed monocytes in some studies [e.g., Ref. (11)]. Since monocytes, once they have arrived in tissue, will start to transform into larger cells and rapidly lose their monocyte characteristics, others have called these recently emigrated cells “small macrophages” (12).
More detailed studies in the mouse have demonstrated tissue cells with characteristics close to blood monocytes (13, 14). However, these cells in the lymph node show a gene expression pattern that distinguishes them from the blood cells (14) and in the skin they show increased expression of lysozyme and CD68, markers typical of mature macrophages (13). Therefore, more data are required in the mouse model and obviously also in man before a consensus can be reached whether we use the term tissue monocyte or whether we continue to call these cells macrophages. Until these issues have been resolved, the term monocyte should be restricted to cells in the blood compartment and the bone marrow and spleen reservoirs that can replenish the blood monocyte pool.
As explained above, monocytes initially had been identified by function and morphology and these criteria have been misleading especially when disease processes altered these features. Therefore, attempts have been made to define unequivocal criteria for monocytes. Here, monoclonal antibodies against cell surface molecules have been proposed. In man, CD14 has been used as a marker (15), and in the mouse, CD115 is often employed (16). CD115 identifies the M-CSF receptor and has the main drawback that in the mouse, it is downregulated on blood monocytes with inflammation (17). Also, the question is whether such markers are sufficiently specific and do not react with other cell types like dendritic cells (DCs). In fact, part of the CD1c+ blood DCs in man can express low-level CD14 (18) and also human B cells have been reported to express some CD14 (19). Therefore, monocytes can be identified with markers like CD14 and CD115, but this should be supported by additional markers and by functional studies. Interestingly, when searching for macrophage-specific transcripts in the mouse, CD64 and MerTK have emerged (20). While CD64 is absent from non-classical monocytes in man, MerTK is a molecule that might prove informative for blood monocytes in different species. In addition, staining for CD16, which is used for monocyte subset definition (see below), will at the same time help to exclude DCs in human blood.
Dendritic cells were first described by Steinman and Cohn as stellate cells isolated from mouse spleen (21). Over the years, there have been debates as to whether these cells are a distinct lineage or part of the mononuclear phagocyte system. A common precursor for monocytes and DCs was described in the mouse (22), but the existence of this cell was later disputed (23) suggesting that DCs and monocytes may diverge at an earlier multi-potent progenitor stage (24).
However, the demonstration that monocytes can be used to generate DCs in vitro by adding GM-CSF and IL-4 suggested a close relationship between monocytes and DCs (25). Later, transcriptome analysis demonstrated that such monocyte-derived DCs rather resemble macrophages than DCs from lymphoid tissue (26). Therefore, these in vitro generated monocyte-derived cells are potent antigen-presenting cells, but they do not represent bona fide DCs; they rather belong to the monocyte/macrophage lineage. Still not resolved is the question whether in tissue the monocyte-derived cells with high levels of class II expression and with high antigen-presenting capacity should be termed monocyte-derived DC (13, 27, 28) or activated macrophages.
In addition to DCs in tissue, cells with DC properties have been described in blood based on the expression of CD68, CD1c, or CD141 (29, 30). Transcriptome analysis has demonstrated that these cells and the monocytes belong to different clusters (26, 31). These data suggest that blood DCs can be segregated from monocytes and macrophages as a separate lineage.
The data also demonstrate the power of transcriptomic analysis in defining and dissecting leukocyte populations like monocytes and DCs. Ontogeny can help in such a definition, but in men, adoptive transfer is limited to strategies like transfer of bone marrow stem cells, and informative mutations are rare. Also, the ontogeny approach needs to be used with caution since a defined progenitor cell can give rise to clearly distinct cell populations. An informative example is the megakaryocyte–erythrocyte progenitor (MEP) cell, which gives rise to either megakaryocytes and their platelet progeny or to erythroblasts and their red blood cell progeny (32, 33). Megakaryocytes and erythroblasts have a distinct transcriptome (34), and they are involved in distinct functions, i.e., in blood clotting and oxygen transport, respectively. Therefore, although having a common ontogeny, these cells belong to clearly separate lineages. This example illustrates that ontogeny can provide a framework, but a comprehensive analyses like transcriptomics and the analysis of cell function are required for dissecting cell types and for developing a nomenclature. Therefore, in order to assign a novel leukocyte population in blood or tissue to either monocytes or DCs, a straight-forward approach is to analyze the transcriptome (and other omics like the proteome, lipidome, glycome, or metabolome) of these cells in comparison to typical monocytes and DCs and to then ask whether the novel cell type co-clusters with either prototypic monocytes or DCs (26).
Evidence for monocyte subpopulations has come from experiments using differential flotation in counter-current elutriation (35) and from differential binding to antibody-coated red blood cells, which has defined populations with different functions (36). With the use of monoclonal antibodies and flow cytometry, tools have become available to clearly define, enumerate, and isolate monocyte subsets based on the differential expression of CD14 and CD16 cell-surface markers (37).
In 2010, an international consortium under the auspices of the IUIS and the WHO has proposed a nomenclature for monocyte subpopulations (38). The proposal defined the major population of CD14high cells found in human blood as classical monocytes and the minor population of cells with low CD14 and high CD16 as non-classical monocytes. A population in between these two subsets was termed intermediate monocytes (see Figure 1).
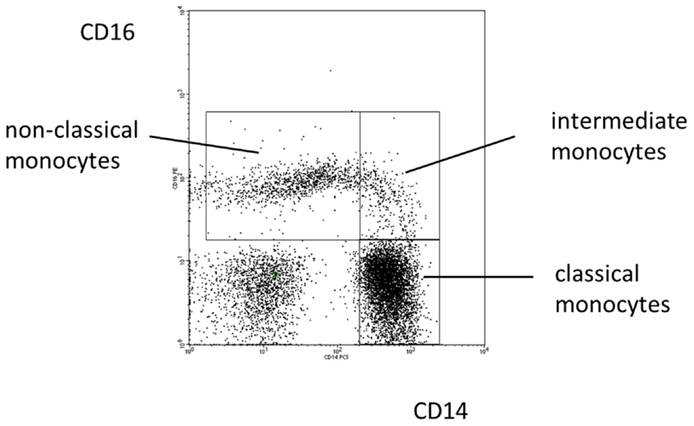
Figure 1. Blood monocyte subsets in man. Illustration of the definition of human monocyte subsets in health based on a typical distribution of events in a CD14 CD16 staining.
While an unequivocal approach to defining the intermediate monocytes has not been developed, as yet (39), a host of studies on intermediate monocytes has been published since the 2010 proposal. In fact, a search for the term “intermediate monocyte” under Google Scholar has revealed more than 100 studies on these cells since 2010. These reports have described an expansion of intermediate monocytes in various inflammatory diseases and these cells have been shown to be of prognostic relevance in cardiovascular disease (40). The use of additional markers for delineation of intermediate monocytes has been suggested (41) and it remains to be shown whether markers, such as CCR2 or slan, will improve the definition of these cells.
The same nomenclature as proposed for man can be used in other species [reviewed in Ref. (42)]. The respective cells can be very similar to men as seen for non-human primates (43, 44). In species like the mouse, the classical and non-classical monocyte subsets can be identified as well, but different markers like CD115, Ly6C, and CD43 are used (16, 45). Also in species like rat, pig, cow, and horse, classical and non-classical monocytes can be defined and even intermediate monocytes have been described in some animals (42). It is predicted that the nomenclature of monocyte subsets will be applicable to all mammalian species.
In human blood, a population of slan-positive cells has been described as DCs, but phenotypic analysis has shown that these cells are CD14-low and CD16-high (46), functional studies demonstrated a high capacity to produce TNF (47), and clinical studies showed that these cells are depleted by glucocorticoid treatment (48). These features are identical to what has been reported as characteristics of non-classical CD14+CD16++ monocytes (37, 49, 50). Also, the increased absolute numbers of slan-positive monocytes and of non-classical monocytes show a clear correlation in HIV-infected patients (51), and part of the non-classical monocytes has been shown to be slan-positive (52–54). Collectively, these findings suggest that the slan-positive cells belong to the non-classical monocytes.
There may be additional monocyte subsets including Fcepsilon-RI-positive cells (55), which were found with a median of 2.5% among CD14-positive blood monocytes in a pediatric cohort (56) and these cells may be involved in IgE clearance (57). Also, proliferating monocytes have been described (58) as well as precursors for fibrocytes (59) and osteoclasts (60). For all of these cell types, further characterization is awaited.
Monocyte numbers as defined in the hematology lab using light scatter properties have not contributed much to diagnosis and monitoring of disease, but with the definition of monocyte subsets by flow cytometry, informative patterns have emerged. For example, severe infection will increase the number of non-classical and intermediate monocytes (61–63). Here, it remains to be analyzed whether such an increase can predict prognosis, as has been suggested (64). Furthermore, therapy with glucocorticoids leads to a decrease of non-classical monocytes, which appears to be due to a selective induction of apoptosis in the non-classical monocytes while classical monocytes even increase in number under glucocorticoids (50, 65). Also, blockade of the M-CSF pathway can lead to depletion of non-classical monocytes (66–68). A likely explanation is that M-CSF signaling via the CD115 M-CSF receptor is required for the classical monocytes to mature into non-classical monocytes. Again still to be determined is whether such a drug-induced depletion can be used to predict therapeutic response in inflammatory diseases. Still unresolved is the mechanism of depletion of non-classical monocytes in three siblings within one family (69). Here, more families with this type of defect need to be analyzed in order to identify the gene and the mechanisms involved. Finally, the absolute count of intermediate monocytes was shown to predict cardiovascular events (70, 71). Hence, analysis of monocyte subsets by flow cytometry now provides clinically useful parameters in various settings. What remains to be established in this context is an unequivocal dissection of the non-classical and the intermediate monocytes.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. van Furth R, Cohn ZA. The origin and kinetics of mononuclear phagocytes. J Exp Med (1968) 128:415–35. doi: 10.1084/jem.128.3.415
2. Tucker SB, Pierre RV, Jordon RE. Rapid identification of monocytes in a mixed mononuclear cell preparation. J Immunol Methods (1977) 14:267–9. doi:10.1016/0022-1759(77)90137-5
3. Uphoff CC, Drexler HG. Biology of monocyte-specific esterase. Leuk Lymphoma (2000) 39:257–70. doi:10.3109/10428190009065825
4. Goud TJ, Schotte C, van Furth R. Identification and characterization of the monoblast in mononuclear phagocyte colonies grown in vitro. J Exp Med (1975) 142:1180–99. doi:10.1084/jem.142.5.1180
5. Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol (2013) 14:821–30. doi:10.1038/ni.2638
6. Klonz A, Wonigeit K, Pabst R, Westermann J. The marginal blood pool of the rat contains not only granulocytes, but also lymphocytes, NK-cells and monocytes: a second intravascular compartment, its cellular composition, adhesion molecule expression and interaction with the peripheral blood pool. Scand J Immunol (1996) 44:461–9. doi:10.1046/j.1365-3083.1996.d01-334.x
7. Steppich B, Dayyani F, Gruber R, Lorenz R, Mack M, Ziegler-Heitbrock HWL. Selective mobilization of CD14(+)CD16(+) monocytes by exercise. Am J Physiol Cell Physiol (2000) 279:C578–86.
8. Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science (2009) 325:612–6. doi:10.1126/science.1175202
9. Shi C, Jia T, Mendez-Ferrer S, Hohl TM, Serbina NV, Lipuma L, et al. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity (2011) 34:590–601. doi:10.1016/j.immuni.2011.02.016
10. Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, Margalit R, et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med (2007) 204:171–80. doi:10.1084/jem.20061011
11. Alexis N, Soukup J, Ghio A, Becker S. Sputum phagocytes from healthy individuals are functional and activated: a flow cytometric comparison with cells in bronchoalveolar lavage and peripheral blood. Clin Immunol (2000) 97:21–32. doi:10.1006/clim.2000.4911
12. Frankenberger M, Eder C, Hofer TP, Heimbeck I, Skokann K, Kassner G, et al. Chemokine expression by small sputum macrophages in COPD. Mol Med (2011) 17:762–70. doi:10.2119/molmed.2010.00202
13. Tamoutounour S, Guilliams M, Montanana Sanchis F, Liu H, Terhorst D, Malosse C, et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity (2013) 39:925–38. doi:10.1016/j.immuni.2013.10.004
14. Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity (2013) 39:599–610. doi:10.1016/j.immuni.2013.08.007
15. Ziegler-Heitbrock HWL, Ulevitch RJ. CD14: cell surface receptor and differentiation marker. Immunol Today (1993) 14:121–5. doi:10.1016/0167-5699(93)90212-4
16. Sunderkotter C, Nikolic T, Dillon MJ, Van Rooijen N, Stehling M, Drevets DA, et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol (2004) 172:4410–7. doi:10.4049/jimmunol.172.7.4410
17. Drevets DA, Schawang JE, Mandava VK, Dillon MJ, Leenen PJ. Severe Listeria monocytogenes infection induces development of monocytes with distinct phenotypic and functional features. J Immunol (2010) 185:2432–41. doi:10.4049/jimmunol.1000486
18. Schwarz H, Schmittner M, Duschl A, Horejs-Hoeck J. Residual endotoxin contaminations in recombinant proteins are sufficient to activate human CD1c+ dendritic cells. PLoS One (2014) 9:e113840. doi:10.1371/journal.pone.0113840
19. Ziegler-Heitbrock HWL, Pechumer H, Petersmann I, Durieux JJ, Vita N, Labeta MO, et al. CD14 is expressed and functional in human B cells. Eur J Immunol (1994) 24:1937–40. doi:10.1002/eji.1830240835
20. Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol (2012) 13:1118–28. doi:10.1038/ni.2419
21. Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med (1973) 137:1142–62. doi:10.1084/jem.137.5.1142
22. Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, Littman DR, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science (2006) 311:83–7. doi:10.1126/science.1117729
23. Sathe P, Metcalf D, Vremec D, Naik SH, Langdon WY, Huntington ND, et al. Lymphoid tissue and plasmacytoid dendritic cells and macrophages do not share a common macrophage-dendritic cell-restricted progenitor. Immunity (2014) 41:104–15. doi:10.1016/j.immuni.2014.05.020
24. Onai N, Ohteki T. Bipotent or oligopotent? A macrophage and DC progenitor revisited. Immunity (2014) 41:5–7. doi:10.1016/j.immuni.2014.07.004
25. Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med (1994) 179:1109–18. doi:10.1084/jem.179.4.1109
26. Robbins SH, Walzer T, Dembele D, Thibault C, Defays A, Bessou G, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol (2008) 9:R17. doi:10.1186/gb-2008-9-1-r17
27. Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity (1999) 11:753–61. doi:10.1016/S1074-7613(00)80149-1
28. Landsman L, Varol C, Jung S. Distinct differentiation potential of blood monocyte subsets in the lung. J Immunol (2007) 178:2000–7. doi:10.4049/jimmunol.178.4.2000
29. Strobl H, Scheinecker C, Riedl E, Csmarits B, Bello-Fernandez C, Pickl WF, et al. Identification of CD68+lin- peripheral blood cells with dendritic precursor characteristics. J Immunol (1998) 161:740–8.
30. Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, et al. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol (2000) 165:6037–46. doi:10.4049/jimmunol.165.11.6037
31. Frankenberger M, Hofer TP, Marei A, Dayyani F, Schewe S, Strasser C, et al. Transcript profiling of CD16-positive monocytes reveals a unique molecular fingerprint. Eur J Immunol (2012) 42:957–74. doi:10.1002/eji.201141907
32. Debili N, Coulombel L, Croisille L, Katz A, Guichard J, Breton-Gorius J, et al. Characterization of a bipotent erythro-megakaryocytic progenitor in human bone marrow. Blood (1996) 88:1284–96.
33. Klimchenko O, Mori M, Distefano A, Langlois T, Larbret F, Lecluse Y, et al. A common bipotent progenitor generates the erythroid and megakaryocyte lineages in embryonic stem cell-derived primitive hematopoiesis. Blood (2009) 114:1506–17. doi:10.1182/blood-2008-09-178863
34. Macaulay IC, Tijssen MR, Thijssen-Timmer DC, Gusnanto A, Steward M, Burns P, et al. Comparative gene expression profiling of in vitro differentiated megakaryocytes and erythroblasts identifies novel activatory and inhibitory platelet membrane proteins. Blood (2007) 109:3260–9. doi:10.1182/blood-2006-07-036269
35. Norris DA, Morris RM, Sanderson RJ, Kohler PF. Isolation of functional subsets of human peripheral blood monocytes. J Immunol (1979) 123:166–72.
36. Zembala M, Uracz W, Ruggiero I, Mytar B, Pryjma J. Isolation and functional characteristics of FcR+ and FcR- human monocyte subsets. J Immunol (1984) 133:1293–9.
37. Passlick B, Flieger D, Ziegler-Heitbrock HWL. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood (1989) 74:2527–34.
38. Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, et al. Nomenclature of monocytes and dendritic cells in blood. Blood (2010) 116:e74–80. doi:10.1182/blood-2010-02-258558
39. Ziegler-Heitbrock L, Hofer TP. Toward a refined definition of monocyte subsets. Front Immunol (2013) 4:23. doi:10.3389/fimmu.2013.00023
40. Rogacev KS, Cremers B, Zawada AM, Seiler S, Binder N, Ege P, et al. CD14++CD16+ monocytes independently predict cardiovascular events: a cohort study of 951 patients referred for elective coronary angiography. J Am Coll Cardiol (2012) 60:1512–20. doi:10.1016/j.jacc.2012.07.019
41. Wong KL, Yeap WH, Tai JJ, Ong SM, Dang TM, Wong SC. The three human monocyte subsets: implications for health and disease. Immunol Res (2012) 53:41–57. doi:10.1007/s12026-012-8297-3
42. Ziegler-Heitbrock L. Monocyte subsets in man and other species. Cell Immunol (2014) 289:135–9. doi:10.1016/j.cellimm.2014.03.019
43. Kim WK, Sun Y, Do H, Autissier P, Halpern EF, Piatak M Jr, et al. Monocyte heterogeneity underlying phenotypic changes in monocytes according to SIV disease stage. J Leukoc Biol (2010) 87:557–67. doi:10.1189/jlb.0209082
44. Kwissa M, Nakaya HI, Oluoch H, Pulendran B. Distinct TLR adjuvants differentially stimulate systemic and local innate immune responses in nonhuman primates. Blood (2012) 119:2044–55. doi:10.1182/blood-2011-10-388579
45. Ingersoll MA, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R, et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood (2010) 115:e10–9. doi:10.1182/blood-2009-07-235028
46. de Baey A, Mende I, Riethmueller G, Baeuerle PA. Phenotype and function of human dendritic cells derived from M-DC8(+) monocytes. Eur J Immunol (2001) 31:1646–55. doi:10.1002/1521-4141(200106)31:6<1646::AID-IMMU1646>3.0.CO;2-X
47. Schakel K, Kannagi R, Kniep B, Goto Y, Mitsuoka C, Zwirner J, et al. 6-Sulfo LacNAc, a novel carbohydrate modification of PSGL-1, defines an inflammatory type of human dendritic cells. Immunity (2002) 17:289–301. doi:10.1016/S1074-7613(02)00393-X
48. Thomas K, Dietze K, Wehner R, Metz I, Tumani H, Schultheiss T, et al. Accumulation and therapeutic modulation of 6-sulfo LacNAc(+) dendritic cells in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm (2014) 1:e33. doi:10.1212/NXI.0000000000000033
49. Belge KU, Dayyani F, Horelt A, Siedlar M, Frankenberger M, Frankenberger B, et al. The proinflammatory CD14+CD16+DR++ monocytes are a major source of TNF. J Immunol (2002) 168:3536–42. doi:10.4049/jimmunol.168.7.3536
50. Fingerle-Rowson G, Angstwurm M, Andreesen R, Ziegler-Heitbrock HWL. Selective depletion of CD14+ CD16+ monocytes by glucocorticoid therapy. Clin Exp Immunol (1998) 112:501–6. doi:10.1046/j.1365-2249.1998.00617.x
51. Dutertre CA, Amraoui S, DeRosa A, Jourdain JP, Vimeux L, Goguet M, et al. Pivotal role of M-DC8(+) monocytes from viremic HIV-infected patients in TNFalpha overproduction in response to microbial products. Blood (2012) 120:2259–68. doi:10.1182/blood-2012-03-418681
52. Siedlar M, Frankenberger M, Ziegler-Heitbrock LH, Belge KU. The M-DC8-positive leukocytes are a subpopulation of the CD14+ CD16+ monocytes. Immunobiology (2000) 202:11–7. doi:10.1016/S0171-2985(00)80047-9
53. Cros J, Cagnard N, Woollard K, Patey N, Zhang SY, Senechal B, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity (2010) 33:375–86. doi:10.1016/j.immuni.2010.08.012
54. Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH, et al. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood (2011) 118:e16–31. doi:10.1182/blood-2010-12-326355
55. Maurer D, Fiebiger E, Reininger B, Wolff-Winiski B, Jouvin MH, Kilgus O, et al. Expression of functional high affinity immunoglobulin E receptors (Fc epsilon RI) on monocytes of atopic individuals. J Exp Med (1994) 179:745–50. doi:10.1084/jem.179.2.745
56. Dehlink E, Baker AH, Yen E, Nurko S, Fiebiger E. Relationships between levels of serum IgE, cell-bound IgE, and IgE-receptors on peripheral blood cells in a pediatric population. PLoS One (2010) 5:e12204. doi:10.1371/journal.pone.0012204
57. Greer AM, Wu N, Putnam AL, Woodruff PG, Wolters P, Kinet JP, et al. Serum IgE clearance is facilitated by human FcepsilonRI internalization. J Clin Invest (2014) 124:1187–98. doi:10.1172/JCI68964
58. Clanchy FI, Holloway AC, Lari R, Cameron PU, Hamilton JA. Detection and properties of the human proliferative monocyte subpopulation. J Leukoc Biol (2006) 79:757–66. doi:10.1189/jlb.0905522
59. Bucala R, Spiegel LA, Chesney J, Hogan M, Cerami A. Circulating fibrocytes define a new leukocyte subpopulation that mediates tissue repair. Mol Med (1994) 1:71–81.
60. Komano Y, Nanki T, Hayashida K, Taniguchi K, Miyasaka N. Identification of a human peripheral blood monocyte subset that differentiates into osteoclasts. Arthritis Res Ther (2006) 8:R152. doi:10.1186/ar2046
61. Fingerle G, Pforte A, Passlick B, Blumenstein M, Strobel M, Ziegler-Heitbrock HWL. The novel subset of CD14+/CD16+ blood monocytes is expanded in sepsis patients. Blood (1993) 82:3170–6.
62. Horelt A, Belge KU, Steppich B, Prinz J, Ziegler-Heitbrock L. The CD14+CD16+ monocytes in erysipelas are expanded and show reduced cytokine production. Eur J Immunol (2002) 32:1319–27. doi:10.1002/1521-4141(200205)32:5<1319::AID-IMMU1319>3.0.CO;2-2
63. Shalova IN, Kajiji T, Lim JY, Gomez-Pina V, Fernandez-Ruiz I, Arnalich F, et al. CD16 regulates TRIF-dependent TLR4 response in human monocytes and their subsets. J Immunol (2012) 188:3584–93. doi:10.4049/jimmunol.1100244
64. Fingerle-Rowson G, Auers J, Kreuzer E, Fraunberger P, Blumenstein M, Ziegler-Heitbrock LH. Expansion of CD14+CD16+ monocytes in critically ill cardiac surgery patients. Inflammation (1998) 22:367–79. doi:10.1023/A:1022316815196
65. Dayyani F, Belge KU, Frankenberger M, Mack M, Berki T, Ziegler-Heitbrock L. Mechanism of glucocorticoid-induced depletion of human CD14+CD16+ monocytes. J Leukoc Biol (2003) 74:33–9. doi:10.1189/jlb.1202612
66. Korkosz M, Bukowska-Strakova K, Sadis S, Grodzicki T, Siedlar M. Monoclonal antibodies against macrophage colony-stimulating factor diminish the number of circulating intermediate and nonclassical (CD14(++)CD16(+)/CD14(+)CD16(++)) monocytes in rheumatoid arthritis patient. Blood (2012) 119:5329–30. doi:10.1182/blood-2012-02-412551
67. MacDonald KP, Palmer JS, Cronau S, Seppanen E, Olver S, Raffelt NC, et al. An antibody against the colony-stimulating factor 1 receptor depletes the resident subset of monocytes and tissue- and tumor-associated macrophages but does not inhibit inflammation. Blood (2010) 116:3955–63. doi:10.1182/blood-2010-02-266296
68. Lenzo JC, Turner AL, Cook AD, Vlahos R, Anderson GP, Reynolds EC, et al. Control of macrophage lineage populations by CSF-1 receptor and GM-CSF in homeostasis and inflammation. Immunol Cell Biol (2012) 90:429–40. doi:10.1038/icb.2011.58
69. Frankenberger M, Ekici AB, Angstwurm MW, Hoffmann H, Hofer TP, Heimbeck I, et al. A defect of CD16-positive monocytes can occur without disease. Immunobiology (2013) 218:169–74. doi:10.1016/j.imbio.2012.02.013
70. Heine GH, Ortiz A, Massy ZA, Lindholm B, Wiecek A, Martinez-Castelao A, et al. Transplant, monocyte subpopulations and cardiovascular risk in chronic kidney disease. Nat Rev Nephrol (2012) 8:362–9. doi:10.1038/nrneph.2012.41
71. Rogacev KS, Zawada AM, Emrich I, Seiler S, Bohm M, Fliser D, et al. Lower Apo A-I and lower HDL-C levels are associated with higher intermediate CD14++CD16+ monocyte counts that predict cardiovascular events in chronic kidney disease. Arterioscler Thromb Vasc Biol (2014) 34:2120–7. doi:10.1161/ATVBAHA.114.304172
Keywords: monocyte subsets, nomenclature, classical monocytes, intermediate monocytes, non-classical monocytes
Citation: Ziegler-Heitbrock L (2015) Blood monocytes and their subsets: established features and open questions. Front. Immunol. 6:423. doi: 10.3389/fimmu.2015.00423
Received: 28 May 2015; Accepted: 03 August 2015;
Published: 17 August 2015
Edited by:
Florent Ginhoux, Singapore Immunology Network, SingaporeReviewed by:
Anne Hosmalin, Cochin Institute, FranceCopyright: © 2015 Ziegler-Heitbrock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Loems Ziegler-Heitbrock, Helmholtz Zentrum München, Asklepios Fachkliniken München-Gauting, Enzianstr.3, 82211 Herrsching, Germany,bHpoQG1vbm9jeXRlLmV1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.