
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 14 July 2015
Sec. Immunological Tolerance and Regulation
Volume 6 - 2015 | https://doi.org/10.3389/fimmu.2015.00352
This article is part of the Research Topic The Role of Aire, microRNAs and Cell-Cell Interactions on Thymic Architecture and Induction of Tolerance. View all 13 articles
Developing thymocytes interact sequentially with two distinct structures within the thymus: the cortex and medulla. Surviving single-positive and double-positive thymocytes from the cortex migrate into the medulla, where they interact with medullary thymic epithelial cells (mTECs). These cells ectopically express a vast set of peripheral tissue antigens (PTAs), a property termed promiscuous gene expression that is associated with the presentation of PTAs by mTECs to thymocytes. Thymocyte clones that have a high affinity for PTAs are eliminated by apoptosis in a process termed negative selection, which is essential for tolerance induction. The Aire gene is an important factor that controls the expression of a large set of PTAs. In addition to PTAs, Aire also controls the expression of miRNAs in mTECs. These miRNAs are important in the organization of the thymic architecture and act as posttranscriptional controllers of PTAs. Herein, we discuss recent discoveries and highlight open questions regarding the migration and interaction of developing thymocytes with thymic stroma, the ectopic expression of PTAs by mTECs, the association between Aire and miRNAs and its effects on central tolerance.
The induction of central immune tolerance is an increasingly complex and intricate process that occurs within the thymus (1, 2). Inside this organ, immature thymocytes interact sequentially and in a three-dimensional architecture with two distinct structures: the cortex and the medulla. In the cortex, the double-negative (DN) and double-positive (DP) thymocytes interact with cortical thymic epithelial cells (cTECs), allowing MHC-mediated self-peptide presentation to DP thymocytes expressing the α/β T cell receptor (α/β TCR), featuring intermediate affinity/avidity. Positive selection is a result of this interaction, which causes DP thymocytes to differentiate into mature single-positive (SP) thymocytes (3, 4).
The DP thymocytes that do not undergo positive selection are eliminated through death by neglect. Thereafter, the surviving SP and DP thymocytes migrate to the thymic medulla, where they interact with medullary thymic epithelial cells (mTECs). These cells are very peculiar because they ectopically express a large set of peripheral tissue antigens (PTAs) (5–8). Therefore, it is possible to find insulin, a PTA that represents pancreatic beta cells, and a myriad of other autoantigens in the thymus.
The immunological significance of this property, which was termed promiscuous gene expression (PGE) (9–12), is associated with the presentation of PTAs by mTECs to SP and DP thymocytes. Thymocyte clones that express α/β TCR with a high affinity for PTAs are eliminated by apoptosis in a process termed negative selection or clonal deletion, which is essential for central tolerance induction (13–16). This process prevents the passage of autoreactive T cell clones to the periphery, which could provoke aggressive autoimmunity.
Therefore, the migration of thymocytes within the thymus enables the physical association of these cells with different thymic microenvironments (15, 17). Immunologists are interested in elucidating which chemotactic factors and/or adhesion molecules are involved in this process (18, 19).
Another very important factor in central tolerance is the autoimmune regulator (Aire) gene that controls the expression of a large set (but not all) of PTAs in mTECs (20, 21). Mutations in these gene that lead to a loss of Aire function can result in autoimmune polyendocrinopathycandidiasis-ectodermal dystrophy (APECED), an autoimmune disease characterized by hypoparathyroidism, candidiasis (yeast infection), and adrenal insufficiency (22–24). The mechanism of the Aire gene as a transcriptional regulator of Aire-dependent PTAs and the effect of point mutations found in the Aire gene sequence on clinical phenotypes (APECED or other autoimmune diseases) have received attention in recent years (25–27).
In addition, researchers have observed that in addition to PTAs, Aire controls the expression of microRNAs (miRNAs) in mTECs (28). In turn, miRNAs are important for the organization of thymic architecture and act as posttranscriptional controllers of PTAs (29, 30).
In this mini-review, we briefly discuss (1) the main aspects of three-dimensional thymus architecture, focusing on the migration and interaction of developing thymocytes with the thymic stroma and positive and negative selection; (2) the ectopic expression of PTAs by mTECs and role of the Aire gene; and (3) the current evidence for the link between Aire and miRNAs in thymic architecture and the induction of central tolerance.
Developing thymocytes interact with the thymic microenvironment while they migrate and differentiate within the organ. This microenvironment is subdivided into two main regions, and each region is composed of different cell types that produce soluble and non-soluble molecules that can modulate thymocyte migration and maturation (31, 32). Thymic lobules are divided into cortical and medullary regions that are connected by a cortico-medullary junction. The cortex microenvironment is filled with cTECs, thymic nurse cells (TECs-thymocyte-forming lymphoepithelial complexes), macrophages, migratory dendritic cells (DCs), and fibroblasts. The medullary region contains mTECs, macrophages, resident and migratory conventional DCs, plasmacytoid DCs, fibroblasts, and B cells (16) (Figure 1A). Both regions are filled with a network of extracellular matrix (ECM) molecules, such as type I and IV collagens, fibronectin, and laminin. Soluble molecules, such as hormones, cytokines, growth factors, chemokines, and sphingolipids, are also found in the thymus and are produced by the lymphoid and non-lymphoid compartments. These soluble moieties can be present in the ECM and mediate cell–ECM and cell–cell interactions (33–35).
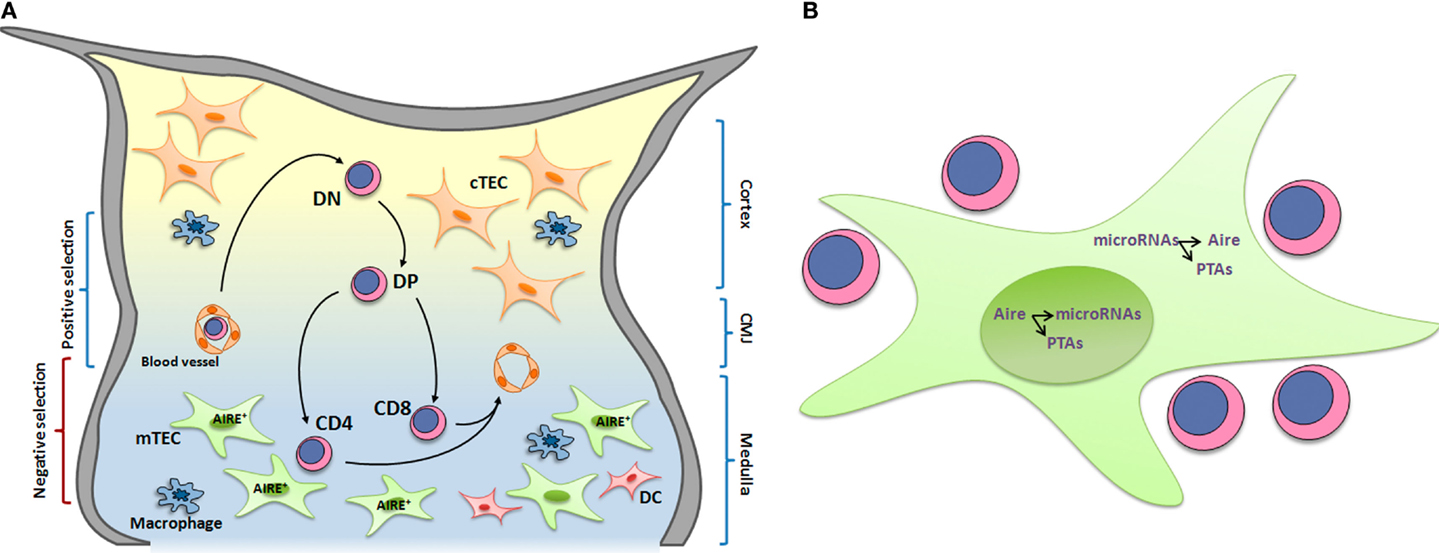
Figure 1. (A) Thymocyte development and interactions with microenvironmental cells. T cell development occurs while cells migrate and interact with thymic microenvironmental components, including thymic epithelial cells, dendritic cells, and macrophages. These interactions are responsible for the selective processes, which lead to the formation of a self-restricted T-cell repertoire. In this context, medullary thymic epithelial cells (mTECs) play an essential role by expressing AIRE, which control the expression of a set of peripheral tissue antigens (PTAs) that are presented to developing thymocytes. High avidity of MHC-PTAs presented by mTECs with TCRs lead to thymocyte death by apoptosis and consequently avoid self-reactive T cell maturation. DN, double-negative CD4−CD8− thymocytes; DP, doublepositive CD4+CD8+; CD4, single-positive CD4+CD8−; CD8, single-positive CD8+CD4− thymocytes; cTEC, cortical thymic epithelial cell; mTEC, medullary thymic epithelial cell. CMJ, cortico-medullary junction. (B) The transcriptional and posttranscriptional control pathway of promiscuous gene expression. The peripheral tissue antigens (PTAs) expressed in medullary thymic epithelial cells (mTECs) are transcriptionally controlled by Aire within the nuclear compartment, which also controls the expression of miRNAs. Within the cytoplasm, the miRNAs control Aire and PTAs.
Thymocyte differentiation and migration occur simultaneously in the thymic microenvironment. T cell progenitors enter the cortico-medullary region via post-capillary venules (36) and rapidly migrate through the cortex toward the subcapsular zone, where DN thymocytes are primarily located. Subsequently, thymocytes migrate to the middle cortex and begin expressing both CD4 and CD8 co-receptors, becoming DP cells. During this stage, cells are selected based on the rearrangement of TCR genes, which leads to the membrane expression of productive TCRs. Cells that do not express productive TCRs undergo apoptosis, whereas cells expressing productive TCRs continue the differentiation process. Then, cells with TCRs that interact with high avidity with MHC-presented self-antigens expressed by mTECs and DCs undergo apoptosis in a process termed negative selection (37). The presentation of self-antigens by mTECs is controlled by Aire and guarantees the deletion of autoreactive T-cell clones, supporting central tolerance (38). In this context, cells with TCRs that interact with low/median avidity with MHC-presented self-antigens survive and continue the maturation process. Survival signals mediated by TCRs and CD4/CD8 co-receptors lead to the down-regulation of a co-receptor, and thymocytes become mature CD4+CD8− or CD8+CD4− SP cells (Figure 1A).
Thymocyte localization and guidance are controlled by ECM molecules and chemokines, among others molecules, and their respective receptors. For example, the entrance of T-cell progenitors in the thymus is controlled by CCL21/CCR7 and CCL25/CCR9 (chemokine/chemokine receptor) interactions (39). Migration of immature cells within the thymus is controlled by CXCL12/CXCR4 and CCL20/CCR6 interactions (40, 41), and CCR7 signaling is essential for the migration of DP thymocytes to the medulla (42). Moreover, CCR7 is involved in thymocyte egress, which is also controlled by sphingosine-1-phosphate receptor 1 signaling (43). The absence of such molecules in the thymus not only abrogates thymocyte development but also induces changes in the histological organization of the organ (44).
Thymic architecture and organization are essential for proper T-cell development and depend on both the lymphoid and non-lymphoid compartments. Alterations in one compartment can affect the other and consequently modify T-cell development and the repertoire of exported mature T cells to peripheral lymphoid organs. For example, Rag mutations substantially impair thymocyte development and consequently affect the distribution and maturation of TECs, diminishing the proportion of mTECs and inducing a lack of AIRE protein expression (45, 46). Lack of AIRE expression can in turn directly affect negative selection and break central tolerance. Interestingly, Aire deficiency can modulate the intrathymic expression of chemokines as a control mechanism of thymocyte development (47). In this context, one can argue that chemokines and other molecules controlling thymocyte migration (such as ECM molecules) could also modulate Aire expression.
During the induction of central tolerance in the thymus, self-reactive regulatory T cells (Treg) are negatively selected, even if these cells play a role in the periphery. In fact, all sets of antigen-presenting cells, including cTECs, mTECs, and thymic DCs, act as self-antigen peptide presenting cells (6–9, 48–53). Thymic DCs present only the PTA peptides that were expressed and processed by mTECs (38).
The expression of PTAs by mTECs is a key process of (auto)immune representation. Due to the wide-ranging diversity of PTAs expressed by these cells, this phenomenon has been termed PGE (5, 9, 12, 48, 54–62).
The primary implication of this type of gene expression, which is heterogeneous and ectopic, is associated with the maintenance of immune homeostasis and controlling the reactivity and self-aggressive autoimmune diseases.
Notably, cTECs and mTECs are essential but not sufficient for these selection events (55). The cTEC-derived signals may regulate the positive selection of thymocytes that recognize the MHC-peptide complexes themselves; however, mTECs that express AIRE help ensure tolerance to self-antigens (63).
A subset of mTECs express the Aire gene (chromosome 10C1 in mice and 21q22.3 in humans) (64) and the claudin proteins (Cld3 or Cld4) on their surface. In these cells, AIRE and the claudin proteins act as adhesion molecules and represent the major proteins that contribute to the molecular architecture of cell junctions. All Cld3+, Cld4+, and Aire+ adult TEC cells strongly express MHC class II and CD80 molecules on their surface (51).
“Immature” CD80−/MHC-II− mTECs express a limited set of PTAs, whereas “mature” CD80+/MHC-II+ mTECs exhibit greater PTA diversity, including PTAs whose expression is Aire dependent (11). These findings have led some researchers to propose “the terminal differentiation model”; i.e., mTECs undergo a continuous process of differentiation similar to the skin or intestinal epithelium, and the full complement of PGE is contingent upon this process (55). mTEC cells are very peculiar due to their unique gene expression pattern. They are capable of expressing more than 19,000 protein-coding genes, including “ectopic” genes that correspond to PTAs. Currently, no other known cell type expresses such a large set of genes (65).
We next sought to determine whether these cells also have unique machinery for gene expression control.
Although the transcriptional control of PGE is partially exerted by Aire, mutations in this gene cause severe autoimmunity that involves various organs and tissues in both mice and humans. In humans, this disease is a syndrome termed APECED, and patients have mutations along the Aire sequence, suggesting that mutations in Aire trigger aggressive autoimmunity (22, 66).
However, the existence of APECED patients who lack Aire mutations (67–69) suggests that other factor(s) may be involved in controlling aggressive autoimmunity. These observations led us to wonder if temporal changes and a slight deregulation of wild-type Aire expression during development could contribute to autoimmunity.
Variation of Aire expression might disturb Aire-dependent PTAs in the thymus and consequently trigger aggressive autoimmunity, a hypothesis that was previously tested by our group (58). This hypothesis is a promising subject for further research and is currently being studied in humans with thymic cells isolated from Down syndrome patients, which feature trisomy of chromosome 21, providing a unique opportunity to evaluate the effect of natural Aire gene dosage in humans (70–72).
The functional role of Aire has been demonstrated using knockout (KO) mice. Aire in mice and humans encodes a protein with affinity for DNA that functions as a positive transcription factor regulating the expression of PTAs mTECs, but AIRE can also act as a negative regulator of other genes (8, 10, 73–76).
The link between Aire expression and the induction of thymocyte apoptosis, a biological process crucial for negative selection, has been demonstrated (37, 77). However, according to our best knowledge, no investigation has assessed this link considering the possible effect of variations in Aire expression in mTECs on adhesion with thymocytes and the induction of apoptosis. Alternatively, Aire-deficient mTEC cells may lose their adhesion ability. This question is still open for further research.
Interestingly, the AIRE targets low-transcribed genes. It interacts with hypomethylated promoter regions in the chromatin through its PHD1 domain (62, 78–82).
However, recent evidence has demonstrated that the AIRE acts indirectly in regulating PTA transcription. According to Giraud et al. (83), AIRE can be considered an unusual transcription factor because it does not appear to function as a typical transactivator. These authors demonstrated that AIRE activates PTA transcription by releasing stalled RNA Pol II from blockage at the promoter region of its target genes. This model suggests that AIRE acts during the elongation stage of transcription rather than at transcription initiation (83). The “promiscuity” of Aire on a large set of downstream PTA genes might be due the unspecific mode of action of RNA Pol II on different promoter regions, but this remains to be determined.
A new exciting possibility for the Aire mechanism is its influence in controlling alternative splicing of PTA genes in mTECs. Aire has been shown to increase the amount of measurable exons per gene and enables the production of PTAs from these exons (84); these properties might significantly increase the diversity of PTA isoforms in mTECs and consequently increase the range of self-representation.
It is possible that aggressive autoimmunity is associated with an imbalance of PTAs isoforms in mTECs.
In our view, not only Aire but also miRNAs may play a role in central tolerance. This hypothesis is plausible considering the vast range of action of miRNAs, which affect more than half of all mRNAs originating from protein-coding genes in human or murine cells (85).
This range of action is expected to reach mRNAs encoding proteins involved in the central tolerance mechanism, including PTA mRNAs and Aire mRNA itself.
First, researchers evaluated the role of the endoribonuclease Dicer, a key enzyme implicated in miRNA maturation, on thymic function. They found that Dicer-KO mice exhibit progressive degeneration in thymic architecture and function, provoking alterations in T cell differentiation and peripheral tolerance, pinpointing miRNA-29a as a specific miRNA participating in this process (29).
Then, mice lacking Dicer expression in the thymic epithelia were found to exhibit a set of abnormalities, including alterations in the expression profiling of cTEC and mTEC mRNAs. T cells obtained from a Dicer-deficient thymus were pathogenic and produced aggressive autoimmunity (86). These finding were instrumental for further research on the role of miRNAs in central tolerance induction.
Moreover, thymic epithelial cells isolated from murine or human thymuses feature overlapping of miRNA signatures, suggesting evolutionary conservation of miRNA expression profiles (87). These authors also demonstrated that Aire expression is associated with maturation-dependent expression of miRNAs.
However, a direct demonstration that Dicer and consequently miRNAs play a role in TEC-thymocyte adhesion, which is crucial for positive and negative selection, is still lacking. This question is open for further investigation.
As discussed above, the AIRE acts in close association with RNA Pol II (83). Because this polymerase transcribes miRNAs in addition to mRNAs (88–92), Aire may affect miRNA expression.
Our group was the first to directly demonstrate this possibility (28). We showed that in murine mTECs, Aire controls the transcription of miRNAs located within a genomic region that encompasses an open-reading frame (ORF of Gm2922 mRNA).
This finding enabled further evaluation of the role played by Aire-dependent miRNAs in the posttranscriptional control of PTAs. Thus, we reconstructed miRNA–mRNA interaction networks from mTECs isolated from BALB/c (non-autoimmune) or non-obese diabetic (NOD) (autoimmune) mice. As expected, dozens of PTA mRNAs interacted with miRNAs. Interestingly, none of the classical Aire-dependent PTAs (e.g., Ins2) interacted with miRNAs, strongly suggesting that they are somewhat resistant to posttranscriptional control (30).
What would be the consequences of a lack of miRNA action on these PTAs? Could this lack of action aid autoantigen synthesis by mTECs, consequently inducing tolerance? What causes these Aire-dependent PTAs to be “resistant” to miRNA action? Could changes in their 3′UTRs (length or mutations) or imbalance in miRNAs expression levels (or both) cause this resistance? We have suggested that there may be changes in length of the 3′UTR sequence of Aire-dependent PTAs expressed in mTECs (30). Researchers including our group and the group of Mathieu Giraud in Paris are now challenged to evaluate the structure of mRNAs in general and/or the 3′UTR of mRNAs of PTAs expressed in mTECs compared with other cell types.
Based on these recent results, is possible to drawn a pathway for the transcriptional and posttranscriptional control of PGE in mTECs (Figure 1B). Within the nuclear compartment, the AIRE controls PTA and miRNA transcription (Aire-dependent PTAs and miRNAs). Once in the cytoplasm, miRNAs play a role in the posttranscriptional control of Aire and PTA mRNAs. Would PTA mRNAs with altered 3′UTRs be refractory to the action of miRNAs?
Although these aspects have only recently begun to be explored, they represent new, exciting questions for present and future research on the molecular genetic basis of immune tolerance.
The molecular genetic control of central tolerance remains an open question in immunology. The identification and cloning of the Aire gene was instrumental in studying the molecular genetics of this process. As the primary controller of PTA expression in mTECs, Aire is the master pillar of central tolerance. Aire expression is common in the thymus; and this observation led to the idea of PGE. However, Aire did not fit well as a classic transcription factor. The AIRE operates in conjunction with various other partner proteins in the release of RNA Pol II shortly after the initiation of PTA gene transcription. This property enabled better understanding of the vast range of AIRE activity. Recently, miRNAs have been found to be the modulators of post-transcriptional controllers in the thymus. Researchers are now challenged with deciphering the transcriptional and post-transcriptional control pathway of PGE involving Aire and miRNAs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the following funding agencies: the São Paulo State Research Foundation (Fapesp) through grant Nos. 13/17481-1 (to GP) and 13/08216-2 (Cepid-Fapesp to Fernando Q. Cunha), the National Council for Research and Technology (CNPq, Brasília) through grant No. 306315/2013-0 (to GP), the CAPES Foundation (Brasília) through grant No. 88881.06105/2014-01 (to GP), the Oswaldo Cruz Foundation and Rio de Janeiro State Research Foundation (Faperj, Rio de Janeiro to DM). EHO was supported by CNPq through post-doctoral fellowship No. 150103/2014-0.
1. Gallegos AM, Bevan MJ. Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. J Exp Med (2004) 200(8):1039–49. doi:10.1084/jem.20041457
2. Mouchess ML, Anderson M. Central tolerance induction. Curr Top Microbiol Immunol (2014) 373:69–86. doi:10.1007/82.2013.321
3. Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol (2009) 9:833–44. doi:10.1038/nri2669
4. Groettrup M, Kirk CJ, Basler M. Proteasomes in immune cells: more than peptide producers? Nat Rev Immunol (2010) 10:73–7. doi:10.1038/nri2687
5. Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes co-localized in chromosomal clusters. J Exp Med (2004) 199(2):155–66. doi:10.1084/jem.20031677
6. Klein L, Kyewski B. Self-antigen presentation by thymic stromal cells: a subtle division of labor. Curr Opin Immunol (2000) 12(2):179–86. doi:10.1016/S0952-7915(99)00069-2
7. Mizuochi T, Kasai M, Kokuho T, Kakiuchi T, Hirokawa K. Medullary but not cortical thymic epithelial cells present soluble antigens to helper T cells. J Exp Med (1992) 175(6):1601–5. doi:10.1084/jem.175.6.1601
8. Tikocinski LO, Sinemus A, Kyewski B. The thymus medulla slowly yields its secrets. Ann N Y Acad Sci (2008) 1143:105–22. doi:10.1196/annals.1443.018
9. Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expressionin medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol (2001) 2(11):1032–9. doi:10.1038/ni723
10. Kyewski B, Derbinski J, Gotter J, Klein L. Promiscuous gene expression and central T-cell tolerance: more than meets the eye. Trends Immunol (2002) 23(7):364–71. doi:10.1016/S1471-4906(02)02248-2
11. Derbinski J, Gäbler J, Brors B, Tierling S, Jonnakuty S, Hergenhan M, et al. Promiscuous gene expression in thymic epithelialcells is regulated at multiple levels. J Exp Med (2005) 202(1):33–45. doi:10.1084/jem.20050471
12. Magalhães DA, Silveira EL, Junta CM, Sandrin-Garcia P, Fachin AL, Donadi EA, et al. Promiscuous gene expression in the thymus: the root of central tolerance. Clin Dev Immunol (2006) 13(2–4):81–99. doi:10.1080/17402520600877091
13. Ma D, Wei Y, Liu F. Regulatory mechanisms of thymus and T cell development. Dev Comp Immunol (2013) 39(1–2):91–102. doi:10.1016/j.dci.2011.12.013
14. Lo WL, Allen PM. Self-peptides in TCR repertoire selection and peripheral T cell function. Curr Top Microbiol Immunol (2014) 373:49–67. doi:10.1007/82.2013.319
15. Anderson G, Baik S, Cowan JE, Holland AM, McCarthy NI, Nakamura K, et al. Mechanisms of thymus medulla development and function. Curr Top Microbiol Immunol (2014) 373:19–47. doi:10.1007/82.2013.320
16. Klein L, Kyewski B, Allen PM, Hoqquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol (2014) 14(6):377–91. doi:10.1038/nri3667
17. Takada K, Ohigashi I, Kasai M, Nakase H, Takahama Y. Development and function of cortical thymic epithelial cells. Curr Top Microbiol Immunol (2014) 373:1–17. doi:10.1007/82.2013.322
18. Scimone ML, Aifantis I, Apostolou I, Von Boehmer H, Von Andrian UH. A multistep adhesion cascade for lymphoid progenitor cell homing to the thymus. Proc Natl Acad Sci USA (2006) 103(18):7006–11. doi:10.1073/pnas.0602024103
19. Zhang SL, Bhandoola A. Trafficking to the thymus. Curr Top Microbiol Immunol (2014) 373:87–111. doi:10.1007/978-3-642-40252-4
20. Mathis D, Benoist C. Aire. Annu Rev Immunol (2009) 27:287–312. doi:10.1146/annurev.immunol.25.022106.141532
21. Laan M, Peterson P. The many faces of aire in central tolerance. Front Immunol (2013) 4:326. doi:10.3389/fimmu.2013.00326
22. Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, et al. Positional cloning of the APECED gene. Nat Genet (1997) 17(4):393–8. doi:10.1038/ng1297-393
23. Consortium FG-A. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet (1997) 17(4):399–403. doi:10.1038/ng1297-399
24. Perheentupa J. Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J Clin Endocrinol Metab (2006) 91(8):2843–50. doi:10.1210/jc.2005-2611
25. Jin P, Zhang Q, Dong CS, Zhao SL, Mo ZH. A novel mutation in autoimmune regulator gene causes autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J Endocrinol Invest (2014) 37(10):941–8. doi:10.1007/s40618-014-0120-7
26. Huibregtse KE, Wolfgram P, Winer KK, Connor EL. Polyglandular autoimmune syndrome type I – a novel AIRE mutation in a North American patient. J Pediatr Endocrinol Metab (2014) 27(11–12):1257–60. doi:10.1515/jpem-2013-0328
27. Gallo V, Giardino G, Capalbo D, Palamaro L, Romano R, Santamaria F. Alterations of the autoimmune regulator transcription factor and failure of central tolerance: APECED as a model. Expert Rev Clin Immunol (2013) 9(1):43–51. doi:10.1586/eci.12.88
28. Macedo C, Evangelista AF, Marques MM, Octacílio-Silva S, Donadi EA, Sakamoto-Hojo ET, et al. Autoimmune regulator (Aire) controls the expression of microRNAs in medullary thymic epithelial cells. Immunobiology (2013) 218(4):554–60. doi:10.1016/j.imbio.2012.06.013
29. Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, Kyewski B, et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat Immunol (2011) 13(2):181–7. doi:10.1038/ni.2193
30. Macedo C, Oliveira EH, Almeida RS, Donate PB, Fornari TA, Pezzi N, et al. Aire-dependent peripheral tissue antigen mRNAs in mTEC cells feature networking refractoriness to microRNA interaction. Immunobiology (2015) 220(1):93–102. doi:10.1016/j.imbio.2014.08.015
31. Savino W, Mendes-da-Cruz DA, Silva JS, Dardenne M, Cotta-de-Almeida V. Intrathymic Tcell migration: a combinatorial interplay of extracellular matrix and chemokines? Trends Immunol (2002) 23(6):305–13. doi:10.1016/S1471-4906(02)02224-X
32. Savino W, Mendes-Da-Cruz DA, Smaniotto S, Silva-Monteiro E, Villa-Verde DM. Molecular mechanisms governing thymocyte migration: combined role of chemokines and extracellular matrix. J Leukoc Biol (2004) 75(6):951–61. doi:10.1189/jlb.1003455
33. Segerer S, Djafarzadeh R, Gröne HJ, Weingart C, Kerjaschki D, Weber C, et al. Selective binding and presentation of CCL5 by discrete tissue microenvironments during renal inflammation. J Am Soc Nephrol (2007) 18(6):1835–44. doi:10.1681/ASN.2006080837
34. Netelenbos T, Zuijderduijn S, Van Den Born J, Kessler FL, Zweegman S, Huijgens PC. Proteoglycans guide SDF-1-induced migration of hematopoietic progenitor cells. J Leukoc Biol (2002) 72(2):353–62.
35. Yanagawa Y, Iwabuchi K, Onoé K. Enhancement of stromal cell-derived factor-1alphainduced chemotaxis for CD4/8 double-positive thymocytes by fibronectin and laminin in mice. Immunology (2001) 104(1):43–9. doi:10.1681/ASN.2006080837
36. Lind EF, Prockop SE, Porritt HE, Petrie HT. Mapping precursor movement through the postnatal thymus reveals specific microenvironments supporting defined stages of early lymphoid development. J Exp Med (2001) 194(2):127–34. doi:10.1084/jem.194.2.127
37. Yamaguchi Y, Kudoh J, Yoshida T, Shimizu N. In vitro co-culture systems for studying molecular basis of cellular interaction between Aire-expressing medullary thymic epithelial cells and fresh thymocytes. Biol Open (2014) 3(11):1071–82. doi:10.1242/bio.201410173
38. Hubert FX, Kinkel SA, Davey GM, Phipson B, Mueller SN, Liston A, et al. Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood (2011) 118(9):2462–72. doi:10.1182/blood-2010-06-286393
39. Liu C, Ueno T, Kuse S, Saito F, Nitta T, Piali L, et al. The role of CCL21 in recruitment of T-precursor cells to fetal thymi. Blood (2005) 105(1):31–9. doi:10.1182/blood-2004-04-1369
40. Kim CH, Pelus LM, White JR, Broxmeyer HE. Differential chemotactic behavior of developing T cells in response to thymic chemokines. Blood (1998) 91(12):4434–43.
41. Bunting MD, Comerford I, Kara EE, Korner H, McColl SR. CCR6 supports migration and differentiation of a subset of DN1 early thymocyte progenitors but is not required for thymic nTreg development. Immunol Cell Biol (2014) 92(6):489–98. doi:10.1038/icb.2014.14
42. Kwan J, Killeen N. CCR7 directs the migration of thymocytes into the thymic medulla. J Immunol (2004) 172(7):3999–4007. doi:10.4049/jimmunol.172.7.3999
43. Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature (2004) 427(6972):355–60. doi:10.1038/nature02284
44. Misslitz A, Pabst O, Hintzen G, Ohl L, Kremmer E, Petrie HT, et al. Thymic T cell development and progenitor localization depend on CCR7. J Exp Med (2004) 200(4):481–91. doi:10.1084/jem.20040383
45. Poliani PL, Facchetti F, Ravanini M, Gennery AR, Villa A, Roifman CM, et al. Early defects in human T-cell development severely affect distribution and maturation of thymic stromal cells: possible implications for the pathophysiology of Omenn syndrome. Blood (2009) 114(1):105–8. doi:10.1182/blood-2009-03-211029
46. Marrella V, Poliani PL, Notarangelo LD, Grassi F, Villa A. Rag defects and thymic stroma: lessons from animal models. Front Immunol (2014) 5:259. doi:10.3389/fimmu.2014.00259
47. Laan M, Kisand K, Kont V, Möll K, Tserel L, Scott HS, et al. Autoimmune regulator deficiency results in decreased expression of CCR4 and CCR7 ligands and in delayed migration of CD4+ thymocytes. J Immunol (2009) 183(12):7682–91. doi:10.4049/jimmunol.0804133
48. Kyewski B, Derbinski J. Self-representation in the thymus: an extended view. Nat Rev Immunol (2004) 4(9):688–98. doi:10.1038/nri1436
49. Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol (2006) 24:571606. doi:10.1146/annurev.immunol.23.021704.115601
50. Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol (2006) 6(2):127–35. doi:10.1038/nri1781
51. Holländer GA. Claudins provide a breath of fresh Aire. Nat Immunol (2007) 8(3):234–6. doi:10.1038/ni0307-234
52. Irla M, Hugues S, Gill J, Nitta T, Hikosaka Y, Williams IR, et al. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity (2008) 29(3):451–63. doi:10.1016/j.immuni.2008.08.007
53. Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci USA (2008) 105(41):15854–9. doi:10.1073/pnas.0808069105
54. Sousa Cardoso R, Magalhães DA, Baião AM, Junta CM, Macedo C, Marques MM, et al. Onset of promiscuous gene expression in murine fetal thymus organ culture. Immunology (2006) 119(3):369–75. doi:10.1111/j.1365-2567.2006.02441.x
55. Gäbler J, Arnold J, Kyewski B. Promiscuous gene expression and the developmental dynamics of medullary thymic epithelial cells. Eur J Immunol (2007) 37(12):3363–72. doi:10.1002/eji.200737131
56. Tykocinski LO, Sinemus A, Kyewski B. The thymus medulla slowly yields its secrets. Ann N Y Acad Sci (2008) 1143:105–22. doi:10.1196/annals.1443.018
57. Irla M, Hollander G, Reith W. Control of central self-tolerance induction by autoreactive CD4+ thymocytes. Trends Immunol (2010) 31(2):71–9. doi:10.1016/j.it.2009.11.002
58. Fornari TA, Donate PB, Macedo C, Marques MM, Magalhães DA, Passos GA. Age-related deregulation of Aire and peripheral tissue antigen genes in the thymic stroma of non-obese diabetic (NOD) mice is associated with autoimmune type 1 diabetes mellitus (DM-1). Mol Cell Biochem (2010) 342(1–2):21–8. doi:10.1007/s11010-010-0464-z
59. Derbinski J, Kyewski B. How thymic antigen presenting cells sample the body’s self-antigens. Curr Opin Immunol (2010) 22(5):592–600. doi:10.1016/j.coi.2010.08.003
60. Donate PB, Fornari TA, Junta CM, Magalhães DA, Macedo C, Cunha TM, et al. Collagen induced arthritis (CIA) in mice features regulatory transcriptional network connecting major histocompatibility complex (MHC H2) with autoantigen genes in the thymus. Immunobiology (2011) 216(5):591–603. doi:10.1016/j.imbio.2010.09.007
61. Pinto S, Michel C, Schmidt-Glenewinkel H, Harder N, Rohr K, Wild S, et al. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc Natl Acad Sci U S A (2013) 110(37):E3497–505. doi:10.1073/pnas.1308311110
62. Ucar O, Rattay K. Promiscuous gene expression in the thymus: a matter of epigenetics, miRNA, and more? Front Immunol (2015) 6:93. doi:10.3389/fimmu.2015.00093
63. Anderson G, Lane PJ, Jenkinson EJ. Generating intrathymic microenvironments to establish T-cell tolerance. Nat Rev Immunol (2007) 7(12):954–63. doi:10.1038/nri2187
64. SOURCE Database. Available from: http://source-search.princeton.edu/
65. Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, et al. Population and single-cell genomics reveal the Aire dependency, relief from polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res (2014) 24(12):1918–31. doi:10.1101/gr.171645.113
66. Capalbo D, De Martino L, Giardino G, Di Mase R, Di Donato I, Parenti G, et al. Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy: insights into genotype-phenotype correlation. Int J Endocrinol (2012) 2012:353250. doi:10.1155/2012/353250
67. Wang CY, Davoodi-Semiromi A, Huang W, Connor E, Shi JD, She JX. Characterization of mutations in patients with autoimmune polyglandular syndrome type 1 (APS1). Hum Genet (1998) 103(6):681–5. doi:10.1007/s004390050891
68. Heino M, Scott HS, Chen Q, Peterson P, Mäebpää U, Papasavvas MP, et al. Mutation analyses of North American APS-1 patients. Hum Mutat (1999) 13(1):69–74. doi:10.1002/(SICI)10981004
69. Björses P, Halonen M, Palvimo JJ, Kolmer M, Aaltonen J, Ellonen P, et al. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am J Hum Genet (2000) 66(2):378–92. doi:10.1086/302765
70. Lima FA, Moreira-Filho CA, Ramos PL, Brentani H, Lima Lde A, Arrais M, et al. Decreased AIRE expression and global thymic hypofunction in Down syndrome. J Immunol (2011) 187(6):3422–30. doi:10.4049/jimmunol.1003053
71. Skogberg G, Lundberg V, Lindgren S, Gudmundsdottir J, Sandström K, Kämpe O, et al. Altered expression of autoimmune regulator in infant down syndrome thymus, a possible contributor to an autoimmune phenotype. J Immunol (2014) 193(5):2187–95. doi:10.4049/jimmunol.1400742
72. Giménez-Barcons M, Casteràs A, Armengol Mdel P, Porta E, Correa PA, Marín A, et al. Autoimmune predisposition in Down syndrome may result from a partial central tolerance failure due to insufficient intrathymic expression of AIRE and peripheral antigens. J Immunol (2014) 193(8):3872–9. doi:10.4049/jimmunol.1400223
73. Taniguchi RT, Anderson MS. The role of Aire in clonal selection. Immunol Cell Biol (2011) 89(1):40–4. doi:10.1038/icb.2010.132
74. Gallegos AM, Bevan MJ. Central tolerance: good but imperfect. Immunol Rev (2006) 209:290–6. doi:10.1111/j.0105-2896.2006.00348.x
76. Mathis D, Benoist C. Levees of immunological tolerance. Nat Immunol (2010) 11(1):3–6. doi:10.1038/ni.1833
77. Yamaguchi Y, Takayanagi A, Chen J, Sakai K, Kudoh J, Shimizu N. Mouse thymic epithelial cell lines expressing “Aire” and peripheral tissue-specific antigens reproduce in vitro negative selection of T cells. Exp Cell Res (2011) 317(14):2019–30. doi:10.1016/j.yexcr.2011.05.00
78. Org T, Chignola F, Hetényi C, Gaetani M, Rebane A, Liiv I, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep (2008) 9(4):370–6. doi:10.1038/sj.embor.2008.11
79. Koh AS, Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, et al. Aire employs a histonebinding module to mediate immunological tolerance, linking chromatin regulation with organspecific autoimmunity. Proc Natl Acad Sci U S A (2008) 105(41):15878–83. doi:10.1073/pnas.080847010
80. Koh AS, Kingston RE, Benoist C, Mathis D. Global relevance of Aire binding to hypomethylated lysine-4 of histone-3. Proc Natl Acad Sci U S A (2010) 107(29):13016–21. doi:10.1073/pnas.1004436107
81. Chignola F, Gaetani M, Rebane A, Org T, Mollica L, Zucchelli C, et al. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res (2009) 37(9):2951–61. doi:10.1093/nar/gkp166
82. Kont V, Murumagi A, Tykocinski LO, Kinkel SA, Webster KE, Kisand K, et al. DNA methylation signatures of the AIRE promoter in thymic epithelial cells, thymomas and normal tissues. Mol Immunol (2011) 49(3):518–26. doi:10.1016/j.molimm.2011.09.022
83. Giraud M, Yoshida H, Abramson J, Rahl PB, Young RA, Mathis D, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A (2012) 109(2):535–40. doi:10.1073/pnas.1119351109
84. Keane P, Ceredig R, Seoighe C. Promiscuous mRNA splicing under the control of AIRE in medullary thymic epithelial cells. Bioinformatics (2015) 31(7):986–90. doi:10.1093/bioinformatics/btu785
85. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem (2010) 79:351–79. doi:10.1146/annurev-biochem-060308103103
86. Zuklys S, Mayer CE, Zhanybekova S, Stefanski HE, Nusspaumer G, Gill J, et al. MicroRNAs control the maintenance of thymic epithelia and their compe-tence for T lineage commitment and thymocyte selection. J Immunol (2012) 189(8):3894–904. doi:10.4049/jimmunol.1200783
87. Ucar O, Tykocinski LO, Dooley J, Liston A, Kyewski B. An evolutionarily conserved mutual interdependence between Aire and microRNAs in promiscuous gene expression. Eur J Immunol (2013) 43(7):1769–78. doi:10.1002/eji.201343343
88. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi:10.1016/S0092-8674(04)00045-5
89. Bui TV, Mendell JT. Myc: maestro of microRNAs. Genes Cancer (2010) 1(6):568–75. doi:10.1177/1947601910377491
90. Fujita S, Iba H. Putative promoter regions of miRNAs genes involved in evolutionarily conserved regulatory systems among vertebrates. Bioinformatics (2008) 24(3):303–8. doi:10.1093/bioinformatics/btm589
91. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J (2002) 21(17):4663–70. doi:10.1093/emboj/cdf476
Keywords: AIRE, miRNA, MTEC, thymus gland, thymocytes, cell adhesion, promiscuous gene expression, central tolerance
Citation: Passos GA, Mendes-da-Cruz DA and Oliveira EH (2015) The thymic orchestration involving Aire, miRNAs, and cell–cell interactions during the induction of central tolerance. Front. Immunol. 6:352. doi: 10.3389/fimmu.2015.00352
Received: 15 May 2015; Accepted: 29 June 2015;
Published: 14 July 2015
Edited by:
Stephen Paul Cobbold, University of Oxford, UKReviewed by:
Hans Acha-Orbea, Université de Lausanne, SwitzerlandCopyright: © 2015 Passos, Mendes-da-Cruz and Oliveira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Geraldo Aleixo Passos, Department of Genetics, Ribeirão Preto Medical School, University of São Paulo, Via Bandeirantes 3900, Ribeirão Preto, São Paulo 14049-900, Brazil,cGFzc29zQHVzcC5icg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.