
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Immunol. , 27 April 2015
Sec. NK and Innate Lymphoid Cell Biology
Volume 6 - 2015 | https://doi.org/10.3389/fimmu.2015.00193
This article mentions parts of:
Human NKG2D-ligands: cell biology strategies to ensure immune recognition
NKG2D is an activating receptor present in all human NK cells, CD8+ T cells, and in certain populations of CD4 T cells, and engagement of this receptor with its ligands is a crucial step in the regulation of both innate and specific immune responses [for review, see Ref. (1)]. NKG2D recognizes two families of MHC-related molecules, whose expression is, in general, upregulated when the cells suffer different types of stress including infection and tumorigenesis. In fact, a large variety of primary tumors and tumor-derived cell lines express NKG2D-ligands and, in the last few years, many reports have established correlations between the presence of NKG2D-ligands in cancer patient serum and disease prognosis [for review, see Ref. (2)]. The presence of NKG2D-ligands in serum is related to another important feature of these molecules: they can be released to the supernatant by either metalloprotease cleavage or incorporated in nanovesicles (exosomes), depending on their biochemical features [for review, see Ref. (3)]. However, it is still not understood why NKG2D-ligands are, in some instances, secreted while in other cases they remain at the cell surface, and understanding the cellular “decisions” on the pathways followed by these molecules will be of crucial importance in the choice of novel therapies based on NKG2D, such as monoclonal antibodies or NKG2D-CARs [for review, see Ref. (4)], since soluble ligands could interfere with these therapies in different manners depending on their biological form. In this sense, detailed study of the differential features of each individual NKG2D-ligand is still needed and investigation of the mechanisms used by different viral gene products to interfere with NKG2D-ligand expression will likely be critical in uncovering these cell biology issues.
NKG2D-ligands belong to two genetic families with multiple members: the major histocompatibility complex class I-related chain (MIC)A/B and UL16 binding protein (ULBP) one to six molecules, also known as retinoic acid early transcripts (RAETs) [for review, see Ref. (5)]. Although both families of NKG2D-ligands are related to MHC molecules and their expression is increased after stress, many differences are observed in their biochemical and cell trafficking properties. Moreover, individual ULBP molecules and MICA/B alleles can share functional characteristics that are not generally conserved within their genetic family. For example, most MICA/B alleles are transmembrane proteins whereas ULBPs have glycosyl-phosphatidyl-inositol (GPI) anchors. However, MICA is a highly polymorphic gene and some of the most frequent alleles, the group known as MICA5.1 whose principal component is MICA*008, contain a frame-shift mutation in the transmembrane region that introduces an early stop codon. These changes mean that MICA5.1 alleles attach to the plasma membrane via a GPI anchor (6), like most of the ULBPs (Figure 1). MICA*008 also differs from other MICA alleles in that it has an extremely slow maturation rate (6, 7) and differs in the N-glycosylation requirements for cell trafficking (8). Functionally, the difference in membrane anchoring of MICA*008 alters its mechanism of release to the extracellular milieu and affects the modulation of the immune response, since multimeric, exosomal MICA*008 are more potent downmodulators of the NKG2D receptor than soluble monomeric MICA molecules (9) and could be more immunogenic (10). Thus, it is important to study the biology of individual NKG2D-ligands separately to understand their involvement in immune recognition and evasion, especially since the use of drugs to modulate immune responses, by regulating NKG2D-ligand release, could also have different outcomes depending on the release mechanism; for example, inhibition of metalloproteases leads to incorporation into exosomes of certain NKG2D-ligands (11).
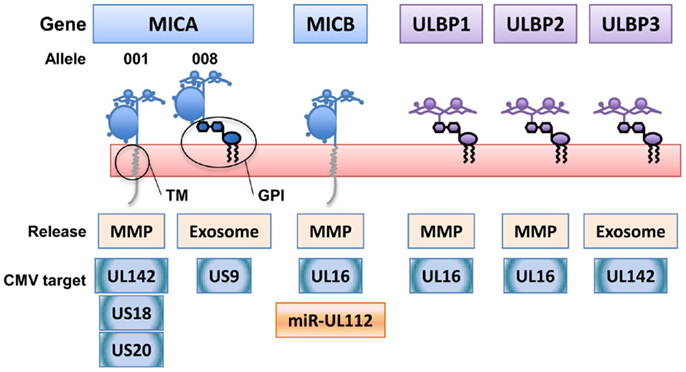
Figure 1. NKG2D-ligands belong to two genetic families with multiple biochemical and cell biology features. The two genetic families of NKG2D-ligands, MICA/B (major histocompatibility complex class I-related chain) and ULBPs (UL16-binding proteins), encode proteins with different membrane attachments [transmembrane (TM) and glycosyl-phosphatidyl- inositol (GPI)] and different mechanisms for release to the extracellular milieu [exosomes versus metalloprotease (MMP) cleavage]. However, not all the members of one of these genetic families of NKG2D-ligands share the same biochemical characteristics. NKG2D-ligands are also differentially recognized by viral proteins and miRNAs that interfere with the immune response. As an example, the human cytomegalovirus evasion molecules targeting NKG2D-ligands (proteins in blue, miRNA in orange) are depicted.
A striking feature of the biology of the MICA*008 allele is its resistance to most of the described viral immune evasion strategies. Human cytomegalovirus (HCMV), for example, encodes several viral proteins and miRNAs that interfere with NKG2D-ligand expression [(12–14) and references therein], yet MICA*008, was resistant to the MICA binding viral protein UL142 (15, 16), and was not downregulated upon infection with HCMV strain AD169VarS (17, 18). Indeed, MICA*008 has been considered to be an HCMV-resistant “escape variant,” and the high prevalence of this allele in multiple populations has been suggested to be the result of an advantage to human NK cells in recognizing infected cells. Recently, however, it has been shown that the US9 gene product of HCMV can selectively downregulate MICA*008 expression in a mechanism that depends on specific interference with the particular cell biology of this MICA allele (7). This observation exemplifies how detailed knowledge of the biochemistry of individual NKG2D-ligands can explain specific functional interactions of these molecules. US9 retention occurs in the ER compartment, in which the MICA*008 protein would be transferred to the GPI moiety (7), while UL142 retains MICA in the Golgi apparatus (16), suggesting different molecular requirements. In fact, UL142 retains inside the cell both the transmembrane MICA alleles and the GPI-anchored protein ULBP3. The use of cell lines susceptible to HCMV infection, but deficient in GPI-transferase enzymes (19, 20), could help to identify whether the GPI anchor is necessary for MICA*008 downregulation by US9 or the molecular mechanism for retention depends on another part of the molecule. Moreover, the information on viral escape and cellular biology of NKG2D-ligands could be applied in other systems in which we still need to understand the role of these molecules, mainly cancer.
That MICA*008 has different biochemical features to transmembrane MICA alleles leads to at least three important consequences: MICA*008 is released preferentially in exosomes, it has a very slow maturation process, and it is not affected by other MICA-modulating pathogen molecules (proteins or miRNAs). It is entirely plausible that in stressed systems, like cancer cells or autoimmunity, certain cellular routes could be blocked while other routes could be enhanced leading to alterations in intracellular protein trafficking. In such a case, NKG2D-ligands could change their expression from intracellular to secreted or appear at the plasma membrane, and this effect will in turn vary depending on the particular ligand. Therefore, it would not be advisable to treat all MICA alleles as biologically equivalent. To interfere intelligently with NKG2D-ligand expression, or to interpret adequately the significance of their expression, will require specific evaluation for each individual patient, determining MICA typing, the amount of ligands released to serum, and the levels of NKG2D receptor on their immune cells. More research is needed, in parallel, to better characterize more differences among MICA alleles and other ligands for NKG2D. In the meantime, it may be worthwhile to revisit the results of previous clinical assays adjusting the data to take into account the MICA typing of the patients, since it might be better to evaluate MICA behavior in function of transmembrane versus GPI-anchored.
The research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author would like to thank H. T. Reyburn for helpful discussions. Work is supported by grants from the Spanish Ministry of Economy and Competitivity (MINECO) [SAF2012-32293] and the Regional Government of Madrid [grant number S2010/BMD-2326 INMUNOTHERCAN].
1. Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol (2013) 31:413–41. doi: 10.1146/annurev-immunol-032712-095951
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene (2008) 27:5944–58. doi:10.1038/onc.2008.272
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Chitadze G, Bhat J, Lettau M, Janssen O, Kabelitz D. Generation of soluble NKG2D ligands: proteolytic cleavage, exosome secretion and functional implications. Scand J Immunol (2013) 78:120–9. doi:10.1111/sji.12072
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Vyas M, Koehl U, Hallek M, Von Strandmann EP. Natural ligands and antibody-based fusion proteins: harnessing the immune system against cancer. Trends Mol Med (2014) 20:72–82. doi:10.1016/j.molmed.2013.10.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Fernandez-Messina L, Reyburn HT, Vales-Gomez M. Human NKG2D-ligands: cell biology strategies to ensure immune recognition. Front Immunol (2012) 3:299. doi:10.3389/fimmu.2012.00299
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Ashiru O, Lopez-Cobo S, Fernandez-Messina L, Pontes-Quero S, Pandolfi R, Reyburn HT, et al. A GPI anchor explains the unique biological features of the common NKG2D-ligand allele MICA*008. Biochem J (2013) 454:295–302. doi:10.1042/BJ20130194
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Seidel E, Le VT, Bar-On Y, Tsukerman P, Enk J, Yamin R, et al. Dynamic co-evolution of host and pathogen: HCMV downregulates the prevalent allele MICA *008 to escape elimination by NK cells. Cell Rep (2015) 10(6):968–82. doi:10.1016/j.celrep.2015.01.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Mellergaard M, Skovbakke SL, Schneider CL, Lauridsen F, Andresen L, Jensen H, et al. N-glycosylation of asparagine 8 regulates surface expression of major histocompatibility complex class I chain-related protein A (MICA) alleles dependent on threonine 24. J Biol Chem (2014) 289:20078–91. doi:10.1074/jbc.M114.573238
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Ashiru O, Boutet P, Fernandez-Messina L, Aguera-Gonzalez S, Skepper JN, Vales-Gomez M, et al. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res (2010) 70:481–9. doi:10.1158/0008-5472.CAN-09-1688
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Tonnerre P, Gerard N, Chatelais M, Poli C, Allard S, Cury S, et al. MICA variant promotes allosensitization after kidney transplantation. J Am Soc Nephrol (2013) 24:954–66. doi:10.1681/ASN.2012080814
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Fernandez-Messina L, Ashiru O, Boutet P, Aguera-Gonzalez S, Skepper JN, Reyburn HT, et al. Differential mechanisms of shedding of the glycosylphosphatidylinositol (GPI)-anchored NKG2D ligands. J Biol Chem (2010) 285:8543–51. doi:10.1074/jbc.M109.045906
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Sutherland CL, Chalupny NJ, Cosman D. The UL16-binding proteins, a novel family of MHC class I-related ligands for NKG2D, activate natural killer cell functions. Immunol Rev (2001) 181:185–92. doi:10.1034/j.1600-065X.2001.1810115.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Fielding CA, Aicheler R, Stanton RJ, Wang EC, Han S, Seirafian S, et al. Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation. PLoS Pathog (2014) 10:e1004058. doi:10.1371/journal.ppat.1004058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Jasinski-Bergner S, Mandelboim O, Seliger B. The role of microRNAs in the control of innate immune response in cancer. J Natl Cancer Inst (2014) 106(10):1–13. doi:10.1093/jnci/dju257
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Chalupny NJ, Rein-Weston A, Dosch S, Cosman D. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun (2006) 346:175–81. doi:10.1016/j.bbrc.2006.05.092
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Ashiru O, Bennett NJ, Boyle LH, Thomas M, Trowsdale J, Wills MR. NKG2D ligand MICA is retained in the cis-Golgi apparatus by human cytomegalovirus protein UL142. J Virol (2009) 83:12345–54. doi:10.1128/JVI.01175-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Zou Y, Bresnahan W, Taylor RT, Stastny P. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J Immunol (2005) 174:3098–104. doi:10.4049/jimmunol.174.5.3098
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, et al. Host immune system gene targeting by a viral miRNA. Science (2007) 317:376–81. doi:10.1126/science.1140956
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Kinoshita T, Maeda Y, Fujita M. Transport of glycosylphosphatidylinositol-anchored proteins from the endoplasmic reticulum. Biochim Biophys Acta (2013) 1833:2473–8. doi:10.1016/j.bbamcr.2013.01.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Kinoshita T. Biosynthesis and deficiencies of glycosylphosphatidylinositol. Proc Jpn Acad Ser B Phys Biol Sci (2014) 90:130–43. doi:10.2183/pjab.90.130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: NKG2D, NKG2D-ligands, MICA/B, ULBP, GPI-anchored proteins, immune evasion, HCMV, exosomes
Citation: Valés-Gómez M (2015) The impact of glycosyl-phosphatidyl-inositol anchored MICA alleles on novel NKG2D-based therapies. Front. Immunol. 6:193. doi: 10.3389/fimmu.2015.00193
Received: 20 March 2015; Accepted: 08 April 2015;
Published: 27 April 2015
Edited by:
Miguel López-Botet, Hospital del Mar Research Institute, SpainReviewed by:
Francisco Borrego, Cruces University Hospital, SpainCopyright: © 2015 Valés-Gómez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mar Valés-Gómez,bXZhbGVzQGNuYi5jc2ljLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.