- 1Integrative Biology Unit, Istituto Nazionale Genetica Molecolare “Romeo ed Enrica Invernizzi”, IRCCS Ospedale Maggiore Policlinico, Milano, Italy
- 2Department of Medical Biotechnology and Translational Medicine, Università degli Studi di Milano, Milano, Italy
Upon recognition of a foreign antigen, CD4+ naïve T lymphocytes proliferate and differentiate into subsets with distinct functions. This process is fundamental for the effective immune system function, as CD4+ T cells orchestrate both the innate and adaptive immune response. Traditionally, this differentiation event has been regarded as the acquisition of an irreversible cell fate so that memory and effector CD4+ T subsets were considered terminally differentiated cells or lineages. Consequently, these lineages are conventionally defined thanks to their prototypical set of cytokines and transcription factors. However, recent findings suggest that CD4+ T lymphocytes possess a remarkable phenotypic plasticity, as they can often re-direct their functional program depending on the milieu they encounter. Therefore, new questions are now compelling such as which are the molecular determinants underlying plasticity and stability and how the balance between these two opposite forces drives the cell fate. As already mentioned, in some cases, the mere expression of cytokines and master regulators could not fully explain lymphocytes plasticity. We should consider other layers of regulation, including epigenetic factors such as the modulation of chromatin state or the transcription of non-coding RNAs, whose high cell-specificity give a hint on their involvement in cell fate determination. In this review, we will focus on the recent advances in understanding CD4+ T lymphocytes subsets specification from an epigenetic point of view. In particular, we will emphasize the emerging importance of non-coding RNAs as key players in these differentiation events. We will also present here new data from our laboratory highlighting the contribution of long non-coding RNAs in driving human CD4+ T lymphocytes differentiation.
The Revolutions of Regulatory Non-Coding RNAs
At the beginning of this century, the results of the human genome project highlighted the complexity of our genome. What emerged was that the fraction of the genome that is informative is higher than we expected. Subsequent analysis revealed that the vast majority of informative sequences does not encode for proteins. Indeed against a total of 62.1% of the human genome covered by processed transcript (74.7% by primary transcripts), exons of protein-coding genes cover only the 2.94% of the genome (1). From an evolutionary point of view, the genome size is in close relationship with coding potential in prokaryotes, which have haploid genomes primarily composed by protein-coding sequences (~88%). Conversely, in eukaryotes, a correlation lacks between protein-coding gene number and organismal complexity. These observations are likely explained by the evolution of a more sophisticated architecture to control gene expression that includes the expansion of non-coding regulatory RNAs (ncRNAs) (2). Thus, we should clearly reassess the centrality of protein-coding RNAs in favor of non-coding ones.
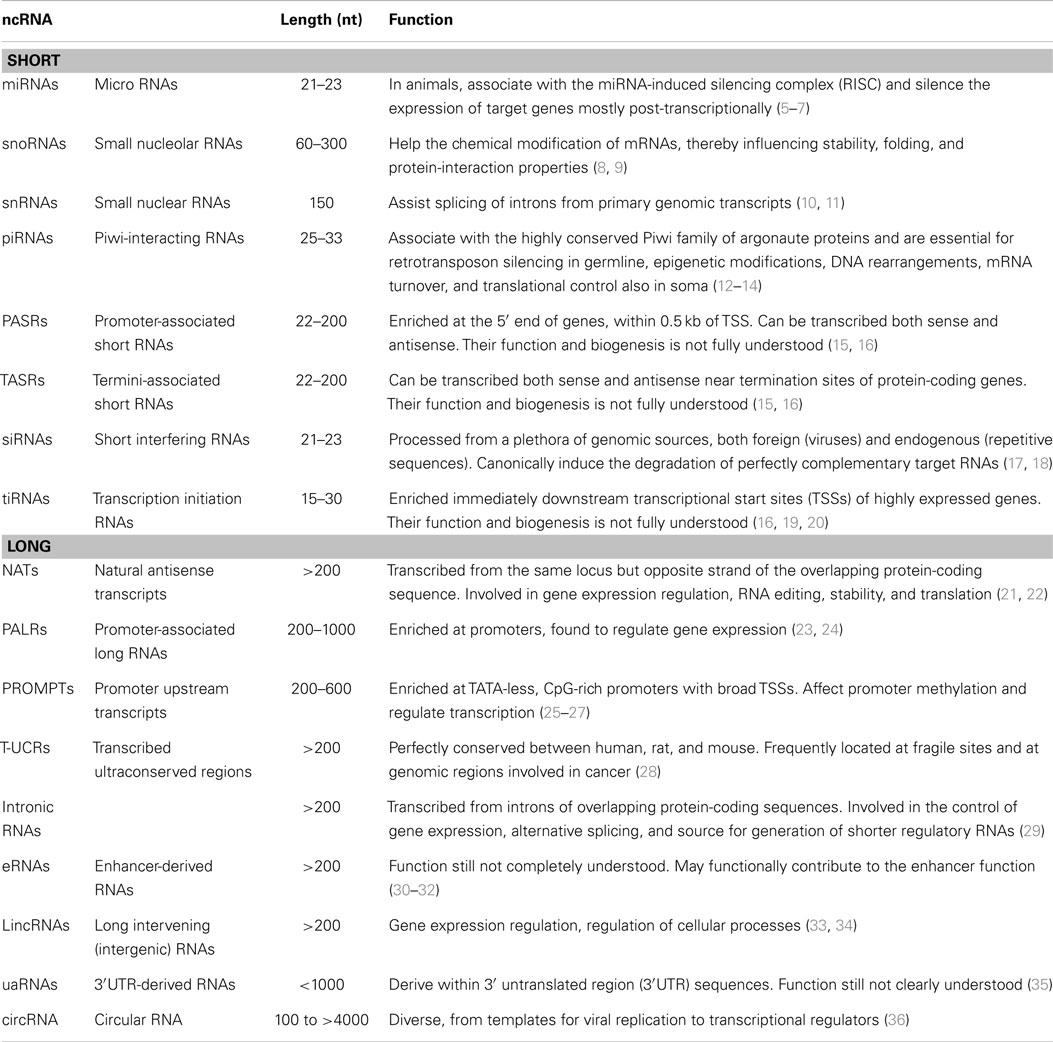
Non-coding RNAs with fundamental functions within cells are known since the discovery of the first transfer RNA (tRNA) (3) and comprise also ribosomal RNAs (rRNAs). Nonetheless, the interest toward non-coding RNAs with regulatory functions arose with the discovery of the first human micro-RNA, let-7 (4). In order to apply a theoretical framework to the transcriptome, regulatory ncRNAs are usually classified based on their dimension: “small” ncRNAs being less than 200 nucleotides in length and “long” or “large” ncRNAs (lncRNAs) ranging from more than 200 to tens of thousands of nucleotides (Table 1).
Further complicating the picture, lncRNAs seem to be the preferred substrate for the generation of small RNAs (21). This maze of non-coding transcripts was revealed also in a genome-wide identification of lncRNAs in mouse CD8+ T lymphocytes, where 18 of the identified lncRNAs appeared to overlap with annotated miRNAs and 21 with snoRNAs (37).
Both classes can be further classified according to their position relative to known sequences of the genome, like in the case of promoter-associated RNAs (PASRs) or transcription initiation small RNAs (tiRNAs). In particular, long non-coding RNAs are usually classified relative to neighboring protein-coding genes. They can be defined as “sense” if they are transcribed from the same strand of the protein-coding gene or “antisense” if the opposite is true. They can be “divergent” if their promoter and the one of the coding transcript are in close proximity and located in a head to head fashion. They can be “exonic” or “intronic” if they overlap one or more exons, or an intron of the protein-coding gene respectively. Instead, they can be “intergenic” (or “intervening”; lincRNAs) if they lie within a sequence between two protein-coding genes (38). In this review, we will focus on this last category, which is probably the most studied given that the location of these lncRNAs avoids complications deriving from the overlap with other genes. The majority of known lncRNAs is generated by the same transcriptional machinery of mRNAs. This means that transcribed lincRNAs genomic sequences are marked by RNA polymerase II occupancy and histone modifications that are shared with active protein-coding genes, such as H3K4me3 at promoters and H3K36me3 within gene bodies (39). They are capped by methylguanosine at their 5′, spliced, and polyadenylated, even if the widespread representation of this last property among known lncRNAs could be partially due to the RNA sequencing strategies used for their identification (15, 40). Indeed, broader analysis identified about 39% of lncRNAs to have at least one of the six most common poly(A) motifs, compared to 51% for coding transcripts (1). These properties imply that there are few distinctive biochemical features that allow the distinction of lncRNAs from protein-coding mRNAs. Among them, lncRNAs have unusual exon structure, with on average 2–5 exons. Intriguingly, lncRNAs are significantly more likely to overlap repetitive elements and particularly RNA-derived transposable elements (TEs). These last account for about 30% of human lncRNAs nucleotides, often in proximity of their transcriptional start site (TSS), which could suggest that TEs could be important drivers of lncRNAs evolution (see below). Nonetheless, the main difference between lncRNAs and protein-coding genes relies by definition on their coding potential: lncRNAs does not possess open reading frames (ORFs), as evaluated based on: the conservation of ORFs codons (41), ORFs length, the presence of known protein domains, in vitro translation (42, 43), and ribosome footprinting (44) assays. However, these conceptual constraints are terribly artificial: short, non-canonical peptides have been found to arise from small ORFs within ncRNA (45–48); lncRNAs genes can also code for proteins and have a double function (49) and ultimately, the coding potential does not necessarily exclude a function as RNA also for known mRNAs (50). Evolution makes boundaries between coding and non-coding genes fainter as ncRNAs can evolve by pseudogenization. This event can follow disruption of the ancestral ORF, but not of the untranslated regulatory regions (UTRs) in protein-coding genes duplicates (50) or can arise without duplication, but from the co-option of ancestral genes to different, non-coding functions (51). This was the case of the long-known Xist RNA, involved in the silencing of the inactive X chromosomes in eutherians. In particular, two exons of the protein-coding gene Lnx3 are homologous to Xist. This gene retained a protein-coding capacity at least in the common ancestor of marsupials and placentals. Conversely, the Xist A-repeat implicated in X-silencing function is not conserved. This sequence likely arose from the insertion of a TE recruited to form a proto-Xist gene (52, 53). Therefore, the difference between dosage compensation in marsupials, eutherians, and monotremes can be ascribed from the presence of a Xist-independent XCI in mammalian ancestor and the peculiar evolution of the proto-Xist gene by pseudogenization in the eutherian ancestor. Intriguingly, other lncRNAs involved in X-inactivation are similarly examples of pseudogenization (54). The boundary between coding and non-coding is even less defined when ncRNAs arise from joining of coding and non-coding exons through alternative splicing (55, 56), from untranslated regions of mRNAs (57, 58) or from the opposite strand of the overlapping protein-coding gene (59). Strikingly, more than a half of protein-coding genes in mammals have a complementary non-coding transcript (60). These findings further challenge our “linear” model of the genome, prompting a re-evaluation of current dogma and genes definitions. Genomic regions indeed are far more complex than previously thought: genes can be used for different purposes and different functional elements can co-locate intermingling coding and non-coding regions.
The interest toward lncRNAs has been rapidly growing and their expressions have been quantitated in many different tissues and cell types by high-throughput sequencing (RNA-seq). These efforts retrieved catalogs with little overlap, so that the number of known lncRNAs is still growing, in contrast with the number of known protein-coding genes that has been remarkably stable over years. Indeed, lncRNAs are far more cell-specific than mRNAs, generally less but also more dynamically expressed at various differentiation stages. For this reason, immune system is an excellent context in which we can deepen our knowledge on lncRNAs. While many excellent reviews cover the recent advances in understanding the role of these molecules within the innate branch (61, 62), little is still known about their importance for the human adaptive immune system. Effector lymphocytes are highly specialized cells that arise from common progenitors through differentiation processes still not completely understood. Besides, lymphocytes can be purified through cell sorting from blood of healthy donors and the existence of in vitro differentiation protocols provide the ideal setting for the identification of lncRNAs expressed in the human immune system and for their functional characterization.
Indeed, the growing interest on lncRNAs and the lack of knowledge on their expression patterns in the human immune system prompted us to perform the RNA-seq analysis on 13 human primary lymphocytes subsets purified by FACS sorting from healthy donors (CD4+ naïve, TH1, TH2, TH17, Treg, TCM, TEM, CD8+ naïve, TCM, TEM, B naïve, B memory, B CD25+) and to develop a bioinformatics pipeline for lincRNAs identification.
Through this analysis, we identified long intergenic ncRNAs genes expressed in these subsets and confirmed that lincRNAs cell-specificity is higher than protein-coding genes even when comparing lincRNAs genes with membrane receptor protein-coding genes, which are generally referred as the most accurate markers for lymphocyte subsets definition. Besides, a major outcome of this analysis is the identification through de novo transcriptome reconstruction of 563 novel, previously unannotated long intergenic ncRNAs genes, increasing by ~12% the number of lincRNAs known to be expressed in human lymphocytes (63). Intriguingly, a fraction of lincRNAs specific for B cells and a fraction of “pan-T” lincRNAs also exist (63). It would be extremely interesting to study these lincRNAs during lymphocytes development in order to understand their likely peculiar role in thymic or bone-marrow-derived cells development.
These observations imply that the little overlap between available catalogs is a direct consequence of lncRNAs specificity and that we could overcome this limitation only assessing lncRNAs expression in every different, highly purified cell type at different developmental stages, instead of considering tissues as a whole. Moreover, due to their specificity of expression, human lymphocytes lincRNAs that are not yet annotated in public resources would have not been identified without performing de novo transcriptome reconstruction. As mentioned before, such tissue-specificity has been linked to the enrichment of TEs in proximity to lincRNAs TSS (64, 65). Moreover, RNA-seq experiments performed in a human CD4+ naïve T cells in vitro differentiation time-course suggest that lincRNA-specific expression in human lymphocyte subsets is acquired during their activation-driven differentiation from naïve to memory cells (63).
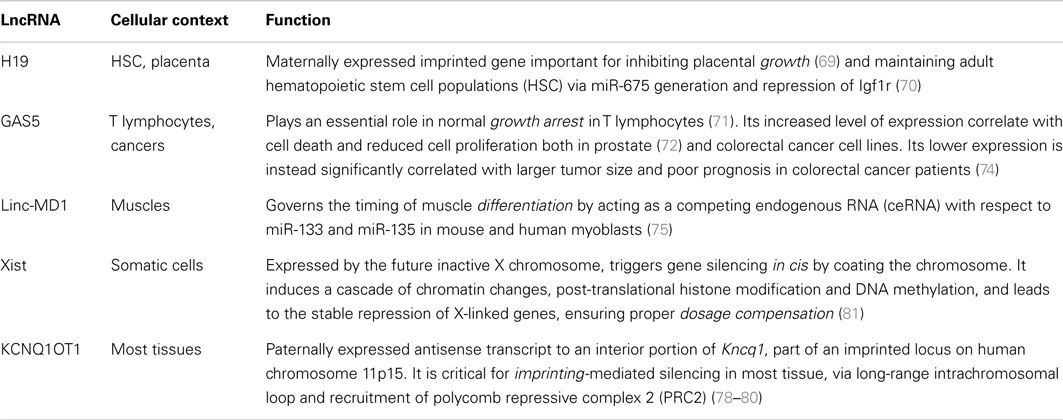
These findings hint to the involvement of lymphocyte-specific lincRNAs as fine-tuners in cell fate commitment, differentiation, and maintenance of cell identity, as demonstrated by many examples in other cell types (66–68). Also, lincRNAs are functionally involved in cell growth (69, 70), apoptosis (71–74), development (75–77), imprinting (78–80), and dosage compensation (81) in almost every cellular context (Table 2).
Long non-coding regulatory RNAs exert their function in these fundamental processes interacting with chromatin or DNA modifiers and transcription factors (TFs) modulating gene expression (82); competing with micro-RNAs acting as sponges (83); modulating subcellular trafficking (84), translation (85), splicing (86), and likely through many other mechanisms still to be discovered (Figure 1).
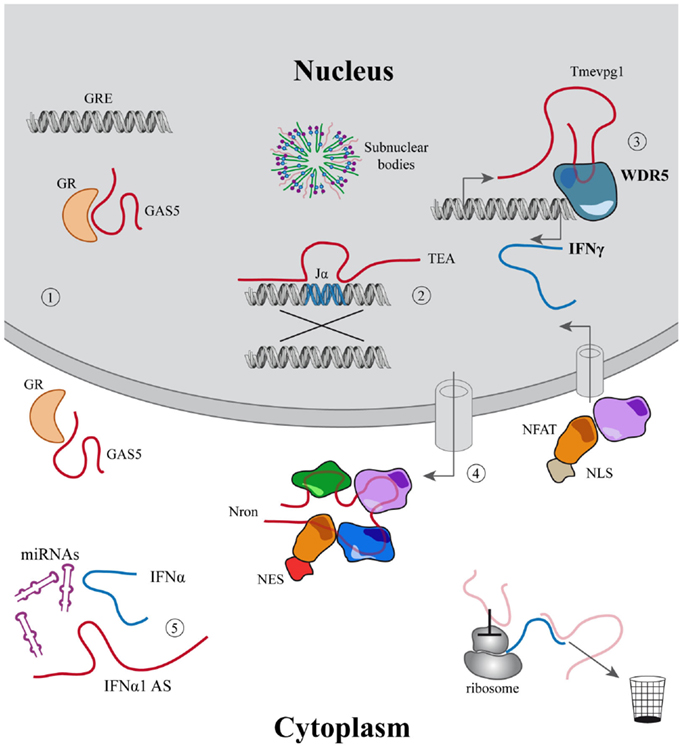
Figure 1. Examples of the main functions associated to lincRNAs. (1–5) LincRNAs described in the immune system. (1) Modulation of cell growth and apoptosis mediated by GAS5 that acts inhibiting glucocorticoid receptors binding to their DNA responsive elements; (2) Jα recombination guided by the PARL TEA; (3) Tmevpg1 recruits WDR5 to induce IFNγ expression; (4) Nron modulates the import–export of NFAT to the nucleus; (5) IFNα1-AS acts as a competing endogenous RNA, releasing IFNα from micro-RNA inhibition. In light red other mechanisms are described for lincRNAs outside the immune system.
Long non-coding regulatory RNAs functional flexibility derive from their intrinsic propensity to fold into thermodynamically stable secondary and higher orders structures that function as interaction modules (87). Each module can fold independently from another, forming bonds at the level of Watson–Crick, Hoogstein, and ribose face (88, 89). These RNAs can rapidly shift between diverse stable structural conformation, allowing allosteric transitions that can act as switches in response to environmental stimuli. They are also processed faster than mRNA, given that they must not be translated, allowing a rapid response to signals. LncRNAs can also be regulated via more than a hundred different nucleotide modifications, like in the case of tRNAs, rRNAs, and snoRNAs (90–92) that modulate their function and probably their structure. RNAs can generate multiple modules within their structure, allowing the interaction with multiple players, the reception of multiple stimuli, and the generation of multiple outputs. The required pairing is likely extremely flexible, such as in the case of micro-RNAs, and allows mismatches, bulges, and wobblings (93). Many of these interaction modules derive from repetitive elements, such as transposons that took advantage of the fewer constraints that lncRNAs sequences have compared to protein-coding genes (1, 94). Indeed, lncRNAs rate of sequence evolution is higher relative to protein-coding genes, even if also these transcripts exhibit evolutionary signatures of functionality. They evolve under modest but detectable selective pressure, accumulating fewer substitutions than neutrally evolving sequences (95, 96). Likely, conservation of relatively small units of lncRNAs sequences (estimated to be less than 5%) could be sufficient to preserve their function, considering their already mentioned modular structure (97). This could be the reason why existing bioinformatic approaches fail to detect low level and scattered selective constraint within these loci (97).
Through such a plastic and versatile structure, lncRNAs can exert their functions binding to proteins, other RNAs (98), and probably also DNA, even if there is still little evidence on the existence of RNA:DNA triplex (99, 100). In particular, lncRNAs can act as scaffolds, bridging together different molecules in a coordinated hub, like in the case of NEAT1: a highly abundant lncRNA that controls sequestration of proteins involved in the formation of paraspeckles, nuclear domains associated with mRNA retention and pathologically enriched in influenza and herpes viruses infections (101, 102). LncRNA can also act as guides, recruiting proteins at specific loci: this has been hypothesized in the case of recombination events that mediate genetic diversity in developing lymphocytes as class switch (CS) and V(D)J recombinations that seem to be mediated by sense and antisense transcripts that dictates the locations of combinatorial events (103–105). Again, lncRNAs can act as control devices or riboswitches in response to extracellular stimuli. For example, they can act as decoys, precluding pre-existing interactions such as GAS5 RNA that detach glucocorticoid receptor (GR) from its responsive elements in conditions of growth arrest (106, 107). Nonetheless, the regulatory potential of lncRNAs has been better characterized in the context of the epigenetic regulation of transcription that ultimately defines the cell transcriptome.
The Role of Long Non-Coding RNAs in Epigenetics
Histones and DNA modifications together with the tridimensional chromosomes conformation within the nucleus define, at least in part, the epigenetic landscape of the cell. This extremely dynamic context modulates gene expression and dictates the final transcriptional output in response to environmental stimuli. By definition, these modifications are then propagated throughout cell divisions. This process is important in every moment of cell life, but particularly during differentiation. Indeed, every cell within our body harbor the same genome, but every cell acquires a particular phenotype according to intrinsic and extrinsic cues that ultimately defines its epigenome and therefore its fate during differentiation. Epigenetics also defines to what extent this fate can be irreversible or plastic (108–110).
As mentioned before, human lymphocytes are an interesting model system for understanding the basis of cell fate specification and plasticity. Indeed, although traditionally the broad range of effector lymphocytes has been referred to as composed by distinct lineages, it has become increasingly clear that these cells also have notable features of plasticity. Differentiation of naïve cells into specific helper subsets requires the integration of extrinsic cues that converge into cell-intrinsic changes in the epigenetic landscape on the genome (111, 112). The interest within the field has been focused on the regulation of prototypical cytokine genes for each subset such as Ifng gene for TH1 or Il-4 for TH2 CD4+ lymphocytes. Much work has been done in both cases to define the complex genetic structure of these loci and the cis regulatory elements bound by TFs and chromatin modifiers promoting or repressing their transcription (113–115). The importance of the setting of epigenetic memory at these fundamental loci was underlined also by treatment with DNA methylation inhibitors (116, 117) or histones deacetylases inhibitors (118–120) and by deletion of DNA methyltransferase (121–123), which caused respectively: constitutive production of IFN-γ, enhanced production of both TH1 and TH2 prototypic cytokines, and inability to activate the proper pattern of expressed cytokines. The same is true for deletion of components of trithorax group (TrxG) or polycomb repressive complex (PRC) that dictates active or repressive epigenetic marks at fundamental loci for proper T-helper cell differentiation, such as Il-4, Il-5, Il-13, and Gata3 (124–129). The pattern of chromatin marks is conventional for signature cytokines: active marks are present at prototypical cytokines whereas repressive marks restrain the expression of antagonistic molecules. However, master regulators and other TFs, usually considered as definers of lineage-specific identity, are characterized by bivalent poised domains, in which both active and repressive chromatin marks are present (130, 131). This histone epigenetic status is peculiar also to promoters in embryonic stem cells, where it poises the expression of key developmental genes thus allowing their timely activation in the presence of differentiation signals and concomitantly precluding expression in their absence (132). Indeed, while the expression of master TFs is quite rapid, cell divisions are required for cytokine loci to become accessible or conversely repressed. Indeed, GATA3 and T-bet/STAT proteins initiate the epigenetic changes at IFN-γ and IL-4 loci that follow the initial activation of naïve T cells and differentiation toward TH1 and TH2 cell fate (133, 134). These observations imply that T-helper cells harbor both clear-cut and plastic epigenetic marks. Nonetheless, we must consider that even cytokines genes that are clearly defined epigenetically, can be expressed or repressed in unexpected context, as reported in TH1 cells converted in IL-4-producing cells during strong TH2-polarizing helminth infections (135) or stable TH1/TH2 hybrid cells derived after parasite infections (136). Therefore, other players must be involved to define the degree of plasticity of lymphocytes in response to these ever-changing environmental conditions during differentiation.
Long non-coding regulatory RNAs have been linked to epigenetic control of gene expression since the first studies regarding the already mentioned Xist transcript, involved in X chromosome inactivation in eutherians. Many other lncRNAs have been associated to chromatin or DNA modifiers and even TFs, thanks to specific mechanistic studies or high-throughput screenings (82, 137–139). This interplay can be observed across a broad range of eukaryotic organisms, suggesting that the epigenetic role of lncRNAs is conserved, even if their mere sequence conservation is often limited (as described previously). It seems that lncRNAs could act as scaffolds, physically associating with proteins that modify chromatin either activating or repressing gene expression. Thanks to the already discussed structural properties of RNA, lncRNAs could organize multiple players in spatially and temporally concerted actions (138). Not only: thanks to their ability to base pair with other nucleic acids, they could recruit these modifiers at specific loci, therefore conferring them specificity of action (98). This property has been an unsolved issue, given that chromatin modifiers do not possess intrinsic bias toward consensus sequences, at least in mammals, while in Drosophila these “docking sites” are well defined (140, 141). Interestingly, while many of these enzymes lack DNA-binding properties, they instead possess RNA-binding motifs (142–144).
The majority of reported lncRNAs are involved in the repression of gene transcription, in particular by interacting with polycomb group (PcG) proteins. The first examples of a direct interaction with PRC2 are the already mentioned Xist (145) and Kcnq1ot1, expressed only in the mammalian paternal chromosome and involved in the silencing of 8–10 protein-coding genes (146). In both these cases, lncRNAs are strictly required for the enrichment of PRC2-associated proteins and for the trimethylation of the lysine 27 of histone H3 at specific loci. Furthermore, other lncRNAs such as NEAT2 and TUG1 promote relocation of growth-control genes at foci of PcG proteins (called PcG bodies), therefore likely facilitating the concerted repression/activation of the transcription units in response to mitogenic signal (147). Many other protein complexes have been found to interact with lncRNAs, the majority targeting histones, either methylases or demethylases, but other involved in DNA methylation (148). Indeed, lncRNAs can bind proteins part of the TrxG (68) that antagonize PcG-mediated silencing (149). Interestingly, an antisense lncRNA has been recently involved in recruiting a regulator of DNA demethylation at a specific promoter (150). This process remains still largely unknown and it has only recently been associated to active enzymatic reactions, via TET family of methylcytosine dioxygenases (151, 152). Even in this case, one of the unsolved questions has been how locus-specificity can be achieved. Particularly, DNA demethylation is often restricted to few dinucleotides at the TSS. The precise mechanism, though, through which lncRNAs could direct DNA or chromatin modification has never been described. Indeed in all reported examples, correlations have been described between lncRNA-modifiers associations and loss of modification after lncRNA gene silencing.
Long non-coding regulatory RNAs are supposed to confer binding specificity to modifiers and recruiting them either in cis or in trans. In the first case, lncRNAs could act directly on sites where they are synthesized without needing to leave DNA. The current hypothesis suggests that the 5′ region of the nascent transcript could bind proteins while the 3′ is transcriptionally lagging, being still tethered to chromatin by RNA polymerase (153). This model is particularly intriguing as through this mechanism, lncRNAs could exert an allele-specific effect, like in the well-studied case of Xist. In trans regulation is instead achieved when lncRNAs act modulating genes across great distances or even on different chromosomes (154). Regarding this dichotomy, we must underline once again its artificiality. Indeed, chromosomes fold into complex, three-dimensional territories together with specialized subnuclear bodies. Proteins that are part of the transcriptional or splicing machinery and regulators of these processes group at these foci (155, 156). These structures are not static, but on the contrary, large-scale chromosomal repositioning is observed in response to environmental stimuli or during differentiation (157, 158). Subnuclear movements are of key importance in regulating events like transcription and rearrangement that occur at immunoglobulin loci during B lymphocytes development (159). The dynamic folding of the genome into higher order structure encompasses loci belonging to the same chromosome, even hundreds of kilobases apart, or different ones, bringing together regions that are distant if we consider the genome as linear. Therefore, in this context, it is extremely difficult to discern what regulations are in cis or in trans, especially when they involve long distance interactions. Intriguingly, lincRNAs have been found to regulate the formation of subnuclear structures, such as NEAT1, required for paraspeckles nucleation (101). LncRNAs can also affect directly the three-dimensional organization of chromosomes enhancing the function of proteins involved in looping formation, like the insulator protein CTCF (160). There are also many examples of lncRNAs involved in three-dimensional local chromatin looping that brings together the ncRNA gene with the region that it regulates within the same chromosome (68, 161). Recently, a lincRNA called Firre has been shown to recruit specific gene loci located on different chromosomes, acting as a docking station for organizing trans-chromosomal associations. Consistently, genetic deletion of Firre leads to a loss of proximity of several trans-interactions (162). A peculiar type of lncRNA has been described that is transcribed from enhancer regions (eRNAs). Classic enhancer elements therefore likely act through transcription of these lncRNAs that upregulate expression at promoters via the recruitment of Mediator complex (163, 164). Finally, there is increasing evidence that even promoters could be transcribed (165), producing lncRNAs probably involved in the enhancer–promoter loop that was hypothesized years ago but never fully resolved (166).
Long Non-Coding RNAs in the Adaptive Immune System
The adaptive immune system is an extraordinary context for the study of the role of lincRNAs in differentiation. Indeed, upon antigen stimulation, naïve CD4+ T cells differentiate into distinct T-helper subsets that were traditionally considered lineages and defined by a prototypic set of expressed cytokines and master TFs. Recently, this relative simple scenario, although useful, has been subjected to debate. CD4+ T cells demonstrated to exhibit substantial plasticity and it has become increasingly clear that they can change the pattern of cytokines and TFs according to the milieu they encounter through their life (167, 168). Not only, in some cases, they can concomitantly express other cytokines and TFs together with their prototypical set. Best examples include IL-10, once thought to specifically identify TH2 and now known to be produced also by TH1, Treg, and TH17 cells (169) and IFN-γ, the classic TH1 cytokine, frequently released by TH17 cells simultaneously with IL-17 (170, 171). Regarding master TFs, Tregs can express Foxp3 (their prototypical TF), but also RORγt (TH17 TF) and Runx3 (172–174); similarly TFH cells can differentiate from FOXP3 positive cells also expressing Bcl6 (their specific TF) (175, 176). In this context, lncRNAs have a fundamental role in governing flexibility and plasticity or maintenance of cell identity, together with lineage-specific TFs and other ncRNAs. In particular, what is emerging from the literature is that ncRNAs typically act as fine-tuners of fate choices and this seems to be true not only in the immune system. Nonetheless, in the case of CD4+ T-cell subsets that are specified but not fully determined, subtle changes in extrinsic signals can reverberate through responsive ncRNAs inducing changes that alter cell phenotype (38, 177, 178). Usually, the stability of lineage identity is achieved through the implementation and inheritance of epigenetic modification, but as mentioned before, lncRNAs can act directly on histone and DNA modifiers redefining this context. Conversely, lncRNAs can also buffer this situation in other conditions, acting as maintainers of cell identity. In the cellular system, lncRNAs can be regarded as minor nodes in a huge interconnected network (179), as they usually interact with few other players. This condition allows them to be more flexible and sensitive to variations without disrupting the whole network integrity (180). This is true both over a very short period, as cells can easily and rapidly adapt to environment, and also over long evolutionary periods, as lncRNAs are among the fastest evolving sequences in the genome (95, 181–183). Conversely, master transcription regulators can be considered highly connected hubs, which confer robustness to the network. Indeed, very few protein-coding genes have been lost from worms to human and mutations are most often pathological (184, 185).
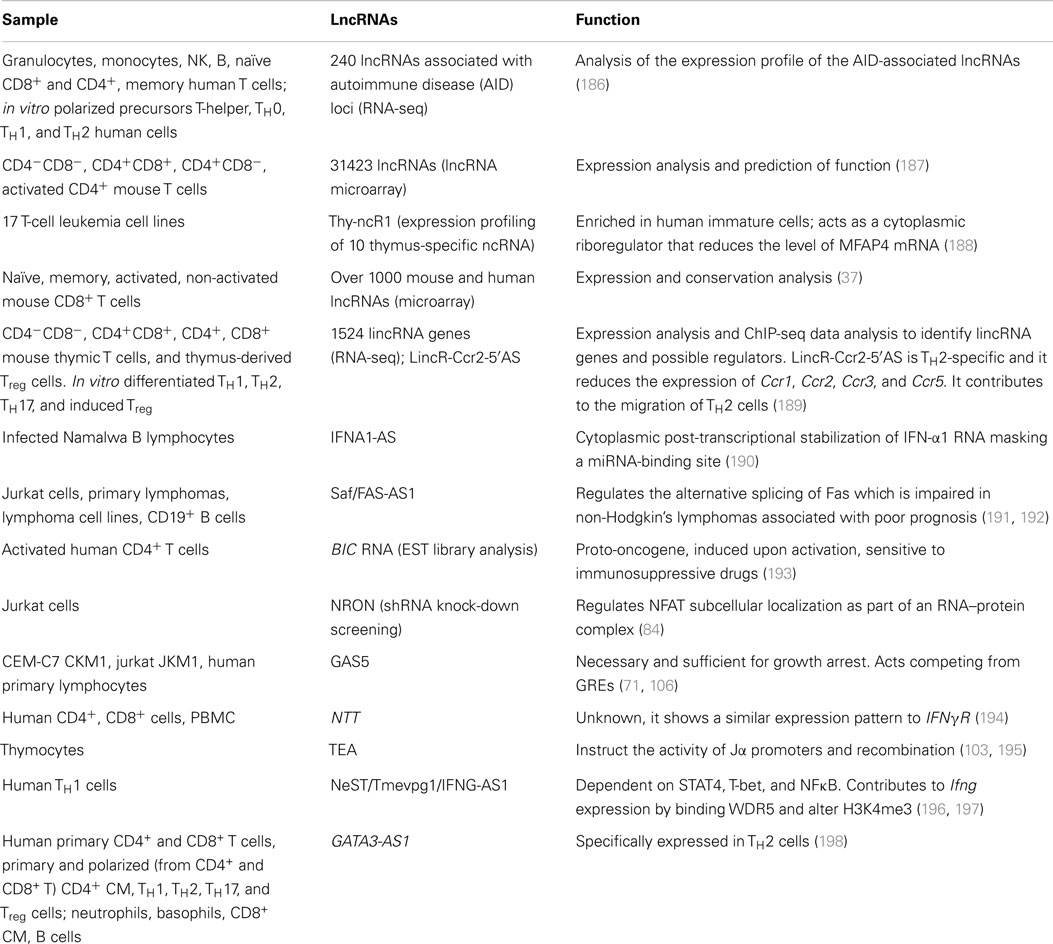
Several single-case or genome-wide studies on lncRNAs in the murine adaptive immune system or cell lines are now available in the literature, whereas only few studies have been conducted until now in the human context. The number of studies that unveiled the function and mechanism of a specific lncRNA is so small that can be counted on one hand (Table 3).
The importance of the studies in the human immune system is underlined by the fact that the differences between experimental animal models and human are still subject of debate in terms of immunologic responses (199–201). Moreover, there are increasing evidences that ncRNAs are poorly conserved between animal models and human (202, 203). In particular, lncRNAs are really fast-evolving elements as demonstrated by the fact that over 80% of the human lncRNAs that arose in the primate lineage, only 3% are conserved across tetrapods and most mammalian lncRNAs lack known orthologs outside vertebrates (97). In detail, even between mouse and human, lncRNAs are poorly conserved (204–206). Despite their rapid evolution, lncRNAs are selected more than neutral sequences and in particular more than intergenic regions, but significantly less than mRNAs (96, 97, 207). It must be underlined that the conservation rate reported could be overestimated: substitution rates are derived from whole-genome alignment and based on the assumption that even segment of homologies imply that that segment belongs to the same RNA class, but this is not necessarily the case. Indeed, it could be that in another genome context a specific lncRNA gene segment is transcribed and processed as part of a protein-coding RNA (208). A striking example is Hotair that is involved in the regulation of the highly conserved cluster of Hox genes (68). The human lincRNA is conserved in the mouse genome (209), nonetheless only the 3′ region is effectively part of the murine homolog (183). The importance of studying lincRNAs specifically within the human immune system derives from these considerations, but this field is still poorly investigated. The majority of the studies focused on the innate immune system (210–212) or analyzed pathological situations, such as cancer-related lncRNAs (192, 213) or responses to specific infections (102, 214–216), mostly in mice. The first functional study that focused on the adaptive immune system, and in particular on TH1 and TH2 lymphocytes, involved a lincRNA, Tmevpg1, that is selectively expressed in TH1 cells via STAT4 and T-bet, both in mouse and human. It participates in the induction of IFN-γ expression strictly in response to TH1 differentiation program and not in other cellular contexts. These results highlight once again the complexity of gene expression regulatory network and the specificity of action of lincRNAs (196). Another paper described a lincRNA, GATA3-AS1, specifically expressed in primary TH2 cells and hypothesized its co-regulation with GATA3 (198). GAS5, expressed in human T lymphocytes, is degraded in optimal growth conditions, but it accumulates contributing to growth arrest in starving conditions (107). In this situation, it competes with GRs DNA-binding sequences, suppressing GR-mediated transcription (106). Broader studies have been performed on the CD8+ T cell transcriptome (37), and recently on CD4+ T lymphocytes (189), but still on mice models. In B cells, chromatin remodeling associated with V(D)J recombination has been potentially linked to a widespread antisense intergenic transcription that occurs in the variable (V) region of the immunoglobulin heavy chain (Igh) locus (104, 105). So far, no studies have been published that performed a deep transcriptomic analysis on human primary lymphocytes from healthy donors, identifying lncRNAs fundamental for differentiation processes. These few examples are just clues of the importance that lincRNA could have for the proper function also of the human immune system and prompt to a deeper analysis of their role in this particularly intriguing context.
Long Non-Coding RNAs as Epigenetic Modulators in Lymphocyte Differentiation
Traditionally, the secretion of IFN-γ and TNF-α characterizes TH1 lymphocytes, whereas IL-4, IL-5, and IL-13 are considered prototypic cytokines secreted by TH2 cells. According to this classic paradigm, these differences underline the different functions exerted by these lymphocytes: TH1 are considered as important to eliminate intracellular bacteria and viruses, whereas TH2 to resist parasitic infections (217). The advantage of solid in vitro differentiation protocols allowed a deep understanding of the genetic mechanisms governing these cells. Since the discovery of this dichotomy, other cell subsets have been identified, but this TH1/TH2 paradigm was undoubtedly useful. Therefore, it is not a case that among the few lncRNAs identified in the immune system, many of those functionally characterized have been described in these two cell subsets. Nevertheless as mentioned before, just one lincRNA, Tmevpg1 (also known as NeST or IFNG-AS1) has been characterized in deep. Tmevpg1 is located proximal to IFN-γ gene both in mice and humans, antisense and convergently transcribed respect to the neighboring gene and plays a role in chromatin remodeling. This transcript is a TH1-specific lincRNA: it requires STAT4 and T-bet for being transcribed and is also bound by CTCF and cohesin during lineage-specific induction (196). Therefore, Tmevpg1 is directly dependent on the activation of a TH1-polarizing transcriptional program, in which the presence of IL-12 leads to the activation of the JAK/STAT pathway via STAT4 (and STAT1) that induces the expression of T-bet. Interestingly, Tmevpg1 gene harbor sequences regulated by histone acetylation and DNase I hypersensitive sites found in TH1 but not TH2 cells (218, 219). Tmevpg1, in its turn, plays a direct part in defining the proper TH1 cytokine expression pattern, influencing Ifng transcription in the presence of T-bet (196), via H3K4 trimethylation by WDR5 binding in mice models (197).
Given the increasing number of lncRNAs described in different cellular contexts and the high number of specific lincRNAs expressed in the different lymphocytes subsets identified with the aforementioned RNA-seq analysis, many more lincRNAs will likely be characterized in the future with a relevant function in the human immune system. A major limitation, though, in the studies on lncRNAs is that there is little biological knowledge on the biochemical or molecular function of lncRNA genes. Compared to classical protein-coding gene studies, hints on their functions cannot be gained simply by the analysis of their primary sequence and application of computational methods to infer lncRNA function are also still in their infancy. As lincRNAs have been reported to influence the expression of neighboring genes (25, 26, 28, 39), one possible approach to investigate their putative function is to focus on lymphocyte lincRNAs proximal to protein-coding genes involved in key cell-functions.
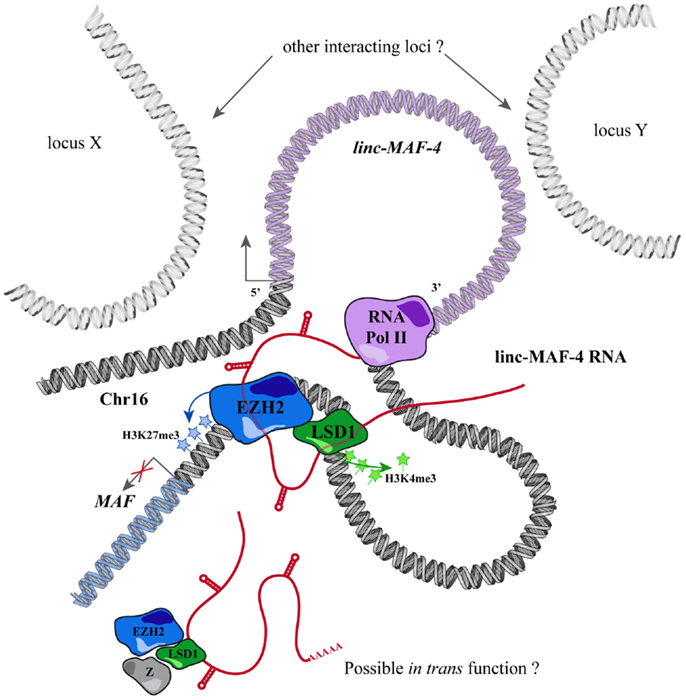
Through this approach, we identified a TH1-specific lincRNA that localized ~140 kb upstream to MAF that was therefore called linc-MAF-4. MAF is a TF involved in TH2 differentiation and required for the efficient secretion of IL-4 by TH17 and the proper development of TFH cells (220–222). Intriguingly, the expression of linc-MAF-4 is negatively correlated with respect to the expression of MAF: linc-MAF-4 expression is high and specific in TH1 lymphocytes, where MAF is lowly expressed whereas in TH2 cells the expression of linc-MAF-4 is extremely low and MAF is highly expressed. Coherently, linc-MAF-4 knock-down in naïve CD4+ T cells increased the expression of MAF and interestingly induced a more general skewing of the whole transcriptomic profile of these cells toward a TH2-like fate (63). The regulation exerted by linc-MAF-4 on MAF gene was analyzed in more detail and this lincRNA proved to modulate MAF expression in cis, as hypothesized by expression analysis. Linc-MAF-4 exerts this regulation by exploiting a chromatin loop that brings its genomic region close to the promoter of MAF gene. Indeed, the chromatin organization of this region allows linc-MAF-4 transcript to recruit chromatin remodelers that inhibit MAF transcription. In particular, linc-MAF-4 was found to associate with EZH2, key enzymatic subunit of the PRC2 complex, and LSD1. These proteins methylate H3K27 and demethylate H3K4, respectively: two histones modifications that code for transcriptional repression. (63). A similar mechanism was described for other lincRNAs, such as HOTAIR and MEG3 (154, 223) but never before for other lncRNAs expressed in the adaptive immune system (Figure 2).
Changes of lincRNAs expression during naïve to memory CD8+ T-cell differentiation (37) and during naïve CD4+ T cells differentiation into distinct helper T-cell lineages (189) have been described in the mouse immune system. linc-MAF-4, though, is, to our knowledge, the first example of a lincRNA playing a role in the proper differentiation of human TH1 cells, suggesting that, besides cytokines and TFs, lncRNAs take part in the TH1 differentiation program as already shown in many other cell types. At this point, an obvious question arises: to what extent are these cells plastic? These findings are evidences that it is possible to re-direct the differentiation path of naïve CD4+ T cells acting on their lincRNA content. Nevertheless, could it be possible to modulate already committed cells? We would expect that the mere down-regulation of a lincRNA would not be sufficient nor a lincRNA knock-out: as mentioned before, lincRNAs are minor nodes in a huge interconnected network composed by feedback mechanisms and epigenetic marks that act stabilizing a pre-existent differentiation status. However, a modulation in lincRNA content may be sufficient to make these cells more responsive to environmental cues that could overcome stabilizing forces, inducing a sort of trans-differentiation event. Functional characterization of other lncRNAs is required to address this crucial issue and to assess the extent of their contribution to cell differentiation and to the maintenance of cell identity in human lymphocytes. Based on what we discussed so far on lncRNA functions and cell-specificity, we believe that future studies will show how these molecules could be capitalized as new molecular targets for the development of novel and highly specific therapies for diseases, such as autoimmunity, immunodeficiencies, allergy, and cancer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank V. Ranzani for support in figures preparation and for discussions and critical revision of the manuscript. Supported by Consiglio Nazionale delle Ricerche – Ministero dell’Istruzione dell’Universita e della Ricerca (EPIGEN), Fondazione Cariplo (2013-0955), AIRC Associazione Italiana per la Ricerca sul Cancro (IG2013-ID14596), the European Research Council (269022 to SA; 617978 to MP), and Fondazione Romeo ed Enrica Invernizzi.
References
1. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res (2012) 22(9):1775–89. doi: 10.1101/gr.132159.111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays (2007) 29(3):288–99. doi:10.1002/bies.20544
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Hoagland MB, Keller EB, Zamecnik PC. Enzymatic carboxyl activation of amino acids. J Biol Chem (1956) 218(1):345–58.
4. Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature (2000) 408(6808):86–9. doi:10.1038/35040556
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol (2013) 13(9):666–78. doi:10.1038/nri3494
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Bronevetsky Y, Ansel KM. Regulation of miRNA biogenesis and turnover in the immune system. Immunol Rev (2013) 253(1):304–16. doi:10.1111/imr.12059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Monticelli S. MicroRNAs in T helper cell differentiation and plasticity. Semin Immunol (2013) 25(4):291–8. doi:10.1016/j.smim.2013.10.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Bratkovič T, Rogelj B. The many faces of small nucleolar RNAs. Biochim Biophys Acta (2014) 1839(6):438–43. doi:10.1016/j.bbagrm.2014.04.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics (2009) 94(2):83–8. doi:10.1016/j.ygeno.2009.05.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Lui L, Lowe T. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem (2013) 54:53–77. doi:10.1042/bse0540053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Valadkhan S, Gunawardane LS. Role of small nuclear RNAs in eukaryotic gene expression. Essays Biochem (2013) 54:79–90. doi:10.1042/bse0540079
12. Bamezai S, Rawat VPS, Buske C. Concise review: the Piwi-piRNA axis: pivotal beyond transposon silencing. Stem Cells (2012) 30(12):2603–11. doi:10.1002/stem.1237
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Ku H-Y, Lin H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl Sci Rev (2014) 1(2):205–18. doi:10.1093/nsr/nwu014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet (2013) 14(8):523–34. doi:10.1038/nrg3495
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science (2007) 316(5830):1484–8. doi:10.1126/science.1138341
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle (2009) 8(15):2332–8. doi:10.4161/cc.8.15.9154
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell (2009) 136(4):642–55. doi:10.1016/j.cell.2009.01.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Svoboda P. Renaissance of mammalian endogenous RNAi. FEBS Lett (2014) 588(15):2550–6. doi:10.1016/j.febslet.2014.05.030
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet (2009) 41(5):572–8. doi:10.1038/ng.312
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol (2010) 17(8):1030–4. doi:10.1038/nsmb.1841
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi AM, et al. Landscape of transcription in human cells. Nature (2012) 489(7414):101–8. doi:10.1038/nature11233
22. Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol (2009) 10(9):637–43. doi:10.1038/nrm2738
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Kurokawa R. Promoter-associated long noncoding RNAs repress transcription through a RNA binding protein TLS. Adv Exp Med Biol (2011) 722:196–208. doi:10.1007/978-1-4614-0332-6_12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Zaphiropoulos PG. A promoter-associated RNA downregulates the oncogenic GLI1 transcription factor in rhabdomyosarcoma cells. RNA Dis (2014) 1(1):e254. doi:10.14800/rd.254
25. Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science (2008) 322(5909):1851–4. doi:10.1126/science.1164096
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Imamura T, Yamamoto S, Ohgane J, Hattori N, Tanaka S, Shiota K. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem Biophys Res Commun (2004) 322(2):593–600. doi:10.1016/j.bbrc.2004.07.159
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Preker P, Almvig K, Christensen MS, Valen E, Mapendano CK, Sandelin A, et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res (2011) 39(16):7179–93. doi:10.1093/nar/gkr370
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Peng JC, Shen J, Ran ZH. Transcribed ultraconserved region in human cancers. RNA Biol (2013) 10(12):1771–7. doi:10.4161/rna.26995
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics (2009) 93(4):291–8. doi:10.1016/j.ygeno.2008.11.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci (2014) 39(4):170–82. doi:10.1016/j.tibs.2014.02.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol (2011) 18(8):956–63. doi:10.1038/nsmb.2085
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet (2012) 46:1–19. doi:10.1146/annurev-genet-110711-155459
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. doi:10.1146/annurev-biochem-051410-92902
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Merry C, Niland C, Khalil A. Diverse functions and mechanisms of mammalian long noncoding RNAs. In: Carmichael GG, editor. Regulatory Non-Coding RNAs: Methods in Molecular Biology. Vol. 1206. New York, NY: Springer (2015). p. 1–14.
35. Mercer TR, Wilhelm D, Dinger ME, Soldà G, Korbie DJ, Glazov EA, et al. Expression of distinct RNAs from 3’ untranslated regions. Nucleic Acids Res (2011) 39(6):2393–403. doi:10.1093/nar/gkq1158
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA (2014) 20(12):1829–42. doi:10.1261/rna.047126.114
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol (2009) 182(12):7738–48. doi:10.4049/jimmunol.0900603
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Pagani M, Rossetti G, Panzeri I, de Candia P, Bonnal RJ, Rossi RL, et al. Role of microRNAs and long-non-coding RNAs in CD4(+) T-cell differentiation. Immunol Rev (2013) 253(1):82–96. doi:10.1111/imr.12055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature (2009) 458(7235):223–7. doi:10.1038/nature07672
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Dieci G, Conti A, Pagano A, Carnevali D. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim Biophys Acta (2013) 1829(3–4):296–305. doi:10.1016/j.bbagrm.2012.09.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene (2007) 26(6):851–8. doi:10.1038/sj.onc.1209846
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol (2007) 5(5):e106. doi:10.1371/journal.pbio.0050106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell (1999) 97(1):17–27. doi:10.1016/S0092-8674(00)80711-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep (2014) 8(5):1365–79. doi:10.1016/j.celrep.2014.07.045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res (2012) 22(9):1646–57. doi:10.1101/gr.134767.111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A (2012) 109(37):E2424–32. doi:10.1073/pnas.1207846109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol (2013) 9(1):59–64. doi:10.1038/nchembio.1120
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet (2014) 15(3):193–204. doi:10.1038/nrg3520
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Ulveling D, Francastel C, Hube F. When one is better than two: RNA with dual functions. Biochimie (2011) 93(4):633–44. doi:10.1016/j.biochi.2010.11.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature (2010) 465(7301):1033–8. doi:10.1038/nature09144
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science (2006) 312(5780):1653–5. doi:10.1126/science.1126316
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Elisaphenko EA, Kolesnikov NN, Shevchenko AI, Rogozin IB, Nesterova TB, Brockdorff N, et al. A dual origin of the Xist gene from a protein-coding gene and a set of transposable elements. PLoS One (2008) 3(6):e2521. doi:10.1371/journal.pone.0002521
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Horvath JE, Sheedy CB, Merrett SL, Diallo AB, Swofford DL, Program NCS, et al. Comparative analysis of the primate X-inactivation center region and reconstruction of the ancestral primate XIST locus. Genome Res (2011) 21(6):850–62. doi:10.1101/gr.111849.110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Romito A, Rougeulle C. Origin and evolution of the long non-coding genes in the X-inactivation center. Biochimie (2011) 93(11):1935–42. doi:10.1016/j.biochi.2011.07.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Bussotti G, Notredame C, Enright A. Detecting and comparing non-coding RNAs in the high-throughput era. Int J Mol Sci (2013) 14(8):15423–58. doi:10.3390/ijms140815423
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell (1991) 64(3):607–13. doi:10.1016/0092-8674(91)90244-S
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Beltran M, Puig I, Peña C, García JM, Álvarez AB, Peña R, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev (2008) 22(6):756–69. doi:10.1101/gad.455708
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature (2008) 454(7206):899–902. doi:10.1038/nature07117
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotech (2012) 30(1):99–104. doi:10.1038/nbt.2024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science (2005) 309(5740):1564–6. doi:10.1126/science.1112009
61. Li Z, Rana TM. Decoding the noncoding: prospective of lncRNA-mediated innate immune regulation. RNA Biol (2014) 11(8):979–85. doi:10.4161/rna.29937
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Imamura K, Akimitsu N. Long non-coding RNAs involved in immune responses. Front Immunol (2014) 5:573. doi:10.3389/fimmu.2014.00573
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Ranzani VRG, Panzeri I, Arrigoni A, Bonnal R, Curti S, Gruarin P, et al. LincRNA landscape in human lymphocytes highlights regulation of T cell differentiation by linc-MAF-4. Nat Immunol (2015) 16(3):318–25. doi:10.1038/ni.3093
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, et al. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet (2013) 9(4):e1003470. doi:10.1371/journal.pgen.1003470
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol (2012) 13(11):R107. doi:10.1186/gb-2012-13-11-r107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell (2014) 53(6):1005–19. doi:10.1016/j.molcel.2014.01.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell (2013) 152(3):570–83. doi:10.1016/j.cell.2013.01.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature (2011) 472(7341):120–4. doi:10.1038/nature09819
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol (2012) 14(7):659–65. doi:10.1038/ncb2521
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature (2013) 500(7462):345–9. doi:10.1038/nature12303
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5). J Cell Sci (2008) 121(7):939–46. doi:10.1242/jcs.024646
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta (2013) 1832(10):1613–23. doi:10.1016/j.bbadis.2013.05.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Rossi MN, Antonangeli F. LncRNAs: new players in apoptosis control. Int J Cell Biol (2014) 2014:7. doi:10.1155/2014/473857
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol (2014) 31(11):253. doi:10.1007/s12032-014-0253-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell (2011) 147(2):358–69. doi:10.1016/j.cell.2011.09.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet (2014) 15(1):7–21. doi:10.1038/nrg3606
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep (2012) 13(11):971–83. doi:10.1038/embor.2012.145
78. Lee Jeannie T, Bartolomei Marisa S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell (2013) 152(6):1308–23. doi:10.1016/j.cell.2013.02.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, et al. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet (1999) 8(7):1209–17. doi:10.1093/hmg/8.7.1209
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Zhang H, Zeitz MJ, Wang H, Niu B, Ge S, Li W, et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J Cell Biol (2014) 204(1):61–75. doi:10.1083/jcb.201304152
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Gendrel AV, Heard E. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol (2014) 30:561–80. doi:10.1146/annurev-cellbio-101512-122415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A (2009) 106(28):11667–72. doi:10.1073/pnas.0904715106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi Pier P. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell (2011) 146(3):353–8. doi:10.1016/j.cell.2011.07.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science (2005) 309(5740):1570–3. doi:10.1126/science.1115901
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature (2012) 491(7424):454–7. doi:10.1038/nature11508
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell (2010) 39(6):925–38. doi:10.1016/j.molcel.2010.08.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Conrad NK. The emerging role of triple helices in RNA biology. Wiley Interdiscip Rev RNA (2014) 5(1):15–29. doi:10.1002/wrna.1194
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell (2009) 136(4):604–9. doi:10.1016/j.cell.2009.02.003
89. Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA (2006) 12(1):83–93. doi:10.1261/rna.2208106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol (2011) 7(12):885–7. doi:10.1038/nchembio.687
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Yang Y, Zhou X, Jin Y. ADAR-mediated RNA editing in non-coding RNA sequences. Sci China Life Sci (2013) 56(10):944–52. doi:10.1007/s11427-013-4546-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res (2012) 40(11):5023–33. doi:10.1093/nar/gks144
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Chi SW, Hannon GJ, Darnell RB. An alternative mode of microRNA target recognition. Nat Struct Mol Biol (2012) 19(3):321–7. doi:10.1038/nsmb.2230
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell (2008) 29(4):499–509. doi:10.1016/j.molcel.2007.12.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet (2012) 8(7):e1002841. doi:10.1371/journal.pgen.1002841
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res (2007) 17(5):556–65. doi:10.1101/gr.6036807
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Marques AC, Ponting CP. Intergenic lncRNAs and the evolution of gene expression. Curr Opin Genet Dev (2014) 27(0):48–53. doi:10.1016/j.gde.2014.03.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Engreitz Jesse M, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell (2014) 159(1):188–99. doi:10.1016/j.cell.2014.08.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell (2011) 44(4):667–78. doi:10.1016/j.molcel.2011.08.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev (2010) 24(20):2264–9. doi:10.1101/gad.590910
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell (2009) 33(6):717–26. doi:10.1016/j.molcel.2009.01.026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell (2014) 53(3):393–406. doi:10.1016/j.molcel.2014.01.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Abarrategui I, Krangel MS. Noncoding transcription controls downstream promoters to regulate T-cell receptor alpha recombination. EMBO J (2007) 26(20):4380–90. doi:10.1038/sj.emboj.7601866
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, et al. Antisense intergenic transcription in V(D)J recombination. Nat Immunol (2004) 5(6):630–7. doi:10.1038/ni1068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci U S A (2012) 109(42):17004–9. doi:10.1073/pnas.1208398109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal (2010) 3(107):ra8. doi:10.1126/scisignal.2000568
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
107. Williams GT, Mourtada-Maarabouni M, Farzaneh F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem Soc Trans (2011) 39(2):482–6. doi:10.1042/BST0390482
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
108. Bird A. Perceptions of epigenetics. Nature (2007) 447(7143):396–8. doi:10.1038/nature05913
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell (2007) 128(4):635–8. doi:10.1016/j.cell.2007.02.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Costa FF. Non-coding RNAs, epigenetics and complexity. Gene (2008) 410(1):9–17. doi:10.1016/j.gene.2007.12.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol (2003) 4(7):616–23. doi:10.1038/ni0703-616
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Tripathi SK, Lahesmaa R. Transcriptional and epigenetic regulation of T-helper lineage specification. Immunol Rev (2014) 261(1):62–83. doi:10.1111/imr.12204
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol (2006) 24:607–56. doi:10.1146/annurev.immunol.23.021704.115821
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Balasubramani A, Mukasa R, Hatton RD, Weaver CT. Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunol Rev (2010) 238(1):216–32. doi:10.1111/j.1600-065X.2010.00961.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
115. Chang S, Aune TM. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol (2007) 8(7):723–31. doi:10.1038/ni1473
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Ballas ZK. The use of 5-azacytidine to establish constitutive interleukin 2-producing clones of the EL4 thymoma. J Immunol (1984) 133(1):7–9.
117. Young HA, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard JR, et al. Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-gamma gene. J Immunol (1994) 153(8):3603–10.
118. Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, et al. Helper T cell differentiation is controlled by the cell cycle. Immunity (1998) 9(2):229–37. doi:10.1016/S1074-7613(00)80605-6
119. Morinobu A, Kanno Y, O’Shea JJ. Discrete roles for histone acetylation in human T helper 1 cell-specific gene expression. J Biol Chem (2004) 279(39):40640–6. doi:10.1074/jbc.M407576200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
120. Valapour M, Guo J, Schroeder JT, Keen J, Cianferoni A, Casolaro V, et al. Histone deacetylation inhibits IL4 gene expression in T cells. J Allergy Clin Immunol (2002) 109(2):238–45. doi:10.1067/mai.2002.121145
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
121. Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell (2002) 10(1):81–91. doi:10.1016/S1097-2765(02)00564-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
122. Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity (2001) 15(5):763–74. doi:10.1016/S1074-7613(01)00227-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
123. Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat Immunol (2003) 4(12):1183–90. doi:10.1038/ni1004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
124. Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, Kimura MY, et al. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity (2006) 24(5):611–22. doi:10.1016/j.immuni.2006.03.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
125. Onodera A, Yamashita M, Endo Y, Kuwahara M, Tofukuji S, Hosokawa H, et al. STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J Exp Med (2010) 207(11):2493–506. doi:10.1084/jem.20100760
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
126. Kimura M, Koseki Y, Yamashita M, Watanabe N, Shimizu C, Katsumoto T, et al. Regulation of Th2 cell differentiation by mel-18, a mammalian polycomb group gene. Immunity (2001) 15(2):275–87. doi:10.1016/S1074-7613(01)00182-0
127. Yamashita M, Kuwahara M, Suzuki A, Hirahara K, Shinnaksu R, Hosokawa H, et al. Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J Exp Med (2008) 205(5):1109–20. doi:10.1084/jem.20072000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
128. Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem (2005) 280(36):31470–7. doi:10.1074/jbc.M504766200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
129. Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell (2005) 121(3):425–36. doi:10.1016/j.cell.2005.02.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
130. Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity (2009) 30(1):155–67. doi:10.1016/j.immuni.2008.12.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
131. Hirahara K, Vahedi G, Ghoreschi K, Yang XP, Nakayamada S, Kanno Y, et al. Helper T-cell differentiation and plasticity: insights from epigenetics. Immunology (2011) 134(3):235–45. doi:10.1111/j.1365-2567.2011.03483.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
132. Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell (2007) 128(4):669–81. doi:10.1016/j.cell.2007.01.033
133. Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity (2001) 14(3):205–15. doi:10.1016/S1074-7613(01)00103-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
134. Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science (2001) 292(5523):1907–10. doi:10.1126/science.1059835
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
135. Panzer M, Sitte S, Wirth S, Drexler I, Sparwasser T, Voehringer D. Rapid in vivo conversion of effector T cells into Th2 cells during helminth infection. J Immunol (2012) 188(2):615–23. doi:10.4049/jimmunol.1101164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
136. Peine M, Rausch S, Helmstetter C, Fröhlich A, Hegazy AN, Kühl AA, et al. Stable T-bet(+)GATA-3(+) Th1/Th2 hybrid cells arise in vivo, can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS Biol (2013) 11(8):e1001633. doi:10.1371/journal.pbio.1001633
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
137. Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell (2010) 40(6):939–53. doi:10.1016/j.molcel.2010.12.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
138. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science (2010) 329(5992):689–93. doi:10.1126/science.1192002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
139. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature (2011) 477(7364):295–300. doi:10.1038/nature10398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
140. Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, et al. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol (2009) 7(1):e1000013. doi:10.1371/journal.pbio.1000013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
141. Kassis JA, Brown JL. Polycomb group response elements in Drosophila and vertebrates. Adv Genet (2013) 81:83–118. doi:10.1016/B978-0-12-407677-8.00003-8
142. Bernstein E, Allis CD. RNA meets chromatin. Genes Dev (2005) 19(14):1635–55. doi:10.1101/gad.1324305
143. Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem (2004) 279(47):49479–87. doi:10.1074/jbc.M409070200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
144. Hiragami-Hamada K, Fischle W. RNAs – physical and functional modulators of chromatin reader proteins. Biochim Biophys Acta (2014) 1839(8):737–42. doi:10.1016/j.bbagrm.2014.03.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
145. Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science (2008) 322(5902):750–6. doi:10.1126/science.1163045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
146. Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell (2008) 32(2):232–46. doi:10.1016/j.molcel.2008.08.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
147. Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell (2011) 147(4):773–88. doi:10.1016/j.cell.2011.08.054
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
148. Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW, et al. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife (2014) 3:e04530. doi:10.7554/eLife.04530
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
149. Steffen PA, Ringrose L. What are memories made of? How polycomb and trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol (2014) 15(5):340–56. doi:10.1038/nrm3789
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
150. Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell (2014) 55(4):604–14. doi:10.1016/j.molcel.2014.06.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
151. Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci (2011) 101:25–104. doi:10.1016/B978-0-12-387685-0.00002-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
152. Ponnaluri VK, Maciejewski JP, Mukherji M. A mechanistic overview of TET-mediated 5-methylcytosine oxidation. Biochem Biophys Res Commun (2013) 436(2):115–20. doi:10.1016/j.bbrc.2013.05.077
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
153. Lee JT. Epigenetic regulation by long noncoding RNAs. Science (2012) 338(6113):1435–9. doi:10.1126/science.1231776
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
154. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell (2007) 129(7):1311–23. doi:10.1016/j.cell.2007.05.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
155. Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet (2001) 2(4):292–301. doi:10.1038/35066075
156. Sleeman JE, Trinkle-Mulcahy L. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol (2014) 28(0):76–83. doi:10.1016/j.ceb.2014.03.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
157. Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature (2008) 452(7184):243–7. doi:10.1038/nature06727
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
158. Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell (2008) 132(6):929–34. doi:10.1016/j.cell.2008.03.004
159. Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science (2002) 296(5565):158–62. doi:10.1126/science.1068768
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
160. Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev (2010) 24(22):2543–55. doi:10.1101/gad.1967810
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
161. Xiang J-F, Yin Q-F, Chen T, Zhang Y, Zhang X-O, Wu Z, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res (2014) 24(5):513–31. doi:10.1038/cr.2014.35
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
162. Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol (2014) 21(2):198–206. doi:10.1038/nsmb.2764
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
163. Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature (2013) 494(7438):497–501. doi:10.1038/nature11884
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
164. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell (2010) 143(1):46–58. doi:10.1016/j.cell.2010.09.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
165. Krivega I, Dean A. Enhancer and promoter interactions-long distance calls. Curr Opin Genet Dev (2012) 22(2):79–85. doi:10.1016/j.gde.2011.11.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
166. Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature (2010) 467(7314):430–5. doi:10.1038/nature09380
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
167. O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science (2010) 327(5969):1098–102. doi:10.1126/science.1178334
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
168. Geginat J, Paroni M, Maglie S, Alfen JS, Kastirr I, Gruarin P, et al. Plasticity of human CD4 T cell subsets. Front Immunol (2014) 5:630. doi:10.3389/fimmu.2014.00630
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
169. Hedrich CM, Bream JH. Cell type-specific regulation of IL-10 expression in inflammation and disease. Immunol Res (2010) 47(1–3):185–206. doi:10.1007/s12026-009-8150-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
170. Chen Z, Tato CM, Muul L, Laurence A, O’Shea JJ. Distinct regulation of interleukin-17 in human T helper lymphocytes. Arthritis Rheum (2007) 56(9):2936–46. doi:10.1002/art.22866
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
171. Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol (2007) 8(9):950–7. doi:10.1038/ni1497
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
172. Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol (2008) 9(11):1297–306. doi:10.1038/ni.1663
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
173. Klunker S, Chong MM, Mantel PY, Palomares O, Bassin C, Ziegler M, et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med (2009) 206(12):2701–15. doi:10.1084/jem.20090596
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
174. Li L, Patsoukis N, Petkova V, Boussiotis VA. Runx1 and Runx3 are involved in the generation and function of highly suppressive IL-17-producing T regulatory cells. PLoS One (2012) 7(9):e45115. doi:10.1371/journal.pone.0045115
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
175. Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med (2011) 17(8):983–8. doi:10.1038/nm.2426
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
176. Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science (2009) 323(5920):1488–92. doi:10.1126/science.1169152
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
177. Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJP, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol (2011) 12(8):796–803. doi:10.1038/ni.2057
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
178. Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol (2014) 15(6):484–91. doi:10.1038/ni.2887
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
179. Barabasi A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet (2004) 5(2):101–13. doi:10.1038/nrg1272
180. Zhang B, Arun G, Mao Yuntao S, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep (2013) 2(1):111–23. doi:10.1016/j.celrep.2012.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
181. Necsulea A, Kaessmann H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat Rev Genet (2014) 15(11):734–48. doi:10.1038/nrg3802
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
182. Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet (2006) 2(10):e168. doi:10.1371/journal.pgen.0020168
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
183. Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet (2011) 7(5):e1002071. doi:10.1371/journal.pgen.1002071
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
184. Gerstein MB, Rozowsky J, Yan KK, Wang D, Cheng C, Brown JB, et al. Comparative analysis of the transcriptome across distant species. Nature (2014) 512(7515):445–8. doi:10.1038/nature13424
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
185. On T, Xiong X, Pu S, Turinsky A, Gong Y, Emili A, et al. The evolutionary landscape of the chromatin modification machinery reveals lineage specific gains, expansions, and losses. Proteins (2010) 78(9):2075–89. doi:10.1002/prot.22723
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
186. Hrdlickova B, Kumar V, Kanduri K, Zhernakova D, Tripathi S, Karjalainen J, et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med (2014) 6(10):88. doi:10.1186/s13073-014-0088-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
187. Xia F, Dong F, Yang Y, Huang A, Chen S, Sun D, et al. Dynamic transcription of long non-coding RNA genes during CD4+ T cell development and activation. PLoS One (2014) 9(7):e101588. doi:10.1371/journal.pone.0101588
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
188. Aoki K, Harashima A, Sano M, Yokoi T, Nakamura S, Kibata M, et al. A thymus-specific noncoding RNA, Thy-ncR1, is a cytoplasmic riboregulator of MFAP4 mRNA in immature T-cell lines. BMC Mol Biol (2010) 11(1):99. doi:10.1186/1471-2199-11-99
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
189. Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol (2013) 14(11):1190–8. doi:10.1038/ni.2712
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
190. Kimura T, Jiang S, Nishizawa M, Yoshigai E, Hashimoto I, Nishikawa M, et al. Stabilization of human interferon-alpha1 mRNA by its antisense RNA. Cell Mol Life Sci (2013) 70(8):1451–67. doi:10.1007/s00018-012-1216-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
191. Yan M-D, Hong C-C, Lai G-M, Cheng A-L, Lin Y-W, Chuang S-E. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet (2005) 14(11):1465–74. doi:10.1093/hmg/ddi156
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
192. Sehgal L, Mathur R, Braun FK, Wise JF, Berkova Z, Neelapu S, et al. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia (2014) 28(12):2376–87. doi:10.1038/leu.2014.126
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
193. Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol (2002) 217(1–2):78–86. doi:10.1016/S0008-8749(02)00506-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
194. Liu AY, Torchia BS, Migeon BR, Siliciano RF. The human NTT gene: identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics (1997) 39(2):171–84. doi:10.1006/geno.1996.4463
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
195. Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nat Immunol (2006) 7(10):1109–15. doi:10.1038/ni1379
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
196. Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol (2012) 189(5):2084–8. doi:10.4049/jimmunol.1200774
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
197. Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell (2013) 152(4):743–54. doi:10.1016/j.cell.2013.01.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
198. Zhang H, Nestor CE, Zhao S, Lentini A, Bohle B, Benson M, et al. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1. J Allergy Clin Immunol (2013) 132(4):1005–8. doi:10.1016/j.jaci.2013.05.033
199. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A (2013) 110(9):3507–12. doi:10.1073/pnas.1222878110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
200. Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A (2015) 112(4):1167–72. doi:10.1073/pnas.1401965111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
201. Tuomela S, Lahesmaa R. Early T helper cell programming of gene expression in human. Semin Immunol (2013) 25(4):282–90. doi:10.1016/j.smim.2013.10.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
202. Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature (2014) 505(7485):635–40. doi:10.1038/nature12943
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
203. Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, et al. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res (2001) 11(5):833–49. doi:10.1101/gr.174901
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
204. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev (2011) 25(18):1915–27. doi:10.1101/gad.17446611
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
205. Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol (2009) 7(5):e1000112. doi:10.1371/journal.pbio.1000112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
206. Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet (2006) 22(1):1–5. doi:10.1016/j.tig.2005.10.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
207. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science (2005) 309(5740):1559–63. doi:10.1126/science.1112014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
208. Ulitsky I, Shkumatava A, Jan Calvin H, Sive H, Bartel David P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell (2011) 147(7):1537–50. doi:10.1016/j.cell.2011.11.055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
209. He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol Biol (2011) 11:102. doi:10.1186/1471-2148-11-102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
210. Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood (2009) 113(11):2526–34. doi:10.1182/blood-2008-06-162164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
211. Wright PW, Huehn A, Cichocki F, Li H, Sharma N, Dang H, et al. Identification of a KIR antisense lncRNA expressed by progenitor cells. Genes Immun (2013) 14(7):427–33. doi:10.1038/gene.2013.36
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
212. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science (2014) 344(6181):310–3. doi:10.1126/science.1251456
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
213. Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell (2014) 159(5):1110–25. doi:10.1016/j.cell.2014.10.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
214. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science (2013) 341(6147):789–92. doi:10.1126/science.1240925
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
215. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge R-M, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife (2013) 2:e00762. doi:10.7554/eLife.00762
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
216. Li Z, Chao T-C, Chang K-Y, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A (2014) 111(3):1002–7. doi:10.1073/pnas.1313768111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
217. Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol (1986) 136(7):2348–57.
218. Chang S, Aune TM. Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci U S A (2005) 102(47):17095–100. doi:10.1073/pnas.0502129102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
219. Zhou W, Chang S, Aune TM. Long-range histone acetylation of the Ifng gene is an essential feature of T cell differentiation. Proc Natl Acad Sci U S A (2004) 101(8):2440–5. doi:10.1073/pnas.0306002101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
220. Ho IC, Lo D, Glimcher LH. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4-dependent and -independent mechanisms. J Exp Med (1998) 188(10):1859–66. doi:10.1084/jem.188.10.1859
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
221. Liu X, Nurieva RI, Dong C. Transcriptional regulation of follicular T-helper (Tfh) cells. Immunol Rev (2013) 252(1):139–45. doi:10.1111/imr.12040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
222. Sato K, Miyoshi F, Yokota K, Araki Y, Asanuma Y, Akiyama Y, et al. Marked induction of c-Maf protein during Th17 cell differentiation and its implication in memory Th cell development. J Biol Chem (2011) 286(17):14963–71. doi:10.1074/jbc.M111.218867
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
223. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature (2010) 464(7291):1071–6. doi:10.1038/nature08975
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: long non-coding RNAs, epigenetic regulation, lymphocyte differentiation
Citation: Panzeri I, Rossetti G, Abrignani S and Pagani M (2015) Long intergenic non-coding RNAs: novel drivers of human lymphocyte differentiation. Front. Immunol. 6:175. doi: 10.3389/fimmu.2015.00175
Received: 22 January 2015; Accepted: 28 March 2015;
Published online: 15 April 2015.
Edited by:
Dragana Jankovic, National Institutes of Health (NIH), USAReviewed by:
Christopher E. Rudd, University of Cambridge, UKJohn J. Miles, Queensland Institute of Medical Research, Australia
Susan Carpenter, University of California San Francisco, USA
Copyright: © 2015 Panzeri, Rossetti, Abrignani and Pagani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Massimiliano Pagani, Integrative Biology Unit, Istituto Nazionale Genetica Molecolare “Romeo ed Enrica Invernizzi”, IRCCS Ospedale Maggiore Policlinico, via Francesco Sforza 35, Milano 20122, Italy e-mail:cGFnYW5pQGluZ20ub3Jn
 Ilaria Panzeri
Ilaria Panzeri Grazisa Rossetti
Grazisa Rossetti Sergio Abrignani
Sergio Abrignani Massimiliano Pagani
Massimiliano Pagani