- Radioisotope Centre, The University of Tokyo, Tokyo, Japan
A large number of human RNA transcripts, which do not encode proteins are defined as non-coding RNAs (ncRNAs). These ncRNAs are divided into two classes of different lengths; short and long ncRNAs. MicroRNAs are a major class of short ncRNAs, ~22 nucleotides in length that regulate gene expression at the post-transcriptional level. Long non-coding RNAs (lncRNAs) are more than 200 nucleotides in length and play roles in various biological pathways. In this review, we summarize the functions of lncRNAs which regulate immune responses.
Introduction
Whole transcriptome analyses of mammalian genomes, such as studies documented in the ENCODE project, have characterized RNA transcripts that have low-protein coding potential, known as non-coding RNAs (ncRNAs) (1). In recent years, it has become increasingly apparent that ncRNAs are involved in diverse biological processes (2–5). Based on their length, ncRNAs are classified into short ncRNAs and long non-coding RNAs (lncRNAs) (6). MicroRNAs (miRNAs) are a major class of short ncRNAs, ~22 nucleotides (nt) in length that regulate post-transcriptional gene silencing by controlling translation and RNA stability (7, 8). miRNAs are involved in the regulation of diverse biological processes, such as proliferation, differentiation, apoptosis, development, and immune responses (9–12). LncRNAs are longer than 200 nt and play roles in diverse biological processes, such as proliferation, differentiation, and development through various modes of action. For instance, several lncRNAs, such as XIST, ANRIL, and HOTAIR, recruit polycomb-repressive complex 2 to target loci for epigenetic regulation (13–18). Recently, it was reported that several lncRNAs regulate post-transcriptional gene regulation through binding to specific RNA-binding proteins (16).
The innate and adaptive immune responses provide immunity to pathogens. The innate immune response is the first line of defense. Cells infected with a pathogen trigger the innate immune response by synthesizing inflammatory mediators or cytokines through the transcriptional and post-transcriptional gene regulations. The innate immune response also activates adaptive immune responses resulting in facilitated elimination of the pathogen. Therefore, the innate immune response is crucial to the host for protection against pathogens, such as bacterium or viruses. A primary challenge for the innate immune system is the discrimination of pathogens by specific receptors. Toll-like receptors (TLRs) are a major receptor type for the recognition of pathogens (19). For example, TLR2 recognizes lipo-protein, TLR3 recognizes double-strand RNAs produced by viruses, TLR4 recognizes lipopolysaccharide, and TLR5 recognizes flagella of bacteria. Following ligand recognition by these receptors, specific signaling cascades are activated that results in the synthesis of inflammatory mediators or cytokines. This involves transcriptional or post-transcriptional gene regulation via transcription factors, such as API and NF-κB, and miRNAs (19, 20). For instance, miR-203 regulates inflammatory cytokines tumor necrosis factor (TNF)-α and interleukin (IL)-24 (21), while miR-26a, -34a, -145, and let-7b regulate IFN-β (22). However, the regulation of inflammatory mediators or cytokines by lncRNAs is still poorly understood. Several hypotheses have recently emerged concerning lncRNA involvement in infectious diseases (23) and some studies found that several lncRNAs might be involved in the regulation of immune responses. In this review, we focus on five lncRNAs, which are clearly related with immune responses. Three of them are involved in bacterial infection, because of stimulation of TLR2 or 4. Two of them are involved in viral infection.
lncRNAs Involved in Bacterial Infection
lincRNA-Cox2
Whole transcriptome analysis (RNA-seq) of mouse bone marrow-derived macrophages (BMDMs) showed that stimulation by the synthetic bacterial lipopeptide Pam3CSK4, a TLR2 ligand, increased the expression of 62 lncRNAs (24). In these lncRNAs, one of the most highly induced lncRNAs was lincRNA-Cox2 (ENSMUSG00000091113, Gm17311). Lipopolysaccharide, a TLR4 ligand, and R848, a synthetic antiviral compound that activates Tlr7/8, also increased lincRNA-Cox2 levels via the Myd88-NF-κB pathway. Conversely, polyinosinic-polycytidylic acid (poly I:C), a synthetic double-stranded RNA that is recognized by TLR3, did not affect lincRNA-Cox2 expression. LincRNA-Cox2 has three variants and is about 2 kb in length. LincRNA-Cox2 variant 1 is the most abundant transcript, but it is unclear, which one acts in immune response and these differences. LincRNA-Cox2 was localized in both the nuclear and the cytosolic compartments of macrophages.
In Pam3CSK4 treatment of BMDMs, silencing of lincRNA-Cox2 induced up-regulation of Ccl5, but down-regulation of Il-6. These results suggest that lincRNA-Cox2 mediates both activation and repression of immune responses. LincRNA-Cox2 bound hnRNP-A/B and hnRNP-A2/B1 in both the nuclear and the cytosolic compartments. These complexes regulated repression of immune genes, including Ccl5, but Il-6. It is unclear how lincRNA-Cox2 regulates Il-6 expression so far.
linc1992/THRIL
Custom lincRNA microarray analysis of THP1-derived macrophages showed that Pam3CSK4 stimulation increased the levels of 127 lincRNAs and decreased the levels of 32 lincRNAs (25). In these lincRNAs, Pam3CSK4 stimulation decreased the expression of linc1992 (NR_110375) and knockdown of linc1992 notably restrained TNFα secretion. Linc 1992 is about 2 kb in length and widely expresses in human tissues. The authors also found other lncRNAs that regulates TNFα or IL6 secretion. Linc0206 (ENST00000450206, RP11-432J24), Linc7190 (ENST00000437190, RP11-296O14), and Linc7705(ENST00000417705, RP11-54O7) regulate TNFα and IL6 secretion under Pam3CSK4 stimulation. Moreover, Linc3995 (ENST00000523995, RP11-37B2) regulate IL6 secretion under Pam3CSK4 stimulation. However, it is unclear how these lncRNAs regulate cytokine expression.
Knockdown of linc 1992 decreased significantly production of TNFα mRNA. Thus, linc1992 regulates TNFα expression through a negative feedback system.
By the analysis of pull-down assay and RIP assay, linc1992 bound hnRNP-L and linc 1992 and hnRNPL formed an RNP complex in vivo. Moreover, ChIP assay revealed that hnRNPL bound to the TNFα promoter region and knockdown of linc1992 reduced binding of hnRNPL to the TNFα promoter region. Thus, this complex may regulate the transcription of downstream genes, including TNFα. Accordingly, Linc1992/THRIL (TNFα ανδ HnRNP-L Related Immunoregulatory LincRNA) is named after this regulation. Furthermore, Linc1992/THRIL regulated expression of IL-8, CXCL10, CCL1, and CSF1, but it is unclear whether hnRNP-L is involved in these expressions.
Moreover, linc1992/THRIL is associated with Kawasaki disease. Kawasaki disease is the most common cause of multisystem vasculitis in childhood. TNF-α is a pleiotropic inflammatory cytokine elevated during the acute phase of Kawasaki disease. In 17 patients with Kawasaki disease, linc1992/THRIL expression was lower during acute phase of disease when TNFα levels are elevated, so linc1992/THRIL could be a new biomarker for immune activation.
lnc-IL7R
LncRNA microarray analysis showed that stimulation of THP-1 cells with lipopolysaccharide, a TLR4 ligand, induced the expression of 443 lncRNAs by more than twofold and decreased the expression 718 lncRNAs more than twofold. Among these lncRNAs, lnc-IL7R (ENST00000303115), one of the most up-regulated genes, is 1427 nt in length and overlaps the 3′-untranslated region (3′-UTR) of the human interleukin-7 receptor α-subunit (IL7R) (26). The majority of lnc-IL7R existed in the nucleus. The increase in lnc-IL7R expression after lipopolysaccharide stimulation indicates that lnc-IL7R is involved in the early immune response. Pam3CSK4 also increased lnc-IL7R expression, but poly I:C did not. In human peripheral blood mononuclear cells, stimulation by lipopolysaccharide or Pam3CSK4 increased lnc-IL7R levels. Lnc-IL7R negatively regulated E-selectin, VCAM-1, IL-8, and IL-6 expression following lipopolysaccharide stimulation. The mechanism by which lncRNA-IL7R regulated E-selectin and VCAM-1 was dependent on the trimethylation of histone H3 at lysine (H3K27me3). Lnc-1L7R knockdown also diminished IL-6 and IL-8 mRNA levels in an as yet unknown way.
lncRNAs Involved in Bacterial Infection
NeST/Tmevpg1
NeST, also known as Tmevpg1 or IFNG-AS1 (GS1-410F4), is an lncRNA located adjacent to the interferon (IFN)-γ-encoding gene in the Tmevp3 locus that is a candidate gene in a susceptibility locus for Theiler’s virus (27, 28) in mouse. NeST RNA is encoded on the DNA strand opposite to that coding for IFN-γ, and the most abundant splice variant has six exons, 914 bp on length, in murine chromosome 10 and expressed in CD4+ T, CD8+ T, and natural killer cells.
By analysis of RIP and ChIP assay, NeST RNA bound WDR5, a subunit of the MLL/SET1 H3K4 methylase complex, and modified the chromatin at the Ifng locus. NeST RNA induced secretion of IFN-γ in CD8+ T cells. Transgenic mice, in which NeST is over-expressed, are protected against Theiler’s virus. NeST expression leaded to persistence of Theiler’s virus, but clearance of Salmonella infection.
NEAT1
NEAT1 has two isoforms, 3.7 kb NEAT1v1 and 23 kb NEAT1v2 (29). NEAT1v2 is an essential factor in the structure of paraspeckles (30–33). Paraspeckles contain about 40 protein factors (34), including NONO/p54nrb and SFPQ/PSF, which function as a transcriptional inhibitor (35, 36).
NEAT1 was originally identified as an inducible lncRNA in mice brains infected with Japanese encephalitis or rabies viruses (37). HIV-I infection also induced NEAT1 in mice and NEAT1 modulates HIV-I replication by affecting the nucleus-to-cytoplasm export of Rev-dependent, instability element-containing HIV-I mRNAs (38). Moreover, it was found that poly I:C, a TLR3 ligand, herpes simplex virus 1 (HSV-1), and measles virus (MV) infection all induce NEAT1v2 (39). Furthermore, in normal cells, SFPQ, a paraspeckle protein, binds the IL-8 promoter to suppress its expression. Poly I:C stimulation, HSV-1, and MV infections caused the relocation of SFPQ from the IL-8 gene promoter to NEAT1, resulting in the formation of excess paraspeckles, which in turn leaded to the transcriptional activation of IL-8.
Conclusion
Several lines of evidence have recently emerged concerning lncRNA involvement in the regulation of inflammatory mediator or cytokine expression. Unlike miRNAs, lncRNAs regulate expression of inflammatory mediators or cytokines by working with RBPs in a number of ways. Some complexes of lncRNAs and RBPs bind to promoters of inflammatory mediator or cytokine genes to regulate transcription or to modify the chromatin (Figures 1A,B). NEAT1 regulates the location of the transcription inhibitor SFPQ from promoter to paraspeckle (Figure 1C). These observations suggest that lncRNAs are factors of the innate immune response. However, there is only evidence that lncRNA induce under TLR ligand stimulation so far and little evidence that lncRNA function under the real bacterial infection, which induces more complicated event in cells. Further studies of this regulation by lncRNAs are likely to reveal novel drug targets for therapy of infectious and inflammatory diseases.
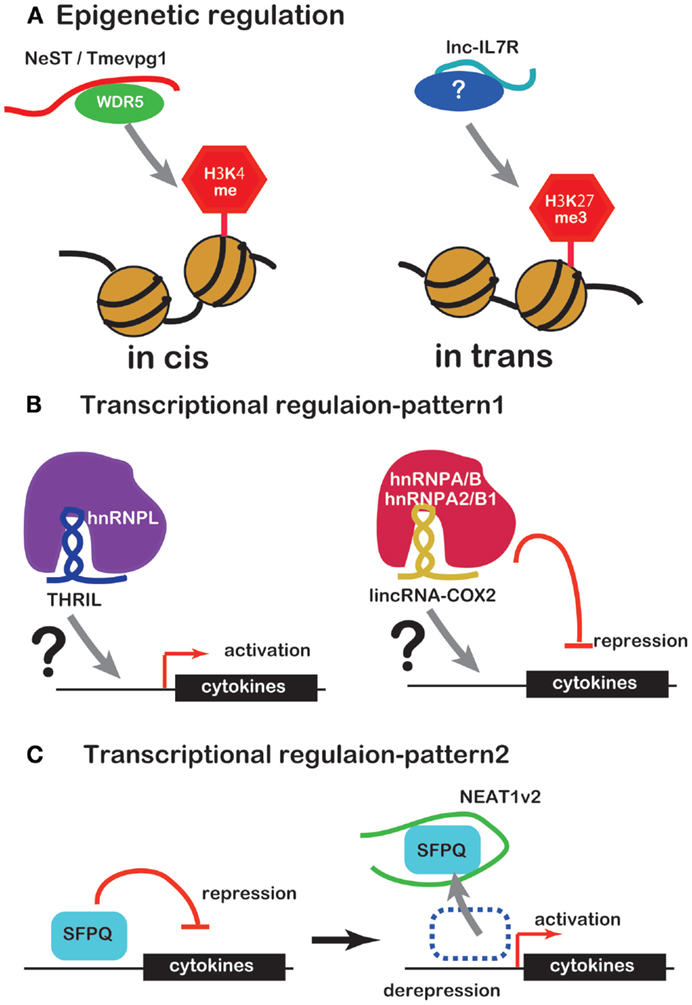
Figure 1. ncRNAs that regulate chromatin structure and transcription factors. (A) NeST/Tmevpg1 activates transcription of genes by recruiting the histone modifier WDR5, which methylates H3K4. Lnc-IL7R activates transcription of genes by H3K27 trimethylation. (B) The complex of THRIL and hnRNPL activates transcription of genes. The complex of lincRNA-Cox2 and hnRNPA/B or hnRNPA2/B1 represses transcription of genes. (C) NEAT1 activates transcription of genes by sequestration of transcriptional factor SFPQ.
Why should ncRNAs be involved in regulating the expression of inflammatory mediators or cytokines? In virus-infected cells, translation is highly inhibited. For this reason, a translation-independent acute response system is required for an effective early immune response. Furthermore, involvement of ncRNA in such a response is biologically meaningful, because non-coding RNAs do not demand translational activity to express their functions. We therefore speculate that many other lncRNAs may play a role in situations where translation is highly repressed, such as viral infection, heat shock, and hypoxia.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature (2012) 489:101–8. doi: 10.1038/nature11233
2. Chen LL, Carmichael GG. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol (2010) 22:357–64. doi:10.1016/j.ceb.2010.03.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature (2010) 464:1071–6. doi:10.1038/nature08975
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell (2009) 136:629–41. doi:10.1016/j.cell.2009.02.006
5. Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol (2012). doi:10.1016/j.jmb.2012.11.024
6. Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci (2013) 14:16010–39. doi:10.3390/ijms140816010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol (2013) 425:3582–600. doi:10.1016/j.jmb.2013.03.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell (2005) 120:623–34. doi:10.1016/j.cell.2004.12.038
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Iyengar BR, Choudhary A, Sarangdhar MA, Venkatesh KV, Gadgil CJ, Pillai B. Non-coding RNA interact to regulate neuronal development and function. Front Cell Neurosci (2014) 8:47. doi:10.3389/fncel.2014.00047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Jeker LT, Bluestone JA. MicroRNA regulation of T-cell differentiation and function. Immunol Rev (2013) 253:65–81. doi:10.1111/imr.12061
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Nagpal N, Kulshreshtha R. miR-191: an emerging player in disease biology. Front Genet (2014) 5:99. doi:10.3389/fgene.2014.00099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Olivieri F, Procopio AD, Montgomery RR. Effect of aging on microRNAs and regulation of pathogen recognition receptors. Curr Opin Immunol (2014) 29C:29–37. doi:10.1016/j.coi.2014.03.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Brockdorff N. Noncoding RNA and polycomb recruitment. RNA (2013) 19:429–42. doi:10.1261/rna.037598.112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett (2013) 339:159–66. doi:10.1016/j.canlet.2013.06.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res (2011) 71:5365–9. doi:10.1158/0008-5472.CAN-10-4379
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell (2011) 147:773–88. doi:10.1016/j.cell.2011.08.054
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog (2013). doi:10.1002/mc.22120
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. He Z, Bammann H, Han D, Xie G, Khaitovich P. Conserved expression of lincRNA during human and macaque prefrontal cortex development and maturation. RNA (2014) 20:1103–11. doi:10.1261/rna.043075.113
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol (2010) 11:373–84. doi:10.1038/ni.1863
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs: novel regulators in skin inflammation. Clin Exp Dermatol (2008) 33:312–5. doi:10.1111/j.1365-2230.2008.02804.x
21. Primo MN, Bak RO, Schibler B, Mikkelsen JG. Regulation of pro-inflammatory cytokines TNFα and IL24 by microRNA-203 in primary keratinocytes. Cytokine (2012) 60:741–8. doi:10.1016/j.cyto.2012.07.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol (2010) 184:2369–76. doi:10.4049/jimmunol.0902712
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Scaria V, Pasha A. Long non-coding RNAs in infection biology. Front Genet (2012) 3:308. doi:10.3389/fgene.2012.00308
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science (2013) 341:789–92. doi:10.1126/science.1240925
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A (2014) 111:1002–7. doi:10.1073/pnas.1313768111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Cui H, Xie N, Tan Z, Banerjee S, Thannickal VJ, Abraham E, et al. The human long noncoding RNA, lnc-IL7R, regulates inflammatory response. Eur J Immunol (2014) 44:2085–95. doi:10.1002/eji.201344126
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol (2003) 77:5632–8. doi:10.1128/JVI.77.10.5632-5638.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell (2013) 152:743–54. doi:10.1016/j.cell.2013.01.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics (2007) 8:39. doi:10.1186/1471-2164-8-39
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell (2009) 35:467–78. doi:10.1016/j.molcel.2009.06.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell (2009) 33:717–26. doi:10.1016/j.molcel.2009.01.026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. Men epsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A (2009) 106:2525–30. doi:10.1073/pnas.0807899106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. Men epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res (2009) 19:347–59. doi:10.1101/gr.087775.108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3’-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J (2012) 31:4020–34. doi:10.1038/emboj.2012.251
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Iacobazzi V, Infantino V, Costanzo P, Izzo P, Palmieri F. Functional analysis of the promoter of the mitochondrial phosphate carrier human gene: identification of activator and repressor elements and their transcription factors. Biochem J (2005) 391:613–21. doi:10.1042/BJ20050776
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc Natl Acad Sci U S A (2005) 102:12189–93. doi:10.1073/pnas.0505179102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Saha S, Murthy S, Rangarajan PN. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol (2006) 87:1991–5. doi:10.1099/vir.0.82107-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio (2013) 4. doi:10.1128/mBio.00596-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell (2014) 53:393–406. doi:10.1016/j.molcel.2014.01.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: long non-coding RNA (lncRNA), innate immune response, NEAT1, translational repression
Citation: Imamura K and Akimitsu N (2014) Long non-coding RNAs involved in immune responses. Front. Immunol. 5:573. doi: 10.3389/fimmu.2014.00573
Received: 14 July 2014; Accepted: 26 October 2014;
Published online: 13 November 2014.
Edited by:
Susan Carpenter, University of Massachusetts Medical School, USAReviewed by:
Paul Quax, Leiden University Medical Center, NetherlandsPeter L. Molloy, Commonwealth Scientific and Industrial Research Organisation, Australia
Maninjay K. Atianand, University of Massachusetts Medical School, USA
Copyright: © 2014 Imamura and Akimitsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nobuyoshi Akimitsu, Radioisotope Centre, The University of Tokyo, 2-11-16 Yayoi, Bunkyo, Tokyo 113-0032, Japan e-mail:YWtpbWl0c3VAcmljLnUtdG9reW8uYWMuanA=
 Katsutoshi Imamura
Katsutoshi Imamura Nobuyoshi Akimitsu
Nobuyoshi Akimitsu