
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 10 November 2014
Sec. T Cell Biology
Volume 5 - 2014 | https://doi.org/10.3389/fimmu.2014.00550
This article is part of the Research TopicRecent Advances in HBV and HCV ImmunologyView all 10 articles
Hepatitis C virus (HCV) remains a global problem, despite advances in treatment. The low cost and high benefit of vaccines have made them the backbone of modern public health strategies, and the fight against HCV will not be won without an effective vaccine. Achievement of this goal will benefit from a robust understanding of virus–host interactions and protective immunity in HCV infection. In this review, we summarize recent findings on HCV-specific antibody responses associated with chronic and spontaneously resolving human infection. In addition, we discuss specific epitopes within HCV’s envelope glycoproteins that are targeted by neutralizing antibodies. Understanding what prompts or prevents a successful immune response leading to viral clearance or persistence is essential to designing a successful vaccine.
Between 130 and 185 million people worldwide are infected with hepatitis C virus (HCV) and are at risk of cirrhosis, hepatocellular carcinoma, and end-stage liver disease (1–3). HCV, a member of the Flaviviridae family, is parenterally transmitted. HCV establishes a persistent infection in 60–80% of individuals infected. The treatment for HCV has long been pegylated interferon alpha co-administrated with ribavirin, but the response rates were unsatisfactory with only 50–60% of patients achieving a sustained virologic response (4, 5). The welcomed discovery of new directly acting antiviral drugs (DAAs) is expected to lead to a dramatic increase in cure rates (6–8). However, it is unlikely that the global HCV problem will be eliminated any time soon. There are numerous challenges that must be overcome first, including the prohibitive cost of treatment and the need for new treatment strategies for patients with advanced liver disease or co-morbidities (9). Another important obstacle is identifying those in need of treatment, since symptoms may be absent or non-specific until after significant liver damage has set in (10).
The development of a protective vaccine is essential in combating the global HCV epidemic. Understanding the immune response in those who spontaneously resolve HCV infections versus those who develop chronic infection is key to the development of prophylactic or therapeutic vaccine (11). So far, developing a HCV vaccine has proven challenging, not least because HCV is genetically highly diverse; there are seven known major genotypes that differ from each other by 30–35%, and over 60 subtypes (12). Indeed, the virus exists as a quasispecies – a swarm of related but distinct sequences – within an infected patient. This diversity is a consequence of HCV’s high replication rate, and an RNA polymerase that lacks any proofreading mechanism. High viral diversity within and between infected individuals poses challenges to vaccine developers: how can we devise a vaccine that will stimulate broadly cross-reactive immune responses to such a changeable foe? The key may well be to target an array of viral epitopes that are functionally constrained, and to enlist both humoral and cellular arms of the adaptive immune response. In particular, it will be important for the vaccine to elicit neutralizing antibodies (nAbs) to block viral access to target cells, and T-cell responses targeting infected cells (13).
Adaptive immune responses are typically delayed during acute HCV infection. HCV RNA can be detected 1–3 weeks following infection, but neither HCV-specific T-cells nor HCV-specific antibodies (Ab) are observed until 1–2 months after infection (14–18). Both CD4+ and CD8+ T-cell responses play essential roles in the outcome of infection. CD8+ T-cells limit HCV replication through cytolytic and non-cytolytic immune mechanisms that are highly dependent on CD4+ T-cell function [reviewed in Ref. (19–23)]. Vigorous and broadly directed anti-HCV T-cell responses are observed in patients who resolve infection (24–27). In patients who progress to chronicity, initial vigorous T-cell responses wane and weaken. Loss of CD4+ T-cell help, a switch to a Treg cell profile, viral epitope escape, and chronic antigen stimulation may all contribute to T-cell exhaustion (23).
It was widely thought that the humoral immune response to HCV played only a peripheral role in HCV infection (24, 28, 29). However, recent studies suggest that B-cells and nAbs may play active roles in the spontaneous resolution of HCV (30–33). Typically, an nAb response would be a component of sterilizing antiviral immunity and has long been a quintessential part of vaccine design (13, 34). An HCV vaccine will need to stimulate strong humoral as well as cellular immune responses. The role of humoral immune system in the both the control of HCV infection and in the pathogenesis of liver disease is still unclear. In this review, we hope to outline our current understanding of the humoral immune system’s roles in acute infection, the progression to chronicity, and the spontaneous resolution of HCV infection, and to highlight some of the pressing questions that need to be addressed.
Antibodies produced during acute HCV infection target epitopes within both structural and non-structural (NS) viral proteins. However, all known nAbs target epitopes within the HCV envelope glycoproteins E1 and E2, or the E1E2 heterodimer. The structural proteins core, E1, and E2 are released from the viral polyprotein by cellular signal peptidases. The viral particle contains the nucleocapsid, formed by the close interaction of the HCV RNA genome and core protein, surrounded by a lipid bilayer envelope into which the glycoproteins E1 and E2 are anchored. E1 and E2 form a heterodimer that mediates viral entry. Determining the structure of the E1E2 heterodimer has proven problematic. E2 is required for the correct folding of E1, so that E1’s structure is still uncertain (35, 36). It is thought that the glycans on both the heavily glycosylated E1 and E2 are involved in folding of the E1E2 heterodimer (37). Interestingly, nAbs (AR4 and AR5) have been found that recognize conformational epitopes on the E1E2 heterodimer with broad neutralizing crossreactivity between diverse HCV genotypes (32).
Most nAbs target E2. E2 plays a key role in HCV entry, directly interacting with two of the cellular proteins needed for viral entry, CD81 and scavenger receptor class B type I (SR-BI) (38, 39). CD81 and SB-RI alone are not sufficient for viral entry; tight junction proteins claudin-1 and occludin are also required (40, 41). Other factors, such as the cholesterol absorption receptor Niemann-Pick C1-like 1, epidermal growth factor receptor, ephrin receptor type A2, and most recently, transferrin receptor 1 enhance viral entry (42–44).
Two recent reports have shed light on the structure of E2 (45, 46). Most surprisingly, E2 did not adopt the expected, highly extended conformation of class II fusion envelope protein like other members of Flaviviridae, such as Tick-borne encephalitis virus or West Nile virus; instead, HCV E2 was found to be compact and globular (Figure 1), with a central beta sandwich surrounded front and back by short alpha helices, loops, beta sheets, and regions lacking organized structure (45, 46).
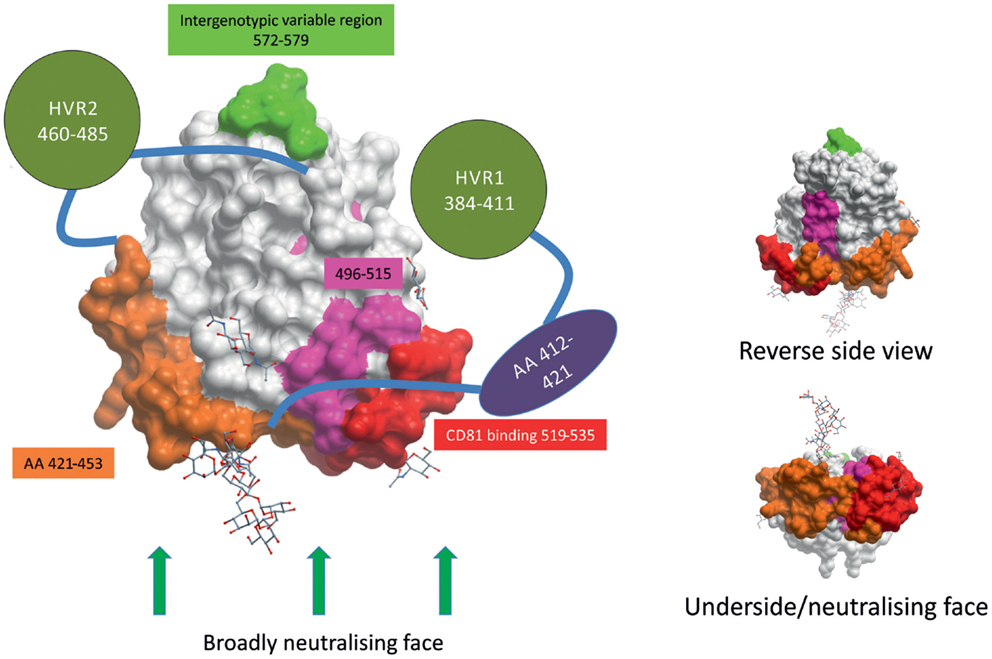
Figure 1. HCV envelope glycoprotein 2 surface representation. E2 is a globular protein with three regions of hypervariablity – HVR1, HVR2, and intergenotypic variable region – shown in green. Domains whose structures are currently unknown are depicted as shapes apart from the structure. HVR1 is predicted to mask a hydrophobic region that is sensitive to nAbs. E2’s broadly neutralizing face, where many broadly neutralizing Abs bind, comprises CD81-binding loop (in red), residues 421–453 (in orange), residues 502–520 (in pink), and residues 412–421 (in purple). The possible positions of some glycans are shown as stick and ball figures. E2 structure obtained from PDB (4MWF) (45).
The ectodomain of E2 contains three regions of variability that are targeted by nAbs: hyper-variable region (HVR) 1, HVR2, and intergenotypic variable region. HVR1 contains dominant neutralizing epitopes, and its variation leads to immunological escape (47–50). HVR1 is a 26–28 amino acid segment located near the amino terminus of E2. HVR1 interacts with SR-BI and is, therefore, likely involved in virus entry, making it an interesting target for nAbs (51, 52). The most-effective nAbs that target HVR1 recognize epitopes found in the C-terminus, where HVR1 interacts with SR-BI (52–54). However, HVR1 mutates rapidly and antibodies that recognize HVR1 demonstrate very poor cross-neutralization across different isolates of the same HCV genotype (50, 55, 56). As of yet, no HVR1 nAbs have been found that display broadly cross-reactive neutralizing activity.
CD81 was the first host receptor identified as being a crucial entry factor for HCV and, therefore, the CD81-binding region of E2 is a likely target for nAbs (38). Indeed, numerous broadly nAbs have been found to target the CD81-binding loop (residues 519–535, numbered according to the HCV H77 consensus sequence accession number AF009606) of E2 (30, 32, 45, 57–60). Kong and colleagues found evidence to suggest CD81 interacts with both the CD81-binding loop (519–535) and residues 421–453 of E2, making this domain in E2 a most intriguing target for vaccine design (Figure 1) (45). nAbs have also been found that target the less variable region directly downstream of HVR1 (residues 412–421) (61–63). However, less than 5% of spontaneous resolvers have nAbs targeting this region, suggesting that this region is not as immunogenic in vivo (64). A highly conserved neutralizing epitope encompassing residues 496–515 is located between the two CD81-binding regions of E2 (Figure 1). Although not directly involved in CD81 binding, binding of nAbs to the 496–515 epitope may cause a conformational change in the CD81-binding region (65). Contrary to what was previously thought, the overlapping residues 502–520 do not contain a fusion peptide (66, 67).
Most of the identified nAbs target regions within E2 that interact with the viral entry factors CD81 and SR-BI. Occludin is also thought to interact directly with E2 (68); perhaps mapping of this interaction will allow the development of novel nAbs. In contrast, claudin-1 has not been shown to interact directly with E2. Would targeting the viral epitope that recognizes claudin-1 prevent entry? Perhaps we know that blocking claudin-1 prevents E2/CD81/claudin-1 interaction, thereby preventing HCV entry (69). Targeting host factors such as SR-BI, CD81, occludin, or claudin-1 may provide novel therapy options that could be used in conjunction with current treatments (70).
Few nAbs have been identified that specifically target E1 (11, 71). Whether this is due to the poor immunogenicity of E1 is still unclear. Anti-E1 Abs are only detected at low levels in HCV patients (72, 73). A recent study reported that patients develop almost sevenfold more antibodies to E2 than to E1 (study in four patients); however, the E1-specific Abs contributed largely to the overall neutralization of HCVpp, despite the relatively low number of E1-specific Abs compared to E2 (74). Anti-E1 Abs have proven difficult to induce in vaccines expressing the E1E2 heterodimer, and are more efficiently induced by expressing E1 alone (75). Whether this is due to the immunodominance of E2 or the masking of E1 neutralizing epitopes by E2 is unknown. Two broadly neutralizing epitopes in E1 encompass residues 192–202 (76) and 264–327 (65, 74, 77). Little is known about the structure, function, and interactions of E1. We know that E1 is involved in E2 folding (78). It is uncertain how E1-specific nAbs mediate their activity. It has been suggested that E1 may contain the element needed for envelope membrane fusion; as such E1 and the (as yet unidentified) fusion epitope it may contain remain an intriguing target for nAbs (79).
The HCV virion forms a complex with low-density and very low-density lipoproteins, forming a lipoviral particle (80). Studies of lipoviral particles showed that host lipoprotein apoE was incorporated into particles with significantly more apoE incorporated than E2 (81, 82). Lipoproteins limit the amount E1E2 to be seen on the surface of the lipoviral particle (82).
Interestingly, many HCV diagnostic assays detect Abs directed against both structural and NS HCV proteins. Why are NS proteins targeted? There is little evidence to suggest they are incorporated into the viral particle, yet NS3-specific Abs are detected before envelope-specific Abs during acute HCV infection (17). It is not yet clear when and in what form the NS proteins are exposed to B-cells. Perhaps NS protein-specific Abs are produced in response to debris from damaged cells. Abs binding to non-neutralizing targets may aid in clearance through opsonization.
The study of Ab responses in HCV infection was long hampered by the lack of a cell culture system or permissive small animal model. Early in vivo studies, conducted in chimpanzees, confirmed the presence of nAbs in plasma from a human being with chronic HCV infection (83). Initial in vitro research on the effect of nAbs in HCV exploited E2/CD81 interaction to perform a neutralization of binding assay with recombinant E2 (38). This system was limited, as it could only evaluate putative neutralizing epitopes overlapping with the E2/CD81-binding region, and likely underestimated the quantity and complexity of nAbs present. Virus-like particles (VLP), produced in a baculovirus system, expressed the E1E2 glycoproteins in a more native conformation (84). The development of retroviral pseudoparticles (HCVpp) expressing unmodified E1E2 glycoproteins that has permitted a more in-depth study of HCV-specific nAbs (50, 85). The HCVpp system is adaptable to allow the expression of E1E2 glycoproteins from diverse HCV genotypes as well as the expression of patient-derived E1E2 (86). This allows patient sera to be screened for neutralizing activity against autologous viral envelope glycoproteins. E1E2 sequences may be cloned from patients’ serial samples, permitting the study of quasispecies and nAb co-evolution over time. Unfortunately, a disadvantage of this system is that the structure and neutralization requirements of HCVpp are still significantly different from those of authentic hepatocyte-derived HCV. The cell cultured derived HCV system (HCVcc) (87–90) may help overcome some of the limitations of the HCVpp system.
While the majority of HCV-infected patients progress to chronic hepatitis with persistent viremia, a significant number (up to 40%) of patients spontaneously clear the infection depending on factors such as, race, sex, and genetics (91–95). It is widely accepted that the cellular immune response can mediate clearance of HCV infection [reviewed in Ref. (23)], but the role of the humoral immune response in acute infection and spontaneous clearance is not fully understood. nAbs are produced in response to HCV infection, but their contribution to control of infection is unclear (83). The acute humoral immune response to HCV has proved challenging to study, as most often patients are asymptomatic and unaware of their infection status; many studies have been retrospective. Typically, IgM is the first immunoglobulin isotype produced by the humoral system in response to infection; however, HCV-specific IgM has not proved a good marker of acute HCV infection as HCV-specific IgM is readily detected in chronically infected patients (96, 97), and HCV-specific IgM and IgG are both almost simultaneously detected in acute infection (98, 99).
There is much evidence to support the theory that Abs have a limited impact on HCV disease outcome as HCV seroconversion is delayed (15, 17, 100, 101), nAbs that target E1E2 are readily detected in the serum of chronically infected patients (29, 102–104), HCV-specific Ab titers wane in patients who have controlled the infection (17, 29, 105, 106), and there are numerous reports of the clearance of HCV infection in the absence of any detected HCV-specific Ab response (15, 28, 107–110).
In contrast, there is striking evidence supporting a role for Abs in control of HCV infection and more interestingly in preventing reinfection. Early induction of cross-reactive nAbs during acute infection strongly correlates with the spontaneous clearance of HCV (31, 33, 50, 73, 111, 112). Conversely, in patients who became persistently infected (>1 year), nAbs were delayed and initially had a narrow neutralizing range, which widened over time (31, 33, 73). In one remarkable case, a patient with established chronic HCV developed a broadly reactive nAb response followed by spontaneous viral clearance (111). Even in chronicity, nAbs may mediate some control of HCV infection as hypogammaglobulinemic patients experience a more rapid and severe progression of disease (113), and patients treated with rituximab show an increase in viral load, which returns to pretreatment levels after completion of treatment (114). Studies in cohorts of intravenous drug users have shown that individuals who spontaneously resolved one episode of HCV infection were more likely to clear a subsequent HCV infection (31, 91, 94, 115), and the time taken to clear the reinfection was significantly shorter (31, 115). Similar to observations in cohorts of intravenous drug users, >80% of chimpanzees that have previously spontaneously cleared HCV, rapidly clear a second infection (16, 109, 116), supporting the hope that protective immunity may be an achievable goal. More work is needed to elucidate the contribution of Abs to the clearance of in HCV in reinfection.
Passive immunization with nAbs can mediate protection: chimpanzees passively immunized with rabbit antisera specific for E1 and E2 were somewhat protected against HCV (117). Recently, HCV1, a human monoclonal Ab targeting E2, has been shown to prevent HCV infection and to reduce the viral load in chronically infected chimpanzees (118). Passive immunity is possible in human beings – it is best highlighted by the Gammagard incident (119). In the early 1990s, the makers of Gammagard (an immunoglobulin product prepared from pooled human plasma), in a move to improve the safety of their product, excluded sera containing anti-HCV Abs from the donor pool; unfortunately, the pooled preparations that removed the HCV seropositive serum lots transmitted HCV to patients (120, 121). Removing HCV-specific Abs from the product removed the protection provided by nAbs previously present. Subsequent screening of the product found HCV RNA (122). HCV-specific Abs provide immunity; however, it is not a sterilizing immunity, as demonstrated by the presence of high-titer nAbs in many patients with persistent HCV infection.
Vaccination has been the most-effective strategy used to control infections that have been a major public health concern. The hepatitis B virus (HBV) vaccine has proven a great success, greatly reducing the number of HBV infections worldwide (123). All successful viral vaccines that have been developed to date induce nAbs (13). However, an HCV vaccine has remained elusive. Most HCV vaccine trials have been conducted in chimpanzees, the best model permitting challenge with infectious HCV. A meta-analysis of HCV vaccine trials in chimpanzees has shown that the vaccines with greatest success contained part or all of the HCV envelope region inducing nAb responses, generating humoral or both humoral and cellular immune responses (124). Recently, a recombinant E1E2 vaccine (derived from HCV 1a) induced protective humoral immune responses in chimpanzees challenged with homologous or heterologous HCV 1a strains (125), and was approved for phase I clinical trial in human beings. In the clinical trial, the recombinant E1E2 vaccine induced antibody and cellular responses in healthy volunteers (126). Further investigation showed the vaccine-induced nAbs against heterologous HCV 1a strains in some healthy volunteers, and one volunteer (out of 16 tested) produced broadly cross-neutralizing Abs against all 7 HCV genotypes (127). A prophylactic vaccine that blocks all infection upon exposure would be ideal. However, preventing the progression of HCV infection to chronicity through a therapeutic vaccine may be a more realistic goal (124, 128, 129).
Neutralizing antibodies are induced during HCV infection, which in some patients contribute to the spontaneous clearance of infection, yet the majority of infected patients progress to chronicity. How does HCV evade the humoral immune response to progress to chronicity? Several mechanisms may contribute to evasion of sterilizing Ab-mediated clearance. These include sequence changes, decoy epitopes, epitope masking, lipid shielding, induction of interfering antibodies, and the ability to move from one cell to another in a neutralization-resistant fashion (Figure 2).
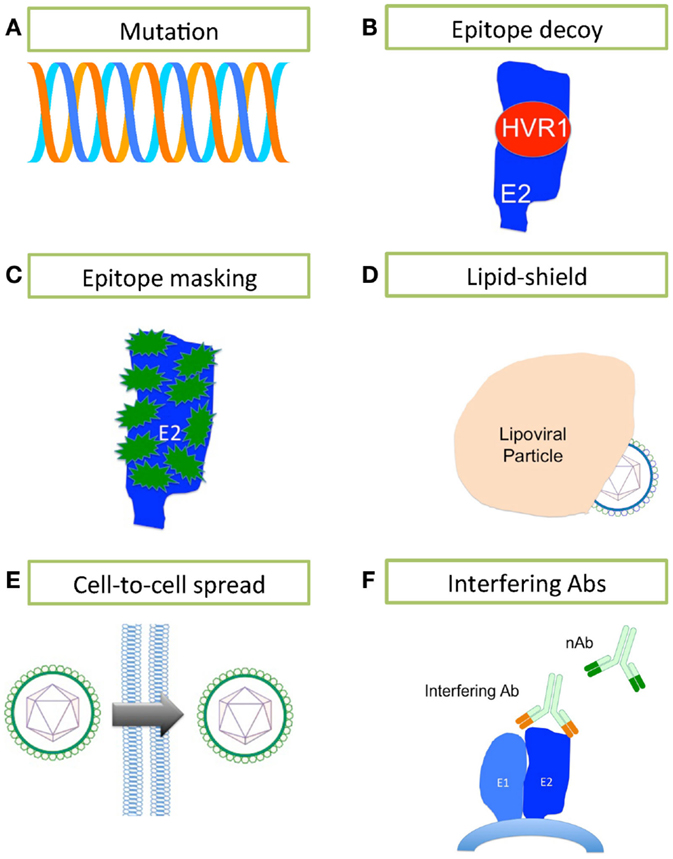
Figure 2. Mechanisms of HCV evasion of the humoral immune system. HCV has developed several strategies for evading the humoral immune system. (A) A high rate of replication using a polymerase that lacks any proofreading mechanism leads to generation of a rapidly changing quasispecies. (B) The highly immunogenic HVR1 masks a hydrophobic region that is sensitive to nAbs. (C) E2 is heavily glycosylated, containing between 9 and 11 N-linked glycans (in green) that mask much of the surface of E2 from nAbs. (D) The close association between HCV and lipoprotein effectively conceals the lipoviral particle from nAbs. (E) HCV may bypass the extracellular space and nAbs by spreading cell-to-cell. (F) Interfering antibodies can disrupt the action of nAbs by binding to non-neutralizing sites and masking neutralizing sites nearby.
Hepatitis C virus’ error-prone replication mechanism permits rapid escape from Ab-mediated and other pressures. Each day, an estimated 1012 new HCV virions are produced in the infected liver (130); it is estimated that thousands of virions bearing each possible single and double nucleotide substitution are made daily in an infected person (131). The resulting quasispecies swarm provides the raw material for selection of nAb-resistant populations (50, 132, 133). In fact, there are numerous reports that link viral sequence evolution, particularly within the E2 glycoprotein, to nAb escape in chronic infection (Figure 2A) (49, 50, 132, 133). Host nAb responses lag behind the rapidly mutating E2 sequences within the quasispecies (50, 132). That nAbs fail to neutralize the dominant viral strain at a given time, yet successfully neutralize previously dominant viral strains in the same patient, clearly demonstrates the continued evolution and escape of the virus under selective pressure from nAbs, with the humoral immune system always, alas, one step behind (132).
It has been suggested that HVR1 of E2 acts as an immunological decoy (Figure 2B) (71, 134). HVR1 is highly immunogenic, but is not essential for viral entry/infection (135); however, HVR1 deletion mutants are far more sensitive to Ab-mediated neutralization, suggesting that HVR1 also acts to conceal epitopes sensitive to neutralization (134–136). nAb selection drives HVR1 sequence evolution in chronically infected patients, while HVR1 remains stable over time in immunoglobulin-deficient patients (137–139). While HVR1 was predicted to be close to the CD81-binding site, Kong and colleagues have suggested that HVR1 lies on the opposite side of E2 molecule, where it masks a hydrophobic surface that is very sensitive to nAbs (45).
E1 and E2 are heavily glycosylated, particularly the immunodominant E2 (Figure 2C). E2 contains up to 11 N-linked glycosylation sites, most of them highly conserved across the different genotypes [reviewed in Ref. (140)]. The N-linked glycans of the ectodomains of E1E2 are reported to contribute almost 50% to the apparent molecular weight of these proteins, and are thought to limit nAbs’ access to key neutralization epitopes (35, 141). These glycans are also essential for the structure and function of E1E2, and play critical roles in viral entry (37, 142–144). Removal of the glycan shield increases the sensitivity of HCVpp to nAb activity (143).
The HCV particle is closely associated with lipoproteins (145), and this association reduces HCV’s buoyant density. Low-density and very low-density virions are more infectious than high-density particles (146, 147). The neutralization of HCVcc by nAbs increased with virion density, suggesting that lipoproteins masked neutralizing epitopes (148). Significantly more host-derived apoE was incorporated into HCV virions than E2, making it far more difficult for the humoral immune system to target E2 (82). Interestingly, apoC-I, the major structural protein of high-density lipoproteins (HDL), is also incorporated in virions (82, 149, 150). SR-BI binds HDL and is a known HCV entry factor, suggesting that HCV has evolved to exploit the normal HDL – SR-BI interaction to avoid the humoral immune system and expedite the virus lifecycle (151). Lipoproteins aid HCV evasion of humoral immunity through two mechanisms: first, the close association of HCV with low-density and very low-density lipoproteins cloaks the virus, thus protecting it from nAbs (Figure 2D) (82, 152) and secondly, HDL expedites virus entry (153).
In vitro and in vivo data suggest that HCV can spread by cell-to-cell transmission (22, 154, 155). Such cell-to-cell spread may enable HCV to bypass extracellular fluids, thereby denying nAbs access to viral particles; indeed, this mechanism appears to be resistant to Ab-mediated neutralization (Figure 2E) (156). CD81, claudin-1, occludin, and SR-BI have pivotal roles in the lateral transmission of HCV, although virions that were not dependent on SR-BI for transmission were significantly more sensitive to nAbs (156, 157). The importance of SR-BI in cell-to-cell spread of HCV would suggest that targeting SR-BI might be valuable for preventing cell-to-cell transmission and avoidance of the humoral immune response. It is, therefore, critical we elucidate the mechanism of cell-to-cell transmission.
Competition between interfering Abs and nAbs can disrupt virus neutralization by nAbs (Figure 2F). Interfering Abs are proposed to work in two ways: first, by directly competing with nAbs for the same epitope; second, by binding an epitope near a neutralizing epitope, thereby masking it. The role of interfering Abs in HCV is controversial (158–163). Abs binding E2 residues 436–447 interfere with nAbs binding to a CD81-binding domain containing E2 residues 412–421 (58). In contrast, a second study failed to find any interfering activity and reported the neutralizing activity was augmented by Abs binding both the 436–447 region and 412–421 region concurrently (162). Further study is needed to tease apart the mechanisms of interference, especially when considering using monoclonal Abs in passive immunizations and vaccine design.
Ideally, an effective HCV vaccine will need to generate a broad and highly reactive immune response at the first signs of HCV infection, before the virus has the chance to unleash its many immune escape mechanisms. A vaccine would need to target multiple antigenic determinants, thus raising the genetic barrier for mutational escape.
The new DAAs will dramatically improve HCV outcome. However, the development of a prophylactic or therapeutic vaccine is needed to control the global HCV problem. Successful vaccine development is dependent on our understanding of the immune response to HCV infection. In particular, it is imperative that we understand why some patients clear the virus naturally and how they are protected from reinfection. The relativity low cost and high benefit of vaccines have made them the cornerstone of modern public health strategies. To date, all successful viral vaccines elicit nAbs (123). The burning question of whether the humoral immune system can mediate or contribute to the clearance of HCV is still unanswered. In the majority of HCV infections, slow development of an nAb response allows HCV to establish widespread and persistent infection. For the lucky minority, the humoral immune system mounts a rapid, broad attack on HCV, contributing to spontaneous clearance of infection. Perhaps the question is far too simple. Our immune system by its very nature is a multifaceted entity, where no one part acts in isolation from the whole. Would the question of the role of B-cells in HCV be best addressed by taking a systems approach to the problem? Is it the failure of CD4+ T helper cells that ultimately leads to the failure of the humoral immune response, and thus the failure to control the infection? Do the other immune cells remain silent or do they engage with B-cells? As we enter the big data era in science, we may be better positioned to answer some of these questions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Seeff LB. The history of the “natural history” of hepatitis C (1968-2009). Liver Int (2009) 29(Suppl 1):89–99. doi: 10.1111/j.1478-3231.2008.01927.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol (2013) 10:553–62. doi:10.1038/nrgastro.2013.107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology (2013) 57:1333–42. doi:10.1002/hep.26141
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Manns MP, Mchutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet (2001) 358:958–65. doi:10.1016/S0140-6736(01)06102-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Hadziyannis SJ, Sette H Jr, Morgan TR, Balan V, Diago M, Marcellin P, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med (2004) 140:346–55. doi:10.7326/0003-4819-140-5-200403020-00010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Heim MH. 25 years of interferon-based treatment of chronic hepatitis C: an epoch coming to an end. Nat Rev Immunol (2013) 13:535–42. doi:10.1038/nri3463
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med (2013) 19:837–49. doi:10.1038/nm.3248
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Tse MT. All-oral HCV therapies near approval. Nat Rev Drug Discov (2013) 12:409–11. doi:10.1038/nrd4036
9. Manns MP, von Hahn T. Novel therapies for hepatitis C – one pill fits all? Nat Rev Drug Discov (2013) 12:595–610. doi:10.1038/nrd4050
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nat Med (2013) 19:850–8. doi:10.1038/nm.3184
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Wang Y, Keck ZY, Foung SK. Neutralizing antibody response to hepatitis C virus. Viruses (2011) 3:2127–45. doi:10.3390/v3112127
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology (2014) 59:318–27. doi:10.1002/hep.26744
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science (2012) 337:183–6. doi:10.1126/science.1225416
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Courouce AM, Bouchardeau F, Girault A, Le Marrec N. Significance of NS3 and NS5 antigens in screening for HCV antibody. Lancet (1994) 343:853–4. doi:10.1016/S0140-6736(94)92054-0
15. Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med (2001) 194:1395–406. doi:10.1084/jem.194.10.1395
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Dahari H, Major M, Zhang X, Mihalik K, Rice CM, Perelson AS, et al. Mathematical modeling of primary hepatitis C infection: noncytolytic clearance and early blockage of virion production. Gastroenterology (2005) 128:1056–66. doi:10.1053/j.gastro.2005.01.049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Netski DM, Mosbruger T, Depla E, Maertens G, Ray SC, Hamilton RG, et al. Humoral immune response in acute hepatitis C virus infection. Clin Infect Dis (2005) 41:667–75. doi:10.1086/432478
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Sagnelli E, Coppola N, Marrocco C, Coviello G, Battaglia M, Messina V, et al. Diagnosis of hepatitis C virus related acute hepatitis by serial determination of IgM anti-HCV titres. J Hepatol (2005) 42:646–51. doi:10.1016/j.jhep.2004.12.027
19. Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature (2005) 436:946–52. doi:10.1038/nature04079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Callendret B, Walker C. A siege of hepatitis: immune boost for viral hepatitis. Nat Med (2011) 17:252–3. doi:10.1038/nm0311-252
21. Thimme R, Binder M, Bartenschlager R. Failure of innate and adaptive immune responses in controlling hepatitis C virus infection. FEMS Microbiol Rev (2012) 36:663–83. doi:10.1111/j.1574-6976.2011.00319.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Dustin LB, Cashman SB, Laidlaw SM. Immune control and failure in HCV infection-tipping the balance. J Leukoc Biol (2014) 96:535–48. doi:10.1189/jlb.4RI0214-126R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Park SH, Rehermann B. Immune responses to HCV and other hepatitis viruses. Immunity (2014) 40:13–24. doi:10.1016/j.immuni.2013.12.010
24. Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, et al. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci U S A (2002) 99:15661–8. doi:10.1073/pnas.202608299
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Lauer GM, Barnes E, Lucas M, Timm J, Ouchi K, Kim AY, et al. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology (2004) 127:924–36. doi:10.1053/j.gastro.2004.06.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest (2009) 119:1745–54. doi:10.1172/JCI39133
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med (2013) 19:859–68. doi:10.1038/nm.3251
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Cooper S, Erickson AL, Adams EJ, Kansopon J, Weiner AJ, Chien DY, et al. Analysis of a successful immune response against hepatitis C virus. Immunity (1999) 10:439–49. doi:10.1016/S1074-7613(00)80044-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Logvinoff C, Major ME, Oldach D, Heyward S, Talal A, Balfe P, et al. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci U S A (2004) 101:10149–54. doi:10.1073/pnas.0403519101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med (2008) 14:25–7. doi:10.1038/nm1698
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Osburn WO, Fisher BE, Dowd KA, Urban G, Liu L, Ray SC, et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology (2010) 138:315–24. doi:10.1053/j.gastro.2009.09.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A (2012) 109:6205–10. doi:10.1073/pnas.1114927109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Osburn WO, Snider AE, Wells BL, Latanich R, Bailey JR, Thomas DL, et al. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology (2014) 59:2140–51. doi:10.1002/hep.27013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol (2013) 31:705–42. doi:10.1146/annurev-immunol-032712-095916
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Michalak JP, Wychowski C, Choukhi A, Meunier JC, Ung S, Rice CM, et al. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol (1997) 78(Pt 9):2299–306.
36. Liu J, Zhu LX, Kong YY, Li GD, Wang Y. Purification and application of C-terminally truncated hepatitis C virus E1 proteins expressed in Escherichia coli. World J Gastroenterol (2005) 11:503–7. Available from: http://www.wjgnet.com/1007-9327/11/503.asp
37. Goffard A, Callens N, Bartosch B, Wychowski C, Cosset FL, Montpellier C, et al. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol (2005) 79:8400–9. doi:10.1128/JVI.79.13.8400-8409.2005
38. Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, et al. Binding of hepatitis C virus to CD81. Science (1998) 282:938–41. doi:10.1126/science.282.5390.938
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J (2002) 21:5017–25. doi:10.1093/emboj/cdf529
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Evans MJ, Von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature (2007) 446:801–5. doi:10.1038/nature05654
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, De Jong YP, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature (2009) 457:882–6. doi:10.1038/nature07684
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med (2011) 17:589–95. doi:10.1038/nm.2341
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Sainz B Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med (2012) 18:281–5. doi:10.1038/nm.2581
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci U S A (2013) 110:10777–82. doi:10.1073/pnas.1301764110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science (2013) 342:1090–4. doi:10.1126/science.1243876
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature (2014) 509:381–4. doi:10.1038/nature13117
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, et al. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol (1993) 67:3923–30.
48. van Doorn LJ, Capriles I, Maertens G, Deleys R, Murray K, Kos T, et al. Sequence evolution of the hypervariable region in the putative envelope region E2/NS1 of hepatitis C virus is correlated with specific humoral immune responses. J Virol (1995) 69:773–8.
49. Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science (2000) 288:339–44. doi:10.1126/science.288.5464.339
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Dowd KA, Netski DM, Wang XH, Cox AL, Ray SC. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology (2009) 136:2377–86. doi:10.1053/j.gastro.2009.02.080
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, et al. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem (2003) 278:41624–30. doi:10.1074/jbc.M305289200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Vieyres G, Dubuisson J, Patel AH. Characterization of antibody-mediated neutralization directed against the hypervariable region 1 of hepatitis C virus E2 glycoprotein. J Gen Virol (2011) 92:494–506. doi:10.1099/vir.0.028092-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, et al. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci U S A (2003) 100:14199–204. doi:10.1073/pnas.2335981100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci U S A (2003) 100:7271–6. doi:10.1073/pnas.0832180100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Shimizu YK, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell RH, et al. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology (1996) 223:409–12. doi:10.1006/viro.1996.0497
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Sabo MC, Luca VC, Ray SC, Bukh J, Fremont DH, Diamond MS. Hepatitis C virus epitope exposure and neutralization by antibodies is affected by time and temperature. Virology (2012) 422:174–84. doi:10.1016/j.virol.2011.10.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, et al. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol (1999) 73:6235–44.
58. Zhang P, Wu CG, Mihalik K, Virata-Theimer ML, Yu MY, Alter HJ, et al. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc Natl Acad Sci U S A (2007) 104:8449–54. doi:10.1073/pnas.0703039104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Dhillon S, Witteveldt J, Gatherer D, Owsianka AM, Zeisel MB, Zahid MN, et al. Mutations within a conserved region of the hepatitis C virus E2 glycoprotein that influence virus-receptor interactions and sensitivity to neutralizing antibodies. J Virol (2010) 84:5494–507. doi:10.1128/JVI.02153-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Keck ZY, Saha A, Xia J, Wang Y, Lau P, Krey T, et al. Mapping a region of hepatitis C virus E2 that is responsible for escape from neutralizing antibodies and a core CD81-binding region that does not tolerate neutralization escape mutations. J Virol (2011) 85:10451–63. doi:10.1128/JVI.05259-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Tarr AW, Owsianka AM, Timms JM, Mcclure CP, Brown RJ, Hickling TP, et al. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology (2006) 43:592–601. doi:10.1002/hep.21088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Potter JA, Owsianka AM, Jeffery N, Matthews DJ, Keck ZY, Lau P, et al. Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J Virol (2012) 86:12923–32. doi:10.1128/JVI.02052-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Keck ZY, Angus AG, Wang W, Lau P, Wang Y, Gatherer D, et al. Non-random escape pathways from a broadly neutralizing human monoclonal antibody map to a highly conserved region on the hepatitis C virus E2 glycoprotein encompassing amino acids 412-423. PLoS Pathog (2014) 10:e1004297. doi:10.1371/journal.ppat.1004297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Tarr AW, Owsianka AM, Jayaraj D, Brown RJ, Hickling TP, Irving WL, et al. Determination of the human antibody response to the epitope defined by the hepatitis C virus-neutralizing monoclonal antibody AP33. J Gen Virol (2007) 88:2991–3001. doi:10.1099/vir.0.83065-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Kachko A, Kochneva G, Sivolobova G, Grazhdantseva A, Lupan T, Zubkova I, et al. New neutralizing antibody epitopes in hepatitis C virus envelope glycoproteins are revealed by dissecting peptide recognition profiles. Vaccine (2011) 30:69–77. doi:10.1016/j.vaccine.2011.10.045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Krey T, D’Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog (2010) 6:e1000762. doi:10.1371/journal.ppat.1000762
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Lavie M, Sarrazin S, Montserret R, Descamps V, Baumert TF, Duverlie G, et al. Identification of conserved residues in hepatitis C virus envelope glycoprotein E2 that modulate virus dependence on CD81 and SRB1 entry factors. J Virol (2014) 88:10584–97. doi:10.1128/JVI.01402-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Sourisseau M, Michta ML, Zony C, Israelow B, Hopcraft SE, Narbus CM, et al. Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies. PLoS Pathog (2013) 9:e1003244. doi:10.1371/journal.ppat.1003244
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology (2010) 139:e951–4. doi:10.1053/j.gastro.2010.05.073
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Xiao F, Fofana I, Heydmann L, Barth H, Soulier E, Habersetzer F, et al. Hepatitis C virus cell-cell transmission and resistance to direct-acting antiviral agents. PLoS Pathog (2014) 10:e1004128. doi:10.1371/journal.ppat.1004128
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Wahid A, Dubuisson J. Virus-neutralizing antibodies to hepatitis C virus. J Viral Hepat (2013) 20:369–76. doi:10.1111/jvh.12094
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Leroux-Roels G, Esquivel CA, Deleys R, Stuyver L, Elewaut A, Philippe J, et al. Lymphoproliferative responses to hepatitis C virus core, E1, E2, and NS3 in patients with chronic hepatitis C infection treated with interferon alfa. Hepatology (1996) 23:8–16. doi:10.1053/jhep.1996.v23.pm0008550052
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A (2007) 104:6025–30. doi:10.1073/pnas.0607026104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Luo K, Li S, Jiang L, Zuo T, Qing J, Shi X, et al. Combinatorial library based profiling of antibody response against hepatitis C virus in human. J Gen Virol (2014). doi:10.1099/vir.0.069278-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Garrone P, Fluckiger AC, Mangeot PE, Gauthier E, Dupeyrot-Lacas P, Mancip J, et al. A prime-boost strategy using virus-like particles pseudotyped for HCV proteins triggers broadly neutralizing antibodies in macaques. Sci Transl Med (2011) 3:94ra71. doi:10.1126/scitranslmed.3002330
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Keck ZY, Sung VM, Perkins S, Rowe J, Paul S, Liang TJ, et al. Human monoclonal antibody to hepatitis C virus E1 glycoprotein that blocks virus attachment and viral infectivity. J Virol (2004) 78:7257–63. doi:10.1128/JVI.78.13.7257-7263.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Meunier JC, Russell RS, Goossens V, Priem S, Walter H, Depla E, et al. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the E1 glycoprotein of hepatitis C virus. J Virol (2008) 82:966–73. doi:10.1128/JVI.01872-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Brazzoli M, Helenius A, Foung SK, Houghton M, Abrignani S, Merola M. Folding and dimerization of hepatitis C virus E1 and E2 glycoproteins in stably transfected CHO cells. Virology (2005) 332:438–53. doi:10.1016/j.virol.2004.11.034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Lavillette D, Pecheur EI, Donot P, Fresquet J, Molle J, Corbau R, et al. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J Virol (2007) 81:8752–65. doi:10.1128/JVI.02642-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol (2013) 11:688–700. doi:10.1038/nrmicro3098
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol (2007) 81:13783–93. doi:10.1128/JVI.01091-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, et al. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci U S A (2013) 110:9505–10. doi:10.1073/pnas.1307527110
83. Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, et al. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A (1994) 91:7792–6. doi:10.1073/pnas.91.16.7792
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Baumert TF, Ito S, Wong DT, Liang TJ. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J Virol (1998) 72:3827–36.
85. Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med (2003) 197:633–42. doi:10.1084/jem.20021756
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Tarr AW, Owsianka AM, Szwejk A, Ball JK, Patel AH. Cloning, expression, and functional analysis of patient-derived hepatitis C virus glycoproteins. Methods Mol Biol (2007) 379:177–97. doi:10.1007/978-1-59745-393-6_13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science (2005) 309:623–6. doi:10.1126/science.1114016
88. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med (2005) 11:791–6. doi:10.1038/nm0805-905b
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A (2005) 102:9294–9. doi:10.1073/pnas.0503596102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, et al. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology (2009) 49:364–77. doi:10.1002/hep.22673
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat (2006) 13:34–41. doi:10.1111/j.1365-2893.2005.00651.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Page K, Hahn JA, Evans J, Shiboski S, Lum P, Delwart E, et al. Acute hepatitis C virus infection in young adult injection drug users: a prospective study of incident infection, resolution, and reinfection. J Infect Dis (2009) 200:1216–26. doi:10.1086/605947
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature (2009) 461:798–801. doi:10.1038/nature08463
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Page K, Osburn W, Evans J, Hahn JA, Lum P, Asher A, et al. Frequent longitudinal sampling of hepatitis C virus infection in injection drug users reveals intermittently detectable viremia and reinfection. Clin Infect Dis (2013) 56:405–13. doi:10.1093/cid/cis921
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Grebely J, Page K, Sacks-Davis R, Van Der Loeff MS, Rice TM, Bruneau J, et al. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology (2014) 59:109–20. doi:10.1002/hep.26639
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Lau GK, Lesniewski R, Johnson RG, Davis GL, Lau JY. Immunoglobulin M and A antibodies to hepatitis C core antigen in chronic hepatitis C virus infection. J Med Virol (1994) 44:1–4. doi:10.1002/jmv.1890440102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Yamaguchi N, Tokushige K, Yamauchi K, Hayashi N. Humoral immune response in Japanese acute hepatitis patients with hepatitis C virus infection. Can J Gastroenterol (2000) 14:593–8.
98. Chen PJ, Wang JT, Hwang LH, Yang YH, Hsieh CL, Kao JH, et al. Transient immunoglobulin M antibody response to hepatitis C virus capsid antigen in posttransfusion hepatitis C: putative serological marker for acute viral infection. Proc Natl Acad Sci U S A (1992) 89:5971–5. doi:10.1073/pnas.89.13.5971
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Nikolaeva LI, Blokhina NP, Tsurikova NN, Voronkova NV, Miminoshvili MI, Braginsky DM, et al. Virus-specific antibody titres in different phases of hepatitis C virus infection. J Viral Hepat (2002) 9:429–37. doi:10.1046/j.1365-2893.2002.00369.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, Choo QL, et al. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med (1989) 321:1494–500. doi:10.1056/NEJM198911303212202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Dustin LB, Rice CM. Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol (2007) 25:71–99. doi:10.1146/annurev.immunol.25.022106.141602
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Chen M, Sallberg M, Sonnerborg A, Weiland O, Mattsson L, Jin L, et al. Limited humoral immunity in hepatitis C virus infection. Gastroenterology (1999) 116:135–43. doi:10.1016/S0016-5085(99)70237-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Steinmann D, Barth H, Gissler B, Schurmann P, Adah MI, Gerlach JT, et al. Inhibition of hepatitis C virus-like particle binding to target cells by antiviral antibodies in acute and chronic hepatitis C. J Virol (2004) 78:9030–40. doi:10.1128/JVI.78.17.9030-9040.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Kaplan DE, Sugimoto K, Newton K, Valiga ME, Ikeda F, Aytaman A, et al. Discordant role of CD4 T-cell response relative to neutralizing antibody and CD8 T-cell responses in acute hepatitis C. Gastroenterology (2007) 132:654–66. doi:10.1053/j.gastro.2006.11.044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Takaki A, Wiese M, Maertens G, Depla E, Seifert U, Liebetrau A, et al. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat Med (2000) 6:578–82. doi:10.1038/75063
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Semmo N, Lucas M, Krashias G, Lauer G, Chapel H, Klenerman P. Maintenance of HCV-specific T-cell responses in antibody-deficient patients a decade after early therapy. Blood (2006) 107:4570–1. doi:10.1182/blood-2005-11-4522
107. Bassett SE, Thomas DL, Brasky KM, Lanford RE. Viral persistence, antibody to E1 and E2, and hypervariable region 1 sequence stability in hepatitis C virus-inoculated chimpanzees. J Virol (1999) 73:1118–26.
108. Razvi S, Schneider L, Jonas MM, Cunningham-Rundles C. Outcome of intravenous immunoglobulin-transmitted hepatitis C virus infection in primary immunodeficiency. Clin Immunol (2001) 101:284–8. doi:10.1006/clim.2001.5132
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Major ME, Mihalik K, Puig M, Rehermann B, Nascimbeni M, Rice CM, et al. Previously infected and recovered chimpanzees exhibit rapid responses that control hepatitis C virus replication upon rechallenge. J Virol (2002) 76:6586–95. doi:10.1128/JVI.76.13.6586-6595.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Post JJ, Pan Y, Freeman AJ, Harvey CE, White PA, Palladinetti P, et al. Clearance of hepatitis C viremia associated with cellular immunity in the absence of seroconversion in the hepatitis C incidence and transmission in prisons study cohort. J Infect Dis (2004) 189:1846–55. doi:10.1086/383279
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Raghuraman S, Park H, Osburn WO, Winkelstein E, Edlin BR, Rehermann B. Spontaneous clearance of chronic hepatitis C virus infection is associated with appearance of neutralizing antibodies and reversal of T-cell exhaustion. J Infect Dis (2012) 205:763–71. doi:10.1093/infdis/jir835
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Esteban-Riesco L, Depaulis F, Moreau A, Bacq Y, Dubois F, Goudeau A, et al. Rapid and sustained autologous neutralizing response leading to early spontaneous recovery after HCV infection. Virology (2013) 444:90–9. doi:10.1016/j.virol.2013.05.037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Bjoro K, Froland SS, Yun Z, Samdal HH, Haaland T. Hepatitis C infection in patients with primary hypogammaglobulinemia after treatment with contaminated immune globulin. N Engl J Med (1994) 331:1607–11. doi:10.1056/NEJM199412153312402
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Ennishi D, Terui Y, Yokoyama M, Mishima Y, Takahashi S, Takeuchi K, et al. Monitoring serum hepatitis C virus (HCV) RNA in patients with HCV-infected CD20-positive B-cell lymphoma undergoing rituximab combination chemotherapy. Am J Hematol (2008) 83:59–62. doi:10.1002/ajh.21022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
115. Sacks-Davis R, Aitken CK, Higgs P, Spelman T, Pedrana AE, Bowden S, et al. High rates of hepatitis C virus reinfection and spontaneous clearance of reinfection in people who inject drugs: a prospective cohort study. PLoS One (2013) 8:e80216. doi:10.1371/journal.pone.0080216
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Bassett SE, Guerra B, Brasky K, Miskovsky E, Houghton M, Klimpel GR, et al. Protective immune response to hepatitis C virus in chimpanzees rechallenged following clearance of primary infection. Hepatology (2001) 33:1479–87. doi:10.1053/jhep.2001.24371
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
117. Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci U S A (1996) 93:15394–9. doi:10.1073/pnas.93.26.15394
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
118. Morin TJ, Broering TJ, Leav BA, Blair BM, Rowley KJ, Boucher EN, et al. Human monoclonal antibody HCV1 effectively prevents and treats HCV infection in chimpanzees. PLoS Pathog (2012) 8:e1002895. doi:10.1371/journal.ppat.1002895
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
119. Yu MY, Bartosch B, Zhang P, Guo ZP, Renzi PM, Shen LM, et al. Neutralizing antibodies to hepatitis C virus (HCV) in immune globulins derived from anti-HCV-positive plasma. Proc Natl Acad Sci U S A (2004) 101:7705–10. doi:10.1073/pnas.0402458101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
120. Yu MW, Mason BL, Guo ZP, Tankersley DL, Nedjar S, Mitchell FD, et al. Hepatitis C transmission associated with intravenous immunoglobulins. Lancet (1995) 345:1173–4. doi:10.1016/S0140-6736(95)91002-6
121. Healey CJ, Sabharwal NK, Daub J, Davidson F, Yap PL, Fleming KA, et al. Outbreak of acute hepatitis C following the use of anti-hepatitis C virus – screened intravenous immunoglobulin therapy. Gastroenterology (1996) 110:1120–6. doi:10.1053/gast.1996.v110.pm8613001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
122. Bresee JS, Mast EE, Coleman PJ, Baron MJ, Schonberger LB, Alter MJ, et al. Hepatitis C virus infection associated with administration of intravenous immune globulin. A cohort study. JAMA (1996) 276:1563–7.
123. Zanetti AR, Van Damme P, Shouval D. The global impact of vaccination against hepatitis B: a historical overview. Vaccine (2008) 26:6266–73. doi:10.1016/j.vaccine.2008.09.056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
124. Dahari H, Feinstone SM, Major ME. Meta-analysis of hepatitis C virus vaccine efficacy in chimpanzees indicates an importance for structural proteins. Gastroenterology (2010) 139:965–74. doi:10.1053/j.gastro.2010.05.077
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
125. Meunier JC, Gottwein JM, Houghton M, Russell RS, Emerson SU, Bukh J, et al. Vaccine-induced cross-genotype reactive neutralizing antibodies against hepatitis C virus. J Infect Dis (2011) 204:1186–90. doi:10.1093/infdis/jir511
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
126. Frey SE, Houghton M, Coates S, Abrignani S, Chien D, Rosa D, et al. Safety and immunogenicity of HCV E1E2 vaccine adjuvanted with MF59 administered to healthy adults. Vaccine (2010) 28:6367–73. doi:10.1016/j.vaccine.2010.06.084
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
127. Law JL, Chen C, Wong J, Hockman D, Santer DM, Frey SE, et al. A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS One (2013) 8:e59776. doi:10.1371/journal.pone.0059776
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
128. Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med (2013) 19:869–78. doi:10.1038/nm.3183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
129. Xue J, Zhu H, Chen Z. Therapeutic vaccines against hepatitis C virus. Infect Genet Evol (2014) 22:120–9. doi:10.1016/j.meegid.2014.01.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
130. Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science (1998) 282:103–7. doi:10.1126/science.282.5386.103
131. Rong L, Dahari H, Ribeiro RM, Perelson AS. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med (2010) 2:30ra32. doi:10.1126/scitranslmed.3000544
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
132. von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology (2007) 132:667–78. doi:10.1053/j.gastro.2006.12.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
133. Liu L, Fisher BE, Dowd KA, Astemborski J, Cox AL, Ray SC. Acceleration of hepatitis C virus envelope evolution in humans is consistent with progressive humoral immune selection during the transition from acute to chronic infection. J Virol (2010) 84:5067–77. doi:10.1128/JVI.02265-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
134. Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, et al. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol (2010) 84:5751–63. doi:10.1128/JVI.02200-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
135. Forns X, Thimme R, Govindarajan S, Emerson SU, Purcell RH, Chisari FV, et al. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc Natl Acad Sci U S A (2000) 97:13318–23. doi:10.1073/pnas.230453597
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
136. Prentoe J, Jensen TB, Meuleman P, Serre SB, Scheel TK, Leroux-Roels G, et al. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J Virol (2011) 85:2224–34. doi:10.1128/JVI.01594-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
137. Booth JC, Kumar U, Webster D, Monjardino J, Thomas HC. Comparison of the rate of sequence variation in the hypervariable region of E2/NS1 region of hepatitis C virus in normal and hypogammaglobulinemic patients. Hepatology (1998) 27:223–7. doi:10.1002/hep.510270134
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
138. Gaud U, Langer B, Petropoulou T, Thomas HC, Karayiannis P. Changes in hypervariable region 1 of the envelope 2 glycoprotein of hepatitis C virus in children and adults with humoral immune defects. J Med Virol (2003) 69:350–6. doi:10.1002/jmv.10296
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
139. Lee WM, Polson JE, Carney DS, Sahin B, Gale M Jr. Reemergence of hepatitis C virus after 8.5 years in a patient with hypogammaglobulinemia: evidence for an occult viral reservoir. J Infect Dis (2005) 192:1088–92. doi:10.1086/432917
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
140. Helle F, Duverlie G, Dubuisson J. The hepatitis C virus glycan shield and evasion of the humoral immune response. Viruses (2011) 3:1909–32. doi:10.3390/v3101909
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
141. Whidby J, Mateu G, Scarborough H, Demeler B, Grakoui A, Marcotrigiano J. Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J Virol (2009) 83:11078–89. doi:10.1128/JVI.00800-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
142. Falkowska E, Kajumo F, Garcia E, Reinus J, Dragic T. Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J Virol (2007) 81:8072–9. doi:10.1128/JVI.00459-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
143. Helle F, Goffard A, Morel V, Duverlie G, Mckeating J, Keck ZY, et al. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J Virol (2007) 81:8101–11. doi:10.1128/JVI.00127-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
144. Helle F, Vieyres G, Elkrief L, Popescu CI, Wychowski C, Descamps V, et al. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J Virol (2010) 84:11905–15. doi:10.1128/JVI.01548-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
145. Bartenschlager R, Penin F, Lohmann V, Andre P. Assembly of infectious hepatitis C virus particles. Trends Microbiol (2011) 19:95–103. doi:10.1016/j.tim.2010.11.005
146. Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, et al. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol (2002) 76:6919–28. doi:10.1128/JVI.76.14.6919-6928.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
147. Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, Mckeating JA, et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A (2006) 103:3805–9. doi:10.1073/pnas.0511218103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
148. Grove J, Nielsen S, Zhong J, Bassendine MF, Drummer HE, Balfe P, et al. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J Virol (2008) 82:12020–9. doi:10.1128/JVI.01569-08
149. Meunier JC, Russell RS, Engle RE, Faulk KN, Purcell RH, Emerson SU. Apolipoprotein c1 association with hepatitis C virus. J Virol (2008) 82:9647–56. doi:10.1128/JVI.00914-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
150. Merz A, Long G, Hiet MS, Brugger B, Chlanda P, Andre P, et al. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem (2011) 286:3018–32. doi:10.1074/jbc.M110.175018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
151. Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, et al. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem (2006) 281:18285–95. doi:10.1074/jbc.M602706200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
152. Di Lorenzo C, Angus AG, Patel AH. Hepatitis C virus evasion mechanisms from neutralizing antibodies. Viruses (2011) 3:2280–300. doi:10.3390/v3112280
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
153. Dreux M, Boson B, Ricard-Blum S, Molle J, Lavillette D, Bartosch B, et al. The exchangeable apolipoprotein ApoC-I promotes membrane fusion of hepatitis C virus. J Biol Chem (2007) 282:32357–69. doi:10.1074/jbc.M705358200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
154. Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology (2008) 47:17–24. doi:10.1002/hep.21959
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
155. Witteveldt J, Evans MJ, Bitzegeio J, Koutsoudakis G, Owsianka AM, Angus AG, et al. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J Gen Virol (2009) 90:48–58. doi:10.1099/vir.0.006700-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
156. Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, et al. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol (2011) 85:596–605. doi:10.1128/JVI.01592-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
157. Catanese MT, Loureiro J, Jones CT, Dorner M, Von Hahn T, Rice CM. Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission. J Virol (2013) 87:8282–93. doi:10.1128/JVI.01102-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
158. Zhang P, Zhong L, Struble EB, Watanabe H, Kachko A, Mihalik K, et al. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci U S A (2009) 106:7537–41. doi:10.1073/pnas.0902749106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
159. Lapierre P, Troesch M, Alvarez F, Soudeyns H. Structural basis for broad neutralization of hepatitis C virus quasispecies. PLoS One (2011) 6:e26981. doi:10.1371/journal.pone.0026981
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
160. Duan H, Kachko A, Zhong L, Struble E, Pandey S, Yan H, et al. Amino acid residue-specific neutralization and nonneutralization of hepatitis C virus by monoclonal antibodies to the E2 protein. J Virol (2012) 86:12686–94. doi:10.1128/JVI.00994-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
161. Sautto G, Mancini N, Diotti RA, Solforosi L, Clementi M, Burioni R. Anti-hepatitis C virus E2 (HCV/E2) glycoprotein monoclonal antibodies and neutralization interference. Antiviral Res (2012) 96:82–9. doi:10.1016/j.antiviral.2012.07.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
162. Tarr AW, Urbanowicz RA, Jayaraj D, Brown RJ, Mckeating JA, Irving WL, et al. Naturally occurring antibodies that recognize linear epitopes in the amino terminus of the hepatitis C virus E2 protein confer noninterfering, additive neutralization. J Virol (2012) 86:2739–49. doi:10.1128/JVI.06492-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
163. Keck Z, Wang W, Wang Y, Lau P, Carlsen TH, Prentoe J, et al. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J Virol (2013) 87:37–51. doi:10.1128/JVI.01941-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: HCV, HCV/E2 glycoprotein, monoclonal antibodies, viral antibodies, chronic infection, acute infection, neutralizing antibodies
Citation: Cashman SB, Marsden BD and Dustin LB (2014) The humoral immune response to HCV: understanding is key to vaccine development. Front. Immunol. 5:550. doi: 10.3389/fimmu.2014.00550
Received: 01 September 2014; Paper pending published: 06 October 2014;
Accepted: 16 October 2014; Published online: 10 November 2014.
Edited by:
Ivo Paul Touw, Erasmus University Medical Center, NetherlandsReviewed by:
Koji Yasutomo, University of Tokushima, JapanCopyright: © 2014 Cashman, Marsden and Dustin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lynn B. Dustin, The Peter Medawar Building for Pathogen Research, South Parks Road, Oxford, OX1 3SY, UK e-mail:bHlubi5kdXN0aW5Aa2VubmVkeS5veC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.