
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 05 September 2014
Sec. Vaccines and Molecular Therapeutics
Volume 5 - 2014 | https://doi.org/10.3389/fimmu.2014.00427
This article is part of the Research TopicUnderstanding the Ontogeny of the Immune System to Promote Immune-Mediated Health for LifeView all 20 articles
Human microbial colonization begins at birth and continues to develop and modulate in species abundance for about 3 years, until the microbiota becomes adult-like. During the same time period, children experience significant developmental changes that influence their health status as well as their immune system. An ever-expanding number of articles associate several diseases with early-life imbalances of the gut microbiota, also referred to as gut microbial dysbiosis. Whether early-life dysbiosis precedes and plays a role in disease pathogenesis, or simply originates from the disease process itself is a question that is beginning to be answered in a few diseases, including IBD, obesity, and asthma. This review describes the gut microbiome structure and function during the formative first years of life, as well as the environmental factors that determine its composition. It also aims to discuss the recent advances in understanding the role of the early-life gut microbiota in the development of immune-mediated, metabolic, and neurological diseases. A greater understanding of how the early-life gut microbiota impacts our immune development could potentially lead to novel microbial-derived therapies that target disease prevention at an early age.
The infant gut undergoes important developmental stages that are entirely dependent upon the colonization with microorganisms, beginning at birth. Experiments in germ-free animal models have shown that microbial colonization induces anatomical development of the intestinal epithelium into the typical microvilli pattern, increases epithelial cell turnover rates, and kick-starts the maturation of the gut-associated lymphoid (immune) tissue (GALT) (1, 2). Functionally, germ-free mice do not develop oral tolerance (3) and mice treated with antibiotics are far more susceptible to intestinal pathogens (4–6). Furthermore, upon antibiotic treatment, several animal models show either an exacerbated or improved phenotype of immune-mediated diseases, including asthma (7) and type 1 diabetes (8), emphasizing the role of microbiota in the development of immune-mediated diseases.
The behavior of children in the first 3 years of life clearly promotes significant exposure to microbes: feeding directly from maternal skin, constant introduction of hands, feet, and other objects to mouth, and contact of hands onto floor surfaces, especially during crawling and early walking stages. Children also suffer more infectious diseases than adults. Not surprisingly, the microbiota in children under 3 years of age fluctuates substantially and is more impressionable to environmental factors than the adult microbiota (9). Modern changes in lifestyle, including improved sanitization, cesarean sections, antibiotic usage, and immunizations are among some of the factors that can shift the microbiota, and are being studied as potential drivers of the sudden increase in immune-mediated diseases in the developed world. It has been hypothesized that there is a “critical window” early in life during which the microbiota can be disrupted in a way that may favor the development of disease later in life (10). This has been shown to be the case in an animal model of asthma, in which antibiotic treatment exclusively during the perinatal period leads to a more severe disease phenotype (7).
The aim of this review is to summarize the latest findings of childhood microbiota studies focusing on the first 3 years of life. In addition, we compile some of the most relevant prenatal, perinatal, and postnatal events that are known to alter the early-life gut microbiome. We also discuss the increasing evidence indicating a role for microbiota changes during early-life impacting the development of intestinal and extra intestinal diseases. As the study of the microbiota has changed drastically with the emergence of culture-independent techniques, we begin this review by describing the high-throughput sequencing methods currently used to analyze microbial communities.
Our concept of the microbiota has profoundly expanded with the emergence of molecular methods to study microbial communities. These methods have eclipsed the small proportion of bacterial groups capable of growing in cultures. Although in other natural environments, the cultivation rates are slightly higher (11, 12), cultivability of human-associated communities remains very low (13, 14). These methods have also led to the discovery of novel taxa, such as the phylum TM7 in the human intestinal tract and other body sites (15–18). It is beyond the scope of this review to discuss all methods to study the microbiota as there are already reviews dedicated to this topic (19–21). Instead, we discuss the most relevant methods used in microbiota research, including targeted approaches like 16S rRNA gene next-generation sequencing (NGS), as well as the large-scale metagenomics approach, also known as shot-gun sequencing.
The current gold standard for microbial community analysis is the amplification of the 16S rRNA gene, although other bacteria-specific targets such as the ribosomal 23s subunit and the internally transcribed spacer (ITS) 16S-23S spacer region (22, 23) have been used. The 16s rRNA gene has the advantages of encoding several conserved regions that are exclusive to all bacteria and hypervariable regions that confer specificity to a large number of bacterial species. Current taxonomy reference databases such as SILVA (24) contain over 3 million aligned 16S sequences. The typical 16S community analysis involves DNA or RNA extraction, amplification, standardization, library construction, sequencing, and subsequent bioinformatic analysis. The choice of primers to amplify the desired region of the 16S gene, as well as the extraction protocol used, may introduce bias to the microbial composition results. For example, primers that target regions V1–V2 fail to amplify important bacterial groups, such as Bifidobacterium. The use of these primers led to the erroneous conclusion that Bifidobacteria were an unimportant bacterial group in the infant intestine (25). Although there is still no consensus on this, comparative analysis with full length Sanger sequencing of the 16S rRNA gene supports the use of primers that amplify the V4 region (26), as they yield the most taxonomically informative sequences (27).
Currently, there are two main sequencing platforms being used by most research groups: the Roche 454 pyrosequencer (GS, FLX, and FLX Titanium) and the Illumina sequencer (MiSeq and HiSeq2000; Illumina Inc., San Diego, CA, USA). In brief, 454 pyrosequencing allows reading DNA fragments of up to 500 bp, allowing the coverage of multiple hypervariable V regions, therefore improving the taxonomic resolution of the dataset. The Illumina platform provides sequencing of DNA fragments of up to 150 bp in length but it sequences 10–100 more samples, at a much higher sequencing depth and lower cost than the 454 pyrosequencer. By choosing the appropriate primers, one can achieve an overlap in the sequences, reducing the sequencing error, and increasing the taxonomic information that can be obtained from the reads. Principal component analysis from a large cohort study that used both 454 and Illumina platforms showed that reads from Illumina clustered very close to the reads obtained from full length Sanger sequencing, whereas reads from 454 pyrosequencing clustered separately (28).
There is not a sequencing technique that is considered gold standard yet, but it is very important to choose the sample preparation protocols and sequencing platforms that will most benefit each study, taking into consideration the environment sampled, the number of samples, cost, and depth of sequencing desired, among other factors.
While 16S rRNA gene analysis of the microbial communities gives a survey of the bacteria present in a particular environment, it fails to provide any functional information. For human clinical studies, this means that one cannot infer a possible mechanism that explains the associations between differences in microbial communities and particular disease states. One approach to obtain functional biochemical information is metagenomics. These large-scale studies aim to provide a gene-based inventory of the microbial community. It involves sequencing of sheared genomic DNA without any previous amplification, also known as shot-gun sequencing. To avoid missing the genetic information from very low abundant species, metagenomic sequencing must be done to a very high depth. This was evidenced in the recent Human Microbiome Project study, in which each sample yielded ~107 reads (29). The same NGS platforms discussed for targeted amplicon studies are used in metagenomic sequencing. The short sequences obtained are assembled into longer contiguous reads (contigs) and compared to databases of known genes, such as the NIH Genbank. There are also databases, such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) (30) that organize the genes into biochemical pathways.
The superiority of nucleic acid-based methods to study the microbiota over culture-based methods is very well established. Although methods like metagenomics remain technically sophisticated and expensive to many laboratories, NGS of the 16S rRNA gene is an affordable and a very informative choice.
Age is the major driver of differences in gut microbiota in several human studies (31). Using sequential fecal sampling from one infant during the first two and a half years of life, and two large cohort human microbiome studies across North America, Africa, South America, and Europe, it is apparent the gut microbiome is highly unstable during the first 3 years of life (9, 32, 33). Children younger than 3 years of age have a significantly lower diversity index compared to adults, with ~1000 operational taxonomic units (OTUs) detected in the first year of life, compared to almost 2000 OTUs after this. However, while the gut microbiota of infants is dominated by fewer bacterial species, the interindividual variability in this age group is significantly higher than in adults (33).
The newborn intestine at birth is an aerobic environment where only facultative anaerobes, such as members of the Enterobacteriaceae family can grow. In a matter of days, however, the intestinal lumen turns anaerobic, allowing for strict anaerobes, such as Bifidobacterium, Clostridium, and Bacteroides to colonize (34). During the first few weeks, the microbiota of the newborn gut resembles the maternal skin and vaginal microbiome, with Enterococcaceae, Streptococcaceae, Lactobacillaceae, Clostridiaceae, and Bifidobacteriaceae being predominant bacterial taxa. During the first few months, the diet of the infant is almost exclusively milk, favoring milk oligosaccharide fermenters, such as Bifidobacterium to thrive. Many bifidobacterial species have been isolated from the infant gut (35–37) and it is considered the most prevalent bacterial group at this stage (31). Weaning and/or introduction of solids foods mark another rapid and important shift in gut microbiota. The introduction of a variety of nutrients, many of which are polysaccharides not digested by host enzymes, triggers an increase in abundance of Bacteroides, Clostridium, Ruminococcus, and a decrease in Bifidobacterium and Enterobacteriaceae (9, 38). In the ensuing 12–30 months, the infant gut microbiota progresses into an adult-like gut microbiota abundant in Ruminococcaceae, Lachnospiraceae, Bacteroidaceae, and Prevotellaceae (31) (Figure 1).
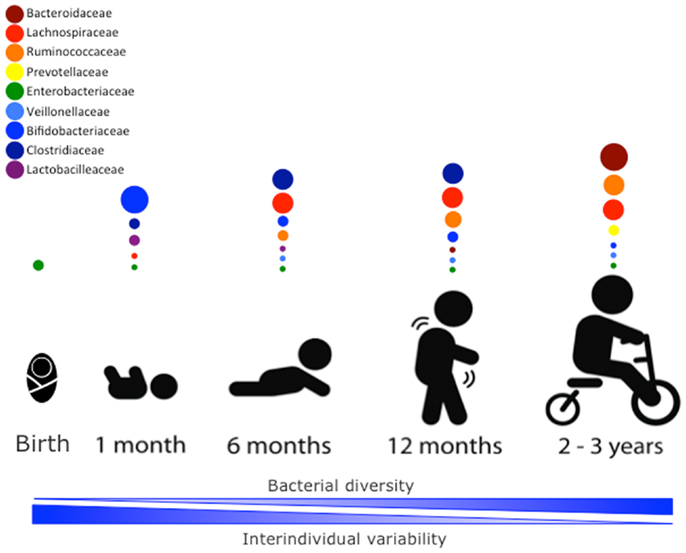
Figure 1. Stages of microbial colonization of the infant and child intestine. Most abundant bacterial families are depicted in circles, where the size of the circle is proportional to the relative abundance of the bacterial taxa at each growth stage. The intestinal microbiota of the newborn is initially colonized by Enterobacteria. In the days after, strict anaerobic bacteria dominate the microbial community. During the first month, bifidobacterial species predominate in the gut, but the introduction of solid foods at around 4–6 months is accompanied by an expansion of clostridial species (Lachnospiracea, Clostridiaceae, and Ruminococcaceae). Members of the Ruminococcaceae family continue to increase in abundance in the following months. By 2–3 years of age, the microbiota composition consists of mainly Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae, which then remains stable into adulthood.
Many factors determine the establishment and composition of microbial communities in all mucose, including the gut. The most important microbial inoculum occurs at birth and shortly afterward. However, the type of diet babies and toddlers ingest, their geographical location, and the use of antibiotics during this period of life can have life-long effects on the composition and function of their gastrointestinal microbiota. Birth was once thought to be the first microbial exposure to the infant, but there is now sufficient evidence to support that prenatal microbial exposure occurs. The following section begins by exploring in utero encounters with microorganisms.
The development of the microbiota begins well before the infant is born. Contrary to what was previously thought, amniotic fluid is not sterile (39, 40). In some cases, bacterial presence in the amniotic fluid is associated with a diseased state. Mycoplasma and Ureaplasma in the amniotic fluid are frequent isolates associated with health detriments such as chorioamnionitis, pre-term delivery, and necrotizing enterocolitis (NEC) (41–43). Also, women with vaginal infections are much more likely to deliver pre-term (PT) babies (44). Aside from this, bacteria are also often detected in the amniotic fluid and placentas of full-term healthy infants (45–47). Other phyla detected in amniotic fluid and placenta overlap with phyla commonly found in the oral microbiota: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Fusobacteria (40, 48). Meconium is also not sterile (49–51), which supports the notion that microbes in the amniotic fluid have access to the unborn fetus. A recent study compared meconium microbiota of pre-term infants to separate datasets of amniotic, vaginal, and oral cavity microbiotas, finding that most overlap between the meconium was from amniotic datasets (52). The bacterial taxa found in meconium using both culture-dependent and independent approaches overlap with adult intestinal microbiota. Enterobacteriaceae (including Escherichia coli and Shigella spp.), Enterococci, Streptococci, Staphylococci (including Staphylococcus epidermidis), and Bifidobacteria have been detected in healthy, full-term infants (50, 53). Furthermore, administration of Enterococcus faecium to pregnant rats allowed for isolation of the same bacteria from the meconium of term pups immediately after birth by caesarian section (53). Thus, while exposure to pathogenic vaginal microbes may be considered infectious events, prenatal exposure to fecal microbes is likely a natural part of in utero development. How these microbes gain access to the uterus remains unknown, although bacterial translocation from the gut into the bloodstream and then to the uterus is one theory that has been proposed, but not tested experimentally yet (54).
Approximately 26% of infants born in Canada are born by cesarean section (55) and this percentage is equal or higher in many other developed countries. The early colonization patterns of cesarean section born infants differ greatly from children born vaginally (34, 56). Knight et al. showed that the first microbiotas of human infants are structured mainly by their mode of delivery, and differences in the bacterial populations within the infant gut are similar to the type of microbiota that the child encounters at birth (31, 56). In fact, post-birth 16S rRNA sequencing data conducted by Knight et al. demonstrate how similar the infant gut microbiota is to the mother’s vaginal or skin microbiota, depending on their mode of birth (56). Additionally, analysis of fecal samples from children 3 days after birth by temporal temperature and denaturing gradient gel electrophoresis (TTGE and DGGE) displayed significant differences in the bacterial populations within the guts of cesarean and vaginally delivered infants (57). Cesarean born infants harbored less Bifidobacterium and Bacteroides species compared to children born vaginally (57–59). Also, the gut microbiota of cesarean delivered infants at 24 months of age is less diverse than those delivered vaginally (60). The authors hypothesize that this drop in diversity may be due to delayed colonization of the gut by Bacteroidetes, as some C-section delivered infants showed no signs of Bacteroidetes colonization until 1 year of age (60). These differences are not, however, apparent in pre-term infants. Although a shorter gestation time is associated with a higher prevalence of Clostridium difficile and Staphylococcus species (59), delivery mode has little effect on the development of the premature infant gut microbiota (61). It may also be that the colonization patterns differ in infants born prematurely due to an increase in antibiotics and various medical treatments administered in the neonatal intensive care unit (NICU) (62, 63).
With the establishment of varying gut microbiotas among infants born vaginally or by C-section comes the subsequent development of various immunological disorders associated with mode of birth. Bager et al. show that cesarean birth is associated with a higher risk of development of inflammatory bowel disease (IBD) between 0 and 14 years of age regardless of parental history of IBD (64). Blustein et al. conducted a study including 10,219 children (926 born by C-section) and found that cesarean delivery was consistently associated with adiposity at 6 weeks of age and this association was even stronger if the children were born from obese mothers (65). Additionally, by age 11 these children were 1.83 times more likely to be overweight or obese (65). Furthermore, Decker et al. found that children born by cesarean section also have an enhanced risk for developing celiac disease (66). Gut microbial dysbiosis has been the most accepted explanation for the association of delivery mode with disease outcome, but more research is needed to adequately support these hypotheses, as these remain as association studies.
Human milk satisfies the nutritional requirements of the infant and confers protection against pathogens through the transmission of maternal antibodies (IgA) and other antimicrobial factors (67–72). The World Health Organization (WHO) recommends exclusive breastfeeding of children up to 6 months of age in order to ensure that the growing infant receives the full nutritional benefits of breast-milk (73). A number of studies have compared the effects of breast versus formula feeding (74–76). Human breast-milk harbors its own microbial consortium that is passed on to the infant along with complex non-digestible human milk oligosaccharides (HMOs) that promote the proliferation of specific gut microbes (77). An infant who consumes approximately 800 mL of human breast-milk per day is thought to ingest between 1 × 105 and 1 × 107 commensal bacteria (78), however, the origin of these commensals remains unclear. Bacterial transfer from the mother’s skin during suckling is essentially unavoidable, but a number of studies also support the entero-mammary pathway hypothesis, wherein bacteria from the maternal gut may reach the mammary glands via maternal dendritic cells and macrophages (79). Pyrosequencing of breast-milk from seven mothers revealed the presence of DNA from a number of major gut-associated bacteria (e.g., Bacteroides and Clostridia) (80) and it also identified a number of gut-associated bacterial genera shared between the maternal feces, breast-milk, and neonatal feces (79). Furthermore, Martin et al. confirmed that breast-milk and infant feces from mother-infant pairs share the same bacterial strains (81). Regardless of the origin of these gut-associated commensals, a number of studies have attempted to identify the mechanisms by which breastfeeding promotes overall immune health via the entero-mammary pathway (67, 81–84). Comparisons between breast-fed and formula-fed infants show that breast-fed infants tend to contain a more uniform population of gut microbes (84). For example, Bifidobacteria and Lactobacillus tend to dominate the guts of breast-fed infants whereas formula-fed infants exhibit higher proportions of Bacteroides, Clostridium, Streptococcus, Enterobacteria, and Veillonella spp. (82–85).
Another area of research regarding formula enrichment is in HMOs and their effects on with the infant gut microbiota. HMOs are considered a type of prebiotic as they promote the growth and proliferation of beneficial commensals and consequently, prevent pathogen colonization of the infant gut and exert positive health effects (86). Certain gut-associated bacterial populations such as Bifidobacterium spp. possess gene clusters dedicated to the metabolism of these substrates (87, 88). An in vitro study assessed HMO consumption by Bifidobacterium spp., E. coli, and Clostridium perfringens and found that only the Bifidobacterium spp. were able to effectively metabolize HMOs (89). Furthermore, the metabolism of these substrates resulted in the production of lactate and short-chain fatty acids (SCFA), which in turn increased the acidity of the surrounding environment, an important factor in preventing pathogen invasion (89). Although Bifidobacteria tend to dominate the guts of breast-fed infants, HMOs are consumed by other bacterial taxa and consequently play a large role in the colonization of the infant gut by various microbial species. Unlike Bifidobacterium spp., Bacteroides spp. (e.g., Bacteroides fragilis and Bacteroides vulgatus) consume a broad range of HMO glycans (90). A piglet model study investigated the role of formula supplementation with an HMO prebiotic mixture on the gut microbiota and overall health after rotavirus infection (91). The group found that piglets fed the supplemented formula versus those fed formula alone had an increased abundance of butyrate-producing bacteria in the Lachnospiraceae family in addition to a reduced duration of rotavirus-induced diarrhea (91). Continued research regarding the role of infant feeding methods in the development of the gut microbiota will likely shed light on the immune and metabolic mechanisms that promote overall infant health.
The introduction of solid foods into the infant gastrointestinal tract (GIT) plays a significant role in the development of the early-life gut microbiota. The gut microbiota conducts a much broader range of metabolic processes than the mammalian cells of the GIT, as it produces a number of degradative enzymes that are not encoded by mammalian genomes (92). Consequently, non-digestible carbohydrates such as plant cell wall polysaccharides, cellulose, and xylans can be broken down and fermented by the gut microbiota (93). SCFAs (the most common being butyrate, acetate, and propionate) are end products of microbial fermentation and are essential energy sources for cells in the mammalian gut in addition to being precursors for gluconeogenesis, liponeogenesis, and protein and cholesterol synthesis (94). The types and amounts of SCFAs produced as well as the prevalence of gut microbial species that produce them are determined by the types of carbohydrates consumed (93–95). Additionally, changes in diet can shift the types and prevalence of microbial species in the gut, as certain microbial species may be better equipped to utilize specific substrates (i.e., inulin and fructo-oligosaccharides promote Bifidobacterium growth) (93). Conversely, some bacterial phyla (e.g., Bacteroidetes) produce numerous carbohydrate-active enzymes that cover a large spectrum of substrates allowing them to switch between energy sources depending on what is available to them (96).
The first introduction of infants to solid foods occurs during weaning (complimentary feeding) when infants are exposed to a much larger array of non-digestible carbohydrates than those present in breast-milk or formula (38). As discussed above, shifts in diet can significantly alter the gut microbiota due to the presence of new substrates that promote the survival and proliferation of varied types of microbial species (38, 93). Additionally, the pancreatic function, small intestinal absorption, and colonic fermentation abilities of the weaning infant mature with the introduction of non-digestible carbohydrates – changing the overall conditions of the digestive tract and the materials that eventually reach the developing colon (97). Fallani et al. used fluorescent in situ hybridization (FISH) to characterize the gut microbiotas of 531 infants across Europe before and after weaning (38). They found that weaning was associated with a significant decrease in proportions of Bifidobacteria and Enterobacteria species as well as in C. difficile and C. perfringens (38). Conversely, there was a significant increase in proportions of Clostridium coccoides and Clostridium leptum (38). In a recent study, 45 exclusively breast-fed 5-month-old infants were randomly assigned to 1 of 3 feeding groups to assess the effect of iron supplementation on the enteric microbiota (98). Children were either fed pureed meats, iron and zinc fortified cereals, or iron-only fortified cereals as the primary complementary food until they were 9–10 months old and fecal samples from 5 to 9 months of age were compared among the children in the three groups (98). Actinobacterial taxa (such as Bifidobacteria and Rothia) as well as Lactobacillales decreased over time with Bacteroides remaining the most abundant in children fed iron-only fortified cereals (98). Clostridium group XIVa (large group of butyrate-producing bacteria) was much more abundant in the microbiotas of the children that were fed meat (98). Koenig et al. found that the introduction of formula and peas to the infant’s diet was associated with an increase in the Bacteroidetes phylum and, after metagenomic analysis, an increase in functional adult-microbiome genes associated with carbohydrate utilization and vitamin biosynthesis (9). The introduction of solid foods to the infant diet seems to initiate the maturation of the infant gut microbiota toward that of an adult. However, more research is needed to clarify what specific components of a solid food diet play the biggest role in developing the infant gut microbiota and how these feeding regimens affect the overall health of the infant.
Intestinal microbiota differs by geographical location for a number of reasons. Microbial and environmental pressures can alter both the repertoire of bacterial species inhabiting the region and their abundance. Different ethnogeographic populations have distinct genetic backgrounds, regional diets, and cultural practices. Of course, resource-replete regions also have access to better sanitation and healthcare than developing nations. Thus, when studies are designed to assess geographical differences in intestinal communities, any trends are attributable to a large body of differences other than geographical separation. Still, comparisons between several developed and developing regions have provided some insight into which geography-associated variables are the strongest drivers of microbial diversity.
A comparison between continentally distinct populations led to the emergence of distinct types of community structures, driven by composition of the Bacteroidetes phylum (28). Danish, Spanish, Italian, French, Japanese, and American adults were distinguished by the dominant genera in the phylum being either Prevotella or Bacteroides, or with a less pronounced Ruminococcus signature. Interestingly, children do not develop a microbial community signature with either Bacteroides or Prevotella until after weaning (32). A longitudinal study on Danish infants revealed that a community structure signature was only detectable after 36 months of life, when the Bacteroidetes phylum, undetectable at 9 or 18 months, expanded in abundance (99).
A sequence-based study compared African children living the rural lifestyle in Burkina Faso (BF) to European urban dwellers in Florence, Italy (EU). These juxtaposed populations were selected because BF represents a society closely resembling the ancestral Neolithic lifestyle, accompanied by high-fiber diet of vegetables, grains, and legumes and absence of processed foods. EU children have a typical western diet saturated in sugars and animal fats, accompanied by a greater caloric intake (32). Compared to EU children, BF children are dominated by Bacteroidetes with a reduced Firmicutes population, along with decreased Proteobacteria and reduced Actinobacteria. The Bacteroidetes-Firmicutes balance has been recently hypothesized to reflect climate, in that Firmicutes dominance is associated with colder climates necessitating increased body fat percentage (100). A community structure signature was found in this study as well, with Prevotella dominant microbiotas being exclusive to BF and Bacteroides exclusive to EU. Besides the differences in microbial species, BF also had raised levels of SCFAs in their stool.
Further studies expanded the developing vs. developed country comparison to include a large cohort of pediatric and adult samples from urban United States and rural villages in Venezuela and Malawi. A Prevotella predominant community signature was found in Malawian and Venezuelan microbiotas, whereas Bacteroides predominated in the North American samples (33). In addition, there were far fewer differences between Venezuelan and Malawian microbiotas, compared to the samples from USA. Taxa discriminating the two rural communities belong predominantly to the Firmicutes phylum with Enterococci being more common to Venezuelan babies and different distributions of the Clostridia class separating adults. Moreover, metagenomics revealed enrichment in glycan and urease metabolic pathways in Venezuelan and Malawian babies, indicative of enhanced ability to forage nitrogen and glycans as an energy source from breast-milk. This enhanced metabolic efficiency of Venezuelan and Malawian infant microbiotas may be an adaptation to decreased volume of nutrition available to these infants compared to North Americans. The Prevotella–Bacteroides split is also observed when comparing children living in a Bangladeshi slum compared to American children living in affluent neighborhoods (101).
Overall, geography is thought to impact the microbiota primarily based on the regional lifestyle. Microbiotas throughout the USA fail to form discrete clusters despite multicultural and multicenter sampling (33, 102, 103). The same could be said for European populations, which are similar to American populations (28, 104). Meanwhile, the microbiotas in developing nations are divergent, but similar between geographically distinct regions with similar dietary habits (32, 33). Importantly, the Prevotella signature is detectable in both Danish (99) and American (33) cohorts, albeit with much more rarity. Thus, the human infant microbiome, irrespective of genetic background, can develop along contrasting trajectories that are dictated by regional lifestyle and diet.
Broad-spectrum antibiotics are often prescribed to infants in the Western world in an attempt to protect the developing child from disease (105). In addition to conferring antibiotic resistance in infancy (106), antibiotic over usage can significantly disrupt the overall ecology of the gut microbiota, alter the abundances of resident gut bacteria, and potentially bias the child toward certain diseases (107–109).
Although the gut microbiota is rather resilient to disruptive factors such as antibiotics (110), the ecology of this dense microbial population can be severely altered if exposed to antibiotics too early in its development and/or for long periods of time (111). This ecological disruption combined with the decreased microbial diversity of the infant gut can provide opportunities for enteric pathogens (112–114). C. difficile is a common infection associated with the antibiotic disturbed gut microbiota (109). A study including 53 infants between the ages of 0 and 13 months linked the onset of C. difficile infections in infancy with alterations in the infant gut microbiota (114).
Antibiotic usage in early-life can also significantly impact the growth of otherwise dominant bacterial phyla in the human gut (111). A study by Fouhy et al. showed that infants exposed to ampicillin and gentamicin shortly after birth tend to harbor higher proportions of Proteobacteria, Actinobacteria, and Lactobacillus than the unexposed children for up to 4 weeks after concluding treatment (111). These sorts of effects are even more visible at the genus level, as seen in a study conducted by Tanaka et al. (105). Terminal restriction fragment length polymorphism (TRFLP) analysis of 26 infants (5 of them received oral broad-spectrum antibiotics) showed that the subjects that received either oral or intravenous antibiotics during the first 4 days of life have less gut microbial diversity as well as an attenuation in colonization with Bifidobacterium and an increase in colonization with Enterococcus (105).
As stated above, antibiotic exposure in early life can render the infant susceptible to numerous diseases later in life (7, 59, 83, 107, 115). Russell et al. showed that vancomycin treatment of ovalbumin (OVA)-challenged mice in early life altered the relative prevalence of microbial populations within the gut microbiota and consequently increased the susceptibility of these mice to asthma (7). A human study associated vancomycin treatment with increased bile acid and glucose metabolism related to the development of obesity (115). Furthermore, antibiotic therapy given to a mouse model of adiposy in early life alters the relative abundances of bacterial populations in the intestine (107). In addition, they showed that this antibiotic therapy in early-life increases colonic short-chain fatty acid levels and increases in adiposy in these mice by altering the regulation of lipid and cholesterol metabolism (107). There is also evidence of antibiotics playing a role in the development of IBD in children (116–118). Shaw et al. found that antibiotic usage during the first year of life was more common in those diagnosed with IBD later in life (116). These results strongly hint at a link between antibiotic usage and disease onset, but research is needed to fully understand the mechanism by which antibiotic-induced microbial dysbiosis influences early-life immune development.
This section reviews the most relevant scientific evidence associating alterations of the intestinal microbiota and pediatric diseases. A summary of this section in included as Table 1.
One of the most lethal threats to a PT infant is NEC (138, 139). With roughly 7% of low birth weight (LBW) infants being diagnosed with NEC in Canada and USA (140, 141) and up to 30% of these infants going on to die from this disease (142), there is an enormous need to understand its etiology in order to develop novel therapeutic interventions. NEC can have different presentations depending on the age of the newborn and the gestational time. NEC in the term infant is often due to structural abnormalities in the intestine and ischemic injury, and often presents within the first week of life (143–145). NEC in the pre-term infant that occurs in the first day of life is often due to intestinal perforation, and is associated with only minor tissue necrosis and lacks the elevated serum cytokine signature seen in classical NEC (146). Classical NEC, which manifests in pre-term infants 1-week after birth or later, is associated with extensive intestinal tissue necrosis (146), elevated serum proinflammatory cytokines (147–149), and bacteremia and endotoxemia (148, 150). The etiology of classical NEC likely involves the intestinal microbiota. The premature neonate gut barrier is leakier than that of term babies (151, 152), a trend that also holds true in animal models of classical NEC (153–155). Increased intestinal permeability promotes bacterial translocation and may account for the endotoxemia and bacteremia associated with NEC. Other risk factors for NEC include antibiotic use (156, 157) and formula feedings (158, 159), both of which are associated with divergent microbial communities. Breastfeeding favors growth of bacteria beneficial to the host while formula feeding might introduce microbes not normally present in the microbiota. The Chronobacter genus is one example of an emerging opportunistic pathogen that can contaminate infant formula and is isolated from NEC patients with higher frequency (160). Thus, the presence of unfavorable bacteria in the leaky pre-term gut is being assessed as another risk factor for NEC.
In general, surveys of the PT infant gut show a microbiota dominated by the Firmicutes and Proteobacteria phyla (119–123, 161). Using culture-based methods, pathogenic bacteria were isolated with more frequency from infants diagnosed with NEC (162) vs. controls, with Coagulase-Negative Staphylococci as a common pathogen. Further, culture-independent case–control comparisons sampled the microbiota within a week of diagnosis and found an outgrowth of Proteobacteria in NEC compared to controls (119, 120).
Several prospective longitudinal studies assessed whether PT infants who would go on to develop NEC followed a distinct trajectory with respect to microbial succession. One such study compared PT infants who did or did not develop NEC to healthy term infants. They found that healthy PT infants began converging to a term-like profile around 6 weeks of age while NEC infants had further outgrowth of Proteobacteria at the expense of Firmicutes, in addition to a reduction of lactose fermenters from the Veillonellaceae family (122). The trend toward a microbiota dominated by Proteobacteria has been observed in further studies of the microbiota in NEC (119–124). Stewart et al. had the unique opportunity to analyze the microbiotas of twins discordant for NEC from 5 days old to over a month in age (162). A sudden outgrowth of E. coli the week before diagnosis was found only in the NEC twin, with a subsequent outgrowth of Klebsiella pneumoniae shortly following diagnosis, but after commencement of antibiotic therapy. A more comprehensive study prospectively evaluated 18 NEC and 35 control PT infants from birth to hospital discharge and found a Proteobacteria bloom 2 weeks before diagnosis, including K. pneumoniae, C. perfringens, and S. epidermidis (124). A gram-negative gut dominated by Proteobacteria should therefore be considered a significant risk factor for NEC development. Mechanistically, the innate sensor for bacterial lipopolysaccharide (LPS) toll-like receptor (TLR)-4, is expressed more by the pre-term than term infant gut in both humans (163, 164) and rodent models of pre-term NEC (165). Moreover, in such animal models, pups lacking TLR-4 are protected from NEC (163, 165). These findings suggest that while prematurity may be the seed cause for dysbiosis, the microbiota of all pre-term infants does not follow the same trajectory and for those that Proteobacteria outcompetes other commensals, NEC becomes a much bigger threat.
Introduction of beneficial commensals, or probiotics, has shown some promise in reducing NEC incidence. Of note, feeding of Bifidobacterium to rat pups improves gut barrier integrity and diminishes NEC incidence (155). This commensal thrives in breast-milk and is found in the term gut a few days after birth, although it is found less in the pre-term gut (166). This, together with other strains of probiotic bacteria, may provide a means to modulate NEC dysbiosis and accelerate the maturation of the prenatal intestine (167).
Inflammatory bowel diseases comprises Crohn’s disease (CD) and ulcerative colitis (UC), two disorders of the GIT whose global prevalence continues to climb. Europe and North America have the highest levels of both CD and UC incidence, with over 300 per 100,000 cases for CD and 90–505 per 100,000 cases for UC (168) and 20–30% of diagnoses are made in the pediatric population (169). Host genotype plays a role in IBD, and several genome-wide association studies (GWAS) have uncovered hundreds of risk alleles associated with both UC and CD. The majority of these are genes related to immune homeostasis in the intestine such as MUC19, involved in gut barrier function, CARD9 (mucosal defense), CCL8 and IL8R (innate cell recruitment), IL23R (mucosal T cell responses), and NOD2, an associated gene involved in bacterial sensing (170). However, it is unlikely that IBD is solely a host-mediated disease, as these risk alleles still only count for a small proportion associated with disease.
The gene products of many polymorphisms tied to disease are also involved with host conversations with intestinal microbes. Moreover, the increasing rates of disease incidence far outpace genetic drift in the human population, which suggests that the environment plays a major role in IBD pathogenesis. Several aspects of the environment have been linked to pediatric IBD, including stress, diet, antibiotic use, and prenatal exposure to infections and smoking (171), and all of these environmental exposures have also been associated with aberrant intestinal microbiotas. Animal model studies have shown that the intestinal microbiota plays a critical role in disease development, as IBD does not occur under germ-free conditions (172, 173). Analyses of fecal microbiota consistently reveals less bacterial diversity in UC and CD patients compared to controls, with an increased density of adherent bacteria in biopsy samples. The loss of diversity in CD has been attributed to fewer members of the Firmicutes phylum (174). Within the phylum, Faecalibacterium and Roseburia are depleted while the Proteobacteria family Enterobacteriaceae increases in abundance (175–177). Some of these findings have used hypothesis-generating approaches to place these associations into causal contexts. For example, the reduction of Faecalibacterium prausnitzii in IBD microbiota has been observed extensively in the literature (178). F. prausnitzii was then shown to induce human T-regulatory cell differentiation in vitro (179) as well as improve gut barrier integrity in a mouse model of colitis (180). Studies such as these, among several others, have provided insight into the dysbiosis of adult IBD and further findings are summarized elsewhere (181, 182). A small collection of studies focus on the pediatric population and their findings are highlighted below.
In a small cohort recruited in Scotland, the microbiota of colonoscopy samples were compared between 25 children with UC or CD and 10 healthy controls. Consistent with the adult literature, diversity was reduced between CD and controls, but this was not true for UC. Also in stark contrast to adult studies, levels of F. prausnitzii were increased in CD compared to controls (125). An earlier study of children with severe UC found reduced diversity in patients compared to healthy controls, as well as reduced abundance of Firmicutes and Verrucomicrobia with an increase in Proteobacteria including E. coli (183). However, while UC children had not received antibiotics for at least 1 month before sampling, they were on corticosteroid therapy, which could have contributed to these trends.
A more comprehensive, sequence-based study by Papa et al. explored the possibility of using stool microbial signatures as a non-invasive diagnostic tool for pediatric IBD. With a cohort of 23 CD, 43 UC patients, and 24 controls, they were able to discriminate not only IBD from controls but also UC and CD, later applying their algorithm to a dataset of adult IBD biopsy samples with some accuracy (126). Bacterial diversity was reduced in IBD patients and this became more apparent during active disease and less so in remission, with an increase in Proteobacteria (including Escherichia and Shigella). During remission, there was an increase in Bifidobacterium abundance. Unique to UC was the complete absence of Verrucomicrobia, which were present in both CD and controls. Interestingly, UC and CD patients were more distinguishable in remission and more similar during active disease.
All of these studies, while showing small trends, suffer from small sample size, confounding effects of treatment, and for some, relying on stool alone instead of biopsy samples. A recent study done by Gevers et al. addressed these pitfalls, using a cohort of 447 children diagnosed with CD alongside 221 controls. They obtained stool together with both ileal and rectal biopsy samples for 16S analysis alongside shot-gun sequencing on a subset of samples (127). Patients were sampled at diagnosis, thus eliminating pharmaceutical treatment as a confounder. Their findings provided several novel insights. Several taxa were negatively associated with CD, including Bacteroides, Bifidobacterium, Blautia, and F. prausnitzii. Taxa positively associated CD included Haemophilus sp., Neiseriaceae, Fusobacterium, Haemophilus influenzae, and E. coli. Veillonellaceae and Pasteurellaceae were specifically associated with deep ulceration as determined by endoscopy. Most of these trends were much stronger in the ileal samples and would not have been detected in the stool, where all the same microbes were present but at much lower abundance. Gene content, predicted by the PICRUSt algorithm for ileal microbiota or shot-gun sequenced for stool samples, revealed an enrichment of pathobiont-promoting pathways in IBD. These include enriched benzoate metabolism in the ileum, by-products of which have been associated with dysbiosis and Enterobacteriaceae virulence (184, 185). Stool samples were enriched for glycerophospholipid and LPS metabolism, products of which have been found in increased levels in CD and UC biopsy tissues (184). Meanwhile, stool microbiome genes involved in complex carbohydrate metabolism were diminished and may result in decreased utilization of these substrates in the gut (186).
Taken together, there is now sufficient evidence that pediatric IBD is associated with aberrant microbial communities even before treatment occurs. It is still unclear, however, whether the dysbiosis observed at the time of diagnosis is a cause for the disease or an early manifestation of it. Regardless, as a dysbiotic disease, it has been treated with probiotics in several clinical trials. There is very minimal evidence that probiotics are effective at treating or preventing remission of CD in children. However, in UC, the probiotic preparation known as VSL-3 has proven as an effective therapy for the ongoing disease and remission prevention (187).
The combination of obesity and malnutrition has had a substantial impact on human health globally. Obesity and diabetes rates have almost doubled since 1980 worldwide, more than 40 million children under 5 were overweight in 2011 (188) and 170 million children are malnourished (189). Diet and nutrition early in life play an important role in these metabolic disorders. However, the prevalence and severity of obesity and malnutrition cannot be attributed to over-eating or food insecurity alone (190). Early-life dietary intake is a strong driver of one’s composition of intestinal microbes (191). In turn, diet driven alteration of the intestinal microbiota can feed back into host metabolism and immunity, with differing consequences dependent on the composition and metabolic potential of the colonizing microbes (192). Early in life, weight gain and height are important measures of human health, and are surrogate markers for nutritional status. In both malnutrition and obesity, the interrelationship between the microbiota, metabolism, and immunity plays an important role in determining outcomes of severity of diseases seen in malnourished and obese humans, such as environmental enteropathy (EE) in malnourished children and early onset of type 2 diabetes in obese children (193).
In obesity, seminal early research implicated the composition of the intestinal microbiota as mediators of weight gain, as the ratio between Bacteroidetes and Firmicutes in the gut correlated with the ability of the host to extract energy from their diet (128). Furthermore, in twins discordant for obesity, transfer of their microbiota into germ-free mice was sufficient to account for their variations in weight, independent of genetics (194). The mechanisms that the microbiota use to signal a shift in host metabolism include signaling through TLR5 (195), glucagon-like peptide (GLP)-1/2 (196), and mediating systemic LPS levels (197). Thus, not only microbiota function, but altered immune function is linked to the pathophysiology of obesity. Obesity has an inflammatory component that can account for the development of metabolic disease, reflected by higher levels of circulating inflammatory proteins, increased adipokine secretion by tissues, and dysregulated activation of leukocytes across various tissue sites, including the liver and brain (198).
It is now clear that early-life changes in microbiota composition can alter susceptibility to developing obesity later in life. As discussed previously, mode of delivery at birth can alter the early-life microbial community, where the gut microbiota of children delivered by C-section is more similar to that of the skin microbiota, rather than vaginal microbiota. In a study of obese Brazilian children, subjects born by C-section had a significantly higher risk for obesity as young adults compared to those born by vaginal delivery (199). Similarly, children fed infant formula rather than breast-milk during the first 6 months of life were more than twice as likely to be obese later in life (200). Aside from nutritional intake early in life, many children are exposed to antibiotics throughout their childhood. Sub-therapeutic doses of several classes of antibiotics have widely been used as early-life growth-promoters in the agriculture industry for decades. Mechanisms of this effect were unclear until recently, where a study by Cho et al. showed these sub-therapeutic doses are sufficient to alter the intestinal microbiota, which resulted in enrichment of key microbial genes involved in carbohydrate metabolism to create SCFAs, as well as systemic changes to hepatic lipid and cholesterol metabolism, leading to increased adiposity in young mice (107). Thus, low levels or exposure to antibiotics can induce subtle shifts in the microbial ecosystem, selecting for an environment more conducive for weight gain and energy harvest. Epidemiological studies show early-life antibiotic use and obesity are correlated, as states in the U.S. with the highest obesity rates also have the highest rates of antibiotic use (188). Many studies have shown presence/absence of specific microbes can modulate and program life-long changes in immunity (201), yet future studies must assess in greater detail how these changes could impact metabolic disease progression. By understanding the differing energy harvest and metabolic capabilities of each child’s gut microbiota, we may be able to create microbiota-based interventions to reverse susceptibility to obesity early in life. A recent study showed that treatment of obese mice with Akkermansia muciniphila reduced high-fat diet-induced metabolic disorders, including fat-mass gain, metabolic endotoxemia, adipose tissue inflammation, and insulin resistance (202). Colonization with Akkermansia mucinophila increased intestinal endocannabinoid levels, controlling inflammation, and also increased the thickness of the inner mucus layer. This treatment required viable A. mucinophila, highlighting the importance of active metabolic output of the bacterium. This study and future efforts hold hope that susceptibility to obesity could be controlled early in life through the microbiota and disease outcomes of obesity could also be improved by probiotics.
Function and composition of the intestinal microbiota also plays a key role in the development and severity of malnutrition. By transferring the feces from Malawian twins discordant for severe protein malnutrition into germ-free mice, Smith et al. were able to show a direct role for the microbiota in mediating symptoms of malnutrition (203). Furthermore, germ-free mice fed a Malawian diet and Malawian microbes lost weight quickly, and this effect could not be fully reversed after feeding with a diet of therapeutic food, implying changes in the microbiota can persistently alter host immunity and metabolism. Since microbiota can mediate the impact of malnutrition, early-life fecal–oral microbial exposure during a critical window of growth and development in children can have a lasting impact on the host’s ability to derive nutrients from the diet, resulting in EE (204). While fecal-oral pathogen exposure can significantly alter the early-life gut microbiota and lead to diarrhea and stunting (205), EE presents as a sub-clinical disorder characterized by villous blunting, crypt hyperplasia, increased permeability, chronic inflammation, and malabsorption in the small intestine (204, 206). It is hypothesized that in regions with poor sanitation, children become exposed to increased levels of fecal-associated bacteria that can colonize the small intestine and lead to EE. Nutritional interventions are not sufficient to reverse symptoms of malnutrition in two-thirds of children, suggesting that the microbiota composition in the small intestine may skew the interactions between immunity and metabolism to a scenario where nutrient uptake and microbe-host mutualism are not favored (193). In this way, EE is thought to have a major impact on outcomes of malnutrition early in life, which can lead to persistent loss of gut function, immune function and cognitive ability (207).
Mechanisms explaining how the microbiota may impact EE and malnutrition are yet to be elucidated, although there are many studies implicating a link between early-life diet and environmental exposures altering immunity and the microbiota, which can drive the pathophysiology and diseases related to malnutrition in a paradoxically similar way to how the microbiota could impact obesity early in life. As the efficacy of nutritional recovery after early-life malnutrition remains low, a seminal study by Trehan et al. noted a significant improvement in nutritional recovery and mortality rates post-antibiotic treatment in children with severe-acute malnutrition (208). The authors did not sequence the microbiota in these children to correlate differences in recovery rate from severe-malnourished, although one can speculate that the antibiotic treatment (amoxicillin) shifted the microbiota into a compositional arrangement more favorable for nutrient uptake.
A critical early window exists in a child’s life where its environmental exposures (including diet and microbes) can shift the immune–metabolism–microbiota interactions to pathophysiological states, which lead to alterations in host growth rates, metabolism, and immunity, ultimately resulting in diseases relating to obesity and malnutrition (such as type 2 diabetes and EE, respectively). A recent study suggests the impact of diet and environmental change stresses on the host can be passed on maternally to children, through epigenetic modulation of the DNA by methylation (209). Thus, maternal dietary and microbial exposures are also crucial to the development of the microbiota early in life, as children may inherit genes with differing potential for predisposition for malnutrition or obesity, based on the diet of their mother. Future studies must focus not only on early-life therapeutic interventions to promote improved intestinal health to combat obesity and malnutrition, but also focus on maternal health to achieve a holistic approach to quelling the impact malnutrition and obesity have on society today.
The atopic diseases (atopic dermatitis, allergic rhinitis, allergic conjunctivitis, anaphylaxis, and asthma) are characterized by IgE-mediated hypersensitivity to an external antigen (210). Strachan’s hygiene hypothesis, proposed in 1989, suggested that exposure of children to infectious agents during infancy will decrease their susceptibility to hyper-inflammatory diseases later in life (211). A recent example of testing this theory was conducted by Ege et al. in Central Europe; they found that children growing up on farms experienced a wide range of microbial exposures and tended to be protected from childhood asthma and other atopic diseases (212).
Significant changes in gut microbiota along with noteworthy changes in immune development and diseases paralleled the industrial revolution, suggesting that environmentally induced changes in the gut microbiota are associated with the development of hypersensitivity diseases. The prevalence of asthma and allergies has continued to rise in industrialized countries and in developing countries where living conditions and hygiene standards are becoming more like those of the Western world (213). Consequently, higher sanitation standards and more readily available antibiotics are likely decreasing our exposure to early childhood microbial antigens at the expense of our immune development (7, 213).
The microflora hypothesis is an extension of Strachan’s hygiene hypothesis in that it argues that there are critical interactions that must occur between our gut microbiota and our immune system in early life in order to circumvent the development of hypersensitivities (214). Studies in germ-free mice show the polarization of the post-birth immune system toward a T-helper 2 cell (TH2) driven immune response (215). With the restoration of the gut microbiota, there is a shift toward a TH1 and TH17 dominated immune phenotype, suggesting that the gut microbiota is important in establishing the balance between the TH1/TH2 subtypes in early life (a balance often disrupted in subjects with atopy) (215–217).
A number of mouse model studies aim to identify that bacterial taxa play a significant role in preventing or promoting the development of atopy. Perinatal antibiotic treatment of OVA-challenged mice (asthma-induced) has been shown to exacerbate the disease potentially by increasing serum and surface bound IgE as well as decreasing T-regulatory cell accumulation in the colon (218). Arnold et al. show that infection of asthma-induced neonate mice versus adult mice with Helicobacter pylori protects these mice from airway hyperresponsiveness, tissue inflammation, and goblet cell metaplasia (common asthma characteristics) (135). Furthermore, Cahenzli et al. provide evidence in mice that a less diverse gut microbiota in early life is associated with elevated mast cell surface bound IgE and exaggerated systemic anaphylaxis (219). Though these studies provide insight for the relationship between gut microbiota and the immune system, researchers are questioning whether specific microbes are actually required to prevent atopy development or whether the metagenomic and metabolomic profiles of the gut microbiota as a whole should be the main focus.
For example, the gut microbiota promotes immune tolerance through the regular stimulation of pattern-recognition receptors (PRRs) and through the production of metabolites (e.g., SCFA) (94, 220). Establishing this immune tolerance to external antigens and host microbes is necessary to prevent the development of hypersensitivity reactions, as T-regulatory cells (+FoxP3) are critical in maintaining the TH1/TH2 balance (94). SCFAs have been shown to play a role in regulating the proliferation of colonic T-regulatory cells (94). These microbial-derived metabolites may be the key to the microbial-host crosstalk that influences systemic inflammation. Low levels of i-butyric, i-valeric, and valeric acids in stool samples from children 1 year of age were associated with the development of food allergy at 4 years of age; however, analysis of the gut microbiota composition was not conducted (221). A recent study showed that the gut microbiota metabolized a high-fiber diet fed to adult mice and this in turn increased the amount of circulating SCFA and consequently decreased allergic inflammation in the lungs of these mice (95). Additional research that focuses on the early-life microbiota is necessary to determine whether these metabolites may be influencing the development of our immune system and thus potentially biasing us toward or away from certain inflammatory diseases.
The majority of human studies regarding the early-life gut microbiota and the development of atopic disease focuses on the role of variables such as mode of delivery (vaginal vs. cesarean birth), feeding methods (breast-milk vs. formula/solid food diet), and early-life antibiotic exposure. Though the infant gut microbiota is influenced significantly by the child’s mode of birth (56), there are conflicting results regarding the association of birth mode with the development of atopic diseases. For example, according to Kolokotroni et al. children born by cesarean section are at an increased risk of developing asthma later in life, yet van Nimwegen et al. argue that mode of birth is less significant than the place of birth (hospital or home) in the development of the intestinal microbiota and asthma (222, 223).
Breastfeeding is a significant contributor to the development of the infant gut microbiota (224). As discussed earlier in this review, breast-fed babies typically have higher numbers of Bifidobacteria and lower numbers of Bacteroides and Atopobium when compared to infants that are exclusively formula-fed (83, 84), whether or not data such as this can be associated with the protection or promotion of atopic disease is still under debate (225–227). However, higher proportions of certain non-digestible HMOs have been associated with a decreased risk of respiratory disease in infants (228).
Antibiotic treatment during the first few years of life has an equally significant impact on the bacterial ecology of the infant gut (107, 109, 111). Epidemiological studies in humans indicate that broad-spectrum antibiotic exposure may play a role in the development of asthma and atopy. Muc et al. conducted a questionnaire-based study and found that antibiotic exposure in the first year of life plays a significant role in the development of asthma and allergic rhinitis in children (229). Additionally, Hoskin-Parr et al. assessed data from 4952 children enrolled in the Avon Longitudinal Study of Parents and Children and found a dose-dependent association between antibiotic usage during the first 2-years of life and the development of asthma at 7.5 years of age (230).
The development of atopic disease in children is characterized by an extremely complex network of environmental and genetic factors, but as the current research shows, the role of the gut microbiota cannot be ignored.
The gut–brain axis is the biochemical signaling that takes place between the GIT and the nervous system. This interaction involves the composition and function of the intestinal microbiota, as it has been shown to alter hormones relating to neurochemical changes in the brain that can modulate the behavior of the host, including anxiety, cognition, stress, mood, and energy-level (231). There are many recent reviews covering this phenomenon extensively (232, 233), so for the purposes of this review, we focus on how early-life changes in the microbiota can alter susceptibility to neurological disease, specifically autism-spectrum disorder (ASD). ASD is a collection of neurodevelopmental changes in children where they exhibit complex behavioral changes in their abilities in social interaction and communication, as well as presence of behaviors similar to obsessive–compulsive disorder, including repetitive and narrow interests. While the disease impacts the brain, gastrointestinal symptoms are commonly described in children with ASD (234, 235), and studies have identified the presence of inflammatory infiltrate and histopathology in biopsies of children with ASD, including increased numbers of cytotoxic T cells, CD19+ B-cells, and increased enteric IgG1/4 (236, 237). Possibly related to these GIT immune changes and dysfunction, some studies have demonstrated an altered composition of intestinal microbiota in young children with ASD compared to healthy control children with typical neurological function (238–242). These data should be interpreted with caution, however, since these studies are all relatively small and ASD children tend to have alternate diets and antibiotic exposures in relation to healthy, typical children. The microbiota composition may be of clinical importance, as a clinical study treated children with ASD with vancomycin and found a high efficacy of recovery from symptoms (243). However, this was a small study and the validity of antibiotic treatment in ASD needs to be assessed with more data.
When identifying a role for the microbiota in ASD, rather than making predictions based on taxonomic sequence data, recent studies have been studying the metabolite secretions of gut microbes and the impact of these microbiotas on host serum metabolites. A potential mechanism behind ASD symptoms could be neuro-active metabolites mediated by or produced by the microbiota, which could disseminate systemically and penetrate the blood–brain barrier. A recent study by Hsiao et al. sought to understand which bacterially produced metabolites modulate progression of ASD by taking advantage of the maternal immune activation (MIA) mouse model, in which mice are known to display features of ASD. They demonstrated that MIA mice exhibit increased intestinal permeability and alterations in the microbiota and serum metabolite profile compared to control mice (137). Remarkably, the authors found that oral treatment with B. fragilis reversed gut permeability and reduced the neurological defects in the MIA mice similar to ASD, including anxiety-like and communicative behavior, while also modulating the serum metabolome. Furthermore, treatment of a single metabolite that is upregulated in MIA mice, restored to normal levels by B. fragilis, induced behavioral abnormalities, highlighting the importance of the serum metabolome profile in ASD (137). This research opens up exciting avenues for future studies in ASD and the microbiome in the early stages of life, and hints at the ability of beneficial microbes to modulate symptoms of ASD. Many studies have suggested early-life exposure to Bifidobacterium or Lactobacillus species modulate behavior in mice (244, 245), and whether these more classical probiotic strains could impact the development of ASD in children remains to be seen. Other studies have altered the hypothesis to argue that it is microbial metabolites meditating the symptoms of ASD. In support of this, elevated SCFA concentrations in the stool of children with ASD have been reported (246). The microbiota early in life produces lower levels of SCFAs compared to adults, since they lack the prevalence of the microbes with the enzymes for their production and generally eat a diet low in complex dietary carbohydrates necessary for the production of SCFAs. Direct exposure of the SCFA propionate to animal brain tissue results in the development of autistic-like behavior (247, 248). Thus, an increase in these metabolites may lead to alterations in brain function.
Once considered in the realms of “pseudo-science,” there has been a groundswell of attention in the role of intestinal microbes and metabolic changes in neurodevelopmental disorders in children, such as ASD. The complexity and heterogeneity of ASD makes it a challenging disease to understand and reverse therapeutically. By taking the new research on the microbiota’s role in the gut-brain access, and the knowledge of importance in the early-life changes, the next-generation of therapies could focus on therapies that increase bacterial homeostasis and barrier function.
Microbiota surveys worldwide have yielded important lessons regarding what influences the microbial composition of the adult intestine, and how this is associated with different diseases. The shift in the Firmicutes/Bacteroidetes ratio is a constant finding among several of the studies looking at disease. Also, the abundance of Prevotella is higher in indigenous regions of the world where there is a much lower incidence of immune-mediated diseases. However, these findings arise from studies done in adults, and the intestinal microbiota during the first 3 years of life differs significantly from adulthood. The microbial community “signatures” associated with the diseases reviewed here do not appear until after 2–3 years, yet it is known that alterations to the early-life microbiota are closely associated the development of these diseases. A recent study in children newly diagnosed with IBD showed that the microbial alterations accompanying disease were different than the changes observed in studies done after diagnosis and during pharmacological treatment (127). This important study suggests that the microbial alterations observed after the disease manifests clinically may not be involved directly in disease pathogenesis and may in fact be, at least in part, a product of the disease manifestations. It is critical to perform additional microbiota studies during the first year of life, a period during which the microbiota composition may give a more accurate portrayal of the microbial conditions that are involved in pathogenesis.
To date, the vast majority of the microbiota studies have focused on the taxonomic profiles of the microbial communities. However, metagenomic studies have shown that different microbes in the gut do not necessarily reflect a different metabolic profile, emphasizing the high degree of redundancy in the biochemical activity among different bacterial taxa (249). Thus, exploring the metagenomic and metabolomic profiles that result from early-life microbial changes associated with disease may provide a better understanding of the role of the microbiota (and their molecules) in the disease process, and may also produce biomarkers of disease before it manifests clinically.
Given that children undergo a drastic succession of microbial changes over a few months, it is clearly important to understand how and when these changes occur. There is a real need for large cohort studies that survey the infant microbiome and metabolome from birth and during at least the first year of life, a period of time most often free of the diseases discussed here yet full of microbial and metabolic changes that may explain the process of later disease emergence. A full understanding of this disease-related changes could allow us to create interventions that rationally shift the microbiota in infants to construct a healthy intestinal environment from a young age. A “critical window” exists early in life where interventions could have more of a profound, long lasting impact on health. Thus, understanding the microbiota changes during this critical window of time will be of great importance for disease prevention.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Brett Finlay lab is funded by grants from the Canadian Institutes of Health Research (CIHR), Genome British Columbia, and Institut Merieux.
1. Abrams GD, Bauer H, Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest (1963) 12:355–64.
2. Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through nod1 regulates intestinal homeostasis. Nature (2008) 456:507–10. doi: 10.1038/nature07450
3. Rask C, Evertsson S, Telemo E, Wold AE. A full flora, but not monocolonization by Escherichia coli or Lactobacilli, supports tolerogenic processing of a fed antigen. Scand J Immunol (2005) 61:529–35. doi:10.1111/j.1365-3083.2005.01598.x
4. Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun (2008) 76:4726–36. doi:10.1128/IAI.00319-08
5. Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, et al. The intestinal microbiota plays a role in salmonella-induced colitis independent of pathogen colonization. PLoS One (2011) 6:e20338. doi:10.1371/journal.pone.0020338
6. Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated citrobacter rodentium-induced colitis. Infect Immun (2011) 79:1536–45. doi:10.1128/IAI.01104-10
7. Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep (2012) 13:440–7. doi:10.1038/embor.2012.32
8. Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN, et al. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol (2012) 189:3805–14. doi:10.4049/jimmunol.1201257
9. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A (2011) 108(Suppl 1):4578–85. doi:10.1073/pnas.1000081107
10. Penders J, Stobberingh EE, Van Den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy (2007) 62:1223–36. doi:10.1111/j.1398-9995.2007.01462.x
11. Bruns A, Nubel U, Cypionka H, Overmann J. Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl Environ Microbiol (2003) 69:1980–9. doi:10.1128/AEM.69.4.1980-1989.2003
12. Kopke B, Wilms R, Engelen B, Cypionka H, Sass H. Microbial diversity in coastal subsurface sediments: a cultivation approach using various electron acceptors and substrate gradients. Appl Environ Microbiol (2005) 71:7819–30. doi:10.1128/AEM.71.12.7819-7830.2005
13. Kovatcheva-Datchary P, Zoetendal EG, Venema K, De Vos WM, Smidt H. Tools for the tract: understanding the functionality of the gastrointestinal tract. Therap Adv Gastroenterol (2009) 2:9–22. doi:10.1177/1756283X09337646
14. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol (2010) 192:5002–17. doi:10.1128/JB.00542-10
15. Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. Investigation of candidate division tm7, a recently recognized major lineage of the domain bacteria with no known pure-culture representatives. Appl Environ Microbiol (2001) 67:411–9. doi:10.1128/AEM.67.1.411-419.2001
16. Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. Prevalence of bacteria of division tm7 in human subgingival plaque and their association with disease. Appl Environ Microbiol (2003) 69:1687–94. doi:10.1128/AEM.69.3.1687-1694.2003
17. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science (2005) 308:1635–8. doi:10.1126/science.1110591
18. Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J (2010) 4:962–74. doi:10.1038/ismej.2010.30
19. Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, et al. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet (2012) 13:47–58. doi:10.1038/nrg3129
20. Lawley B, Tannock GW. Nucleic acid-based methods to assess the composition and function of the bowel microbiota. Gastroenterol Clin North Am (2012) 41:855–68. doi:10.1016/j.gtc.2012.08.010
21. Reigstad CS, Kashyap PC. Beyond phylotyping: understanding the impact of gut microbiota on host biology. Neurogastroenterol Motil (2013) 25:358–72. doi:10.1111/nmo.12134
22. Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol (2009) 75:1534–45. doi:10.1128/AEM.02216-08
23. Gonzales-Marin C, Spratt DA, Allaker RP. Maternal oral origin of Fusobacterium nucleatum in adverse pregnancy outcomes as determined using the 16s-23s rrna gene intergenic transcribed spacer region. J Med Microbiol (2013) 62:133–44. doi:10.1099/jmm.0.049452-0
24. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. Silva: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with arb. Nucleic Acids Res (2007) 35:7188–96. doi:10.1093/nar/gkm864
25. Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol (2007) 5:e177. doi:10.1371/journal.pbio.0050177
26. Liu Z, Desantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16s rrna sequences produced by highly parallel pyrosequencers. Nucleic Acids Res (2008) 36:e120. doi:10.1093/nar/gkn491
27. Claesson MJ, Wang Q, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16s rrna gene regions. Nucleic Acids Res (2010) 38:e200. doi:10.1093/nar/gkq873
28. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature (2011) 473:174–80. doi:10.1038/nature09944
29. Human Microbiome Project Consortium. A framework for human microbiome research. Nature (2012) 486:215–21. doi:10.1038/nature11209
30. Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. Kegg for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res (2010) 38:D355–60. doi:10.1093/nar/gkp896
31. Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vazquez-Baeza Y, et al. Meta-analyses of studies of the human microbiota. Genome Res (2013) 23:1704–14. doi:10.1101/gr.151803.112
32. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from europe and rural africa. Proc Natl Acad Sci U S A (2010) 107:14691–6. doi:10.1073/pnas.1005963107
33. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature (2012) 486:222–7. doi:10.1038/nature11053
34. Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, De La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol (2013) 21:167–73. doi:10.1016/j.tim.2012.12.001
35. Lahtinen SJ, Boyle RJ, Kivivuori S, Oppedisano F, Smith KR, Robins-Browne R, et al. Prenatal probiotic administration can influence bifidobacterium microbiota development in infants at high risk of allergy. J Allergy Clin Immunol (2009) 123:499–501. doi:10.1016/j.jaci.2008.11.034
36. Aires J, Thouverez M, Allano S, Butel MJ. Longitudinal analysis and genotyping of infant dominant bifidobacterial populations. Syst Appl Microbiol (2011) 34:536–41. doi:10.1016/j.syapm.2011.02.007
37. Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, et al. Diversity of Bifidobacteria within the infant gut microbiota. PLoS One (2012) 7:e36957. doi:10.1371/journal.pone.0036957
38. Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five european centres. Microbiology (2011) 157:1385–92. doi:10.1099/mic.0.042143-0
39. DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One (2008) 3:e3056. doi:10.1371/journal.pone.0003056
40. DiGiulio DB. Diversity of microbes in amniotic fluid. Semin Fetal Neonatal Med (2012) 17:2–11. doi:10.1016/j.siny.2011.10.001
41. Goldenberg RL, Culhane JF. Infection as a cause of preterm birth. Clin Perinatol (2003) 30:677–700. doi:10.1016/S0095-5108(03)00110-6
42. Okogbule-Wonodi AC, Gross GW, Sun CC, Agthe AG, Xiao L, Waites KB, et al. Necrotizing enterocolitis is associated with Ureaplasma colonization in preterm infants. Pediatr Res (2011) 69:442–7. doi:10.1203/PDR.0b013e3182111827
43. Kwak DW, Hwang HS, Kwon JY, Park YW, Kim YH. Co-infection with vaginal Ureaplasma urealyticum and Mycoplasma hominis increases adverse pregnancy outcomes in patients with preterm labor or preterm premature rupture of membranes. J Matern Fetal Neonatal Med (2014) 27:333–7. doi:10.3109/14767058.2013.818124
44. Menon R, Dunlop AL, Kramer MR, Fortunato SJ, Hogue CJ. An overview of racial disparities in preterm birth rates: caused by infection or inflammatory response? Acta Obstet Gynecol Scand (2011) 90:1325–31. doi:10.1111/j.1600-0412.2011.01135.x
45. Steel JH, Malatos S, Kennea N, Edwards AD, Miles L, Duggan P, et al. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr Res (2005) 57:404–11. doi:10.1203/01.PDR.0000153869.96337.90
46. Satokari R, Gronroos T, Laitinen K, Salminen S, Isolauri E. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett Appl Microbiol (2009) 48:8–12. doi:10.1111/j.1472-765X.2008.02475.x
47. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med (2014) 6:237–65. doi:10.1126/scitranslmed.3008599
48. Prince AL, Antony KM, Chu DM, Aagaard KM. The microbiome, parturition, and timing of birth: more questions than answers. J Reprod Immunol (2014). doi:10.1016/j.jri.2014.03.006
49. Mshvildadze M, Neu J, Shuster J, Theriaque D, Li N, Mai V. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr (2010) 156:20–5. doi:10.1016/j.jpeds.2009.06.063
50. Gosalbes MJ, Llop S, Valles Y, Moya A, Ballester F, Francino MP. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy (2013) 43:198–211. doi:10.1111/cea.12063
51. Hu J, Nomura Y, Bashir A, Fernandez-Hernandez H, Itzkowitz S, Pei Z, et al. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS One (2013) 8:e78257. doi:10.1371/journal.pone.0078257
52. Ardissone AN, De La Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One (2014) 9:e90784. doi:10.1371/journal.pone.0090784
53. Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Res Microbiol (2008) 159:187–93. doi:10.1016/j.resmic.2007.12.007
54. Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLoS Biol (2013) 11:e1001631. doi:10.1371/journal.pbio.1001631
55. Canadian Institute for Health Information. C-Section Rate: Exclusions [Online]. (2014). Available from: http://www.cihi.ca/CIHI-ext-portal/internet/en/documentfull/health+system+performance/indicators/performance/indicator_ent
56. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A (2010) 107:11971–5. doi:10.1073/pnas.1002601107
57. Biasucci G, Benenati B, Morelli L, Bessi E, Boehm G. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr (2008) 138:1796S–800S.
58. Adlerberth I, Lindberg E, Aberg N, Hesselmar B, Saalman R, Strannegard IL, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res (2006) 59:96–101. doi:10.1203/01.pdr.0000191137.12774.b2
59. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics (2006) 118:511–21. doi:10.1542/peds.2005-2824
60. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed bacteroidetes colonisation and reduced th1 responses in infants delivered by caesarean section. Gut (2014) 63:559–66. doi:10.1136/gutjnl-2012-303249
61. Arboleya S, Binetti A, Salazar N, Fernandez N, Solis G, Hernandez-Barranco A, et al. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol (2012) 79:763–72. doi:10.1111/j.1574-6941.2011.01261.x
62. Berrington JE, Stewart CJ, Embleton ND, Cummings SP. Gut microbiota in preterm infants: assessment and relevance to health and disease. Arch Dis Child Fetal Neonatal Ed (2013) 98:F286–90. doi:10.1136/archdischild-2012-302134
63. Mai V, Torrazza RM, Ukhanova M, Wang X, Sun Y, Li N, et al. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One (2013) 8:e52876. doi:10.1371/journal.pone.0052876
64. Bager P, Simonsen J, Nielsen NM, Frisch M. Cesarean section and offspring’s risk of inflammatory bowel disease: a national cohort study. Inflamm Bowel Dis (2012) 18:857–62. doi:10.1002/ibd.21805
65. Blustein J, Attina T, Liu M, Ryan AM, Cox LM, Blaser MJ, et al. Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. Int J Obes (Lond) (2013) 37:900–6. doi:10.1038/ijo.2013.49
66. Decker E, Hornef M, Stockinger S. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Gut Microbes (2011) 2:91–8. doi:10.4161/gmic.2.2.15414
67. Martin R, Jimenez E, Olivares M, Marin ML, Fernandez L, Xaus J, et al. Lactobacillus salivarius cect 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair. Int J Food Microbiol (2006) 112:35–43. doi:10.1016/j.ijfoodmicro.2006.06.011
68. Palmer AC. Nutritionally mediated programming of the developing immune system. Adv Nutr (2011) 2:377–95. doi:10.3945/an.111.000570
69. Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol (2011) 9:279–90. doi:10.1038/nrmicro2540
70. Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides. Adv Nutr (2012) 3:450S–5S. doi:10.3945/an.112.001859
71. O’Sullivan A, He X, Mcniven EM, Haggarty NW, Lonnerdal B, Slupsky CM. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res (2013) 12:2833–45. doi:10.1021/pr4001702
72. Rogier EW, Frantz AL, Bruno ME, Wedlund L, Cohen DA, Stromberg AJ, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci U S A (2014) 111:3074–9. doi:10.1073/pnas.1315792111
73. WHO. World Health Organization: Breastfeeding [Online]. (2014). Available from: http://www.who.int/topics/breastfeeding/en/
74. Andres A, Cleves MA, Bellando JB, Pivik RT, Casey PH, Badger TM. Developmental status of 1-year-old infants fed breast milk, cow’s milk formula, or soy formula. Pediatrics (2012) 129:1134–40. doi:10.1542/peds.2011-3121
75. Pozo-Rubio T, Capilla A, Mujico JR, De Palma G, Marcos A, Sanz Y, et al. Influence of breastfeeding versus formula feeding on lymphocyte subsets in infants at risk of coeliac disease: the proficel study. Eur J Nutr (2013) 52:637–46. doi:10.1007/s00394-012-0367-8
76. Martin FP, Moco S, Montoliu I, Collino S, Da Silva L, Rezzi S, et al. Impact of breast-feeding and high- and low-protein formula on the metabolism and growth of infants from overweight and obese mothers. Pediatr Res (2014) 75:535–43. doi:10.1038/pr.2013.250
77. Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res (2013) 69:1–10. doi:10.1016/j.phrs.2012.09.001
78. Heikkila MP, Saris PE. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol (2003) 95:471–8. doi:10.1046/j.1365-2672.2003.02002.x
79. Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol (2014) 16:2891–904. doi:10.1111/1462-2920.12238
80. Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr (2013) 110:1253–62. doi:10.1017/S0007114513000597
81. Martin V, Maldonado-Barragan A, Moles L, Rodriguez-Banos M, Campo RD, Fernandez L, et al. Sharing of bacterial strains between breast milk and infant feces. J Hum Lact (2012) 28:36–44. doi:10.1177/0890334411424729
82. Adlerberth I, Wold AE. Establishment of the gut microbiota in western infants. Acta Paediatr (2009) 98:229–38. doi:10.1111/j.1651-2227.2008.01060.x
83. Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, et al. Intestinal microbiota of 6-week-old infants across europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr (2010) 51:77–84. doi:10.1097/MPG.0b013e3181d1b11e
84. Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (fish). Anaerobe (2011) 17:478–82. doi:10.1016/j.anaerobe.2011.03.009
85. Sahl JW, Matalka MN, Rasko DA. Phylomark, a tool to identify conserved phylogenetic markers from whole-genome alignments. Appl Environ Microbiol (2012) 78:4884–92. doi:10.1128/AEM.00929-12
86. German JB, Freeman SL, Lebrilla CB, Mills DA. Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria. Nestle Nutr Workshop Ser Pediatr Program (2008) 62:205–18. doi:10.1159/000146322 discussion 218-222,
87. Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentability of human milk oligosaccharides by several strains of Bifidobacteria. Mol Nutr Food Res (2007) 51:1398–405. doi:10.1002/mnfr.200700150
88. Sela DA, Mills DA. Nursing our microbiota: molecular linkages between Bifidobacteria and milk oligosaccharides. Trends Microbiol (2010) 18:298–307. doi:10.1016/j.tim.2010.03.008
89. Yu ZT, Chen C, Kling DE, Liu B, Mccoy JM, Merighi M, et al. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology (2013) 23:169–77. doi:10.1093/glycob/cws138
90. Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect (2012) 18(Suppl 4):12–5. doi:10.1111/j.1469-0691.2012.03863.x
91. Li M, Monaco MH, Wang M, Comstock SS, Kuhlenschmidt TB, Fahey GC Jr., et al. Human milk oligosaccharides shorten rotavirus-induced diarrhea and modulate piglet mucosal immunity and colonic microbiota. ISME J (2014) 8:1609–20. doi:10.1038/ismej.2014.10
92. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes (2012) 3:289–306. doi:10.4161/gmic.19897
93. Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature (2012) 489:242–9. doi:10.1038/nature11552
94. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science (2013) 341:569–73. doi:10.1126/science.1241165
95. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med (2014) 20:159–66. doi:10.1038/nm.3444
96. Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut bacteroidetes: the food connection. Front Microbiol (2011) 2:93. doi:10.3389/fmicb.2011.00093
97. Parrett AM, Edwards CA. In vitro fermentation of carbohydrate by breast fed and formula fed infants. Arch Dis Child (1997) 76:249–53. doi:10.1136/adc.76.3.249
98. Krebs NF, Sherlock LG, Westcott J, Culbertson D, Hambidge KM, Feazel LM, et al. Effects of different complementary feeding regimens on iron status and enteric microbiota in breastfed infants. J Pediatr (2013) 163:416–23. doi:10.1016/j.jpeds.2013.01.024
99. Bergstrom A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of danish infants. Appl Environ Microbiol (2014) 80:2889–900. doi:10.1128/AEM.00342-14
100. Suzuki TA, Worobey M. Geographical variation of human gut microbial composition. Biol Lett (2014) 10:20131037. doi:10.1098/rsbl.2013.1037
101. Lin A, Bik EM, Costello EK, Dethlefsen L, Haque R, Relman DA, et al. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and The United States. PLoS One (2013) 8:e53838. doi:10.1371/journal.pone.0053838
102. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science (2009) 326:1694–7. doi:10.1126/science.1177486
103. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature (2009) 457:480–4. doi:10.1038/nature07540
104. Lay C, Rigottier-Gois L, Holmstrom K, Rajilic M, Vaughan EE, De Vos WM, et al. Colonic microbiota signatures across five Northern European countries. Appl Environ Microbiol (2005) 71:4153–5. doi:10.1128/AEM.71.7.4153-4155.2005
105. Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol (2009) 56:80–7. doi:10.1111/j.1574-695X.2009.00553.x
106. Mitsou EK, Kirtzalidou E, Pramateftaki P, Kyriacou A. Antibiotic resistance in faecal microbiota of Greek healthy infants. Benef Microbes (2010) 1:297–306. doi:10.3920/BM2010.0007
107. Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature (2012) 488:621–6. doi:10.1038/nature11400
108. Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol (2013) 14:685–90. doi:10.1038/ni.2608
109. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature (2013) 502:96–9. doi:10.1038/nature12503
110. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature (2012) 489:220–30. doi:10.1038/nature11550
111. Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother (2012) 56:5811–20. doi:10.1128/AAC.00789-12
112. Doorduyn Y, Van Den Brandhof WE, Van Duynhoven YT, Wannet WJ, Van Pelt W. Risk factors for salmonella enteritidis and typhimurium (dt104 and non-dt104) infections in the Netherlands: predominant roles for raw eggs in enteritidis and sandboxes in typhimurium infections. Epidemiol Infect (2006) 134:617–26. doi:10.1017/S0950268805005406
113. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16s rRNA sequencing. PLoS Biol (2008) 6:e280. doi:10.1371/journal.pbio.0060280
114. Rousseau C, Levenez F, Fouqueray C, Dore J, Collignon A, Lepage P. Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. J Clin Microbiol (2011) 49:858–65. doi:10.1128/JCM.01507-10
115. Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol (2014) 60:824–31. doi:10.1016/j.jhep.2013.11.034
116. Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterol (2010) 105:2687–92. doi:10.1038/ajg.2010.398
117. Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics and new diagnoses of Crohn’s disease and ulcerative colitis. Am J Gastroenterol (2011) 106:2133–42. doi:10.1038/ajg.2011.304
118. Kronman MP, Zaoutis TE, Haynes K, Feng R, Coffin SE. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics (2012) 130:e794–803. doi:10.1542/peds.2011-3886
119. Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, et al. 16s rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J (2009) 3:944–54. doi:10.1038/ismej.2009.37
120. Mai V, Young CM, Ukhanova M, Wang X, Sun Y, Casella G, et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One (2011) 6:e20647. doi:10.1371/journal.pone.0020647
121. Stewart CJ, Marrs EC, Magorrian S, Nelson A, Lanyon C, Perry JD, et al. The preterm gut microbiota: changes associated with necrotizing enterocolitis and infection. Acta Paediatr (2012) 101:1121–7. doi:10.1111/j.1651-2227.2012.02801.x
122. Claud EC, Keegan KP, Brulc JM, Lu L, Bartels D, Glass E, et al. Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome (2013) 1:20. doi:10.1186/2049-2618-1-20
123. Morrow AL, Lagomarcino AJ, Schibler KR, Taft DH, Yu Z, Wang B, et al. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome (2013) 1:13. doi:10.1186/2049-2618-1-13
124. Torrazza RM, Ukhanova M, Wang X, Sharma R, Hudak ML, Neu J, et al. Intestinal microbial ecology and environmental factors affecting necrotizing enterocolitis. PLoS One (2013) 8:e83304. doi:10.1371/journal.pone.0083304
125. Hansen R, Russell RK, Reiff C, Louis P, Mcintosh F, Berry SH, et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn’s but not in ulcerative colitis. Am J Gastroenterol (2012) 107:1913–22. doi:10.1038/ajg.2012.335
126. Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One (2012) 7:e39242. doi:10.1371/journal.pone.0039242
127. Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe (2014) 15:382–92. doi:10.1016/j.chom.2014.02.005
128. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444:1027–31. doi:10.1038/nature05414
129. Armougom F, Henry M, Vialettes B, Raccah D, Raoult D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and methanogens in anorexic patients. PLoS One (2009) 4:e7125. doi:10.1371/journal.pone.0007125
130. Zuo HJ, Xie ZM, Zhang WW, Li YR, Wang W, Ding XB, et al. Gut bacteria alteration in obese people and its relationship with gene polymorphism. World J Gastroenterol (2011) 17:1076–81. doi:10.3748/wjg.v17.i8.1076
131. Zhang H, Dibaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A (2009) 106:2365–70. doi:10.1073/pnas.0812600106
132. Million M, Angelakis E, Paul M, Armougom F, Leibovici L, Raoult D. Comparative meta-analysis of the effect of Lactobacillus species on weight gain in humans and animals. Microb Pathog (2012) 53:100–8. doi:10.1016/j.micpath.2012.05.007
133. Balamurugan R, George G, Kabeerdoss J, Hepsiba J, Chandragunasekaran AM, Ramakrishna BS. Quantitative differences in intestinal Faecalibacterium prausnitzii in obese Indian children. Br J Nutr (2010) 103:335–8. doi:10.1017/S0007114509992182
134. Hunt JR, Martinelli R, Adams VC, Rook GA, Brunet LR. Intragastric administration of Mycobacterium vaccae inhibits severe pulmonary allergic inflammation in a mouse model. Clin Exp Allergy (2005) 35:685–90. doi:10.1111/j.1365-2222.2005.02239.x
135. Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C, et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory t cells. J Clin Invest (2011) 121:3088–93. doi:10.1172/JCI45041
136. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory t cells by indigenous Clostridium species. Science (2011) 331:337–41. doi:10.1126/science.1198469
137. Hsiao EY, Mcbride SW, Hsien S, Sharon G, Hyde ER, Mccue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell (2013) 155:1451–63. doi:10.1016/j.cell.2013.11.024
138. Lin PW, Stoll BJ. Necrotising enterocolitis. Lancet (2006) 368:1271–83. doi:10.1016/S0140-6736(06)69525-1
139. Obladen M. Necrotizing enterocolitis – 150 years of fruitless search for the cause. Neonatology (2009) 96:203–10. doi:10.1159/000215590
140. Horbar JD, Badger GJ, Carpenter JH, Fanaroff AA, Kilpatrick S, Lacorte M, et al. Trends in mortality and morbidity for very low birth weight infants, 1991-1999. Pediatrics (2002) 110:143–51. doi:10.1542/peds.110.1.143
141. Holman RC, Stoll BJ, Curns AT, Yorita KL, Steiner CA, Schonberger LB. Necrotising enterocolitis hospitalisations among neonates in the united states. Paediatr Perinat Epidemiol (2006) 20:498–506. doi:10.1111/j.1365-3016.2006.00756.x
142. Fitzgibbons SC, Ching Y, Yu D, Carpenter J, Kenny M, Weldon C, et al. Mortality of necrotizing enterocolitis expressed by birth weight categories. J Pediatr Surg (2009) 44:1072–5. doi:10.1016/j.jpedsurg.2009.02.013 discussion 1075-1076,
143. Martinez-Tallo E, Claure N, Bancalari E. Necrotizing enterocolitis in full-term or near-term infants: risk factors. Biol Neonate (1997) 71:292–8. doi:10.1159/000244428
144. Ostlie DJ, Spilde TL, St Peter SD, Sexton N, Miller KA, Sharp RJ, et al. Necrotizing enterocolitis in full-term infants. J Pediatr Surg (2003) 38:1039–42. doi:10.1016/S0022-3468(03)00187-8
145. Al Tawil K, Sumaily H, Ahmed IA, Sallam A, Al Zaben A, Al Namshan M, et al. Risk factors, characteristics and outcomes of necrotizing enterocolitis in late preterm and term infants. J Neonatal Perinatal Med (2013) 6:125–30. doi:10.3233/NPM-1365912
146. Neu J, Walker WA. Necrotizing enterocolitis. N Engl J Med (2011) 364:255–64. doi:10.1056/NEJMra1005408
147. Markel TA, Crisostomo PR, Wairiuko GM, Pitcher J, Tsai BM, Meldrum DR. Cytokines in necrotizing enterocolitis. Shock (2006) 25:329–37. doi:10.1097/01.shk.0000192126.33823.87
148. Sharma R, Tepas JJ III, Hudak ML, Mollitt DL, Wludyka PS, Teng RJ, et al. Neonatal gut barrier and multiple organ failure: role of endotoxin and proinflammatory cytokines in sepsis and necrotizing enterocolitis. J Pediatr Surg (2007) 42:454–61. doi:10.1016/j.jpedsurg.2006.10.038
149. Maheshwari A, Schelonka RL, Dimmitt RA, Carlo WA, Munoz-Hernandez B, Das A, et al. Cytokines associated with necrotizing enterocolitis in extremely-low-birth-weight infants. Pediatr Res (2014) 76:100–8. doi:10.1038/pr.2014.48
150. Bizzarro MJ, Ehrenkranz RA, Gallagher PG. Concurrent bloodstream infections in infants with necrotizing enterocolitis. J Pediatr (2014) 164:61–6. doi:10.1016/j.jpeds.2013.09.020
151. Weaver LT, Laker MF, Nelson R. Intestinal permeability in the newborn. Arch Dis Child (1984) 59:236–41. doi:10.1136/adc.59.3.236
152. van Elburg RM, Fetter WP, Bunkers CM, Heymans HS. Intestinal permeability in relation to birth weight and gestational and postnatal age. Arch Dis Child Fetal Neonatal Ed (2003) 88:F52–5. doi:10.1136/fn.88.1.F52
153. Clark JA, Doelle SM, Halpern MD, Saunders TA, Holubec H, Dvorak K, et al. Intestinal barrier failure during experimental necrotizing enterocolitis: protective effect of EGF treatment. Am J Physiol Gastrointest Liver Physiol (2006) 291:G938–49. doi:10.1152/ajpgi.00090.2006
154. Shiou SR, Yu Y, Chen S, Ciancio MJ, Petrof EO, Sun J, et al. Erythropoietin protects intestinal epithelial barrier function and lowers the incidence of experimental neonatal necrotizing enterocolitis. J Biol Chem (2011) 286:12123–32. doi:10.1074/jbc.M110.154625
155. Bergmann KR, Liu SX, Tian R, Kushnir A, Turner JR, Li HL, et al. Bifidobacteria stabilize claudins at tight junctions and prevent intestinal barrier dysfunction in mouse necrotizing enterocolitis. Am J Pathol (2013) 182:1595–606. doi:10.1016/j.ajpath.2013.01.013
156. Alexander VN, Northrup V, Bizzarro MJ. Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. J Pediatr (2011) 159:392–7. doi:10.1016/j.jpeds.2011.02.035
157. Greenwood C, Morrow AL, Lagomarcino AJ, Altaye M, Taft DH, Yu Z, et al. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of enterobacter. J Pediatr (2014) 165:23–9. doi:10.1016/j.jpeds.2014.01.010
158. Sullivan S, Schanler RJ, Kim JH, Patel AL, Trawoger R, Kiechl-Kohlendorfer U, et al. An exclusively human milk-based diet is associated with a lower rate of necrotizing enterocolitis than a diet of human milk and bovine milk-based products. J Pediatr (2010) 156:562.e–7.e. doi:10.1016/j.jpeds.2009.10.040
159. Quigley M, McGuire W. Formula versus donor breast milk for feeding preterm or low birth weight infants. Cochrane Database Syst Rev (2014) 4:CD002971. doi:10.1002/14651858.CD002971.pub3
160. Hunter CJ, Bean JF. Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J Perinatol (2013) 33:581–5. doi:10.1038/jp.2013.26
161. Torrazza RM, Neu J. The altered gut microbiome and necrotizing enterocolitis. Clin Perinatol (2013) 40:93–108. doi:10.1016/j.clp.2012.12.009
162. Stewart CJ, Marrs EC, Nelson A, Lanyon C, Perry JD, Embleton ND, et al. Development of the preterm gut microbiome in twins at risk of necrotising enterocolitis and sepsis. PLoS One (2013) 8:e73465. doi:10.1371/journal.pone.0073465
163. Leaphart CL, Cavallo J, Gribar SC, Cetin S, Li J, Branca MF, et al. A critical role for tlr4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. J Immunol (2007) 179:4808–20. doi:10.4049/jimmunol.179.7.4808
164. Chan KL, Wong KF, Luk JM. Role of lps/cd14/tlr4-mediated inflammation in necrotizing enterocolitis: pathogenesis and therapeutic implications. World J Gastroenterol (2009) 15:4745–52. doi:10.3748/wjg.15.4745
165. Jilling T, Simon D, Lu J, Meng FJ, Li D, Schy R, et al. The roles of bacteria and tlr4 in rat and murine models of necrotizing enterocolitis. J Immunol (2006) 177:3273–82. doi:10.4049/jimmunol.177.5.3273
166. Butel MJ, Suau A, Campeotto F, Magne F, Aires J, Ferraris L, et al. Conditions of bifidobacterial colonization in preterm infants: a prospective analysis. J Pediatr Gastroenterol Nutr (2007) 44:577–82. doi:10.1097/MPG.0b013e3180406b20
167. Jakaitis BM, Denning PW. Commensal and probiotic bacteria may prevent nec by maturing intestinal host defenses. Pathophysiology (2014) 21:47–54. doi:10.1016/j.pathophys.2013.11.012
168. Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology (2012) 142:46.e–54.e. doi:10.1053/j.gastro.2011.10.001
169. Heyman MB, Kirschner BS, Gold BD, Ferry G, Baldassano R, Cohen SA, et al. Children with early-onset inflammatory bowel disease (ibd): analysis of a pediatric ibd consortium registry. J Pediatr (2005) 146:35–40. doi:10.1016/j.jpeds.2004.08.043
170. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature (2011) 474:307–17. doi:10.1038/nature10209
171. Aujnarain A, Mack DR, Benchimol EI. The role of the environment in the development of pediatric inflammatory bowel disease. Curr Gastroenterol Rep (2013) 15:326. doi:10.1007/s11894-013-0326-4
172. Madsen KL, Malfair D, Gray D, Doyle JS, Jewell LD, Fedorak RN. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm Bowel Dis (1999) 5:262–70. doi:10.1097/00054725-199911000-00004
173. Kennedy RJ, Hoper M, Deodhar K, Erwin PJ, Kirk SJ, Gardiner KR. Interleukin 10-deficient colitis: new similarities to human inflammatory bowel disease. Br J Surg (2000) 87:1346–51. doi:10.1046/j.1365-2168.2000.01615.x
174. Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut (2006) 55:205–11. doi:10.1136/gut.2005.073817
175. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A (2008) 105:16731–6. doi:10.1073/pnas.0804812105
176. Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, et al. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis (2010) 16:2034–42. doi:10.1002/ibd.21319
177. Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology (2010) 139:1844.e–54.e. doi:10.1053/j.gastro.2010.08.049
178. Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Gastroenterol Res Pract (2014) 2014:872725. doi:10.1155/2014/872725
179. Qiu X, Zhang M, Yang X, Hong N, Yu C. Faecalibacterium prausnitzii upregulates regulatory t cells and anti-inflammatory cytokines in treating tnbs-induced colitis. J Crohns Colitis (2013) 7:e558–68. doi:10.1016/j.crohns.2013.04.002
180. Carlsson AH, Yakymenko O, Olivier I, Hakansson F, Postma E, Keita AV, et al. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice dss colitis. Scand J Gastroenterol (2013) 48:1136–44. doi:10.3109/00365521.2013.828773
181. Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in ibd. Nat Rev Gastroenterol Hepatol (2012) 9:599–608. doi:10.1038/nrgastro.2012.152
182. Shim JO. Gut microbiota in inflammatory bowel disease. Pediatr Gastroenterol Hepatol Nutrition (2013) 16:17–21. doi:10.5223/pghn.2013.16.1.17
183. Michail S, Durbin M, Turner D, Griffiths AM, Mack DR, Hyams J, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis (2012) 18:1799–808. doi:10.1002/ibd.22860
184. Morita H, Nakanishi K, Dohi T, Yasugi E, Oshima M. Phospholipid turnover in the inflamed intestinal mucosa: arachidonic acid-rich phosphatidyl/plasmenyl-ethanolamine in the mucosa in inflammatory bowel disease. J Gastroenterol (1999) 34:46–53. doi:10.1007/s005350050215
185. Eloe-Fadrosh EA, Rasko DA. The human microbiome: from symbiosis to pathogenesis. Annu Rev Med (2013) 64:145–63. doi:10.1146/annurev-med-010312-133513
186. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol (2012) 13:R79. doi:10.1186/gb-2012-13-9-r79
187. Guandalini S. Update on the role of probiotics in the therapy of pediatric inflammatory bowel disease. Expert Rev Clin Immunol (2010) 6:47–54. doi:10.1586/eci.09.70
188. Riley LW, Raphael E, Faerstein E. Obesity in the united states – dysbiosis from exposure to low-dose antibiotics? Front Public Health (2013) 1:69. doi:10.3389/fpubh.2013.00069
189. Lutter CK, Daelmans BM, De Onis M, Kothari MT, Ruel MT, Arimond M, et al. Undernutrition, poor feeding practices, and low coverage of key nutrition interventions. Pediatrics (2011) 128:e1418–27. doi:10.1542/peds.2011-1392
190. Gordon JI, Dewey KG, Mills DA, Medzhitov RM. The human gut microbiota and undernutrition. Sci Transl Med (2012) 4:137s12. doi:10.1126/scitranslmed.3004347
191. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature (2014) 505:559–63. doi:10.1038/nature12820
192. Sommer F, Backhed F. The gut microbiota – masters of host development and physiology. Nat Rev Microbiol (2013) 11:227–38. doi:10.1038/nrmicro2974
193. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature (2011) 474:327–36. doi:10.1038/nature10213
194. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (2013) 341:1241214. doi:10.1126/science.1241214
195. Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science (2010) 328:228–31. doi:10.1126/science.1179721
196. Madsbad S. The role of glucagon-like peptide-1 impairment in obesity and potential therapeutic implications. Diabetes Obes Metab (2014) 16:9–21. doi:10.1111/dom.12119
197. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes (2007) 56:1761–72. doi:10.2337/db06-1491
198. Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol (2011) 29:415–45. doi:10.1146/annurev-immunol-031210-101322
199. Mesquita DN, Barbieri MA, Goldani HA, Cardoso VC, Goldani MZ, Kac G, et al. Cesarean section is associated with increased peripheral and central adiposity in young adulthood: Cohort study. PLoS One (2013) 8:e66827. doi:10.1371/journal.pone.0066827
200. Gibbs BG, Forste R. Socioeconomic status, infant feeding practices and early childhood obesity. Pediatr Obes (2014) 9:135–46. doi:10.1111/j.2047-6310.2013.00155.x
201. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (2012) 336:1268–73. doi:10.1126/science.1223490
202. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A (2013) 110:9066–71. doi:10.1073/pnas.1219451110
203. Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, et al. Gut microbiomes of malawian twin pairs discordant for kwashiorkor. Science (2013) 339:548–54. doi:10.1126/science.1229000
204. Humphrey JH. Child undernutrition, tropical enteropathy, toilets, and handwashing. Lancet (2009) 374:1032–5. doi:10.1016/S0140-6736(09)60950-8
205. Guerrant RL, Oria RB, Moore SR, Oria MO, Lima AA. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev (2008) 66:487–505. doi:10.1111/j.1753-4887.2008.00082.x
206. Korpe PS, Petri WA Jr. Environmental enteropathy: critical implications of a poorly understood condition. Trends Mol Med (2012) 18:328–36. doi:10.1016/j.molmed.2012.04.007
207. Guerrant RL, Deboer MD, Moore SR, Scharf RJ, Lima AA. The impoverished gut – a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol (2013) 10:220–9. doi:10.1038/nrgastro.2012.239
208. Trehan I, Goldbach HS, Lagrone LN, Meuli GJ, Wang RJ, Maleta KM, et al. Antibiotics as part of the management of severe acute malnutrition. N Engl J Med (2013) 368:425–35. doi:10.1056/NEJMoa1202851
209. Dominguez-Salas P, Moore SE, Baker MS, Bergen AW, Cox SE, Dyer RA, et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun (2014) 5:3746. doi:10.1038/ncomms4746
210. Kay AB. Allergy and allergic diseases. First of two parts. N Engl J Med (2001) 344:30–7. doi:10.1056/NEJM200101043440106
211. Strachan DP. Hay fever, hygiene, and household size. BMJ (1989) 299:1259–60. doi:10.1136/bmj.299.6710.1259
212. Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med (2011) 364:701–9. doi:10.1056/NEJMoa1007302
214. Shreiner A, Huffnagle GB, Noverr MC. The “microflora hypothesis” of allergic disease. Adv Exp Med Biol (2008) 635:113–34. doi:10.1007/978-0-387-09550-9_10
215. Smith PM, Garrett WS. The gut microbiota and mucosal t cells. Front Microbiol (2011) 2:111. doi:10.3389/fmicb.2011.00111
216. Oyama N, Sudo N, Sogawa H, Kubo C. Antibiotic use during infancy promotes a shift in the t(h)1/t(h)2 balance toward t(h)2-dominant immunity in mice. J Allergy Clin Immunol (2001) 107:153–9. doi:10.1067/mai.2001.111142
217. Smith K, Mccoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol (2007) 19:59–69. doi:10.1016/j.smim.2006.10.002
218. Russell SL, Gold MJ, Willing BP, Thorson L, Mcnagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes (2013) 4:158–64. doi:10.4161/gmic.23567
219. Cahenzli J, Koller Y, Wyss M, Geuking MB, Mccoy KD. Intestinal microbial diversity during early-life colonization shapes long-term ige levels. Cell Host Microbe (2013) 14:559–70. doi:10.1016/j.chom.2013.10.004
220. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell (2004) 118:229–41. doi:10.1016/j.cell.2004.07.002
221. Sandin A, Braback L, Norin E, Bjorksten B. Faecal short chain fatty acid pattern and allergy in early childhood. Acta Paediatr (2009) 98:823–7. doi:10.1111/j.1651-2227.2008.01215.x
222. van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol (2011) 128:948–55. doi:10.1016/j.jaci.2011.07.027
223. Kolokotroni O, Middleton N, Gavatha M, Lamnisos D, Priftis KN, Yiallouros PK. Asthma and atopy in children born by caesarean section: effect modification by family history of allergies – a population based cross-sectional study. BMC Pediatr (2012) 12:179. doi:10.1186/1471-2431-12-179
224. Chan ES, Cummings C, Feldman M, Grimes RB, Gruiger B, Pancer L. Dietary exposures and allergy prevention in high-risk infants: a joint statement with the canadian society of allergy and clinical immunology. Paediatr Child Health (2013) 18:545–54.
225. Gdalevich M, Mimouni D, David M, Mimouni M. Breast-feeding and the onset of atopic dermatitis in childhood: a systematic review and meta-analysis of prospective studies. J Am Acad Dermatol (2001) 45:520–7. doi:10.1067/mjd.2001.114741
226. Sears MR, Greene JM, Willan AR, Taylor DR, Flannery EM, Cowan JO, et al. Long-term relation between breastfeeding and development of atopy and asthma in children and young adults: a longitudinal study. Lancet (2002) 360:901–7. doi:10.1016/S0140-6736(02)11025-7
227. Garcia-Marcos L, Mallol J, Sole D, Brand PL, The EISL Study Group. International study of wheezing in infants: risk factors in affluent and non-affluent countries during the first year of life. Pediatr Allergy Immunol (2010) 21:878–88. doi:10.1111/j.1399-3038.2010.01035.x
228. Stepans MBF, Wilhelm SL, Hertzog M, Rodehorst TKC, Blaney S, Clemens B, et al. Early consumption of human milk oligosaccharides is inversely related to subsequent risk of respiratory and enteric disease in infants. Breastfeed Med (2006) 1:207–15. doi:10.1089/bfm.2006.1.207
229. Muc M, Padez C, Pinto AM. Exposure to paracetamol and antibiotics in early life and elevated risk of asthma in childhood. Adv Exp Med Biol (2013) 788:393–400. doi:10.1007/978-94-007-6627-3_53
230. Hoskin-Parr L, Teyhan A, Blocker A, Henderson AJ. Antibiotic exposure in the first two years of life and development of asthma and other allergic diseases by 7.5 yr: a dose-dependent relationship. Pediatr Allergy Immunol (2013) 24:762–71. doi:10.1111/pai.12153
231. Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol (2009) 6:306–14. doi:10.1038/nrgastro.2009.35
232. Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci (2012) 13:701–12. doi:10.1038/nrn3346
233. Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Microbiology. Animal behavior and the microbiome. Science (2012) 338:198–9. doi:10.1126/science.1227412
234. Valicenti-McDermott M, Mcvicar K, Rapin I, Wershil BK, Cohen H, Shinnar S. Frequency of gastrointestinal symptoms in children with autistic spectrum disorders and association with family history of autoimmune disease. J Dev Behav Pediatr (2006) 27:S128–36. doi:10.1097/00004703-200604002-00011
235. de Theije CG, Wu J, Da Silva SL, Kamphuis PJ, Garssen J, Korte SM, et al. Pathways underlying the gut-to-brain connection in autism spectrum disorders as future targets for disease management. Eur J Pharmacol (2011) 668(Suppl 1):S70–80. doi:10.1016/j.ejphar.2011.07.013
236. Furlano RI, Anthony A, Day R, Brown A, Mcgarvey L, Thomson MA, et al. Colonic cd8 and gamma delta t-cell infiltration with epithelial damage in children with autism. J Pediatr (2001) 138:366–72. doi:10.1067/mpd.2001.111323
237. Torrente F, Ashwood P, Day R, Machado N, Furlano RI, Anthony A, et al. Small intestinal enteropathy with epithelial igg and complement deposition in children with regressive autism. Mol Psychiatry (2002) 7(375–382):334. doi:10.1038/sj.mp.4001079
238. Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe (2010) 16:444–53. doi:10.1016/j.anaerobe.2010.06.008
239. Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism-comparisons to typical children and correlation with autism severity. BMC Gastroenterol (2011) 11:22. doi:10.1186/1471-230X-11-22
240. Williams BL, Hornig M, Parekh T, Lipkin WI. Application of novel pcr-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio (2012) 3:e261–211. doi:10.1128/mBio.00261-11
241. De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One (2013) 8:e76993. doi:10.1371/journal.pone.0076993
242. Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One (2013) 8:e68322. doi:10.1371/journal.pone.0068322
243. Sandler RH, Finegold SM, Bolte ER, Buchanan CP, Maxwell AP, Vaisanen ML, et al. Short-term benefit from oral vancomycin treatment of regressive-onset autism. J Child Neurol (2000) 15:429–35. doi:10.1177/088307380001500701
244. Bercik P, Park AJ, Sinclair D, Khoshdel A, Lu J, Huang X, et al. The anxiolytic effect of bifidobacterium longum ncc3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil (2011) 23:1132–9. doi:10.1111/j.1365-2982.2011.01796.x
245. Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central gaba receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A (2011) 108:16050–5. doi:10.1073/pnas.1102999108
246. Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig Dis Sci (2012) 57:2096–102. doi:10.1007/s10620-012-2167-7
247. MacFabe DF, Cain NE, Boon F, Ossenkopp KP, Cain DP. Effects of the enteric bacterial metabolic product propionic acid on object-directed behavior, social behavior, cognition, and neuroinflammation in adolescent rats: relevance to autism spectrum disorder. Behav Brain Res (2011) 217:47–54. doi:10.1016/j.bbr.2010.10.005
248. Thomas RH, Meeking MM, Mepham JR, Tichenoff L, Possmayer F, Liu S, et al. The enteric bacterial metabolite propionic acid alters brain and plasma phospholipid molecular species: further development of a rodent model of autism spectrum disorders. J Neuroinflammation (2012) 9:153. doi:10.1186/1742-2094-9-153
Keywords: child microbiota, intestinal microbiota, immune-mediated disease, pediatric disease, intestinal dysbiosis
Citation: Arrieta M-C, Stiemsma LT, Amenyogbe N, Brown EM and Finlay B (2014) The intestinal microbiome in early life: health and disease. Front. Immunol. 5:427. doi: 10.3389/fimmu.2014.00427
Received: 19 June 2014; Accepted: 22 August 2014;
Published online: 05 September 2014.
Edited by:
Arnaud Marchant, Université Libre de Bruxelles, BelgiumReviewed by:
Rae Ritchie, Bioscience Vaccines Inc., USACopyright: © 2014 Arrieta, Stiemsma, Amenyogbe, Brown and Finlay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brett Finlay, Michael Smith Laboratories, University of British Columbia, #301 – 2185 East Mall, Vancouver, BC V6T 1Z4, Canada e-mail:YmZpbmxheUBtYWlsLnViYy5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.