- Department of Immunology, University of Connecticut Health Center, Farmington, CT, USA
Mounting a protective immune response is critically dependent on the orchestrated movement of cells within lymphoid tissues. The structure of secondary lymphoid organs regulates immune responses by promoting optimal cell–cell and cell–extracellular matrix interactions. Naïve T cells are initially activated by antigen presenting cells in secondary lymphoid organs. Following priming, effector T cells migrate to the site of infection to exert their functions. Majority of the effector cells die while a small population of antigen-specific T cells persists as memory cells in distinct anatomical locations. The persistence and location of memory cells in lymphoid and non-lymphoid tissues is critical to protect the host from re-infection. The localization of memory T cells is carefully regulated by several factors including the highly organized secondary lymphoid structure, the cellular expression of chemokine receptors and compartmentalized secretion of their cognate ligands. This balance between the anatomy and the ordered expression of cell surface and soluble proteins regulates the subtle choreography of T cell migration. In recent years, our understanding of cellular dynamics of T cells has been advanced by the development of new imaging techniques allowing in situ visualization of T cell responses. Here, we review the past and more recent studies that have utilized sophisticated imaging technologies to investigate the migration dynamics of naïve, effector, and memory T cells.
Imaging Technology
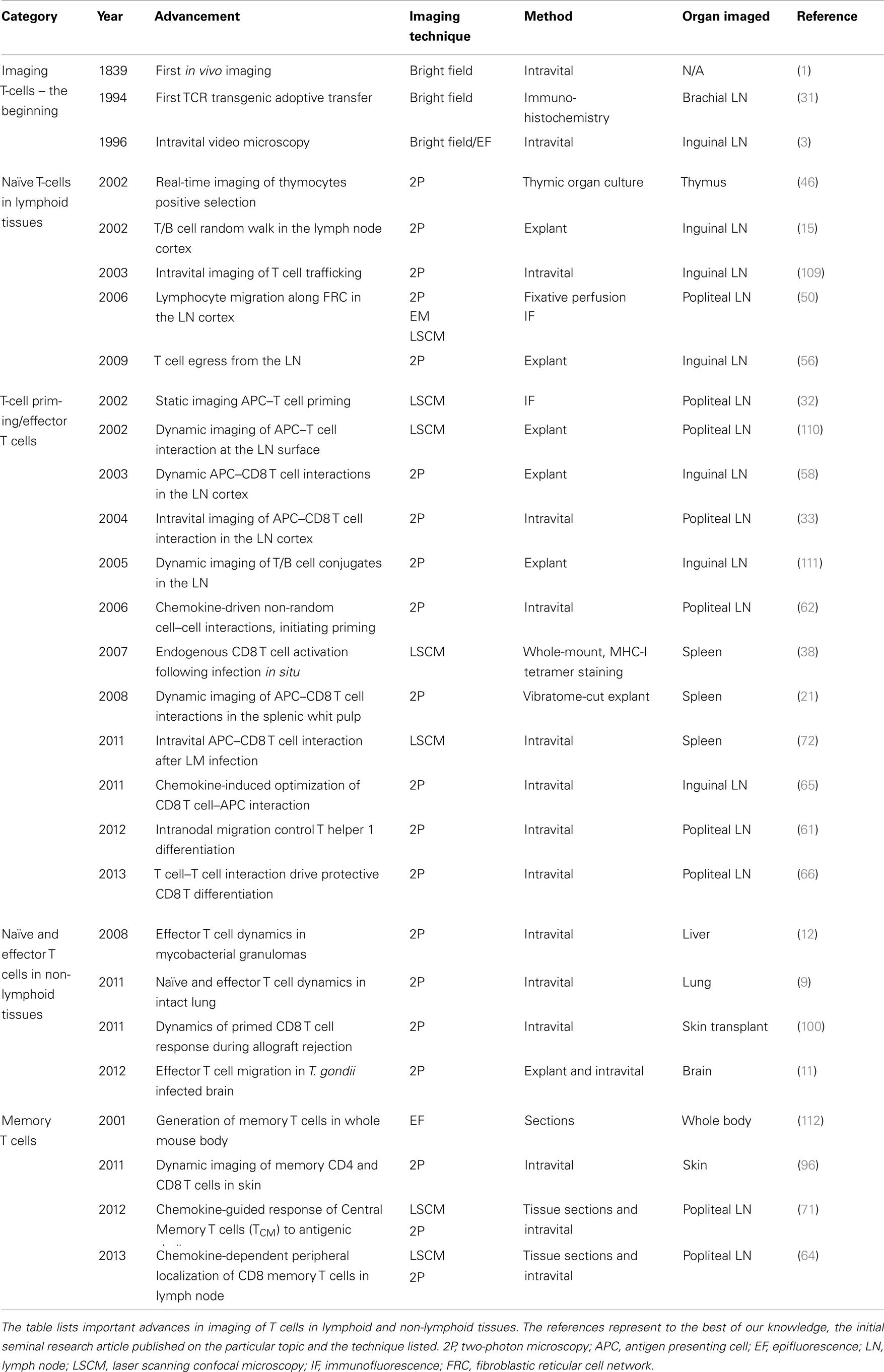
The ability to image the dynamics of T cell immune responses in situ has undergone significant advances over the past decade. For over a century, bright field transillumination or epifluoresecence microscopy was the only technology utilized to image excised organ sections or to visualize cellular processes in vivo. These techniques were useful for visualizing leukocyte interactions with the endothelium (1–3). The introduction of immunohistochemistry and immunofluorescence coupled with the use of monoclonal antibodies introduced specificity to the staining of lymphocytes. More recently, the advent of integrated fluorescent probes (e.g., CFSE) or natural fluorescent proteins (e.g., green fluorescent protein) permitted investigators to tag specific cell populations in vivo. These fluorescently labeled cells could now be tracked in real-time by directly imaging organs in explant preparations or directly intravitally in live animals. An overview of the techniques used for dynamic imaging of T cells is shown in Figure 1.
Figure 1. Overview of surgical techniques for studying T cell dynamics. (A) Explant system, the organ is kept in a heated chamber. Constant flow of warm media (bubble with 95% oxygen and 5% carbon dioxide) is maintained by a peristaltic pump. (B) Examples of intravital imaging methods previously used to observed T cell dynamic in vivo. (C,D) Examples of custom built stages used to immobilize the mouse for intravital imaging. (C) This stage has been used to image the popliteal lymph node; for a detailed description of this method, please refer to a publication by Murooka and Mempel (4). (D) The second stage is designed for imaging the spleen (5).
A significant technological advance was achieved with the laser scanning confocal microscope (LSCM). This type of microscope uses a lens to focus the laser excitation light on the specimen and the emitted light from the focal plane is refocused trough the same lens to the center of an open detector aperture (pinhole). This innovation obstructs the light coming from above and below the focal plane and thus increases the resolution. Sharp optical sectioning through a specimen at different depths can be performed to produce a 3 dimensional reconstruction of the sample. However, single photon confocal microcopy does not allow imaging at great depth (>100 μm) due to light scattering, photobleaching of stained tissue that is outside of the focal plane, and slow speed of data acquisition. Thus, it is very suitable for imaging thin tissues sections. Real-time dynamic imaging using LSCM is limited to the surface of the organ and for shorter periods of time. However, recent modifications to the standard single photon confocal microscope such as the addition of a microlens high speed spinning disk prevents cell damage and allows for rapid acquisition of imaging data of very large surfaces (approximately 870 μm × 660μm) (6). Thus, if deep tissue imaging is not required, the spinning disk confocal microscope can be very effective for performing dynamic imaging of large areas of various tissues. Several groups have recently used this technology for in vivo imaging, since it allows superior resolution (7). In a recent study, Cockburn and colleagues described the antigen-specific CD8+ T cell mediated killing of liver stage malaria parasites using a high speed spinning disk confocal microscope (7). In this case, even a superficial penetration of the laser beam was sufficient to observe the morphology of the liver parenchyma.
Compared to conventional lower wavelength and single photon excitation, the use of near-infrared two-photon (2P) excitation permits imaging of tissues at substantially greater depth (>300 μm). Moreover, the fact that the excitation of fluorescent proteins is confined to the focal plane significantly minimizes the problem of photobleaching. Consequently, by using 2P microscopy it is now possible to visualize the dynamics of immune cells in real-time, and at greater depths in intact explanted tissues or in live animals without causing overt cellular damage (8). Readily available tissues like the skin and the associated draining lymph nodes (dLN) were among the first tissues that were imaged intravitally using elegant surgical techniques (Figure 1). More recently, 2P microscopes have been modified and used to image several non-lymphoid tissues such as the lung, the intestines, the brain, and the liver (Figure 1) (9–12). 2P microscopy can also be used to visualize non-centrosymmetric structures such as collagen fibers (13). Non-linear optical effect called second harmonic generation (SHG) can be used to image collagen bundles in muscle and in bone tissues. When using a 2P laser, the emission of the SHG signal is exactly half of the excitation wavelength and can be very useful for providing structural reference of most tissues being imaged in vivo (14).
T cells are constantly moving inside and between organs, they are among the most motile cells in the body (an average of 10 μm/min, with peak velocity as high as 25 μm/min in the LN) (15). For this reason, the use of 2P microscopy has been a critical tool that has significantly increased our understanding of the dynamics of T cell responses in vivo (8, 16, 17). The disadvantages of this technique are the cost, and the limitation of the available fluorescent reporter mice or fluorescent probes.
Surgical Techniques to Study T Cell Dynamics in situ
Among the first techniques used for observing T cell dynamics in situ was the organ explant system (Figure 1A) (18). It consists of a heated imaging chamber in which an organ such as a LN is immobilized and the chamber is then perfused with heated oxygenated media. This method offers greater stability and is suitable for imaging number of lymphoid and non-lymphoid tissues (11, 15, 19–21). However, excised organs that are submerged in a media filled chamber lack major vascular innervations such lymphatics and blood vessels. Moreover, chemokine production and distribution within the organ may be completely disrupted, and thus, the milieu in the excise organ may not reflect the tissue environment that exists in vivo in live animals. Moreover, in certain situations the dynamics of T cell behavior depends on the forces exerted by the fluid circulation. The best example is leukocytes extravasation from blood circulation into the underlying tissues where shear forces play an important role (22). Thus, intravital microscopic techniques to image myriad of different organs have been developed by several investigators (an overview is shown in Figure 1B) (23–25). As noted earlier, any studies that investigate the role of chemokines in regulating T cell migration will benefit from intravital microcopy since chemokine and the cytokine milieu can change drastically after an organ is removed. However, intravital microscopy involves complicated surgical techniques that can be invasive and cause vascular damage. As a result, several controls have to be performed and the experiments have to be repeated many times. In addition, other issues associated with intravital imaging must be considered; for example, the protracted anesthesia induced unconsciousness can decrease the heart rate impacting normal levels of blood circulation and ambient body temperature (26). However, it is possible to detect vascular leakage within the tissue being imaged by the systemic injection of fluorescent quantum dots. Local body temperature can be measured by the use of portable thermometers and constant temperature can be maintained by the use of a heated stage (Figures 1C,D) (5, 24). Certain organs (i.e., thymus) within their normal bodily context simply cannot be accessed for intravital imaging. For this reason, several groups have developed transplantation methods to provide better access of the organ for imaging (27, 28). For instance, a recent report described a thymic transplant on the kidney capsule of a nude mouse (that lacked an endogenous thymus) enabling better access of the organ for intravital imaging (27). Another group designed a thoracic suction window that stabilized the lung tissue in a live mouse without overtly disrupting the function (9). Although these surgical approaches for in situ imaging of lymphocytes are technically demanding, they have greatly facilitated our ability to observe T cell behavior directly in vivo.
Visualizing Antigen-Specific T Cells in situ
The interaction of the T cell receptor (TCR) expressed on CD4+ and CD8+ T cells with a cognate peptide bound to a major histocompatibility complex (MHC) on an antigen presenting cell (APC) is essential to initiate the signaling cascade that eventually leads to T cell activation. Since, at steady state the precursor frequency of a naïve antigen-specific T cell population for a given epitope is extremely low (29, 30), adoptive transfer of labeled or congenically mismatched antigen-specific T cells isolated from TCR transgenic mice into a wild-type host has been a very useful tool for visualizing T cell dynamics in vivo (31–33). However, studies using adoptive transfer of TCR transgenic T cells are associated with certain caveats. Transfer of large numbers of naïve TCR transgenic T cells do not reflect the physiological precursor frequency, and will likely fail to mimic normal T cell responses (34). In addition, transferred TCR transgenic T cells express TCRs that exhibit identical affinity or avidity for a particular antigen, and thus, may not reflect a more physiological polyclonal endogenous T cell response to a pathogen (35). Nevertheless, in the absence of alternative technologies, in order to visualize the initial T cell activation (within minutes to hours after immunization), transfer of large numbers of TCR transgenic T cells is required.
Another major advance in detecting antigen-specific T cells was the development of MHC-multimers (36). An MHC monomer binds poorly to a specific TCR, while a multimeric MHC molecule binds stably to TCRs expressed on T cells and thus can be used effectively to stain antigen-specific TCRs allowing the detection of endogenous antigen-specific T cells. MHC class I tetramers have largely been used in flow-cytometric analysis, however, our group and others have effectively used in situ MHC-I tetramer staining for static imaging studies (37–39). Using this technique, we have previously visualized the anatomical program followed by endogenous antigen-specific CD8 T cells during a primary and a memory immune response against Listeria monocytogenes (LM) in the spleen (38). For a list of seminal publications that have contributed to the advancement of T cell imaging in situ please refer to Table 1.
Analysis and Data Interpretation
Observing the orchestrated movement of immune cells within intact organs without disrupting intricate organ structure is a powerful benefit of using 2P microscopy. However, imaging techniques described above offer a full spectrum of parameters that have to be effectively analyzed to obtain physiologically relevant and reliable data. Real-time imaging requires the acquisition of four dimensional data (x, y, z, t; time), which can be used effectively to quantify cellular dynamics such as cell–cell interactions, cellular velocity, cellular contact time, chemotactic and shape index, and much more. For a more thorough review of this topic readers should refer to previously published reviews (8, 40). However, it is noteworthy that two groups recently combined flow cytometry and in situ imaging (41, 42) to develop a novel way to analyze imaging data. The first group published the “histo-cytometry” method, which was applied to investigate dendritic cell (DC) subset localization in the LN. DCs represent a highly heterogeneous population of cells and thus it is necessary to stain for at least five markers to identify several specific subsets. Using this novel method they were able to gate on a specific DC subset and simultaneously analyze the localization of the particular population directly within the LN section (42). The second group, Moreau et al. developed the so-called “dynamic in situ cytometry” (DISC), by combining 2P imaging with direct in vivo staining by injecting Fab fragments of antibodies against cell surface molecules of interest. By converting files that represent imaging data into the regular FCS format, data were easily analyzed using a flow-cytometric software. As a consequence, cell phenotype was effectively linked to in situ cell behavior (41). In addition, another elegant method termed, the “intravital dynamics-immunosignal correlative microscopy” linked dynamic behavior of T cells with static antibody stained imaging. Chodaczek and colleagues fixed the whole tissue immediately after dynamic imaging and proceeded to stain the fixed tissue with antibodies to specific proteins. By using tissue landmarks they were able to realign the T cell movements with static immunofluorescent images, and thus, single cell dynamic behavior was effectively linked to the location of TCRs and signaling molecules in situ (43).
Visualization of T Cell Responses in Lymphoid Organs
Thymus
Early histological studies using fixed thymic sections revealed the geographical location of developing thymocytes in situ (44). It was demonstrated that double-negative (DN) thymocytes spent an average period of 14 days before becoming double-positive (DP) cells at the corticomedullary junction. In the next 3–5 days these DP cells were shown to migrate to the cortex where they interacted with the cortical thymic epithelial cells (cTEC) and underwent positive selection and matured into single-positive (SP) thymocytes. The final process of negative selection occurred in the thymic medulla where SP thymocytes interacted with the medullary thymic epithelial cells (mTECs), before exiting to the periphery (45).
Recent studies using 2P dynamic imaging have significantly increased our understanding of the T cell developmental process by defining thymocyte–APC interactions and trafficking patterns within the thymus. The first real-time imaging study utilized reaggregate thymic organ cultures to characterize the dynamics of thymocyte behavior during the process of positive selection (46). The same group further extended their findings by using thymic explants and 2P microscopy (19). They showed that T cells located in the thymic cortex exhibited the same stochastic migration pattern previously observed in the LN cortex (15), but the cells moved at a relatively low speed of 3–8 μm/min. However, after undergoing positive selection, T cells exhibited significantly higher motility of 10–25 μm/min as they migrated toward the medulla. Since the thymic medulla is located at greater depth, dynamic 2P imaging of this region required the use of vibratome-cut thymic explants (47). Intriguingly, SP thymocytes undergoing negative selection were confined to a specific area of the thymic medullary region and exhibited an average velocity of 10 μm/min. By using a mouse model that expressed the ovalbumin (OVA) antigen in mTECs, Le Borgne et al. visualized the process of negative selection of OVA specific TCR transgenic CD8 T cells (OTI) in situ, and showed that negatively selected T cells surprisingly continued to stay motile but were confined to a restricted area of the medulla. This observation implied that SP thymocytes needed continuous cellular interactions and integrations of signals before undergoing apoptosis (47).
Lymph Node
The emerging data indicate that mounting a protective immune response against pathogens or tumors is critically dependent on the orchestrated movement of cells within lymphoid organs. The lymph node structure is one of the underlying regulators of immune responses against mucosal infections or following vaccination by promoting interactions between different cell types. Thus, understanding the dynamics of T cell behavior in situ within the LN is essential. Skin draining LN can be accessed for intravital imaging, therefore, several previous studies have reported intranodal T cell behavior in several different contexts (48).
Naïve T cell trafficking
T cell trafficking patterns even under steady state conditions is a highly regulated process and several recent elegant studies have helped illuminate the processes that control this complex behavior of T cells in situ. Naïve T cells access the LN via the blood and enter the LN cortex through the high endothelial venules (HEV). Once in the cortex, T cells scan the DC networks (49) for antigen as they follow the fibroblastic reticular network within the LN (50). Factors that govern this migration pattern are not completely understood (48); however, G-protein coupled receptors (GPCRs) are very important in regulating this process. By abrogating global GPCRs signaling with pertussis toxin (PTX) treatment, Cyster and colleagues showed that PTX treated T cells showed a 50% reduction in median velocity, and a 90% decrease in mean motility coefficient in the LNs when compared to untreated T cells (51). Among the different GPCR, CCR7 was shown to be important for the localization of T cells in the paracortex. Indeed, modification of the CCR7 ligand (CCL19 and CCL21) distribution by subcutaneous injection distracted the lymphocytes from the T cell zone (52). The absence of the CCR7 signaling on naïve T cells significantly reduced the intranodal normal T cell velocity. However, this deficiency did not introduce any directional biases (52, 53), and thus, the “random walk” behavior exhibited by T cells was unchanged. Although both ligands for CCR7, CCL21, and CCL19 are produced by FRCs (54) only surface bound CCL21 is require for the random T cell motility (55) within the LN. These observations suggest that T cells follow a haptotactic (immobilized ligand) instead of a chemotactic (soluble ligand) gradient.
Egress of naïve T cells is also regulated by GPCRs. Upon LN entry, naïve T cells spend on average 6–12 h in the LN cortex, before using the cortical lymphatic sinuses to exit the LNs. Cyster and colleagues elegantly visualized this process, and showed that naïve T cells first probed the cortical lymphatic sinuses, and subsequently entered the lymphatic vessels by a sphingosine-1-phosphate receptor-1 (S1PR1) dependent mechanism (56). The S1PR1 ligand shingosine-1-phosphate (S1P) is present at high concentrations in the blood and in the lymph, but virtually absent in the tissues due to the degradation by the enzyme S1P lyase (57). S1PR1 is rapidly desensitized after S1P ligation, thus newly arrived T cells in the LN cortex fail to express the receptor on their surface. Once S1PR1 is recycled back to the surface, T cells are able to respond to the S1P gradient and return back to the circulation following exit from the LNs.
Naïve T cell priming
Visualizing T cell activation in situ has considerably enhanced our understanding of the mechanisms that regulate T cell and APC interactions in vivo. Three major experimental models have been utilized to image T cell priming; subcutaneous antigen delivery coupled with adjuvant, transfer of antigen pulsed DCs or direct infection of animals with pathogens.
Early 2P microscopy studies using antigen pulsed DCs to prime T cells revealed the dynamics of T cells–DC interactions in situ (33, 58) during antigen presentation. Antigen-specific T cells formed protracted interactions with DCs that lasted not minutes but hours (33, 58). Mempel et al. described a more complex process that characterized the dynamics of T cell activation. They demonstrated that T cell priming occurred in distinct phases where initial repeated brief encounters with DCs was followed by long-lived stable DC–T cell conjugates that in some instances lasted for more than 20 h in the LNs (33). The stability of these interactions depends on the antigen dose and TCR–MHC affinity (59, 60). Chemokines are also important for promoting T cell–APC interactions. CD4 T cell that are deficient in CXCR3 display fewer and shorter interactions with DCs that expressed the cognate ligand CXCL10, which resulted in poor Th1 differentiation as well as misplaced intranodal migration of primed CD4 T cells (61). In addition, collaboration between lymphocyte subsets was shown to facilitate antigen recognition of rare antigen presenting DCs at early stages of an immune response in the LN, resulting in non-random cell–cell interactions (62). Early during an immune response, the interaction of CD4 T cells with antigen bearing DCs resulted in the production of chemokines CCL3 and CCL4. This in turn lead to the recruitment of CCR5 expressing CD8 T cells to these rare sites of antigen depot in the LN, allowing for optimized T cell priming and memory cell generation (62).
Thus, the emerging data suggest that the “dwell time” (the length of initial T cell–DC conjugate formation) and the subsequent T cell motility and migration within defined compartments of secondary lymphoid organs are important parameters directing optimal T cell activation in vivo and these parameters are sensitive in situ indicators of antigen recognition.
In limited number of cases T cell dynamics in secondary lymphoid organs has been investigated following an infection. Although imaging studies using simple antigens have improved our knowledge regarding the mechanics of T cell activation, observing T cell responses to live replicating pathogens adds new layers of complexity. Since naïve T cells are largely located in the LN cortex where they continuously scan DCs, it was tempting to assume that following infection, T cell priming would occur deep in the LN cortex. However, in vivo visualization of early T cell–APC interactions immediately following viral infections revealed that T cell priming in fact occurred near the cortical ridge or at the interfollicular region of the dLN (32, 63–65). Subcutaneous infection with vaccinia virus or vesicular stomatitis virus (VSV) resulted in the infection of macrophages and DCs present in the LN subcapsular sinus (SCS), however, only DCs that expressed virus encoded protein appeared to present antigen directly to transferred TCR transgenic CD8 T cells (32, 63, 65). Interestingly, acquisition of effector functions and proper differentiation of primed CD8 T cells during the later phases of the T cell priming process depends on T cell–T cell interactions rather than just the early DC–T cell interactions (66). Integrin (CD11a) mediated T cell homotypic interactions was shown to be essential for the ability of antigen-specific CD8 T cells to secrete interferon-γ and subsequently differentiate into memory T cells following infection.
Following infection, effector T cells proliferate briefly in the LNs before exiting via the efferent lymphatics into the circulation. Most studies have focused on early events using TCR transgenic adoptive transfer methods but the subsequent intranodal migration of newly primed T cells, or the mechanisms that drive the egress kinetics from the LN following a localized infection have not been investigated adequately. However, a recent study used antigen pulsed DCs to immunize mice and demonstrated that stromal cells and DCs in the interfollicular area express CXCL9 and CXCL10, respectively, and attract newly primed CXCR3 expressing CD4 T cells to the interfollicular and medullary areas of the LN (61). In this case, cell–cell interactions in the periphery of the LN were important for the proper intranodal T cell positioning as well as adequate Th1 cell differentiation.
Thus, these observations clearly demonstrate the complexity of the T cell activation process that requires several types of dynamic interactions between immune as well as stromal cells. Clarification of these elaborate processes has only been made possible by the use of in situ imaging methods.
Central memory T cells
Following the resolution of an acute infection, a heterogeneous population of memory T cells are generated that exhibit differential tissue tropism (67–70). Central memory T cells (TCM) that express the lymphoid homing receptors CCR7 and CD62L preferentially localize to the secondary lymphoid organs, while effector memory T cells (TEM) that fail to express lymph node homing receptors migrate to non-lymphoid tissues where they may upregulate CD69 and CD103 and form a stable resident memory T cell population. Central memory CD4+ and CD8+ T cells (64, 71) are largely located in the periphery of the LNs in close proximity to the lymphatic sinuses where a potential re-infection may occur. This peripheralization was shown to be dependent on the expression of the chemokine receptor CXCR3. Thus, this “pre-positioning” of memory T cells allowed for a more rapid response to a challenge infection.
Spleen
Following a blood borne infection, the spleen plays an essential role in the initiation of an anti-microbial immune response. A systemic infection with the intracellular Gram-positive bacteria LM is a widely used model to study the immune responses in this important lymphoid organ. Contrary to the LN, the spleen is more difficult to access for intravital imaging. Moreover, light scattering and absorption by red blood cells makes multi-photon microscopy of the spleen challenging. Additionally, the splenic white pulp, which plays host to T and B cells, is too deep to be imaged directly by 2P microscopy. However, using a standard confocal microscope the splenic red pulp was imaged intravitally following LM infection (72). During early time points after infection, DCs in the splenic red pulp established static LM depots, which were subsequently swarmed by neutrophils and monocytes, as well as CD8+ T cells (72). At later times after infection, infected DCs migrate to the splenic white pulp where they form stable interactions with CD8+ T cells, resulting in T cell activation (21, 73).
Using in situ tetramer staining and static whole-mount confocal microscopy with thick spleen sections, we have previously mapped the entire anatomical program followed by endogenous antigen-specific CD8+ T cells in the spleen after a primary and secondary LM infection (38). At day 3 after infection tetramer positive (tet+) cells were readily detected in small clusters at the T/B cell border and in the splenic marginal zone in close contact with DCs. After a brief period of expansion in the T cell zones, by 6 days after infection, virtually all antigen-specific effector CD8+ T cells had exited the white pulp using the bridging channels (lymphatic vessels are absent in the spleen). Similar to what was observed in the LN (64, 71), we found that early memory T cells were preferentially localized to the periphery of the splenic T cell zones and surprisingly even in the B cell follicles. Interestingly, another study showed that memory CD8 T cells beyond 55 days post LCMV infection were primarily located in splenic T cells zones (74), suggesting that as memory T cells mature their anatomical localization may change. This process will likely depend on the differential expression of homing and chemokine receptors that are involved in memory cell localization and migration.
Imaging T Cell Responses in Non-Lymphoid Organs
Following an infection, effector T cells, apart from proliferating in lymphoid organs, also migrate to peripheral inflamed tissues via the vasculature. Several adhesion molecules and chemokines are involved in the migration and entry of effector T cells into peripheral tissues. Receptors like E-selectin and P-selectin expressed on endothelial cells within tissues and molecules like CD44, P-selectin glycoprotein ligand 1 (PSGL1) and α4β1 integrin on antigen-challenged T cells enable non-selective localization of T cells to various peripheral organs (75). However, tissue-specific chemokines and receptors expressed on T cells specifically direct the homing of activated T cells to a particular non-lymphoid organ such as the skin, brain, or the gut (76). E-selectin ligands and the chemokine receptors CCR4 and CCR10 direct T cell homing specifically to the skin, while CXCL10 and CXCR3 regulate recruitment to the brain (11). At mucosal sites such as the intestinal mucosa, DCs in the Peyer’s patches or mesenteric LNs imprint T cells to specifically migrate to the intestinal tissue (77). These mucosal activated T cells upregulate CCR9 and α4β7, which allow the T cells to home to the gut mucosa. α4β7 binds to the mucosal addressin cell adhesion molecule-1 (MADCAM-1), which is expressed heavily on vascular endothelial cells in the intestinal tissue (78), while CCL25 (CCR9 ligand) is produced by the intestinal epithelial cells (79).
Effector T Cells
Once effector T cells enter peripheral tissues, they search for their cognate antigen, which is presented in context with MHC molecules by APCs or tissue stromal cells. Advanced imaging techniques like intravital imaging and multi-photon microscopy have attempted to shed light on the behavior of T cells navigating peripheral tissues (11, 16, 80). 2P imaging in the brain showed CD8+ T cells undergoing a non-random, directed mode of migration (11). The authors speculated that this unusual pattern of migration called a “Levy walk” enabled T cells to find rare APCs faster. In this particular study, the observed Levy walk helped T cells control Toxoplasma gondii infection in the brains of infected mice. Analysis of cell motility in non-lymphoid tissues revealed that T cells move at an average velocity that is significantly lower than their counterparts in the LNs. Intravital imaging in the lung was used to image naïve and activated T cells. Naïve T cells maintained an average track speed of around 2.5 μm/s, whereas activated T cells moved at a substantially lower speed of 0.4 μm/s (9). In the liver, granuloma formation following infection with Mycobacterium bovis (BCG) has been used as a model system to image the dynamics of innate and adaptive immune cells (12, 81). The study revealed fast but restricted movement of CD4+ T cells inside granulomas (12). The same group later demonstrated that antigen presentation within granulomas determined T cell motility and cytokine production (81), which in turn influenced pathogen clearance. Thus, it is clear that tissue microenvironment likely influences T cell motility. There are several factors that can influence the differential dynamic behavior of T cells in non-lymphoid tissues. These factors include but not limited to: (i) nature of the replicating pathogen with regards to cellular tropism and the inflammatory milieu the pathogen may induce; (ii) the production of chemokines and their receptors expressed on T cells (62, 82); and (iii) the tissue resident cells including stromal or APCs that T cells may interact with (83). Determining the role each of these factors play in regulating T cell dynamics in non-lymphoid tissues will be essential for closing the large gaps that exist in our knowledge regarding T cell behavior in non-lymphoid tissues.
Non-T cell populations like neutrophils and DCs in the lung and DCs in the gut lumen have also been studied using 2P imaging (9, 84–88). However, as the focus here is on T cells, a detailed review of non-T cell populations is beyond the scope of this article.
Memory T Cells
After effector T cells perform their function of antigen recognition, interaction, and clearance, a subpopulation of T cells persist as memory cells. A hallmark of protective immunity is that these cells help mount a rapid immune response following a secondary infection. As noted earlier, memory cells can be divided into TCM and TEM cells. TCM re-circulate predominantly between secondary lymphoid organs and the blood, whereas TEM re-circulate through peripheral non-lymphoid tissues (68). A third category of memory cells are called resident memory cells (TRM) that remain in peripheral tissues and fail to re-circulate (89–93). Understanding the receptors, chemokines, and other factors that influence formation of a TRM population in non-lymphoid tissues is an area of active research. Intra-epithelial lymphocytes (IELs), the TRM cells in the gut epithelium, downregulate expression of homing receptors α4β7 and CCR9 and upregulate CD69 and CD103 to establish residence (89, 94). Decrease in the expression of KLF2, the transcriptional activator of the gene that codes for S1PR1 was recently shown to be important for establishment of resident memory CD8+ T cells (95) in non-lymphoid tissues. Since S1PR1 plays an important role in naïve T cell egress from lymph nodes (56), transcriptional downregulation of this gene for the generation of resident memory population is an important and interesting observation. 2P microscopy has helped address previously unappreciated questions about the nuances of memory T cell dynamics in non-lymphoid tissues following infection. After HSV infection in the skin, memory CD4+ and CD8+ T cell populations adopt different rates of migration based on tissue localization. CD8+ memory T cells remain in the epidermis close to the initial site of infection and migrate at a slower rate (2–3 μm/min) than dermis-localized CD4+ T memory cells (5–6 μm/min) (96). Interestingly, CD4+ T cells could re-enter the blood circulation, whereas CD8+ cells did not, suggesting that CD4 and CD8 T cells are regulated differently with regards to tissue residence. The mechanisms responsible for these differences are not known. As a follow-up, recent published work using multi-photon imaging and computer modeling showed that the slower velocities of CD8+ TRM cells in the epidermis enabled these cells to remain on site throughout the life of the mouse (97). The observed cell velocities obtained in vivo over 2–3 h were used to model migration patterns over longer periods of time. The mathematical modeling showed that percentages of CD8+ T cells within a 5-μm region of skin at day 100 (49.8%) and day 365 (49.4%) after infection were not significantly different from day 0 (50%). Furthermore, two-photon imaging also demonstrated that the CD8+ TRM population interacted with and were influenced by other cell populations like Langerhans cells (LCs) and dendritic epidermal γδ T cells in the epidermis.
Autoimmunity, Tissue Rejection, and Tumor Immunity
The immune system can sometimes backfire, leading to autoimmunity and medical complications like graft rejection. Using mouse models and novel imaging tools, efforts are being made to understand these processes better. In a diabetic mouse model, 2P microscopic study showed that cytotoxic T lymphocytes (CTLs) underwent random walks in the exocrine tissues of the pancreas. On encountering β-cells in the pancreatic islets, the CTL motility slowed and led to the eventual death of β-cells, causing diabetes (98). In experimental autoimmune encephalomyelitis (EAE), a mouse model for multiple sclerosis, a molecular sensor that combines fluorescent nuclear factor of activated T cells (NFAT) with the histone protein H2B was used to detect intravital T cell activation (99). In activated T cells, NFAT is dephosphorylated and translocates into the nucleus. Tagging NFAT with fluorescent histone 2B enables simultaneously tracking NFAT nuclear translocation and cell mitosis, a powerful tool to follow activation of individual T cells in vivo following CNS invasion during development of EAE. Initially, T cells that traversed the leptomeningeal blood vessels in the CNS were activated following transient contacts with resident macrophages. During disease progression, activated T cells spread to the CNS parenchyma. In another study, the step-wise host tissue destruction was studied in a mouse model of allograft rejection, where ear skin grafts were imaged using intravital 2P microscopy (100). Donor dermal DCs were destroyed within 3–5 days after transplant. Rapid infiltration of host CD11b+ cells, initially neutrophils replaced by monocytes, could act as APCs that transported antigen from graft to the dLN, thereby activating CD8+ T cells. The above-mentioned two studies, therefore, demonstrated how intravital imaging and multi-photon microscopy could provide crucial temporal information on important processes like disease progression and graft rejection.
T cell homing and infiltration of tumors play an important role in improved cancer prognosis. Receptors like CXCR3, CCR5, CCR4, and adhesion molecules like intercellular adhesion molecule-1 (ICAM-1) have been implicated in T cell infiltration to various types of tumors (101, 102). Cytotoxic T lymphocytes (CTLs) infiltrate tumors and mediate tumor cell destruction. Multi-photon microscopy has been used effectively to image the dynamics of CTLs within solid tumors. In a mouse thymoma model, E.G7-OVA tumor cells (modified to express CD8 T cell antigen OVA) were injected into mice subcutaneously (s.c), and 2P microscopy was utilized to assess OVA specific CD8 T cells (OTI) dynamics during early and late phases of tumor rejection. OTI arrest coefficient was higher, while the mean velocities were decreased in the OVA-expressing tumors during early stage of rejection, when compared to non-OVA bearing tumors, suggesting antigen-specific recognition by CD8 T cells was necessary for T cell arrest and the eventual destruction of tumor cells. (103). To better understand if CD8 T cells directly mediate tumor cell lysis, a combination of 2P microscopy and a fluorescence resonance energy transfer (FRET) based caspase-3 activity reporter system was used to track apoptosis of tumor cells following adoptive transfer of activated CD8 T cells (104). Indeed, this study revealed that CD8 T cells directly kill individual tumor cells, however, the time required for tumor cell lysis was unusually lengthy (6 h on average), which may explain why T cell mediated tumor regression is often inefficient. Other immune cell types that target tumor antigens, like natural killer (NK) cells, have also been imaged using multi-photon microscopy and were shown to have very different dynamics as compared to CTLs (105).
Real-time in situ imaging of tumor tissue has also shed light on potential mechanisms that can restrict CTLs from carrying out their antitumor function within solid tumors. Myeloid-derived suppressor cells (MDSCs) are heterogeneous populations of APCs such as tumor dendritic cells (TuDCs) or tumor-associated macrophages (TAMs), which can subvert the antitumor activity of CTLs (106). 2P imaging of tumor tissue showed that following chemotherapy, tumor-infiltrating lymphocytes (TILs) made increased contacts with TuDCs but were trapped in TuDC-rich areas in the tumor parenchyma, restricting their infiltration deeper into tumor tissue (107). Furthermore, in a mouse model of adoptive T cell transfer, intratumoral regulatory T cells (Tregs) in an antigen-dependent manner, induced a functional anergic state in tumor-infiltrating CTLs, which resulted in poor tumor regression (108). Thus, using 2P microscopy to image solid tumors has been a valuable tool for gaining critical information regarding the dynamic behavior of TILs in vivo.
Conclusion
Two-photon microscopy and intravital imaging have helped make significant strides in increasing our understanding of the spatial and temporal behavior of T cells and APCs in situ during an immune response and subsequent steps of memory cell generation. With lesser restraints on the type and thickness of peripheral live tissues that can be imaged in real-time, the possibilities are limitless to address further unanswered questions regarding the biology of the immune response to a tumor or following an infection, and thus the development of effective vaccines and therapeutics.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors’ work is supported by the National Institutes of Health (grants RO1AI097375 and RO1AI041576). The authors apologize to those colleagues whose work was not cited as a result of space constraints.
References
1. Wagner R. Erläuterungstafeln zur Physiologie und Entwicklungsgeschichte. Leipzig: Leopold, Voss Leipzig (1839).
3. von Andrian UH. Intravital microscopy of the peripheral lymph node microcirculation in mice. Microcirculation (1996) 3:287–300. doi: 10.3109/10739689609148303
4. Murooka TT, Mempel TR. Multiphoton intravital microscopy to study lymphocyte motility in lymph nodes. Methods Mol Biol (2012) 757:247–57. doi:10.1007/978-1-61779-166-6_16
5. Arnon TI, Horton RM, Grigorova IL, Cyster JG. Visualization of splenic marginal zone B-cell shuttling and follicular B-cell egress. Nature (2013) 493:684–8. doi:10.1038/nature11738
6. Thiberge S, Blazquez S, Baldacci P, Renaud O, Shorte S, Menard R, et al. In vivo imaging of malaria parasites in the murine liver. Nat Protoc (2007) 2:1811–8. doi:10.1038/nprot.2007.257
7. Cockburn IA, Amino R, Kelemen RK, Kuo SC, Tse SW, Radtke A, et al. In vivo imaging of CD8+ T cell-mediated elimination of malaria liver stages. Proc Natl Acad Sci U S A (2013) 110:9090–5. doi:10.1073/pnas.1303858110
8. Cahalan MD, Parker I. Choreography of cell motility and interaction dynamics imaged by two-photon microscopy in lymphoid organs. Annu Rev Immunol (2008) 26:585–626. doi:10.1146/annurev.immunol.24.021605.090620
9. Looney MR, Thornton EE, Sen D, Lamm WJ, Glenny RW, Krummel MF. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods (2011) 8:91–6. doi:10.1038/nmeth.1543
10. McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature (2012) 483:345–9. doi:10.1038/nature10863
11. Harris TH, Banigan EJ, Christian DA, Konradt C, Tait Wojno ED, Norose K, et al. Generalized Levy walks and the role of chemokines in migration of effector CD8+ T cells. Nature (2012) 486:545–8. doi:10.1038/nature11098
12. Egen JG, Rothfuchs AG, Feng CG, Winter N, Sher A, Germain RN. Macrophage and T cell dynamics during the development and disintegration of mycobacterial granulomas. Immunity (2008) 28:271–84. doi:10.1016/j.immuni.2007.12.010
13. Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods (2005) 2:932–40. doi:10.1038/nmeth818
14. Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol (2003) 21:1369–77. doi:10.1038/nbt899
15. Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science (2002) 296:1869–73. doi:10.1126/science.1070051
16. Germain RN, Robey EA, Cahalan MD. A decade of imaging cellular motility and interaction dynamics in the immune system. Science (2012) 336:1676–81. doi:10.1126/science.1221063
17. Munoz MA, Biro M, Weninger W. T cell migration in intact lymph nodes in vivo. Curr Opin Cell Biol (2014) 30C:17–24. doi:10.1016/j.ceb.2014.05.002
18. Dzhagalov IL, Melichar HJ, Ross JO, Herzmark P, Robey EA. Two-photon imaging of the immune system. Curr Protoc Cytom (2012) Chapter 12:Unit12. doi:10.1002/0471142956.cy1226s60
19. Witt CM, Raychaudhuri S, Schaefer B, Chakraborty AK, Robey EA. Directed migration of positively selected thymocytes visualized in real time. PLoS Biol (2005) 3:e373. doi:10.1371/journal.pbio.0030373
20. Veres TZ, Voedisch S, Spies E, Tschernig T, Braun A. Spatiotemporal and functional behavior of airway dendritic cells visualized by two-photon microscopy. Am J Pathol (2011) 179:603–9. doi:10.1016/j.ajpath.2011.04.039
21. Aoshi T, Zinselmeyer BH, Konjufca V, Lynch JN, Zhang X, Koide Y, et al. Bacterial entry to the splenic white pulp initiates antigen presentation to CD8+ T cells. Immunity (2008) 29:476–86. doi:10.1016/j.immuni.2008.06.013
22. Rosen SD. Ligands for L-selectin: homing, inflammation, and beyond. Annu Rev Immunol (2004) 22:129–56. doi:10.1146/annurev.immunol.21.090501.080131
23. Schramm R, Schafers HJ, Harder Y, Schmits R, Thorlacius H, Menger MD. The cervical lymph node preparation: a novel approach to study lymphocyte homing by intravital microscopy. Inflamm Res (2006) 55:160–7. doi:10.1007/s00011-006-0066-0
24. Mempel TR. Single-cell analysis of cytotoxic T cell function by intravital multiphoton microscopy. Methods Mol Biol (2010) 616:181–92. doi:10.1007/978-1-60761-461-6_12
25. Sellers SL, Payne GW. Intravital microscopy of the inguinal lymph node. J Vis Exp (2011) 50:2551. doi:10.3791/2551
26. Hart CY, Burnett JC Jr, Redfield MM. Effects of avertin versus xylazine-ketamine anesthesia on cardiac function in normal mice. Am J Physiol Heart Circ Physiol (2001) 281:H1938–45.
27. Caetano SS, Teixeira T, Tadokoro CE. Intravital imaging of the mouse thymus using 2-photon microscopy. J Vis Exp (2012) 59:e3504. doi:10.3791/3504
28. Lawton JC, Benson RA, Garside P, Brewer JM. Using lymph node transplantation as an approach to image cellular interactions between the skin and draining lymph nodes during parasitic infections. Parasitol Int (2014) 63:165–70. doi:10.1016/j.parint.2013.07.010
29. Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity (2007) 27:203–13. doi:10.1016/j.immuni.2007.07.007
30. Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity (2008) 28:859–69. doi:10.1016/j.immuni.2008.04.010
31. Kearney ER, Pape KA, Loh DY, Jenkins MK. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity (1994) 1:327–39. doi:10.1016/1074-7613(94)90084-1
32. Norbury CC, Malide D, Gibbs JS, Bennink JR, Yewdell JW. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat Immunol (2002) 3:265–71. doi:10.1038/ni762
33. Mempel TR, Henrickson SE, von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature (2004) 427:154–9. doi:10.1038/nature02238
34. Marzo AL, Klonowski KD, Le Bon A, Borrow P, Tough DF, Lefrancois L. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat Immunol (2005) 6:793–9. doi:10.1038/ni1227
35. Hao Y, Legrand N, Freitas AA. The clone size of peripheral CD8 T cells is regulated by TCR promiscuity. J Exp Med (2006) 203:1643–9. doi:10.1084/jem.20052174
36. Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science (1996) 274:94–6. doi:10.1126/science.274.5284.94
37. Skinner PJ, Daniels MA, Schmidt CS, Jameson SC, Haase AT. Cutting edge: in situ tetramer staining of antigen-specific T cells in tissues. J Immunol (2000) 165:613–7. doi:10.4049/jimmunol.165.2.613
38. Khanna KM, McNamara JT, Lefrancois L. In situ imaging of the endogenous CD8 T cell response to infection. Science (2007) 318:116–20. doi:10.1126/science.1146291
39. Khanna KM, Bonneau RH, Kinchington PR, Hendricks RL. Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity (2003) 18:593–603. doi:10.1016/S1074-7613(03)00112-2
40. Matheu MP, Cahalan MD, Parker I. Immunoimaging: studying immune system dynamics using two-photon microscopy. Cold Spring Harb Protoc (2011) 2011(2): pdb.top99. doi:10.1101/pdb.top99
41. Moreau HD, Lemaitre F, Terriac E, Azar G, Piel M, Lennon-Dumenil AM, et al. Dynamic in situ cytometry uncovers T cell receptor signaling during immunological synapses and kinapses in vivo. Immunity (2012) 37:351–63. doi:10.1016/j.immuni.2012.05.014
42. Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity (2012) 37:364–76. doi:10.1016/j.immuni.2012.07.011
43. Chodaczek G, Papanna V, Zal MA, Zal T. Body-barrier surveillance by epidermal gammadelta TCRs. Nat Immunol (2012) 13:272–82. doi:10.1038/ni.2240
44. Anderson G, Moore NC, Owen JJ, Jenkinson EJ. Cellular interactions in thymocyte development. Annu Rev Immunol (1996) 14:73–99. doi:10.1146/annurev.immunol.14.1.73
45. Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol (2007) 25:649–79. doi:10.1146/annurev.immunol.23.021704.115715
46. Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science (2002) 296:1876–80. doi:10.1126/science.1070945
47. Le BM, Ladi E, Dzhagalov I, Herzmark P, Liao YF, Chakraborty AK, et al. The impact of negative selection on thymocyte migration in the medulla. Nat Immunol (2009) 10:823–30. doi:10.1038/ni.1761
48. Worbs T, Bernhardt G, Forster R. Factors governing the intranodal migration behavior of T lymphocytes. Immunol Rev (2008) 221:44–63. doi:10.1111/j.1600-065X.2008.00580.x
49. Lindquist RL, Shakhar G, Dudziak D, Wardemann H, Eisenreich T, Dustin ML, et al. Visualizing dendritic cell networks in vivo. Nat Immunol (2004) 5:1243–50. doi:10.1038/ni1139
50. Bajenoff M, Egen JG, Koo LY, Laugier JP, Brau F, Glaichenhaus N, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity (2006) 25:989–1001. doi:10.1016/j.immuni.2006.10.011
51. Cyster JG. Specifying the patterns of immune cell migration. Novartis Found Symp (2007) 281:54–61. doi:10.1002/9780470062128.ch6
52. Worbs T, Mempel TR, Bolter J, von Andrian UH, Forster R. CCR7 ligands stimulate the intranodal motility of T lymphocytes in vivo. J Exp Med (2007) 204:489–95. doi:10.1084/jem.20061706
53. Sperti-Boursin F, Real E, Bismuth G, Trautmann A, Donnadieu E. CCR7 ligands control basal T cell motility within lymph node slices in a phosphoinositide 3-kinase-independent manner. J Exp Med (2007) 204:1167–79. doi:10.1084/jem.20062079
54. Malhotra D, Fletcher AL, Astarita J, Lukacs-Kornek V, Tayalia P, Gonzalez SF, et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat Immunol (2012) 13:499–510. doi:10.1038/ni.2262
55. Woolf E, Grigorova I, Sagiv A, Grabovsky V, Feigelson SW, Shulman Z, et al. Lymph node chemokines promote sustained T lymphocyte motility without triggering stable integrin adhesiveness in the absence of shear forces. Nat Immunol (2007) 8:1076–85. doi:10.1038/ni1499
56. Grigorova IL, Schwab SR, Phan TG, Pham TH, Okada T, Cyster JG. Cortical sinus probing, S1P1-dependent entry and flow-based capture of egressing T cells. Nat Immunol (2009) 10:58–65. doi:10.1038/ni.1682
57. Schwab SR, Cyster JG. Finding a way out: lymphocyte egress from lymphoid organs. Nat Immunol (2007) 8:1295–301. doi:10.1038/ni1545
58. Bousso P, Robey E. Dynamics of CD8(+) T cell priming by dendritic cells in intact lymph nodes. Nat Immunol (2003) 4:579–85. doi:10.1038/ni928
59. Henrickson SE, Mempel TR, Mazo IB, Liu B, Artyomov MN, Zheng H, et al. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat Immunol (2008) 9:282–91. doi:10.1038/ni1559
60. Shakhar G, Lindquist RL, Skokos D, Dudziak D, Huang JH, Nussenzweig MC, et al. Stable T cell-dendritic cell interactions precede the development of both tolerance and immunity in vivo. Nat Immunol (2005) 6:707–14. doi:10.1038/ni1210
61. Groom JR, Richmond J, Murooka TT, Sorensen EW, Sung JH, Bankert K, et al. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity (2012) 37:1091–103. doi:10.1016/j.immuni.2012.08.016
62. Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature (2006) 440:890–5. doi:10.1038/nature04651
63. Hickman HD, Takeda K, Skon CN, Murray FR, Hensley SE, Loomis J, et al. Direct priming of antiviral CD8+ T cells in the peripheral interfollicular region of lymph nodes. Nat Immunol (2008) 9:155–65. doi:10.1038/ni1557
64. Kastenmuller W, Brandes M, Wang Z, Herz J, Egen JG, Germain RN. Peripheral prepositioning and local CXCL9 chemokine-mediated guidance orchestrate rapid memory CD8+ T cell responses in the lymph node. Immunity (2013) 38:502–13. doi:10.1016/j.immuni.2012.11.012
65. Hickman HD, Li L, Reynoso GV, Rubin EJ, Skon CN, Mays JW, et al. Chemokines control naive CD8+ T cell selection of optimal lymph node antigen presenting cells. J Exp Med (2011) 208:2511–24. doi:10.1084/jem.20102545
66. Gerard A, Khan O, Beemiller P, Oswald E, Hu J, Matloubian M, et al. Secondary T cell-T cell synaptic interactions drive the differentiation of protective CD8+ T cells. Nat Immunol (2013) 14:356–63. doi:10.1038/ni.2547
67. Hamann D, Baars PA, Rep MH, Hooibrink B, Kerkhof-Garde SR, Klein MR, et al. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med (1997) 186:1407–18. doi:10.1084/jem.186.9.1407
68. Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature (1999) 401:708–12. doi:10.1038/44385
69. Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, Woodland DL. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med (2007) 204:1625–36. doi:10.1084/jem.20070322
70. Masopust D, Vezys V, Marzo AL, Lefrançois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science (2001) 291:2413–7. doi:10.1126/science.1058867
71. Sung JH, Zhang H, Moseman EA, Alvarez D, Iannacone M, Henrickson SE, et al. Chemokine guidance of central memory T cells is critical for antiviral recall responses in lymph nodes. Cell (2012) 150:1249–63. doi:10.1016/j.cell.2012.08.015
72. Waite JC, Leiner I, Lauer P, Rae CS, Barbet G, Zheng H, et al. Dynamic imaging of the effector immune response to Listeria infection in vivo. PLoS Pathog (2011) 7:e1001326. doi:10.1371/journal.ppat.1001326
73. Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, KC W, et al. CD8alpha(+) dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity (2011) 35:236–48. doi:10.1016/j.immuni.2011.06.012
74. Dauner JG, Williams IR, Jacob J. Differential microenvironment localization of effector and memory CD8 T cells. J Immunol (2008) 180:291–9. doi:10.4049/jimmunol.180.1.291
75. Woodland DL, Kohlmeier JE. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol (2009) 9:153–61. doi:10.1038/nri2496
76. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol (2014) 32:659–702. doi:10.1146/annurev-immunol-032713-120145
77. Mora JR, Bono MR, Manjunath N, Weninger W, Cavanagh LL, Rosemblatt M, et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature (2003) 424:88–93. doi:10.1038/nature01726
78. Streeter PR, Berg EL, Rouse BT, Bargatze RF, Butcher EC. A tissue-specific endothelial cell molecule involved in lymphocyte homing. Nature (1988) 331:41–6. doi:10.1038/331041a0
79. Wurbel MA, Malissen M, Guy-Grand D, Malissen B, Campbell JJ. Impaired accumulation of antigen-specific CD8 lymphocytes in chemokine CCL25-deficient intestinal epithelium and lamina propria. J Immunol (2007) 178:7598–606. doi:10.4049/jimmunol.178.12.7598
80. Mueller SN. Effector T-cell responses in non-lymphoid tissues: insights from in vivo imaging. Immunol Cell Biol (2013) 91:290–6. doi:10.1038/icb.2012.75
81. Egen JG, Rothfuchs AG, Feng CG, Horwitz MA, Sher A, Germain RN. Intravital imaging reveals limited antigen presentation and T cell effector function in mycobacterial granulomas. Immunity (2011) 34:807–19. doi:10.1016/j.immuni.2011.03.022
82. Castellino F, Germain RN. Cooperation between CD4+ and CD8+ T cells: when, where, and how. Annu Rev Immunol (2006) 24:519–40. doi:10.1146/annurev.immunol.23.021704.115825
83. Heath WR, Carbone FR. The skin-resident and migratory immune system in steady state and memory: innate lymphocytes, dendritic cells and T cells. Nat Immunol (2013) 14:978–85. doi:10.1038/ni.2680
84. Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med (2006) 203:2841–52. doi:10.1084/jem.20061884
85. GeurtsvanKessel CH, Willart MA, van Rijt LS, Muskens F, Kool M, Baas C, et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. J Exp Med (2008) 205:1621–34. doi:10.1084/jem.20071365
86. McGill J, van RN, Legge KL. Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. J Exp Med (2008) 205:1635–46. doi:10.1084/jem.20080314
87. Kreisel D, Nava RG, Li W, Zinselmeyer BH, Wang B, Lai J, et al. In vivo two-photon imaging reveals monocyte-dependent neutrophil extravasation during pulmonary inflammation. Proc Natl Acad Sci U S A (2010) 107:18073–8. doi:10.1073/pnas.1008737107
88. Thornton EE, Looney MR, Bose O, Sen D, Sheppard D, Locksley R, et al. Spatiotemporally separated antigen uptake by alveolar dendritic cells and airway presentation to T cells in the lung. J Exp Med (2012) 209:1183–99. doi:10.1084/jem.20112667
89. Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med (2010) 207:553–64. doi:10.1084/jem.20090858
90. Mackay LK, Stock AT, Ma JZ, Jones CM, Kent SJ, Mueller SN, et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci U S A (2012) 109:7037–42. doi:10.1073/pnas.1202288109
91. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol (2013) 14:1294–301. doi:10.1038/ni.2744
92. Carbone FR, Mackay LK, Heath WR, Gebhardt T. Distinct resident and recirculating memory T cell subsets in non-lymphoid tissues. Curr Opin Immunol (2013) 25:329–33. doi:10.1016/j.coi.2013.05.007
93. Wu T, Hu Y, Lee YT, Bouchard KR, Benechet A, Khanna K, et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J Leukoc Biol (2014) 95:215–24. doi:10.1189/jlb.0313180
94. Woodberry T, Suscovich TJ, Henry LM, August M, Waring MT, Kaur A, et al. Alpha E beta 7 (CD103) expression identifies a highly active, tonsil-resident effector-memory CTL population. J Immunol (2005) 175:4355–62. doi:10.4049/jimmunol.175.7.4355
95. Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol (2013) 14:1285–93. doi:10.1038/ni.2745
96. Gebhardt T, Whitney PG, Zaid A, Mackay LK, Brooks AG, Heath WR, et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature (2011) 477:216–9. doi:10.1038/nature10339
97. Zaid A, Mackay LK, Rahimpour A, Braun A, Veldhoen M, Carbone FR, et al. Persistence of skin-resident memory T cells within an epidermal niche. Proc Natl Acad Sci U S A (2014) 111:5307–12. doi:10.1073/pnas.1322292111
98. Coppieters K, Amirian N, von HM. Intravital imaging of CTLs killing islet cells in diabetic mice. J Clin Invest (2012) 122:119–31. doi:10.1172/JCI59285
99. Lodygin D, Odoardi F, Schlager C, Korner H, Kitz A, Nosov M, et al. A combination of fluorescent NFAT and H2B sensors uncovers dynamics of T cell activation in real time during CNS autoimmunity. Nat Med (2013) 19:784–90. doi:10.1038/nm.3182
100. Celli S, Albert ML, Bousso P. Visualizing the innate and adaptive immune responses underlying allograft rejection by two-photon microscopy. Nat Med (2011) 17:744–9. doi:10.1038/nm.2376
101. Musha H, Ohtani H, Mizoi T, Kinouchi M, Nakayama T, Shiiba K, et al. Selective infiltration of CCR5(+)CXCR3(+) T lymphocytes in human colorectal carcinoma. Int J Cancer (2005) 116:949–56. doi:10.1002/ijc.21135
102. Buckanovich RJ, Facciabene A, Kim S, Benencia F, Sasaroli D, Balint K, et al. Endothelin B receptor mediates the endothelial barrier to T cell homing to tumors and disables immune therapy. Nat Med (2008) 14:28–36. doi:10.1038/nm1699
103. Boissonnas A, Fetler L, Zeelenberg IS, Hugues S, Amigorena S. In vivo imaging of cytotoxic T cell infiltration and elimination of a solid tumor. J Exp Med (2007) 204:345–56. doi:10.1084/jem.20061890
104. Breart B, Lemaitre F, Celli S, Bousso P. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J Clin Invest (2008) 118:1390–7. doi:10.1172/JCI34388
105. Deguine J, Breart B, Lemaitre F, Di Santo JP, Bousso P. Intravital imaging reveals distinct dynamics for natural killer and CD8(+) T cells during tumor regression. Immunity (2010) 33:632–44. doi:10.1016/j.immuni.2010.09.016
106. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol (2009) 9:162–74. doi:10.1038/nri2506
107. Boissonnas A, Licata F, Poupel L, Jacquelin S, Fetler L, Krumeich S, et al. CD8+ tumor-infiltrating T cells are trapped in the tumor-dendritic cell network. Neoplasia (2013) 15:85–94.
108. Bauer CA, Kim EY, Marangoni F, Carrizosa E, Claudio NM, Mempel TR. Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. J Clin Invest (2014) 124:2425–40. doi:10.1172/JCI66375
109. Miller MJ, Wei SH, Cahalan MD, Parker I. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proc Natl Acad Sci U S A (2003) 100:2604–9. doi:10.1073/pnas.2628040100
110. Stoll S, Delon J, Brotz TM, Germain RN. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science (2002) 296:1873–6. doi:10.1126/science.1071065
111. Okada T, Miller MJ, Parker I, Krummel MF, Neighbors M, Hartley SB, et al. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol (2005) 3:e150. doi:10.1371/journal.pbio.0030150
Keywords: CD8 T cells, imaging techniques, intravital microscopy, migration, T cells, infections
Citation: Benechet AP, Menon M and Khanna KM (2014) Visualizing T cell migration in situ. Front. Immunol. 5:363. doi: 10.3389/fimmu.2014.00363
Received: 31 May 2014; Accepted: 14 July 2014;
Published online: 29 July 2014.
Edited by:
Kimberly Sue Schluns, University of Texas MD Anderson Cancer Center, USAReviewed by:
Tomasz Zal, University of Texas MD Anderson Cancer Center, USAShahram Salek-Ardakani, University of Florida, USA
Copyright: © 2014 Benechet, Menon and Khanna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kamal M. Khanna, Department of Immunology, University of Connecticut Health Center, L3057, 263 Farmington Avenue, Farmington, CT 06030, USA e-mail:a2toYW5uYUB1Y2hjLmVkdQ==
 Alexandre P. Benechet
Alexandre P. Benechet Manisha Menon
Manisha Menon Kamal M. Khanna
Kamal M. Khanna