- 1Department of Medicine, Immunology Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA
- 2Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA
Cytosolic NOD-like receptors (NLRs) have been associated with human diseases including infections, cancer, and autoimmune and inflammatory disorders. These innate immune pattern recognition molecules are essential for controlling inflammatory mechanisms through induction of cytokines, chemokines, and anti-microbial genes. Upon activation, some NLRs form multi-protein complexes called inflammasomes, while others orchestrate caspase-independent nuclear factor kappa B (NF-κB) and mitogen activated protein kinase (MAPK) signaling. Moreover, NLRs and their downstream signaling components engage in an intricate crosstalk with cell death and autophagy pathways, both critical processes for cancer development. Recently, increasing evidence has extended the concept that chronic inflammation caused by abberant NLR signaling is a powerful driver of carcinogenesis, where it abets genetic mutations, tumor growth, and progression. In this review, we explore the rapidly expanding area of research regarding the expression and functions of NLRs in different types of cancers. Furthermore, we particularly focus on how maintaining tissue homeostasis and regulating tissue repair may provide a logical platform for understanding the liaisons between the NLR-driven inflammatory responses and cancer. Finally, we outline novel therapeutic approaches that target NLR signaling and speculate how these could be developed as potential pharmaceutical alternatives for cancer treatment.
Introduction
Over the past two decades, immunologists have begun to appreciate the complexity of the innate immune system, its importance as the first wave of defensive action against perceived harmful microbes or foreign particles and its functions in triggering antigen-specific responses by engaging the adaptive immune system. Innate immune responses are orchestrated by germline-encoded pattern recognition receptors (PRRs) (1). PRRs recognize conserved pathogen-derived and damaged self-derived molecular components, commonly referred to as pathogen associated molecular patterns (PAMPs) and danger associated molecular patterns (DAMPs), respectively (2, 3). PRR superfamilies are broadly classified based upon structural homology and the requirement of different adaptor proteins that ensure their function and downstream signal transduction (4). The PRRs include members of the Toll-like receptors (TLRs) (3), nucleotide-binding, and oligomerization domain containing receptors [NOD-like receptors (NLRs)] (5, 6), retinoic acid-inducible gene (RIG) I-like RNA helicases (7), C-type lectins (8), and AIM2 like receptors (ALRs) (9). Evidence in the field points to a paramount importance of NLRs in human diseases with increasing interest in translating this knowledge toward clinical benefits. Due to the active role of NLRs in regulating pro-inflammatory signals and recruiting the adaptive arm of the immune system, dysregulation of microbial sensing has been reported to influence disease outcomes and tumorigenesis (10). In this review, we will describe the crucial roles of NLRs in cancer development and progression, and discuss the possibility of NLRs as targets for tumor therapy.
Factors That Influence Tumorigenesis
Observations by Rudolf Virchow in the nineteenth century indicated a link between inflammation and cancer, and suggested that immune and inflammatory cells are frequently present within tumors. Indeed, chronic inflammation plays critical roles in various stages of cancer development and progression (11–13). Many cancer risk factors are associated with a source of inflammation or act through inflammatory mechanisms such as those evoked by bacterial and viral infections (14), tobacco smoke (15), obesity (16, 17), and aging or cell senescence (18, 19). While some cancers arise from chronic inflammation or after immune deregulation and autoimmunity, solid malignancies elicit intrinsic immune mechanisms that guide the construction of a tumorigenic microenvironment (12, 13, 20). Although the exact mechanism of how inflammation leads to neoplastic transformation is not fully known, it is suggested that inflammatory immune cells like macrophages and T cells are the main orchestrators of inflammation-mediated tumor progression. These cells secrete cytokines and chemokines that cause DNA damage, generate mutagenic reactive oxygen species (ROS), and supply cancer cells with growth factors (13). In addition, inflammatory mechanisms were shown to promote genetic instability by impairing DNA repair mechanisms, altering cell cycle checkpoints, and often facilitating epigenetic silencing of anti-tumor genes, thus contributing to the high degree of genetic heterogeneity in tumors (21). Oncogenic mutations prompted by an inflammatory microenvironment frequently cause neoplastic transformation by promoting excessive proliferation and resistance to cell death (22). Indeed, impaired expression and activity of proteins that control cell survival, such as the inhibitor of apoptosis proteins (IAPs) and the BCL2 family of proteins, is a common occurrence in many cancers (23, 24). Typically known to exert strong anti-apoptotic functions, IAPs neutralize pro-apoptotic second mitochondrial activator of caspases (SMAC) and inhibit activation of apoptotic caspases, thereby promoting cell survival during both physiological stresses and pathogenic stimulations (25–29). Owing to their strong pro-survival potency, enhanced expression of IAPs has been correlated with several human cancers (22). Unlike IAPs, the BCL2 family of proteins consists of both pro- and anti-apoptotic proteins that control critical checkpoints of intrinsic apoptosis by regulating mitochondrial integrity and release of cytochrome c into the cytosol (30). Deregulation of the functions of BCL2 proteins, i.e., down-regulation of pro-apoptotic members and over expression of pro-survival members, has been strongly correlated with tumorigenesis and resistance to chemotherapy (31). Interestingly, the pro-apoptotic BID, PUMA, and NOXA are transcriptional targets of the tumor suppressor gene p53 and loss of their expression enhances tumorigenesis and morbidity of MYC overexpressing transgenic mice (32, 33). It was described that the transcription factor p53 senses physiological stresses and is critical for restraining tumor growth. Indeed, loss of p53 expression or function in both humans and mice has been proven to promote sporadic tumorigenesis (34, 35). Induction of target genes that inhibit cancer progression is generally considered to be the canonical mechanism of p53-mediated tumor-suppression. These target genes directly modulate cellular programs involving induction of apoptosis, cell cycle arrest, and promotion of cellular senescence and DNA repair (36). Recently, non-canonical functions of p53 have come to light, like the regulation of cellular metabolism, cell-to-cell communication, autophagy, tumor invasion, and metastasis, making p53 an attractive pharmaceutical target for treating cancers [reviewed in Ref. (37)]. Early detection of rogue tumor cells by the innate immune cells and their rapid removal is a key host defense strategy for evading tumorigenesis. In particular, natural killer (NK) cells are primary sentinels that guarantee such immune surveillance by differentiating normal cells from stressed or tumor cells via the expression of specific NK receptors (38). Indeed, increased presence of NK cells at tumor sites has been reported to improve remission, whereas decreased NK cell anti-tumor activity has been correlated with a greater likelihood for developing cancer (39).
Nod-Like Receptors in Cancer
Overview of NLRs
NOD-like receptors are a relatively recent addition to the PRR superfamily (40–42). All NLRs contain a central NACHT domain that facilitates oligomerization, and bear multiple leucine-rich repeats (LRRs) on their C-terminal for ligand sensing (5, 43). The 22 human NLRs can be distinguished into five subfamilies by their N-terminal effector domains that bestow unique functional characteristics to each NLR (43) (Figure 1). NLRs with an N-terminal acidic transactivation domain are termed NLRA (CIITA) and serve as transcriptional regulators of MHC class II antigen presentation (44). NLRB (NAIP) proteins have an N-terminal baculoviral inhibition of apoptosis repeat (BIR) domain and are largely recognized for their roles in host defense and cell survival. For instance, NAIP5 is known to induce host defense against bacterial infections by curtailing macrophage permissiveness to Legionella pneumophila, the causative agent of the Legionnaires’ disease (45–47). N-terminal caspase activation and recruitment domain (CARD) distinguishes the NLRC subfamily (NLRC 1–5) and allows direct interaction between members of this family and other CARD carrying adaptor proteins. NOD1 (NLRC1) and NOD2 (NLRC2), the founding members of the NLRs, are key sensors of bacterial peptidoglycan (PGN) and are crucial for tissue homeostasis and host defense against bacterial pathogens (48). Notably, single-nucleotide polymorphisms (SNPs) in the NOD2 (CARD15) gene are among the most significant genetic risk factors associated with Crohn’s disease (CD) susceptibility (49, 50), hence the rising interest in unraveling the functions of NOD1 and NOD2 receptors in microbial sensing, intestinal homeostasis, and disease. Members of the pyrin domain (PYD) containing NLRP subfamily (NLRP 1–14) are best known for their role in inducing the formation of the oligomeric inflammatory complex “Inflammasome” (51). NLRX1, the only described member of the NLRX subfamily contains an N-terminal mitochondria-targeting sequence required for its trafficking to the mitochondrial membrane (Figure 1). Mechanistically, NLRX1 was shown to down-regulate mitochondrial anti-viral signaling protein (MAVS)-mediated type I interferon (IFN) production (52), interfere with the TLR-TRAF6-NF-κB pathways (53, 54), and enhance virus induced-autophagy (55, 56). On the other hand, NLRX1 was implicated in the generation of ROS induced by TNFα and Shigella infection magnifying the JNK and NF-κB signaling (57). Interestingly, NLRX1-mediated ROS generation was involved in promoting Chlamydia trachomatis replication in epithelial cells (58). However, recent data from Soares et al. revealed that bone marrow macrophages (BMMs) and mouse embryonic fibroblasts (MEFs) from Wild type (WT) or Nlrx1−/− mice respond equally to in vitro infection with Sendai virus or following in vivo challenge with influenza A virus and TLR3 ligand Poly I:C (59). Additionally, Rebsamen et al. reported no significant contribution of NLRX1 in RLR–MAVS signaling both in vitro and in vivo (60). Overall, the precise role of NLRX1 remains controversial and further research is required to validate its pro or anti-inflammatory properties.
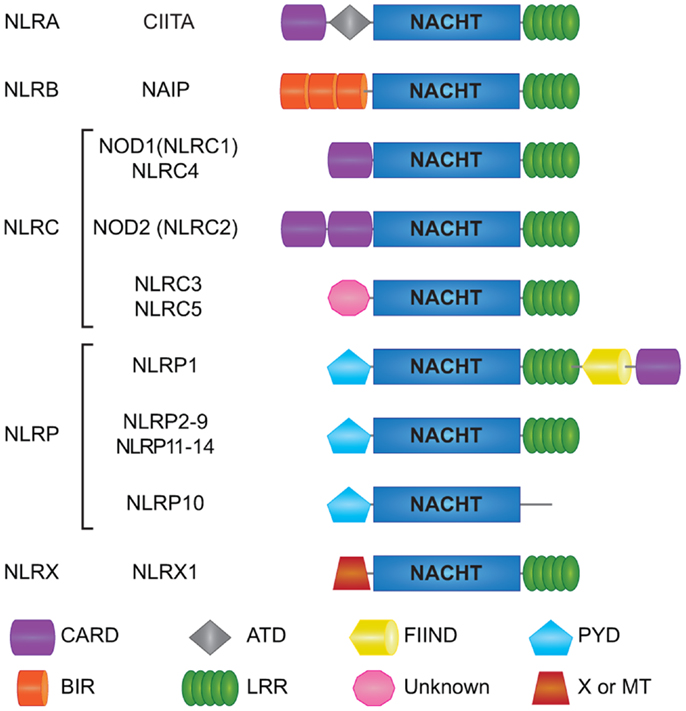
Figure 1. Schematic representation of individual NLR domains. Domain architecture of human NLRs is depicted here. Human NLRs are sub-classified into five categories: NLRA, NLRB, NLRC, NLRP, and NLRX. All 22 human NLRs contain a central NACHT domain and a C-terminal ligand sensing LRR domain, with the exception of NLRP10. The N-terminal domains ascribe functional properties to the NLRs; however, the function of some of the domains is still unclear like for the N-terminal domain of NLRC3 and NLRC5, as well as the C-terminal FIIND in NLRP1. CARD; caspase association and recruitment domain, ATD; acidic transactivation domain, FIIND; function to find domain, PYD; pyrin domain, BIR; Baculoviral inhibition of apoptosis protein repeat domain, LRR; leucine-rich repeats, MT; targets NLRX1 to the mitochondria but no sequence homology with traditional mitochondrial targeting sequence has been reported.
Dysregulated apoptosis and autophagy pathways, as well as excessive chronic inflammation are major drivers of carcinogenesis. NLRs are innate immune sensors that actively communicate with a myriad of cell death regulators. Hence, these PRRs are well-positioned to influence tumor development and progression particularly at sites with high host-microbiome interactions like the gut. One of the mysteries of the innate immune system is how do NLRs sense molecular patterns from both commensal and pathogenic microorganisms and manage to tolerate one while help eradicate the other (5, 61). This disparity in NLR functions is particularly useful in the intestinal epithelia where host cells are in constant contact with millions of microbes. Consequently, it came as little surprise when common variants in the NLR genes were correlated with the incidence of CD and susceptibility to cancers (50, 62–64). Due to these correlations, most of the studies have been focused on understanding the mechanisms by which NODs and inflammasome NLRs regulate intestinal inflammation and tumorigenesis.
NOD1 and NOD2 in Cancer
NOD-Dependent Signaling Cascades
NOD1 and NOD2 are cytosolic proteins that sense intracellular bacterial PGN and trigger signal transduction via NF-κB and MAPK activation. NOD1 is expressed in both hematopoietic and non-hematopoietic cells and responds to intracellular gamma-d-glutamyl-meso-diaminopimelic acid (iE-DAP) mostly present on Gram-negative bacteria and only on some select Gram-positive bacteria, like Listeria and Bacillus species (65–67). Unlike NOD1, NOD2 expression is largely restricted to hematopoietic cells and certain specialized epithelial cells such as the small intestinal Paneth cells (68). NOD2 recognizes cytosolic muramyl dipeptide (MDP) found in the PGN of all bacteria (69). Besides providing immunity against intracellular bacteria, NODs were revealed to be critical for host defense against non-invasive Gram-negative bacteria like Helicobacter pylori, following delivery of its PGN into the host cells through the bacterial type IV secretion system (70). Moreover, NOD1 and NOD2 ligands were also described to gain access to the cytosol by endocytosis with the help of transporter proteins like SLC15A3 and SLC15A4 (71–73). Notably, NOD1 and NOD2 have been reported to localize to the plasma membrane at the sites of infection; however, the biological relevance of this translocation remains elusive (74, 75). Interestingly, a recent report accentuated the importance of NOD proteins in monitoring the activation state of small Rho GTPases (e.g., RAC1, CDC42, and RHOA) and inducing unusual immune responses in the host in response to bacterial infection (76). Upon activation by their cognate ligands both NOD1 and NOD2 self-oligomerize, undergo a conformational change, and through homotypic CARD–CARD interactions allow the recruitment of the CARD containing adaptor Receptor-interacting protein kinase 2 (RIP2 or RIPK2) (41, 42, 77, 78) (Figure 2). This event facilitates the formation of a multi-protein signaling complex termed “Nodosome,” which leads to downstream NF-κB and MAPK-mediated inflammatory and anti-microbial output. Indeed, cells or mice lacking RIP2 do not respond to NOD agonists and fail to produce pro-inflammatory and anti-microbial molecules (78–80). Initially, it was thought that NOD oligomerization initiated RIP2 aggregation and activation by “induced proximity” (81). While this model still stands true, over the years new body of research has contributed a wealth of data regarding specific sequence of events that leads to RIP2 activation. In contrast to the earlier studies (82–85), recent in vitro data using pharmacological inhibitors as well as in vivo evidence using a knock-in mouse with kinase-dead RIP2 (K47A) have highlighted the key role of the kinase activity of RIP2 in NOD-mediated immune responses (86, 87).
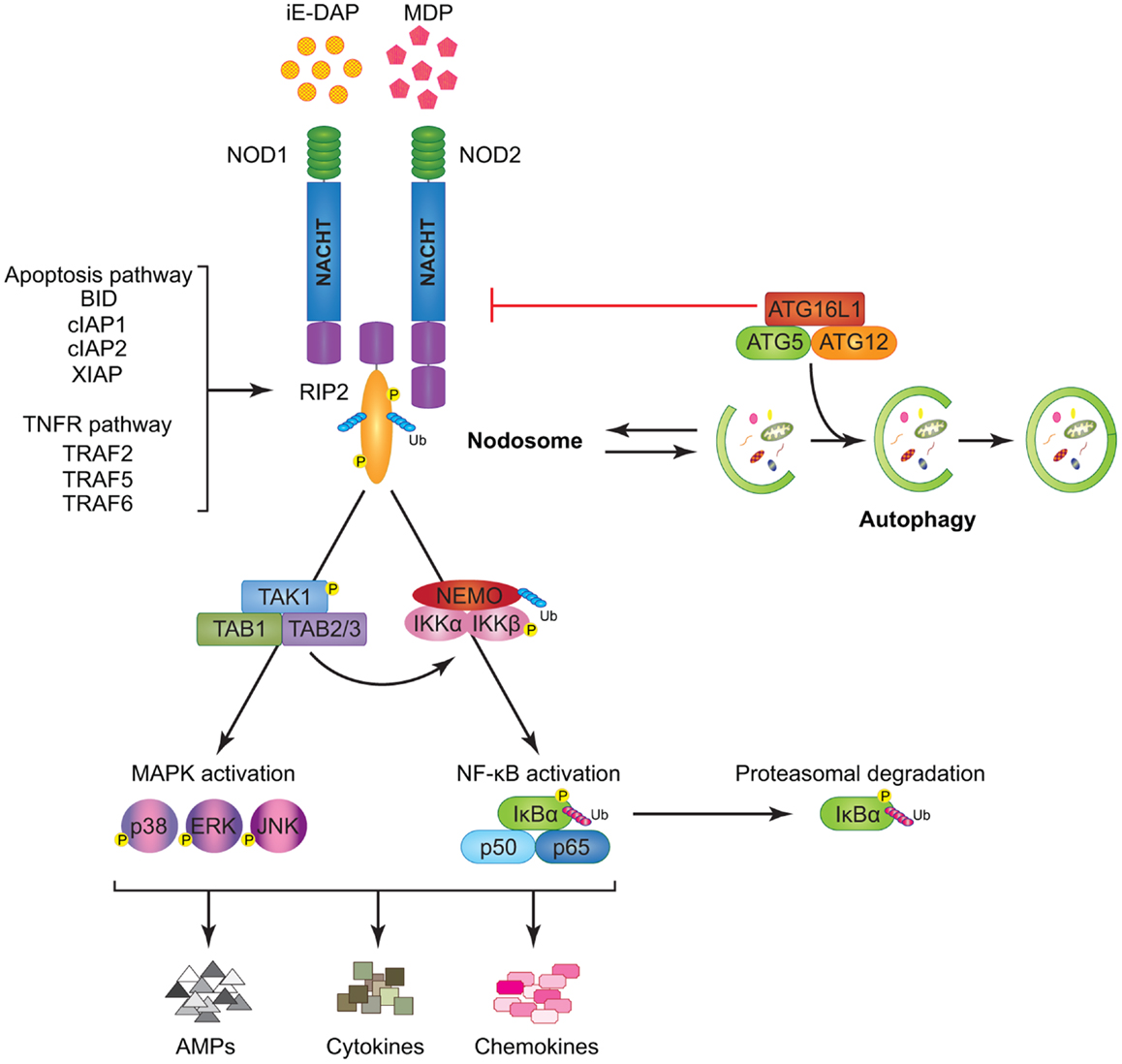
Figure 2. Model of NOD1 and NOD2 signaling cascades. NOD1 and NOD2 recognize bacterial PGNs, iE-DAP, and MDP, respectively. Following ligand sensing the NODs recruit their common adaptor RIP2 by CARD–CARD interactions and induce RIP2 to undergo phosphorylation. The members of the TRAF family (TRAF2, TRAF5, and TRAF6), the IAP family (XIAP, cIAP1, and cIAP2), and the BCL2 family (BID) bind to RIP2 and facilitate its ubiquitination allowing the recruitment of TAK1 and ubiquitinated NEMO to the Nodosome. On one hand, NEMO instigates activation of the canonical NF-κB pathway by phosphorylating IKKα and IKKβ, by inducing IκBα phosphorylation and proteasomal degradation, and by freeing p50 and p65 NF-κB subunits. On the other hand, TAK1 recruits TAB1 and TAB2/3 inducing both (p38, ERK, and JNK) MAPK and NF-κB activation. Stimulation of both arms culminates in the induction of anti-microbial peptides (AMPs), cytokines, and chemokines. The formation of the Nodosome promotes autophagy and conversely, a fully functional autophagy machinery helps in signal transduction through the Nodosome. ATG16L1 along with ATG5 and ATG12 is required for autophagosome formation, however, independently of its autophagy functions, ATG16L1 negatively regulates NOD/RIP2 signaling.
Lately, it was described that the pathways activated downstream of NOD proteins are closely related to those activated by death receptors, notably TNF receptor 1 (TNFR1). For instance, hierarchical recruitment of selective TNFR-associated factors (TRAF2, TRAF5, or TRAF6) facilitates Lys63 poly-ubiquitination and activation of RIP2 (88–90). Activated RIP2 facilitates ubiquitination of NEMO (also called IKKγ) leading to the recruitment of tumor growth factor β-activated kinase 1 (TAK1) and TAK1 binding proteins (TAB) 1, TAB2, or TAB3 (91, 92). Following this complex formation, IKKs (IKKα and IKKβ) get phosphorylated eventually driving the phosphorylation and degradation of IκBα and subsequent transcription of NF-κB target genes (5, 89, 92) (Figure 2). RIP2 activation also constitutes a key event that links the NOD–RIP2 cascade with the p38, extracellular signal-regulated kinase (ERK), and c-Jun N-terminal kinase (JNK) MAPK pathways (93).
In addition to TRAFs, members of the IAP family including X-linked IAP (XIAP) and cellular IAP1 (cIAP1) and cIAP2 were described to physically interact with RIP2 and facilitate NOD-mediated immunity (94–98). Both in vitro and in vivo studies suggest a strong role for cIAP1 and cIAP2 in promoting NOD signaling (Figure 2); however, the mechanism for such positive regulation is still not fully understood (94, 99–101). Similarly, XIAP was reported to recruit a linear ubiquitin chain assembly complex (LUBAC) for RIP2 ubiquitination and this step was proven critical for downstream NF-κB regulation (96, 97). Upon microbial sensing another E3 ubiquitin ligase, ITCH, also ubiquitinates RIP2, and it is speculated that ITCH-mediated ubiquitination acts like a molecular switch dictating the fate of the signaling circuit to NF-κB or p38 and JNK activation (102). Pathogen-mediated NOD1 activation has also been shown to elicit protective immune responses via RIP2-TRAF3-IRF7-mediated transcription of IFNβ (79). Overall, it is tempting to speculate that similar to pro-survival association of RIP1 with cIAP1 and cIAP2 (103), interactions between RIP2 and the IAPs may also lead to modulation of cellular apoptosis. However, neither NODs nor RIP2 has been demonstrated to exploit these associations to affect cell survival. Similarly, several studies have alluded to NODs as being regulators of caspase-mediated apoptosis (82, 104, 105); yet, no direct link has so far been reported. Recently, the pro-apoptotic BH3 only BCL2 family protein BID (BH3 interacting-domain death agonist) was identified in a genome wide siRNA screen as a positive regulator of NOD signaling (101). BID was demonstrated to bind to RIP2 bridging both NOD and IKK complexes to specifically transduce NF-κB and ERK signaling events (101). Notably, BID was phosphorylated upon activation with NOD agonists and these innate immune functions of BID were found to be independent of its pro-apoptotic processing by caspase-8 (101). The discovery involving a classical pro-apoptotic protein, such as BID, in NOD–RIP2 signaling strengthens the concept that inflammatory and cell death pathways do not function as discrete mechanisms but share common adaptors. Such adaptors can exert multiple functions depending upon the nature of the stimuli (5, 106–108) (Figure 2). One recent study have reported that BID-deficient mice exhibit a normal NOD-mediated immunity (109), suggesting that further investigations are still needed to clearly decipher the implication of BID in NOD signaling.
Similar to NOD2, a SNP encoding a missense variant in the autophagy gene ATG16L1 was strongly associated with the incidence of CD, raising a possible common role of both genes in host defense mechanisms (110, 111). Intriguingly, it has been described that NOD1 and NOD2 stimulation enhances autophagy, either directly by interacting with ATG16L1 (112) or indirectly (112–115). Conversely, pharmacological inhibition of both early and late autophagy has been proven to down-regulate MDP-mediated NF-κB and MAPK activation, suggesting that autophagocytic trafficking of MDP may be required for efficient NOD2 signaling (114). Surprisingly, ATG16L1 was recently shown to negatively regulate NOD1- and NOD2-mediated inflammatory signaling by interfering with RIP2 ubiquitination and recruitment to the Nodosome (116) (Figure 2). Taken together, this information suggests that different NLRs can have opposing regulatory effects on autophagy and cell death, yet the molecular triggers that dictate these actions are not fully understood.
NOD Proteins and Cancer
Three mutations within the LRR region of the NOD2 gene have been associated with increased CD susceptibility. Interestingly, these same mutations have also been found to directly interfere with NOD2’s bacterial sensing faculties and downstream NF-κB activation (49, 50). Notably, such inactivation of NOD2 immunity has been indicated to enhance the risk of bacterial infections following chemotherapy in patients with acute myeloid leukemia (117). In addition, NOD2 polymorphisms have been correlated with modifications in gastric mucosa and increased risk for H. pylori induced gastric cancer (118). Apart from intestinal disorders, mutations in NOD2 have been linked with increased prevalence of early onset breast (119) and lung cancers (120, 121). However, how NOD2 contributes to the initiation and the progression of cancer remains ill defined. Although no mutations in the NOD1 gene have been so far associated with the incidence of intestinal inflammation or even colorectal cancer (CRC), murine models clearly designate a central anti-tumorigenic function for NOD1 in the pathophysiology of disease. For instance, Nod1−/− mice have been described to be susceptible to dextran sulfate sodium (DSS), a sulfated polysaccharide highly toxic to enterocytes (122). Upon combination of a single hit of the carcinogen, azoxymethane (AOM), with DSS (123), NOD1-deficient mice were found to develop significantly more and larger colonic tumors as compared to WT mice (122). This experimental CRC model is particularly applicable when the focus is on understanding colitis-driven tumor initiation and progression. The ApcMin/+ mouse is a N-Ethyl-N-Nitrosourea (ENU) mutant model carrying the multiple intestinal neoplasia (Min/+) mutation and recapitulates many aspects of human hereditary or sporadic CRCs with mutations in the adenomatous polyposis coli (Apc) gene (124–127). Intriguingly, it has been reported that treatment with low doses of DSS leads to increased colonic tumors in ApcMin/+Nod1−/−mice suggesting that NOD1 serves as a negative regulator of the tumor-promoting Wnt/β-catenin cascade (128, 129). Further analysis revealed that absence of NOD1 exacerbated NF-κB-mediated inflammation early during colitis causing gut barrier damage and prompted a second wave of microbiota driven inflammation and intestinal epithelial cell (IEC) proliferation, thus initiating tumor development. These conclusions are supported by the observation that antibiotic treatment of Nod1−/− mice ameliorated DSS-induced CRC (122). While most investigations have been focused on the role of NOD1 in models of intestinal tumorigenesis, one report provided experimental evidence for the protective role of NOD1 in breast cancer (104). Herein, it was shown that NOD1-deficient MCF-7 breast cancer cells were resistant to iE-DAP and cycloheximide mediated cell death. Interestingly, SCID mice grafted with NOD1 overexpressing cells exhibited rapid tumor regression. In sharp contrast, mice grafted with NOD1-deficient MCF-7 cells displayed increased and continued tumor growth (104).
Like Nod1−/− mice, NOD2-deficient mice have been revealed to be highly susceptible to DSS-induced colitis by inheritance of dysbiotic microbiota that markedly sensitizes mice to injury (130). Furthermore, Nod2−/− mice have been found to display worse disease outcome with increased epithelial dysplasia, heightened tumor burden, and elevated expression of the pro-inflammatory cytokine IL-6 when subjected to AOM–DSS treatment. This transmissible phenotype was significantly ameliorated upon treatment with broad-spectrum antibiotics or using the neutralizing IL-6 receptor antibody (130). Altogether, these findings reinforce the idea that aberrant NOD signaling gives rise to dysbiosis that in an inflammatory setting ultimately causes mucosal injury and drives CRC. So far, the translational value of this knowledge is limited but with the recent technological advances in the microbiome research it is predicted that modulation of dysbiosis could be used as a therapeutic strategy for patients with CD as well as CRC.
Contrary to the protective role for NODs in intestinal tumorigenesis, increased expression of both NOD1 and NOD2 has been reported in the head and neck squamous cell carcinoma biopsies as compared to the healthy nasal biopsies. These findings implicate NODs in enhancing head and neck cancers; however, thus far no corroborating experimental evidence has been reported (131). Furthermore, iE-DAP stimulation of human pharyngeal squamous carcinoma cell lines (Detroit 562 and Fadu) has been determined to augment the production of β-defensins, which can serve as chemoattractants, thus fostering an inflammatory and pro-tumorigenic environment (131).
Inflammasome NLRs in Cancer
Inflammasome NLRs: NLRP3-Mediated Signaling Cascades
While NOD1 and NOD2 form the Nodosome, other NLRs assemble macromolecular inflammasome complexes. To date, various inflammasome platforms have been described (132), but the NLRP3 inflammasome is the most commonly studied. The reason behind this could be the initial discovery of mutations in the NLRP3 (CIAS1) gene implicating this PYD containing protein in both familial cold auto-inflammatory syndrome (FCAS) and Muckle–Wells Syndrome (MWS) (133). Thus, the NLRP3 inflammasome will be described here as a prototype for these NLRs (Figure 3). Classically, the inflammasome has been described to consist of an NLRP, the inflammatory protease caspase-1, and the apoptosis-associated speck like protein (ASC) (51). ASC contains an N-terminal PYD and a C-terminal CARD making it uniquely suited for bringing into close proximity the two key components, caspase-1 and NLRPs (134, 135). Upon activation, NLRP3 recruits ASC and caspase-1, which is a prerequisite for the cleavage and maturation of the inflammatory cytokines IL-1β and IL-18 and consequent inflammatory cell death named pyroptosis (136–141). Lately, a more complex model for NLRP3-inflammasome activation has been proposed where two adaptors, ASC and mitochondrial MAVS, are required for optimal inflammasome triggering (142).
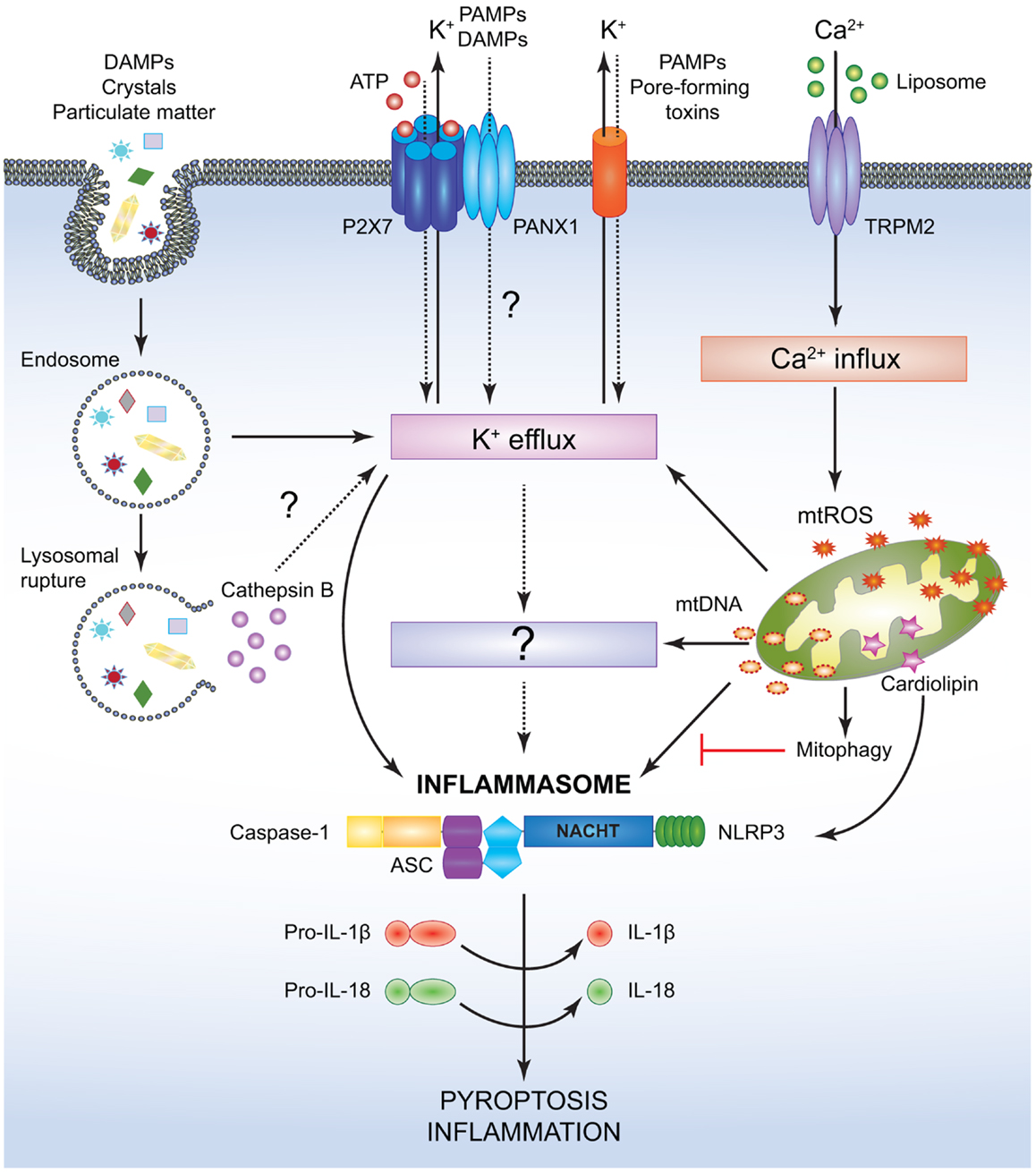
Figure 3. Simplified mechanisms for the canonical NLRP3- inflammasome activation. Various PAMPs and DAMPs provide the signal 2 required to assemble and activate the NLRP3 inflammasome comprised of NLRP3, ASC, and caspase-1. Although the precise mechanism leading to NLRP3 activation is still controversial, it is speculated that K+ efflux may be the common cellular response that triggers inflammasome activation. However, this notion has not been fully verified and it is possible that an unidentified or intermediate adaptor may be required for transmitting signals between K+ efflux and the NLRP3 inflammasome. Crystals and particulate DAMPs enter the cell via endocytosis directly inducing K+ efflux and NLRP3-inflammasome formation. In addition, the endo- lysosomes carrying these DAMPs undergo lysosomal rupture and release cathepsin B, which acts as an intracellular DAMP and can induce K+ efflux. However, contradicting studies indicate that lysosomal rupture may cause K+ efflux and inflammasome activation even in the absence of cathepsin B. ATP binds to the P2X7 receptor on the cell membrane and causes opening of the PANX1 channels allowing K+ efflux and influx of any PAMPs and DAMPs present in the extracellular space. PAMPs such as pore-forming toxins activate the NLRP3 inflammasome and facilitate K+ efflux. Liposomes instigate Ca2+ influx through opening of the TRPM2 channels. Accumulation of excessive Ca2+ in the cytosol causes mitochondrial dysfunction and release of mtROS and oxidized mtDNA, which may activate the NLRP3 inflammasome either directly or by inducing K+ efflux. Clearance of distressed mitochondria by mitophagy serves to evade such inflammasome activation. Mitochondrial Cardiolipin binds to NLRP3 and is required for the NLRP3-inflammasome activation. Following NLRP3-inflammasome assembly, caspase-1 undergoes proximity driven proteolytic cleavage and further processes pro-IL-18 and pro-IL-1β into their mature active forms. Activation of the NLRP3-caspase-1 axis results in inflammation and pyroptotic cell death.
Owing to its widespread expression in numerous cell types such as neutrophils, monocytes, DCs, epithelial cells, and T cells (140, 143, 144), NLRP3 is exposed to a wide array of PAMPs and DAMPs that instigate the assembly and activation of the inflammasome [reviewed in Ref. (5, 132, 145–148)]. The NLRP3-inflammasome formation requires a two-step process (149). The priming step (or signal 1) involves TLR-NF-κB-driven induction of inflammasome components, as basal expression of NLRP3 in resting cells is insufficient for effective inflammasome activation (149, 150). However, certain cells like the human blood monocytes and murine macrophages appear to activate the NLRP3 inflammasome in response to LPS stimulation alone (151, 152). It is noteworthy that a transcriptionally silent mechanism for TLR4-mediated inflammasome priming has been lately discovered (153, 154). This mechanism involves mitochondrial ROS (mtROS)-driven deubiquitination of NLRP3, suggesting that constitutive ubiquitination of NLRs may be a homeostatic mechanism to prevent overt inflammasome activity (154). The second activation step (or signal 2) promotes the NLRs to undergo homotypic oligomerization and assemble the inflammasome.
While several models have been proposed to define the signals behind NLRP3 activation, the precise mechanisms remain hitherto unresolved. Various bacterial pathogens induce potassium efflux and activate the NLRP3 inflammasome via the action of secreted pore-forming toxins (e.g., nigericin from Streptomyces hygroscopicus, listeriolysin O from Listeria monocytogenes, pneumolysin from Streptococcus pneumoniiae, alpha-hemolysin, etc.) (138, 155, 156) (Figure 3). In addition, NLRP3 inflammasomes have been known to assemble in response to cytosolic bacterial and viral RNA both in vivo and in vitro (137, 157–160). Extracellular adenosine tri-phosphate (ATP) released from dying or damaged cells also causes NLRP3-inflammasome activation through either paracrine or autocrine sensing of ATP by the purinergic receptor P2X7 (138, 161–163). Besides, it has been defined that ATP released from phagocytosed dying cells acts similarly on P2X7 and prompts pannexin-1 (PANX1) channels to open, thus resulting in potassium (K+) efflux and allowing other agonists to further engage and activate NLRP3 (164) (Figure 3).
Monosodium urate (MSU) and calcium pyrophosphate dehydrate crystals, alum, amyloid-β fibrils, as well as environmental pollutants like asbestos and silica strongly activate the NLRP3 inflammasome (139, 165–170). According to one model for this mode of activation, uptake of the crystalline and particulate matters into the cell causes lysosomal destabilization and release of cathepsin B, which is sensed by NLRP3 (168, 169). Interestingly, however, opposing results were obtained when cathepsin B-deficient BMMs were used to test this hypothesis, as no differences in IL-1β or caspase-1 cleavage were observed in response to several inflammasome activators such as hemozoin, MSU, or alum (171). Another model suggests that these activators prompt generation of mtROS and mitochondrial DNA, both of which are responsible for NLRP3-inflammasome activation (172–174). Evidently, pharmacological inhibition of mtROS production has been shown to prevent NLRP3-inflammasome formation indicating that ROS generation is an upstream event for NLRP3 activation (165, 166) (Figure 3). Liposomes have been found to induce mtROS and NLRP3-inflammasome activation by triggering calcium (Ca2+) influx via the transient receptor potential melastatin 2 (TRPM2), although the exact mechanism linking ROS production to TRPM2 channel opening is still not well-characterized (175). On the other hand, the mitochondrial protein cardiolipin has been shown to directly bind and activate NLRP3 in a ROS-independent manner suggesting that ROS may not be the common denominator engaging the NLRP3 inflammasome (176). Recent advances have put forward additional mechanisms underlying NLRP3-inflammasome activation. In BMMs stimulated with PAMPs, extracellular calcium has been shown to activate the calcium sensing receptor (CASR) mediating signal transduction pathways that culminate in the release of calcium stores from the endoplasmic reticulum (ER), eventually activating the NLRP3 inflammasome (177–179). The diverse nature of the NLRP3-inflammasome agonists allude to the likelihood that, instead of directly sensing PAMPs and DAMPs, NLRP3 may be activated by converging pathways with a final common ligand for NLRP3. Guanylate binding protein 5 (GBP5) has been recently proposed as one such component that directly participates in NLRP3-inflammasome activation; however, further investigation is needed to decipher how the GBP5 is activated and why it is required for select inflammasome assembly (180). Finally, studies by Munoz-Planillo et al. suggest that potassium efflux may perhaps be the sole intracellular event necessary for NLRP3 activation in response to a wide array of stimuli arguing for a unifying model for the NLRP3-inflammasome activation (181) (Figure 3).
Production of mtROS often culminates in mitophagy, an autophagic clearance of dysfunctional mitochondria. It has been demonstrated that inhibition of mitophagy enhances NLRP3-caspase-1-mediated secretion of IL-1β and IL-18 in response to LPS and ATP (172). In addition, deletion of ATG16L1 was found to promote IL-1β release in response to ATP, MSU, or LPS alone (182). Moreover, it has been recently suggested that autophagy may restrict NLRP3 activity by directly sequestering and targeting inflammasome components for degradation (183, 184). Overall, it is reasonable to speculate that autophagy could serve as a mechanism for preventing excessive NLRP3-inflammasome activation (172, 173, 183–185).
Mitochondrial dysfunction plays a central role in regulating the mechanisms involved in both inflammasome and apoptosis pathways. Loss of mitochondrial membrane potential is a pivotal event in intrinsic apoptosis and is tightly regulated by the BCL2 family of proteins through a system of checks and balances (30). Interestingly, anti-apoptotic BCL2 and BCL-XL proteins have been reported to directly interact with NLRP1 (CARD and PYD domain containing NLRP) to negatively regulate caspase-1 activation (186, 187). Similarly, BCL2 overexpression was shown to limit NLRP3-inflammasome activation (173, 174). In addition to BCL2 proteins, cIAP1, cIAP2, and XIAP have also been linked with inflammasome activation. Unlike their role in NOD signaling, initial studies have proposed that expression of these proteins might prevent caspase-1-dependent cell death (188). However, more recently cIAP1 and cIAP2 along with TRAF2 were found to enhance inflammasome activation seemingly by ubiquitinating and stabilizing caspase-1 and consequently prompting IL-1β release (189). In another report, genetic ablation of cIAP1 or cIAP2 had no effect on NLRP3-inflammasome activation, but concurrent pharmacological degradation of XIAP, cIAP1, and cIAP2 using SMAC mimetics was shown to limit caspase-1 activation (190). Interestingly, further inquiries revealed that in the absence of XIAP, cIAP1, and cIAP2, cell death in response to LPS was primarily incited by RIP3 activation causing NLRP3-caspase-1- as well as caspase-8-dependent IL-1β secretion (190). Lately, the concept of non-canonical inflammasome has been defined, which requires activation of caspase-11 in response to Gram-negative bacteria to facilitate either caspase-1-mediated IL-1β secretion or caspase-1-independent pyroptosis (191–194). Interestingly, apoptosis mediators FADD and caspase-8 have been involved in canonical and non-canonical NLRP3-inflammasome signaling. Indeed, FADD and caspase-8 facilitate the priming in “signal 1” by instigating both, LPS-TLR-MyD88-triggered induction of pro-IL-1β and NLRP3, as well as TLR-TRIF-mediated upregulation of pro-caspase-11 (195). Upon infection with Citrobacter rodentium or Escherichia coli, FADD and caspase-8 have been found to promote the “signal 2” by interacting with the NLRP3-inflammasome complex, thus influencing both canonical (caspase-1-dependent IL-1β maturation) and non-canonical (caspase-11-dependent pyropotosis) inflammasomes (194, 195). Conversely, it has been exhibited that caspase-8-deficient murine DCs are hyper-responsive to LPS-induced NLRP3-inflammasome assembly and activation (196). Overall, these studies place caspase-11 and caspase-8 at the center of inflammasome activation; however, a general lack of consensus in the field makes it hard to aptly judge their contribution in inflammasome-induced inflammation.
Inflammasome NLRs and Cancer
NLRP3, previously associated with rare and severe auto-inflammatory disorders, has been lately implicated in CD susceptibility and correlated with decreased NLRP3 expression and IL-1β production (62). Indeed, mice lacking NLRP3 have been shown to display exacerbated colonic inflammation upon DSS-induced colitis characterized by greater gut barrier damage, inflammatory immune cell infiltration, and cytokine production (197, 198). In accord, a central role has been ascribed for caspase-1 and ASC in intestinal epithelial repair after DSS-injury (199). Specifically, caspase-1, ASC, or NLRP3 deficiency in mice has been shown to be detrimental in DSS-induced intestinal inflammation, a mechanism attributed to the lack of IL-18 production by IECs (198, 199). Concomitantly, the increased colitogenic phenotype was completely reversed when mice were exogenously administered with the recombinant IL-18 cytokine (198, 199). The same lack of inflammatory regulation was found to render Nlrp3−/− and Casp1−/− mice more susceptible to AOM–DSS carcinogenesis (197, 200). The heightened tumor growth in the caspase-1 deficient mice was accompanied with drastically low levels of colonic IL-18. Overall, NLRP3 was shown to be important for IL-18 secretion, which in turn through IFNγ production induces STAT1 (Signal transducers and activators of transcription 1) phosphorylation and thus promotes an anti-tumorigenic environment (200). Moreover, it has been shown that Il18−/− or Il18r−/− mice are more susceptible to DSS-induced colitis and CRC, mimicking the increased tumor burdens observed in NLRP3 and caspase-1 deficient mice (201). Recent findings have put forward a novel concept for the dual function of IL-18 in intestinal inflammation and colitis-driven CRC (202, 203). For instance, during acute injury IEC-derived IL-18 triggers repair and restitution of the ulcerated epithelial barrier, whereas under chronic inflammatory settings the excessive release of IL-18 both from IECs and lamina propria macrophages and DCs is deleterious (203, 204). A protective role for NLRP3 has also been described in hepatocellular carcinoma (HCC) (205). This correlation is primarily based on mRNA and protein expression data showing reduced levels of NLRP3 and other related inflammasome components seen in hepatic parenchymal cells derived from HCC tissue specimens as compared to non-cancerous liver sections (205). On the other hand, a gain of function SNP (Q705K) within the NLRP3 gene has been associated with increased mortality in CRC patients (206). Significantly, the same SNP was also found to be more prevalent in patients with malignant melanoma (207). Human monocytic THP-1 cells overexpressing a mutant variant of NLRP3 bearing the Q705K SNP have been reported to greatly respond to the inflammasome agonist alum and to trigger the production of IL-1β and IL-18, implying that overt NLRP3 activation could be detrimental for certain types of cancer (208). Similarly, another group implicated constitutive NLRP3-inflammasome signaling in the development and progression of melanomas (209).
Loss of function in the tumor suppressor gene p53 has been associated with a large number of sporadic cancers (36). One of the mechanisms for p53-induced clearance of potentially carcinogenic cells has been found to be via transcriptional up regulation of cell death activators (210). In light of this knowledge, the discovery of NLRC4 as a downstream transcriptional target of p53 was a promising evidence for the anti-tumorigenic functions of this NLR (211). Moreover, lack of NLRC4 inflammasome has been associated with the attenuation of p53-mediated cell death, indicative of a protective role of NLRC4 during tumor development (211). Several groups have investigated the role of NLRC4 in colitis and CRC. However, lack of consensus in the susceptibility of Nlrc4−/− mice to DSS as well as AOM–DSS treatment makes it difficult to gage the protective effect of NLRC4 in these models (197, 212). It has been demonstrated that mice deficient in NLRC4 develop higher tumor burdens than WT mice when subjected to DSS-induced CRC (212). In addition, bone marrow chimera experiments verified that NLRC4 expression within the radioresistant compartment was the major driver of CRC protection (212). Surprisingly, similar colitic phenotypes have been observed between WT and Nlrc4−/− mice following DSS administration, suggesting that tumor regulation by NLRC4 is mostly cell intrinsic and not through down-regulation of inflammation (213). Given the unique capacity of NLRC4 to sense and differentiate between commensal and pathogenic microbes in the gut (214), it is surprising that the tumor restraining roles of NLRC4 have been ruled to be independent of its immune regulatory functions. One unifying theory addressing these discrepancies could be that anti-tumor functions of NLRC4 are attributed to the cells of non-hematopoietic origin, whereas intestinal mononuclear phagocytes are the primary source of NLRC4 for microbial sensing and pathogen clearance (213, 214). Overall, these assumptions warrant deeper inquiries to clearly elucidate the mechanisms by which NLRC4 exerts protective functions during CRC and to decipher the relevance of p53-mediated role of NLRC4 in tumorigenesis.
Akin to NLRP3, both NLRP6 and NLRP12 have been recently described to use ASC-caspase-1 molecular platforms and assemble inflammasomes. A first hint of NLRP6 being an inflammasome NLR was gleaned from in vitro experiments showing increased caspase-1 cleavage when ASC and NLRP6 were co-expressed (215). Further in vivo evidence emphasized a protective role for NLRP6 in intestinal inflammation and tumorigenesis as Nlrp6−/− mice showed high susceptibility to DSS-induced colitis and AOM–DSS-induced CRC (216–218). Unlike NLRC4, dampening of inflammation is purported to be one of the primary mechanisms for NLRP6-mediated protection and tissue homeostasis. NLRP6 has been shown to promote a gut microbiome that limits chronic inflammation. In fact, it has been evidenced that Nlrp6−/− mice display a distinct transmissible pro-colitogenic microbiome with increased prevalence of the bacterial genus Prevotellaceae (217). These mice presented a steady state colitic phenotype and an enhanced sensitivity to DSS colitis (217). Overall, a mechanism has been suggested wherein dysbiosis in the gut, caused by aberrant NLRP6 inflammasome signaling, drives excessive CCL5-mediated IL-6 production, barrier damage, and inflammation (217). In agreement with the findings in Casp1−/− mice (199), NLRP6-deficient mice had impaired IL-18 production mainly from the intestinal epithelial compartment further diminishing the capacity of these mice to recover from colitis. Likewise, overt inflammation and lack of IL-18 in the Nlrp6−/− mice has been associated with increased colonic tumor development (216), however, as seen for Nlrp3−/− mice it is still unknown whether administration of IL-18 is capable of rescuing the susceptibility phenotype. Interestingly, gene expression profiling of colorectal tumors derived from WT and Nlrp6−/− mice revealed an increased expression of paracrine factors of the Wnt and NOTCH signaling cascades, underscoring a novel function of NLRP6 in controlling intestinal proliferation (218). Sensing of damaged or dying cells by NLRP6 and NLRP3 inflammasomes has lately been hypothesized to prevent CRC through maintaining the balance between IL-22 and IL-22 binding protein (IL22-BP) (219). It has been speculated that sensing of DAMPs by both NLRs instigates IL-18-dependent down-regulation of the inhibitory molecule IL-22BP, thus allowing IL-22 to repair the injured tissue. However, dysregulated NLRP6 or NLRP3 signaling could potentially lead to inappropriate IL-22BP expression, thus creating a pro-tumorigenic environment caused by either excessive cell proliferation or lack of tissue repair (219). Although the dual function of IL-22 in CRC has been well-described, further experimental validation is needed to pinpoint the exact mode by which NLRP3 or NLRP6 regulate IL-22/IL-22BP ratio during colon tumorigenesis.
NLRP12 was originally defined as an inflammasome NLR due to its co-localization with ASC and caspase-1, induction of IL-1β and IL-18 secretion as well as NF-κB activation (220, 221). SNPs within the NLRP12 gene have been associated with increased susceptibility to atopic dermatitis and periodic fever syndromes accompanied mostly with caspase-1 activation and IL-1β release (222–225). It has been observed that NLRP12 can negatively regulate both canonical and non-canonical NF-κB pathways by targeting the IL-1R-associated kinase 1 (IRAK1) and NF-κB inducing kinase (NIK) for proteasomal degradation (226–228). Two independent studies proposed that NLRP12 acts as a tumor suppressive molecule ex vivo and in in vivo animal models of colitis and colitis-induced CRC (229, 230). Mice lacking NLRP12 have been found to be more susceptible to DSS-injury with increased body weight loss, enhanced pathology scores coupled with massive infiltration of inflammatory cells and high inflammatory cytokine production (229, 230). Furthermore, AOM–DSS treatment of Nlrp12−/− mice has been shown to further provoke colonic tumor development and progression (229, 230). In the first study, it was clearly demonstrated that lack of NLRP12 increases NIK-dependent non-canonical NF-κB signaling and drives the regulation of cancer promoting genes like CXCL12 and CXCL13 (230). In the second report, the enhanced tumorigenicity in knockout mice was traced to excessive canonical NF-κB activation due to lack of NLRP12 in hematopoietic cells. Indeed, enhanced LPS-induced canonical NF-κB activation was exhibited in Nlrp12−/− macrophages ex vivo, suggesting that microbial sensing and negative regulation of inflammation may account for NLRP12-mediated tumor suppression (229). Altogether, these results underscore the importance of anti-inflammatory signals provided by NLRP12 in maintaining colonic homeostasis and protecting from colitis and colon tumorigenesis.
Therapeutic Strategies and Conclusion
It has been suggested that the strong immunomodulatory properties of NLRs could be exploited for mounting potent anti-tumorigenic responses. In fact, mice injected with B16 melanoma cells or EL4 thymoma cells expressing flagellin from Salmonella typhimurium were shown to display dramatic resistance to tumor establishment in NLRC4 dependent manner (231). In addition, immunization with flagellin expressing cancer cells lead to impressive antigen-specific CD4 and CD8 T cell responses via NLRC4 and NAIP5 signaling and bestowed anti-tumor immunity against a secondary inoculation with tumor cells (231). Similarly, activation of NODs, in particular NOD2, to elicit robust cell-based anti-tumor immunity has been under scrutiny for several years. Indeed, instillation of MDP in patients with lung cancer has been reported to enhance expression of inflammatory cytokines and neutrophils in the pleural fluid (232). Relatedly, it has been suggested that the local immune-modulatory activity of MDP helps improve prognosis in hamsters suffering from osteosarcoma (233) and significantly reduces tumor metastasis in several murine cancer models, such as B16–BL6 melanoma, colon 26-M#1 carcinoma, and L5178Y-ML25T T lymphoma (234, 235).
Overt activation of the NLRP3 inflammasome has been demonstrated to elicit cancer progression. For instance, in mouse models of methylcholanthrene (MCA, a highly potent carcinogen) induced fibrosarcoma, NLRP3 was demonstrated to promote cancer progression. Moreover, NLRP3 expression in myeloid cells was shown to interfere with the suppression of cancer metastasis by inhibiting recruitment of anti-tumor NK cells to the site of carcinogenesis (236). Besides interfering with natural tumor control, NLRP3 inflammasome-mediated IL-1β has been described to attenuate anti-tumor effects of chemotherapeutic agents, gemcitabine (Gem), and 5-fluorouracil (5FU) (237). Mice lacking NLRP3 were far more receptive to thymoma regression upon treatment with Gem or 5FU as compared to WT mice. Furthermore, enhanced NLRP3-driven IL-1β release was linked with the induction of T helper 17 (Th17) cells that promoted chemo-resistance in WT mice (237). Keeping these observations in view, several studies support the use of specific inhibitors, antagonists, and monoclonal antibodies against components of the inflammasome, e.g., caspase-1, IL-1β, and IL-18, as therapeutic approaches beneficial for controlling inflammation and improving cancer prognosis (238).
An early phase clinical study suggests that administration of the IL-1R antagonist, Anakinra, alone or in combination with dexamethasone could potentially impede human multiple myeloma progression (239). Furthermore, it was demonstrated that IL-18 derived from tumor cells had the ability to subvert the NK cell-mediated tumor immunosurveillance and to promote tumor progression in a programed death receptor 1 (PD1)-dependent manner (240, 241). These findings suggest the potential of using IL-18 as well as PD1 neutralization for cancer immunotherapy. Overall, selective attenuation of the activities of certain NLRs could potentially boost regression and improve responsiveness to chemotherapy. The variability in NLRP3- and IL-18-mediated effects in different cancers highlights the complexity in NLR circuits and suggests that any broad implications regarding NLR intervention in tumorigenesis should be carefully investigated.
Microbial environment, diet, mouse strain, tumor ontogeny, etc. are all part of the complex network that dictates how an NLR influences inflammation and tumorigenesis. Sensitivity to these factors has lead to conflicting disease phenotypes in genetically modified mice lacking specific NLRs. Furthermore, NLR expression in hematopoietic or non-hematopoietic cellular compartments appears to have distinct influence on inflammatory regulation and tumorigenesis. Due to such discrepancies, it is still uncertain how dysregulation of these innate immune sensors incites inflammation that leads to carcinogenic transformation of cells. Although several mechanisms have been suggested like control of NF-κB signaling, regulation of tissue repair factors, and IL-18 secretion, no unifying hypothesis exists. In addition, interaction of NLRs with different members of the TNFR pathway, BCL2 family of proteins, IAPs, apoptotic caspases, and autophagy regulators point toward more intricate mechanisms for NLR regulation than currently acknowledged. Future studies focusing on the biochemistry of interactions between cell death regulators and NLRs are required to delineate the co-integration of NLR-cell death mechanisms so as to facilitate implementation of NLR modifying therapeutic strategies for inflammatory diseases and cancer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the Yeretssian lab members for the insightful and critical reading of the manuscript. This work was supported by the Helmsley Foundation.
References
1. Janeway CA Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol (2002) 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359
2. Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol (2007) 81:1–5. doi:10.1189/jlb.0306164
3. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity (2011) 34:637–50. doi:10.1016/j.immuni.2011.05.006
4. Hansen JD, Vojtech LN, Laing KJ. Sensing disease and danger: a survey of vertebrate PRRs and their origins. Dev Comp Immunol (2011) 35:886–97. doi:10.1016/j.dci.2011.01.008
5. Yeretssian G. Effector functions of NLRs in the intestine: innate sensing, cell death, and disease. Immunol Res (2012) 54:25–36. doi:10.1007/s12026-012-8317-3
6. Wen H, Miao EA, Ting JP. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity (2013) 39:432–41. doi:10.1016/j.immuni.2013.08.037
7. Goubau D, Deddouche S, Reis E, Sousa C. Cytosolic sensing of viruses. Immunity (2013) 38:855–69. doi:10.1016/j.immuni.2013.05.007
8. Hardison SE, Brown GD. C-type lectin receptors orchestrate antifungal immunity. Nat Immunol (2012) 13:817–22. doi:10.1038/ni.2369
9. Ratsimandresy RA, Dorfleutner A, Stehlik C. An update on PYRIN domain-containing pattern recognition receptors: from immunity to pathology. Front Immunol (2013) 4:440. doi:10.3389/fimmu.2013.00440
10. Janowski AM, Kolb R, Zhang W, Sutterwala FS. Beneficial and detrimental roles of NLRs in carcinogenesis. Front Immunol (2013) 4:370. doi:10.3389/fimmu.2013.00370
11. Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet (2001) 357:539–45. doi:10.1016/S0140-6736(00)04046-0
12. Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature (2008) 454:436–44. doi:10.1038/nature07205
13. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell (2010) 140:883–99. doi:10.1016/j.cell.2010.01.025
14. de Martel C, Franceschi S. Infections and cancer: established associations and new hypotheses. Crit Rev Oncol Hematol (2009) 70:183–94. doi:10.1016/j.critrevonc.2008.07.021
15. Takahashi H, Ogata H, Nishigaki R, Broide DH, Karin M. Tobacco smoke promotes lung tumorigenesis by triggering IKKbeta- and JNK1-dependent inflammation. Cancer Cell (2010) 17:89–97. doi:10.1016/j.ccr.2009.12.008
16. Khasawneh J, Schulz MD, Walch A, Rozman J, Hrabe De Angelis M, Klingenspor M, et al. Inflammation and mitochondrial fatty acid beta-oxidation link obesity to early tumor promotion. Proc Natl Acad Sci U S A (2009) 106:3354–9. doi:10.1073/pnas.0802864106
17. Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell (2010) 140:197–208. doi:10.1016/j.cell.2009.12.052
18. Ershler WB, Keller ET. Age-associated increased interleukin-6 gene expression, late-life diseases, and frailty. Annu Rev Med (2000) 51:245–70. doi:10.1146/annurev.med.51.1.245
19. Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol (2009) 11:973–9. doi:10.1038/ncb1909
21. Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell (2014) 54:716–27. doi:10.1016/j.molcel.2014.05.015
22. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi:10.1016/j.cell.2011.02.013
23. Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J (2011) 30:3667–83. doi:10.1038/emboj.2011.307
24. Bai L, Smith DC, Wang S. Small-molecule SMAC mimetics as new cancer therapeutics. Pharmacol Ther (2014). doi:10.1016/j.pharmthera.2014.05.007
25. Suzuki Y, Nakabayashi Y, Nakata K, Reed JC, Takahashi R. X-linked inhibitor of apoptosis protein (XIAP) inhibits caspase-3 and -7 in distinct modes. J Biol Chem (2001) 276:27058–63. doi:10.1074/jbc.M102415200
26. Busca A, Saxena M, Kryworuchko M, Kumar A. Anti-apoptotic genes in the survival of monocytic cells during infection. Curr Genomics (2009) 10:306–17. doi:10.2174/138920209788920967
27. Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer (2010) 10:561–74. doi:10.1038/nrc2889
28. Saxena M, Busca A, Pandey S, Kryworuchko M, Kumar A. CpG protects human monocytic cells against HIV-Vpr-induced apoptosis by cellular inhibitor of apoptosis-2 through the calcium-activated JNK pathway in a TLR9-independent manner. J Immunol (2011) 187:5865–78. doi:10.4049/jimmunol.1100115
29. Busca A, Saxena M, Kumar A. Critical role for antiapoptotic Bcl-xL and Mcl-1 in human macrophage survival and cellular IAP1/2 (cIAP1/2) in resistance to HIV-Vpr-induced apoptosis. J Biol Chem (2012) 287:15118–33. doi:10.1074/jbc.M111.312660
30. Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol (2008) 9:47–59. doi:10.1038/nrm2308
31. Kelly PN, Strasser A. The role of Bcl-2 and its pro-survival relatives in tumourigenesis and cancer therapy. Cell Death Differ (2011) 18:1414–24. doi:10.1038/cdd.2011.17
32. Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol (2002) 4:842–9. doi:10.1038/ncb866
33. Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ (2009) 16:684–96. doi:10.1038/cdd.2008.195
34. Donehower LA, Harvey M, Slagle BL, Mcarthur MJ, Montgomery CA Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature (1992) 356:215–21. doi:10.1038/356215a0
35. Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol (2010) 2:a001008. doi:10.1101/cshperspect.a001008
36. Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer (2014) 14:359–70. doi:10.1038/nrc3711
37. Hager KM, Gu W. Understanding the non-canonical pathways involved in p53-mediated tumor suppression. Carcinogenesis (2014) 35:740–6. doi:10.1093/carcin/bgt487
38. Chretien AS, Le Roy A, Vey N, Prebet T, Blaise D, Fauriat C, et al. Cancer-induced alterations of NK-mediated target recognition: current and investigational pharmacological strategies aiming at restoring NK-mediated anti-tumor activity. Front Immunol (2014) 5:122. doi:10.3389/fimmu.2014.00122
39. Vivier E, Ugolini S, Blaise D, Chabannon C, Brossay L. Targeting natural killer cells and natural killer T cells in cancer. Nat Rev Immunol (2012) 12:239–52. doi:10.1038/nri3174
40. Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, et al. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J Biol Chem (1999) 274:12955–8. doi:10.1074/jbc.274.19.12955
41. Inohara N, Koseki T, Del Peso L, Hu Y, Yee C, Chen S, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem (1999) 274:14560–7. doi:10.1074/jbc.274.21.14560
42. Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem (2001) 276:4812–8. doi:10.1074/jbc.M008072200
43. Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, et al. The NLR gene family: a standard nomenclature. Immunity (2008) 28:285–7. doi:10.1016/j.immuni.2008.02.005
44. Nickerson K, Sisk TJ, Inohara N, Yee CS, Kennell J, Cho MC, et al. Dendritic cell-specific MHC class II transactivator contains a caspase recruitment domain that confers potent transactivation activity. J Biol Chem (2001) 276:19089–93. doi:10.1074/jbc.M101295200
45. Diez E, Lee SH, Gauthier S, Yaraghi Z, Tremblay M, Vidal S, et al. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat Genet (2003) 33:55–60. doi:10.1038/ng1065
46. Wright EK, Goodart SA, Growney JD, Hadinoto V, Endrizzi MG, Long EM, et al. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol (2003) 13:27–36. doi:10.1016/S0960-9822(02)01359-3
47. Lightfield KL, Persson J, Brubaker SW, Witte CE, Von Moltke J, Dunipace EA, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol (2008) 9:1171–8. doi:10.1038/ni.1646
48. Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol (2014) 14:9–23. doi:10.1038/nri3565
49. Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature (2001) 411:599–603. doi:10.1038/35079107
50. Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature (2001) 411:603–6. doi:10.1038/35079114
51. Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell (2002) 10:417–26. doi:10.1016/S1097-2765(02)00599-3
52. Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature (2008) 451:573–7. doi:10.1038/nature06501
53. Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity (2011) 34:854–65. doi:10.1016/j.immuni.2011.03.026
54. Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q, et al. NLRX1 negatively regulates TLR-induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity (2011) 34:843–53. doi:10.1016/j.immuni.2011.02.022
55. Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity (2012) 36:933–46. doi:10.1016/j.immuni.2012.03.025
56. Lei Y, Wen H, Ting JP. The NLR protein, NLRX1, and its partner, TUFM, reduce type I interferon, and enhance autophagy. Autophagy (2013) 9:432–3. doi:10.4161/auto.23026
57. Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep (2008) 9:293–300. doi:10.1038/sj.embor.7401161
58. Abdul-Sater AA, Said-Sadier N, Lam VM, Singh B, Pettengill MA, Soares F, et al. Enhancement of reactive oxygen species production and chlamydial infection by the mitochondrial Nod-like family member NLRX1. J Biol Chem (2010) 285:41637–45. doi:10.1074/jbc.M110.137885
59. Soares F, Tattoli I, Wortzman ME, Arnoult D, Philpott DJ, Girardin SE. NLRX1 does not inhibit MAVS-dependent antiviral signalling. Innate Immun (2013) 19:438–48. doi:10.1177/1753425912467383
60. Rebsamen M, Vazquez J, Tardivel A, Guarda G, Curran J, Tschopp J. NLRX1/NOD5 deficiency does not affect MAVS signalling. Cell Death Differ (2011) 18:1387. doi:10.1038/cdd.2011.64
61. Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol (2012) 13:325–32. doi:10.1038/ni.2231
62. Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, et al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat Genet (2009) 41:71–6. doi:10.1038/ng.285
63. Kutikhin AG. Role of NOD1/CARD4 and NOD2/CARD15 gene polymorphisms in cancer etiology. Hum Immunol (2011) 72:955–68. doi:10.1016/j.humimm.2011.06.003
64. Jostins L, Ripke S, Weersma RK, Duerr RH, Mcgovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature (2012) 491:119–24. doi:10.1038/nature11582
65. Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol (2003) 4:702–7. doi:10.1038/ni945
66. Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science (2003) 300:1584–7. doi:10.1126/science.1084677
67. Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, Philpott DJ, et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem (2003) 278:41702–8. doi:10.1074/jbc.M307198200
68. Ogura Y, Lala S, Xin W, Smith E, Dowds TA, Chen FF, et al. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut (2003) 52:1591–7. doi:10.1136/gut.52.11.1591
69. Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem (2003) 278:8869–72. doi:10.1074/jbc.C200651200
70. Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol (2004) 5:1166–74. doi:10.1038/ni1131
71. Lee J, Tattoli I, Wojtal KA, Vavricka SR, Philpott DJ, Girardin SE. pH-dependent internalization of muramyl peptides from early endosomes enables Nod1 and Nod2 signaling. J Biol Chem (2009) 284:23818–29. doi:10.1074/jbc.M109.033670
72. Marina-Garcia N, Franchi L, Kim YG, Hu Y, Smith DE, Boons GJ, et al. Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J Immunol (2009) 182:4321–7. doi:10.4049/jimmunol.0802197
73. Nakamura N, Lill JR, Phung Q, Jiang Z, Bakalarski C, De Maziere A, et al. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature (2014) 509:240–4. doi:10.1038/nature13133
74. Fukazawa A, Alonso C, Kurachi K, Gupta S, Lesser CF, Mccormick BA, et al. GEF-H1 mediated control of NOD1 dependent NF-kappaB activation by Shigella effectors. PLoS Pathog (2008) 4:e1000228. doi:10.1371/journal.ppat.1000228
75. Kufer TA, Kremmer E, Adam AC, Philpott DJ, Sansonetti PJ. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol (2008) 10:477–86.
76. Keestra AM, Winter MG, Auburger JJ, Frassle SP, Xavier MN, Winter SE, et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature (2013) 496:233–7. doi:10.1038/nature12025
77. Chin AI, Dempsey PW, Bruhn K, Miller JF, Xu Y, Cheng G. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature (2002) 416:190–4. doi:10.1038/416190a
78. Park JH, Kim YG, Mcdonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol (2007) 178:2380–6. doi:10.4049/jimmunol.178.4.2380
79. Watanabe T, Asano N, Fichtner-Feigl S, Gorelick PL, Tsuji Y, Matsumoto Y, et al. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Invest (2010) 120:1645–62. doi:10.1172/JCI39481
80. Magalhaes JG, Lee J, Geddes K, Rubino S, Philpott DJ, Girardin SE. Essential role of Rip2 in the modulation of innate and adaptive immunity triggered by Nod1 and Nod2 ligands. Eur J Immunol (2011) 41:1445–55. doi:10.1002/eji.201040827
81. Inohara N, Koseki T, Lin J, Del Peso L, Lucas PC, Chen FF, et al. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem (2000) 275:27823–31.
82. Inohara N, Del Peso L, Koseki T, Chen S, Nunez G. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J Biol Chem (1998) 273:12296–300. doi:10.1074/jbc.273.20.12296
83. McCarthy JV, Ni J, Dixit VM. RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J Biol Chem (1998) 273:16968–75. doi:10.1074/jbc.273.27.16968
84. Thome M, Hofmann K, Burns K, Martinon F, Bodmer JL, Mattmann C, et al. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr Biol (1998) 8:885–8. doi:10.1016/S0960-9822(07)00352-1
85. Eickhoff J, Hanke M, Stein-Gerlach M, Kiang TP, Herzberger K, Habenberger P, et al. RICK activates a NF-kappaB-dependent anti-human cytomegalovirus response. J Biol Chem (2004) 279:9642–52. doi:10.1074/jbc.M312893200
86. Windheim M, Lang C, Peggie M, Plater LA, Cohen P. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem J (2007) 404:179–90. doi:10.1042/BJ20061704
87. Nembrini C, Kisielow J, Shamshiev AT, Tortola L, Coyle AJ, Kopf M, et al. The kinase activity of Rip2 determines its stability and consequently Nod1- and Nod2-mediated immune responses. J Biol Chem (2009) 284:19183–8. doi:10.1074/jbc.M109.006353
88. Yang Y, Yin C, Pandey A, Abbott D, Sassetti C, Kelliher MA. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J Biol Chem (2007) 282:36223–9. doi:10.1074/jbc.M703079200
89. Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J (2008) 27:373–83. doi:10.1038/sj.emboj.7601962
90. Tigno-Aranjuez JT, Asara JM, Abbott DW. Inhibition of RIP2’s tyrosine kinase activity limits NOD2-driven cytokine responses. Genes Dev (2010) 24:2666–77. doi:10.1101/gad.1964410
91. Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol (2004) 14:2217–27. doi:10.1016/j.cub.2004.12.032
92. Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol (2007) 27:6012–25. doi:10.1128/MCB.00270-07
93. Navas TA, Baldwin DT, Stewart TA. RIP2 is a Raf1-activated mitogen-activated protein kinase kinase. J Biol Chem (1999) 274:33684–90. doi:10.1074/jbc.274.47.33684
94. Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity (2009) 30:789–801. doi:10.1016/j.immuni.2009.04.011
95. Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z, et al. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci U S A (2009) 106:14524–9. doi:10.1073/pnas.0907131106
96. Damgaard RB, Nachbur U, Yabal M, Wong WW, Fiil BK, Kastirr M, et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell (2012) 46:746–58. doi:10.1016/j.molcel.2012.04.014
97. Tokunaga F, Iwai K. Linear ubiquitination: a novel NF-kappaB regulatory mechanism for inflammatory and immune responses by the LUBAC ubiquitin ligase complex. Endocr J (2012) 59:641–52. doi:10.1507/endocrj.EJ12-0148
98. Damgaard RB, Fiil BK, Speckmann C, Yabal M, Zur Stadt U, Bekker-Jensen S, et al. Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Mol Med (2013) 5:1278–95. doi:10.1002/emmm.201303090
99. Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, et al. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest (2008) 118:545–59. doi:10.1172/JCI33145
100. Reardon C, Mak TW. cIAP proteins: keystones in NOD receptor signal transduction. Immunity (2009) 30:755–6. doi:10.1016/j.immuni.2009.06.005
101. Yeretssian G, Correa RG, Doiron K, Fitzgerald P, Dillon CP, Green DR, et al. Non-apoptotic role of BID in inflammation and innate immunity. Nature (2011) 474:96–9. doi:10.1038/nature09982
102. Tao M, Scacheri PC, Marinis JM, Harhaj EW, Matesic LE, Abbott DW. ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr Biol (2009) 19:1255–63. doi:10.1016/j.cub.2009.06.038
103. Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell (2008) 30:689–700. doi:10.1016/j.molcel.2008.05.014
104. da Silva Correia J, Miranda Y, Austin-Brown N, Hsu J, Mathison J, Xiang R, et al. Nod1-dependent control of tumor growth. Proc Natl Acad Sci U S A (2006) 103:1840–5. doi:10.1073/pnas.0509228103
105. da Silva Correia J, Miranda Y, Leonard N, Hsu J, Ulevitch RJ. Regulation of Nod1-mediated signaling pathways. Cell Death Differ (2007) 14:830–9. doi:10.1038/sj.cdd.4402070
106. Yeretssian G, Labbe K, Saleh M. Molecular regulation of inflammation and cell death. Cytokine (2008) 43:380–90. doi:10.1016/j.cyto.2008.07.015
107. Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. Non-apoptotic functions of apoptosis-regulatory proteins. EMBO Rep (2012) 13:322–30. doi:10.1038/embor.2012.19
108. Yeretssian G, Correa RG, Doiron K, Fitzgerald P, Dillon CP, Green DR, et al. Is BID required for NOD signalling? reply. Nature (2012) 488:E6–8. doi:10.1038/nature11367
109. Nachbur U, Vince JE, O’Reilly LA, Strasser A, Silke J. Is BID required for NOD signalling? Nature (2012) 488:E4–6. doi:10.1038/nature11366
110. Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet (2007) 39:207–11. doi:10.1038/ng1954
111. Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet (2007) 39:596–604. doi:10.1038/ng2032
112. Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol (2010) 11:55–62. doi:10.1038/ni.1823
113. Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med (2010) 16:90–7. doi:10.1038/nm.2069
114. Homer CR, Richmond AL, Rebert NA, Achkar JP, Mcdonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology (2010) 139:1630–41. doi:10.1053/j.gastro.2010.07.006
115. Homer CR, Kabi A, Marina-Garcia N, Sreekumar A, Nesvizhskii AI, Nickerson KP, et al. A dual role for receptor-interacting protein kinase 2 (RIP2) kinase activity in nucleotide-binding oligomerization domain 2 (NOD2)-dependent autophagy. J Biol Chem (2012) 287:25565–76. doi:10.1074/jbc.M111.326835
116. Sorbara MT, Ellison LK, Ramjeet M, Travassos LH, Jones NL, Girardin SE, et al. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity (2013) 39:858–73. doi:10.1016/j.immuni.2013.10.013
117. Yomade O, Spies-Weisshart B, Glaser A, Schnetzke U, Hochhaus A, Scholl S. Impact of NOD2 polymorphisms on infectious complications following chemotherapy in patients with acute myeloid leukaemia. Ann Hematol (2013) 92:1071–7. doi:10.1007/s00277-013-1734-0
118. Hnatyszyn A, Szalata M, Stanczyk J, Cichy W, Slomski R. Association of c.802C >T polymorphism of NOD2/CARD15 gene with the chronic gastritis and predisposition to cancer in H. pylori infected patients. Exp Mol Pathol (2010) 88:388–93. doi:10.1016/j.yexmp.2010.03.003
119. Huzarski T, Lener M, Domagala W, Gronwald J, Byrski T, Kurzawski G, et al. The 3020insC allele of NOD2 predisposes to early-onset breast cancer. Breast Cancer Res Treat (2005) 89:91–3. doi:10.1007/s10549-004-1250-y
120. Lener MR, Oszutowska D, Castaneda J, Kurzawski G, Suchy J, Nej-Wolosiak K, et al. Prevalence of the NOD2 3020insC mutation in aggregations of breast and lung cancer. Breast Cancer Res Treat (2006) 95:141–5. doi:10.1007/s10549-005-9057-z
121. Rigoli L, Di Bella C, Fedele F, Procopio V, Amorini M, Lo Giudice G, et al. TLR4 and NOD2/CARD15 genetic polymorphisms and their possible role in gastric carcinogenesis. Anticancer Res (2010) 30:513–7.
122. Chen GY, Shaw MH, Redondo G, Nunez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res (2008) 68:10060–7. doi:10.1158/0008-5472.CAN-08-2061
123. Tanaka T, Kohno H, Suzuki R, Yamada Y, Sugie S, Mori H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci (2003) 94:965–73. doi:10.1111/j.1349-7006.2003.tb01386.x
124. Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science (1990) 247:322–4. doi:10.1126/science.2296722
125. Itzkowitz S. Colon carcinogenesis in inflammatory bowel disease: applying molecular genetics to clinical practice. J Clin Gastroenterol (2003) 36:S70–4. doi:10.1097/00004836-200305001-00012
126. Salinger AP, Justice MJ. Mouse mutagenesis using N-Ethyl-N-nitrosourea (ENU). CSH Protoc (2008) 2008:pdb.prot4985. doi:10.1101/pdb.prot4985
127. Nassiri M, Kooshyar MM, Roudbar Z, Mahdavi M, Doosti M. Genes and SNPs associated with non-hereditary and hereditary colorectal cancer. Asian Pac J Cancer Prev (2013) 14:5609–14. doi:10.7314/APJCP.2013.14.10.5609
128. Maruyama K, Ochiai A, Akimoto S, Nakamura S, Baba S, Moriya Y, et al. Cytoplasmic beta-catenin accumulation as a predictor of hematogenous metastasis in human colorectal cancer. Oncology (2000) 59:302–9. doi:10.1159/000012187
129. Yamada Y, Mori H. Multistep carcinogenesis of the colon in Apc(Min/+) mouse. Cancer Sci (2007) 98:6–10. doi:10.1111/j.1349-7006.2006.00348.x
130. Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest (2013) 123:700–11. doi:10.1172/JCI62236
131. Millrud CR, Kvarnhammar AM, Tajti J, Munck-Wikland E, Uddman R, Cardell LO. Nod-like receptors in head and neck squamous cell carcinoma. Acta Otolaryngol (2013) 133:1333–44. doi:10.3109/00016489.2013.831476
132. Schroder K, Tschopp J. The inflammasomes. Cell (2010) 140:821–32. doi:10.1016/j.cell.2010.01.040
133. Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet (2001) 29:301–5. doi:10.1038/ng756
134. Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature (2004) 430:213–8. doi:10.1038/nature02664
135. Mariathasan S, Monack DM. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol (2007) 7:31–40. doi:10.1038/nri1997
136. Agostini L, Martinon F, Burns K, Mcdermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity (2004) 20:319–25. doi:10.1016/S1074-7613(04)00046-9
137. Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature (2006) 440:233–6. doi:10.1038/nature04517
138. Mariathasan S, Weiss DS, Newton K, Mcbride J, O’Rourke K, Roose-Girma M, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature (2006) 440:228–32. doi:10.1038/nature04515
139. Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature (2006) 440:237–41. doi:10.1038/nature04516
140. Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity (2006) 24:317–27. doi:10.1016/j.immuni.2006.02.004
141. Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol (2009) 7:99–109. doi:10.1038/nrmicro2070
142. Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell (2013) 153:348–61. doi:10.1016/j.cell.2013.02.054
143. Kummer JA, Broekhuizen R, Everett H, Agostini L, Kuijk L, Martinon F, et al. Inflammasome components NALP 1 and 3 show distinct but separate expression profiles in human tissues suggesting a site-specific role in the inflammatory response. J Histochem Cytochem (2007) 55:443–52. doi:10.1369/jhc.6A7101.2006
144. Guarda G, Zenger M, Yazdi AS, Schroder K, Ferrero I, Menu P, et al. Differential expression of NLRP3 among hematopoietic cells. J Immunol (2011) 186:2529–34. doi:10.4049/jimmunol.1002720
145. Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol (2011) 29:707–35. doi:10.1146/annurev-immunol-031210-101405
146. Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol (2012) 28:137–61. doi:10.1146/annurev-cellbio-101011-155745
147. Muniz LR, Knosp C, Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front Immunol (2012) 3:310. doi:10.3389/fimmu.2012.00310
148. Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol (2013) 13:397–411. doi:10.1038/nri3452
149. Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, Macdonald K, Speert D, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol (2009) 183:787–91. doi:10.4049/jimmunol.0901363
150. Ghonime MG, Shamaa OR, Das S, Eldomany RA, Fernandes-Alnemri T, Alnemri ES, et al. Inflammasome priming by lipopolysaccharide is dependent upon ERK signaling and proteasome function. J Immunol (2014) 192:3881–8. doi:10.4049/jimmunol.1301974
151. Schumann RR, Belka C, Reuter D, Lamping N, Kirschning CJ, Weber JR, et al. Lipopolysaccharide activates caspase-1 (interleukin-1-converting enzyme) in cultured monocytic and endothelial cells. Blood (1998) 91:577–84.
152. Netea MG, Nold-Petry CA, Nold MF, Joosten LA, Opitz B, Van Der Meer JH, et al. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood (2009) 113:2324–35. doi:10.1182/blood-2008-03-146720
153. Bauernfeind F, Bartok E, Rieger A, Franchi L, Nunez G, Hornung V. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J Immunol (2011) 187:613–7. doi:10.4049/jimmunol.1100613
154. Juliana C, Fernandes-Alnemri T, Kang S, Farias A, Qin F, Alnemri ES. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem (2012) 287:36617–22. doi:10.1074/jbc.M112.407130
155. Craven RR, Gao X, Allen IC, Gris D, Bubeck Wardenburg J, Mcelvania-Tekippe E, et al. Staphylococcus aureus alpha-hemolysin activates the NLRP3-inflammasome in human and mouse monocytic cells. PLoS One (2009) 4:e7446. doi:10.1371/journal.pone.0007446
156. McNeela EA, Burke A, Neill DR, Baxter C, Fernandes VE, Ferreira D, et al. Pneumolysin activates the NLRP3 inflammasome and promotes proinflammatory cytokines independently of TLR4. PLoS Pathog (2010) 6:e1001191. doi:10.1371/journal.ppat.1001191
157. Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem (2006) 281:36560–8. doi:10.1074/jbc.M607594200
158. Allen IC, Scull MA, Moore CB, Holl EK, Mcelvania-Tekippe E, Taxman DJ, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity (2009) 30:556–65. doi:10.1016/j.immuni.2009.02.005
159. Thomas PG, Dash P, Aldridge JR Jr., Ellebedy AH, Reynolds C, Funk AJ, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity (2009) 30:566–75. doi:10.1016/j.immuni.2009.02.006
160. Rajan JV, Warren SE, Miao EA, Aderem A. Activation of the NLRP3 inflammasome by intracellular poly I:C. FEBS Lett (2010) 584:4627–32. doi:10.1016/j.febslet.2010.10.036
161. Kanneganti TD, Lamkanfi M, Kim YG, Chen G, Park JH, Franchi L, et al. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity (2007) 26:433–43. doi:10.1016/j.immuni.2007.03.008
162. Piccini A, Carta S, Tassi S, Lasiglie D, Fossati G, Rubartelli A. ATP is released by monocytes stimulated with pathogen-sensing receptor ligands and induces IL-1beta and IL-18 secretion in an autocrine way. Proc Natl Acad Sci U S A (2008) 105:8067–72. doi:10.1073/pnas.0709684105
163. Gombault A, Baron L, Couillin I. ATP release and purinergic signaling in NLRP3 inflammasome activation. Front Immunol (2012) 3:414. doi:10.3389/fimmu.2012.00414
164. Ayna G, Krysko DV, Kaczmarek A, Petrovski G, Vandenabeele P, Fesus L. ATP release from dying autophagic cells and their phagocytosis are crucial for inflammasome activation in macrophages. PLoS One (2012) 7:e40069. doi:10.1371/journal.pone.0040069
165. Cassel SL, Eisenbarth SC, Iyer SS, Sadler JJ, Colegio OR, Tephly LA, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci U S A (2008) 105:9035–40. doi:10.1073/pnas.0803933105
166. Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science (2008) 320:674–7. doi:10.1126/science.1156995
167. Eisenbarth SC, Colegio OR, O’Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature (2008) 453:1122–6. doi:10.1038/nature06939
168. Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol (2008) 9:857–65. doi:10.1038/ni.1636
169. Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol (2008) 9:847–56. doi:10.1038/ni.1631
170. Kingsbury SR, Conaghan PG, Mcdermott MF. The role of the NLRP3 inflammasome in gout. J Inflamm Res (2011) 4:39–49. doi:10.2147/JIR.S11330
171. Dostert C, Guarda G, Romero JF, Menu P, Gross O, Tardivel A, et al. Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PLoS One (2009) 4:e6510. doi:10.1371/journal.pone.0006510
172. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol (2011) 12:222–30. doi:10.1038/ni.1980
173. Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature (2011) 469:221–5. doi:10.1038/nature09663
174. Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity (2012) 36:401–14. doi:10.1016/j.immuni.2012.01.009
175. Zhong Z, Zhai Y, Liang S, Mori Y, Han R, Sutterwala FS, et al. TRPM2 links oxidative stress to NLRP3 inflammasome activation. Nat Commun (2013) 4:1611. doi:10.1038/ncomms2608
176. Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity (2013) 39:311–23. doi:10.1016/j.immuni.2013.08.001
177. Lee GS, Subramanian N, Kim AI, Aksentijevich I, Goldbach-Mansky R, Sacks DB, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature (2012) 492:123–7. doi:10.1038/nature11588
178. Murakami T, Ockinger J, Yu J, Byles V, Mccoll A, Hofer AM, et al. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci U S A (2012) 109:11282–7. doi:10.1073/pnas.1117765109
179. Rossol M, Pierer M, Raulien N, Quandt D, Meusch U, Rothe K, et al. Extracellular Ca2+ is a danger signal activating the NLRP3 inflammasome through G protein-coupled calcium sensing receptors. Nat Commun (2012) 3:1329. doi:10.1038/ncomms2339
180. Shenoy AR, Wellington DA, Kumar P, Kassa H, Booth CJ, Cresswell P, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science (2012) 336:481–5. doi:10.1126/science.1217141
181. Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity (2013) 38:1142–53. doi:10.1016/j.immuni.2013.05.016
182. Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature (2008) 456:264–8. doi:10.1038/nature07383
183. Harris J, Hartman M, Roche C, Zeng SG, O’Shea A, Sharp FA, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem (2011) 286:9587–97. doi:10.1074/jbc.M110.202911
184. Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol (2012) 13:255–63. doi:10.1038/ni.2215
185. Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol (2013) 14:480–8. doi:10.1038/ni.2563
186. Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, Xu C, et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell (2007) 129:45–56. doi:10.1016/j.cell.2007.01.045
187. Faustin B, Chen Y, Zhai D, Le Negrate G, Lartigue L, Satterthwait A, et al. Mechanism of Bcl-2 and Bcl-X(L) inhibition of NLRP1 inflammasome: loop domain-dependent suppression of ATP binding and oligomerization. Proc Natl Acad Sci U S A (2009) 106:3935–40. doi:10.1073/pnas.0809414106
188. Hawkins CJ, Uren AG, Hacker G, Medcalf RL, Vaux DL. Inhibition of interleukin 1 beta-converting enzyme-mediated apoptosis of mammalian cells by baculovirus IAP. Proc Natl Acad Sci U S A (1996) 93:13786–90. doi:10.1073/pnas.93.24.13786
189. Labbe K, Mcintire CR, Doiron K, Leblanc PM, Saleh M. Cellular inhibitors of apoptosis proteins cIAP1 and cIAP2 are required for efficient caspase-1 activation by the inflammasome. Immunity (2011) 35:897–907. doi:10.1016/j.immuni.2011.10.016
190. Vince JE, Wong WW, Gentle I, Lawlor KE, Allam R, O’Reilly L, et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity (2012) 36:215–27. doi:10.1016/j.immuni.2012.01.012
191. Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, et al. Non-canonical inflammasome activation targets caspase-11. Nature (2011) 479:117–21. doi:10.1038/nature10558
192. Gurung P, Malireddi RK, Anand PK, Demon D, Vande Walle L, Liu Z, et al. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-beta (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J Biol Chem (2012) 287:34474–83. doi:10.1074/jbc.M112.401406
193. Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell (2012) 150:606–19. doi:10.1016/j.cell.2012.07.007
194. Sagulenko V, Thygesen SJ, Sester DP, Idris A, Cridland JA, Vajjhala PR, et al. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ (2013) 20:1149–60. doi:10.1038/cdd.2013.37
195. Gurung P, Anand PK, Malireddi RK, Vande Walle L, Van Opdenbosch N, Dillon CP, et al. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J Immunol (2014) 192:1835–46. doi:10.4049/jimmunol.1302839
196. Kang TB, Yang SH, Toth B, Kovalenko A, Wallach D. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity (2013) 38:27–40. doi:10.1016/j.immuni.2012.09.015
197. Allen IC, Tekippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med (2010) 207:1045–56. doi:10.1084/jem.20100050
198. Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity (2010) 32:379–91. doi:10.1016/j.immuni.2010.03.003
199. Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, Mcintire CR, Leblanc PM, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity (2010) 32:367–78. doi:10.1016/j.immuni.2010.02.012
200. Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol (2010) 185:4912–20. doi:10.4049/jimmunol.1002046
201. Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai RM, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med (2010) 207:1625–36. doi:10.1084/jem.20100199
202. Reuter BK, Pizarro TT. Commentary: the role of the IL-18 system and other members of the IL-1R/TLR superfamily in innate mucosal immunity and the pathogenesis of inflammatory bowel disease: friend or foe? Eur J Immunol (2004) 34:2347–55. doi:10.1002/eji.200425351
203. Siegmund B. Interleukin-18 in intestinal inflammation: friend and foe? Immunity (2010) 32:300–2. doi:10.1016/j.immuni.2010.03.010
204. Pastorelli L, De Salvo C, Mercado JR, Vecchi M, Pizarro TT. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: lessons learned from animal models and human genetics. Front Immunol (2013) 4:280. doi:10.3389/fimmu.2013.00280
205. Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X, et al. Deregulation of the NLRP3 inflammasome in hepatic parenchymal cells during liver cancer progression. Lab Invest (2014) 94:52–62. doi:10.1038/labinvest.2013.126
206. Ungerback J, Belenki D, Jawad Ul-Hassan A, Fredrikson M, Fransen K, Elander N, et al. Genetic variation and alterations of genes involved in NFkappaB/TNFAIP3- and NLRP3-inflammasome signaling affect susceptibility and outcome of colorectal cancer. Carcinogenesis (2012) 33:2126–34. doi:10.1093/carcin/bgs256
207. Verma D, Sarndahl E, Andersson H, Eriksson P, Fredrikson M, Jonsson JI, et al. The Q705K polymorphism in NLRP3 is a gain-of-function alteration leading to excessive interleukin-1beta and IL-18 production. PLoS One (2012) 7:e34977. doi:10.1371/journal.pone.0034977
208. Verma D, Bivik C, Farahani E, Synnerstad I, Fredrikson M, Enerback C, et al. Inflammasome polymorphisms confer susceptibility to sporadic malignant melanoma. Pigment Cell Melanoma Res (2012) 25:506–13. doi:10.1111/j.1755-148X.2012.01008.x
209. Okamoto M, Liu W, Luo Y, Tanaka A, Cai X, Norris DA, et al. Constitutively active inflammasome in human melanoma cells mediating autoinflammation via caspase-1 processing and secretion of interleukin-1beta. J Biol Chem (2010) 285:6477–88. doi:10.1074/jbc.M109.064907
210. Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ (2012) 19:1992–2002. doi:10.1038/cdd.2012.89
211. Sadasivam S, Gupta S, Radha V, Batta K, Kundu TK, Swarup G. Caspase-1 activator Ipaf is a p53-inducible gene involved in apoptosis. Oncogene (2005) 24:627–36. doi:10.1038/sj.onc.1208201
212. Hu B, Elinav E, Flavell RA. Inflammasome-mediated suppression of inflammation-induced colorectal cancer progression is mediated by direct regulation of epithelial cell proliferation. Cell Cycle (2011) 10:1936–9. doi:10.4161/cc.10.12.16008
213. Hu B, Elinav E, Huber S, Booth CJ, Strowig T, Jin C, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci U S A (2010) 107:21635–40. doi:10.1073/pnas.1016814108
214. Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, et al. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol (2012) 13:449–56. doi:10.1038/ni.2263
215. Grenier JM, Wang L, Manji GA, Huang WJ, Al-Garawi A, Kelly R, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett (2002) 530:73–8. doi:10.1016/S0014-5793(02)03416-6
216. Chen GY, Liu M, Wang F, Bertin J, Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol (2011) 186:7187–94. doi:10.4049/jimmunol.1100412
217. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell (2011) 145:745–57. doi:10.1016/j.cell.2011.04.022
218. Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci U S A (2011) 108:9601–6. doi:10.1073/pnas.1100981108
219. Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature (2012) 491:259–63. doi:10.1038/nature11535
220. Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem (2002) 277:29874–80. doi:10.1074/jbc.M203915200
221. Vladimer GI, Weng D, Paquette SW, Vanaja SK, Rathinam VA, Aune MH, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity (2012) 37:96–107. doi:10.1016/j.immuni.2012.07.006
222. Macaluso F, Nothnagel M, Parwez Q, Petrasch-Parwez E, Bechara FG, Epplen JT, et al. Polymorphisms in NACHT-LRR (NLR) genes in atopic dermatitis. Exp Dermatol (2007) 16:692–8. doi:10.1111/j.1600-0625.2007.00589.x
223. Jeru I, Duquesnoy P, Fernandes-Alnemri T, Cochet E, Yu JW, Lackmy-Port-Lis M, et al. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci U S A (2008) 105:1614–9. doi:10.1073/pnas.0708616105
224. Arthur JC, Lich JD, Ye Z, Allen IC, Gris D, Wilson JE, et al. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J Immunol (2010) 185:4515–9. doi:10.4049/jimmunol.1002227
225. Borghini S, Tassi S, Chiesa S, Caroli F, Carta S, Caorsi R, et al. Clinical presentation and pathogenesis of cold-induced autoinflammatory disease in a family with recurrence of an NLRP12 mutation. Arthritis Rheum (2011) 63:830–9. doi:10.1002/art.30170
226. Williams KL, Lich JD, Duncan JA, Reed W, Rallabhandi P, Moore C, et al. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem (2005) 280:39914–24. doi:10.1074/jbc.M502820200
227. Arthur JC, Lich JD, Aziz RK, Kotb M, Ting JP. Heat shock protein 90 associates with monarch-1 and regulates its ability to promote degradation of NF-kappaB-inducing kinase. J Immunol (2007) 179:6291–6. doi:10.4049/jimmunol.179.9.6291
228. Lich JD, Williams KL, Moore CB, Arthur JC, Davis BK, Taxman DJ, et al. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J Immunol (2007) 178:1256–60. doi:10.4049/jimmunol.178.3.1256
229. Zaki MH, Vogel P, Malireddi RK, Body-Malapel M, Anand PK, Bertin J, et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell (2011) 20:649–60. doi:10.1016/j.ccr.2011.10.022
230. Allen IC, Wilson JE, Schneider M, Lich JD, Roberts RA, Arthur JC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity (2012) 36:742–54. doi:10.1016/j.immuni.2012.03.012
231. Garaude J, Kent A, Van Rooijen N, Blander JM. Simultaneous targeting of toll- and nod-like receptors induces effective tumor-specific immune responses. Sci Transl Med (2012) 4:120ra116. doi:10.1126/scitranslmed.3002868
232. Yanagawa H, Haku T, Takeuchi E, Suzuki Y, Nokihara H, Sone S. Intrapleural therapy with MDP-Lys (L18), a synthetic derivative of muramyl dipeptide, against malignant pleurisy associated with lung cancer. Lung Cancer (2000) 27:67–73. doi:10.1016/S0169-5002(99)00090-2
233. Nitta Y, Sugita T, Ikuta Y, Murakami T. Inhibitory effect of liposomal MDP-Lys on lung metastasis of transplantable osteosarcoma in hamster. Oncol Res (2000) 12:25–31.
234. Yoo YC, Saiki I, Sato K, Azuma I. MDP-Lys(L18), a lipophilic derivative of muramyl dipeptide, inhibits the metastasis of haematogenous and non-haematogenous tumours in mice. Vaccine (1994) 12:175–260. doi:10.1016/0264-410X(94)90057-4
235. Fujimura T, Yamasaki K, Hidaka T, Ito Y, Aiba S. A synthetic NOD2 agonist, muramyl dipeptide (MDP)-Lys (L18) and IFN-beta synergistically induce dendritic cell maturation with augmented IL-12 production and suppress melanoma growth. J Dermatol Sci (2011) 62:107–15. doi:10.1016/j.jdermsci.2011.02.002
236. Chow MT, Sceneay J, Paget C, Wong CS, Duret H, Tschopp J, et al. NLRP3 suppresses NK cell-mediated responses to carcinogen-induced tumors and metastases. Cancer Res (2012) 72:5721–32. doi:10.1158/0008-5472.CAN-12-0509
237. Bruchard M, Mignot G, Derangere V, Chalmin F, Chevriaux A, Vegran F, et al. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat Med (2013) 19:57–64. doi:10.1038/nm.2999
238. Zitvogel L, Kepp O, Galluzzi L, Kroemer G. Inflammasomes in carcinogenesis and anticancer immune responses. Nat Immunol (2012) 13:343–51. doi:10.1038/ni.2224
239. Lust JA, Lacy MQ, Zeldenrust SR, Dispenzieri A, Gertz MA, Witzig TE, et al. Induction of a chronic disease state in patients with smoldering or indolent multiple myeloma by targeting interleukin 1{beta}-induced interleukin 6 production and the myeloma proliferative component. Mayo Clin Proc (2009) 84:114–22. doi:10.4065/84.2.114
240. Terme M, Ullrich E, Aymeric L, Meinhardt K, Desbois M, Delahaye N, et al. IL-18 induces PD-1-dependent immunosuppression in cancer. Cancer Res (2011) 71:5393–9. doi:10.1158/0008-5472.CAN-11-0993
Keywords: apoptosis, autophagy, colorectal cancer, innate immunity, intestinal inflammation, inflammasome, nod-like receptors, nodosome
Citation: Saxena M and Yeretssian G (2014) NOD-like receptors: master regulators of inflammation and cancer. Front. Immunol. 5:327. doi: 10.3389/fimmu.2014.00327
Received: 27 April 2014; Paper pending published: 02 May 2014;
Accepted: 27 June 2014; Published online: 14 July 2014.
Edited by:
Anton G. Kutikhin, Russian Academy of Medical Sciences, RussiaReviewed by:
Dario S. Zamboni, Universidade de São Paulo, BrazilMaya Saleh, McGill University, Canada
Arseniy E. Yuzhalin, University of Oxford, UK
Copyright: © 2014 Saxena and Yeretssian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Garabet Yeretssian, Department of Medicine, Clinical Immunology, Immunology Institute, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, 12-20E, New York, NY 10029, USA e-mail:Z2FyYWJldC55ZXJldHNzaWFuQG1zc20uZWR1
 Mansi Saxena
Mansi Saxena Garabet Yeretssian
Garabet Yeretssian