- Department of Pathology and Microbiology, University of Nebraska Medical Center, Omaha, NE, USA
Staphylococcus aureus and Staphylococcus epidermidis are notable for their propensity to form biofilms on implanted medical devices. Staphylococcal biofilm infections are typified by their recalcitrance to antibiotics and ability to circumvent host immune-mediated clearance, resulting in the establishment of chronic infections that are often recurrent in nature. Indeed, the immunomodulatory lifestyle of biofilms seemingly shapes the host immune response to ensure biofilm engraftment and persistence in an immune competent host. Here, we provide a brief review of the mechanisms whereby S. aureus and S. epidermidis biofilms manipulate host–pathogen interactions and discuss the concept of microenvironment maintenance in infectious outcomes, as well as speculate how these findings pertain to the challenges of staphylococcal vaccine development.
Introduction
Staphylococcus aureus and Staphylococcus epidermidis are highly opportunistic pathogens and a major cause of nosocomial and community-associated infections (1–4). While both staphylococcal species harbor multiple virulence factors, they are also capable of biofilm formation, which represents a communal virulence determinant to circumvent immune-mediated clearance and establish persistent infection (5–8). Most medical device-associated biofilm infections are caused by S. epidermidis and S. aureus, and both species can also establish biofilms on native host surfaces, such as infective endocarditis and osteomyelitis (9–11). Biofilms are heterogeneous bacterial communities encased within a self-produced matrix composed of eDNA (12), proteins (13), and polysaccharides (14), and their composition is often dependent on environmental factors, such as nutrient availability and mechanical stress (15). Biofilms represent a spatially diverse population of cells in terms of gene expression and metabolic activity, which is thought to arise in response to differences in nutrient, pH, and oxygen gradients within the structure (5, 16). Biofilm infections are recalcitrant to antibiotics (17, 18), which often necessitates removal of the infected device or native tissue and is associated with significant morbidity and economic impact (10, 19, 20). Indeed, it has been estimated that approximately $1.8 billion is spent annually in the US for the treatment and clinical management of orthopedic implant-related infections (21, 22).
Neutrophils and macrophages are the main innate immune effectors against acute planktonic staphylococcal infections (i.e., abscess, sepsis) (23–29). Neutrophil antimicrobial activity is mediated by defensins, cathelicidins, and lysozymes, as well as reactive oxygen species catalyzed by NADPH oxidase (30, 31). Recently, neutrophil and macrophage extracellular traps (NETs and METs, respectively) have been identified as another means of antimicrobial action (31–34). This “beyond the grave” mechanism is typified by an extracellular net of DNA released from dying phagocytes that contains localized islands of lytic enzymes that kill ensnared extracellular bacteria. Classically activated (M1) macrophages are key microbicidal effectors that exert their effector functions, in part, through the production of reactive oxygen and nitrogen species, in addition to proinflammatory cytokines (35–38). Macrophages also clear extracellular bacteria by phagocytosis, which is tightly linked with proinflammatory cytokine and chemokine production to initiate adaptive immune responses (39–41). Collectively, these innate leukocyte responses, coupled with complement activation, usually ensure the successful clearance of planktonic staphylococcal infections in an immune competent host.
In contrast, a very different story has emerged regarding biofilm infections (Figure 1). Recent studies have demonstrated that staphylococcal biofilms actively skew host immunity toward an anti-inflammatory, pro-fibrotic response that favors bacterial persistence (6, 42–44). This is typified by alternatively activated (M2) macrophages and arginase-1 (Arg1) activity, resulting in urea and ornithine production, which are involved in collagen formation and tissue remodeling (35, 45, 46). Our laboratory has shown that macrophages associated with S. aureus biofilms both in vitro and in vivo have decreased inducible nitric oxide synthase (iNOS) concomitant with increased Arg1 expression, as well as attenuated cytokine and chemokine production (6, 43, 44). Similar findings have been reported in response to S. epidermidis biofilms (42, 47–49) and biofilms from other bacterial species (50–53), suggesting a conserved mechanism exists to thwart host immunity to ensure biofilm persistence (54). However, recent data suggest a protective role of anti-inflammatory Th2/Treg cells, as opposed to proinflammatory Th1/Th17 signaling, during early S. aureus biofilm development (55), highlighting the complexity of staphylococcal biofilm infections and perhaps distinctions between innate and adaptive immune mechanisms. For in-depth discussions on host immunity to staphylococcal infections, the reader is referred to recent reviews (7, 56–58). This Perspective is focused on how changes in biofilm gene expression/secretion impact host immune mechanisms and potential implications regarding novel therapeutic strategies.
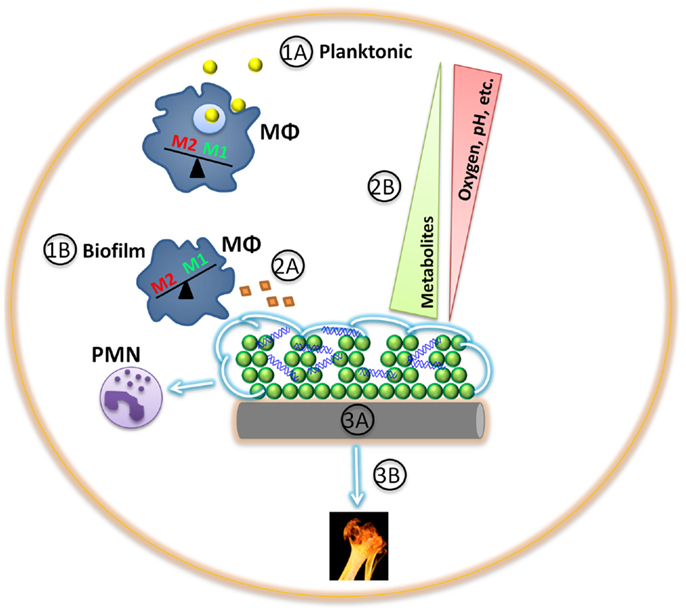
Figure 1. Game of hide-and-seek between staphylococcal biofilms and host innate immunity. (1A) Planktonic staphylococci (yellow) are more readily phagocytosed and cleared by innate immune cells, such as macrophages (MΦ; blue) and neutrophils (PMNs), triggering a M1 proinflammatory response. (1B) Differential gene expression between planktonic and biofilm staphylococci provide survival benefits in harsh environments, such as the presence of an extracellular matrix (green). In addition, S. aureus biofilms exhibit differential gene expression in response to macrophages or neutrophils (violet), shielding the biofilm from detection by the former while facilitating cytotoxicity of the latter. (2A) Staphylococcal biofilm protein secretion in vivo is relatively unknown and may represent fertile ground for clinical intervention and/or vaccine development. (2B) Staphylococcal biofilms are composed of a large biomass, of which some cells are metabolically active and transform the surrounding microenvironment by establishing various metabolic gradients. (3A) Implanted medical devices (gray) not only provide a scaffold for bacterial adhesion and biofilm formation that is facilitated through host protein deposition (tan), but can also induce a persistent, anti-inflammatory immune response that favors bacterial persistence. (3B). Tissue damage provoked by staphylococcal nutrient procurement may further incite an overall anti-inflammatory milieu.
Differential Gene Expression in Staphylococcal Biofilms: A Game of Hide-and-Seek
Staphylococcal biofilms exhibit distinct gene expression profiles compared to planktonic cells, demonstrating unique growth differences between the two lifestyles (16, 59, 60). One major disparity in staphylococcal biofilms is the induction of genes associated with potential acid tolerance response pathways (i.e., arginine deiminase and urease), reflecting anaerobic growth and the need for pH homeostasis (5, 16). In addition, an increased demand for pyrimidine nucleotide biosynthesis has been associated with the biofilm lifestyle of staphylococci (16) and other bacterial species (61, 62). The DNA-binding regulatory protein SarA is critical for biofilm formation and virulence factor expression in both S. aureus and S. epidermidis. SarA acts, in part, through the accessory gene regulator agr, which encodes a quorum-sensing system responsible for transitioning the synthesis of surface and adhesive molecules to secreted factors and is an important regulatory switch between planktonic and biofilm lifestyles (63–66).
In addition to unique genetic changes during biofilm growth, we have recently reported that innate immune cells (i.e., macrophages and neutrophils) alter biofilm gene expression profiles (67). Transcriptome analysis of S. aureus biofilms following macrophage exposure revealed a rapid down-regulation of over 550 staphylococcal genes within a 1 h period. This effect occurred despite the fact that macrophages did not significantly invade the biofilm or exhibit phagocytic activity. This transcriptional repression was mitigated after 24 h, which was likely explained by the significant degree of macrophage death observed at this late interval. In essence, the biofilm was no longer required to cloak itself, since the threat of macrophage attack had dissipated. In contrast, although neutrophils displayed a much higher propensity for biofilm invasion and phagocytosis, they did not significantly alter the biofilm transcriptome over similar time periods (67). This data supports previous reports demonstrating that biofilms are not inherently protected from neutrophil assault (44, 68, 69).
These results suggest that staphylococcal biofilms “hide” in response to macrophages while they elect to tackle neutrophil challenge head on. But why does the biofilm down-regulate gene expression in the face of a seemingly inactive macrophage challenge and barely break a transcriptional sweat when confronted with an assault of phagocytic neutrophils? One possibility involves the reported ability of S. aureus to survive intracellularly in neutrophils (70, 71). Another is that S. aureus secretes a number of neutrophil-lytic toxins, including α-toxin (Hla) (72) and phenol-soluble modulins (PSMs) (73–77). Interestingly, both Hla and PSMs are under agr control, and agr expression was significantly increased in S. aureus biofilms following neutrophil exposure (67, 74). Notably, we have found few neutrophil infiltrates in either S. aureus catheter-associated or orthopedic implant models of biofilm infection in vivo (7, 44). Likewise, the exogenous introduction of neutrophils at the site of S. aureus biofilm infection is not capable of limiting bacterial growth, whereas M1-activated macrophages can significantly prevent or clear established biofilms (44). Collectively, these findings highlight the well-adapted nature of S. aureus biofilms to neutrophils. Although both neutrophils and macrophages are phagocytic, macrophages are recognized as a signaling hub during bacterial infections to coordinate evolving immune responses through cytokine and chemokine release (78–80). While it is possible that staphylococcal biofilms are more responsive to this secreted milieu than physical disruption via phagocytosis, it is clear that they respond differently to challenges from distinct leukocyte populations in regards to their transcriptional profile.
Microenvironment Modulation
Previous work has characterized numerous secreted virulence factors used by staphylococci that target specific host cell populations. S. aureus, in particular, utilizes numerous hemolysins (81), leukocidins (82, 83), and proteases to evade the host immune system (84). While staphylococci certainly divert resources from secreted to structural proteins during early biofilm growth, protein secretion is still maintained in biofilms. For example, several reports have demonstrated that various bacterial biofilm-forming species secrete peptides and full-length proteins (i.e., Esp) that interfere with biofilm development of competing organisms, presumably to eliminate competition for a biofilm-friendly niche (85–87). Additionally, studies have demonstrated evidence of host-biofilm cross-talk involving the quorum-sensing molecules N-Acyl homoserine lactones (AHL) (88, 89). Therefore, it is likely that staphylococcal biofilms also secrete factors in vivo, perhaps under quorum-sensing system regulation, in an attempt to evade immune recognition and clearance. However, the identity of such molecules remains to be elucidated but could represent future attractive anti-biofilm agents.
Besides virulence factors, staphylococci also secrete molecules for nutrient procurement and cell signaling. For example, siderophores are critical for iron acquisition (90), while several signaling molecules are important for biofilm remodeling and dispersal, such as auto-inducing peptide (AIP) (63), nuclease (91), and PSMs (74). While the primary roles of these molecules are apparently unrelated to immune interactions, recent evidence suggests the potential for alternative functions. For example, nuclease, which mediates biofilm dispersal (65, 91), can also participate in NET degradation (32). Recently, nuclease in combination with adenosine synthase has been implicated in the conversion of NETs to leukotoxic deoxyadenosine, highlighting a clever means whereby S. aureus turns the immune response against itself (92). In addition, S. aureus PSM expression is induced following neutrophil phagocytosis, resulting in neutrophil lysis and bacterial escape (74, 76, 93). This process is regulated by the stringent response characterized by the synthesis of the intracellular signaling alarmone (p)ppGpp (93). Therefore, the stringent response provides yet another means of staphylococcal adaptation in response to select immune pressures, something that has previously been well-established in relation to nutrient availability and metabolism (94–99). Indeed, recent evidence has implicated (p)ppGpp and di-cyclic NTPs as critical signals in the switch from planktonic to biofilm growth (100).
However, secreted factors are not the only means whereby staphylococcal biofilms could regulate the host response in an exogenous manner. While bacteria have a plethora of environmental sensory mechanisms at their disposal, host immune cells are also very sensitive to their surrounding environment (101–103). Because biofilms represent a large biomass, the sum metabolic activity of the bacteria themselves would be expected to have an impact on pH and oxygen levels in the surrounding tissue microenvironment. Indeed, even subtle changes in pH (101, 104) or oxygen (102) can significantly alter the nature of the immune response. Furthermore, novel research with fungal and bacterial biofilms has identified a coordinated system of ROS signaling for biofilm maturation (105). It will be interesting to see what, if any, impact this biofilm ROS gradient has on the host immune response and whether host-generated ROS can also act as a signaling molecule within the biofilm to influence target pathways, such as biofilm formation via quorum-sensing (105).
Hijacking the Host Response
Most implanted medical devices are at risk for biofilm colonization (4, 10, 11, 15). Upon placement, the device is conditioned by host proteins, such as fibronectin and collagen, which can serve as ligands for bacterial attachment via microbial surface components recognizing adhesive matrix molecules (MSCRAMMS) (4, 9, 11, 15, 106, 107). Therefore, the wound healing responses elicited after medical device implantation can inadvertently provide a rich environment for bacterial colonization; however, they can also set the stage for a very specific immune response that is incapable of mediating bacterial clearance, as described below (108). Staphylococcal biofilm-associated protein (Bap) can facilitate adherence to host epithelial cells through the inhibition of MSCRAMM-mediated attachment (109, 110). Following adherence, bacteria begin to proliferate and accumulate by producing adherence factors and matrix components, resulting in biofilm formation. Bacterial-derived accumulation-associated protein (Aap) and polysaccharide intercellular adhesin (PIA) are important accumulation molecules in S. epidermidis.
Our laboratory has reported that S. aureus biofilms promote their growth and expansion by manipulating the host immune response (6, 43, 44, 111). Several studies demonstrated that staphylococci have developed strategies to interfere with M1 macrophage polarization either by directly inhibiting antimicrobial activity or hindering M1 cytokine production (6, 42, 44, 48). In addition, the expression of numerous proinflammatory mediators is significantly attenuated during S. aureus biofilm infection (6, 7, 43, 44). Since many of these molecules are also associated with wound healing responses, their active repression by staphylococcal biofilms is likely one mechanism to explain the paucity of neutrophil and M1-activated macrophage infiltrates into biofilms in vivo (6, 43, 44).
Besides dampening the local inflammatory environment, numerous mechanisms likely exist to explain the chronicity of staphylococcal biofilm infections in an immune competent host. For example, we have demonstrated the preferential accumulation of M2-activated macrophages in S. aureus biofilms in vivo and in vitro typified by Arg1 concomitant with minimal iNOS expression (6, 43, 44). Increased Arg1 activity during staphylococcal infection can regulate T cell proliferation and effector functions via arginine depletion from the environment, resulting in defective TCR signaling from inhibiting CD3ζ expression, cell cycle, and cytokine production (112–116). Indeed, our laboratory has found few T cell infiltrates associated with S. aureus biofilm infections, whereas another group reported Th2/Treg involvement in biofilm clearance (7, 55). The reasons for these discrepancies are not clear but may arise from differences in experimental models and/or S. aureus strains tested. Impaired adaptive immunity or memory responses could compromise the success of vaccine strategies against staphylococcal biofilms.
Future Directions
As ongoing staphylococcal biofilm research elucidates mechanisms contributing to the persistence and virulence of these infections, it is becoming clear that novel strategies are needed to combat the complex biofilm lifestyle. The inherent ability of S. aureus to circumvent immune-mediated clearance has complicated staphylococcal vaccine development. Indeed, previous studies have shown that S. aureus infection alone is not sufficient to induce protective immunity (117). Moreover, recent vaccine development efforts have stalled in clinical trials, likely due to the organism’s immunomodulatory capabilities coupled with the differential expression of targeted antigens in vivo (118–120). Another challenge is to develop a vaccine efficacious against both planktonic and biofilm infections that provides protection across a wide range of S. aureus clonal complex groups (121).
Recent staphylococcal vaccine efforts have utilized new approaches to increase efficacy. Vaccination with attenuated S. aureus strains (122) has led to the discovery of protective antigens with vaccine potential in a process termed “genetic vaccinology” (123, 124). In addition, multivalent vaccines targeting multiple secreted and cell surface-associated virulence factors from both planktonic (125), and biofilm (126, 127) modes of growth have been employed with some success in vivo (121, 128). Furthermore, the use of Toll-like receptor agonists as adjuvants and novel DNA vaccines are currently under investigation (129, 130). While the efforts of our laboratory and others have implicated a subset of putative factors responsible for the immunosuppressive and chronic nature of staphylococcal biofilms, significant obstacles remain for developing a successful vaccine. In our opinion, future anti-staphylococcal therapeutics for the biofilm lifestyle will likely require a combinatorial approach of bacteriocidal and immunostimulatory treatments.
In this context, a successful staphylococcal vaccine may be facilitated by a more comprehensive understanding of the staphylococcal biofilm transcriptome and secretome, particularly in response to relevant host phagocytes, which will be an important next step toward replicating complex in vivo interactions. Additional insights into staphylococcal biofilm metabolism, in particular, how the biofilm metabolome changes once immune cells are encountered, may be beneficial as it could facilitate the identification of virulence determinants for vaccine development. A recent analogy is the use of metabolic stimuli to induce aminoglycoside susceptibility in otherwise recalcitrant S. aureus cell populations, such as biofilms (131). Also, characterizing how the natural wound healing response promotes bacterial colonization could provide invaluable information on key immune cell populations for vaccine targeting. A better understanding of how biofilms manipulate the host immune response should provide valuable insights to direct the continued evolution of staphylococcal vaccines. Until, we better define how staphylococci circumvent host immunity, they will likely continue to have the upper hand.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) P01 AI083211 to Tammy Kielian and an American Heart Association predoctoral fellowship to Cortney E. Heim (13PRE16910040).
References
1. Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev (2002) 15:167–93. doi:10.1128/CMR.15.2.167-193.2002
2. Fitzpatrick F, Humphreys H, O’Gara JP. The genetics of staphylococcal biofilm formation – will a greater understanding of pathogenesis lead to better management of device-related infection? Clin Microbiol Infect (2005) 11:967–73. doi:10.1111/j.1469-0691.2005.01274.x
3. Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol (2008) 322:207–28. doi:10.1007/978-3-540-75418-3_10
4. Arciola CR, Campoccia D, Speziale P, Montanaro L, Costerton JW. Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials. Biomaterials (2012) 33:5967–82. doi:10.1016/j.biomaterials.2012.05.031
5. Fey PD, Olson ME. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol (2010) 5:917–33. doi:10.2217/fmb.10.56
6. Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol (2011) 186:6585–96. doi:10.4049/jimmunol.1002794
7. Hanke ML, Kielian T. Deciphering mechanisms of staphylococcal biofilm evasion of host immunity. Front Cell Infect Microbiol (2012) 2:62. doi:10.3389/fcimb.2012.00062
8. Watkins RR, David MZ, Salata RA. Current concepts on the virulence mechanisms of meticillin-resistant Staphylococcus aureus. J Med Microbiol (2012) 61:1179–93. doi:10.1099/jmm.0.043513-0
9. McCann MT, Gilmore BF, Gorman SP. Staphylococcus epidermidis device-related infections: pathogenesis and clinical management. J Pharm Pharmacol (2008) 60:1551–71. doi:10.1211/jpp/60.12.0001
10. Montanaro L, Speziale P, Campoccia D, Ravaioli S, Cangini I, Pietrocola G, et al. Scenery of Staphylococcus implant infections in orthopedics. Future Microbiol (2011) 6:1329–49. doi:10.2217/fmb.11.117
11. Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med (2013) 64:175–88. doi:10.1146/annurev-med-042711-140023
12. Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A (2007) 104:8113–8. doi:10.1073/pnas.0610226104
13. Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog (2012) 8:e1002744. doi:10.1371/journal.ppat.1002744
14. Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials (2007) 28:1711–20. doi:10.1016/j.biomaterials.2006.11.046
15. Kiedrowski MR, Horswill AR. New approaches for treating staphylococcal biofilm infections. Ann N Y Acad Sci (2011) 1241:104–21. doi:10.1111/j.1749-6632.2011.06281.x
16. Beenken KE, Dunman PM, Mcaleese F, Macapagal D, Murphy E, Projan SJ, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol (2004) 186:4665–84. doi:10.1128/JB.186.14.4665-4684.2004
17. Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol (1999) 37:1771–6.
18. Anderl JN, Zahller J, Roe F, Stewart PS. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother (2003) 47:1251–6. doi:10.1128/AAC.47.4.1251-1256.2003
19. Zimmerli W, Trampuz A, Ochsner PE. Prosthetic-joint infections. N Engl J Med (2004) 351:1645–54. doi:10.1056/NEJMra040181
20. Lora-Tamayo J, Murillo O, Iribarren JA, Soriano A, Sanchez-Somolinos M, Baraia-Etxaburu JM, et al. A large multicenter study of methicillin-susceptible and methicillin-resistant Staphylococcus aureus prosthetic joint infections managed with implant retention. Clin Infect Dis (2013) 56:182–94. doi:10.1093/cid/cis746
21. Darouiche RO. Treatment of infections associated with surgical implants. N Engl J Med (2004) 350:1422–9. doi:10.1056/NEJMra035415
22. Haenle M, Skripitz C, Mittelmeier W, Skripitz R. Economic impact of infected total knee arthroplasty. ScientificWorldJournal (2012) 2012:196515. doi:10.1100/2012/196515
23. Verdrengh M, Tarkowski A. Role of neutrophils in experimental septicemia and septic arthritis induced by Staphylococcus aureus. Infect Immun (1997) 65:2517–21.
24. Molne L, Verdrengh M, Tarkowski A. Role of neutrophil leukocytes in cutaneous infection caused by Staphylococcus aureus. Infect Immun (2000) 68:6162–7. doi:10.1128/IAI.68.11.6162-6167.2000
25. Verdrengh M, Tarkowski A. Role of macrophages in Staphylococcus aureus-induced arthritis and sepsis. Arthritis Rheum (2000) 43:2276–82. doi:10.1002/1529-0131(200010)43:10<2276::AID-ANR15>3.0.CO;2-C
26. Kielian T, Barry B, Hickey WF. CXC chemokine receptor-2 ligands are required for neutrophil-mediated host defense in experimental brain abscesses. J Immunol (2001) 166:4634–43.
27. Asai A, Tsuda Y, Kobayashi M, Hanafusa T, Herndon DN, Suzuki F. Pathogenic role of macrophages in intradermal infection of methicillin-resistant Staphylococcus aureus in thermally injured mice. Infect Immun (2010) 78:4311–9. doi:10.1128/IAI.00642-10
28. Cheng AG, Dedent AC, Schneewind O, Missiakas D. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol (2011) 19:225–32. doi:10.1016/j.tim.2011.01.007
29. Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol (2012) 34:237–59. doi:10.1007/s00281-011-0295-3
30. Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol (2006) 6:173–82. doi:10.1038/nri1785
31. Nauseef WM. How human neutrophils kill and degrade microbes: an integrated view. Immunol Rev (2007) 219:88–102. doi:10.1111/j.1600-065X.2007.00550.x
32. Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, Von Kockritz-Blickwede M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun (2010) 2:576–86. doi:10.1159/000319909
33. Brinkmann V, Zychlinsky A. Neutrophil extracellular traps: is immunity the second function of chromatin? J Cell Biol (2012) 198:773–83. doi:10.1083/jcb.201203170
34. Goldmann O, Medina E. The expanding world of extracellular traps: not only neutrophils but much more. Front Immunol (2012) 3:420. doi:10.3389/fimmu.2012.00420
35. Mosser DM. The many faces of macrophage activation. J Leukoc Biol (2003) 73:209–12. doi:10.1189/jlb.0602325
36. Martinez FO. Regulators of macrophage activation. Eur J Immunol (2011) 41:1531–4. doi:10.1002/eji.201141670
37. Mege JL, Mehraj V, Capo C. Macrophage polarization and bacterial infections. Curr Opin Infect Dis (2011) 24:230–4. doi:10.1097/QCO.0b013e328344b73e
38. Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol (2011) 11:762–74. doi:10.1038/nri3070
39. Aderem A. Phagocytosis and the inflammatory response. J Infect Dis (2003) 187(Suppl 2):S340–5. doi:10.1086/374747
40. Underhill DM, Gantner B. Integration of Toll-like receptor and phagocytic signaling for tailored immunity. Microbes Infect (2004) 6:1368–73. doi:10.1016/j.micinf.2004.08.016
41. Underhill DM, Goodridge HS. Information processing during phagocytosis. Nat Rev Immunol (2012) 12:492–502. doi:10.1038/nri3244
42. Schommer NN, Christner M, Hentschke M, Ruckdeschel K, Aepfelbacher M, Rohde H. Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A.1. Infect Immun (2011) 79:2267–76. doi:10.1128/IAI.01142-10
43. Hanke ML, Angle A, Kielian T. MyD88-dependent signaling influences fibrosis and alternative macrophage activation during Staphylococcus aureus biofilm infection. PLoS One (2012) 7:e42476. doi:10.1371/journal.pone.0042476
44. Hanke ML, Heim CE, Angle A, Sanderson SD, Kielian T. Targeting macrophage activation for the prevention and treatment of Staphylococcus aureus biofilm infections. J Immunol (2013) 190:2159–68. doi:10.4049/jimmunol.1202348
45. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol (2008) 8:958–69. doi:10.1038/nri2448
46. Comalada M, Yeramian A, Modolell M, Lloberas J, Celada A. Arginine and macrophage activation. Methods Mol Biol (2012) 844:223–35. doi:10.1007/978-1-61779-527-5_16
47. Kristian SA, Birkenstock TA, Sauder U, Mack D, Gotz F, Landmann R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J Infect Dis (2008) 197:1028–35. doi:10.1086/528992
48. Cerca F, Andrade F, Franca A, Andrade EB, Ribeiro A, Almeida AA, et al. Staphylococcus epidermidis biofilms with higher proportions of dormant bacteria induce a lower activation of murine macrophages. J Med Microbiol (2011) 60:1717–24. doi:10.1099/jmm.0.031922-0
49. Spiliopoulou AI, Kolonitsiou F, Krevvata MI, Leontsinidis M, Wilkinson TS, Mack D, et al. Bacterial adhesion, intracellular survival and cytokine induction upon stimulation of mononuclear cells with planktonic or biofilm phase Staphylococcus epidermidis. FEMS Microbiol Lett (2012) 330:56–65. doi:10.1111/j.1574-6968.2012.02533.x
50. Yu H, Head NE. Persistent infections and immunity in cystic fibrosis. Front Biosci (2002) 7:d442–57.
51. Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, et al. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol (2003) 171:4329–39.
52. Mittal R, Sharma S, Chhibber S, Harjai K. Effect of macrophage secretory products on elaboration of virulence factors by planktonic and biofilm cells of Pseudomonas aeruginosa. Comp Immunol Microbiol Infect Dis (2006) 29:12–26. doi:10.1016/j.cimid.2005.11.002
53. Chandra J, Mccormick TS, Imamura Y, Mukherjee PK, Ghannoum MA. Interaction of Candida albicans with adherent human peripheral blood mononuclear cells increases C. albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect Immun (2007) 75:2612–20. doi:10.1128/IAI.01841-06
54. Otto M. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr Top Microbiol Immunol (2006) 306:251–8. doi:10.1007/3-540-29916-5_10
55. Prabhakara R, Harro JM, Leid JG, Keegan AD, Prior ML, Shirtliff ME. Suppression of the inflammatory immune response prevents the development of chronic biofilm infection due to methicillin-resistant Staphylococcus aureus. Infect Immun (2011) 79:5010–8. doi:10.1128/IAI.05571-11
56. DeLeo FR, Diep BA, Otto M. Host defense and pathogenesis in Staphylococcus aureus infections. Infect Dis Clin North Am (2009) 23:17–34. doi:10.1016/j.idc.2008.10.003
57. Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence (2011) 2:445–59. doi:10.4161/viru.2.5.17724
58. Kim HK, Thammavongsa V, Schneewind O, Missiakas D. Recurrent infections and immune evasion strategies of Staphylococcus aureus. Curr Opin Microbiol (2012) 15:92–9. doi:10.1016/j.mib.2011.10.012
59. Resch A, Rosenstein R, Nerz C, Gotz F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol (2005) 71:2663–76. doi:10.1128/AEM.71.5.2663-2676.2005
60. Yao Y, Sturdevant DE, Otto M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis (2005) 191:289–98. doi:10.1086/426945
61. Garavaglia M, Rossi E, Landini P. The pyrimidine nucleotide biosynthetic pathway modulates production of biofilm determinants in Escherichia coli. PLoS One (2012) 7:e31252. doi:10.1371/journal.pone.0031252
62. Yadav MK, Kwon SK, Cho CG, Park SW, Chae SW, Song JJ. Gene expression profile of early in vitro biofilms of Streptococcus pneumoniae. Microbiol Immunol (2012) 56:621–9. doi:10.1111/j.1348-0421.2012.00483.x
63. Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci U S A (1995) 92:12055–9. doi:10.1073/pnas.92.26.12055
64. Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun (2003) 71:4206–11. doi:10.1128/IAI.71.7.4206-4211.2003
65. Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog (2008) 4:e1000052. doi:10.1371/journal.ppat.1000052
66. Mrak LN, Zielinska AK, Beenken KE, Mrak IN, Atwood DN, Griffin LM, et al. saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS One (2012) 7:e38453. doi:10.1371/journal.pone.0038453
67. Scherr TD, Roux CM, Hanke ML, Angle A, Dunman PD, Kielian T. Global transcriptome analysis of Staphylococcus aureus biofilms in response to innate immune cells. Infect Immun (2013) 81:4363–76. doi:10.1128/IAI.00819-13
68. Gunther F, Wabnitz GH, Stroh P, Prior B, Obst U, Samstag Y, et al. Host defence against Staphylococcus aureus biofilms infection: phagocytosis of biofilms by polymorphonuclear neutrophils (PMN). Mol Immunol (2009) 46:1805–13. doi:10.1016/j.molimm.2009.01.020
69. Graves SF, Kobayashi SD, Deleo FR. Community-associated methicillin-resistant Staphylococcus aureus immune evasion and virulence. J Mol Med (Berl) (2010) 88:109–14. doi:10.1007/s00109-009-0573-x
70. Gresham HD, Lowrance JH, Caver TE, Wilson BS, Cheung AL, Lindberg FP. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J Immunol (2000) 164:3713–22.
71. Anwar S, Prince LR, Foster SJ, Whyte MK, Sabroe I. The rise and rise of Staphylococcus aureus: laughing in the face of granulocytes. Clin Exp Immunol (2009) 157:216–24. doi:10.1111/j.1365-2249.2009.03950.x
72. Spaan AN, Surewaard BG, Nijland R, Van Strijp JA. Neutrophils versus Staphylococcus aureus: a biological tug of war. Annu Rev Microbiol (2013) 67:629–50. doi:10.1146/annurev-micro-092412-155746
73. Clarke SR. Phenol-soluble modulins of Staphylococcus aureus lure neutrophils into battle. Cell Host Microbe (2010) 7:423–4. doi:10.1016/j.chom.2010.05.015
74. Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, et al. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A (2012) 109:1281–6. doi:10.1073/pnas.1115006109
75. Chatterjee SS, Joo HS, Duong AC, Dieringer TD, Tan VY, Song Y, et al. Essential Staphylococcus aureus toxin export system. Nat Med (2013) 19:364–7. doi:10.1038/nm.3047
76. Chatterjee SS, Otto M. How can Staphylococcus aureus phenol-soluble modulins be targeted to inhibit infection? Future Microbiol (2013) 8:693–6. doi:10.2217/fmb.13.37
77. Tsompanidou E, Denham EL, Van Dijl JM. Phenol-soluble modulins, hellhounds from the staphylococcal virulence-factor pandemonium. Trends Microbiol (2013) 21:313–5. doi:10.1016/j.tim.2013.04.007
78. Silva MT. When two is better than one: macrophages and neutrophils work in concert in innate immunity as complementary and cooperative partners of a myeloid phagocyte system. J Leukoc Biol (2010) 87:93–106. doi:10.1189/jlb.0809549
79. Silva MT. Macrophage phagocytosis of neutrophils at inflammatory/infectious foci: a cooperative mechanism in the control of infection and infectious inflammation. J Leukoc Biol (2011) 89:675–83. doi:10.1189/jlb.0910536
80. Silva MT, Correia-Neves M. Neutrophils and macrophages: the main partners of phagocyte cell systems. Front Immunol (2012) 3:174. doi:10.3389/fimmu.2012.00174
81. Kebaier C, Chamberland RR, Allen IC, Gao X, Broglie PM, Hall JD, et al. Staphylococcus aureus alpha-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J Infect Dis (2012) 205:807–17. doi:10.1093/infdis/jir846
82. Dumont AL, Nygaard TK, Watkins RL, Smith A, Kozhaya L, Kreiswirth BN, et al. Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol Microbiol (2011) 79:814–25. doi:10.1111/j.1365-2958.2010.07490.x
83. Alonzo F III, Torres VJ. Bacterial survival amidst an immune onslaught: the contribution of the Staphylococcus aureus leukotoxins. PLoS Pathog (2013) 9:e1003143. doi:10.1371/journal.ppat.1003143
84. Oogai Y, Matsuo M, Hashimoto M, Kato F, Sugai M, Komatsuzawa H. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl Environ Microbiol (2011) 77:8097–105. doi:10.1128/AEM.05316-11
85. Otto M, Echner H, Voelter W, Gotz F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun (2001) 69:1957–60. doi:10.1128/IAI.69.3.1957-1960.2001
86. Qazi S, Middleton B, Muharram SH, Cockayne A, Hill P, O’Shea P, et al. N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect Immun (2006) 74:910–9. doi:10.1128/IAI.74.2.910-919.2006
87. Qin Z, Yang L, Qu D, Molin S, Tolker-Nielsen T. Pseudomonas aeruginosa extracellular products inhibit staphylococcal growth, and disrupt established biofilms produced by Staphylococcus epidermidis. Microbiology (2009) 155:2148–56. doi:10.1099/mic.0.028001-0
88. Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology (2007) 153:3923–38. doi:10.1099/mic.0.2007/012856-0
89. Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol (2008) 6:111–20. doi:10.1038/nrmicro1836
90. Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, Skaar EP. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog (2006) 2:e87. doi:10.1371/journal.ppat.0020087
91. Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One (2009) 4:e5822. doi:10.1371/journal.pone.0005822
92. Thammavongsa V, Missiakas DM, Schneewind O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science (2013) 342:863–6. doi:10.1126/science.1242255
93. Geiger T, Francois P, Liebeke M, Fraunholz M, Goerke C, Krismer B, et al. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog (2012) 8:e1003016. doi:10.1371/journal.ppat.1003016
94. Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol (2003) 48:1429–49. doi:10.1046/j.1365-2958.2003.03526.x
95. Somerville GA, Said-Salim B, Wickman JM, Raffel SJ, Kreiswirth BN, Musser JM. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect Immun (2003) 71:4724–32. doi:10.1128/IAI.71.8.4724-4732.2003
96. Seidl K, Goerke C, Wolz C, Mack D, Berger-Bachi B, Bischoff M. Staphylococcus aureus CcpA affects biofilm formation. Infect Immun (2008) 76:2044–50. doi:10.1128/IAI.00035-08
97. Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev (2009) 73:233–48. doi:10.1128/MMBR.00005-09
98. Sadykov MR, Zhang B, Halouska S, Nelson JL, Kreimer LW, Zhu Y, et al. Using NMR metabolomics to investigate tricarboxylic acid cycle-dependent signal transduction in Staphylococcus epidermidis. J Biol Chem (2010) 285:36616–24. doi:10.1074/jbc.M110.152843
99. Nuxoll AS, Halouska SM, Sadykov MR, Hanke ML, Bayles KW, Kielian T, et al. CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog (2012) 8:e1003033. doi:10.1371/journal.ppat.1003033
100. Valle J, Solano C, Garcia B, Toledo-Arana A, Lasa I. Biofilm switch and immune response determinants at early stages of infection. Trends Microbiol (2013) 21:364–71. doi:10.1016/j.tim.2013.05.008
102. Nizet V, Johnson RS. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol (2009) 9:609–17. doi:10.1038/nri2607
103. Leemans JC, Cassel SL, Sutterwala FS. Sensing damage by the NLRP3 inflammasome. Immunol Rev (2011) 243:152–62. doi:10.1111/j.1600-065X.2011.01043.x
104. Rajamaki K, Nordstrom T, Nurmi K, Akerman KE, Kovanen PT, Oorni K, et al. Extracellular acidosis is a novel danger signal alerting innate immunity via the NLRP3 inflammasome. J Biol Chem (2013) 288:13410–9. doi:10.1074/jbc.M112.426254
105. Cap M, Vachova L, Palkova Z. Reactive oxygen species in the signaling and adaptation of multicellular microbial communities. Oxid Med Cell Longev (2012) 2012:976753. doi:10.1155/2012/976753
106. Joh D, Wann ER, Kreikemeyer B, Speziale P, Hook M. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol (1999) 18:211–23. doi:10.1016/S0945-053X(99)00025-6
107. Fowler T, Wann ER, Joh D, Johansson S, Foster TJ, Hook M. Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell beta1 integrins. Eur J Cell Biol (2000) 79:672–9. doi:10.1078/0171-9335-00104
108. Rochford ET, Richards RG, Moriarty TF. Influence of material on the development of device-associated infections. Clin Microbiol Infect (2012) 18:1162–7. doi:10.1111/j.1469-0691.2012.04002.x
109. Cucarella C, Tormo MA, Knecht E, Amorena B, Lasa I, Foster TJ, et al. Expression of the biofilm-associated protein interferes with host protein receptors of Staphylococcus aureus and alters the infective process. Infect Immun (2002) 70:3180–6. doi:10.1128/IAI.70.6.3180-3186.2002
110. Valle J, Latasa C, Gil C, Toledo-Arana A, Solano C, Penades JR, et al. Bap, a biofilm matrix protein of Staphylococcus aureus prevents cellular internalization through binding to GP96 host receptor. PLoS Pathog (2012) 8:e1002843. doi:10.1371/journal.ppat.1002843
111. Cheatle J, Aldrich A, Thorell WE, Boska MD, Kielian T. Compartmentalization of immune responses during Staphylococcus aureus cranial bone flap infection. Am J Pathol (2013) 183:450–8. doi:10.1016/j.ajpath.2013.04.031
112. Taheri F, Ochoa JB, Faghiri Z, Culotta K, Park HJ, Lan MS, et al. l-Arginine regulates the expression of the T-cell receptor zeta chain (CD3zeta) in Jurkat cells. Clin Cancer Res (2001) 7:958s–65s.
113. Bronte V, Serafini P, Mazzoni A, Segal DM, Zanovello P. l-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol (2003) 24:302–6. doi:10.1016/S1471-4906(03)00132-7
114. Rodriguez PC, Zea AH, Desalvo J, Culotta KS, Zabaleta J, Quiceno DG, et al. l-arginine consumption by macrophages modulates the expression of CD3 zeta chain in T lymphocytes. J Immunol (2003) 171:1232–9.
116. Rodriguez PC, Quiceno DG, Ochoa AC. l-arginine availability regulates T-lymphocyte cell-cycle progression. Blood (2007) 109:1568–73. doi:10.1182/blood-2006-06-031856
117. Daum RS, Spellberg B. Progress toward a Staphylococcus aureus vaccine. Clin Infect Dis (2012) 54:560–7. doi:10.1093/cid/cir828
118. Bagnoli F, Bertholet S, Grandi G. Inferring reasons for the failure of Staphylococcus aureus vaccines in clinical trials. Front Cell Infect Microbiol (2012) 2:16. doi:10.3389/fcimb.2012.00016
119. Botelho-Nevers E, Verhoeven P, Paul S, Grattard F, Pozzetto B, Berthelot P, et al. Staphylococcal vaccine development: review of past failures and plea for a future evaluation of vaccine efficacy not only on staphylococcal infections but also on mucosal carriage. Expert Rev Vaccines (2013) 12:1249–59. doi:10.1586/14760584.2013.840091
120. Jansen KU, Girgenti DQ, Scully IL, Anderson AS. Vaccine review: “Staphyloccocus aureus vaccines: problems and prospects”. Vaccine (2013) 31:2723–30. doi:10.1016/j.vaccine.2013.04.002
121. Harro JM, Peters BM, O’May GA, Archer N, Kerns P, Prabhakara R, et al. Vaccine development in Staphylococcus aureus: taking the biofilm phenotype into consideration. FEMS Immunol Med Microbiol (2010) 59:306–23. doi:10.1111/j.1574-695X.2010.00708.x
122. Kim HK, Kim HY, Schneewind O, Missiakas D. Identifying protective antigens of Staphylococcus aureus, a pathogen that suppresses host immune responses. FASEB J (2011) 25:3605–12. doi:10.1096/fj.11-187963
123. Kim HK, Emolo C, Dedent AC, Falugi F, Missiakas DM, Schneewind O. Protein A-specific monoclonal antibodies and prevention of Staphylococcus aureus disease in mice. Infect Immun (2012) 80:3460–70. doi:10.1128/IAI.00230-12
124. Kim HK, Emolo C, Missiakas D, Schneewind O. A monoclonal antibody that recognizes the E domain of staphylococcal protein A. Vaccine (2014) 32:464–9. doi:10.1016/j.vaccine.2013.11.054
125. Stranger-Jones YK, Bae T, Schneewind O. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci U S A (2006) 103:16942–7. doi:10.1073/pnas.0606863103
126. Arrecubieta C, Matsunaga I, Asai T, Naka Y, Deng MC, Lowy FD. Vaccination with clumping factor A and fibronectin binding protein A to prevent Staphylococcus aureus infection of an aortic patch in mice. J Infect Dis (2008) 198:571–5. doi:10.1086/590210
127. Brady RA, O’May GA, Leid JG, Prior ML, Costerton JW, Shirtliff ME. Resolution of Staphylococcus aureus biofilm infection using vaccination and antibiotic treatment. Infect Immun (2011) 79:1797–803. doi:10.1128/IAI.00451-10
128. Anderson AS, Miller AA, Donald RG, Scully IL, Nanra JS, Cooper D, et al. Development of a multicomponent Staphylococcus aureus vaccine designed to counter multiple bacterial virulence factors. Hum Vaccin Immunother (2012) 8:1585–94. doi:10.4161/hv.21872
129. Ingolotti M, Kawalekar O, Shedlock DJ, Muthumani K, Weiner DB. DNA vaccines for targeting bacterial infections. Expert Rev Vaccines (2010) 9:747–63. doi:10.1586/erv.10.57
130. Levitz SM, Golenbock DT. Beyond empiricism: informing vaccine development through innate immunity research. Cell (2012) 148:1284–92. doi:10.1016/j.cell.2012.02.012
Keywords: Staphylococcus aureus, Staphylococcus epidermidis, biofilm, macrophage, neutrophil
Citation: Scherr TD, Heim CE, Morrison JM and Kielian T (2014) Hiding in plain sight: interplay between staphylococcal biofilms and host immunity. Front. Immunol. 5:37. doi: 10.3389/fimmu.2014.00037
Received: 31 October 2013; Accepted: 22 January 2014;
Published online: 05 February 2014.
Edited by:
Fabio Bagnoli, Novartis Vaccines and Diagnostics, ItalyReviewed by:
Dane Parker, Columbia University, USAAndrea Guido Oreste Manetti, Novartis Vaccines and Diagnostics, Italy
Copyright: © 2014 Scherr, Heim, Morrison and Kielian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tammy Kielian, Department of Pathology and Microbiology, University of Nebraska Medical Center, 985900 Nebraska Medical Center, Omaha, NE 68198-5900, USA e-mail:dGtpZWxpYW5AdW5tYy5lZHU=
 Tyler D. Scherr
Tyler D. Scherr Cortney E. Heim
Cortney E. Heim John M. Morrison
John M. Morrison Tammy Kielian
Tammy Kielian