
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 16 December 2013
Sec. Antigen Presenting Cell Biology
Volume 4 - 2013 | https://doi.org/10.3389/fimmu.2013.00464
This article is part of the Research Topic Processing, editing, and presenting the MHC class II peptidome View all 21 articles
The standard model of Major Histocompatibility Complex class II (MHCII)-restricted antigen processing depicts a straightforward, linear pathway: internalized antigens are converted into peptides that load in a chaperone dependent manner onto nascent MHCII in the late endosome, the complexes subsequently trafficking to the cell surface for recognition by CD4+ T cells (TCD4+). Several variations on this theme, both moderate and radical, have come to light but these alternatives have remained peripheral, the conventional pathway generally presumed to be the primary driver of TCD4+ responses. Here we continue to press for the conceptual repositioning of these alternatives toward the center while proposing that MHCII processing be thought of less in terms of discrete pathways and more in terms of a network whose major and minor conduits are variable depending upon many factors, including the epitope, the nature of the antigen, the source of the antigen, and the identity of the antigen-presenting cell.
In the early twentieth century delayed-type hypersensitivity (DTH) established itself as the mainstay for cellular immunologists, providing the launching point for many of the antigen systems that remain in use today (1) (Figure 1). A point of emphasis is that many of these proteins (ovalbumin, lysozyme, myoglobin, …) shared the properties of being plentiful and sturdy, and therefore amenable to the early protein purification schemes, which were relatively harsh and inefficient. These properties also facilitated structure determination, reinforcing their popularity as immunologists sought greater mechanistic insight into immune recognition.
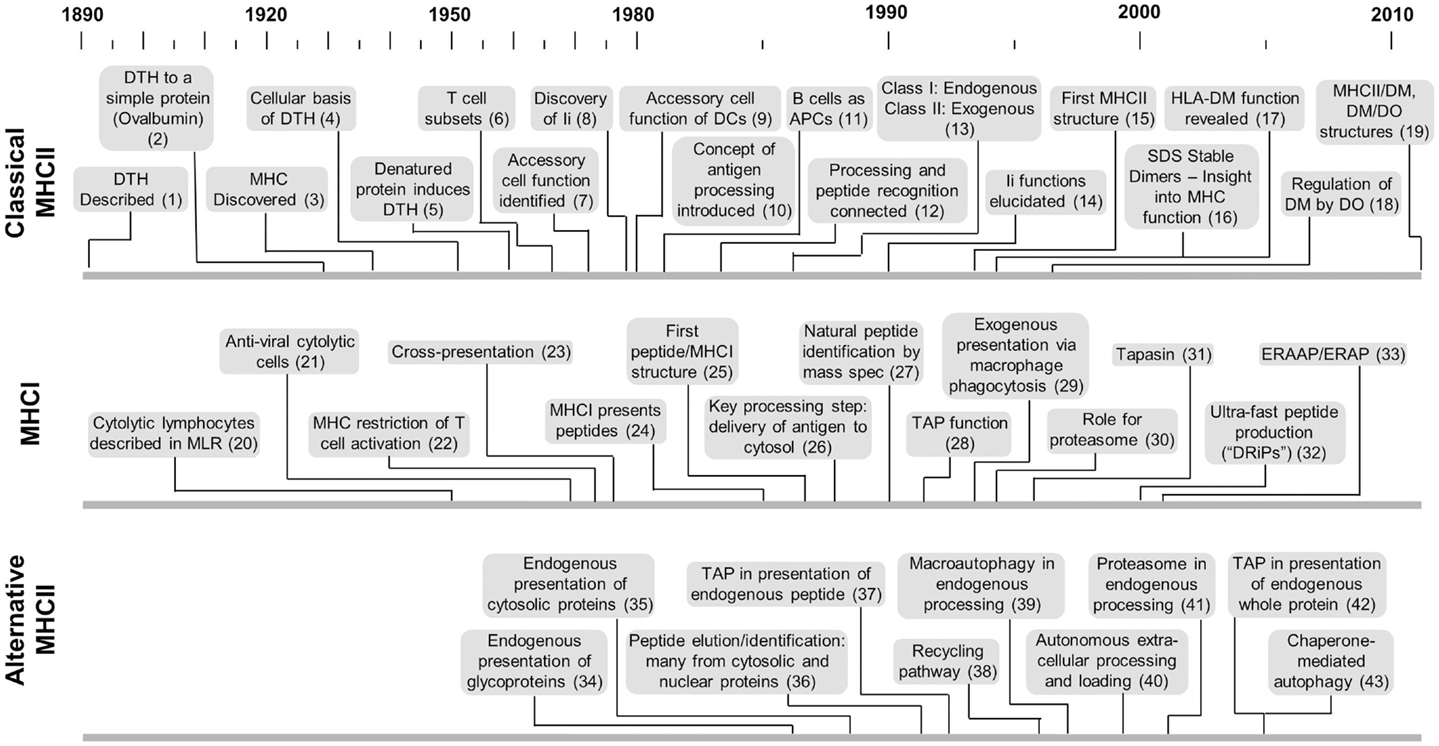
Figure 1. Timelines of key developments with respect to MHCII-classical processing, MHCI processing, and MHCII-alternative processing. This is not intended to provide a comprehensive listing but more a sense of the genesis and evolution of each area with respect to the others. Citation key: (1) Koch (117), (2) Dienes and Schoenheit (118), (3) MHC discovered by Gorer (119), (4) Gell and Hinde (120), (5) Gell and Benacerraf (2), (6) Claman et al. (121), (7) Shevach and Rosenthal (5), Rosenthal and Shevach (122), (8) Jones et al. (123), (9) Nussenzweig (124), Steinman and Nussenzweig (125) after earlier identification by Steinman and Cohn (126) and later demonstration of antigen processing by Sunshine et al. (23), Van Voorhis et al. (24), (10) Ziegler and Unanue (8), (11) Chesnut and Grey (20), Chesnut et al. (99, 100), Lanzavecchia (21), (12) Shimonkevitz et al. (11), Babbitt et al. (13), (13) Germain (54), (14) Bakke and Dobberstein (27), Peterson and Miller (29), Roche and Cresswell (30), Teyton et al. (31), Roche et al. (32), (15) Brown et al. (127), (16) Germain and Hendrix (35), Sadegh-Nasseri and Germain (36), Amigorena et al. (37), Qui et al. (38), Tulp et al. (39), West et al. (40), Riberdy et al. (128), (17) Fling et al. (25), Morris et al. (26), (18) Denzin et al. (129), (19) Pos et al. (130), Guce et al. (131), (20) Govaerts (132), (21) Lundstedt (133), (22) Zinkernagel and Doherty (134), (23) Bevan (55), (24) Townsend et al. (135), (25) Bjorkman et al. (136), (26) Moore et al. (47), Yewdell et al. (48), (27) Falk et al. (137), Rötzschke et al. (138), Van Bleek and Nathenson (139), (28) Powis et al. (49), (29) Kovacsovics-Bankowski et al. (140), (30) Rock et al. (52), (31) Sadasivan et al. (141), (32) Reits et al. (142), Schubert et al. (143), (33) Brouwenstijn et al. (144), Serwold et al. (145), (34) Bikoff and Bershtein (75), Eisenlohr and Hackett (77), Weiss and Bogen (78), (35) Jacobson et al. (81, 82), (36) Rudensky et al. (87), (37) Malnati et al. (83), (38) Pinet et al. (70), (39) Brazil et al. (146), (40) Santambrogio et al. (62, 63), (41) Mukherjee et al. (92), (42) Tewari et al. (94), (43) Zhou et al. (147).
By the mid-1970s it was known that denatured proteins and synthetic peptides could induce DTH (2, 3) and that DTH is mediated by the “helper” T cell subset (4). This foundation provided the springboard for two key subsequent discoveries. First, the Rosenthal laboratory demonstrated that in vitro T cell activation requires an MHCII-compatible “accessory cell,” later renamed antigen-presenting cell (APC) (5–7). At the time, the accessory cell was synonymous with the macrophage, whose longstanding reputation for phagocytosis further reinforced focus on exogenously provided antigens. Dendritic cells (DCs) and B cells would only later be identified as “professional” APC (Figure 1). Subsequently Unanue and co-workers pioneered the concept of “processing” in which antigen is taken up by the APC and handled internally for a defined period of time before emerging on the cell surface in a form capable of activating TCD4+ (8). A fragmentation step was implied by the observation that presentation is inhibitable by weak bases that prevent activation of the endosomal proteases (9).
While Listeria monocytogenes was utilized in initial experiments (8, 10), the nominal DTH antigens were far more suitable for experiments that sought greater mechanistic insight, due in large part to the challenge of epitope identification. At the time, the standard approach entailed chemical and/or proteolytic fragmentation of whole antigen, identification of the active fragment with in vitro assays, and confirmation and fine mapping with synthetic peptides. Thus, it was demonstrated with the ovalbumin system that the same TCD4+ hybridoma could be activated by whole antigen or proteolytic OVA fragments provided to the APC, or synthetic peptide pulsed onto gluteraldehyde-fixed APCs (11). An epitope within hen egg lysozyme (12) was used to demonstrate that the immunogenic peptide binds directly to MHCII (13) and, later, to map the residues of the peptide that contact MHCII and those that contact the T cell receptor (14). Subsequent key insights were made using the same or similar globular protein antigen systems, including the identification of specific endosomal proteases that participate in antigen processing (15–19), the impact that surface immunoglobulin has on the efficiency and specificity of processing by B cells (20–22), the antigen processing abilities of DCs (23, 24), and the critical role that the chaperone HLA-DM (in humans, H2-M in mice, referred to collectively as “DM”) plays in late endosomal (“classical”) peptide loading (25, 26).
Concurrent biochemical experiments served to reinforce the classical pathway. Efforts by several groups, most notably the Cresswell laboratory, elucidated the role of the transient MHCII binding partner, invariant chain (Ii), in delivering MHCII to the late endosome where Ii is removed by the combined actions of proteases and DM, and high affinity peptides are loaded (27–34). Germain and co-workers subsequently demonstrated that acquisition of high affinity peptide correlates with a discernible change in MHCII conformation, the so-called SDS-resistant “compact dimer” (35, 36). Several groups exploited this property to demonstrate via subcellular fractionation that compact dimer formation occurs in the late endosome (37–40), and is generally dependent upon DM (41, 42). Direct imaging studies tracing the fates of MHCII and Ii generally provided corroboration (43–46).
The processing of antigen for recognition by CD8+ T cells (TCD8+) had long been viewed as fundamentally different. This is because most MHC class I (MHCI) processing begins with delivery of antigen to the cytosol (47, 48), usually via infection, which allows for access to the proteasome and the transporter associated with antigen processing (TAP), both being critical for the production and delivery of most peptides to nascent MHCI in the ER (49–52). The apparent dichotomy – MHCI for endogenous antigen and MHCII from exogenous antigen – was reinforced by Morrison et al. who reported that inactivation of influenza virus obviates TCD8+ but not TCD4+ cell line activation while expression of influenza protein by a recombinant vaccinia virus results in TCD8+ but not TCD4+ activation (53). Thus, MHCI and MHCII appeared to be fundamentally different in terms of where the peptides come from (54).
The distinction stood for many years until the concept of cross-presentation, essentially MHCI-restricted presentation of exogenous antigen, gained traction. First observed by Bevan as the development of a host response to allogeneic cells (55), cross-presentation was eventually demonstrated to apply to virus infection, and attributed in most cases to the ability of the DC to take up material released from the antigen bearing cell and transfer it to the cytosol for conventional processing via mechanisms that are still under investigation (56).
Cross-presentation expands the potential for TCD8+ activation in two ways. First, it ensures delivery of antigen to DCs, generally considered essential for T cell priming (57), under circumstances when the invading organism does not infect DCs. Second, it short circuits many of the strategies that pathogens have developed to thwart MHCI antigen processing (58). For the similar purpose of expanding the potential for TCD4+ activation, one might expect that additional mechanisms exist for the generation of MHCII-peptide complexes. Indeed, there is longstanding evidence for alternative MHCII processing pathways, although they have yet to take hold like cross-presentation. These pathways fall into three general categories.
Exceptions to the need for intracellular antigen processing were provided by reports that location of the epitope within a disordered region of the antigen (59), denaturation of the antigen (60), and catabolism of some antigens by serum proteases (61), precludes the need for internalization. Subsequently it was demonstrated that immature DCs secrete lysosomal proteases into the extracellular space and express abundant amounts of empty (peptide receptive) MHC, allowing for autonomous extracellular processing (62, 63) and peptide loading at the cell surface independent of DM (64–66). More recently, extracellular processing of B cell receptor-captured antigen has been demonstrated to occur within the synaptic space between B cell and TCD4+ (67). Despite providing a potential explanation for the presentation of antigens that preclude internalization (large parasites, for example), extracellular processing has thus far been vastly understudied.
Experiments demonstrating an active peptide exchange mechanism in live APCs at relatively neutral pH (68) and the ability of mature MHCII to be internalized (69) suggested that MHCII could be recycled for a second round of peptide loading in an early endosomal compartment. Functional operation of this pathway was first demonstrated by the laboratory of Eric Long (70), and subsequently demonstrated to operate independent of DM (71). Influenza hemagglutinin, the antigen they focused upon, represents a group of proteins, including other viral glycoproteins and many bacterial toxins, that may be particularly relevant to the recycling pathway as they unfold in the early endosome as part of their biological programs (72–74).
Beginning in the mid-1980s, reports emerged that efficient, and in some cases any detectable presentation of a specific MHCII epitope depends upon synthesis of the antigen within the APC (75–80). Because the first antigens were glycoproteins, the suspicion remained, despite many controls, that the antigens trafficked in some fashion to the endosomal compartment for conventional processing. This concern was obviated by the Long lab, who demonstrated efficient presentation of epitopes within cytosolically located measles virus matrix protein and signal sequence-deleted influenza hemagglutinin (81–83). Further, expression of matrix by a recombinant vaccinia virus, where the antigen is not incorporated into the virion, also resulted in presentation (84). Of note, presentation from cytosolic HA was independent of TAP expression, but presentation from a cytosolic “minigene” construct was TAP-dependent (83).
Numerous subsequent reports have provided additional evidence of endogenous presentation (85, 86), including demonstrations that a substantial portion of peptides eluted from MHCII are derived from cytosolic and nuclear proteins (87, 88). Particularly notable is the variety of underlying mechanisms compared to the classical pathway. The Münz laboratory has focused upon a role for macroautophagy, the process in which cytosolic contents are enveloped in a bilayer membrane that subsequently fuses with the lysosome (89). The Blum laboratory has demonstrated that chaperone mediated autophagy, which involves translocation of individual proteins bearing the KFERQ motif, is critical in the presentation of some self-antigens (90). A third form of autophagy, microautophagy, in which cytosolic proteins are delivered to the late endosome during multivesicular body formation (91), is also likely to contribute to MHCII processing although a direct connection remains to be made. We and others have shown roles for the proteasome (92–95) and even TAP (94) in endogenous processing of some proteins. Like cross-presentation, these observations blur the line between the MHCI and MHCII systems.
Several key questions concerning MHCII processing remain.
(1) How common are the alternative pathways? As recounted elsewhere (86), our laboratory has addressed this question with influenza and ectromelia viruses, and results suggest that most of the TCD4+ responses to both viruses is driven by alternative, mainly endogenous processing. It will be of great interest to determine the extent to which this holds for other viruses and other types of pathogens.
(2) Where are MHCII molecules loaded in the cell? An intriguing alternative to the endosomal compartment is the endoplasmic reticulum, the site of Ii loading. There is no obvious reason why incompletely folded proteins could not compete with Ii. Indeed, there are longstanding reports of ER-resident MHCII binding proteins other than Ii (96). The greater challenge has been to connect this event with TCD4+ activation. This is exacerbated by the extreme sensitivity of T cells, allowing for the possibility that ER loading is a far greater contributor to TCD4+ activation than is appreciable by biochemical assays. Compared to the endocytic compartment there may be limited competition for MHCII in the ER, particularly during a virus infection that shuts off host synthesis. Thus, the bulk of MHCII may be directed to the endosomal compartment due to less efficient acquisition of epitopes derived from exogenous antigen.
(3) How many components of the antigen processing machinery remain to be discovered? Probably many. Components of any metabolic pathway are generally not appreciable until they are disabled, often unintentionally. For example, the role of DM in MHCII processing was revealed by a mutagenesis screen intended to identify MHCII structural mutants (97). A recent antibody-based genome-wide siRNA screen by the Neefjes lab revealed 276 genes that contribute to peptide presentation, only 10% of which had been previously implicated (98). The screen was performed on uninfected cells, and it is possible that infection and the ensuing innate activation enlists additional cellular components.
The data in aggregate indicate that MHCII processing and presentation extends well beyond the classical pathway. Following are additional considerations leading up to our proposal for a different way of thinking about MHCII processing:
(1) A cellular component can contribute to different processing schemes. For example, H-2M can contribute to loading of both proteasome-dependent and -independent epitopes (94).
(2) The same epitope can be produced via multiple pathways. We were first made aware of this possibility by an epitope within the influenza hemagglutinin that is presentable from exogenous antigen via the recycling pathway and from endogenous sources by a proteasome-dependent pathway (73, 94). Since that time we have expanded the analysis, finding that most of the influenza epitopes we have examined, are presentable from both exogenous and endogenous sources of the parent antigen, just much more efficiently in most cases via endogenous sources (unpublished). We speculate that an important factor is the proteolytic activity of various subcellular compartments (the cytosol being relatively hospitable and the endosomal compartment being relatively inhospitable) and the resistance of particular linear sequences to attack by resident proteases.
(3) Different APC types have different antigen processing capabilities. This has been apparent since the first comparisons of DCs, macrophages and B cells (99–104) and has been attributed to a variety of factors including internalization capabilities, protease profile, and activation state (105–112). We recently compared the abilities of three distinct primary APC types (bone marrow-derived DCs, splenic DCs, and peritoneal macrophages) to present six different influenza epitopes from exogenous and endogenous sources. We observed clear differences that were more or less accentuated depending upon the epitope (unpublished observation). An important ramification is that the presentation characteristics of APCs in vitro will not necessarily reflect antigen presentation in vivo, which in many cases may be an aggregate of several APC types.
(4) Different pathogens will likely demonstrate different processing signatures, both in terms of the processing pathways that are utilized and the extent to which each one is utilized. The various categories of pathogen interact with APCs in profoundly different ways. Large parasites, such as helminths, are presumably processed exogenously, via either the classical or extracellular pathways following some degree of extracellular digestion. This may also be the case for extracellular bacteria although, as noted above, membrane crossing bacterial toxins may be an exception. Viral proteins will have access to endogenous pathways but pathway usage may also be dramatically different depending upon replication scheme, the extent to which vesicular trafficking is disrupted, and the selective activation of cellular processes (e.g., autophagy).
(5) MHCII processing diversity – facilitation and benefits. The open-ended nature of the MHCII-peptide binding groove ensures that many epitope-containing forms can be presented. In contrast, closed-ended MHCI molecules generally require peptides of specific lengths and, thus, more focused and coordinated processing events. A driving force for the existence of multiple pathways is likely the greater diversity of peptides displayed, thereby ensuring sufficient TCD4+ engagement, whose importance in resolving infections is becoming increasingly apparent (113–116). In addition, processing diversity reduces the prospect that MHCII-restricted antigen presentation can be thwarted by an infectious agent. This redundancy may be one reason why there are many more reports of pathogens that attack components of the class I vs. class II processing machinery.
The physical properties of the DTH proteins considerably narrow processing scenarios, resulting in the detailed mechanistic insights that have been gained. In contrast, the robust diversity in alternative MHCll processing leads to superficially conflicting data and a diffusion of effort, both of which impede validation by concerted mechanistic studies. A way forward might be facilitated by reframing the problem. Therefore, based upon the points that have been made, we propose a network model of MHCII antigen processing (Figure 2), in which each epitope is produced via several pathways, the composition and balance of which are unique for each epitope, and strongly modulated by factors such as APC type and nature of the infectious agent. In vitro experimentation may validate the general tenets of the model, but its true scope will only be appreciated in vivo, a far more complex landscape that will be tackled only with the development of additional tools. This framework could catalyze insight into host/pathogen interplay and, consequently, new strategies for rational vaccine design. A critical first step will be increased numbers of investigators who use infectious organisms to investigate MHCII antigen processing and presentation. The systems are admittedly more cumbersome but the territory that they open up more than compensate for the extra effort.
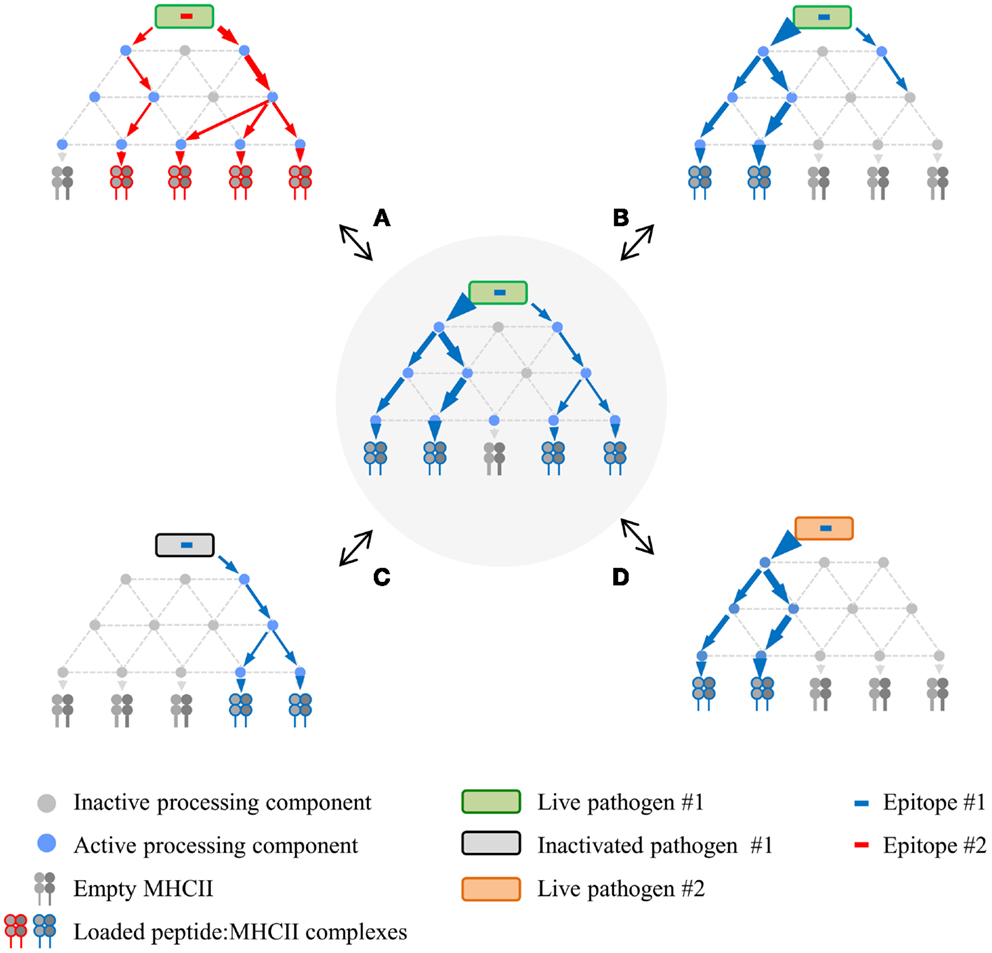
Figure 2. Proposed network model of MHCII antigen processing. The network in the center represents a hypothetical processing scheme for epitope #1 derived from live pathogen #1 following infection of APC #1. Processing variations surround this central network, demonstrating altered processing and presentation depending on different epitopes from the same live pathogen (A), different APC for the same epitope (B), same epitope from inactivated pathogen (C), or same epitope expressed via recombination by a different live pathogen (D). Processing components can be either active or inactive depending on APC type and/or APC activation state in response to infection.
Laurence C. Eisenlohr wrote the first draft. Laurence C. Eisenlohr and Michael A. Miller worked on revisions and the figures together.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Drs. Elizabeth Mellins, Jonathan Yewdell, and Peter Cresswell for helpful comments and suggestions.
2. Gell PGH, Benacerraf B. Studies on hypersensitivity. II. Delayed hypersensitivity to denatured proteins in guinea pigs. Immunology (1959) 2:64–70.
3. Kantor FS, Ojeda A, Benacerraf B. Studies of artificial antigens I. Antigenicity of DNP-polylysine and DNP copolymer of lysine and glutamic acid in guinea pigs. J Exp Med (1963) 117:55–69. doi: 10.1084/jem.117.1.55
4. Vadas MA, Miller JF, McKenzie IF, Chism SE, Shen FW, Boyse EA, et al. Ly and Ia antigen phenotypes of T cells involved in delayed-type hypersensitivity and in suppression. J Exp Med (1976) 144:10–9. doi:10.1084/jem.144.1.10
5. Shevach EM, Rosenthal AS. Function of macrophages in antigen recognition by guinea pig T lymphocytes. II. Role of the macrophage in the regulation of genetic control of the immune response. J Exp Med (1973) 138:1213–29. doi:10.1084/jem.138.5.1213
6. Waldron JA, Horn RG, Rosenthal AS. Antigen-induced proliferation of guinea pig lymphocytes in vitro: obligatory role of macrophages in the recognition of antigen by immune T-lymphocytes. J Immunol (1973) 111:58–64.
7. Rosenthal AS. Determinant selection and macrophage function in genetic control of the immune response. Immunol Rev (1978) 40:135–52. doi:10.1111/j.1600-065X.1978.tb00404.x
8. Ziegler K, Unanue ER. Identification of a macrophage antigen-processing event required for I-region-restricted antigen presentation to T lymphocytes. J Immunol (1981) 127:1869–75.
9. Ziegler HK, Unanue ER. Decrease in macrophage antigen catabolism caused by ammonia and chloroquine is associated with inhibition of antigen presentation to T cells. Proc Natl Acad Sci U S A (1982) 79:175–8. doi:10.1073/pnas.79.1.175
10. Ziegler K, Unanue ER. The specific binding of Listeria monocytogenes-immune T lymphocytes to macrophages. I. Quantitation and role of H-2 gene products. J Exp Med (1979) 150:1143–60. doi:10.1084/jem.150.5.1143
11. Shimonkevitz R, Kappler J, Marrack P, Grey H. Antigen recognition by H-2-restricted T cells. I. Cell-free antigen processing. J Exp Med (1983) 158:303–16. doi:10.1084/jem.158.2.303
12. Allen PM, Strydom DJ, Unanue ER. Processing of lysozyme by macrophages: identification of the determinant recognized by two T-cell hybridomas. Proc Natl Acad Sci U S A (1984) 81:2489–93. doi:10.1073/pnas.81.8.2489
13. Babbitt BP, Allen PM, Matsueda GR, Haber E, Unanue ER. Binding of immunogenic peptides to Ia histocompatibility molecules. Nature (1985) 317:359–61. doi:10.1038/317359a0
14. Allen PM, Matsueda GR, Evans RJ, Dunbar JB Jr, Marshall GR, Unanue ER. Identification of the T-cell and Ia contact residues of a T-cell antigenic epitope. Nature (1987) 327:713–5. doi:10.1038/327713a0
15. Takahashi H, Cease KB, Berzofsky JA. Identification of proteases that process distinct epitopes on the same protein. J Immunol (1989) 142:2221–9.
16. van Noort JM, Boon J, Van Der Drift ACM, Wagenaar JPA, Boots AMH, Boog CJP. Antigen processing by endosomal proteases determines which sites of sperm-whale myoglobin are eventually recognized by T cells. Eur J Immunol (1991) 21:1989–96. doi:10.1002/eji.1830210904
17. Bennett K, Levine T, Ellis JS, Peanasky RJ, Samloff IM, Kay J, et al. Antigen processing for presentation by class II major histocompatibility complex requires cleavage by cathepsin E. Eur J Immunol (1992) 22:1519–24. doi:10.1002/eji.1830220626
18. Williams KP, Smith JA. Isolation of a membrane-associated cathepsin D-like enzyme from the model antigen presenting cell, A20, and its ability to generate antigenic fragments from a protein antigen in a cell-free system. Arch Biochem Biophys (1993) 305:298–306. doi:10.1006/abbi.1993.1426
19. Manoury B, Hewitt EW, Morrice N, Dando PM, Barrett AJ, Watts C. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature (1998) 396:695–9. doi:10.1038/25379
20. Chesnut RW, Grey HM. Studies on the capacity of B cells to serve as antigen-presenting cells. J Immunol (1981) 126:1075–9.
21. Lanzavecchia A. Antigen-specific interaction between T and B cells. Nature (1985) 314:537–9. doi:10.1038/314537a0
22. Simitsek PD, Campbell DG, Lanzavecchia A, Fairweather N, Watts C. Modulation of antigen processing by bound antibodies can boost or suppress class II major histocompatibility complex presentation of different T cell determinants. J Exp Med (1995) 1957:1963.
23. Sunshine GH, Gold DP, Wortis HH, Marrack P, Kappler JW. Mouse spleen dendritic cells present soluble antigens to antigen-specific T cell hybridomas. J Exp Med (1983) 158:1745–50. doi:10.1084/jem.158.5.1745
24. Van Voorhis WC, Valinsky J, Hoffman E, Luban J, Hair LS, Steinman RM. Relative efficacy of human monocytes and dendritic cells as accessory cells for T cell replication. J Exp Med (1983) 158:174–91. doi:10.1084/jem.158.1.174
25. Fling SP, Arp B, Pious D. HLA-DMA and -DMB genes are both required for MHC class II/peptide complex formation in antigen-presenting cells. Nature (1994) 368:554–8. doi:10.1038/368554a0
26. Morris P, Shaman J, Attaya M, Amaya M, Goodman S, Bergman C, et al. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature (1994) 368:551–4. doi:10.1038/368551a0
27. Bakke O, Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell (1990) 63:707–16. doi:10.1016/0092-8674(90)90137-4
28. Marks MS, Blum JS, Cresswell P. Invariant chain trimers are sequestered in the rough endoplasmic reticulum in the absence of association with HLA class II antigens. J Cell Biol (1990) 111:839–55. doi:10.1083/jcb.111.3.839
29. Peterson M, Miller J. Invariant chain influences the immunological recognition of MHC class II molecules. Nature (1990) 345:172–4. doi:10.1038/345172a0
30. Roche PA, Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature (1990) 345:615–8. doi:10.1038/345615a0
31. Teyton L, O’Sullivan D, Dickson PW, Lotteau V, Sette A, Fink P, et al. Invariant chain distinguishes between the exogenous and endogenous antigen presentation pathways. Nature (1990) 348:39–44. doi:10.1038/348039a0
32. Roche PA, Marks MS, Cresswell P. Formation of a nine-subunit complex by HLA class II glycoproteins and the invariant chain. Nature (1991) 354:392–4. doi:10.1038/354392a0
33. Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol (1994) 12:259–93. doi:10.1146/annurev.iy.12.040194.001355
34. Cresswell P. Invariant chain structure and MHC class II function. Cell (1996) 84:505–7. doi:10.1016/S0092-8674(00)81025-9
35. Germain RN, Hendrix LR. MHC class II structure, occupancy and surface expression determined by post-endoplasmic reticulum antigen binding. Nature (1991) 353:134–9. doi:10.1038/353134a0
36. Sadegh-Nasseri S, Germain RN. A role for peptide in determining MHC class II structure. Nature (1991) 353:167–70. doi:10.1038/353167a0
37. Amigorena S, Drake JR, Webster P, Mellman I. Transient accumulation of new class II MHC molecules in a novel endocytic compartment in B lymphocytes. Nature (1994) 369:113–20. doi:10.1038/369113a0
38. Qui Y, Xu X, Wandinger-Ness A, Dalke DP, Pierce SK. Separation of subcellular compartments containing distinct functional forms of MHC class II. J Cell Biol (1994) 125:595–605. doi:10.1083/jcb.125.3.595
39. Tulp A, Verwoerd D, Dobberstein B, Ploegh HL, Pieters J. Isolation and characterization of the intracellular MHC class II compartment. Nature (1994) 369:120–6. doi:10.1038/369120a0
40. West MA, Lucocq JM, Watts C. Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells. Nature (1994) 369:147–51. doi:10.1038/369147a0
41. Pious D, Dixon L, Levine F, Cotner T, Johnson R. HLA class II regulation and structure. Analysis with HLA-DR3 and HLA-DP point mutants. J Exp Med (1985) 162:1193–207. doi:10.1084/jem.162.4.1193
42. Stebbins CC, Loss GE Jr, Elias CG, Chervonsky A, Sant AJ. The requirement for DM in class II-restricted antigen presentation and SDS-stable dimer formation is allele and species dependent. J Exp Med (1995) 184:1747–53.
43. Guagliardi LE, Koppelman B, Blum JS, Marks MS, Cresswell P, Brodsky FM. Co-localization of molecules involved in antigen processing and presentation in an early endocytic compartment. Nature (1990) 343:133–9. doi:10.1038/343133a0
44. Sanderson F, Kleijmeer MJ, Kelly A, Verwoerd D, Tulp A, Neefjes JJ, et al. Accumulation of HLA-DM, a regulator of antigen presentation, in MHC class II compartments. Science (1994) 266:1566–9. doi:10.1126/science.7985027
45. Peters PJ, Raposo G, Neefjes JJ, Oorschot V, Leijendedder RL, Geuze HJ, et al. Major histocompatibility complex class II compartments in human B lymphoblastoid cells are distinct from early endosomes. J Exp Med (1995) 182:325–34. doi:10.1084/jem.182.2.325
46. Kleijmeer MJ, Morkowski S, Griffith JM, Rudensky AY, Geuze HJ. Major histocompatibility complex class II compartments in human and mouse B lymphoblasts represent conventional endocytic compartments. J Cell Biol (1997) 139:639–49. doi:10.1083/jcb.139.3.639
47. Moore MW, Carbone FR, Bevan MJ. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell (1988) 54:777–85. doi:10.1016/S0092-8674(88)91043-4
48. Yewdell JW, Bennink JR, Hosaka Y. Cells process exogenous proteins for recognition by cytotoxic T lymphocytes. Science (1988) 239:637–40. doi:10.1126/science.3257585
49. Powis SJ, Townsend ARM, Deverson EV, Bastin J, Butcher GW, Howard JC. Restoration of antigen presentation to the mutant cell line RMA-S by an MHC-linked transporter. Nature (1991) 354:528–31. doi:10.1038/354528a0
50. Spies T, Cerundolo V, Colonna M, Cresswell P, Townsend A, Demars R. Presentation of endogenous viral antigen by major histocompatibility class I molecules dependent on putative peptide transporter heterodimer. Nature (1992) 355:644–6. doi:10.1038/355644a0
51. Spies T, Cerundolo V, Colonna M, Cresswell P, Townsend A, Demars R. Presentation of viral antigen by MHC class I molecules is dependent on a putative peptide transporter heterodimer. Nature (1992) 355:644–6. doi:10.1038/355644a0
52. Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell (1994) 78:761–71. doi:10.1016/S0092-8674(94)90462-6
53. Morrison LA, Lukacher AE, Braciale VL, Fan DP, Braciale TJ. Differences in antigen presentation to MHC class I- and class II-restricted influenza virus-specific cytolytic T lymphocyte clones. J Exp Med (1986) 163:903–21. doi:10.1084/jem.163.4.903
54. Germain RN. The ins and outs of antigen processing and presentation. Nature (1986) 322:687–9. doi:10.1038/322687a0
55. Bevan MJ. Cross-priming for a secondary cytotoxic response to minor H antigens with H-2 congenic cells which do not cross-react in the cytotoxic assay. J Exp Med (1976) 143:1283. doi:10.1084/jem.143.5.1283
56. Segura E, Villadangos JA. A modular and combinatorial view of the antigen cross-presentation pathway in dendritic cells. Traffic (2011) 12:1677–85. doi:10.1111/j.1600-0854.2011.01254.x
57. Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol (2005) 23:975–1028. doi:10.1146/annurev.immunol.22.012703.104538
58. Zhou F. Molecular mechanisms of viral immune evasion proteins to inhibit MHC class I antigen processing and presentation. Int Rev Immunol (2009) 28:376–93. doi:10.1080/08830180903013034
59. Lee P, Matsueda GR, Allen PM. T cell recognition of fibrinogen. A determinant on the Aα-chain does not require processing. J Immunol (1988) 140:1063–8.
60. Hampl J, Gradehandt G, Kalbacher H, Rude E. In vitro processing of insulin for recognition by murine T cells results in the generation of A chains with free CysSH. J Immunol (1992) 148:2664–71.
61. Accapezzato D, Nisini R, Paroli M, Bruno G, Bonino F, Houghton M, et al. Generation of an MHC class II-restricted T cell epitope by extracellular processing of hepatitis delta antigen. J Immunol (1998) 160:5262–6.
62. Santambrogio L, Sato AK, Carven GJ, Belyanskaya SL, Strominger JL, Stern LJ. Extracellular antigen processing and presentation by immature dendritic cells. Proc Natl Acad Sci U S A (1999) 96:15056–61. doi:10.1073/pnas.96.26.15056
63. Santambrogio L, Sato AK, Fischer FR, Dorf ME, Stern LJ. Abundant empty class II MHC molecules on the surface of immature dendritic cells. Proc Natl Acad Sci U S A (1999) 96:15050–5. doi:10.1073/pnas.96.26.15050
64. Rabinowitz JD, Vrljic M, Kasson PM, Liang MN, Busch R, Boniface JJ, et al. Formation of a highly peptide-receptive state of class II MHC. Immunity (1998) 9:699–709. doi:10.1016/S1074-7613(00)80667-6
65. Vacchino JF, McConnell HM. Peptide binding to active class II MHC protein on the cell surface. J Immunol (2001) 166:6680–5.
66. Venkatraman P, Nguyen TT, Sainlos M, Bilsel O, Chitta S, Imperiali B, et al. Fluorogenic probes for monitoring peptide binding to class II MHC proteins in living cells. Nat Chem Biol (2007) 3:222–8. doi:10.1038/nchembio868
67. Yuseff MI, Reversat A, Lankar D, Diaz J, Fanget I, Pierobon P, et al. Polarized secretion of lysosomes at the B cell synapse couples antigen extraction to processing and presentation. Immunity (2011) 35:361–74. doi:10.1016/j.immuni.2011.07.008
68. Adorini L, Apella E, Doria G, Cardinaux F, Nagy ZA. Competition for antigen presentation in living cells involves exchange of peptides bound by class II MHC molecules. Nature (1989) 342:800–3. doi:10.1038/342800a0
69. Harding CV, Roof RW, Unanue ER. Turnover of Ia-peptide complexes is facilitated in viable antigen-presenting cells: biosynthetic turnover of Ia vs. peptide exchange. Proc Natl Acad Sci U S A (1989) 86:4230–4. doi:10.1073/pnas.86.11.4230
70. Pinet V, Vergelli M, Martin R, Bakke O, Long EO. Antigen presentation mediated by recycling of surface HLA-DR molecules. Nature (1995) 375:603–6. doi:10.1038/375603a0
71. Chianese-Bullock KA, Russell HI, Moller C, Gerhard W, Monaco JJ, Eisenlohr LC. Antigen processing of two H2-IEd restricted epitopes is differentially influenced by the structural changes in a viral glycoprotein. J Immunol (1998) 161:1599–607.
72. Falnes PO, Sandvig K. Penetration of protein toxins into cells. Curr Opin Cell Biol (2000) 12:407–13. doi:10.1016/S0955-0674(00)00109-5
73. Sinnathamby G, Eisenlohr LC. Presentation by recycling MHC class II molecules of an influenza hemagglutinin-derived epitope that is revealed in the early endosome by acidification. J Immunol (2003) 170:3504–13.
74. Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol (2008) 9:543–56. doi:10.1038/nrm2417
75. Bikoff E, Bershtein BK. T cell clones specific for Ig G2a of the α allotype: direct evidence for presentation of endogenous antigen. J Immunol (1986) 137:28–34.
76. Bikoff EK, Eckhardt LA. Presentation of IgG2a antigens to class II-restricted T cells by stably transfected B lymphoma cells. Eur J Immunol (1989) 19:1903–9. doi:10.1002/eji.1830191022
77. Eisenlohr LC, Hackett CJ. Class II major histocompatibility complex-restricted T cells specific for a virion structural protein that do not recognize exogenous influenza virus. J Exp Med (1989) 169:921–31. doi:10.1084/jem.169.3.921
78. Weiss S, Bogen B. B-lymphoma cells process and present their endogenous immunoglobulin to major histocompatibility complex-restricted T cells. Proc Natl Acad Sci U S A (1989) 86:282–6. doi:10.1073/pnas.86.1.282
79. Weiss S, Bogen B. MHC class II-restricted presentation of intracellular antigen. Cell (1991) 64:767–76. doi:10.1016/0092-8674(91)90506-T
80. Loss GE Jr, Elias CG, Fields PE, Ribaudo RK, McKisic M, Sant AJ. Major histocompatibility complex class II-restricted presentation of an internally synthesized antigen displays cell-type variability and segregates from the exogenous class II and endogenous class I presentation pathways. J Exp Med (1993) 178:73–85. doi:10.1084/jem.178.1.73
81. Jacobson S, Sekaly RP, Bellini WJ, Johnson CL, McFarland HF, Long EO. Recognition of intracellular measles virus antigens by HLA class II restricted measles virus-specific cytotoxic T lymphocytes. Ann N Y Acad Sci (1988) 540:352–3. doi:10.1111/j.1749-6632.1988.tb27096.x
82. Jacobson S, Sekaly RP, Jacobson CL, McFarland HF, Long EO. HLA class II restricted presentation of cytoplasmic measles virus antigens to cytotoxic T cells. J Virol (1989) 63:1756–62.
83. Malnati MS, Marti M, Lavaute T, Jaraquemada D, Biddison W, Demars R, et al. Processing pathways for presentation of cytosolic antigen to MHC class II-restricted T cells. Nature (1992) 357:702–4. doi:10.1038/357702a0
84. Jaraquemada D, Marti M, Long EO. An endogenous processing pathway in vaccinia virus-infected cells for presentation of cytoplasmic antigens to class II restricted T cells. J Exp Med (1990) 172:947–54. doi:10.1084/jem.172.3.947
85. Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol (2013) 31:443–73. doi:10.1146/annurev-immunol-032712-095910
86. Eisenlohr LC. Alternative generation of MHC class II-restricted epitopes: not so exceptional? Mol Immunol (2013) 55:169–71. doi:10.1016/j.molimm.2012.10.020
87. Rudensky AY, Preston-Hulburt P, Hong S-C, Barlow A, Janeway CJ. Sequence analysis of peptides bound to MHC class II molecules. Nature (1991) 353:32767. doi:10.1038/353622a0
88. Yeager M, Carrington M, Hughes AL. Class I and class II MHC bind self peptide sets that are strikingly different in their evolutionary characteristics. Immunogenetics (2000) 51:8–15. doi:10.1007/s002510050002
89. Gannage M, Münz C. Autophagy in MHC class II presentation of endogenous antigens. Curr Top Microbiol Immunol (2009) 335:123–40. doi:10.1007/978-3-642-00302-8_6
90. Crotzer VL, Blum JS. Autophagy and adaptive immunity. Immunology (2010) 131:9–17. doi:10.1111/j.1365-2567.2010.03321.x
91. Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell (2011) 20:131–9. doi:10.1016/j.devcel.2010.12.003
92. Mukherjee P, Dani A, Bhatia S, Singh N, Rudensky AY, George A, et al. Efficient presentation of both cytosolic and endogenous transmembrane protein antigens on MHC class ii is dependent on cytoplasmic proteolysis. J Immunol (2001) 167:2632–41.
93. Li P, Gregg JL, Wang N, Zhou D, O’Donnell P, Blum JS, et al. Compartmentalization of class II antigen presentation: contribution of cytoplasmic and endosomal processing. Immunol Rev (2005) 207:206–17. doi:10.1111/j.0105-2896.2005.00297.x
94. Tewari MK, Sinnathamby G, Rajagopal D, Eisenlohr LC. A cytosolic pathway for MHC class II-restricted antigen processing that is proteasome and TAP dependent. Nat Immunol (2005) 6:287–94. doi:10.1038/ni0405-420
95. Comber JD, Robinson TM, Siciliano NA, Snook AE, Eisenlohr LC. Functional macroautophagy induction by influenza A virus without a contribution to major histocompatibility complex class II-restricted presentation. J Virol (2011) 85:6453–63. doi:10.1128/JVI.02122-10
96. Lechler R, Aichinger G, Lightstone L. The endogenous pathway of MHC class II antigen presentation. Immunol Rev (1996) 151:51–79. doi:10.1111/j.1600-065X.1996.tb00703.x
97. Mellins E, Smith L, Arp B, Cotner T, Celis E, Pious D. Defective processing and presentation of exogenous antigens in mutants with normal HLA class II genes. Nature (1990) 343:71–4. doi:10.1038/343071a0
98. Paul P, Van Den Hoorn T, Jongsma ML, Bakker MJ, Hengeveld R, Janssen L, et al. A Genome-wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell (2011) 145:268–83. doi:10.1016/j.cell.2011.03.023
99. Chesnut RW, Colon SM, Grey HM. Antigen presentation by normal B cells, B cell tumors, and macrophages: functional and biochemical comparison. J Immunol (1982) 128:1764–8.
100. Chesnut RW, Colon SM, Grey HM. Requirements for the processing of antigens by antigen-presenting B cells. I. Functional comparison of B cell tumors and macrophages. J Immunol (1982) 129:2382–8.
101. Grey HM, Colon SM, Chesnut RW. Requirements for the processing of antigen by antigen-presenting B cells. II. Biochemical comparison of the fate of antigen in B cell tumors and macrophages. J Immunol (1982) 129:2389–95.
102. Guidos C, Wong M, Lee K-C. A comparison of the stimulatory activities of lymphoid dendritic cells and macrophages in T proliferative responses to various antigens. J Immunol (1984) 133:1179–84.
103. Lee KC, Guidos C. Heterogeneity of macrophages and dendritic cells as accessory cells. Immunobiology (1984) 168:172–81. doi:10.1016/S0171-2985(84)80108-4
104. Bjercke S, Gaudernack G. Dendritic cells and monocytes as accessory cells in T-cell responses in man. II. Function as antigen-presenting cells. Scand J Immunol (1985) 21:501–8. doi:10.1111/j.1365-3083.1985.tb01839.x
105. Scheerlinck JP, Burssens G, Brys L, Michel A, Hauser P, De Baetselier P. Differential presentation of hepatitis B S-preS(2) particles and peptides by macrophages and B-cell like antigen-presenting cells. Immunology (1991) 73:88–94.
106. Garrigan K, Moroni-Rawson P, McMurray C, Hermans I, Abernethy N, Watson J, et al. Functional comparison of spleen dendritic cells and dendritic cells cultured in vitro from bone marrow precursors. Blood (1996) 88:3508–12.
107. Milich DR, Chen M, Schodel F, Peterson DL, Jones JE, Hughes JL. Role of B cells in antigen presentation of the hepatitis B core. Proc Natl Acad Sci U S A (1997) 94:14648–53. doi:10.1073/pnas.94.26.14648
108. Gluckman JC, Canque B, Rosenzwajg M. Dendritic cells: a complex cellular system. Transfus Clin Biol (1998) 5:47–55. doi:10.1016/S1246-7820(98)80110-9
109. Lennon-Dumenil AM, Bakker AH, Wolf-Bryant P, Ploegh HL, Lagaudriere-Gesbert C. A closer look at proteolysis and MHC-class-II-restricted antigen presentation. Curr Opin Immunol (2002) 14:15–21. doi:10.1016/S0952-7915(01)00293-X
110. Holmdahl M, Grubb A, Holmdahl R. Cysteine proteases in Langerhans cells limits presentation of cartilage derived type II collagen for autoreactive T cells. Int Immunol (2004) 16:717–26. doi:10.1093/intimm/dxh079
111. Pereira SR, Faca VM, Gomes GG, Chammas R, Fontes AM, Covas DT, et al. Changes in the proteomic profile during differentiation and maturation of human monocyte-derived dendritic cells stimulated with granulocyte macrophage colony stimulating factor/interleukin-4 and lipopolysaccharide. Proteomics (2005) 5:1186–98. doi:10.1002/pmic.200400988
112. Duclos S, Clavarino G, Rousserie G, Goyette G, Boulais J, Camossetto V, et al. The endosomal proteome of macrophage and dendritic cells. Proteomics (2011) 11:854–64. doi:10.1002/pmic.201000577
113. Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med (2000) 191:1499–512. doi:10.1084/jem.191.9.1499
114. Shoukry NH, Cawthon AG, Walker CM. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol (2004) 58:391–424. doi:10.1146/annurev.micro.58.030603.123836
115. Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med (2012) 18:274–80. doi:10.1038/nm.2612
116. Zhou Y, Callendret B, Xu D, Brasky KM, Feng Z, Hensley LL, et al. Dominance of the CD4+ T helper cell response during acute resolving hepatitis A virus infection. J Exp Med (2012) 209:1481–92. doi:10.1084/jem.20111906
117. Koch R. Mittheilungen über ein Heilmittel gegen Tuberculose. Dt med Wochenschr (1891) 17:101–2. doi:10.1055/s-0029-1206198
118. Dienes L, Schoenheit EW. The reproduction of tuberculin hypersensitiveness in guinea pigs with various protein substances. Am Rev Tubercul (1929) 20:92–105.
119. Gorer P. The genetic and antigenic basis of tumor transplantation. J Pathol Bacteriol (1937) 44:691–7. doi:10.1002/path.1700440313
120. Gell PG, Hinde IT. The histology of the tuberculin reaction and its modification by cortisone. Br J Exp Pathol (1951) 32:516–29.
121. Claman HN, Chaperon EA, Triplett RF. Thymus-marrow cell combinations. Synergism in antibody production. Proc Soc Exp Biol Med (1966) 122:1167–71. doi:10.3181/00379727-122-31353
122. Rosenthal AS, Shevach EM. Function of macrophages in antigen recognition by guinea pig T lymphocytes. I. Requirement for histocompatible macrophages and lymphocytes. J Exp Med (1973) 138:1194–212. doi:10.1084/jem.138.5.1194
123. Jones PP, Murphy DB, Hewgill D, McDevitt HO. Detection of a common polypeptide chain in I – A and I – E sub-region immunoprecipitates. Mol Immunol (1979) 16:51–60. doi:10.1016/0161-5890(79)90027-0
124. Nussenzweig MC, Steinman RM, Gutchinov B, Cohn ZA. Dendritic cells are accessory cells for the development of anti-trinitrophenyl cytotoxic T lymphocytes. J Exp Med (1980) 152:1070–84. doi:10.1084/jem.152.4.1070
125. Steinman RM, Nussenzweig MC. Dendritic cells: features and functions. Immunol Rev (1980) 53:125–47. doi:10.1111/j.1600-065X.1980.tb01042.x
126. Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med (1973) 137:1142–62. doi:10.1084/jem.137.5.1142
127. Brown JH, Jardetsky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature (1993) 364:33–9. doi:10.1038/364033a0
128. Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR Molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature (1992) 360:474–6. doi:10.1038/360474a0
129. Denzin LK, Sant’angelo DB, Hammond C, Surman MJ, Cresswell P. Negative regulation by HLA-DO of MHC class II-restricted antigen processing. Science (1997) 278:106–9. doi:10.1126/science.278.5335.106
130. Pos W, Sethi DK, Call MJ, Schulze MS, Anders AK, Pyrdol J, et al. Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell (2012) 151:1557–68. doi:10.1016/j.cell.2012.11.025
131. Guce AI, Mortimer SE, Yoon T, Painter CA, Jiang W, Mellins ED, et al. HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism. Nat Struct Mol Biol (2013) 20:90–8. doi:10.1038/nsmb.2460
133. Lundstedt C. Interaction between antigenically different cells. Virus-induced cytotoxicity by immune lymphoid cells in vitro. Acta Pathol Microbiol Scand (1969) 75:139–52.
134. Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature (1974) 248:701–2. doi:10.1038/248701a0
135. Townsend ARM, Gotch FM, Davey J. Cytotoxic T cells recognize fragments of the influenza nucleoprotein. Cell (1985) 42:457–67. doi:10.1016/0092-8674(85)90103-5
136. Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature (1987) 329:506–12. doi:10.1038/329506a0
137. Falk K, Rötzschke O, Rammensee H-G. Cellular peptide composition governed by major histocompatibility complex class I molecules. Nature (1990) 348:248–51. doi:10.1038/348248a0
138. Rötzschke O, Falk K, Deres K, Schild H, Norda M, Metzger J, et al. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature (1990) 348:252–4. doi:10.1038/348252a0
139. Van Bleek GM, Nathenson SG. Isolation of an endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature (1990) 348:213–6. doi:10.1038/348213a0
140. Kovacsovics-Bankowski M, Clark K, Benacerraf B, Rock KL. Efficient major histocompatibility complex class I presentation of exogenous antigen upon phagocytosis by macrophages. Proc Natl Acad Sci U S A (1993) 90:4942–6. doi:10.1073/pnas.90.11.4942
141. Sadasivan B, Lahner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity (1996) 5:103–14. doi:10.1016/S1074-7613(00)80487-2
142. Reits EA, Vos JC, Gromme M, Neefjes J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature (2000) 404:774–8. doi:10.1038/35008103
143. Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature (2000) 404:770–4. doi:10.1038/35008096
144. Brouwenstijn N, Serwold T, Shastri N. MHC class I molecules can direct proteolytic cleavage of antigenic precursors in the endoplasmic reticulum. Immunity (2001) 15:95–104. doi:10.1016/S1074-7613(01)00174-1
145. Serwold T, Gaw S, Shastri N. ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat Immunol (2001) 2:644–51. doi:10.1038/89800
146. Brazil MI, Weiss S, Stockinger B. Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur J Immunol (1997) 27:1506–14. doi:10.1002/eji.1830270629
Keywords: antigen processing, antigen presentation, MHC class II, alternative processing, endogenous, review
Citation: Miller MA, Ganesan APV and Eisenlohr LC (2013) Toward a network model of MHC class II-restricted antigen processing. Front. Immunol. 4:464. doi: 10.3389/fimmu.2013.00464
Received: 25 September 2013; Accepted: 03 December 2013;
Published online: 16 December 2013.
Edited by:
Laura Santambrogio, Albert Einstein College of Medicine, USAReviewed by:
Laura Santambrogio, Albert Einstein College of Medicine, USACopyright: © 2013 Miller, Ganesan and Eisenlohr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laurence C. Eisenlohr, Thomas Jefferson University, Department of Microbiology and Immunology, Bluemle Life Sciences Building, Room 730, 233 South 10th Street, Philadelphia, PA 19107, USA e-mail:bGF1cmVuY2UuZWlzZW5sb2hyQGplZmZlcnNvbi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.