
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 21 October 2013
Sec. Molecular Innate Immunity
Volume 4 - 2013 | https://doi.org/10.3389/fimmu.2013.00348
This article is part of the Research TopicNLR-protein functions in immunityView all 18 articles
Nucleotide-binding domain leucine-rich repeat (NLR) proteins play a central role in the innate immune systems of plants and vertebrates. In plants, NLR proteins function as intracellular receptors that detect pathogen effector proteins directly, or indirectly by recognizing effector-induced modifications to other host proteins. NLR activation triggers a suite of defense responses associated with programed cell death (PCD). The molecular mechanisms underlying NLR activation, and how activation is translated into defense responses, have been particularly challenging to elucidate in plants. Recent reports, however, are beginning to shed some light. It is becoming clear that plant NLR proteins are targeted to diverse sub-cellular locations, likely depending on the locations where the effectors are detected. These reports also indicate that some NLRs re-localize following effector detection, while others do not, and such relocalization may reflect differences in signaling pathways. There have also been recent advances in understanding the structure of plant NLR proteins, with crystal structures now available for the N-terminal domains of two well-studied NLRs, a coiled-coil (CC) domain and a Toll-interleukin Receptor (TIR). Significant improvements in molecular modeling have enabled more informed structure-function studies, illuminating roles of intra- and inter-molecular interactions in NLR activation regulation. Several independent studies also suggest that intracellular trafficking is involved in NLR-mediated resistance. Lastly, progress is being made on identifying transcriptional regulatory complexes activated by NLRs. Current models for how plant NLR proteins are activated and how they induce defenses are discussed, with an emphasis on what remains to be determined.
Plants do not have an adaptive immune system like that found in vertebrate animals. Instead, plants depend solely on an innate immune system that bears intriguing similarities to animal innate immune systems, but is likely independently evolved [see review by Jacob et al. (1)]. Plant innate immunity is a two-tier resistance system (2). The first tier consists of plasma-membrane (PM) localized pattern recognition receptors (PRRs) that mediate detection of conserved microbial molecules referred to as pathogen associated molecular patterns (PAMPs). This type of resistance is known as PAMP triggered immunity (PTI). Most plant PRRs are transmembrane receptor kinases, with the majority containing extracellular leucine-rich repeats (LRR), thus have functional and structural similarity to the Toll-like Receptors of animals. The second tier system consists of intracellular receptors that detect the presence of pathogen proteins inside the host cell. Pathogen proteins that get inside host cells are commonly referred to as effector proteins, thus this second tier is usually referred to as effector triggered immunity (ETI).
Effector triggered immunity is mostly mediated by nucleotide-binding leucine-rich repeat (NLR) proteins. Plant NLR proteins usually contain a C-terminal LRR domain and a central NB-ARC domain (nucleotide-binding adaptor shared by Apaf-1, Resistance proteins, and CED-4) (3).The NB-ARC proteins form a subclass in the STAND super family (signal transduction ATPases with numerous domains) and function as molecular switches regulating many processes, including immunity and apoptosis (4, 5). Plant NLRs are roughly divided into two groups, depending on their N-terminal structures, CNL (CC-NB-LRR) with an N-terminal coiled-coil domain and TNL (TIR-NB-LRR) with an N-terminal Toll/interleukin-1 receptor domain (TIR) (6). Plant NLR proteins recognize the presence of pathogens either directly by binding to pathogen effectors, or indirectly by sensing effector-induced modification of other host proteins. The activation of ETI usually results in localized cell death at the infection site, which is referred to as a hypersensitive response (HR). The HR is commonly used as a read-out for the activation of NLR proteins in plants. The first NLR proteins, N and RPS2, were cloned in 1994 based on their ability to confer resistance to specific diseases in plants (7–9). However, the molecular mechanisms that control NLR activation and signaling remain poorly understood.
Here, we focus on the advances made in the last 2 years toward understanding how plant NLRs are activated and how signaling is initiated and transduced. We highlight the compartmentalization of plant NLRs, intra-/inter-molecular interactions before and after activation, and structural and genetic insights into NLR downstream signaling.
The activation of NLR proteins is commonly associated with significant transcriptional reprograming. Consistent with this observation, several plant NLRs have been shown to accumulate in the nucleus upon effector-induced activation (10–13)(Figure 1A). For example, in the presence of the cognate powdery mildew effector AvrA10, the barley CNL, MLA10, translocates into the nucleus and interacts with both WRKY transcriptional repressors and MYB6, a transcriptional activator, to activate defense responses (10, 14). Similarly, nuclear accumulation of the Arabidopsis TNL, RPS4, is required for RPS4-mediated resistance in the presence of its cognate effector AvrRps4 (11, 15, 16). However, a number of recent studies have demonstrated that coordinated nucleo-cytoplasmic trafficking of plant NLRs is required for the full activation of defense responses, suggesting that a single NLR protein may activate distinct signaling pathways in the cytoplasm and nucleus. For example, the RPS4 protein of Arabidopsis, a TNL that mediates recognition of the effector protein AvrRps4 from P. syringae, appears to localize to both the nucleus and cytoplasm and activate different pathways in each. Forced nuclear accumulation of AvrRps4 is sufficient to activate RPS4-mediated bacterial growth inhibition, but blocks RPS4-mediated HR (16). On the other hand, sequestration of AvrRps4 in the cytosol using a nuclear export signal significantly impairs RPS4-mediated resistance but only moderately reduces RPS4-mediated HR. Therefore, nucleo-cytoplasmic partitioning of plant NLR proteins seems to be a regulatory mechanism for differential activation of downstream signaling. These studies also point out that activation of host cell death (HR) can be separated from activation of resistance.
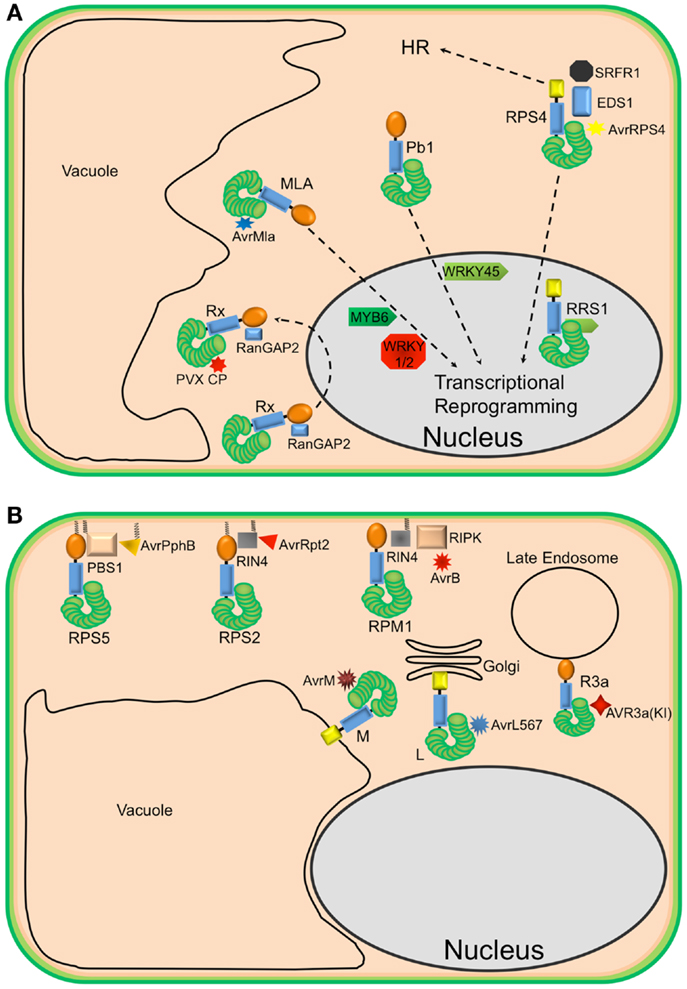
Figure 1. Diverse localization of plant NLR proteins. (A) Nuclear-localized plant NLRs. The barley MLA proteins reside in the cytoplasm but, in the presence of corresponding AvrMla effectors, translocate into the nucleus where they interact with both WRKY transcription repressors and MYB6, a transcriptional activator, to activate defense responses. Similarly, the rice CNL Pb1 also accumulates in the nucleus where it interacts with and stabilizes rice WRKY45 to activate defense responses. RPS4 also translocates into the nucleus, upon recognition of AvrRps4, to activate defense responses in conjunction with RRS1, an atypical TNL in Arabidopsis that contains a WRKY domain. At the same time, a subset of RPS4 complexes stays in the cytoplasm to activate HR. The potato CNL, Rx, interacts with the cytosolic Ran GTPase Activating Protein 2 (RanGAP2) and actively shuttles between the nucleus and the cytoplasm. However, the recognition of PVX CP and activation of signaling seem to occur in the cytoplasm. (B) Endomembrane associated plant NLRs and their corresponding “guardees” and pathogen effectors. RPS5 (an NLR), PBS1 (guardee), and AvrPphB (P. syringae effector) localize to the plasma membrane (PM). This is mediated by N-terminal acylation (myristoylation and/or palmitoylation). Similarly, RPS2 (an NLR) is PM-associated via a predicted N-terminal palmitoylation signal while RIN4 (guardee) localizes to the PM via a C-terminal prenylation or palmitoylation signal. RPM1 (NLR) also localizes to the PM, but lacks a predicted acylation signal. These three well-studied Arabidopsis NLR proteins are activated on the PM and initiate signaling on the PM. Relocalization following activation does not appear to occur. The flax rust resistance proteins L6 and M are respectively targeted to the Golgi apparatus and vacuolar membrane. Re-directing L6 to the vacuolar membrane, however, does not affect its function. The potato resistance protein, R3a, relocalizes from the cytoplasm to late endosomes in the presence of its corresponding effector AVR3a(KI), which also relocalizes to late endosomes in the presence of R3a.
Not all plant NLRs require nuclear localization for activation of resistance, and in fact, this may be the exception rather than the rule. The CNL protein, Rx, which mediates recognition of the Potato Virus X coat protein (CP), localizes to both the nucleus and cytosol (17, 18). Sequestration of Rx in the nucleus impairs its function, but forced cytosolic accumulation enhances Rx function (19). Moreover, Rx is not activated in the presence of forced nuclear PVX CP accumulation (20). Taken together, these results suggest that both pathogen recognition and resistance signaling by Rx need to take place in the cytoplasm. Thus, nuclear accumulation of Rx may represent a form of negative regulation. Alternatively, but not exclusively, Rx may have to traffic to the nucleus to form a functional complex and then back to the cytosol, where it surveys the presence of the cognate virus CP.
In contrast to Rx, RPS4, and MLA10, the CNL proteins RPS5 and RPM1 from Arabidopsis require PM localization to function (Figure 1B). This requirement likely reflects the localization of the pathogen effector proteins detected by each. RPS5 detects the P. syringae effector AvrPphB, which is a cysteine protease that targets the protein kinase PBS1.(21). AvrPphB autoprocesses upon entry into host cells, revealing an N-terminal motif that becomes myristoylated by host cell enzymes, which then targets AvrPphB to the PM (22). RPS5 is also acylated on its N-terminus and localizes to the PM (23). Mutation of the predicted acylation sites of RPS5 (Glycine 2 and Cysteine 4) disrupts RPS5-mediated HR and PM localization (23). Although RPM1 does not possess predicted acylation motifs at its N-terminus, it also localizes to the PM, where its corresponding effector AvrB and co-activators RIPK and RIN4 also localize (24–26). An auto-active RPM1 mutant, T166E, also localizes to the PM, indicating that RPM1 does not move following activation. Furthermore, sequestration of RPM1 on the PM does not affect RPM1-mediated resistance (25). Together, these observations indicate that activation of RPS5 and RPM1 and subsequent signaling occurs on the PM.
Plant NLR proteins have also been localized to other endomembrane locations. For example, the flax rust resistance proteins L6 and M localize to the Golgi apparatus and the tonoplast, respectively (27). Swapping the N-terminal sequences between L6 and M swapped their localization, indicating that the localization signals reside at the N-termini of these proteins, which are predicted to function as signal anchor sequences. Signal anchors are hydrophobic N-terminal sequences that direct nascent polypeptides to the endoplasmic reticulum, but unlike signal peptides, are not removed by a signal peptidase. Interestingly, changing the localization of L6 from the Golgi to the tonoplast did not affect its ability to detect its corresponding effector protein, nor activate resistance. Deletion of the signal anchor sequence, however, destabilized L6 protein accumulation, rendering it non-functional (27). A similar loss of protein stability was observed for RPS5 when its acylation motif was mutated (23), suggesting that at least a subset of NLR proteins require membrane localization for function and stability.
Plant NLRs can also move between the cytosol and endomembrane system. The potato resistance protein, R3a, relocates from the cytoplasm to endosomal compartments in the presence of the Phytophthora infestans effector AVR3a(KI) but not an unrecognized derivative AVR3a(EM) (28). Moreover, AVR3a(KI), but not AVR3a(EM), relocalizes to endosomes in the presence of R3a. Treatment with inhibitors of endocytic trafficking affects both the relocalization of R3a and its function. These observations suggest that the recognition of AVR3a(KI) by R3a and signal initiation occur in endocytic vesicles.
The LRR domains are the most polymorphic part of plant NLR proteins, which likely reflects their role in effector recognition. Direct interaction between NLR proteins and pathogen effectors has been demonstrated for only a subset of plant NLRs, however. The best characterized of these is the flax L protein, in which allele specific interactions between L and its corresponding fungal effector AvrL567 have been demonstrated for the C-terminal LRR regions using yeast-two-hybrid analysis (29, 30). More recently, race-specific interactions between the Arabidopsis RPP1 LRR domain and the oomycete ATR1 effector have been demonstrated using co-immunoprecipitation analysis (31). However, race-specific physical interactions have also been shown between the coiled-coil (CC) domains of a rice NLR, Pik, and corresponding Avr-Pik effectors from the fungus Magnaporthe oryzae (32).
In addition to its role in effector recognition, the LRR domain also plays an important role in keeping NLR proteins in the “off” state. Studies of Bs2, RPS5, and Rx have demonstrated that the LRR domain physically associates with the NB-ARC domain (33–35). Furthermore, deletion of the LRR domain typically results in auto-activation (20, 35). A recent study on RPS5 established that only the first four LRRs are required to inhibit this auto-activation (23). Auto-activation has also been reported for the potato NLR Rx when its CC-NB-ARC region was co-expressed with RanGAP2 in tobacco plants (20). Auto-activation is also frequently observed when LRR domains are swapped between NLR proteins, suggesting that the LRR and NB-ARC domains co-evolve with each other (23, 36, 37). Consistent with this hypothesis, a highly acidic loop region in the Rx ARC2 domain has been shown to associate with basic patches in the N-terminal end of the Rx LRR domain (37).
The recently solved crystal structure of the mouse NLRC4 protein [NLR family, Caspase activation and recruitment domain (CARD) containing 4] provides additional insights into the physical interactions between the NB-ARC domain and the LRR domain that function to inhibit NLR auto-activation (38). NLRC4 displays an inverted “question-mark” structure, where the N-terminal region of the horse-shoe shaped LRR interacts with the NB subdomain of the NB-ARC. This interaction sterically restricts the accessibility of the side of the NB that is required for oligomerization. Deletion of the LRR domain, or point mutations in the NB/LRR interaction surface, result in constitutive activation of NLRC4 (38). The N-terminal region of the NLRC4 LRR domain also interacts with the ARC3 subdomain (also known as the helix domain 2), with this interaction playing an important role in the overall positioning of the LRR domain relative to the NB-ARC. Plant NLRs, however, do not contain an ARC3 subdomain (39), making it difficult to predict whether the LRRs of plant NLRs will be similarly positioned.
An open question in plant NLR studies is which domain(s) of plant NLRs is/are directly responsible for downstream signaling. In mammalian cells, the NLR activation usually results in the recruitment and activation of pro-caspase-1 through homotypic interaction with the N-terminal CARD (40). This leads to the formation of inflammasomes, which is linked to pyroptosis. By analogy, it is reasonable to assume that activation of plant NLRs exposes the N-terminal domain for downstream signaling. Indeed, overexpression of the N-terminal CC or TIR domains from two plant NLRs causes effector-independent HR, supporting a signaling role (41, 42). Crystal structures of the CC domain of the barley MLA10 CNL and the TIR domain of the flax L6 TNL indicate that homodimerization is necessary for downstream signaling activity (41, 42). In both studies, mutations at the dimer interface disrupted dimerization and signaling activity. However, mutations in the BB loop and αC helix of the L6 TIR domain did not affect homodimerization, but did disable downstream signaling, indicating the presence of discrete interfaces for self-association and engaging other unidentified signaling molecules (41). In addition, the presence of the L6 NB-ARC domain inhibited the dimerization of the L6 TIR and prevented signaling.
The above studies on L6 and MLA10 provide strong support for the CC and TIR domains functioning as the sole domains engaging downstream signaling components. However, conflicting data have been obtained from studies on the potato Rx and Arabidopsis RPS5 proteins. In the case of Rx, overexpression of the NB subdomain alone was found to be sufficient for inducing cell death, suggesting that this domain plays a roll in engaging downstream signaling components (43). For RPS5, overexpression of the CC or NB-ARC domains alone did not induce cell death, while overexpression of a CC-NB-ARC construct did, suggesting that the two domains function together to engage downstream components (35). It is not yet clear whether these conflicting data reflect fundamental differences between NLR proteins in terms of their signaling mechanisms, or are due to differences in how the experiments were conducted (e.g., different levels of overexpression, different epitope tags, etc.).
In addition to signaling, the N-terminal domains of plant NLRs may also function in effector recognition. For example, many effector targets, such as Pto, RIN4, PBS1, and NRIP, are found to interact with the N-terminal domains of their corresponding NLRs (44). Thus, the N-terminal domains of these NLRs may be responsible for directly monitoring effector-induced modifications of these target proteins or, alternatively, place their LRR domains in appropriate proximity for optimal surveillance. As mentioned above, race-specific interactions are reported to occur between the CC domains of the rice NLR, Pik, and corresponding Avr-Pik effectors (32). Similarly, L6 and L7 from the flax L locus recognize different effectors, but their amino acid sequences differ only in the N-terminal TIR domains (45).
Although direct association with pathogen effector proteins has been documented for some plant NLRs, many appear to detect pathogen effectors indirectly via sensing effector-induced modifications of other host proteins (46, 47). As mentioned above, the Arabidopsis CNL RPS5 detects the presence of the cysteine protease effector AvrPphB by monitoring the integrity of PBS1. In addition, insertion of seven amino acids at the AvrPphB cleavage site of PBS1 activates RPS5 as strongly as PBS1 cleavage, suggesting that RPS5 senses subtle conformational changes in PBS1 associated with its cleavage (48). Sensing of these structural changes by RPS5 is likely mediated by the LRR domain, as partial deletions as small as one LRR abolished activation by PBS1 cleavage, but did not abolish auto-activation by mutations in the NB-ARC domain (23). As a second example of indirect recognition, the Arabidopsis CNL, RPM1, detects modification of the Arabidopsis RIN4 protein induced by the P. syringae effector proteins AvrB and AvrRpm1. Current data indicate that AvrB physically associates with the Arabidopsis protein kinase RIPK, which then phosphorylates RIN4 (26). The phosphorylation of specific RIN4 residues then leads to the activation of RPM1 (24, 26). AvrRpm1 appears to induce modification of other RIN4 residues, but the specific residue(s) modified, and whether it is by phosphorylation, is not yet clear. A third example of indirect recognition of pathogen effectors is recognition of the P. syringae effector AvrRpt2 (a cysteine protease) by Arabidopsis RPS2. In this example, RPS2 is activated by the degradation of RIN4 following cleavage by AvrRpt2 (49). Thus RIN4 is required to keep RPS2 in an off state, and rin4 null mutations are lethal in Arabidopsis if RPS2 is functional.
The NB-ARC domain is conserved among plant and animal NLRs, and in the animal literature is often referred to as the nucleotide binding and oligomerization domain (NOD). NB-ARC domains form a deep nucleotide-binding pocket. In the “off” state, the NB-ARC domain adopts a “closed” structure where ADP is preferentially bound and coordinates intramolecular interactions to stabilize this structure (38, 50, 51). Activation is thought to require release of the ADP to be replaced by ATP and adoption of an “open” structure. This structural change is then thought to promote homo-oligomerization via the NB-ARC domain, which in turn enables the N-terminal domains to engage in downstream signaling. Thus, the NB-ARC domain is thought to function as a molecular switch that determines the “on” and “off” state of NLR signaling with ADP bound form for “off” and ATP bound form for “on.” Due to difficulties in purifying soluble plant NLR proteins, however, this long-standing model was not tested until recently. Biochemical studies using the CC-NB-ARC region of tomato I-2 and Mi-1 demonstrated that ATP is bound by these proteins (52). This binding activity depends on a functional P-loop (Phosphate-binding loop), also known as the Walker A motif, which is a glycine-rich flexible loop containing a highly conserved lysine residue that interacts with the phosphates of the nucleotide and with a magnesium cation that coordinates the β- and γ-phosphates (53). In addition, these proteins display ATPase activity. A follow-up study reported that mutations within the NB-ARC domain that cause an auto-activation phenotype impair the ATPase activity of I-2, supporting the model that the ATP bound form represents the “on” state (54). The first direct experimental evidence that a full-length plant NLR protein preferentially binds ADP in its resting state was reported in 2011. In that study, the barley CNL, MLA27, co-purified specifically with ADP but not ATP (42). Also in 2011, the flax rust resistance protein M was found to co-purify with ADP, while an auto-active mutant form (D555V in the conserved MHD motif) co-purified with more ATP than ADP (55), supporting the model that nucleotide exchange is required for switching from the off state to the “on” state.
Apaf-1 and CED-4 are known to form oligomers through NB-ARC-mediated interaction (51, 56). The crystal structure of the mouse NLRC4 protein mentioned above revealed that ADP coordinates interactions between the central NB subdomain and the ARC2 subdomain (also called the winged-helix domain) to stabilize a closed conformation. A second interaction surface between the ARC3 subdomain and the NB masks an α-helix of the NB subdomain that participates in oligomerization. This α-helix is part of a conserved structure within the STAND family of ATPases (38). Specific point mutations within these interaction surfaces also result in an auto-active phenotype. A structure for NLRC4 in an active confirmation is not yet available, but it is predicted that ligand binding leads to the disengagement of the ARC2, ARC3, and LRR domains from the NB simultaneously, allowing the oligomerization of NLRC4 mediated by the NB subdomain.
There are several reports that indicate plant NLR proteins can also self-associate. For example, tomato Prf forms a dimer, which then incorporates into a complex containing two accessory molecules of the Pto protein kinase (57, 58). Similarly, co-immunoprecipitation assays have demonstrated that Arabidopsis RPS5 self-associates prior to activation (35). Oligomer formation has also been reported for the tobacco TNL protein, N, but only its N-terminal TIR domain has been associated with oligomerization (59). The crystal structures of the MLA10 CC and L6 TIR domains revealed that both form homodimers (41, 42), and the MLA1 protein (allelic to MLA10) was observed to self-associate in planta (42). Whether the NB-ARC domain also plays a role in plant NLR oligomerization remains unclear, however. So far, only the RPS5 NB-ARC domain is known to self-associate, and this was shown under conditions of transient overexpression (35). In contrast, the L6 NB-ARC domain was shown to inhibit the homodimerization of its TIR domain and the activation of defense responses (41). ATP binding seems to be a must, however, for oligomerization because P-loop mutations disrupted the formation of N oligomers (59).
In addition to self-association, different NLRs may interact with each other to form heterodimers or hetero-oligomers. Interaction between NLRs has been reported between mouse NLRC4 and NAIP2, and between NLRC4 and NAIP5 (60, 61). NLRC4-containing oligomers assemble in response to two distinct PAMPs, flagellin and PrgJ, a component of type III secretion systems (40, 62). However, NLRC4 does not directly interact with flagellin or PrgJ. Instead, these PAMPs bind to NAIP2 and NAIP5, respectively, which then bind to NLRC4 (60, 61), triggering formation of a functional NLRC4 inflammasome. Like NAIP2 and NAIP5, some plant NLRs appear to require a second NLR for signaling. The Arabidopsis NLR RPS4 requires a second NLR, RRS1, to recognize the AvrRps4 effector protein from P. syringae (63, 64). Interestingly, RRS1 is encoded by a gene immediately adjacent to RPS4, in a head-to-head orientation (63, 64). This pair of NLRs is also involved in recognition of the PopP2 effector protein from the bacterial pathogen Ralstonia solanacearum and an unidentified effector(s) from the fungus Colletotrichum higginsianum. It is not yet known whether RRS1 and RPS4 physically associate, but both at least partially localize to the nucleus in the presence of effectors (11, 65).
A second example of a “helper” NLR is the Arabidopsis ADR1 (Activated Disease Resistance 1) family, which contributes to defense responses activated by Arabidopsis RPS2 (a CNL), RPP2, and RPP4 (TNLs) (66). There are three copies of ADR1 in the Arabidopsis genome and all three must be knocked out to affect RPS2 signaling. Interestingly, ADR1 does not rely on an intact P-loop motif for this function, suggesting that ATP binding is not required for signaling. ADR1 family members are also required for basal resistance and PTI, suggesting that this family of CNLs may function more generally in regulating defense responses, rather than functioning specifically in effector detection. Consistent with this, attempts at showing direct physical interactions between ADR1 and other NLRs have not been successful (66). In addition, mutations in ADR1 family members suppress runaway cell death triggered by loss of the LSD1 gene, and autoactivating mutations in ADR1-L2 cause large increases in the defense hormone salicylic acid (67). Together, these data suggest that ADR1 may be part of an amplification loop that leads to elevated levels of salicylic acid during defense responses.
An ADR1 homolog has also been described in tobacco, and has been named NRG1. The NRG1 protein is required for resistance mediated by the TNL protein N, which mediates recognition of tobacco mosaic virus (TMV) (68). Consistent with their proposed role in signaling, transient expression of the CC domains of NRG1 and ADR1 induces HR in tobacco plants (69). NRG1 and ADR1 belong to an ancient clade of CNLs that is unusually conserved relative to other plant NLR proteins. Phylogenetic analyses have revealed the correlated absence of both NRG1 homologs and TIR-NB-LRR-encoding genes from the dicot Aquilegia caerulea and the dicotyledonous order Lamiales, as well as from the grass family (Poaceae), suggesting that the TNL family may be dependent on ADR1 family members for activating resistance (69). Since grasses contain numerous CNL family members, this functional requirement appears not to be true for CNLs in general. Indeed, resistance mediated by the Arabidopsis RPM1 protein is not affected by loss of ADR1 function (66).
It has been almost 20 years since the cloning of the first plant NLR gene. During this period, major advances have been made in our understanding of NLR structure, activation, and localization. However, little is known about the signal transduction steps following plant NLR activation. Forward genetic screens have been mostly unsuccessful at identifying downstream components, likely due to redundancy of signaling pathways. One exception was the identification of EDS1 (enhanced disease susceptibility 1) in Arabidopsis, which is required for resistance mediated by TNLs but not CNLs (70, 71). EDS1 has recently been shown to form protein complexes with the Arabidopsis TNLs RPS4, RPS6, and SNC1 (72). These complexes also contain an unrelated protein named SRFR1, which was identified in a screen for mutations that restored resistance to rps4 mutant Arabidopsis (73, 74). Furthermore, the bacterial effectors recognized by RPS4 and RPS6 (AvrRps4 and HopA1) bind to EDS1 and disrupt EDS1-SRFR1 interactions (72). This study suggests that EDS1 may be a “guardee” of RPS4 and RPS6 (and possibly other TNLs) and that these TNLs are activated by effector mediated disruption of the EDS1-SRFR1 complex. More recent work, however, found that the C-terminal half of AvrRps4, which is necessary and sufficient for activating RPS4, does not interact with EDS1 in co-immunoprecipitation or yeast two-hybrid assays (75). This finding suggests that physical association between AvrRps4 and EDS1 is not required for activation of RPS4, thus the molecular mechanism underlying AvrRps4 recognition remains unclear.
Regardless of whether EDS1 is a true target of AvrRps4, it is clear that EDS1 and SRFR1 represent a signaling complex that is employed by multiple TNLs. SRFR1 contains a tetratricopeptide repeat domain and displays similarities to transcriptional repressors in Saccharomyces cerevisiae and Caenorhabditis elegans (74). Consistent with SRFR1 possibly functioning as a transcriptional repressor, two independent studies reported that loss of SRFR1 function activates the expression of SNC1, an Arabidopsis TNL, resulting in constitutive defense responses (76, 77). Furthermore, bifluorescence complementation analyses showed that SRFR1 interacts with RPS4 and SNC1 in the nucleus (72), suggesting that TNLs may directly regulate SRFR1 activity.
Other transcriptional regulators have also been shown to directly interact with TNLs. For example, Topless-related 1 (TPR1) interacts with SNC1, a TNL protein, and knocking out TPR1 compromises immunity mediated by SNC1 (78). Significantly, TPR1 represses the expression of two well-known negative regulators of immunity, Defense no Death 1 (DND1) and Defense no Death 2 (DND2). Therefore, the SNC1-mediated immune responses are activated by TPR1 through its repression of negative regulators. SPL6, a squamosa promoter binding protein (SBP)-domain transcription factor interacts with the N protein of tobacco within distinct nuclear compartments (79). The Arabidopsis ortholog of SPL6 is required for the RPS4-mediated resistance, indicating that this transcription factor plays a conserved role in activating TNL-mediated defenses. Also, as described above, the CNL protein MLA 10 translocates into the nucleus upon activation and interacts with both WRKY transcriptional repressors and MYB6, a transcriptional activator, to activate defense responses (10, 14). Most recently, the rice CNL Pb1, which confers resistance to rice blast (Magnaporthe oryzae), was shown to interact with the WRKY45 transcription factor in the nucleus (80). This interaction is mediated by the CC domain of Pb1, and mutations in the CC domain that disrupt the interaction compromise Pb1-mediated resistance. Thus both CNLs and TNLs have the capacity to impact gene expression by direct interaction with transcriptional repressors and activators, making these NLR signal transduction pathways quite short.
It is unlikely, however, that all NLR proteins regulate gene expression by direct interaction with transcription factors. As described above, the CNL proteins RPS2, RPS5, and RPM1 are localized to the PM. Activation of defenses by PM-localized NLRs appears to require an influx of extracellular Ca2+, as cell death induced by RPS2 and RPM1 can be eliminated by the calcium channel blocker LaCl3 (81). Recent reverse genetic studies indicate that RPS2- and RPM1-mediated resistance is at least partially dependent on calcium dependent protein kinases (CPKs) (82, 83), with different CPKs being involved in different aspects of resistance (83). Specifically CPK1 and CPK2 contribute to HR development, while CPK4/5/6/11 all contribute to transcriptional reprograming by phosphorylating the transcription factors WRKY8/28/48. Additionally, CPK1/2/4/11 also contribute to production of reactive oxygen species via phosphorylation of PM-associated NADPH oxidases. Based on these observations, the authors proposed a model in which NLR activation triggers a sustained influx of calcium, which then triggers multiple CPK signaling pathways that lead to ROS production, defense gene activation, and cell death. In addition to cytoplasmic calcium signaling, RPS2 and RPM1 activation has been shown to elicit specific Ca2+ signatures inside chloroplasts (84). These calcium transients are dependent on a chloroplast-localized protein named CAS for calcium-sensing receptor. Mutations in the CAS gene compromise both PTI and HR development during ETI. This study thus provides a possible link between NLR activation and chloroplast functions such as the production of the defense-related hormones jasmonic acid and salicylic acid.
Although it is clear that different plant NLRs employ different signaling pathways, these signaling pathways appear to be broadly conserved across plant species, as evidenced by functional transfer of NLRs between species. For example, the RRS1-RPS4 gene pair described above has been functionally transferred from Arabidopsis to five different plant species from three different families (Brassica rapa and Brassica napus (Brassicaceae); Nicotiana benthamiana and Solanum lycopersicum (tomato) (Solanaceae), and Cucumis sativus (cucumber, Cucurbitaceae) (85). In addition, cell death can be activated in N. benthamiana and/or N. tabacum (tobacco) by transient expression of several different TNL and CNL proteins from diverse plant species, including Arabidopsis, flax, and barley (35, 86–88). Particularly noteworthy is the recent demonstration that the MLA1 protein from barely can function in transgenic Arabidopsis to confer resistance against the barley powdery mildew fungus, Blumeria graminis f. sp. hordei (89). Interestingly, this resistance remains effective in Arabidopsis mutants defective in ethylene, jasmonic acid, and salicylic acid signaling, indicating the presence of a hormone independent NLR-mediated defense mechanism that is conserved between barley (a monocot and member of the grass family) and Arabidopsis (a dicot and member of the mustard family).
The HR is usually associated with NLR-activated immunity in plants. However, the HR can be genetically uncoupled from restriction of pathogen growth, at least in the case of resistance to P. syringae (16, 90). In addition, it remains unclear how cell death is executed, or indeed, whether different classes of NLRs share the same cell death pathway. For the PM-localized NLRs, RPM1, and RPS2, cell death is preceded by fusion of the vacuolar membrane with the PM, resulting in release of vacuolar proteins to the apoplast (extracellular space) (91). The resulting extracellular fluid possesses both antibacterial activity and cell death-inducing activity. This membrane fusion process depends on the activity of the proteasome subunit PBA1, suggesting that there may be an “HR inhibitor” protein that must be degraded to enable HR activation.
Plants lack canonical caspase proteases that are associated with apoptosis in mammalian cells. However, they do contain proteins with weak structural similarities to caspases called metacaspases that have recently been implicated in regulating HR cell death (90). Knockout of the Arabidopsis metacaspase AtMC1 reduces, but does not eliminate, RPM1-mediated HR, but has no effect on RPM1-mediated growth restriction of P. syringae. Conversely, knockout of a second Arabidopsis metacaspase AtMC2, enhances RPM1-mediated HR, but again has no effect on restricting bacterial growth. These observations suggest that metacaspases play an accessory role in regulating HR, but are not a central trigger.
A second type of protease associated with HR regulation in plants has recently been identified and named phytaspase (90, 92). Phytaspases are structurally unrelated to animal caspases, but like caspases, catalyze cleavage following aspartate residues. RNAi-mediated silencing of phytaspase in tobacco reduced N-gene mediated HR triggered by TMV infection and reduced resistance to TMV, indicating that phytaspases may play a central role in resistance mediated by N, a TNL family member (90, 92). Interestingly, tobacco phytaspase is constitutively expressed and secreted to the extracellular space, but during the HR, partially relocalizes to the cytoplasm (90, 92), raising the possibility that it is actively transported back into the cell during the HR, where it must cleave specific substrates to activate cell death. Although phytaspase has also been purified from rice, there are not yet any reports on whether it is required for NLR-mediated resistance in other plant species.
Several recent studies indicate that secretion may play an important role in NLR-mediated defense. For example, Arabidopsis AtMIN7, an ADP ribosylation factor-guanine nucleotide exchange factor (ARF-GEF) protein, has recently been shown to be required for RPS2- and RPS5-mediated resistance, but not for HR cell death (93). ARF-GEF proteins regulate the activity of small GTPases involved in endomembrane trafficking. AtMIN7 is a target of the P. syringae effector, HopM1, which promotes proteasome-dependent degradation of AtMIN7 (94). Activation of RPS2 and RPS5 somehow prevents HopM1-mediated degradation of AtMIN7 (93). Consistent with AtMIN7 playing a role in endomembrane trafficking, confocal microscopy showed that MIN7 and HopM1 localize to the trans-Golgi network/early endosomes. Further evidence that endomembrane trafficking/secretion plays a role in RPS2-mediated resistance comes from quantitative proteomic analysis of PMs following RPS2 activation (95). In this study, a transgenic Arabidopsis line expressing a dexamethasone-inducible AvrRpt2 gene was used to activate RPS2. Comparison of activated to unactivated samples uncovered 235 proteins that were significantly up-regulated. This set of up-regulated proteins was highly enriched in proteins involved in endocytosis and exocytosis, including Syntaxin of plants 122 (SYP122) and N-ethylmaleimide-sensitive factor vesicle fusing ATPase, and soluble N-ethylmaleimide-sensitive factor adaptor protein 33 (SNAP33). RPS2 has also been shown to upregulate production of miR393b, a microRNA that targets at least three different genes likely involved in endomembrane trafficking (MEMB12, a golgi-localized SNARE protein; VPS54, homologous to a yeast protein involved in retrograde transport from late endosomes to the Golgi, and EXO70H3, a subunit of the exocyst complex thought to be required for exocytosis (96). Knockout of MEMB12 enhances secretion of the defense protein PR-1 that is induced by RPS2 activation. Thus MEMB12 appears to function as negative regulator of exocytosis, with RPS2 inducing production of a miRNA that inhibits translation of the MEMB12 protein. Lower MEMB12 protein levels then enable an increase in defense protein secretion. Consistent with this model, the MEMB12 knockout line displays enhanced basal resistance in the absence of RPS2 activation (96). The endomembrane trafficking system is thus emerging as important arm of the NLR-mediated defense system that is also targeted by pathogen effectors.
As should be apparent from the discussion above, plant NLRs have evolved diverse mechanisms for recognizing pathogens and diverse mechanisms for activating resistance. However, a feature that is likely shared among all “sensor” NLRs in plants (as opposed to “helper” NLRs) is the dual role of the LRR domain in keeping the NLR in the “off state” in the absence of pathogen, and promoting the switch to the “on state” in the presence of pathogen (via binding to effectors or effector-modified host proteins). We have very little insight, however, into how the LRR domain accomplishes either of these roles. The recent structure of mouse NLRC4 indicates that in animal NLRs, the LRR folds back across NB-ARC domain with the N-terminal portion of the LRR making multiple contacts with the NB and ARC3 domains, effectively placing a lid over the ADP/ATP binding pocket. The absence of the ARC3 domain in plant NLRs makes it a certainty that the contacts between the LRR and NB-ARC will differ from NLRC4, but based on the locations of autoactivating mutations and on deletion analyses, the general structure is likely to be similar, with just the N-terminal portion of the LRR (approximately four repeats) required to form the lid (23). The C-terminal portion of the LRR appears to be where specificity for effector recognition generally lies, but how effector binding alters NB-ARC:LRR interaction is unknown. What remains a holy grail for the field, in both plants and animals, is obtaining the structure of an NLR complexed with its activating protein. The insolubility of NLRs when overexpressed in bacteria or insect cells has been a major barrier to progress on this front. Surmounting this barrier for plant NLRs may require purification from plant systems in which the necessary chaperones should be present.
A second holy grail is identifying the immediate downstream interacting proteins for PM associated CNLs. Although several transcription factors have now been identified that interact with nuclear-localized TNLs and CNLs, we still lack good candidates for downstream signaling proteins for NLRs that signal from the PM such as RPM1, RPS2, and RPS5. Proteomic approaches hold some promise for shedding light on this unknown (95, 97), but face the additional challenge of rapid turnover of NLR proteins following activation. The finding that extracellular calcium influx is required for RPM1- and RPS2-mediated HR suggests that there may be a fairly direct link between NLR activation and calcium channels (81), which merits further exploration.
A third holy grail is a better understanding of how cell death is executed during NLR-mediated HR. Although cell death is apparently not required for resistance, at least to P. syringae, the HR is still a hallmark of NLR activation. One study has implicated fusion of the vacuolar and PM as the primary event leading to cell death (91). If this is true, the question becomes how NLR activation triggers such membrane fusion events. More generally, accumulating data have implicated endomembrane trafficking as playing a central role in NLR-mediated resistance, presumably to increase secretion of antimicrobial compounds. How does NLR activation regulate this process?
In summary, although much has been learned in the nearly 20 years since the first NLR was identified, major questions remain. Providing answers to these questions will require both creativity and improvements in technology, but will no doubt come.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank the National Institute of General Medical Sciences of the National Institutes of Health for their financial support (grant numbers R01 GM046451 and R01 GM063761 to Roger W. Innes).
1. Jacob F, Vernaldi S, Maekawa T. Evolution and conservation of plant NLR functions. Front Immunol (2013) 4:297. doi: 10.3389/fimmu.2013.00297
3. van der Biezen EA, Jones JD. The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol (1998) 8:R226–7. doi:10.1016/S0960-9822(98)70145-9
4. Inohara N, Chamaillard M, Mcdonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem (2005) 74:355–83. doi:10.1146/annurev.biochem.74.082803.133347
5. Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of life, wheel of death: a mechanistic insight into signaling by STAND proteins. Structure (2009) 17:172–82. doi:10.1016/j.str.2009.01.001
6. Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell (2003) 15:809–34. doi:10.1105/tpc.009308
7. Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, et al. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science (1994) 265:1856–60. doi:10.1126/science.8091210
8. Mindrinos M, Katagiri F, Yu GL, Ausubel FM. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell (1994) 78:1089–99. doi:10.1016/0092-8674(94)90282-8
9. Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell (1994) 78:1101–15. doi:10.1016/0092-8674(94)90283-6
10. Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science (2007) 315:1098–103. doi:10.1126/science.1136372
11. Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol (2007) 17:2023–9. doi:10.1016/j.cub.2007.10.042
12. Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell (2008) 132:449–62. doi:10.1016/j.cell.2007.12.031
13. Cheng YT, Germain H, Wiermer M, Bi D, Xu F, Garcia AV, et al. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell (2009) 21:2503–16. doi:10.1105/tpc.108.064519
14. Chang C, Yu D, Jiao J, Jing S, Schulze-Lefert P, Shen QH. Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell (2013) 25:1158–73. doi:10.1105/tpc.113.109942
15. Garcia AV, Blanvillain-Baufume S, Huibers RP, Wiermer M, Li G, Gobbato E, et al. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog (2010) 6:e1000970. doi:10.1371/journal.ppat.1000970
16. Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, Parker JE. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science (2011) 334:1401–4. doi:10.1126/science.1211641
17. Sacco MA, Mansoor S, Moffett P. A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J (2007) 52:82–93. doi:10.1111/j.1365-313X.2007.03213.x
18. Tameling WI, Baulcombe DC. Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell (2007) 19:1682–94. doi:10.1105/tpc.107.050880
19. Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell (2010) 22:4195–215. doi:10.1105/tpc.110.077537
20. Tameling WI, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, et al. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell (2010) 22:4176–94. doi:10.1105/tpc.110.077461
21. Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science (2003) 301:1230–3. doi:10.1126/science.1085671
22. Dowen RH, Engel JL, Shao F, Ecker JR, Dixon JE. A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J Biol Chem (2009) 284:15867–79. doi:10.1074/jbc.M900519200
23. Qi D, Deyoung BJ, Innes RW. Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol (2012) 158:1819–32. doi:10.1104/pp.112.194035
24. Chung EH, Da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, et al. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe (2011) 9:125–36. doi:10.1016/j.chom.2011.01.009
25. Gao Z, Chung EH, Eitas TK, Dangl JL. Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci U S A (2011) 108:7619–24. doi:10.1073/pnas.1104410108
26. Liu J, Elmore JM, Lin ZJ, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe (2011) 9:137–46. doi:10.1016/j.chom.2011.01.010
27. Takemoto D, Rafiqi M, Hurley U, Lawrence GJ, Bernoux M, Hardham AR, et al. N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol Plant Microbe Interact (2012) 25:379–92. doi:10.1094/MPMI-11-10-0272
28. Engelhardt S, Boevink PC, Armstrong MR, Ramos MB, Hein I, Birch PR. Relocalization of late blight resistance protein R3a to endosomal compartments is associated with effector recognition and required for the immune response. Plant Cell (2012) 24:5142–58. doi:10.1105/tpc.112.104992
29. Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A (2006) 103:8888–93. doi:10.1073/pnas.0602577103
30. Ravensdale M, Bernoux M, Ve T, Kobe B, Thrall PH, Ellis JG, et al. Intramolecular interaction influences binding of the Flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Pathog (2012) 8:e1003004. doi:10.1371/journal.ppat.1003004
31. Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell (2010) 22:2444–58. doi:10.1105/tpc.110.075358
32. Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, Alaux L, et al. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J (2012) 72:894–907. doi:10.1111/j.1365-313X.2012.05110.x
33. Moffett P, Farnham G, Peart J, Baulcombe DC. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J (2002) 21:4511–9. doi:10.1093/emboj/cdf453
34. Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell (2005) 17:1268–78. doi:10.1105/tpc.104.029637
35. Ade J, Deyoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A (2007) 104:2531–6. doi:10.1073/pnas.0608779104
36. Rairdan GJ, Moffett P. Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell (2006) 18:2082–93. doi:10.1105/tpc.106.042747
37. Slootweg EJ, Spiridon LN, Roosien J, Butterbach P, Pomp R, Westerhof L, et al. Structural determinants at the interface of the ARC2 and LRR domains control the activation of the NB-LRR plant immune receptors Rx1 and Gpa2. Plant Physiol (2013) 162:1510–28. doi:10.1104/pp.113.218842
38. Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science (2013) 341:172–5. doi:10.1126/science.1236381
39. Takken FL, Albrecht M, Tameling WI. Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol (2006) 9:383–90. doi:10.1016/j.pbi.2006.05.009
40. Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci USA (2010) 107:3076–80. doi:10.1073/pnas.0913087107
41. Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe (2011) 9:200–11. doi:10.1016/j.chom.2011.02.009
42. Maekawa T, Cheng W, Spiridon LN, Toller A, Lukasik E, Saijo Y, et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe (2011) 9:187–99. doi:10.1016/j.chom.2011.02.008
43. Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P. The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell (2008) 20:739–51. doi:10.1105/tpc.107.056036
44. Lukasik E, Takken FL. Standing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol (2009) 12:427–36. doi:10.1016/j.pbi.2009.03.001
45. Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell (1999) 11:495–506. doi:10.2307/3870876
46. Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature (2001) 411:826–33. doi:10.1038/35081161
47. van der Hoorn RA, Kamoun S. From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell (2008) 20:2009–17. doi:10.1105/tpc.108.060194
48. DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW. Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol (2012) 14:1071–84. doi:10.1111/j.1462-5822.2012.01779.x
49. Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell (2003) 112:369–77. doi:10.1016/S0092-8674(03)00036-9
50. Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature (2005) 434:926–33. doi:10.1038/nature03465
51. Yan N, Chai JJ, Lee ES, Gu LC, Liu Q, He JQ, et al. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature (2005) 437:831–7. doi:10.1038/nature04002
52. Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, et al. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell (2002) 14:2929–39. doi:10.1105/tpc.005793
53. Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J (1982) 1:945–51.
54. Tameling WIL, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, et al. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol (2006) 140:1233–45. doi:10.1104/pp.105.073510
55. Williams SJ, Sornaraj P, Decourcy-Ireland E, Menz RI, Kobe B, Ellis JG, et al. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact (2011) 24:897–906. doi:10.1094/MPMI-03-11-0052
56. Shiozaki EN, Chai JJ, Shi Y. Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc Natl Acad Sci USA (2002) 99:4197–202. doi:10.1073/pnas.072544399
57. Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, et al. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell (2006) 18:2792–806. doi:10.1105/tpc.106.044016
58. Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Jones AM, et al. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J (2010) 61:507–18. doi:10.1111/j.1365-313X.2009.04078.x
59. Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell (2006) 18:491–501. doi:10.1105/tpc.105.037234
60. Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature (2011) 477:592–5. doi:10.1038/nature10394
61. Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature (2011) 477:596–600. doi:10.1038/nature10510
62. Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol (2006) 7:569–75. doi:10.1038/ni1344
63. Birker D, Heidrich K, Takahara H, Narusaka M, Deslandes L, Narusaka Y, et al. A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J (2009) 60:602–13. doi:10.1111/j.1365-313X.2009.03984.x
64. Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, et al. RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J (2009) 60:218–26. doi:10.1111/j.1365-313X.2009.03949.x
65. Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A (2003) 100:8024–9. doi:10.1073/pnas.1230660100
66. Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci U S A (2011) 108:16463–8. doi:10.1073/pnas.1113726108
67. Roberts M, Tang S, Stallmann A, Dangl JL, Bonardi V. Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet (2013) 9:e1003465. doi:10.1371/journal.pgen.1003465
68. Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol (2005) 15:968–73. doi:10.1016/j.cub.2005.04.053
69. Collier SM, Hamel LP, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact (2011) 24:918–31. doi:10.1094/MPMI-03-11-0050
70. Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell (1996) 8:2033–46. doi:10.2307/3870410
71. Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci U S A (1998) 95:10306–11. doi:10.1073/pnas.95.17.10306
72. Bhattacharjee S, Halane MK, Kim SH, Gassmann W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science (2011) 334:1405–8. doi:10.1126/science.1211592
73. Kwon SI, Koczan JM, Gassmann W. Two Arabidopsis srfr (suppressor of rps4-RLD) mutants exhibit avrRps4-specific disease resistance independent of RPS4. Plant J (2004) 40:366–75. doi:10.1111/j.1365-313X.2004.02213.x
74. Kwon SI, Kim SH, Bhattacharjee S, Noh JJ, Gassmann W. SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J (2009) 57:109–19. doi:10.1111/j.1365-313X.2008.03669.x
75. Sohn KH, Hughes RK, Piquerez SJ, Jones JD, Banfield MJ. Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc Natl Acad Sci U S A (2012) 109:16371–6. doi:10.1073/pnas.1212332109
76. Kim SH, Gao F, Bhattacharjee S, Adiasor JA, Nam JC, Gassmann W. The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog (2010) 6:e1001172. doi:10.1371/journal.ppat.1001172
77. Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y. SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog (2010) 6:e1001111. doi:10.1371/journal.ppat.1001111
78. Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, et al. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci U S A (2010) 107:13960–5. doi:10.1073/pnas.1002828107
79. Padmanabhan MS, Ma S, Burch-Smith TM, Czymmek K, Huijser P, Dinesh-Kumar SP. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog (2013) 9:e1003235. doi:10.1371/journal.ppat.1003235
80. Inoue H, Hayashi N, Matsushita A, Xinqiong L, Nakayama A, Sugano S, et al. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc Natl Acad Sci U S A (2013) 110:9577–82. doi:10.1073/pnas.1222155110
81. Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerstrom M. Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J (2006) 47:947–59. doi:10.1111/j.1365-313X.2006.02844.x
82. Boudsocq M, Willmann MR, Mccormack M, Lee H, Shan L, He P, et al. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature (2010) 464:418–22. doi:10.1038/nature08794
83. Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, et al. Bifurcation of Arabidopsis NLR immune signaling via Ca(2)(+)-dependent protein kinases. PLoS Pathog (2013) 9:e1003127. doi:10.1371/journal.ppat.1003127
84. Nomura H, Komori T, Uemura S, Kanda Y, Shimotani K, Nakai K, et al. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat Commun (2012) 3:926. doi:10.1038/ncomms1926
85. Narusaka M, Kubo Y, Hatakeyama K, Imamura J, Ezura H, Nanasato Y, et al. Interfamily transfer of dual NB-LRR genes confers resistance to multiple pathogens. PLoS ONE (2013) 8:e55954. doi:10.1371/journal.pone.0055954
86. Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell (2004) 16:755–68. doi:10.1105/tpc.020040
87. Day B, Dahlbeck D, Huang J, Chisholm ST, Li D, Staskawicz BJ. Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell (2005) 17:1292–305. doi:10.1105/tpc.104.030163
88. Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, et al. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog (2012) 8:e1002752. doi:10.1371/journal.ppat.1002752
89. Maekawa T, Kracher B, Vernaldi S, Ver Loren Van Themaat E, Schulze-Lefert P. Conservation of NLR-triggered immunity across plant lineages. Proc Natl Acad Sci U S A (2012) 109:20119–23. doi:10.1073/pnas.1218059109
90. Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, et al. Arabidopsis type I metacaspases control cell death. Science (2010) 330:1393–7. doi:10.1126/science.1194980
91. Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, et al. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev (2009) 23:2496–506. doi:10.1101/gad.1825209
92. Chichkova NV, Shaw J, Galiullina RA, Drury GE, Tuzhikov AI, Kim SH, et al. Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J (2010) 29:1149–61. doi:10.1038/emboj.2010.1
93. Nomura K, Mecey C, Lee YN, Imboden LA, Chang JH, He SY. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc Natl Acad Sci U S A (2011) 108:10774–9. doi:10.1073/pnas.1103338108
94. Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science (2006) 313:220–3. doi:10.1126/science.1129523
95. Elmore JM, Liu J, Smith B, Phinney B, Coaker G. Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol Cell Proteomics (2012) 11:M111014555. doi:10.1074/mcp.M111.014555
96. Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, et al. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell (2011) 42:356–66. doi:10.1016/j.molcel.2011.04.010
Keywords: plant innate immunity, leucine-rich repeats, disease resistance, hypersensitive response, Pseudomonas syringae, pathogen effectors
Citation: Qi D and Innes RW (2013) Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 4:348. doi: 10.3389/fimmu.2013.00348
Received: 13 July 2013; Accepted: 09 October 2013;
Published online: 21 October 2013.
Edited by:
Jorg Hermann Fritz, McGill University, CanadaCopyright: © 2013 Qi and Innes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roger W. Innes, Department of Biology, Indiana University, 915 East, Third Street, Bloomington, IN 47405, USA e-mail:cmlubmVzQGluZGlhbmEuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.