
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 10 October 2013
Sec. Mucosal Immunity
Volume 4 - 2013 | https://doi.org/10.3389/fimmu.2013.00310
This article is part of the Research Topic Factors that promote T helper and regulatory T cell responses to Helicobacter pylori infection of the gastric mucosa View all 6 articles
The bacterium Helicobacter pylori (H. pylori), has evolved to survive in the highly acidic environment of the stomach and colonize on the epithelial surface of the gastric mucosa. Its pathogenic effects are well known to cause gastritis, peptic ulcers, and gastric cancer. In order to infect the stomach and establish colonies on the mucus epithelial surface, the bacterium has to move across the gel-like gastric mucus lining of the stomach under acidic conditions. In this review we address the question of how the bacterium gets past the protective mucus barrier from a biophysical perspective. We begin by reviewing the molecular structure of gastric mucin and discuss the current state of understanding concerning mucin polymerization and low pH induced gelation. We then focus on the viscoelasticity of mucin in view of its relevance to the transport of particles and bacteria across mucus, the key first step in H. pylori infection. The second part of the review focuses on the motility of H. pylori in mucin solutions and gels, and how infection with H. pylori in turn impacts the viscoelastic properties of mucin. We present recent microscopic results tracking the motion of H. pylori in mucin solutions and gels. We then discuss how the biochemical strategy of urea hydrolysis required for survival in the acid is also relevant to the mechanism that enables flagella-driven swimming across the mucus gel layer. Other aspects of the influence of H. pylori infection such as, altering gastric mucin expression, its rate of production and its composition, and the influence of mucin on factors controlling H. pylori virulence and proliferation are briefly discussed with references to relevant literature.
As is well known, Helicobacter pylori (H. pylori), the most abundant pathogen in the stomach causes gastritis, peptic ulcers, and gastric cancer by establishing colonies on the epithelial surface of the stomach that generate a host-immune response. It has three major pathogenic effects on its host: gastric inflammation, disruption of the gastric mucosal barrier, and alteration of gastric physiology (1–4). The virulence factors that are responsible for the pathogenic effects also enable the bacteria to manipulate the host-immune response and support its long-term survival in the stomach (5–9). The question of how the bacterium initially breaches the protective mucus barrier to reach the epithelial cell surface and colonize in the extreme acidic environment of the stomach (10) is particularly intriguing from a physical perspective. Bacterial motility in aqueous solutions is well understood (11, 12), and motility through viscous polymeric solutions has been investigated for many years (13–18). However, much less is understood about how a bacterium moves through a gel, and how the motion depends on, and in turn, influences the structure and dynamic properties of the mucus gel. Recently, some theoretical advances have been made on the related problem of motility of sperm [for a recent review see Ref. (19)], and on the swimming of helical objects in a viscoelastic medium (20). The presence of an elastic network with fluid filled pores in a gel raises questions such as: (i) “Can a bacterium move through a gel, and if so, how does it move?” (ii) “Is it possible for the bacterium flagella motors to exert sufficient force and torque to deform a gel and enable it to move?” (iii) How do the speed and torque depend on the rheological parameters of the gel? Conversely, does the bacterium alter the physical and chemical properties of the gel? In this review we describe the physical characteristics of the gastric mucus lining, and discuss the structure and gelation of gastric mucin, which is the gel forming component of mucus. We then address the question of the motility of H. pylori in the acidic environment of the stomach, and its influence on the structure and mechanical properties of the mucus gel.
The gastrointestinal (GI) tract, like other physiological systems with a cavity open to the environment, is lined with a protective mucus layer [for reviews see Ref. (21, 22)]. The high molecular weight glycoprotein, mucin, secreted by cells in the lining of these organs is responsible for giving mucus the physical characteristics of a viscoelastic fluid and a hydrogel (21). This physical state presents a unique environment to the more than a trillion bacteria that live or move in the GI tract. In the stomach, like the intestine, bacteria are found in the viscous fluid-like outer mucus layer but not in the dense mucus gel layer that adheres to the cell surface (23, 24). Compared to the intestine, relatively few bacteria inhabit the mucus layer of the stomach, as most are unable to survive in the acidic environment of the stomach (25). H. pylori, the most abundant, and the most long-term inhabitant of gastric mucosa, copes with the acidic environment by secreting urease to hydrolyze urea and produce ammonia to neutralize the acid (26, 27). How it gets across the mucosal barrier in the stomach to colonize on the gastric epithelium (10, 28, 29) is a largely unsolved problem. The commonly held view that it bores its way through the mucus gel-like a corkscrew (10, 30) is probably not valid, in light of observations showing that H. pylori are immobile in porcine gastric mucin (PGM) gels under acidic conditions, although their flagella rotate and they wiggle in-place. Instead, we show that motility across the gel is achieved due to the same biochemical mechanism that H. pylori uses for surviving in the acid, namely urea hydrolysis to elevate the pH of its environment. The elevation of pH to neutral transforms the viscoelastic mucin gel to a viscous liquid, enabling the bacterium to swim in the viscous solution (31). Additionally, the helical cell-shape may enable it to swim faster in the viscous solution, as implied by the theoretical prediction of (20). The helical cell-shape has also been shown to be of importance in colonization, as rod- and C-shaped mutants of H. pylori, although motile in soft agar, are not as effective in establishing colonies (32, 33).
Helicobacter pylori infection is also known to impair mucin production, and alter the composition of mucins in the gastric mucus (34). Byrd et al. (35) show that MUC 6, normally associated with gastric gland mucous cells, is expressed in surface mucous cells of H. pylori infected patients, while the MUC5 component of surface mucous cells decreases. Navabi et al. (36) report that MUC1 turnover and level is decreased upon H. pylori infection in mice. Chronic infection leads to intestinal metaplasia, with the stomach mucus developing characteristics of intestinal mucus (37). Newton et al. (38) report an 18% reduction in the amount of gel forming, high molecular weight mucin, although they noted that thickness of the mucus layer is uncompromised. On the other hand Henriksnäs et al. (39), observed a reduction in the thickness of the adherent mucus layer in mice. Mucins also bind to H. pylori (24, 40), the binding is pH dependent (41, 42) and the bacterium is chemotactic toward mucin. Conversely, mucins influence the proliferation, gene expression, and virulence of H. pylori, implying a dynamic interplay between the bacterium and its host (43). These aspects will not be further addressed in this review; the reader is referred McGuckin et al. (24) for the complex interplay between mucins and bacterial pathogens. In the remainder of this review we focus on the physical properties of mucus relevant to the transport of H. pylori across the mucus barrier, and provide some insight into the mechanism of H. pylori motility. We also address the impact of H. pylori infection on mucus structure and rheology.
Of all the organs, it is in the stomach that mucus faces its severest challenges from secreted HCl, digestive enzymes, alcohol, drugs, and bacteria such as H. pylori (10, 44). Gastric mucus is a highly hydrated (swollen to ∼95% water), viscoelastic substance containing 3% of mucin glycoprotein mixed with about 2% low molecular weight lipids, electrolytes, other small molecules, and other proteins such as trefoil factors (21, 45). The mucin glycoprotein is responsible for the remarkable hydration, viscoelastic, and mucoadhesive properties of the protective mucus layer (21, 46). These properties are primarily related to the ability of mucin to polymerize to high molecular weight [for reviews of biophysical properties of mucin see Ref. (47, 48)]. At the typical concentrations found in mucus secretions in mammalian stomachs, mucin further aggregates, and gels under acidic pH.
The gastric mucosal surface is coated with mucus, about 200–400 μm in thickness, consisting of an adherent layer of mucus on the epithelial surface covered with loosely attached, mobile mucus on the luminal side (49). Atuma et al. (50) reported in vivo measurements of the thickness of the mucus lining from stomach to colon in anesthetized rats. They observed that the mucus lining is continuous and consists of two layers, a loosely adherent outer layer which can be easily removed by suction and a firmly adherent layer attached to the epithelial surface. In the rat stomach the loose layer varies from 100 to 120 μm and the firmly adherent layer ranges from 80 to about 150 μm.
Several investigators have used scanning electron microscopy (SEM) to visualize the stomach mucosal surface [see for e.g., Ref. (49, 51–53)], and numerous images and beautiful illustrations of the microstructure of the inner surface of the stomach, with and without bacteria, are readily available see for e.g., http://katierosejohnston.blogspot.com/2011/09/research-images.html. These pictures reveal a highly convoluted, self-similar, or fractal surface with numerous folds of stomach epithelia forming gastric glands (also called gastric pits) which open on the luminal surface of the stomach. Mucus secretions can be seen as wispy, fibrous material on the surface on many of these images. Forte (52) reported low-resolution images of bullfrog mucosa where the residual mucus secretion left on the SEM specimen was visible as bright white, coagulated strands. In this review, we reproduce an image from Nunn et al. (51) showing surface mucous cells, covered with sheets of mucus (Figure 1).
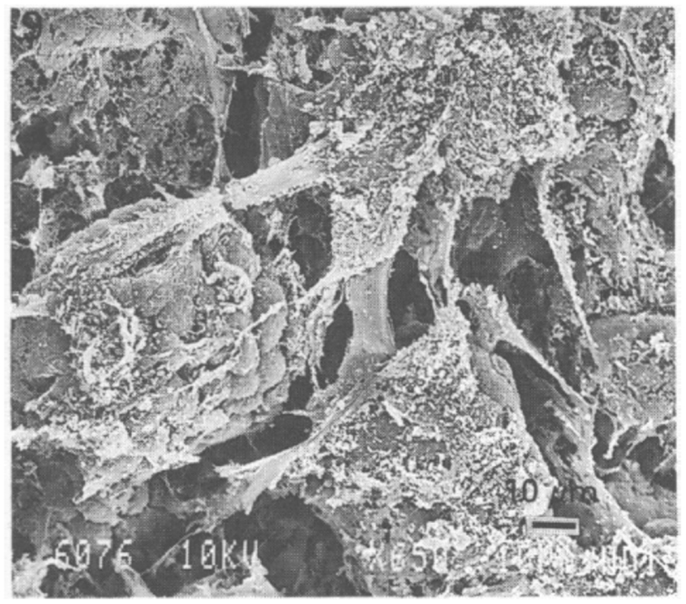
Figure 1. Scanning electron microscopy image of rabbit stomach mucosa showing surface cells and numerous bundles of fiber-like mucus strands. Scale bar 10 μm. Reproduced from Nunn et al. (51) with permission from Wiley.
The shiny, translucent film of mucus, visible to the naked eye, can be removed by gentle scraping and further processed to prepare purified mucin (54). To avoid the perturbative effects of SEM preparation we examined the structure of hydrated mucin and mucus on the sub-micron length scale in vitro by atomic force microscopy (AFM) in a liquid cell under appropriate buffers. A tapping mode AFM measurement of a wet sample of human mucus taken from the discarded material obtained in the lavage following a gastric biopsy reveals a swollen network (Figure 2) formed by the glycoprotein mucin (55). The mucin appears to form aggregates that are connected as in a “pearl-necklace” and enclose aqueous pores of about 200–300 nm in diameter.
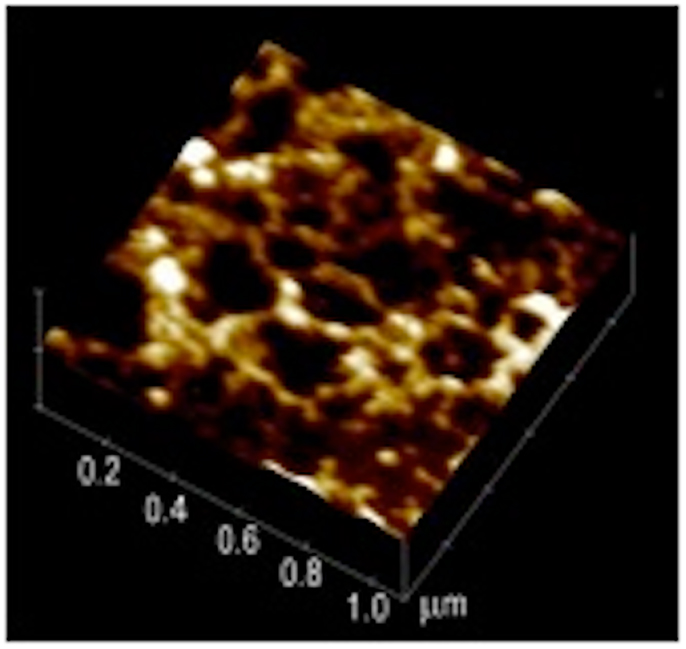
Figure 2. Atomic force microscopy image of endoscopic specimen of human mucus gel. This 1 μm× 1 μm image reveals a network with a “pearl-necklace” morphology formed by mucin aggregates. Reproduced from Hong et al. (55) with permission from American Chemical Society.
For completeness we include a brief description of mucin composition, although this topic has been reviewed extensively. Gastric mucin, like other mucins is a very high molecular weight glycoprotein (2–20 million g/mol) with about 70–80% polysaccharides. The protein, which is quite unlike normal globular proteins, forms the linear core of the molecule on which the polysaccharide chains are radially arrayed similar to the bristles of a bottle-brush, as shown in Figure 3 [adapted from (48)].
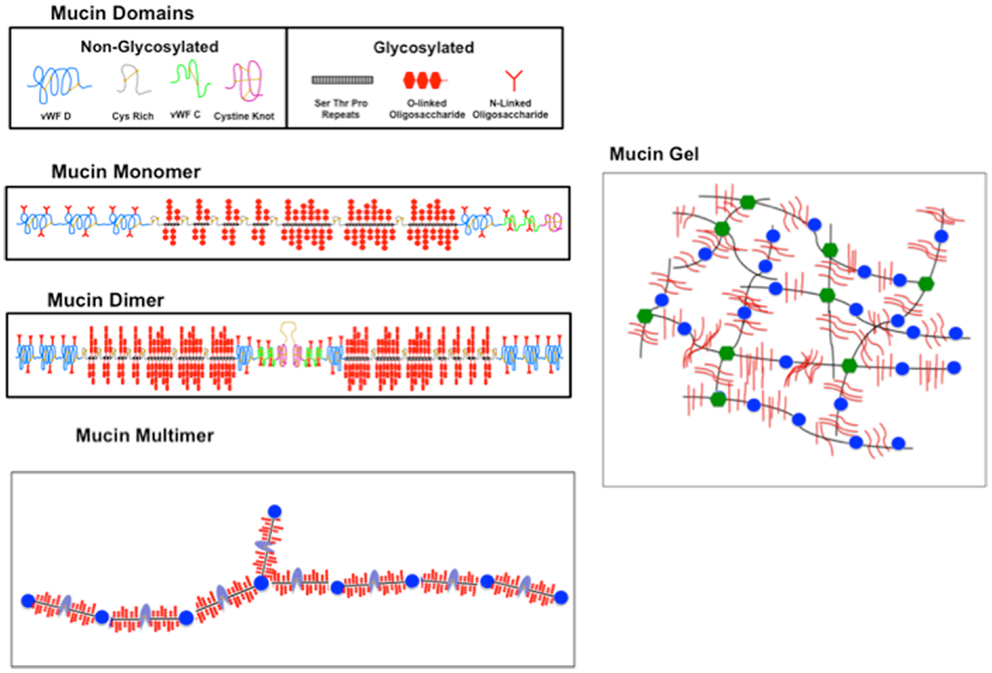
Figure 3. Sketch illustrating the domain structure, polymerization, and gelation of mucin. (Topmost panel) The constituent glycosylated (g) and non- (or weakly) N-glycosylated (ng) domains of mucin represented by icons as labeled. (Second panel) The g and ng domains are linked as shown to form the mucin monomer, with vWF domains at the N terminal and cysteine rich and cysteine knot domains at the C terminal, interspersed with the O-glycosylated STP repeats forming the g domain. (Third panel) A dimer formed by two monomers linked with a disulfide (S–S) bond involving the C-terminal cysteine knot domains. (Bottom panel) A multimer with alternating g (black with red brush) and ng (blue) domains. An N-terminal branch is also indicated. (Right hand panel) A sketch of a mucin gel formed by hydrophobic association of unfolded ng domains of the multimer. The crosslinking is represented by changing the color of ng domains from blue to green. New sketch based in part on Ref. (48) with permission from Elsevier.
Of the ∼20 mucin genes that have been identified with a similar sequence structure (56, 57), gastric mucus contains only two secretory mucins MC5AC and MUC6, in addition to the ubiquitous cell surface mucin MUC1 (58). Dekker et al. (59) suggest that the mucin family be separated into two groups on the basis of sequence homology, one group representing those similar to the cell surface mucin MUC 1, and the other group comprising the secretory mucins similar to MUC2. Both MUC1 and MUC2 mucins contain large domains comprised of tandem repeating sequences of serine (S), threonine (T), and proline (P), located in the heavily O-glycosylated portion of the molecule. The S and T amino acids provide the O-glycosylation sites for the covalent attachment of the polysaccharide. The length and number of the STP repeat varies for the different mucins coded by different genes, and also varies between species. For example, MUC5AC contains 66–124 repeats of 8 amino acids with the consensus sequence TTSTTSAP (60) and MUC6 contains 15–30 repeats of 169 amino acids (61). In all MUC2 type secretory mucins, this glycosylated (g) domain occupies the central region of the apoprotein. It is flanked by cysteine rich domains and a Cystine Knot domain, with non-repeating sequences at the C-terminal (see Figure 3), and by domains similar to the von Willebrand Factor (vWF) C, D domains involved in blood clotting pathways, at the N-terminal. These weakly or non-glycosylated (ng) regions resemble typical secreted globular proteins in their amino acid composition and contain small amount of isolated N-glycosylated oligosaccharides, but do not form a bottle-brush.
Early studies of mucin were interpreted in terms of tetrameric wind-mill like structure (45). However, transmission electron microscopy (TEM) studies clearly established that gastric, cervical, and respiratory mucins are all linear polymers (62, 63). AFM and dynamic light scattering (DLS) studies further show that even in solution, mucins are highly elongated, rod-like, or worm-like polymers [for detailed review and references see Ref. (47, 48)]. AFM imaging of individual molecules of PGM in aqueous solution reveals long, curvilinear filaments ranging from 500 nm to 4 μm in length (55) and ∼1 nm in height, reflecting the height of the hydrated brush probably flattened down due to interactions with the AFM tip. The height and diameter reported by Hong et al. (55) were made in an aqueous environment, and thus provide a better estimate of the dimensions of the hydrated, native PGM molecule than that obtained by TEM measurements (63) or earlier AFM measurements (64) which were done on dried films. Hong et al. (55) also examined the pH dependence of PGM, observing in situ aggregation at pH < 4 with the formation of large, spherical, or oblong aggregates and a 5- to 10-fold increase in height.
The formation of large glycoproteins with molecular weight ranging from 2 to 20 million g/mol is generally believed to involve C-terminal dimer formation of the apoprotein via disulfide (S-S) linkages of the Cysteine knot domains (57, 65). These dimers then further polymerize to form large multimers, as illustrated in Figure 3. There is also the possibility of the C-linked dimers to form trimers via N-terminal S-S linkages involving the vWF D domains. This has been observed in MUC2 (66, 67) and in porcine submaxillary mucin (57) predominantly composed of MUC5B. This would imply the occurrence of tri-functional branches in mucin. It is not clear whether vWF D linked trimers are formed in MUC5AC, although other cleavage sites at the C-terminal have been noted (68, 69). Since TEM and AFM images show predominantly linear polymers, it is possible that trimers, if there, are present in small quantities, and the weak branching at the molecular level is not resolvable by TEM and AFM techniques.
The molecular mechanism of crosslinking in PGM gels is not fully understood. Using fluorescent dye binding (70) suggested that aggregation/gelation at low pH involves a complex interplay between electrostatic and hydrophobic interactions (48) with the formation of non-covalent crosslinks via the hydrophobic association of specific regions of ng domains, modulated by pH dependent changes in the electrostatic interactions of charged amino acids in the N- and C-terminal regions of the apoprotein (48). This network is schematically illustrated in Figure 3, which shows the mucin polymer as having long glycosylated (g), hydrophilic domains, alternating with short, and somewhat hydrophobic ng domains. The differential affinity for water of the alternating domains will stabilize non-covalent crosslinks formed by association of hydrophobic amino acids exposed at low pH in the ng domains in the C- and N-terminal regions, as illustrated in Figure 3. Exactly which domains are involved in these hydrophobic interactions, and whether it involves single domains from dimers or whether it involves the N-terminal tri-functional vWF units has not been explored. Discrete molecular dynamics simulations show pH induced changes in the folding of the PGM 2X domains that are not seen in vWF C domains (71). Further work, both theoretical and experimental, concerning the folding and association of the ng domains, and the swelling of the gel due to the electrostatic interactions of the negatively charged polysaccharide brush of the glycosylated domains, would be valuable in developing a detailed molecular model of pH induced gelation of mucin.
The formation of a network in gastric mucin at low pH has a profound influence on its rheological properties, i.e., its fluid-like flow behavior and response to mechanical deformation and shear forces. In view of the key role that these properties play in the transport of particles and bacteria across mucus, we discuss the underlying concepts before discussing the results. Like many other soft biological materials, the mucus layer exhibits a viscoelastic response to deformation that represents the combined effect of both its liquid-like and its solid-like features, arising from the presence of a polymer network filled with liquid. As illustrated in Figure 4A, a normal viscous solution, like water or glycerin, flows when subjected to a tangential shear force with a viscosity η, reflecting the resistance to flow, i.e., the dissipation or loss of energy. In contrast, an elastic solid stretches (or compresses) like a spring when subjected to tensile forces, and its shape deforms, like the spine of a hardcover book, when subjected to shear forces, but it does not flow in either case. A viscoelastic material exhibits both liquid-like flow and solid-like elasticity, as summarized in Figure 4B (72, 73). One of the hallmarks of a viscoelastic material is that its response to deformation depends on the time for which the material is deformed, as illustrated by the familiar Silly Putty which bounces like a ball when dropped quickly (reflecting the solid-like response on short time scales), but flows like a liquid when stretched slowly (reflecting the large time scale liquid-like flow). The text box lists the basic relations of viscoelasticity.
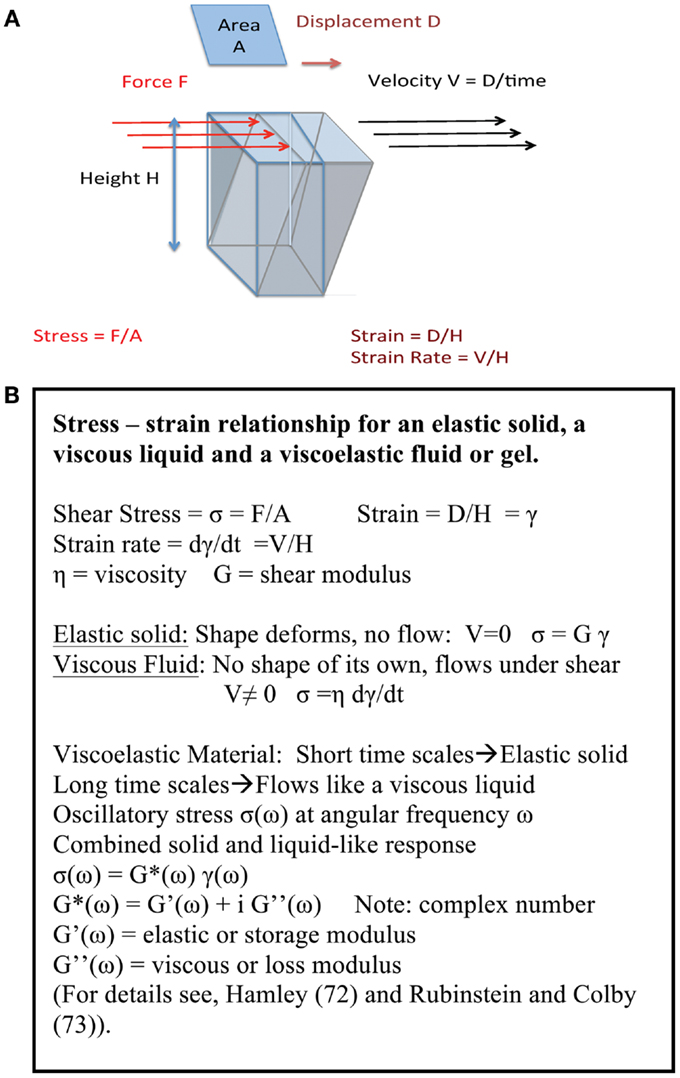
Figure 4. (A) Key concepts of viscoelasticity. A rectangular object with a surface area A and height H, fixed at the bottom surface, is subjected to a shear force F by pushing along the top surface. If the object is a regular solid then the top surface displaces by an amount D deforming the object. If the object is a liquid between two plates, with the bottom plate fixed and the top plate pulled at a speed V, it flows with speed increasing from 0 at the bottom to V at the top, so the deformation is given by V/H. If it is viscoelastic, both solid-like and liquid-like deformations occur simultaneously, albeit on different time scales. The origin of viscoelasticity lies in the ease with which polymer chains can be deformed, and the fact that their motion is controlled by transient entanglements and long-lived crosslinks between chains. (B) Summary of stress – strain relationship. The basic definitions for an elastic solid, a viscous liquid and a viscoelastic fluid or gel are summarized here.
A very simple assay of pH dependence of viscosity of mucin was reported by Bhaskar et al. (54) by measuring the terminal speed1 of a micron sized steel ball falling under gravity in purified PGM mucin solutions. The ball falls slower as the pH is reduced, and does not fall at all in mucin at pH 2, indicting the formation of a gel network at pH 2 (54).
Further insight into the rheological properties of a gel can be obtained from techniques such as oscillatory shear rheology which provides a direct measurement of the frequency dependent bulk viscoelastic moduli of a material. Oscillatory shear measurements clearly show that viscoelastic properties of gastric mucin are highly pH dependent, as shown in Figure 5 (74). At elevated pH (>4) gastric mucin flows like a viscous polymer solution, in which mucin glycoprotein macromolecules are in solution phase, with only transient intermolecular entanglement. In these conditions response to deformation is dominated by viscous flow (and viscous modulus G′′(ω) > elastic modulus G′(ω), over a wide range of values of ω) with a small (but measurable) elasticity. Conversely, at pH < 4, associations between mucin domains give rise to a connected intermolecular network with significant elasticity and minimal flow [elastic modulus G′(ω) > viscous modulus G′′(ω)] (see text box for definitions). Further insight is gained by examining the scaling relationship in plots of G′(ω) and G′′(ω) versus ω and identifying the frequency at which the moduli cross over as described previously (74, 75). This behavior is paralleled in a wide range of biopolymer systems driven by polymer concentration, solvent interactions, pH, salt concentration. Similar results on the pH dependence of PGM have been reported by Maleki et al. (76) using rheo-SALS, a rheometric technique coupled to small angle light scattering.
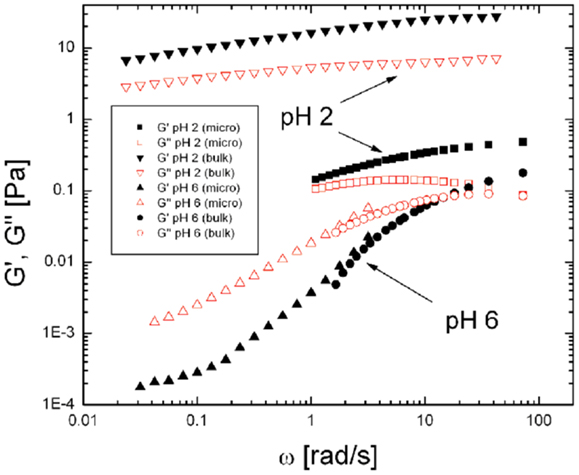
Figure 5. Comparison of viscoelastic moduli, G′(ω) and G′′(ω) for PGM at pH 2 and pH 6 probed by bulk rheology and particle tracking microrheology. In the solution state (pH 6 PGM) bulk and microrheology measurements of both components, G′(ω) and G′′(ω) (labeled as indicated in the Figure) are in agreement over the frequency range accessible. In contrast, the viscoelastic moduli obtained by bulk and microrheology differ significantly in the gel phase (pH 2) suggesting the presence of microstructural heterogeneity and length-scale dependent rheology in the gel state. Reproduced from Celli (80) with permission of author.
Bulk rheology, as described above, provides the average viscoelastic response of a material. However, in a swollen gel such as mucin, the elastic, and viscous response is likely to be length scale dependent due to the inherently heterogeneous structure of a gel consisting of fluid filled pores and network strands. The local, microrheological properties can be probed by tracking the hindered, Brownian motion of micron sized polystyrene latex particles in the medium of interest and calculating the complex elastic modulus G*(ω) from the mean square displacement ⟨Δr2⟩of the particle (77–79). Using this method, Celli (80) found that both G′(ω) and G′′(ω), the elastic and viscous moduli, at pH 6 have similar magnitudes to those obtained in bulk rheology, as expected for particles moving in a viscous solution (80). In contrast, at pH 2 the moduli obtained by microrheology were significantly lower than those obtained by bulk rheology (Figure 5), indicating that, on small length scales, the particles sample a less viscous environment than the bulk gel. Similar results were obtained in a microscopic DLS study (81) in which the much smaller, 100 nm latex particles, appeared to probe two microenvironments; with some beads moving freely in large pores in the pH 2 gel, and others displaying the same slow relaxation as the signal from the dynamics of the gel itself. Lieleg et al. (82) also observed heterogeneities in their investigation of particle translocation through mucin hydrogels.
Time-resolved microscopic particle tracking measurements (80) show that at pH 2 the probe particles move in an inhomogeneous micro-environment consisting of water filled pores in a gel and thus encounter reduced viscous damping. This is illustrated in Figure 6 showing the trajectory of the center of mass (c.m) of a 1 μm latex bead trapped in a pore in the pH 2 mucin network, and only moving at most a total distance of ∼0.1 μm. Rheological studies under non-linear deformations reveal an apparent yield stress, the stress at which the elasticity breaks down, that is also highly influenced by pH. The gel begins to flow just above 10 Pa (74). PGM exhibits a highly non-Newtonian shear thinning behavior, viscosity decreasing with increasing stress in steady shear flow tests, as was previously observed in commercially made PGM (83). The lower pH samples are dramatically shear thinning, decreasing in viscosity by about three orders of magnitude over four decades increase in shear rate and approaching a constant yield stress at low shear rates. These findings on purified mucin are consistent with bulk and microrheological measurements of mucus rheology (84), and provide a molecular basis for understanding the rheological properties of mucus. The shear thinning and yield stress may be relevant to mucus shedding during peristalsis, and could provide a physical mechanism for washing away bacteria that are not adhered to the epithelial surface. It would be interesting to investigate the rheological properties of the mucin from the two distinct mucus layers discussed earlier in this review (50).
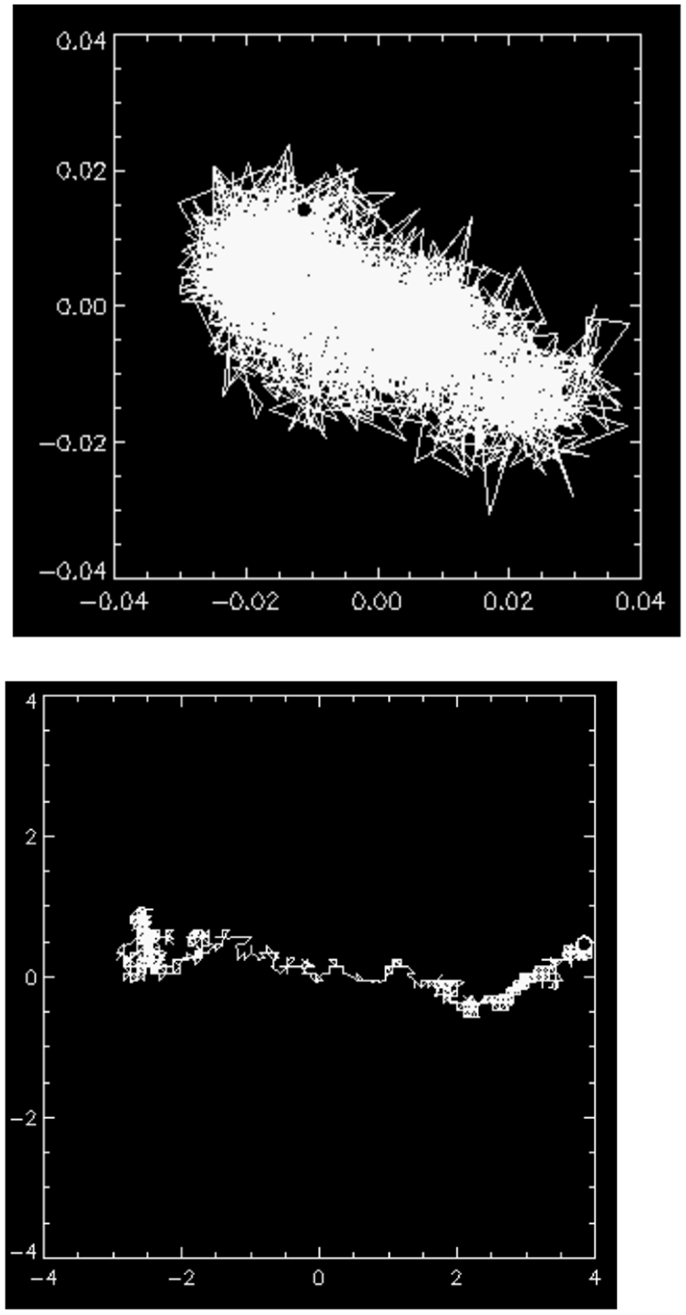
Figure 6. (Top) the motion of the center of mass (c.m) of a 1 μm latex bead trapped in a pore in the mucin gel at pH 2. The maximal excursion of the particle’s c.m <0.1 μm, indicating that it is essentially immobile in the gel network. (Bottom) occasionally, a particle finds a channel in the gel network indicating that the network structure is heterogeneous. The length unit in both images is micrometer. Reproduced from Celli (80) with author’s permission.
We end this section by a cautionary remark concerning the use of commercial mucin preparations such as those obtained by Sigma Aldrich. These do not form a pH dependent gel, because the mucin has been proteolytically digested during purification (85). Such a reduced mucin does not gel upon lowering pH (54). The rheological behavior also depends on which mucin is the predominant component of the preparation. For example, the Orthana MUC6 mucin investigated by Yakubov et al. (86) and Di Cola et al. (87) has a linear dumbbell structure with a central glycosylated portion flanked by hydrophobic, ng regions, suggesting that it consists of monomeric apoprotein, and not the polymerized, gel forming mucin (c.f. Figure 3).
The work described above on particle diffusion in gels shows that the microstructure of mucin and mucus gels restricts the diffusional motion of micron sized particles in such gels. It is not a priori clear how the flagella-driven motion of H. pylori would be impacted by the size restriction due to the movement in a confined geometry. In the next part of this review we address how the helical shape of H. pylori and it’s unique biochemical adaptation to survive in an acidic environment also enables it to go across the mucus gel.
Like most other bacteria that colonize the GI mucosa, H. pylori, too have evolved to be adapted to their unique niche of the stomach mucosa. These Gram-negative, helical-shaped bacteria, 2.5–5.0 μm in length and 0.5–1.0 μm wide (88) have four to six unipolar-sheathed flagella, which are essential for bacterial motility. Each flagellum, ∼3 μm long and 2.5 nm thick, has a characteristic terminal bulb, which is an extension of the flagellar sheath (88). The motility and the helical shape have both been shown to be of importance to the survival of these organisms. Eaton et al. (89) showed that flagellar mutants of H. pylori were unable to colonize the gastric mucosa of gnotobiotic piglets. More recent work by Ottemann and Lowenthal (90) establishes that mutants with non-motile but otherwise intact flagella also do not colonize.
Helicobacter pylori has also evolved to survive in the acidic environment of the stomach (91). It is well established that the ability of H. pylori to hydrolyze urea and elevate the pH of its surroundings is important in enabling it to escape the acidity of the gastric lumen (26, 27, 92, 93), penetrate the thick mucosal gel and reach the surface epithelium (91). H. pylori, exhibits chemotaxis toward urea present in the epithelial cell surface and a pH tactic response toward elevated pH, both of which may also be crucial for survival in the stomach (94). H. pylori survival in acidic conditions is also stated conversely, namely that acidic conditions are required for H. pylori survival in the presence of urea because the subsequent rise in pH to highly alkaline levels is also toxic to the bacterium. To help avoid overproduction of ammonia, the urea channel is regulated by protons to open at low pH and close at high pH. H. pylori’s TlpB receptors enable pH taxis (95, 96). It uses the mucosal pH gradient which varies from low on the luminal surface to neutral on the cell surface (28, 97) to move away from the lumen toward the mucosal surface (98) where it attaches itself with adhesins (10, 99). Mutants lacking either the TlpA or TlpB receptors also show altered extent of inflammation (100). The growth of H. pylori in culture is also pH dependent (101).
The motility and chemotaxis of H. pylori and the related Campylobacter has been investigated by using microscopic tracking [for a recent review see Ref. (102)]. By detailed comparison of the motions of straight-rod E. coli and helical bacteria H. pylori and Campylobacter, in liquid cultures Karim et al. (103) showed that the helical bacteria swim faster than E. coli, presumably due to their helical body shape. Their finding is consistent with the idea of Berg and Turner (16) that a helical shape would result in additional screw-like propulsion for bacteria moving in viscous environments such as those faced by H. pylori in its native environment. However, differences between species cannot be ruled out in this comparison. Yoshiyama et al. (30, 104) and Worku et al. (105) investigated the motility and chemotactic response of H. pylori in viscous synthetic polymer solutions, and found that swimming speeds decrease with increasing viscosity of the polymer solution, and the bacteria became immobile at very high viscosities.
We have examined the motility of H. pylori in purified gastric mucin at different pH’s mucin (31) using phase contrast, digital video microscopy to image live bacteria. Some typical results from analysis of the movies published by Celli et al. (31) are shown and discussed here. We observed that in PGM at neutral pH of 6 or 7 the bacteria swam considerable distances, along almost linear or curved tracks as shown in Figure 7. They exhibit a large spread in their speeds, reflecting both the variation in the size/shape and number of flagella of individual bacteria, as well as the variation in their speed as flagella motors fire asynchronously, and flagella bundle and unbundle. The mean speed averaged over all bacteria is about 16 μm/s which is qualitatively comparable to the speeds of about 20–30 μm/s reported by Worku et al. (105) in methylcellulose.
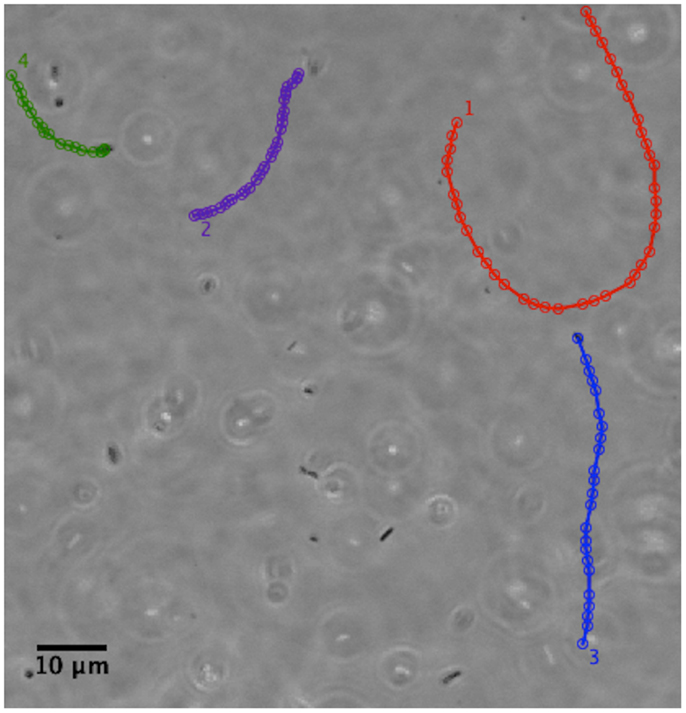
Figure 7. Phase contrast image showing the tracks of a few H. pylori bacteria in a PGM solution 15 mg/ml at pH 6. Adapted from the movies included as supplementary material in Celli et al. (31). The tracks have been colored for ease of visualization. Permission not required for author’s re-use of their own work in PNAS after January 2009. Similar tracks are also shown in Celli (80).
In contrast to the swimming behavior observed in PGM solution at pH 6–7, we noted that when bacteria were added to PGM gels buffered at low pH of 4 or 2 and deprived of urea, they were immobilized and did not move over any measurable distance. Rotation of the flagella and wiggling of the bacteria in-place could be observed at 60–100× magnification, but this did not displace the c.m of the bacterium. Using theoretical models based on resistive force theory of Magariyama et al. (106–108) we obtained the motor torque as 3.6 × 10−18 Nm (31), which is about three times the torque of E. coli swimming in an aqueous environment. The higher torque maybe needed to rotate the flagella in a medium.
We also noticed that, although low pH impairs the flagella motors (109, 110), the bacteria were alive at low pH over the duration of the experiment, as confirmed by increasing the pH back to neutral (see below) and observing the renewed motion of bacteria that were stuck in the low pH environment. In our experiments with motility in PGM gels, we observed a dramatic difference depending on the presence of urea. In the absence of urea the bacteria remained immobile in the low pH gel. However, if urea was present and the sample was not buffered, then bacteria that were immobile in a low pH PGM gel, became mobile within a few minutes. Using two-photon fluorescence microscopy to image the bacterial in an initially low pH mucin gel containing urea we showed that the onset of bacterial movement is directly correlated with a rise in pH to near neutral values, as indicated by the pH sensitive fluorescent dye, BCECF (31). These observations in purified PGM gels are consistent with observations in anesthesized gerbils, which showed that H. pylori became immotile in <1 min at lumen pH values of 2 and 3, and in 2 min at pH 4, but remained motile for more than 15 min at pH 6 (111).
The results above show that the motility of H. pylori depends on the physical state of the mucin gel. To address the converse question, whether the infection by H. pylori impacts the physical properties of mucin or mucus, we performed measurements of the frequency dependent bulk viscoelasticity of purified PGM infected with the bacteria using oscillatory shear methods (31). In earlier studies with purified PGM (shown in Figure 5) we had shown that at pH 4 and 2 purified PGM is a gel with an elastic modulus that dominates over the viscous response, while at pH 6 it is a solution (74). A similar experiment on a 15 mg/ml PGM sample initially at pH 4 incubated with H. pylori for 24 h (Figure 8) clearly shows that the rheological parameters of the infected sample were close to those of pure PGM at pH 6–7 and it remained a solution even at the lowest stress values with a greatly reduced viscosity (31). The concurrent gel to solution transition with the onset of mobility in PGM containing urea suggests that H. pylori gets across the mucus gel using urease secretion to neutralize the acid, raising the pH, and triggering a gel-sol transition. This is illustrated in Figure 9.
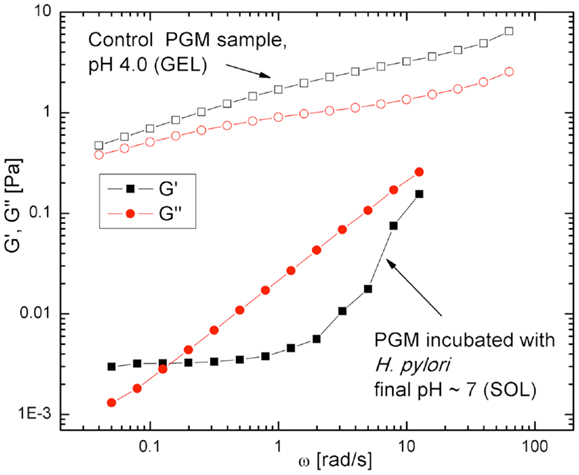
Figure 8. Frequency dependent viscoelastic moduli, for H. pylori infected PGM and control sample, both at pH 4 initially. The elastic response G′ (open black symbols) and viscous response G′′ (filled red symbols) data show that the control sample is a gel (G′ > G′′) while the infected sample is a solution (G′′ > G′). From Celli et al. (31). Copyright permission not required per PNAS policy effective January 2009.
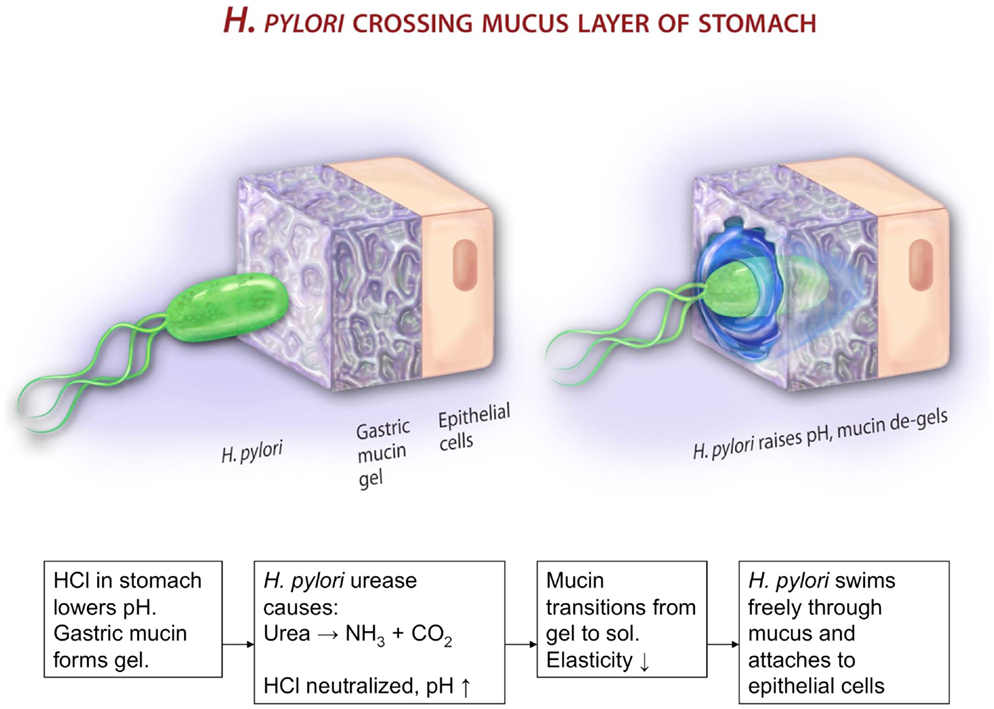
Figure 9. This cartoon illustrates a possible mechanism by which H. pylori gets across the mucus gel. According to Celli et al. (31), gastric mucin forms a gel at low pH < 4. The bacterium cannot move in mucin gels. It secretes urease that metabolizes urea, producing ammonia and elevating the pH. This de-gels the mucin, and enables the bacterium to swim in the resulting polymer solution. The picture, not copyrighted, is reproduced from an NSF press release 09-149. http://www.nsf.gov/news/news_summ.jsp?cntn_id=115409&org=NSF&from=news
Some cautionary remarks about motility and rheology measurements are in order here. Motility measurements in a population of bacteria are inherently polydisperse. Thus, characterizing the behavior by a single average velocity provides limited information and could lead to erroneous predictions; either several different statistical measures such as median, mean, maximal, and minimal speeds should be reported or better yet, the entire distribution. It is also important to analyze hundreds or thousands of tracks, as bacteria move out of the focal plane, and sometimes get stuck in air bubbles and similar defects. By the same token, measurements made very close to the surface of the slide are likely to be influenced by substrate interactions. We also note that bulk rheology provides the average dynamical response, which may not be the same as observed in microrheology, particularly in microstructured environments. The bacteria can also find channels and swim through other heterogeneities in the medium. Bulk rheology experiments require a large sample volume (1–5 ml), while microrheology done on a microscope slide requires only a few microliters. This is a great advantage when analyzing purified samples, mutants, or expressed proteins that are available only in small quantities.
The results discussed above suggest that other factors that can de-gel the mucin would also enable H. pylori to get across the mucus barrier. Worku et al. (105), had observed that H. pylori which were immobile in biopsy samples of mucus, became motile when saline was added to the mucus gel. On the basis of our experiments showing that mucin does not gel at low pH in high salt concentrations (54, 70, 74) we suggest that saline restores motility because it too triggers a gel to solution transition. Whether these observations have implications for identifying factors such as high salt diet, that may promote H. pylori infection in certain populations, remains to be seen.
The review presented here focuses narrowly on the interplay between mucus structure and rheology and the motility of H. pylori. Among the many topics that we have not discussed are motility in a chemotactic environment, possible chemical interactions of the bacterium with mucin or other factors present in mucus, the binding of mucin to H. pylori, the role of H. pylori in altering mucin production or proteolytic digestion of mucin, the effect of the bacterium on mucous producing cells, the mucosal factors that are involved in the adhesion of the bacterium to the epithelial surface, and cell signaling in the mucus environment to form a colony. Some references on these topics have been provided.
The work discussed here however, does show the importance of physical limitations of the mucus gel microstructure and its pH dependence on the motility of this important pathogen. It also shows the usefulness of microscopic bacterial and particle tracking tools for examining the initial stage of bacterial penetration. We hope that this review encourages further theoretical investigations of the fundamental problem of addressing how a helical-shaped bacterium with helical flagella might propel itself in a viscoelastic medium. Further experimental investigations using microfluidics for investigating stomach mucus barrier (112) and single molecule imaging methods, combined with advances in molecular biology, genetics, immunology, and the availability of mutants with specific molecular and functional alterations, will enable deeper understanding of this fascinating problem.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors acknowledge support from NSF (PHY 1058648, PI: Rama Bansil) and NIH (R00CA155045, PI: Jonathan P. Celli).
1. Marshall BJ. Helicobacter Pioneers: Firsthand Accounts from the Scientists who Discovered Helicobacters, 1892-1982. New York: Wiley-Blackwell (2002).
2. Mobley HL, Mendz GL, Hazell SL, editors. Restriction and modification systems – Helicobacter pylori: Physiology Genetics. Washington, DC: ASM press (2001).
3. Lee A. The microbiology and epidemiology of Helicobacter pylori infection. Scand J Gastroenterol Suppl (1994) 201:2–6. doi: 10.3109/00365529409105352
4. Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev (2006) 19(3):449–90. doi:10.1128/CMR.00054-05
5. Peek RM Jr. Events at the host-microbial interface of the gastrointestinal tract IV. The pathogenesis of Helicobacter pylori persistence. Am J Physiol Gastrointest Liver Physiol (2005) 289(1):G8–12. doi:10.1152/ajpgi.00086.2005
6. Tummala S, Keates S, Kelly CP. Update on the immunologic basis of Helicobacter pylori gastritis. Curr Opin Gastroenterol (2004) 20(6):592–7. doi:10.1097/00001574-200411000-00015
7. Mobley HL. Helicobacter pylori factors associated with disease development. Gastroenterology (1997) 113(6 Suppl):S21–8. doi:10.1016/S0016-5085(97)80006-6
8. Blaser MJ, Kirschner D. Dynamics of Helicobacter pylori colonization in relation to the host response. Proc Natl Acad Sci U S A (1999) 96(15):8359–64. doi:10.1073/pnas.96.15.8359
9. Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol (2013) 11:385–99. doi:10.1038/nrmicro3016
10. Montecucco C, Rappuoli R. Living dangerously: how H. pylori survives in the human stomach. Nat Rev Mol Cell Biol (2001) 2:457–66. doi:10.1038/35073084
13. Schneider WR, Doetsch RN. Effect of viscosity on bacterial motility. J Bacteriol (1974) 117:696–701.
14. Greenberg EP, Canale-Parola E. Relationship between cell coiling and motility of spirochetes in viscous environments. J Bacteriol (1977) 131:960–9.
15. Greenberg EP, Canale-Parola E. Motility of flagellated bacteria in viscous environments. J Bacteriol (1977) 132:356–8.
16. Berg HC, Turner L. Movement of microorganisms in viscous environments. Nature (1979) 278:349–51. doi:10.1038/278349a0
17. Chaudhury TK. Swimming in a viscoelastic liquid. J Fluid Mech (1979) 95:189–97. doi:10.1017/S0022112079001415
18. Leshansky AM. Enhanced low-Reynolds-number propulsion in heterogeneous viscous environments. Phys Rev E Stat Nonlin Soft Matter Phys (2009) 80(5 Pt 1):051911. doi:10.1103/PhysRevE.80.051911
19. Lauga E, Powers TR. The hydrodynamics of swimming microorganisms. Rep Prog Phys (2009) 72:096601. doi:10.1088/0034-4885/72/9/096601
20. Spagnolie SE, Liu B, Powers TR. Locomotion of helical bodies in viscoelastic fluids: enhanced swimming at large helical amplitudes. Phys Rev Lett (2013) 111(6):068101. doi:10.1103/PhysRevLett.111.068101
21. Allen A. Structure and function of gastrointestinal mucus. In: Johnson LR editor. Physiology of the Gastrointestinal Tract. New York: Raven Press (1981). p. 617–39.
22. Guth PH. Pathogenesis of gastric mucosal injury. Annu Rev Med (1982) 33:183–96. doi:10.1146/annurev.me.33.020182.001151
23. Hansson GC, Johansson MEV. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes (2010) 1:51–4. doi:10.4161/gmic.1.1.10470
24. McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol (2011) 9(4):265–78. doi:10.1038/nrmicro2538
25. Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A (2006) 103(3):732–7. doi:10.1073/pnas.0506655103
26. Sachs G, Weeks DL, Wen Y, Marcus EA, Scott DR, Melchers K. Acid acclimation by Helicobacter pylori. Physiology (2005) 20:429–38. doi:10.1152/physiol.00032.2005
27. Sidebotham RL, Worku ML, Karim QN, Dhir NK, Baron JH. How Helicobacter pylori urease may affect external pH and influence growth and motility in the mucus environment: evidence from in-vitro studies. Eur J Gastroenterol Hepatol (2003) 15(4):395–401. doi:10.1097/00042737-200304000-00010
28. Hollander F. Two-component mucous barrier: its activity in protecting gastroduodenal mucosa against peptic ulceration. AMA Arch Intern Med (1954) 93:107–29. doi:10.1001/archinte.1954.00240250117009
29. Slomiany BL, Slomiany A. Mechanism of Helicobacter pylori pathogenesis: focus on mucus. J Clin Gastroenterol (1992) 14(Suppl 1):S114–21. doi:10.1097/00004836-199206001-00020
30. Yoshiyama H, Nakazawa T. Unique mechanism of Helicobacter pylori for colonizing the gastric mucus. Microbes Infect (2000) 2(1):55–60. doi:10.1016/S1286-4579(00)00285-9
31. Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci U S A (2009) 106(34):14321–6. doi:10.1073/pnas.0903438106
32. Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, et al. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell (2010) 141(5):822–33. doi:10.1016/j.cell.2010.03.046
33. Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, et al. Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog (2012) 8(3):e1002603. doi:10.1371/journal.ppat.1002603
34. Linden S. Helicobacter pylori Binding to Gastric Mucins and Host Glycosylation Changes After Inoculation. Doctoral Thesis, Lund University (2004). Available from: http://www.lub.lu.se/luft/diss/med_883/med_883_kappa.pdf
35. Byrd JC, Yan P, Sternberg L, Yunker CK, Scheiman JM, Bresalier RS. Aberrant expression of gland-type gastric mucin in the surface epithelium of Helicobacter pylori-infected patients. Gastroenterology (1997) 113(2):455–64. doi:10.1053/gast.1997.v113.pm9247464
36. Navabi N, Johansson ME, Raghavan S, Lindén SK. Helicobacter pylori infection impairs the mucin production rate and turnover in the murine gastric mucosa. Infect Immun (2013) 81(3):829–37. doi:10.1128/IAI.01000-12
37. Hidaka E, Ota H, Hidaka H, Hayama M, Matsuzawa K, Akamatsu T, et al. Helicobacter pylori and two ultrastructurally distinct layers of gastric mucous cell mucins in the surface mucous gel layer. Gut (2001) 49(4):474–80. doi:10.1136/gut.49.4.474
38. Newton JL, Jordan N, Oliver L, Strugala V, Pearson J, James OFW, et al. Helicobacter pylori in vivo causes structural changes in the adherent gastric mucus layer but barrier thickness is not compromised. Gut (1998) 43(4):470–5. doi:10.1136/gut.43.4.470
39. Henriksnäs J, Phillipson M, Storm M, Engstrand L, Soleimani M, Holm L. Impaired mucus-bicarbonate barrier in Helicobacter pylori-infected mice. Am J Physiol Gastrointest Liver Physiol (2006) 291(3):G396–403. doi:10.1152/ajpgi.00017.2006
40. Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol (2008) 1(3):183–97. doi:10.1038/mi.2008.5
41. Nordman H, Borén T, Davies JR, Engstrand L, Carlstedt I. pH-dependent binding of Helicobacter pylori to pig gastric mucins. FEMS Immunol Med Microbiol (1999) 24(2):175–81. doi:10.1111/j.1574-695X.1999.tb01279.x
42. Linden S, Boren T, Mahdavi J, Hedenbro J, Carlstedt I. Effects of pH on Helicobacter pylori binding to human gastric mucins: identification of binding to non-MUC5AC mucins. Biochem J (2004) 384:263–70. doi:10.1042/BJ20040402
43. Skoog EC, Sjöling Å, Navabi N, Holgersson J, Lundin SB, Lindén SK. Human gastric mucins differently regulate Helicobacter pylori proliferation, gene expression and interactions with host cells. PLoS One (2012) 7(5):e36378. doi:10.1371/journal.pone.0036378
44. Blaser MJ. The bacteria behind ulcers. Sci Am (1996) 274(2):104–7. doi:10.1038/scientificamerican0296-104
45. Allen A, Garner A. Mucus and bicarbonate secretion in the stomach and their possible role in mucosal protection. Gut (1980) 21(3):249–62. doi:10.1136/gut.21.3.249
46. Strous GJ, Dekker J. Mucin-type glycoproteins. Crit Rev Biochem Mol Biol (1992) 27(1–2):57–92. doi:10.3109/10409239209082559
47. Bansil R, Stanley HE, LaMont JT. Mucin biophysics. Ann Rev Physiol (1995) 57(1):635–57. doi:10.1146/annurev.ph.57.030195.003223
48. Bansil R, Turner B. Mucin structure, aggregation, physiological functions and biomedical applications. Curr Opin Colloid Interface Sci (2006) 11(2–3):164–70. doi:10.1016/j.cocis.2005.11.001
49. Allen A, Flemstrom G, Garner A, Kivilaakso E. Gastroduodenal mucosal protection. Physiol Rev (1993) 73(4):823–57.
50. Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol (2001) 280:G922–9.
51. Nunn S, Gilmore RS, Dodge JA, Carr KE. Exudate variation in the rabbit gastrointestinal tract: a scanning electron microscope study. J Anat (1990) 170:87.
52. Forte JG. Gastric function. In: Greger R, Windhorst U editors. Comprehensive Human Physiology. Berlin: Springer Verlag (1996). p. 1239–57.
53. Hernández DR, Gianeselli PM, Domitrovic HA. Morphology, histology and histochemistry of the digestive system of South American catfish (Rhamdia quelen). Int J Morphol (2009) 27(1):105–11. doi:10.4067/S0717-95022009000100019
54. Bhaskar KR, Gong D, Bansil R, Pajevic S, Hamilton JA, Turner BS, et al. Profound increase in viscosity and aggregation of pig gastric mucin at low pH. Am J Physiol (1991) 261:G827–32.
55. Hong Z, Chasan B, Bansil R, Turner B, Bhaskar K, Afdhal N. Atomic force microscopy reveals aggregation of gastric mucin at low pH. Biomacromolecules (2005) 6:3458–66. doi:10.1021/bm0505843
56. Gum JR Jr. Mucin genes and the proteins they encode: structure, diversity, and regulation. Am J Respir Cell Mol Biol (1992) 7(6):557–64. doi:10.1165/ajrcmb/7.6.557
57. Perez-Vilar J, Hill RL. The structure and assembly of secreted mucins. J Biol Chem (1999) 274(45):31751–4. doi:10.1074/jbc.274.45.31751
58. Gendler SJ, Spicer AP. Epithelial mucin genes. Ann Rev Physiol (1995) 57(1):607–34. doi:10.1146/annurev.ph.57.030195.003135
59. Dekker J, Rossen JW, Büller HA, Einerhand AW. The MUC family: an obituary. Trends Biochem Sci (2002) 27(3):126–31. doi:10.1016/S0968-0004(01)02052-7
60. Escande F, Aubert JP, Porchet N, Buisine MP. Human mucin gene MUC5AC: organization of its 5’-region and central repetitive region. Biochem J (2001) 358:763–72. doi:10.1042/0264-6021:3580763
61. Toribara NW, Robertson AM, Ho SB, Kuo WL, Gum E, Hicks JW, et al. Human gastric mucin. Identification of a unique species by expression cloning. J Biol Chem (1993) 268:5879–85.
62. Sheehan JK, Oates K, Carlstedt I. Electron microscopy of cervical, gastric and bronchial mucus glycoproteins. Biochem J (1986) 239:147–53.
63. Fiebrig I, Harding S, Rowe A, Hyman S, Davis S. Transmission electron microscopy studies on pig gastric mucin and its interactions with chitosan. Carbohydr Polym (1995) 28(3):239–44. doi:10.1016/0144-8617(95)00105-0
64. Deacon M, Mcgurk S, Roberts C, Williams P, Tendler S, Davies M, et al. Atomic force microscopy of gastric mucin and chitosan mucoadhesive systems. Biochem J (2000) 348:557–63. doi:10.1042/0264-6021:3480557
65. Sheehan JK, Kirkham S, Howard M, Woodman P, Kutay S, Brazeau C, et al. Identification of molecular intermediates in the assembly pathway of the MUC5AC mucin. J Biol Chem (2004) 279:15698–705. doi:10.1074/jbc.M313241200
66. Godl K, Johansson ME, Lidell ME, Mörgelin M, Karlsson H, Olson FJ, et al. The N terminus of the MUC2 mucin forms trimers that are held together within a trypsin-resistant core fragment. J Biol Chem (2002) 277(49):47248–56. doi:10.1074/jbc.M208483200
67. Lidell ME, Moncada DM, Chadee K, Hansson GC. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc Natl Acad Sci U S A (2006) 103(24):9298–303. doi:10.1073/pnas.0600623103
68. Lidell M, Hansson G. Cleavage in the GDPH sequence of the C-terminal cysteine-rich part of the human MUC5AC mucin. Biochem J (2006) 399:121–9. doi:10.1042/BJ20060443
69. Lidell M. Assembly and Proteolytic Cleavages of Gel-Forming Mucins. Doctoral Thesis, University of Gothenburg (2006). Available from: http://hdl.handle.net/2077/16769
70. Cao X, Bansil R, Bhaskar KR, Turner BS, LaMont JT, Niu N, et al. pH-dependent conformational change of gastric mucin leads to sol-gel transition. Biophys J (1999) 76:1250–8. doi:10.1016/S0006-3495(99)77288-7
71. Barz B, Turner BS, Bansil R, Urbanc B. Folding of pig gastric mucin non-glycosylated domains: a discrete molecular dynamics study. J Biol Phys (2012) 38(4):681–703. doi:10.1007/s10867-012-9280-x
72. Hamley IW. Introduction to Soft Matter: Synthetic and Biological Self-Assembling Materials. England: Wiley (2007).
74. Celli JP, Turner BS, Afdhal NH, Ewoldt RH, McKinley GH, Bansil R, et al. Rheology of gastric mucin exhibits a pH-dependent sol-gel transition. Biomacromolecules (2007) 8(5):1580–6. doi:10.1021/bm0609691
75. Winter HH, Chambon F. Analysis of linear viscoelasticity of a crosslinking polymer at the gel point. J Rheol (1986) 30:367. doi:10.1122/1.549853
76. Maleki A, Lafitte G, Kjøniksen AL, Thuresson K, Nyström B. Effect of pH on the association behavior in aqueous solutions of pig gastric mucin. Carbohydr Res (2008) 343(2):328–40. doi:10.1016/j.carres.2007.10.005
77. Mason TG, Weitz DA. Optical measurements of frequency-dependent linear viscoelastic moduli of complex fluids. Phys Rev Lett (1995) 74(7):1250–3. doi:10.1103/PhysRevLett.74.1250
78. Mason TG. Estimating the viscoelastic moduli of complex fluids using the generalized Stokes-Einstein equation. Rheol Acta (2000) 39:371–8. doi:10.1007/s003970000094
79. Tseng Y, Kole TP, Wirtz D. Micromechanical mapping of live cells by multiple-particle-tracking microrheology. Biophys J (2002) 83(6):3162–76. doi:10.1016/S0006-3495(02)75319-8
80. Celli JP. Gastric Mucin and Helicobacter pylori: Rheology, Microrheology and Microscopy Studies [Ph.D. Dissertation]. Boston, MA: Boston University (2007).
81. Celli J, Gregor B, Turner B, Afdhal N, Bansil R, Erramilli S. Viscoelastic properties and dynamics of porcine gastric mucin. Biomacromolecules (2005) 6(3):1329–33. doi:10.1021/bm0493990
82. Lieleg O, Vladescu I, Ribbeck K. Characterization of particle translocation through mucin hydrogels. Biophys J (2010) 98(9):1782–9. doi:10.1016/j.bpj.2010.01.012
83. Waigh T, Papagiannopoulos A, Voice A, Bansil R, Unwin A, Dewhurst C, et al. Entanglement coupling in porcine stomach mucin. Langmuir (2002) 18(19):7188–95. doi:10.1021/la025515d
84. Lai SK, Wang YY, Wirtz D, Hanes J. Micro- and macrorheology of mucus. Adv Drug Deliv Rev (2009) 61(2):86–100. doi:10.1016/j.addr.2008.09.012
85. Kocevar-Nared J, Kristl J, Smid-Korbar J. Comparative rheological investigation of crude gastric mucin and natural gastric mucus. Biomaterials (1997) 18(9):677–81. doi:10.1016/S0142-9612(96)00180-9
86. Yakubov GE, Papagiannopoulos A, Rat E, Easton RL, Waigh TA. Molecular structure and rheological properties of short-side-chain heavily glycosylated porcine stomach mucin. Biomacromolecules (2007) 8(11):3467–77. doi:10.1021/bm700721c
87. Di Cola E, Yakubov GE, Waigh TA. Double-globular structure of porcine stomach mucin: a small-angle X-ray scattering study. Biomacromolecules (2008) 9(11):3216–22. doi:10.1021/bm800799u
88. O’Rourke J, Bode G. Morphology and ultrastructure. In: Mobley H, Mendz G, Hazell S editors. Helicobacter pylori: Physiology and Genetics. Washington, DC: ASM Press (2001). p. 53–67.
89. Eaton KA, Morgan DR, Krakowka S. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol (1992) 37(2):123–7. doi:10.1099/00222615-37-2-123
90. Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun (2002) 70(4):1984–90. doi:10.1128/IAI.70.4.1984-1990.2002
91. Hazell SL, Lee A. Campylobacter pyloridis, urease, hydrogen ion back diffusion, and gastric ulcers. Lancet (1986) 2(8497):15–7. doi:10.1016/S0140-6736(86)92561-4
92. Scott DR, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology (1998) 114(1):58–70. doi:10.1016/S0016-5085(98)84400-4
93. Athmann C, Zeng N, Kang T, Marcus EA, Scott DR, Rektorschek M, et al. Local pH elevation mediated by the intrabacterial urease of Helicobacter pylori cocultured with gastric cells. J Clin Invest (2000) 106(3):339–47. doi:10.1172/JCI9351
94. Nakamura H, Yoshiyama H, Takeuchi H, Mizote T, Okita K, Nakazawa T. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect Immun (1998) 66(10):4832–7.
95. Croxen MA, Sisson G, Melano R, Hoffman PS. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol (2006) 188(7):2656–65. doi:10.1128/JB.188.7.2656-2665.2006
96. Schweinitzer T, Mizote T, Ishikawa N, Dudnik A, Inatsu S, Schreiber S, et al. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J Bacteriol (2008) 190:3244–55. doi:10.1128/JB.01940-07
97. Davenport HW. Fluid produced by the gastric mucosa during damage by acetic and salicylic acids. Gastroenterology (1966) 50:487–99.
98. Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, et al. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci U S A (2004) 101(14):5024–9. doi:10.1073/pnas.0308386101
99. el-Shoura SM. Helicobacter pylori: I. Ultrastructural sequences of adherence, attachment, and penetration into the gastric mucosa. Ultrastruct Pathol (1995) 19(4):323–33. doi:10.3109/01913129509064237
100. Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun (2007) 75(8):3747–57. doi:10.1128/IAI.00082-07
101. Sjostrom JE, Larsson H. Factors affecting growth and antibiotic susceptibility of Helicobacter pylori: effect of pH and urea on the survival of a wild-type strain and a urease-deficient mutant. J Med Microbiol (1996) 44(6):425–33. doi:10.1099/00222615-44-6-425
102. Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol (2011) 65:389–410. doi:10.1146/annurev-micro-090110-102908
103. Karim QN, Logan RPH, Puels J, Karnholz A, Worku ML. Measurement of motility of Helicobacter pylori, Campylobacter jejuni, and Escherichia coli by real time computer tracking using the Hobson BacTracker. J Clin Pathol (1998) 51:623–8. doi:10.1136/jcp.51.8.623
104. Yoshiyama H, Nakamura H, Kimoto M, Okita K, Nakazawa T. Chemotaxis and motility of Helicobacter pylori in a viscous environment. J Gastroenterol (1999) 34(Suppl 11):18–23.
105. Worku ML, Sidebotham RL, Baron JH, Misiewicz JJ, Logan RP, Keshavarz T, et al. Motility of Helicobacter pylori in a viscous environment. Eur J Gastroenterol Hepatol (1999) 11(10):1143–50. doi:10.1097/00042737-199910000-00012
106. Magariyama Y, Sugiyama S, Muramoto K, Kawagishi I, Imae Y, Kudo S. Simultaneous measurement of bacterial flagellar rotation rate and swimming speed. Biophys J (1995) 69:2154–62. doi:10.1016/S0006-3495(95)80089-5
107. Magariyama Y, Sugiyama S, Kudo S. Bacterial swimming speed and rotation rate of bundled flagella. FEMS Microbiol Lett (2001) 199:125–9. doi:10.1111/j.1574-6968.2001.tb10662.x
108. Magariyama Y, Kudo S. A mathematical explanation of an increase in bacterial swimming speed with viscosity in linear-polymer solutions. Biophys J (2002) 83:733–9. doi:10.1016/S0006-3495(02)75204-1
109. Meyer-Rosberg K, Scott DR, Rex D, Melchers K, Sachs G. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology (1996) 111(4):886–900. doi:10.1016/S0016-5085(96)70056-2
110. Nakamura S, Kami-ike N, Yokota JP, Kudo S, Minamino T, Namba K. Effect of intracellular pH on the torque-speed relationship of bacterial proton-driven flagellar motor. J Mol Biol (2009) 386:332–8. doi:10.1016/j.jmb.2008.12.034
111. Schreiber S, Bucker R, Groll C, Azevedo-Vethacke M, Garten D, Scheid P, et al. Rapid loss of motility of Helicobacter pylori in the gastric lumen in vivo. Infect Immun (2005) 73:1584–9. doi:10.1128/IAI.73.3.1584-1589.2005
Keywords: H. pylori, gastric mucosa, mucins, rheology, motility, atomic force microscopy, particle tracking microrheology, bacterial infections
Citation: Bansil R, Celli JP, Hardcastle JM and Turner BS (2013) The influence of mucus microstructure and rheology in Helicobacter pylori infection. Front. Immunol. 4:310. doi: 10.3389/fimmu.2013.00310
Received: 17 May 2013; Paper pending published: 19 July 2013;
Accepted: 16 September 2013; Published online: 10 October 2013.
Edited by:
Thomas Blanchard, University of Maryland School of Medicine, USAReviewed by:
Glen C. Ulett, Griffith University, AustraliaCopyright: © 2013 Bansil, Celli, Hardcastle and Turner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rama Bansil, Department of Physics, Boston University, 590 Commonwealth Avenue, Boston, MA 02215, USA e-mail:cmJAYnUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.