
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 28 February 2013
Sec. NK and Innate Lymphoid Cell Biology
Volume 4 - 2013 | https://doi.org/10.3389/fimmu.2013.00055
This article is part of the Research Topic Biology of NK cells and NK cell receptors View all 45 articles
Upon maturation, natural killer (NK) cells acquire effector functions and regulatory receptors. New insights suggest a considerable functional heterogeneity and dynamic regulation of receptor expression in mature human NK cell subsets based on different developmental axes. Such processes include acquisition of lytic granules as well as regulation of cytokine production in response to exogenous cytokine stimulation or target cell interactions. One axis is regulated by expression of inhibitory receptors for self-MHC class I molecules, whereas other axes are less well defined but likely are driven by different activating receptor engagements or cytokines. Moreover, the recent identification of long-lived NK cell subsets in mice that are able to expand and respond rapidly following a secondary viral challenge suggest previously unappreciated plasticity in the programming of NK cell differentiation. Here, we review advances in our understanding of mature NK cell development and plasticity with regards to regulation of cellular function. Furthermore, we highlight some of the major questions that remain pertaining to the epigenetic changes that underlie the differentiation and functional specialization of NK cells and the regulation of their responses.
Epigenetics refers to functionally relevant modifications to the genome that do not involve a change in the nucleotide sequence, but may impact gene expression, cellular phenotype, and function. Examples of such modifications are acetylation, methylation or phosphorylation of the N-terminal tails of histone proteins. In general, acetylation marks are enriched in open chromatin regions, while histone methylation associates with either open or closed chromatin depending on the degree of methylation and the specific amino acid residues that are modified on the histone proteins (Suganuma and Workman, 2011). Changes in chromatin structure also occur through DNA methylation and demethylation, nucleosome repositioning and the generation of DNase hypersensitive sites. Conclusive evidence has demonstrated that epigenetic changes that take place during cell differentiation can persist through multiple cell divisions (Greer and Shi, 2012).
Natural killer (NK) cells constitute an arm of the innate immune system, as their effector functions are controlled by a repertoire of germline–encoded receptors that do not undergo somatic recombination (Lanier, 2008; Bryceson et al., 2011). However, similar to other lymphocyte subsets such as T and B cells, NK cells may manifest adaptive features including enhanced longevity and recall responses (Sun and Lanier, 2011). NK cells produce interferon (IFN)-γ in response to exogenous cytokines, display immunoregulatory activity in regards to perforin-dependent killing of activated immune cells and mediate immunosurveillance and elimination of intracellular pathogens and tumors through both IFN-γ secretion and target cell killing (Lodoen and Lanier, 2006; Vesely et al., 2011). From a clinical perspective, NK cells can kill allogeneic cells in the setting of hematopoietic stem cell transplantation and are utilized in immunotherapy for the treatment of certain malignancies (Geller and Miller, 2011).
Considerable heterogeneity exists among human peripheral blood NK cells with regards to phenotype and function, and a hierarchy of activating stimuli controls the induction of various effector functions (Bjorkstrom et al., 2010; Fauriat et al., 2010; Juelke et al., 2010) (Figure 1). Although many details are lacking, experimental evidence suggests that NK cell precursors develop in primary lymphoid organs, circulate through the bloodstream, and collect in secondary lymphoid tissues. During the maturation process, NK cells differentiate into highly proliferative CD56bright NK cells. Terminal differentiation involves a downregulation of CD56, changes in the receptor profile and acquisition of cytotoxic function (Freud et al., 2005). In parallel to the continuous maturation process, recognition of major histocompatibility complex (MHC) class I molecules by inhibitory receptors potentiate NK cell responses through a phenomenon known as education (Kim et al., 2005; Anfossi et al., 2006). The expression levels of several surface proteins on CD56dim NK cells including CD57, CD62L, and CD94 correlate either positively or negatively with differentiation as defined by cytotoxic potential (Bjorkstrom et al., 2010; Juelke et al., 2010; Yu et al., 2010), but no cellular marker has been identified to distinguish between educated and non-educated NK cells. Further highlighting cellular plasticity, phenotypically and functionally distinct NK cell subsets are found in the uterus, liver, mucosa, and lymph nodes (Koopman et al., 2003; Cella et al., 2009; Luci et al., 2009). Defining NK cell subsets and understanding the acquisition of function is particularly difficult since superimposing differentiation and education processes on NK cell functionality does not fully explain the heterogeneity in NK cell responses. Here, we provide an overview of current insights into the transcriptional and epigenetic regulation of mature NK cell development and differentiation.
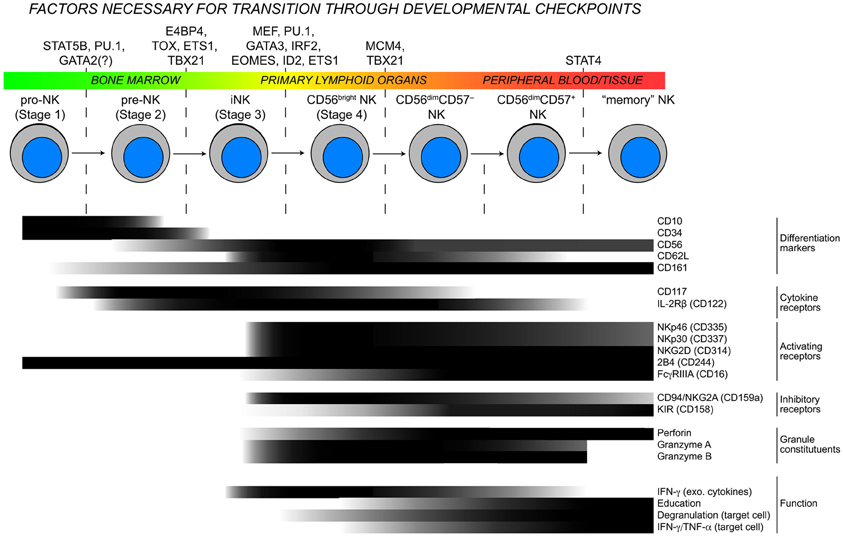
Figure 1. Checkpoints in NK cell development. As NK cells develop, they transit through the body and advance through checkpoints that are controlled by DNA-binding factors. Throughout this process, the expression of developmental markers, cytokine receptors, natural cytotoxicity receptors, and functional competencies are all dynamically regulated resulting in considerable heterogeneity within the NK cell population. Protein expression data are modified from (Freud et al., 2005; Freud and Caligiuri, 2006) and for “memory” NK cells extrapolated from (Sun et al., 2009; Hwang et al., 2012).
Cellular identity is formed and often maintained by a network of transcription factors that influence the chromatin state and drive gene expression patterns. Mouse models have been instrumental for identifying several key transcription factors that are necessary for commitment to the NK cell lineage. Signaling through the interleukin-15 receptor common beta and gamma chains (CD122/132) is essential for NK cell development, and conditional knockout of Stat5a and Stat5b in NK cells results in an almost complete developmental block at the NK progenitor (NKP) to immature NK (iNK) transition (Eckelhart et al., 2011). Moreover, autosomal recessive STAT5B mutations in humans are associated with defective NK cell development and severe herpes virus infections (Kofoed et al., 2003). STAT5 proteins can enhance cell survival by driving expression of important anti-apoptotic genes such as Bcl-2, Bcl-xL, and Mcl-1 (Debierre-Grockiego, 2004) or by inducing activation of the phosphoinostol 3-kinase (PI3K)/Akt and Ras/MAPK pathways (Nyga et al., 2005). While these pathways may contribute to NK cell development, Gascoyne et al. recently discovered an axis in which IL-15R signaling is necessary, specifically in NK cells, for the expression of the transcription factor E4bp4 (Gascoyne et al., 2009). As NKP cells transition to the iNK stage, E4bp4 induces transcription of Id2 and Gata3, which are critical for further NK cell development. The generation of mice with targeted mutations in putative Stat5 binding sites within the regulatory regions E4bp4 are needed to definitively determine whether the requirement for Stat5 proteins in NK cell development is through direct induction of E4bp4 expression. A second axis involved in the control of early NK cell development is dependent upon the transcription factor thymocyte selection-associated high-mobility group box protein (TOX). NK cells in Tox-deficient mice fail to develop from iNK to mature NK (mNK) cells (Aliahmad et al., 2010). Although Id2 expression is slightly lower in Tox-deficient NK cells, overexpression of Id2 in these cells cannot rescue the developmental block. Tox-deficient NKPs also express normal levels of IL-15R, suggesting that TOX acts in a separate pathway to promote early NK cell development.
The homologous transcription factors Tbx21 (T-bet) and Eomes are essential components of another early pathway that regulates NK cell development. NK cell-specific deletion of Tbx21 leads to a block in development at the transition between the NKP and iNK stages of development, while deletion of Eomes causes defects in subsequent maturation events including the acquisition of a diverse Ly49 receptor repertoire and expression of other developmental markers including DX5. Interestingly, sustained expression of both T-bet and Eomes appears to be essential for maintaining the identity/maturity of NK cells (Gordon et al., 2012), though the epigenetic basis for these observations is yet to be explored. Knockout studies in mice have demonstrated that several other transcription factors including Ets1, Spi1, and Irf2 have cell-intrinsic requirements for early NK cell development (Scott et al., 1994; Barton et al., 1998; Lohoff et al., 2000). Spi1−/− mice, which lack expression of the Ets-family transcription factor PU.1, die in utero. Fetal liver transfer experiments have revealed that PU.1-deficiency abrogates T and B cell development and severely impairs the development of NK cell precursors (pre-NK) (Colucci et al., 2001). Similarly, Gata3−/− mice die in utero, but display normal NK cell numbers in the spleen. Gata3−/− NK cells are more immature, suggesting a defect in iNK to mNK transition (Samson et al., 2003). Furthermore, Id2−/− mice are viable and display a defect in iNK to mNK transition (Boos et al., 2007). Mutations in GATA2 were recently identified in patients with a syndrome known as dendritic cell, monocyte, B and natural killer lymphoid (DCML)-deficiency (Dickinson et al., 2011; Hsu et al., 2011), but whether NK cells have a cell-intrinsic requirement for GATA2 expression or whether the NK cell-deficiency is secondary to the loss of monocytes and dendritic cells still needs to be clarified.
While considerable progress has been made in identifying transcription factor networks that control early developmental checkpoints, much less is known with regards to how the terminal maturation stages are regulated in NK cells. Targeted exome sequencing of individuals with a variant of familial glucocorticoid deficiency (FGD) associated with severe herpes virus infections uncovered an essential role for the minichromosome maintenance (MCM) 4 gene NK cell terminal differentiation (Gineau et al., 2012). MCM4 is a component of a protein complex with DNA helicase activity that acts in separating DNA strands during replication. NK cells from patients with partial MCM4-deficiency failed to efficiently differentiate beyond the CD56bright stage and did not proliferate in response to IL-2 or IL-15, suggesting that robust proliferation is necessary for the CD56bright to CD56dim NK cell transition. It is remarkable that the proliferation of B and T cells was not affected in patients with MCM4-deficiency, and more studies are required to explain why the observed proliferation defects are specific to NK cells, whether MCM4-deficiency affects differentiation or survival and to rule out the possibility that the adrenal insufficiency in these patients is impacting NK cell development. Regardless of the mechanistic details, these studies highlight the uniqueness of the CD56bright NK cell subset and demonstrate the importance of proliferation in NK cell development and differentiation.
Although NK cell activation has been studied intensively for several decades, the mechanisms underlying the generation and maintenance of functional NK cells remains only partially understood. The effector molecule that is most highly associated with NK cell cytotoxicity is perforin, which creates pores in the phospholipid bilayer of target cells, facilitating entry of apoptosis-inducing granzymes. Expression of the Ets family transcription factor myeloid Elf1-like factor (MEF) directly binds to two sites within the PRF1 promoter and is obligatory for perforin expression in NK cells but not CD8+ T cells, demonstrating that PRF1 expression is differentially regulated at the transcriptional level in cytotoxic lymphocytes. The 5′ regulatory region of PRF1 contains two enhancers, located at −15 and −1 kb that bind Stat5 and are responsive to IL-2R-activated signal transduction (Zhang et al., 1999). The importance of these enhancers is evident in NK cells from Il2Rb or Stat5b knockout mice that have significantly lower levels of perforin transcription (Imada et al., 1998). Interestingly, the −1 kb enhancer is also responsive to IL-6 and IL-12 and can bind STAT1a and STAT4, respectively, upon cytokine stimulation (Yu et al., 1999; Yamamoto et al., 2002). How these enhancers and other known regulatory elements within PRF1 are regulated during NK cell development and whether they contribute to DNA demethylation within the locus remains to be determined.
Granzymes constitute a family of serine proteases that are found in cytolytic granules of cytotoxic T cells and NK cells. One of the most thoroughly studied members of the family is granzyme B, which can mediate target cell death through the cleavage of caspases or through damage to the mitochondria (Lieberman, 2003). Studies on transcriptional control of granzyme B expression in CD8+ T cells have demonstrated that several transcription factors including AP1, IKAROS, RUNX1, CREB1, and Eomes bind within a large DNase-hypersensitive region comprising the granzyme B promoter and facilitate transcription (Wargnier et al., 1995; Babichuk et al., 1996; Araki et al., 2008). Little is known with regards to epigenetic control of granzyme B expression outside of the observation that histone H3K9 acetylation within the granzyme B promoter correlates with high levels of gene expression (Araki et al., 2008). Signaling through the NF-κ B pathway induces granzyme B expression, and NF-κ B binds to a putative enhancer element approximately 10 kb downstream of the transcriptional start site (Huang et al., 2006). Further studies are needed to test whether this enhancer promotes expression by influencing the chromatin environment surrounding GZMB.
The mechanisms that regulate IFN-γ production are of interest given that a tight balance must be maintained to ensure that enough IFN-γ is produced to bolster immune responses without an overproduction that can lead to severe and widespread pathology. In this regard, CD56bright NK cells produce IFN-γ mainly in response to exogenous cytokines, whereas CD56dim NK cells do so mainly in response to target cells. Furthermore, IFN-γ production by donor NK cells is also impaired following transplantation (Foley et al., 2011). Rapid production of IFN-γ by NK cells upon stimulation appears to be due, at least in part, to the establishment of long-range histone acetylation patterns at distal conserved non-coding sequences (CNS) within the IFNG locus early during NK cell development. Several CNS elements have been identified in both the human and mouse genes. Of note, a CNS element ~22 kb upstream of murine Ifng binds T-bet and serves as an obligate enhancer for opening the locus and allowing expression in NK cells (Hatton et al., 2006).
The binding of several cytokines, including IL-2, IL-15, IL-12, and IL-18, to their cognate receptor complexes on NK cells strongly induces IFN-γ expression. In primary human NK cells, IL-2 treatment enhances histone acetylation at a DNase-I hypersensitivity region containing a Stat5 binding site ~3.5–4.0 kb upstream of the transcriptional start site. Thus, this distal regulatory element is a target of chromatin remodeling in the IFNG locus and acts as an IL-2-responsive transcriptional enhancer (Bream et al., 2004). Since CD56bright NK cells constitutively express both the high- and intermediate-affinity IL-2R chains (Matos et al., 1993), the presence of the ~3.5–4.0 kb upstream element provides a presumptive explanation for the enhanced ability of CD56bright NK cells to produce IFN-γ in response to IL-2 relative to the CD56dim subset.
Changes in nucleosome composition or position are also important for the establishment of open chromatin and active transcription. So far, three distinct chromatin-remodeling complexes have been identified in mammals. These include the Mi-2-NuRD complex, the ISWI complex, and a mammalian Switch (Swi)-sucrose non-fermenter (SNF) homologue complex (Narlikar et al., 2002). The mammalian Swi-SNF complex contains a homologue of Brahma-related gene 1 (Brg1) or Brahma and includes Brahma/Brg1-associated factors that are capable of binding DNA (Khavari et al., 1993). In Th1 cells, Stat4-dependent recruitment of Brg1 is necessary for nucleosome remodeling within the first 600 bases of the Ifng promoter (Zhang and Boothby, 2006). Whether Stat4 recruits Brg1 through a direct interaction or indirectly by promoting histone acetylation and creating favorable conditions for Brg1 binding is still unknown. A similar mechanism presumably controls IFN-γ expression in CD56dim NK cells, which express high levels of both Brg1 and STAT4 (Cichocki and Bryceson, unpublished observations). How the IFNG locus may be distinctly regulated in CD56bright and CD56dim NK cells to control IFN-γ production induced by exogenous cytokines or target cell recognition may provide interesting insights into the mechanisms regulating functional specialization of NK cell subsets.
In addition to IFN-γ, CD56dim NK cells produce high levels of tumor necrosis factor (TNF)-α in response to target cell stimulation, which has a major role in initiating inflammatory responses, promoting the clearance of intracellular pathogens, and killing tumors. TNF-α belongs to the TNF superfamily, which also includes the proinflammatory cytokines lymphotoxin (LT)-α and LT-β. The TNF/LT genes are clustered together within the MHC class III region on human chromosome 6. DNA within the 5′ regulatory region of TNF is densely methylated in embryonic and hematopoietic precursor cells. Studies using monocytes show that lineage commitment is associated with DNA demethylation followed by the acquisition of histone acetylation and the establishment of open chromatin. In mouse monocyte cell lines, histones within the Tnf promoter associate constitutively with Brg1 (Ramirez-Carrozzi et al., 2006). Therefore, a common mechanism involving histone modification and nucleosome repositioning is responsible for creating an open chromatin conformation within the regulatory regions of multiple inflammatory cytokine genes. Figure 2 summarizes the current state of knowledge with regard to transcription factor binding to PRF1, GZMB, IFNG, and TNF and epigenetic control of their expression.
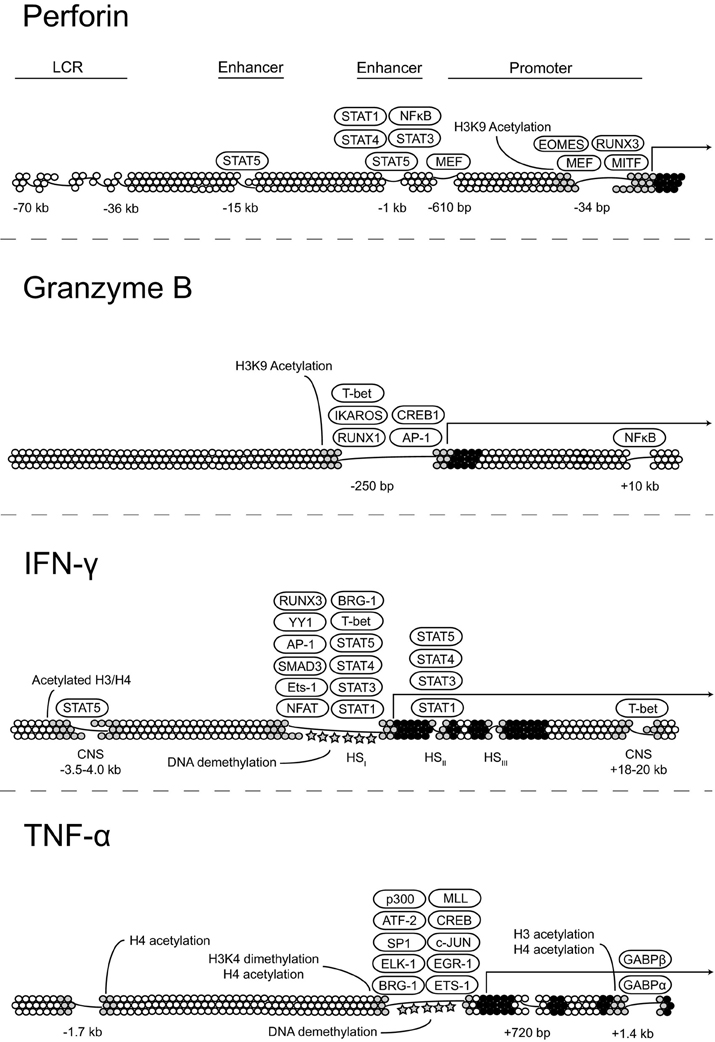
Figure 2. Transcriptional and epigenetic regulation of key effector molecules. Proposed models for the regulation of perforin, granzyme B, IFN-γ, and TNF-α expression at the transcriptional level through transcription factor binding and changes in the chromatin state.
The term “memory” NK cell was first coined after the observation of long-lived NK cells following CMV infection in mice (Sun et al., 2009). Such adaptive NK cells express self-specific inhibitory Ly49 receptors and their expansion is Dap12-dependent (Sun et al., 2009). Similarly, in humans, NK cells expressing the DAP12-coupled activating receptor NKG2C are found at high frequencies in CMV seropositive donors (Lopez-Vergès et al., 2011; Foley et al., 2012). Moreover, in the context of severe viral infections, NKG2C+ NK cells with self-specific inhibitory KIR receptors expand (Bjorkstrom et al., 2011; Beziat et al., 2012).
The formation of B and T cell memory involves antigen recognition by the B cell receptor and T cell receptor complexes, respectively, along with cytokine signaling and clonal expansion. At the molecular level, programming of B and T cell memory is dependent upon transcription factor networks that promote differentiation and survival. Of note, the transcription factors B cell lymphoma 6 (BCL-6) and PR domain-containing protein 1 (PRDM1) act within a regulatory axis to control effector and memory differentiation in CD4+ T cells, CD8+ T cells, and B cells by reinforcing cell-fate decisions (Crotty et al., 2010). The shared requirement for BCL-6 and PRDM1 in the generation of adaptive immune memory make these transcription factors attractive candidates for study in the emerging field of NK cell memory formation.
The mechanisms that control NK cell proliferation and survival are highly relevant to “memory” NK cells. Studies of murine NK cell responses to mouse cytomegalovirus (MCMV) revealed that virus-specific NK cells proliferate 100-fold in the spleen and 1000-fold in the liver after infection. After the initial expansion and contraction phases, a pool of self-renewing “memory” NK cells with the ability to rapidly respond to secondary viral challenges persists for several months (Sun et al., 2009). This adaptive NK cell population can develop in Ifng-deficient mice, but fails to develop in either Il12r or Stat4-deficient mice (Sun et al., 2012). The precise role of STAT4 in the generation of “memory” NK cells remains to be elucidated, but studies of memory formation in CD8+ T cells may provide some clues. STAT4 activation downstream of IL-12 promotes clonal expansion of antigen-specific CD8+ T cells by enhancing expression of the anti-apoptotic genes Bcl2 and Bcl-xL and increasing sensitivity to IL-7 and IL-15 (Li et al., 2006). Moreover, “memory” NK cells in mice vigorously produce IFN-γ and TNF-α in response to ITAM-receptor stimulation. The same phenomenon is observed in a subset of human NK cells with down-regulated FcεRγ-expression (Hwang et al., 2012). At present, data suggest that only educated cells proliferate in response to cytokines and signals from DAP12-coupled receptors (Figure 3). However, many open questions remain with respect to how NK cell function relates to development and the extent to which NK cell differentiation is linear vs. branched. The application of high-throughput sequencing technologies to study distinct subsets of NK cells could be very useful in elucidating the processes that regulate differentiation and the acquisition of function.
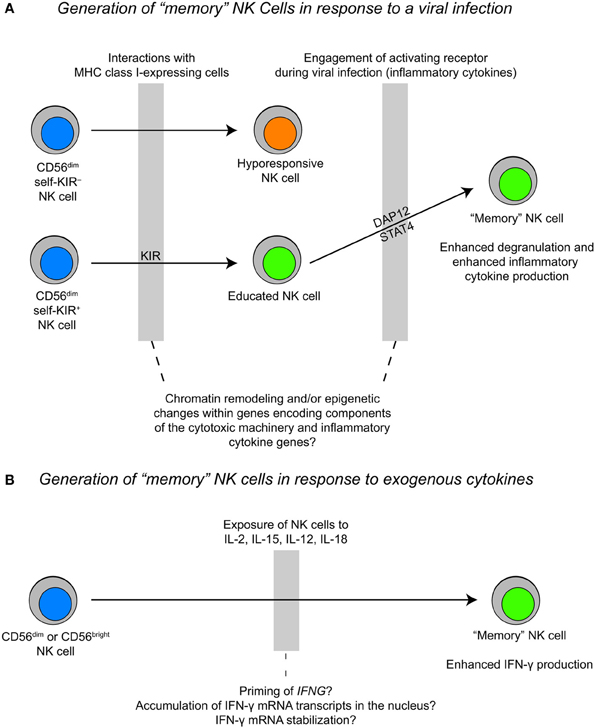
Figure 3. Proposed models for the generation of “memory” NK cells during a viral infection vs. cytokine priming. (A) The generation of “memory” NK cells during a viral infection likely happens within the context of NK cell education, where an “educated” NK cell expands during a viral infection and exhibits rapid cytotoxicity and inflammatory cytokine production upon a secondary challenge. (B) The generation of “memory” NK cells as a result of cytokine stimulation may result in epigenetic priming of the IFNG locus and/or accumulation of IFN-γ transcripts in the nucleus. This process may be fundamentally different from the generation of “memory” NK cells during a viral infection.
Also pertaining to stimulation induced changes in NK cell responsiveness, two recent reports described the generation of “memory” NK cells after in vitro stimulation with combinations of IL-12, IL-18, and IL-15 (Cooper et al., 2009; Romee et al., 2012). In both mouse and human systems, NK cells were primed with cytokines and restimulated at later time points. Cells that were primed demonstrated a more robust IFN-γ response without any enhancement in cytotoxicity upon restimulation with either exogenous cytokines or through activating receptors. These adaptive responses were identified primarily in human CD56bright NK cells and CD56dim NK cells lacking the expression of killer immunoglobulin-like receptors (KIR) and CD57, and might simply reflect epigenetic changes at the IFNG locus. An alternative explanation for these results is that the priming of NK cells with IL-12, IL-18, and IL-15 selectively affects the post-transcriptional regulation of IFN-γ mRNA. Previous studies have shown that co-stimulation of NK cells with IL-2 and IL-12 leads to enhanced mRNA stabilization (Ye et al., 1995). However, IL-12 stimulation is unique in that it causes an elevation in nuclear IFN-γ mRNA retention after transcriptional activity of the gene has ceased, which is likely directed by specific sequence elements within the IFN-γ mRNA transcript. The nuclear retention of IFN-γ mRNA transcripts can be overcome by co-stimulation with IL-2, which leads to transcription-independent transport of IFN-γ mRNA out of the nucleus and into the cytoplasm where it can be translated (Hodge et al., 2002). Enhanced cytokine responses in memory T cells correlate with changes in the chromatin state of cytokine genes. These changes are stable and can be inherited through cell divisions, forming the basis for memory. Future studies should be done to compare the epigenetic changes associated with “memory” NK cells generated from cytokine priming to “memory” NK cells generated from receptor engagement during a viral infection, which exhibit more robust IFN-γ responses and enhanced degranulation upon secondary challenge.
A deeper understanding of NK cell differentiation and the specialization of function will ultimately require high-resolution, genome-wide analyses. The development of the chromatin immunoprecipitation (ChIP)-seq technique, which combines ChIP and high-throughput sequencing methods, provides the opportunity to directly assess histone modifications and DNA-binding proteins. Combining ChIP-seq data with global gene expression results from techniques such as microarrays or RNA-seq makes it possible to produce a genome-wide view that links chromatin state with transcriptional activity within a defined population of cells. These technologies have been used extensively to study T cell differentiation and the chromatin states of genes that are selectively expressed in distinct populations of memory cells. Limited numbers relative to other lymphocyte subsets makes the application of high-throughput technologies to primary NK cells difficult, but improvements in reagents and sequencing sensitivity should soon overcome these obstacles. This will provide new views of how NK cells differentiate and undergo functional specialization.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the Swedish Research Council, Swedish Cancer Foundation, Swedish Children's Cancer Foundation, Åke Olsson Foundation for Hematological Research, Histiocytosis Association, Jeansson's Foundation, Åke Wiberg's Foundation, Clas Groschinsky's Memorial Fund, the Karolinska Institute Research Foundation, and the Frontiers in Biomedical Research Fellowship.
Aliahmad, P., de la Torre, B., and Kaye, J. (2010). Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue-inducer cell and NK cell lineages. Nat. Immunol. 11, 945–952.
Anfossi, N., André, P., Guia, S., Falk, C. S., Roetynck, S., Stewart, C. S., et al. (2006). Human NK cell eduation by inhibitory receptors for MHC class I. Immunity 25, 331–342.
Araki, Y., Fann, M., Wersto, R., and Weng, N. P. (2008). Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B). J. Immunol. 180, 8102–8108.
Babichuk, C. K., Duggan, B. L., and Bleackley, R. C. (1996). In vivo regulation of murine granzyme B gene transcription in activated primary T cells. J. Biol. Chem. 271, 16485–16493.
Barton, K., Muthusamy, N., Fischer, C., Ting, C. N., Walunas, T. L., Lanier, L. L., et al. (1998). The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity 9, 555–563.
Beziat, V., Dalgard, O., Asselah, T., Halfon, P., Bedossa, P., Boudifa, A., et al. (2012). CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur. J. Immunol. 42, 447–457.
Bjorkstrom, N. K., Lindgren, T., Stoltz, M., Fauriat, C., Braun, M., Evander, M., et al. (2011). Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus. J. Exp. Med. 208, 13–21.
Bjorkstrom, N. K., Riese, P., Heuts, F., Andersson, S., Fauriat, C., Ivarsson, M. A., et al. (2010). Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 116, 3853–3864.
Boos, M. D., Yokota, J., Eberl, G., and Kee, B. L. (2007). Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 204, 1119–1130.
Bream, J. H., Hodge, D. L., Gonsky, R., Spolski, R., Leonard, W. J., Krebs, S., et al. (2004). A distal region in the interferon-gamma gene is a site of epigenetic remodeling and transcriptional regulation by interleukin-2. J. Biol. Chem. 279, 41249–41257.
Bryceson, Y. T., Chiang, S. C. C., Darmanin, S., Schlums, H., Theorell, J. T., and Wood, S. M. (2011). Molecular mechanisms of NK cell activation. J. Innate Immun. 3, 216–226.
Cella, M., Fuchs, A., Vermi, W., Facchetti, F., Otero, K., Lennerz, J. K., et al. (2009). A human natural killer cell subset provides and innate source of IL-22 for mucosal immunity. Nature 457, 722–725.
Colucci, F., Samson, S. I., DeKoter, R. P., Lantz, O., Singh, H., and Di Santo, J. P. (2001). Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood 97, 2625–2632.
Cooper, M. A., Elliott, J. M., Keyel, P. A., Yang, L., Carrero, J. A., and Yokoyama, W. M. (2009). Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 1915–1919.
Crotty, S., Johnston, R. J., and Schoenberger, S. P. (2010). Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11, 114–120.
Debierre-Grockiego, F. (2004). Anti-apoptotic role of STAT5 in haematopoietic cells and in the pathogenesis of malignancies. Apoptosis 9, 717–728.
Dickinson, R. E., Griffin, H., Bigley, V., Reynard, L. N., Hussain, R., Haniffa, M., et al. (2011). Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 118, 2656–2658.
Eckelhart, E., Warsch, W., Zebedin, E., Simma, O., Stoiber, D., Kolbe, T., et al. (2011). A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood 117, 1565–1573.
Fauriat, C., Long, E. O., Ljunggren, H. G., and Bryceson, Y. T. (2010). Regulation of human NK cell cytokine and chemokine production by target cell recognition. Blood 115, 2167–2176.
Foley, B., Cooley, S., Verneris, M. R., Curtsinger, J., Luo, X., Waller, E. K., et al. (2011). NK cell education after allogeneic transplantation: dissociation between recovery of cytokine-producing and cytotoxic functions. Blood 118, 2784–2792.
Foley, B., Cooley, S., Verneris, M. R., Pitt, M., Curtsinger, J., Luo, X., et al. (2012). Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 119, 2665–2674.
Freud, A. G., Becknell, B., Roychowdhury, S., Mao, H. C., Ferketich, A. K., Nuovo, G. J., et al. (2005). A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity 22, 295–304.
Freud, A. G., and Caligiuri, M. A. (2006). Human natural killer cell development. Immunol. Rev. 214, 56–72.
Gascoyne, D. M., Long, E., Veiga-Fernandes, H., de Boer, J., Williams, O., Seddon, B., et al. (2009). The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10, 1118–1124.
Geller, M. A., and Miller, J. S. (2011). Use of allogeneic NK cells for cancer immunotherapy. Immunotherapy 3, 1445–1459.
Gineau, L., Cognet, C., Kara, N., Lach, F. P., Dunne, J., Veturi, U., et al. (2012). Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J. Clin. Invest. 122, 821–832.
Gordon, S. M., Chaix, J., Rupp, L. J., Wu, J., Madera, S., Sun, J. C., et al. (2012). The transcripiton factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67.
Greer, E. L., and Shi, Y. (2012). Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357.
Hatton, R. D., Harrington, L. E., Luther, R. J., Wakefield, T., Janowski, K. M., Oliver, J. R., et al. (2006). A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity 25, 717–729.
Hodge, D. L., Martinez, A., Julias, J. G., Taylor, L. S., and Young, H. A. (2002). Regulation of nuclear gamma interferon gene expression by interleukin 12 (IL-12) and iL-2 represents a novel form of posttranscriptional control. Mol. Cell. Biol. 22, 1742–1753.
Hsu, A. P., Sampaio, E. P., Khan, J., Calvo, K. R., Lemieux, J. E., Patel, S. Y., et al. (2011). Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 118, 2653–2655.
Huang, C., Bi, E., Hu, Y., Deng, W., Tian, Z., Dong, C., et al. (2006). A novel NF-kappaB binding site controls human granzyme B gene transcription. J. Immunol. 176, 4173–4181.
Hwang, I., Zhang, T., Scott, J. M., Kim, A. R., Lee, T., Kakarla, T., et al. (2012). Identification of human NK cells that are deficient for signaling adaptor FcRgamma and specialized for antibody-dependent immune functions. Int. Immunol. 24, 793–802.
Imada, K., Bloom, E. T., Nakajima, H., Horvath-Arcidiacono, J. A., Udy, G. B., Davey, H. W., et al. (1998). Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J. Exp. Med. 188, 2067–2074.
Juelke, K., Killig, M., Luetke-Eversloh, M., Parente, E., Gruen, J., Morandi, B., et al. (2010). CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood 116, 1299–1307.
Khavari, P. A., Peterson, C. L., Tamkun, J. W., Mendel, D. B., and Crabtree, G. R. (1993). BRG1 contains a conserved domain of the SWI/SNF2 family necessary for normal mitotic growth and transcription. Nature 366, 170–174.
Kim, S., Poursine-Laurent, J., Truscott, S. M., Lybarger, L., Song, Y. J., Yang, L., et al. (2005). Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436, 709–713.
Kofoed, E. M., Hwa, V., Little, B., Woods, K. A., Buckway, C. K., Tsubaki, J., et al. (2003). Growth hormone insensitivity associated with a STAT5b mutation. N. Engl. J. Med. 349, 1139–1147.
Koopman, L. A., Kopcow, H. D., Rybalov, B., Boyson, J. E., Orange, J. S., Schatz, F., et al. (2003). Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med. 198, 1201–1212.
Lanier, L. L. (2008). Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9, 495–502.
Li, Q., Eppolito, C., Odunsi, K., and Shrikant, P. A. (2006). IL-12-programmed long-term CD8+ T cell responses require STAT4. J. Immunol. 177, 7618–7625.
Lieberman, J. (2003). The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat. Rev. Immunol. 3, 361–370.
Lodoen, M. B., and Lanier, L. L. (2006). Natural killer cells as an initial defense against pathogens. Curr. Opin. Immunol. 18, 391–398.
Lohoff, M., Duncan, G. S., Ferrick, D., Mittrücker, H. W., Bischof, S., Prechtl, S., et al. (2000). Deficiency in the transcription factor interferon regulatory factor (IRF)-2 leads to severely compromosed development of natural killer and T helper type 1 cells. J. Exp. Med. 192, 325–336.
Lopez-Vergès, S., Milush, J. M., Schwartz, B. S., Pando, M. J., Jarjoura, J., York, V. A., et al. (2011). Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 14725–14732.
Luci, C., Reynders, A., Ivanov, I. I., Cognet, C., Chiche, L., Chasson, L., et al. (2009). Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 10, 75–82.
Matos, M. E., Schnier, G. S., Beecher, M. S., Ashman, L. K., William, D. E., and Caligiuri, M. A. (1993). Expression of a functional c-kit receptor on a subset of natural killer cells. J. Exp. Med. 178, 1079–1084.
Narlikar, G. J., Fan, H. Y., and Kingston, R. E. (2002). Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487.
Nyga, R., Pecquet, C., Harir, N., Gu, H., Dhennin-Duthille, I., Regnier, A., et al. (2005). Activated STAT5 proteins induce activation of the PI 3-kinase/Akt and Ras/MAPK pathways via the Gab2 scaffolding adapter. Biochem. J. 390, 359–366.
Ramirez-Carrozzi, V. R., Nazarian, A. A., Li, C. C., Gore, S. L., Sridharan, R., Imbalzano, A. N., et al. (2006). Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 20, 282–296.
Romee, R., Schneider, S. E., Leong, J. W., Chase, J. M., Keppel, C. R., Sullivan, R. P., et al. (2012). Cytokine activation induces human memory-like NK cells. Blood 120, 4751–4760.
Samson, S. I., Richard, O., Tavian, M., Ranson, T., Vosshenrich, C. A., Colucci, F., et al. (2003). GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity 19, 701–711.
Scott, E. W., Simon, M. C., Anastasi, J., and Singh, H. (1994). Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 264, 1573–1577.
Suganuma, T., and Workman, J. L. (2011). Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 80, 473–499.
Sun, J. C., Beilke, J. N., and Lanier, L. L. (2009). Adaptive immune features of natural killer cells. Nature 457, 557–561.
Sun, J. C., and Lanier, L. L. (2011). NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat. Rev. Immunol. 11, 645–657.
Sun, J. C., Madera, S., Bezman, N. A., Beilke, J. N., Kaplan, M. H., and Lanier, L. L. (2012). Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med. 209, 947–954.
Vesely, M. D., Kershaw, M. H., Schreiber, R. D., and Smyth, M. J. (2011). Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 29, 235–271.
Wargnier, A., Legros-Maida, S., Bosselut, R., Bourge, J. F., Lafaurie, C., Ghysdael, C. J., et al. (1995). Identification of human granzyme B promoter regulatory elements interacting with activated T-cell-specific proteins: implication of Ikaros and CBF binding sites in promoter activation. Proc. Natl. Acad. Sci. U.S.A. 92, 6930–6934.
Yamamoto, K., Shibata, F., Miyasaka, N., and Miura, O. (2002). The human perforin gene is a direct target of STAT4 activated by IL-12 in NK cells. Biochem. Biophys. Res. Commun. 297, 1245–1252.
Ye, J., Ortaldo, J. R., Conlon, K., Winkler-Pickett, R., and Young, H. A. (1995). Cellular and molecular mechanisms of IFN-gamma production induced by IL-2 and IL-12 in a human NK cell line. J. Leukoc. Biol. 58, 225–233.
Yu, C. R., Ortaldo, J. R., Curiel, R. E., Young, H. A., Anderson, S. K., and Gosselin, P. (1999). Role of a STAT binding site in the regulation of the human perforin promoter. J. Immunol. 162, 2785–2790.
Yu, J., Mao, H. C., Wei, M., Hughs, T., Zhang, J., Park, I. K., et al. (2010). CD94 surface density identifies a functional intermediary between CD56bright and CD56dim human NK-cell subsets. Blood 115, 274–281.
Zhang, F., and Boothby, M. (2006). T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J. Exp. Med. 203, 1493–1505.
Keywords: NK cell, epigenetics, transcription factors, development, memory
Citation: Cichocki F, Miller JS, Anderson SK and Bryceson YT (2013) Epigenetic regulation of NK cell differentiation and effector functions. Front. Immunol. 4:55. doi: 10.3389/fimmu.2013.00055
Received: 02 November 2012; Accepted: 11 February 2013;
Published online: 28 February 2013.
Edited by:
Hans-Gustaf Ljunggren, Karolinska Institute, SwedenReviewed by:
Christine S. Falk, Hannover Medical School MHH, GermanyCopyright © 2013 Cichocki, Miller, Anderson and Bryceson. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Frank Cichocki, Department of Medicine, Center for Infectious Medicine, F59, Karolinska Institute, Karolinska University Hospital, Huddinge, S-141 86 Stockholm, Sweden. e-mail:ZnJhbmsuY2ljaG9ja2lAa2kuc2U=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.