
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 21 May 2012
Sec. Microbial Immunology
Volume 3 - 2012 | https://doi.org/10.3389/fimmu.2012.00121
This article is part of the Research Topic Immunity to Infection with Leishmania: parasitism from an immunological perspective. View all 16 articles
Intense research efforts so far have not been sufficient to reduce leishmaniasis burden worldwide. This disease is transmitted by bites of infected sand flies, which inject saliva in the host skin in an attempt to obtain a blood meal. Sand fly saliva has an array of proteins with diverse pharmacological properties that modulates the host homeostatic and immune responses. Some of these proteins are also immunogenic and can induce both cellular and humoral immune responses. Recently, the use of sand fly salivary proteins to estimate exposure to sand fly bites and consequently the risk of infection has emerged. Here, we review evidence that supports the use of the host immune responses against sand fly salivary proteins to estimate risk of infection. We also discuss how the use of recombinant salivary proteins can optimize serological surveys and provide guidance for the implementation of specific measures for disease control in endemic areas.
Leishmaniasis is caused by infection with Leishmania parasites transmitted by bites of infected sand flies. The infection can result in a wide range of clinical manifestations varying from self-healing localized skin lesions to lethal visceral disease, and major determinants of the clinical outcome rely on the parasite strain and the host immune response (Murray et al., 2005). Despite ample basic and applied research, there is no effective vaccine to prevent leishmaniasis. As a consequence, the prophylactic strategies proposed by public health authorities are restricted mainly to vector control and consistent screening and elimination of potential reservoirs. In this scenario, understanding the nuances of the host–vector–parasite interactions becomes critical for the development of more reliable tools to adequately control leishmaniasis.
A critical event in Leishmania transmission is the sand fly bite. Female sand flies require hematophagy for nutrition, egg development, and survival. During blood feeding, sand fly saliva containing a number of pharmacologically active molecules with diverse effects on the host’s hemostatic responses is delivered into the host skin (Andrade et al., 2005). There is strong evidence that components of the sand fly saliva play a major role driving both susceptibility to Leishmania infection and disease severity (Titus and Ribeiro, 1988; Belkaid et al., 1998; Morris et al., 2001; de Moura et al., 2007). Indeed, sand fly saliva can exacerbate lesions in experimental models of cutaneous leishmaniasis (CL) and this effect is considered, at least in part, a consequence of its immunomodulatory properties. Salivary proteins are also immunogenic and can elicit specific immune responses that can be detrimental for Leishmania establishment (Kamhawi, 2000; Valenzuela et al., 2001; Thiakaki et al., 2005; de Moura et al., 2007; Gomes et al., 2008; Oliveira et al., 2008; Collin et al., 2009; Rohousova et al., 2011; Tavares et al., 2011; Xu et al., 2011).
The idea of using antibodies against saliva from bloodsucking arthropods as markers of exposure has been proposed for different arthropod vectors. There is extensive work showing that humans and other vertebrates can develop antibodies against salivary components of different bloodsucking vectors like mosquitoes (Brummer-Korvenkontio et al., 1994; Palosuo et al., 1997; Remoue et al., 2006; Orlandi-Pradines et al., 2007; Andrade et al., 2009), ticks (Schwartz et al., 1990, 1991; Sanders et al., 1998), and triatomines (Volf et al., 1993; Nascimento et al., 2001; Schwarz et al., 2009, 2010, 2011). Specific antibodies and their dynamics in vertebrate hosts were described also against fleas and louse (Volf, 1991). As expected, exposure to sand fly bites or saliva also induces antibody production in humans and animal models (Barral et al., 2000; Volf and Rohousova, 2001; Gomes et al., 2002, 2007, 2008; Rohousova et al., 2005; Silva et al., 2005; Clements et al., 2010; Souza et al., 2010; Teixeira et al., 2010; Vlkova et al., 2011). In mice, the antibody isotype most induced by sand fly saliva is IgG1 (Silva et al., 2005) whereas IgG1, IgG2, IgG4, and IgE are more frequent in humans (Vinhas et al., 2007; Marzouki et al., 2011). Specific IgG1 and IgG2 isotypes are also highly induced in dogs bitten by sand flies (Hostomska et al., 2008; Vlkova et al., 2011). Thus, these antibody isotypes are obvious candidates for use as targets in serological surveys in endemic areas. Interestingly, two major patterns of clinical and serological responses to sand fly saliva are identified in human volunteers repeatedly exposed to Lutzomyia longipalpis, the vector of visceral leishmaniasis (VL) in South America (Vinhas et al., 2007). Individuals who develop delayed-type hypersensitivity (DTH)-like skin reactions after exposure usually display higher IgG/IgE ratios than those evolving mild erythematous reactions (Vinhas et al., 2007; Figure 1). These observations argue that the host response to sand fly saliva may present some degree of divergence resulting from genetic variations that could influence susceptibility to Leishmania establishment. Furthermore, characterization of the humoral response against saliva can provide evidence regarding susceptibility to Leishmania infection in humans. In this case, characterizing the antibody profile of an exposed individual may be useful in predicting susceptibility to disease. Whether these immunological aspects are reproducible in individuals exposed to other species of sand flies, including those that transmit the cutaneous form of the disease is still unknown.
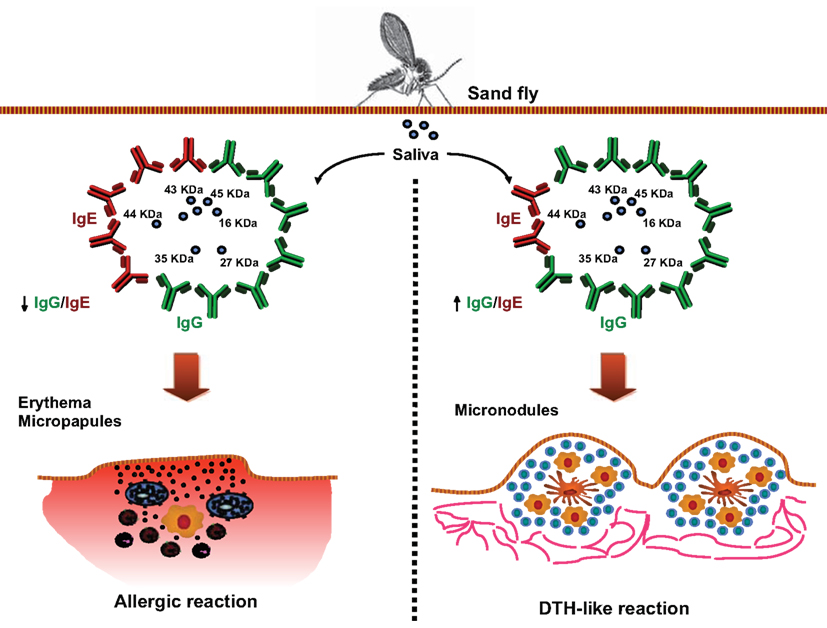
Figure 1. Associations between the pattern of host humoral responses against saliva from L. longipalpis and skin reactions upon exposure to bites. Human anti-saliva responses following exposure to uninfected L. longipalpis sand flies, using an in vivo model in which normal volunteers were exposed four times to 30 laboratory-reared Lutzomyia longipalpis (Vinhas et al., 2007). Following third exposure, volunteers developed diverse dermatological reactions at the site of insect bite. Serum from volunteers displayed high levels of IgG1, IgG4, and IgE anti-saliva and recognized several salivary gland proteins. Interestingly, volunteers who developed immediate skin reactions (left panel) presented higher IgE levels and lower IgG levels than those with delayed skin responses (right panel), as demonstrated by the IgG/IgE ratio. Whether this variability in the host responses against saliva is associated with different degree of susceptibility to Leishmania infection is still unknown.
Several field studies in highly endemic areas for leishmaniasis indicate that natural exposure to non-infected sand fly bites can impact the epidemiology of this disease (Barral et al., 2000; Gomes et al., 2002; Rohousova et al., 2005; de Moura et al., 2007; Aquino et al., 2010; Marzouki et al., 2011; Vlkova et al., 2011). In these regions, residents are frequently bitten by both non-infected and infected sand flies. It is hypothesized that these frequently exposed individuals usually present mild and chronic disease manifestations while travelers from non-endemic areas, not previously exposed to sand flies or Leishmania, present a higher risk of developing severe clinical forms of leishmaniasis. The idea is that frequent exposure to sand fly bites leads to the production of neutralizing antibodies against salivary proteins and also to activation of cellular mechanisms that may have an adverse effect on Leishmania establishment. In this perspective, characterization of immune responses against sand fly saliva can help estimate both risk of infection and to some degree anti-parasite immunity. Although this hypothesis has been proven in animal models, additional large-scale clinical studies are necessary to validate it in humans. Individuals living in leishmaniasis endemic areas that are exposed to bites from uninfected sand flies over a long term still get infected and develop disease. Interestingly, protection against leishmaniasis in mice has been recently shown to be limited to short-term exposure to sand flies immediately before infection (Rohousova et al., 2011). This finding may explain the persistence of leishmaniasis in endemic areas, as individuals might experience periods without exposure to sand fly bites. Moreover, other factors besides exposure to sand fly bites might also be driving susceptibility to infection and disease severity in human leishmaniasis and the interaction between these determinants are still unknown. The characterization of the host’s immune responses against sand fly saliva can lead to the identification of key salivary proteins with at least two remarkable potentials: (i) its use as surrogates for exposure to sand fly bites and the risk of infection and (ii) the identification of candidate vaccines against leishmaniasis. In fact, estimation of exposure to sand fly bites to infer risk of infection has emerged as an important tool to help drive strategic interventions to reduce disease transmission in endemic areas. Here, we review the current advances in the use of sand fly saliva as a helpful tool to estimate exposure to vector bites and risk for leishmaniasis. We also summarize the development of reliable tools to assess vector-exposure in endemic areas.
Visceral leishmaniasis in Latin America is transmitted by the bite of L. longipalpis sand flies infected with Leishmania infantum chagasi. One of the first reports showing the immunogenicity of sand fly saliva in exposed individuals was carried out in a highly endemic VL area in Brazil, where inhabitants differentially recognize salivary gland proteins (Barral et al., 2000). Interestingly, a positive correlation was observed between recognition of salivary proteins and anti-Leishmania DTH reaction to parasite antigens and protection, but not to anti-Leishmania serologic status. Furthermore, children who experienced anti-Leishmania DTH conversion had an increase in anti-L. longipalpis saliva antibodies, suggesting that development of anti-parasite DTH coincides and is directly related to the development of anti-saliva antibodies (Gomes et al., 2002). More recently, in the same endemic area, the rate of a positive anti-Leishmania DTH was higher in individuals that had anti-L. longipalpis antibodies at the baseline of a 2-year follow-up study (Aquino et al., 2010). On the other hand, these results could simply suggest that DTH is also dependent on parasite challenges which was not specifically measured in either studies. Other factors that might contribute to the susceptibility to infection in endemic areas such as vector density and reservoir infection rates still need to be addressed.
The presence of antibodies to sand fly salivary proteins was also demonstrated in canids (dogs and foxes), important reservoirs that serve as a source of parasites to sand flies in endemic regions. In dogs experimentally exposed to L. longipalpis, the induced anti-saliva antibodies can recognize up to six different sand fly salivary proteins (Hostomska et al., 2008). Comparable results were also found in naturally exposed animals captured in a highly endemic area of VL in Brazil. Anti-L. longipalpis saliva antibodies were detected in naturally infected dogs and foxes, suggesting that these animals were in close contact with the vector (Bahia et al., 2007; Gomes et al., 2007). In a follow-up study of dogs experimentally exposed to Phlebotomus perniciosus, the main vector of canine VL in Europe, a positive correlation between the production of specific antibodies against vector saliva and the number of blood fed sand flies was shown. Notably, dogs naturally exposed to bites during the sand fly season that tested positive for Leishmania had decreased systemic levels of anti-P. perniciosus saliva IgG2 antibodies compared to healthy dogs (Vlkova et al., 2011). Such studies suggest that the detection of anti-saliva antibodies can be valuable to evaluate vector control programs. Indeed, in a recent study in VL areas in India and Nepal, the efficacy of insecticide treated nets (ITN) was evaluated in terms of vector-exposure by measuring anti-P. argentipes saliva antibody concentrations in the inhabitants before and after the intervention. This approach showed that exposure to P. argentipes was reduced by up to 12% in people living in villages where nets were used compared to controls (Clements et al., 2010; Gidwani et al., 2011). In summary, both observational and interventional field studies in VL areas strongly suggest that the production of antibodies to specific sand fly salivary proteins could be directly associated to correlates of protection.
Cutaneous leishmaniasis is transmitted by the bite of a number of sand flies species with diverse geographic distributions. There is also evidence suggesting that individuals that live in endemic areas for CL also develop specific antibodies to saliva of sand fly species that transmit the disease.
In an endemic area for Leishmania tropica in Turkey, the antibody response against saliva of P. sergenti (the vector of L. tropica) and P. papatasi (the vector of L. major elsewhere) was compared between healthy individuals and patients with active CL lesions (Rohousova et al., 2005). It was observed that approximately 40% of the study population carried IgG antibodies to the saliva of both sand fly species. Interestingly, individuals with active lesions had higher anti-P. sergenti IgG levels compared to healthy individuals, strongly suggesting that production of anti-saliva antibodies could be used as a marker for risk of developing CL (Rohousova et al., 2005). In a recent study performed in Tunisia, sera from children living in a CL endemic area of L. major showed a consistent production of anti-P. papatasi saliva antibodies after two transmission seasons. Anti-saliva IgG antibodies were significantly higher and correlated with lesion development, arguing that anti-saliva antibodies were strongly associated with the disease development (Marzouki et al., 2011). A similar finding was described for Lutzomyia intermedia, an important vector of CL in South America (de Moura et al., 2007). Residents from an endemic area in Brazil presenting active CL displayed higher titers of antibodies against L. intermedia saliva compared to healthy individuals from the same area, indicating that in CL a humoral immune response to L. intermedia saliva is a marker of risk of disease (de Moura et al., 2007). Similarly to the studies performed in VL areas, these conclusions need to take into account other important determinants such as percentage of infected vectors, the biting rate and the presence of potential reservoirs in the area that could also influence the outcome of the host’s immune response.
These observations indicate that the detection of specific humoral immune response against salivary proteins from different species of sand flies should be carefully evaluated. Altogether, these findings indicate that exposure to saliva of distinct species of sand flies can be correlated with two different outcomes: susceptibility to disease or protection. In VL, the findings were observed in cross-sectional and longitudinal prospective studies showing that the development of both types of immunity occurs simultaneously (Barral et al., 2000; Gomes et al., 2002; Aquino et al., 2010). In CL, however, the development of anti-saliva antibodies and susceptibility to disease are closely related and recent work shows an association with a Th2 phenotype (Rohousova et al., 2005; de Moura et al., 2007; Marzouki et al., 2011). Although production of antibodies against vector saliva suggests an increased chance of encountering an infected bite and developing disease, constant exposure to non-infected bites in endemic areas could possibly modulate the host’s immune response to affect a successful settlement of the parasite. Whereas production of antibodies against sand fly saliva can be used as biomarker of disease susceptibility more studies are required to identify salivary proteins that can affect the development of anti-Leishmania immunity.
The work done so far with detection of anti-sand fly saliva antibodies have shown enough evidence to prove the relevance of using salivary proteins as tools to address the level of exposure to vector bites bringing a new perspective to the field. Although the use of antibody response to sand fly saliva is promising, an approach in which a large number of samples from different vertebrate hosts could be quickly and specifically tested is highly desired. The majority of work evaluating saliva of sand flies as potential biomarkers of vector-exposure used whole salivary extracts. While whole salivary extract has advantages as it represents the complete repertoire of crude secreted salivary proteins, it also has limitations that could restrict their application. Technical limitations to the use of large quantities of whole salivary extracts in population surveys include restrictions on the number of sand flies available for salivary gland dissection, homogeneity of the salivary content, and conservation of the proteins without degradation. These limitations impact the size and extent of field studies. Additionally, the use of total saliva reduces the specificity of detection due to a higher likelihood of cross-reactivity with saliva from other sympatric non-vector sand fly species. For instance, salivary antigens do not cross-react between L. longipalpis and Phlebotomus species but some cross-reactions were demonstrated between diverse Phlebotomus species in both mice (Volf and Rohousova, 2001) and humans (Rohousova et al., 2005). Hence, the identification of specific and highly immunogenic salivary proteins that can also be produced in large quantities as quality-controlled recombinant proteins is of utmost importance for larger epidemiological studies.
The first attempt to identify, produce, and test salivary recombinant proteins for serological surveys was recently demonstrated for L. longipalpis sand flies (Souza et al., 2010; Teixeira et al., 2010). These work describe a practical and functional approach for identification of salivary proteins most recognized by sera from humans and canids residing in endemic areas for VL in Brazil and, more importantly, how to obtain highly pure and immunogenic recombinant proteins. Initially, sera from humans, dogs, and foxes from VL endemic areas were screened against the whole salivary extract from L. longipalpis. The most recognized proteins were in the range of 15–65 kDa. Based on a L. longipalpis salivary transcriptome, nine transcripts that corresponded to proteins within the predicted molecular weight in the desired range were selected as potential candidates. To characterize and confirm the antigenicity of the candidates, each recombinant salivary protein was produced in a mammalian expression system, purified, and tested individually for their recognition by sera from individuals residing in a VL endemic area. Two of the nine salivary recombinant proteins, LJM11 and LJM17, were highly recognized by humans exposed to L. longipalpis, demonstrating their potential as markers for epidemiological studies. To verify their specificity, the two recombinant proteins were further tested against sera from individuals exposed to L. intermedia. Both recombinant proteins were only recognized by sera from individuals exposed to L. longipalpis but not to L. intermedia bites, confirming their potential as specific markers for L. longipalpis exposure. Finally, to validate LJM11 and LJM17 as markers for exposure, they were tested in a large cohort with over a thousand people from a VL area (Souza et al., 2010). Both recombinant proteins (LJM11 and LJM17) were assessed individually and in combination and were able to detect anti-saliva seroconversion but the use of a combination of these two proteins showed a more powerful discrimination equal to the seroconversion observed against whole saliva extract. A summary of the approach used in these two studies is illustrated in Figure 2. Notably, this method used for identification of salivary proteins applied in ELISA assays could also be assessed to identify potential markers from the saliva of other arthropod vectors. The use of ELISA has shown to be a practical tool in different studies where whole vector’s saliva was used. In several epidemiological studies, this approach has been used to determine the levels of specific anti-vector saliva antibodies including Ixodes pacificus, P. argentipes, Aedes caspius, and Aedes albopictus (Lane et al., 1999; Clements et al., 2010; Fontaine et al., 2011; Gidwani et al., 2011; Vlkova et al., 2011; Doucoure et al., 2012), clearly demonstrating its potential to support control strategies programs for vector-borne diseases.
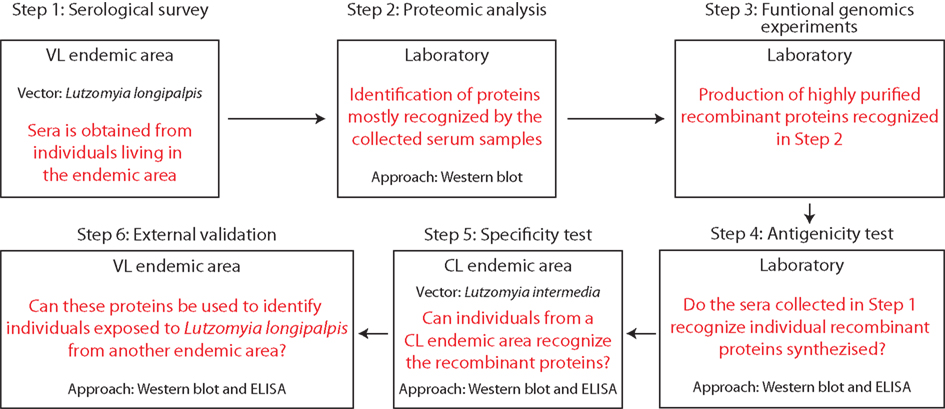
Figure 2. Key steps to implement the use of recombinant proteins from sand fly saliva in serological surveys to estimate exposure to bites and risk of Leishmania infection.
Research focused in the interplay between vector saliva and host immune responses has been very successful in the identification of molecules with diverse biological and pharmacological activities and has contributed impressively to the understanding of the immunopathogenesis of an array of vector-borne diseases. Regarding leishmaniasis, different groups have shown that sand fly saliva plays a critical role in the establishment of Leishmania infection and have also described host immune responses against salivary proteins in mice, dogs, and humans. Interest in the field is empowered by recent observations arguing that it is possible to track human exposure to vectors using salivary recombinant proteins. The use of recombinant proteins in serological surveys performed in leishmaniasis endemic areas is critical to standardize the quality and reproducibility of results from different studies and also to optimize procedures (Figure 3). Unfortunately, although extensive data regarding the quantification of host antibody responses against sand fly saliva has been published, few studies provide validation of their results using large cohorts or multi-center approaches. Moreover, multi-center approaches are also critical to identify salivary proteins that have conserved expression in different endemic areas, as variation in salivary content among the same species has been described in different geographical regions (Lanzaro et al., 1999). An expanded effort for studying salivary content of species from different parts of the world will certainly increase the chances to find common molecules that could function as markers or as candidates for a wide-ranging vaccine. This approach is critical as specificity to saliva-mediated protection against Leishmania infection was demonstrated in the mouse model arguing that immunization with salivary components from one species of sand fly can be ineffective to induce protection against other species (Thiakaki et al., 2005), This finding also suggests that immunity against Leishmania is elicited by unique salivary antigens and not conserved molecules between various species of sand flies. Definitively, coordinated initiatives are necessary to design robust validation studies toward generating standardized guidelines. Ultimately, the use of sand fly salivary proteins in epidemiological studies can provide a fundamental tool to help public health authorities in planning key strategies to reduce the burden of leishmaniasis.
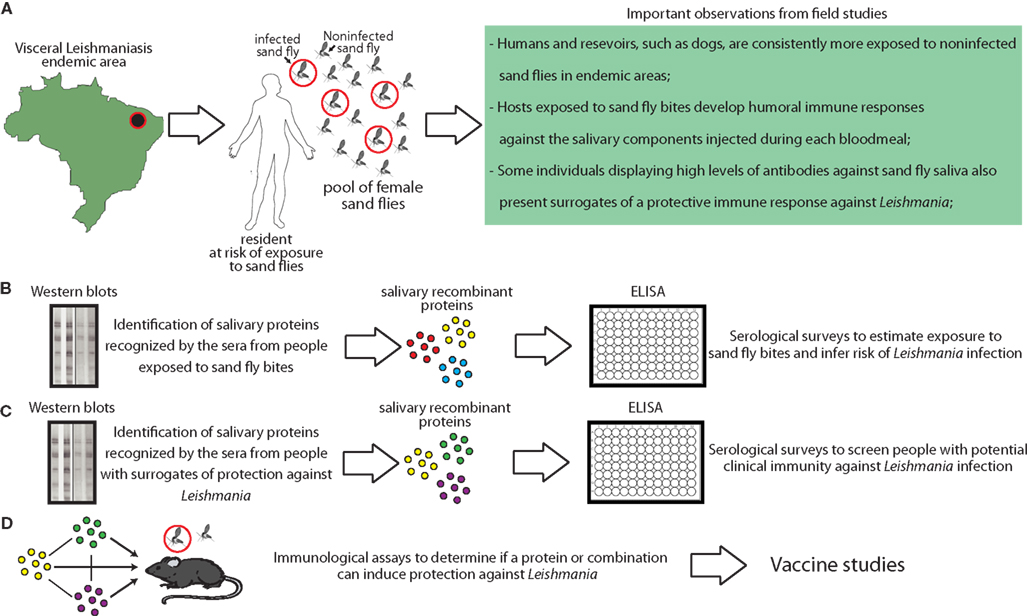
Figure 3. Combining clinical studies with basic proteomic research to generate tools based on sand fly salivary proteins to aid the control of visceral leishmaniasis. (A) Epidemiological and entomological studies have provided critical knowledge to guide development of advanced research studies using sand fly saliva. Proteomic and transcriptome studies made possible to identify and isolate the salivary proteins that are immunogenic and elicit host antibody responses. In addition, we have recently shown an approach to produce large amounts of recombinant versions of these proteins in large-scale (Teixeira et al., 2010). (B) Some proteins are globally identified by the host humoral responses. Such proteins can be used in serological surveys to estimate exposure to sand fly bites and an overall risk to become infected in endemic areas. (C) Other salivary proteins are more frequently recognized by antibodies from people presenting surrogates of clinical immunity against leishmaniasis, such as delayed-type hypersensitivity againstLeishmania antigens. These proteins can also be used in serological surveys in order to identify people with potential protection against infection (Souza et al., 2010). (D) Ultimately, recombinant proteins can be used in experimental models to define potential use in vaccination strategies against leishmaniasis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The National Institutes of Health, NIAID, and Intramural Research Program support the work developed by the authors. We thank Dr. Jesus Valenzuela, Dr. Shaden Kamhawi, and Dr. Luiz Fabiano Oliveira for critical revision of this manuscript.
Andrade, B. B., Rocha, B. C., Reis-Filho, A., Camargo, L. M., Tadei, W. P., Moreira, L. A., Barral, A., and Barral-Netto, M. (2009). Anti-Anopheles darlingi saliva antibodies as marker of Plasmodium vivax infection and clinical immunity in the Brazilian Amazon. Malar. J. 8, 121.
Andrade, B. B., Teixeira, C. R., Barral, A., and Barral-Netto, M. (2005). Haematophagous arthropod saliva and host defense system: a tale of tear and blood. An. Acad. Bras. Cienc. 77, 665–693.
Aquino, D. M., Caldas, A. J., Miranda, J. C., Silva, A. A., Barral-Netto, M., and Barral, A. (2010). Epidemiological study of the association between anti-Lutzomyia longipalpis saliva antibodies and development of delayed-type hypersensitivity to Leishmania antigen. Am. J. Trop. Med. Hyg. 83, 825–827.
Bahia, D., Gontijo, N. F., Leon, I. R., Perales, J., Pereira, M. H., Oliveira, G., Correa-Oliveira, R., and Reis, A. B. (2007). Antibodies from dogs with canine visceral leishmaniasis recognise two proteins from the saliva of Lutzomyia longipalpis. Parasitol. Res. 100, 449–454.
Barral, A., Honda, E., Caldas, A., Costa, J., Vinhas, V., Rowton, E. D., Valenzuela, J. G., Charlab, R., Barral-Netto, M., and Ribeiro, J. M. (2000). Human immune response to sand fly salivary gland antigens: a useful epidemiological marker? Am. J. Trop. Med. Hyg. 62, 740–745.
Belkaid, Y., Kamhawi, S., Modi, G., Valenzuela, J., Noben-Trauth, N., Rowton, E., Ribeiro, J., and Sacks, D. L. (1998). Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J. Exp. Med. 188, 1941–1953.
Brummer-Korvenkontio, H., Lappalainen, P., Reunala, T., and Palosuo, T. (1994). Detection of mosquito saliva-specific IgE and IgG4 antibodies by immunoblotting. J. Allergy Clin. Immunol. 93, 551–555.
Clements, M. F., Gidwani, K., Kumar, R., Hostomska, J., Dinesh, D. S., Kumar, V., Das, P., Muller, I., Hamilton, G., Volfova, V., Boelaert, M., Das, M., Rijal, S., Picado, A., Volf, P., Sundar, S., Davies, C. R., and Rogers, M. E. (2010). Measurement of recent exposure to Phlebotomus argentipes, the vector of Indian visceral leishmaniasis, by using human antibody responses to sand fly saliva. Am. J. Trop. Med. Hyg. 82, 801–807.
Collin, N., Gomes, R., Teixeira, C., Cheng, L., Laughinghouse, A., Ward, J. M., Elnaiem, D. E., Fischer, L., Valenzuela, J. G., and Kamhawi, S. (2009). Sand fly salivary proteins induce strong cellular immunity in a natural reservoir of visceral leishmaniasis with adverse consequences for Leishmania. PLoS Pathog. 5, e1000441. doi:10.1371/journal.ppat.1000441
de Moura, T. R., Oliveira, F., Novais, F. O., Miranda, J. C., Clarencio, J., Follador, I., Carvalho, E. M., Valenzuela, J. G., Barral-Netto, M., Barral, A., Brodskyn, C., and De Oliveira, C. I. (2007). Enhanced Leishmania braziliensis infection following pre-exposure to sandfly saliva. PLoS Negl. Trop. Dis. 1, e84. doi:10.1371/journal.pntd.0000084
Doucoure, S., Mouchet, F., Cornelie, S., Dehecq, J. S., Rutee, A. H., Roca, Y., Walter, A., Herve, J. P., Misse, D., Favier, F., Gasque, P., and Remoue, F. (2012). Evaluation of the human IgG antibody response to Aedes albopictus saliva as a new specific biomarker of exposure to vector bites. PLoS Negl. Trop. Dis. 6, e1487. doi:10.1371/journal.pntd.0001487
Fontaine, A., Pascual, A., Orlandi-Pradines, E., Diouf, I., Remoue, F., Pages, F., Fusai, T., Rogier, C., and Almeras, L. (2011). Relationship between exposure to vector bites and antibody responses to mosquito salivary gland extracts. PLoS ONE 6, e29107. doi:10.1371/journal.pone.0029107
Gidwani, K., Picado, A., Rijal, S., Singh, S. P., Roy, L., Volfova, V., Andersen, E. W., Uranw, S., Ostyn, B., Sudarshan, M., Chakravarty, J., Volf, P., Sundar, S., Boelaert, M., and Rogers, M. E. (2011). Serological markers of sand fly exposure to evaluate insecticidal nets against visceral leishmaniasis in India and Nepal: a cluster-randomized trial. PLoS Negl. Trop. Dis. 5, e1296. doi:10.1371/journal.pntd.0001296
Gomes, R., Teixeira, C., Teixeira, M. J., Oliveira, F., Menezes, M. J., Silva, C., De Oliveira, C. I., Miranda, J. C., Elnaiem, D. E., Kamhawi, S., Valenzuela, J. G., and Brodskyn, C. I. (2008). Immunity to a salivary protein of a sand fly vector protects against the fatal outcome of visceral leishmaniasis in a hamster model. Proc. Natl. Acad. Sci. U.S.A. 105, 7845–7850.
Gomes, R. B., Brodskyn, C., De Oliveira, C. I., Costa, J., Miranda, J. C., Caldas, A., Valenzuela, J. G., Barral-Netto, M., and Barral, A. (2002). Seroconversion against Lutzomyia longipalpis saliva concurrent with the development of anti-Leishmania chagasi delayed-type hypersensitivity. J. Infect. Dis. 186, 1530–1534.
Gomes, R. B., Mendonca, I. L., Silva, V. C., Ruas, J., Silva, M. B., Cruz, M. S., Barral, A., and Costa, C. H. (2007). Antibodies against Lutzomyia longipalpis saliva in the fox Cerdocyon thous and the sylvatic cycle of Leishmania chagasi. Trans. R. Soc. Trop. Med. Hyg. 101, 127–133.
Hostomska, J., Rohousova, I., Volfova, V., Stanneck, D., Mencke, N., and Volf, P. (2008). Kinetics of canine antibody response to saliva of the sand fly Lutzomyia longipalpis. Vector Borne Zoonotic Dis. 8, 443–450.
Kamhawi, S. (2000). The biological and immunomodulatory properties of sand fly saliva and its role in the establishment of Leishmania infections. Microbes Infect. 2, 1765–1773.
Lane, R. S., Moss, R. B., Hsu, Y. P., Wei, T., Mesirow, M. L., and Kuo, M. M. (1999). Anti-arthropod saliva antibodies among residents of a community at high risk for Lyme disease in California. Am. J. Trop. Med. Hyg. 61, 850–859.
Lanzaro, G. C., Lopes, A. H., Ribeiro, J. M., Shoemaker, C. B., Warburg, A., Soares, M., and Titus, R. G. (1999). Variation in the salivary peptide, maxadilan, from species in the Lutzomyia longipalpis complex. Insect Mol. Biol. 8, 267–275.
Marzouki, S., Ben Ahmed, M., Boussoffara, T., Abdeladhim, M., Ben Aleya-Bouafif, N., Namane, A., Hamida, N. B., Ben Salah, A., and Louzir, H. (2011). Characterization of the antibody response to the saliva of Phlebotomus papatasi in people living in endemic areas of cutaneous leishmaniasis. Am. J. Trop. Med. Hyg. 84, 653–661.
Morris, R. V., Shoemaker, C. B., David, J. R., Lanzaro, G. C., and Titus, R. G. (2001). Sandfly maxadilan exacerbates infection with Leishmania major and vaccinating against it protects against L. major infection. J. Immunol. 167, 5226–5230.
Murray, H. W., Berman, J. D., Davies, C. R., and Saravia, N. G. (2005). Advances in leishmaniasis. Lancet 366, 1561–1577.
Nascimento, R. J., Santana, J. M., Lozzi, S. P., Araujo, C. N., and Teixeira, A. R. (2001). Human IgG1 and IgG4: the main antibodies against Triatoma infestans (Hemiptera: Reduviidae) salivary gland proteins. Am. J. Trop. Med. Hyg. 65, 219–226. doi:10.1371/journal.pntd.0000226
Oliveira, F., Lawyer, P. G., Kamhawi, S., and Valenzuela, J. G. (2008). Immunity to distinct sand fly salivary proteins primes the anti-Leishmania immune response towards protection or exacerbation of disease. PLoS Negl. Trop. Dis. 2, e226.
Orlandi-Pradines, E., Almeras, L., Denis De Senneville, L., Barbe, S., Remoue, F., Villard, C., Cornelie, S., Penhoat, K., Pascual, A., Bourgouin, C., Fontenille, D., Bonnet, J., Corre-Catelin, N., Reiter, P., Pages, F., Laffite, D., Boulanger, D., Simondon, F., Pradines, B., Fusai, T., and Rogier, C. (2007). Antibody response against saliva antigens of Anopheles gambiae and Aedes aegypti in travellers in tropical Africa. Microbes Infect. 9, 1454–1462.
Palosuo, K., Brummer-Korvenkontio, H., Mikkola, J., Sahi, T., and Reunala, T. (1997). Seasonal increase in human IgE and IgG4 antisaliva antibodies to Aedes mosquito bites. Int. Arch. Allergy Immunol. 114, 367–372.
Remoue, F., Cisse, B., Ba, F., Sokhna, C., Herve, J. P., Boulanger, D., and Simondon, F. (2006). Evaluation of the antibody response to Anopheles salivary antigens as a potential marker of risk of malaria. Trans. R. Soc. Trop. Med. Hyg. 100, 363–370.
Rohousova, I., Hostomska, J., Vlkova, M., Kobets, T., Lipoldova, M., and Volf, P. (2011). The protective effect against Leishmania infection conferred by sand fly bites is limited to short-term exposure. Int. J. Parasitol. 41, 481–485.
Rohousova, I., Ozensoy, S., Ozbel, Y., and Volf, P. (2005). Detection of species-specific antibody response of humans and mice bitten by sand flies. Parasitology 130, 493–499.
Sanders, M. L., Jaworski, D. C., Sanchez, J. L., Defraites, R. F., Glass, G. E., Scott, A. L., Raha, S., Ritchie, B. C., Needham, G. R., and Schwartz, B. S. (1998). Antibody to a cDNA-derived calreticulin protein from Amblyomma americanum as a biomarker of tick exposure in humans. Am. J. Trop. Med. Hyg. 59, 279–285.
Schwartz, B. S., Ford, D. P., Childs, J. E., Rothman, N., and Thomas, R. J. (1991). Anti-tick saliva antibody: a biologic marker of tick exposure that is a risk factor for Lyme disease seropositivity. Am. J. Epidemiol. 134, 86–95.
Schwartz, B. S., Ribeiro, J. M., and Goldstein, M. D. (1990). Anti-tick antibodies: an epidemiologic tool in Lyme disease research. Am. J. Epidemiol. 132, 58–66.
Schwarz, A., Helling, S., Collin, N., Teixeira, C. R., Medrano-Mercado, N., Hume, J. C., Assumpcao, T. C., Marcus, K., Stephan, C., Meyer, H. E., Ribeiro, J. M., Billingsley, P. F., Valenzuela, J. G., Sternberg, J. M., and Schaub, G. A. (2009). Immunogenic salivary proteins of Triatoma infestans: development of a recombinant antigen for the detection of low-level infestation of triatomines. PLoS Negl. Trop. Dis. 3, e532. doi:10.1371/journal.pntd.0000532
Schwarz, A., Juarez, J. A., Richards, J., Rath, B., Machaca, V. Q., Castro, Y. E., Malaga, E. S., Levy, K., Gilman, R. H., Bern, C., Verastegui, M., and Levy, M. Z. (2011). Anti-triatomine saliva immunoassays for the evaluation of impregnated netting trials against Chagas disease transmission. Int. J. Parasitol. 41, 591–594.
Schwarz, A., Medrano-Mercado, N., Billingsley, P. F., Schaub, G. A., and Sternberg, J. M. (2010). IgM-antibody responses of chickens to salivary antigens of Triatoma infestans as early biomarkers for low-level infestation of triatomines. Int. J. Parasitol. 40, 1295–1302.
Silva, F., Gomes, R., Prates, D., Miranda, J. C., Andrade, B., Barral-Netto, M., and Barral, A. (2005). Inflammatory cell infiltration and high antibody production in BALB/c mice caused by natural exposure to Lutzomyia longipalpis bites. Am. J. Trop. Med. Hyg. 72, 94–98.
Souza, A. P., Andrade, B. B., Aquino, D., Entringer, P., Miranda, J. C., Alcantara, R., Ruiz, D., Soto, M., Teixeira, C. R., Valenzuela, J. G., De Oliveira, C. I., Brodskyn, C. I., Barral-Netto, M., and Barral, A. (2010). Using recombinant proteins from Lutzomyia longipalpis saliva to estimate human vector exposure in visceral leishmaniasis endemic areas. PLoS Negl. Trop. Dis. 4, e649. doi:10.1371/journal.pntd.0000649
Tavares, N. M., Silva, R. A., Costa, D. J., Pitombo, M. A., Fukutani, K. F., Miranda, J. C., Valenzuela, J. G., Barral, A., De Oliveira, C. I., Barral-Netto, M., and Brodskyn, C. (2011). Lutzomyia longipalpis saliva or salivary protein LJM19 protects against Leishmania braziliensis and the saliva of its vector, Lutzomyia intermedia. PLoS Negl. Trop. Dis. 5, e1169. doi:10.1371/journal.pntd.0001169
Teixeira, C., Gomes, R., Collin, N., Reynoso, D., Jochim, R., Oliveira, F., Seitz, A., Elnaiem, D. E., Caldas, A., De Souza, A. P., Brodskyn, C. I., De Oliveira, C. I., Mendonca, I., Costa, C. H., Volf, P., Barral, A., Kamhawi, S., and Valenzuela, J. G. (2010). Discovery of markers of exposure specific to bites of Lutzomyia longipalpis, the vector of Leishmania infantum chagasi in Latin America. PLoS Negl. Trop. Dis. 4, e638. doi:10.1371/journal.pntd.0000638
Thiakaki, M., Rohousova, I., Volfova, V., Volf, P., Chang, K. P., and Soteriadou, K. (2005). Sand fly specificity of saliva-mediated protective immunity in Leishmania amazonensis-BALB/c mouse model. Microbes Infect. 7, 760–766.
Titus, R. G., and Ribeiro, J. M. (1988). Salivary gland lysates from the sand fly Lutzomyia longipalpis enhance Leishmania infectivity. Science 239, 1306–1308.
Valenzuela, J. G., Belkaid, Y., Rowton, E., and Ribeiro, J. M. (2001). The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J. Exp. Biol. 204, 229–237.
Vinhas, V., Andrade, B. B., Paes, F., Bomura, A., Clarencio, J., Miranda, J. C., Bafica, A., Barral, A., and Barral-Netto, M. (2007). Human anti-saliva immune response following experimental exposure to the visceral leishmaniasis vector, Lutzomyia longipalpis. Eur. J. Immunol. 37, 3111–3121.
Vlkova, M., Rohousova, I., Drahota, J., Stanneck, D., Kruedewagen, E. M., Mencke, N., Otranto, D., and Volf, P. (2011). Canine antibody response to Phlebotomus perniciosus bites negatively correlates with the risk of Leishmania infantum transmission. PLoS Negl. Trop. Dis. 5, e1344. doi:10.1371/journal.pntd.0001344
Volf, P. (1991). Polyplax spinulosa infestation and antibody response in various strains of laboratory rats. Folia Parasitol. 38, 355–362.
Volf, P., Grubhoffer, L., and Hosek, P. (1993). Characterisation of salivary gland antigens of Triatoma infestans and antigen-specific serum antibody response in mice exposed to bites of T. infestans. Vet. Parasitol. 47, 327–337.
Volf, P., and Rohousova, I. (2001). Species-specific antigens in salivary glands of phlebotomine sandflies. Parasitology 122(Pt 1), 37–41.
Xu, X., Oliveira, F., Chang, B. W., Collin, N., Gomes, R., Teixeira, C., Reynoso, D., My Pham, V., Elnaiem, D. E., Kamhawi, S., Ribeiro, J. M., Valenzuela, J. G., and Andersen, J. F. (2011). Structure and function of a “yellow” protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection. J. Biol. Chem. 286, 32383–32393.
Keywords: leishmaniasis, sand fly, biomarkers, salivary proteins, vectors
Citation: Andrade BB and Teixeira CR (2012) Biomarkers for exposure to sand flies bites as tools to aid control of leishmaniasis. Front. Immun. 3:121. doi: 10.3389/fimmu.2012.00121
Received: 31 January 2012; Accepted: 27 April 2012;
Published online: 21 May 2012.
Edited by:
Nathan Peters, National Institute of Allergy and Infectious Diseases, USAReviewed by:
Petr Volf, Charles University in Prague, Czech RepublicCopyright: © 2012 Andrade and Teixeira. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Clarissa R. Teixeira, Vector Molecular Biology Section, Laboratory of Malaria and Vector Research, National Institutes of Health, 12735 Twinbrook Parkway, Building Twinbrook III, Room 2E22A, Rockville, MD 20852, USA. e-mail:dGVpeGVpcmFjQG5pYWlkLm5paC5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.