
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 09 May 2012
Sec. T Cell Biology
Volume 3 - 2012 | https://doi.org/10.3389/fimmu.2012.00115
This article is part of the Research Topic How does T cell antigen receptor signaling begin? View all 14 articles
Increasing evidence favors the notion that, before triggering, the T cell antigen receptor (TCR) forms nanometer-scale oligomers that are called nanoclusters. The organization of the TCR in pre-existing oligomers cannot be ignored when analyzing the properties of ligand (pMHC) recognition and signal transduction. As with other membrane receptors, the existence of TCR oligomers points out to cooperativity phenomena. We review the data in support of conformational changes in the TCR as the basic principle to transduce the activation signal to the cytoplasm and the incipient data suggesting cooperativity within nanoclusters.
The molecular mechanisms underlying the initiation of TCR signaling are not completely understood. As for other membrane receptors (Lemmon and Schlessinger, 2010), the predominant view of a purely crosslinking model as the mechanism for signal transmission across the membrane is slowly changing to accept the existence of conformational changes in the TCR (van der Merwe and Dushek, 2011). Conformational changes are widespread both in soluble and membrane-bound proteins (Anyatonwu et al., 2010; Cederholm et al., 2010; Wade et al., 2011). The archetypical example is this of G-protein-coupled receptors (GPCRs) which was originally proposed to oscillate between an inactive and an active state stabilized by ligand binding. However, more recent evidence supports the notion that there is a variability in receptor conformations according to the nature of the ligands (Kahsai et al., 2011). The change from inactive to active conformation initiates in the ectodomains of the receptors and is somehow transmitted to the cytoplasmic tails leading to the unmasking of an intracellular loop that forms a G-protein binding site (Wess, 1997; Wess et al., 1997). More recently, it has been shown that transmission of the active conformation requires extensive rearrangement of the transmembrane domains of the GPCRs (Altan-Bonnet and Germain, 2005; Adler et al., 2008). Many receptors with intrinsic tyrosine kinase activity also undergo ligand-induced conformational changes (Jiang and Hunter, 1999), as well as several receptors of immunological interest, including: integrins, the erythropoietin receptor, the tumor necrosis factor receptors, Fas, the interleukin 6 receptor, IFN receptors (Banner et al., 1993; Walter et al., 1995; Remy et al., 1999; Chan et al., 2000; Siegel et al., 2000; Krause et al., 2002; Murali et al., 2005; Strunk et al., 2008), and most important, the B cell receptor (Cambier et al., 1994; Reth, 2002; Tolar et al., 2005; Yang and Reth, 2010).
The studies on the mechanisms of signal initiation by the TCR are perverted by models which try to explain some of the properties of antigen recognition by T cells, such as the discrimination between MHC loaded with self-peptides and MHC loaded with antigen peptides and the exquisite sensitivity of T cells to low doses of antigen, but that do not provide mechanistic insights into how signal is transmitted from the outside to the cytoplasm. These models are summarized in Figure 1 and include the kinetic proofreading model, the serial triggering model, the kinetic segregation model and the dissociation of the TCR in its composing subunits. The kinetic proofreading model is based on a correlation between the dissociation constant of MHC loaded with different peptides and T cell response, but it has not progressed beyond a mere correlation (McKeithan, 1995). The serial triggering model proposes that an engaged TCR is internalized and leaves the same triggering pMHC molecule accessible to a second non-triggered TCR (Valitutti et al., 1995). This model does not provide mechanistic insights and has not provided evidence as to how and where the disengaged TCR transmits signals once the ligand has dissociated. The kinetic segregation model proposes that T cell activation is made possible by the segregation of proteins with large ectodomains such as the tyrosine phosphatase CD45 from areas of the membrane where the TCR and other proteins with smaller ectodomains are accumulating (Anton van der Merwe et al., 2000; Davis and van der Merwe, 2006). In our view, this model is a variation of the crosslinking model. Likewise, the dissociation in composing subunits that proposes that TCR triggering results in the release of the signaling CD3 subunits from an inhibitory contact with the TCRα/β subunits is a variation of the conformational change model.
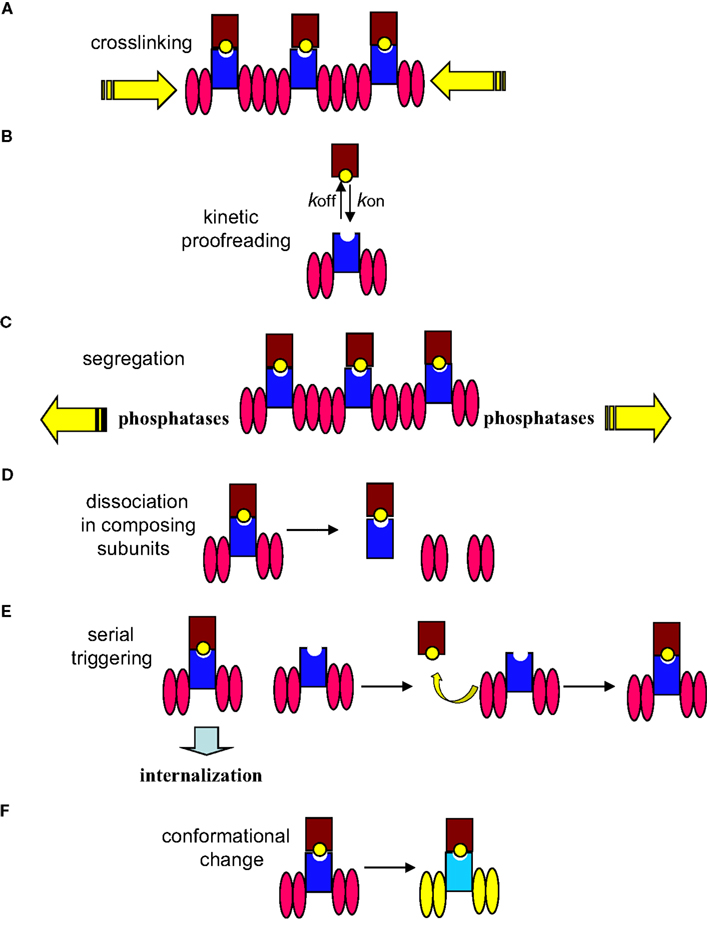
Figure 1. Mechanisms of signal initiation by the TCR. The TCR complex is represented as the TCR α/β heterodimer (shown as a dark blue rectangle) associated with two CD3 dimers depicted as pink ellipses (CD3ε-CD3γ and CD3ε-CD3δ). The MHC is represented as a brown rectangle bound to a yellow ball (peptide antigen). (A) The mere aggregation of TCR complexes following TCR engagement promotes ITAM’s phosphorylation. (B) The degree of response depends on the half-life of the TCR–pMHC interaction that itself relies on the association/dissociation constants (kon/koff) of the interaction. (C) The kinetic segregation model proposes that TCR–pMHC binding segregates the TCR complex from the inhibitory tyrosine phosphatases; leading to sustained phosphorylation of the CD3 ITAMs by Lck. (D) Upon triggering of the TCR, CD3 subunits are released from the inhibitory contact with the TCR α/β subunits. (E) The contacted TCR is internalized leaving the same pMHC available to engage other TCR complexes. (F) Engagement of TCR by pMHC molecules induces conformational changes (represented as change of colors in this cartoon) in the TCR complex that result in a change of conformation of the CD3 cytoplasmic domains, allowing ITAMs phosphorylation.
Nevertheless, crosslinking, kinetic segregation, and conformational changes are not mutually excluding. Indeed, we have evidence that both crosslinking of the TCR and conformational changes are required for T cell activation (Minguet et al., 2007).
The initial evidence for the existence of ligand-induced conformational changes in the TCR complex came from co-capping experiments performed by Janeway and colleagues (Janeway, 1995). They realized that stimulation of T cells with monovalent Fab fragments of anti-TCR antibodies promoted co-capping with the CD4 co-receptor, suggesting that anti-TCR antibodies may promote changes in the T cell in the absence of TCR crosslinking (Yoon et al., 1994). Later, we found that TCR triggering in the presence of the src kinase inhibitor PP2 or even in cell-free purified preparations led to its induced binding to the N-terminal SH3 domain of Nck (Gil et al., 2002). The TCR binds Nck through the proline-rich sequence (PRS) of CD3ε. Therefore the induced binding indicated the existence of a conformational change in the TCR that was manifested in the exposure of the PRS for Nck binding (Figure 2). Induction of PRS exposure was first demonstrated with stimulatory anti-CD3 antibodies (Gil et al., 2002) and later with a panel of pMHC ligands (Gil et al., 2005). The finding that mAb APA1/1 binds the PRS of CD3ε in the active conformation of the TCR, added further support to the conformational change model and served to demonstrate that the TCR adopted the active conformation when stimulated by antigen in vivo (Risueno et al., 2005). However, both Nck binding and APA1/1 recognition reflect the same molecular event, i.e. exposure of the PRS in the tail of CD3ε. The first demonstration that conformational changes affect TCR subunits other than CD3ε came from a protease sensitivity assay in which the cytoplasmic tails of both CD3ε and CD3ζ were shown to adopt a trypsin-resistant conformation when the TCR was triggered (Risueno et al., 2008a). This resistance was proposed to derive from the tight packing of the CD3 tails in the active conformation that contrasted with a loose or floppy state for the cytoplasmic tails in the inactive conformation. Interestingly, a protein chimera containing the cytoplasmic tail of CD3ε was found by Forster’s resonance energy transfer (FRET) to have the two key tyrosine residues of the ITAM inserted in the plasma membrane where they are inaccessible to kinases, suggesting that in the inactive conformation the tail of CD3ε was tightly associated to the plasma membrane’s phospholipids (Xu et al., 2008). These FRET results were in line with the idea of floppy tails resulting from the protease resistance experiments. Nevertheless, it has to be pointed out that the FRET experiments were performed with a CD3ε chimera and therefore it remains to be determined whether the tight association with the plasma membrane holds true in the context of a complete TCR.
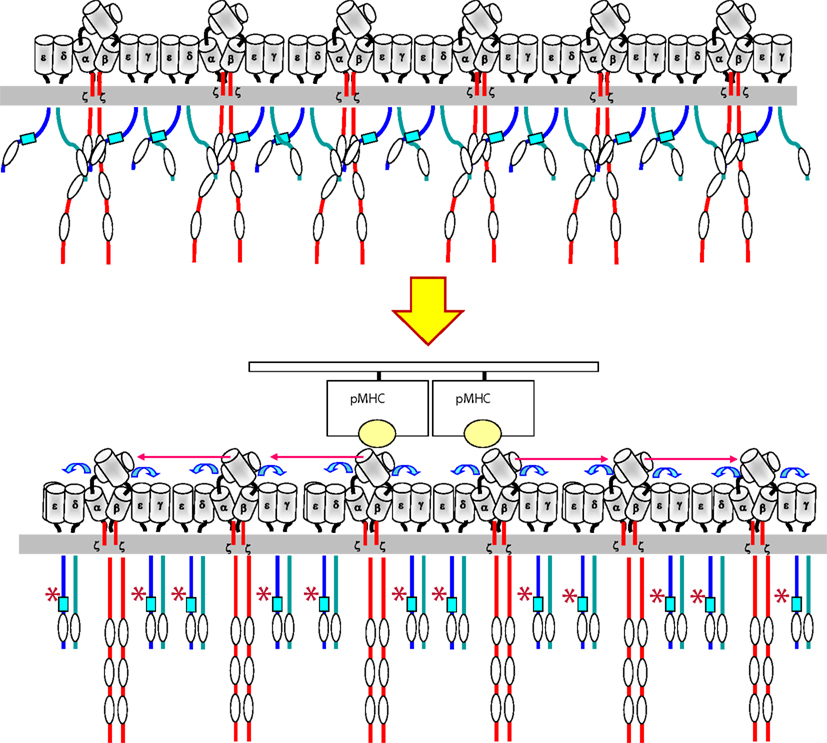
Figure 2. Model for transmission of the ligand-induced conformational change within a TCR cluster. Upon binding of pMHC (a dimer or an oligomer) to a TCR nanocluster, a torque in the contacted TCRα/β heterodimers is produced that is transmitted to the CD3 ectodomains (blue arrows). The CD3 ectodomains adopts the active conformation that is transmitted to their cytoplasmic tails through the transmembrane domains. The adoption of the active conformation by the CD3 tails result in the exposure (represented by an asterisk) of the polyproline sequence (blue square) in the tail of CD3ε and the adoption of a compact structure in contrast to its loose state before engagement. This compact structure facilitates the phosphorylation of ITAMs by Lck. The conformational changes are transmitted to all other TCR complexes within a nanocluster.
Although the ectodomains of the TCRα/β heterodimer and of the CD3γ-CD3ε and CD3δ–CD3ε dimers, as well as the transmembrane domain of CD3ζ, have been structurally determined by X-ray crystallography and NMR (Sun et al., 2001, 2004; Arnett et al., 2004; Kjer-Nielsen et al., 2004; Call et al., 2006; Garcia et al., 2009), the entire TCR complex has not been purified in sufficient amount and purity as to generate atom-scale information (Arechaga et al., 2010). This has not made possible to visualize changes that might occur at the TCRα/β-CD3 ectodomain interfaces or in the transmembrane domains. The TCRα/β ectodomains of several TCRs have been crystallized with or without its pMHC ligand and no detectable changes have been identified in most of them (Ding et al., 1999; Rudolph et al., 2006), that could result in transmission of conformational changes to the CD3 subunits. The exception is the LC13 TCR, where a ligand-induced shift of the Cα AB loop was demonstrated first by X-ray crystallography (Kjer-Nielsen et al., 2003) and later by site-directed fluorescence labeling (Beddoe et al., 2009). Mutation of the AB loop in the LC13 TCR was shown to affect T cell activation, but whether the conformational change of the AB loop is a peculiarity of the LC13 TCR or is a general phenomenon not visualized in other TCRs due to poor resolution of the loop or to the tight packing imposed by the crystal, remains to be determined (Beddoe et al., 2009).
Using existing structural data of CD3γ–CD3ε and CD3δ–CD3ε dimers bound and unbound to stimulatory anti-CD3 antibodies, a molecular dynamics model was generated that predicted a stiffening effect in the structures of the antibody-bound dimers. Two amino acid residues (Lys76 and Cys80) of CD3ε that were particularly changing their position in the bound vs. the unbound dimers were mutated to assess their impact on signal transmission. Mutation of both residues inhibited or abrogated the adoption of the active conformation by the PRS of CD3ε and prevented T cell activation upon antibody or pMHC triggering (Martinez-Martin et al., 2009). These results demonstrated that Lys76 and Cys80 were required for transmission of outside-in information in the TCR and validated the importance of conformational changes as primary signal transducing mechanisms in the TCR. Interestingly, Cys80 forms part of the CxxC motif of CD3ε. It should be pointed out that some of the conformational mutants of CD3ε showed a dominant negative effect in the induction of the conformational change, hence the inhibition of the conformational change took place even in the presence of an excess of wild type CD3ε. In a different study, replacement of the two cysteine residues from the CxxC motif by serine was shown to abrogate T cell differentiation in the thymus (Wang et al., 2009).
As for other membrane receptors (Risueno et al., 2008b), the mechanism proposed for the transmission of the conformational change across the membrane involves piston-like, rotational, and sliding movements (Kuhns et al., 2006; Risueno et al., 2008b). The paired G beta strands of CD3ε and CD3γ form a rigid rod-like connector that could produce a displacement of the transmembrane helices in a piston-like movement (Sun et al., 2001). However it is not clear how a piston-like movement could be transmitted to the ITAMs. The receptor deformation model is a variation of the piston mechanism whereby forces created at the T cell-APC contact site “pull” the TCR when it is engaged by pMHC and induce a conformational change in the TCR (Ma and Finkel, 2010). The idea of the TCR being a mechanosensor is gaining acceptance (Kim et al., 2009). By using immobilized non-stimulatory anti-CD3 mAb it has been shown that calcium fluxes can be induced if a tangential force, but not a normal force, is applied. The physiological source of this force includes oscillatory movements of the cytoskeleton, forces induced during T cell detachment and migration and tensional forces generated by larger molecules in the vicinity of the engaged TCR (Kim et al., 2009). According to this, it is the deformation of the TCRα/β heterodimer induced by a tangential force what causes a push or a pull on the CD3 subunits and allows the transmission of the conformational change to the cytoplasm. However, this model does not explain how soluble anti-CD3 or anti-TCRα/β V region antibodies and pMHC multimers stimulate T cells (Rojo and Janeway, 1988; Rosette et al., 2001; Campanelli et al., 2002).
Early studies using sucrose density gradients (Exley et al., 1995) and on double TCR-transgenic mice, using FRET, co-modulation and co-precipitation techniques, showed that TCRs of different antigen specificity are physically associated (Fernandez-Miguel et al., 1999). This led to the notion that the TCR is at least bivalent. This idea has been recently reinforced using immunoprecipitation-flow cytometry (Schrum et al., 2011). However, in addition to dimers, the TCR seems to form higher-order oligomers independent of stimulation. A number of independent techniques have been used to study this issue, including blue native gels (BN-PAGE) and immuno-gold EM after label-fracture. Immuno-gold EM indicated that the TCR is expressed as a mixture of oligomers of different size in non-stimulated resting T cells, ranging from single complexes to clusters of 20 or more TCR complexes (Schamel et al., 2005). It is unlikely that the oligomeric complexes observed are an artifact of the technique because the cells are fixed with aldehydes before labeling with antibodies, the complexes can be detected by antibodies specific for different chains of the TCR complex, depletion of cholesterol before fixing and labeling with antibodies disrupts TCR oligomers, and the size of the TCR oligomers differs between naïve and antigen-experienced T cells. Even when detected with the same antibody, the change in oligomeric TCR size can also be detected by an independent method (BN-PAGE) and finally, a single mutation in the transmembrane domain of CD3ζ can impair oligomeric TCR formation (Kumar et al., 2011).
Analysis of membrane sheets by EM has shown that many membrane receptors are non-randomly associated forming the so called “protein islands” supported by cholesterol and the underlying cytoskeleton (Lillemeier et al., 2006). The TCR is one of such receptors, as demonstrated in addition by high-speed photo activated localization microscopy, and dual-color fluorescence cross-correlation spectroscopy (Lillemeier et al., 2009). Hence our data, along with studies using alternative techniques (Lillemeier et al., 2009; Sherman et al., 2011), show that the TCR is expressed as pre-existing oligomers before the T cell is engaged by its pMHC ligand. Furthermore, the effect of the CD3ζ mutation on sensitivity of antigen-experienced T cells suggests that the TCR is functionally multivalent in this population. Interestingly, other membrane receptors that undergo conformational changes are being demonstrated to be organized as oligomers. This is the case for GPCRs which were long suspected to form dimers and are now being shown to form higher-order oligomers (Kaczor and Selent, 2011).
Preassembled oligomeric TCR complexes could be a platform for the transmission of the conformational change. Therefore, binding of the pMHC begins with a monovalent interaction between one pMHC and one TCR within a cluster. This initial event favors the interaction of a second pMHC–TCR. Binding of two or more agonist ligands promotes a conformational change in all the TCR complexes of the same oligomer (Schamel et al., 2006). Such a cooperative effect has been proposed for amino acid receptors in bacteria, explaining their high sensitivity to very low concentrations of ligand (Duke et al., 2001) and more recently for GPCRs (Canals et al., 2012). The cooperative effect in the TCR is strongly suggested by the dominant negative effect in the transmission of the conformational change exerted by CD3ε conformational mutants expressed within an excess of endogenous wild type chain (Martinez-Martin et al., 2009). We envision the preassembled TCR oligomers as tightly packed structures in which all TCR complex units are coupled, shifting simultaneously from an inactive to an active conformation, such as the lamellas of a venetian blind open and close. The surface of the pMHC complex always contacts the surface of TCRα/β in a diagonal orientation (Rudolph et al., 2006). Simultaneous binding of two or more pMHC complexes to a TCR oligomer would provoke a rotation of the contacted TCRα/β heterodimers, such as it has been proposed in the “permissive geometry model” (Minguet and Schamel, 2008). This rotation imposed upon the contacted TCRα/β heterodimers could be transmitted to the rest of the TCRα/β heterodimers in the TCR oligomer even if they are not directly contacted by pMHC, thus making all TCR units in the oligomer participants in the signaling process (Figure 3). Interestingly, it has just been published that disruption of the TCR–pMHC binding geometry by using a protruding antigen peptide impedes signal transduction even though the affinity of the interaction in solution is as high as this of other pMHC complexes binding with a normal angle (Adams et al., 2011). Despite the high 3D affinity (in solution) for the TCR of the abnormal pMHC, the affinity in 2D (membrane-bound) is lower than this of normal pMHC ligands. It would be interesting to determine if the incapacity to activate T cells is derived from an inability to induce the conformational change in the TCR and if the diminished 2D affinity is derived from an impossibility to induce the twist or torque in two or more TCRα/β heterodimers within a TCR nanocluster. Finally, the conformational change in the TCR allows the exposure of the CD3 cytoplasmic tails and the recruitment of direct effectors such as Nck and regulates the accessibility of their tyrosine residues to the priming kinase Lck. Recent data indicate that TCR engagement does not increase the activity of Lck (Nika et al., 2010). If this is confirmed, regulation of CD3 tyrosine phosphorylation would occur just at the level of the substrate (tyrosine residues) and not of the kinase, suggesting that the TCR conformational change is the only mechanism regulating the most initial events of TCR signaling.
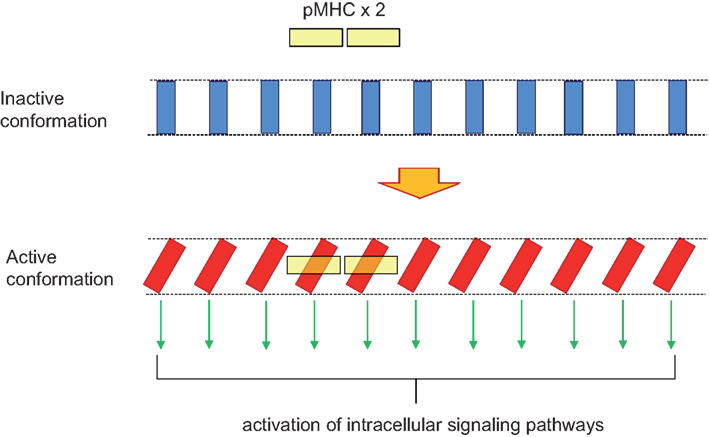
Figure 3. Domino effect explaining TCR cooperativity within a TCR nanocluster. A TCR nanocluster (here a 11mer) has all its TCR complex units in the inactive conformation (blue rectangles). The binding of a pMHC oligomer (here depicted as a yellow dimer) to two of the TCR units within the nanocluster provokes a twist in the two contacted TCR units in order to accommodate the pMHC dimer with an approximately 30° angle. This twist is transmitted along the TCR nanocluster in such a way that all TCR units adopt the active conformation; a movement reminiscent of a venetian blind.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The laboratory of Balbino Alarcón is supported by grants SAF2010-14912 from “Comisión Interministerial de Ciencia y Tecnología” and FP7/2007-2013 “Sybilla” from the European Union.
EM, electron microscopy; FRET, Forester’s resonance energy transfer; GPCRs, G-protein-coupled receptors; ITAM, immunoreceptor tyrosine-based activation motif; NMR, nuclear magnetic resonance; pMHC, major histocompatibility complex loaded with antigen peptide; PRS, proline-rich sequence; TCR, T cell antigen receptor.
Adams, J. J., Narayanan, S., Liu, B., Birnbaum, M. E., Kruse, A. C., Bowerman, N. A., Chen, W., Levin, A. M., Connolly, J. M., Zhu, C., Kranz, D. M., and Garcia, K. C. (2011). T Cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 16, 16.
Adler, J., Pagakis, S. N., and Parmryd, I. (2008). Replicate-based noise corrected correlation for accurate measurements of colocalization. J. Microsc. 230(Pt 1), 121–133.
Altan-Bonnet, G., and Germain, R. N. (2005). Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 3, e356. doi: 10.1371/journal.pbio.0030356
Anton van der Merwe, P., Davis, S. J., Shaw, A. S., and Dustin, M. L. (2000). Cytoskeletal polarization and redistribution of cell-surface molecules during T cell antigen recognition. Semin. Immunol. 12, 5–21.
Anyatonwu, G., Khan, M. T., Schug, Z. T., da Fonseca, P. C., Morris, E. P., and Joseph, S. K. (2010). Calcium-dependent conformational changes in inositol trisphosphate receptors. J. Biol. Chem. 285, 25085–25093.
Arechaga, I., Swamy, M., Abia, D., Schamel, W. A., Alarcon, B., and Valpuesta, J. M. (2010). Structural characterization of the TCR complex by electron microscopy. Int. Immunol. 22, 897–903.
Arnett, K. L., Harrison, S. C., and Wiley, D. C. (2004). Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment. Proc. Natl. Acad. Sci. U.S.A. 101, 16268–16273.
Banner, D. W., D’Arcy, A., Janes, W., Gentz, R., Schoenfeld, H. J., Broger, C., Loetscher, H., and Lesslauer, W. (1993). Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell 73, 431–445.
Beddoe, T., Chen, Z., Clements, C. S., Ely, L. K., Bushell, S. R., Vivian, J. P., Kjer-Nielsen, L., Pang, S. S., Dunstone, M. A., Liu, Y. C., Macdonald, W. A., Perugini, M. A., Wilce, M. C., Burrows, S. R., Purcell, A. W., Tiganis, T., Bottomley, S. P., McCluskey, J., and Rossjohn, J. (2009). Antigen ligation triggers a conformational change within the constant domain of the alphabeta T cell receptor. Immunity 30, 777–788.
Call, M. E., Schnell, J. R., Xu, C., Lutz, R. A., Chou, J. J., and Wucherpfennig, K. W. (2006). The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell 127, 355–368.
Cambier, J. C., Pleiman, C. M., and Clark, M. R. (1994). Signal transduction by the B cell antigen receptor and its coreceptors. Annu. Rev. Immunol. 12, 457–486.
Campanelli, R., Palermo, B., Garbelli, S., Mantovani, S., Lucchi, P., Necker, A., Lantelme, E., and Giachino, C. (2002). Human CD8 co-receptor is strictly involved in MHC-peptide tetramer-TCR binding and T cell activation. Int. Immunol. 14, 39–44.
Canals, M., Lane, J. R., Wen, A., Scammells, P. J., Sexton, P. M., and Christopoulos, A. (2012). A Monod-Wyman-Changeux mechanism can explain G protein-coupled receptor (GPCR) allosteric modulation. J. Biol. Chem. 287, 650–659.
Cederholm, J. M., Absalom, N. L., Sugiharto, S., Griffith, R., Schofield, P. R., and Lewis, T. M. (2010) Conformational changes in extracellular loop 2 associated with signal transduction in the glycine receptor. J. Neurochem. 115, 1245–1255.
Chan, F. K., Chun, H. J., Zheng, L., Siegel, R. M., Bui, K. L., and Lenardo, M. J. (2000). A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288, 2351–2354.
Davis, S. J., and van der Merwe, P. A. (2006). The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 7, 803–809.
Ding, Y. H., Baker, B. M., Garboczi, D. N., Biddison, W. E., and Wiley, D. C. (1999). Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity 11, 45–56.
Duke, T. A., Le Novere, N., and Bray, D. (2001). Conformational spread in a ring of proteins: a stochastic approach to allostery. J. Mol. Biol. 308, 541–553.
Exley, M., Wileman, T., Mueller, B., and Terhorst, C. (1995). Evidence for multivalent structure of T-cell antigen receptor complex. Mol. Immunol. 32, 829–839.
Fernandez-Miguel, G., Alarcon, B., Iglesias, A., Bluethmann, H., Alvarez-Mon, M., Sanz, E., and de la Hera, A. (1999). Multivalent structure of an alphabetaT cell receptor. Proc. Natl. Acad. Sci. U.S.A. 96, 1547–1552.
Garcia, K. C., Adams, J. J., Feng, D., and Ely, L. K. (2009). The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat. Immunol. 10, 143–147.
Gil, D., Schamel, W. W., Montoya, M., Sanchez-Madrid, F., and Alarcon, B. (2002). Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell 109, 901–912.
Gil, D., Schrum, A. G., Alarcon, B., and Palmer, E. (2005). T cell receptor engagement by peptide-MHC ligands induces a conformational change in the CD3 complex of thymocytes. J. Exp. Med. 201, 517–522.
Janeway, C. A. Jr. (1995). Ligands for the T-cell receptor: hard times for avidity models. Immunol. Today 16, 223–225.
Jiang, G., and Hunter, T. (1999). Receptor signaling: when dimerization is not enough. Curr. Biol. 9, R568–R571.
Kaczor, A. A., and Selent, J. (2011). Oligomerization of G protein-coupled receptors: biochemical and biophysical methods. Curr. Med. Chem. 18, 4606–4634.
Kahsai, A. W., Xiao, K., Rajagopal, S., Ahn, S., Shukla, A. K., Sun, J., Oas, T. G., and Lefkowitz, R. J. (2011). Multiple ligand-specific conformations of the beta2-adrenergic receptor. Nat. Chem. Biol. 7, 692–700.
Kim, S. T., Takeuchi, K., Sun, Z. Y., Touma, M., Castro, C. E., Fahmy, A., Lang, M. J., Wagner, G., and Reinherz, E. L. (2009). The alphabeta T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 284, 31028–31037.
Kjer-Nielsen, L., Clements, C. S., Purcell, A. W., Brooks, A. G., Whisstock, J. C., Burrows, S. R., McCluskey, J., and Rossjohn, J. (2003). A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity 18, 53–64.
Kjer-Nielsen, L., Dunstone, M. A., Kostenko, L., Ely, L. K., Beddoe, T., Mifsud, N. A., Purcell, A. W., Brooks, A. G., McCluskey, J., and Rossjohn, J. (2004). Crystal structure of the human T cell receptor CD3 epsilon gamma heterodimer complexed to the therapeutic mAb OKT3. Proc. Natl. Acad. Sci. U.S.A. 101, 7675–7680.
Krause, C. D., Mei, E., Xie, J., Jia, Y., Bopp, M. A., Hochstrasser, R. M., and Pestka, S. (2002). Seeing the light: preassembly and ligand-induced changes of the interferon gamma receptor complex in cells. Mol. Cell Proteomics 1, 805–815.
Kuhns, M. S., Davis, M. M., and Garcia, K. C. (2006). Deconstructing the form and function of the TCR/CD3 complex. Immunity 24, 133–139.
Kumar, R., Ferez, M., Swamy, M., Arechaga, I., Rejas, M. T., Valpuesta, J. M., Schamel, W. W., Alarcon, B., and van Santen, H. M. (2011). Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity 35, 375–387.
Lemmon, M. A., and Schlessinger, J. (2010). Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134.
Lillemeier, B. F., Mortelmaier, M. A., Forstner, M. B., Huppa, J. B., Groves, J. T., and Davis, M. M. (2009). TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96.
Lillemeier, B. F., Pfeiffer, J. R., Surviladze, Z., Wilson, B. S., and Davis, M. M. (2006). Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 103, 18992–18997.
Martinez-Martin, N., Risueno, R. M., Morreale, A., Zaldivar, I., Fernandez-Arenas, E., Herranz, F., Ortiz, A. R., and Alarcon, B. (2009). Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3epsilon. Sci. Signal. 2, ra43.
McKeithan, T. W. (1995). Kinetic proofreading in T-cell receptor signal transduction. Proc. Natl. Acad. Sci. U.S.A. 92, 5042–5046.
Minguet, S., and Schamel, W. W. (2008). A permissive geometry model for TCR-CD3 activation. Trends Biochem. Sci. 33, 51–57.
Minguet, S., Swamy, M., Alarcon, B., Luescher, I. F., and Schamel, W. W. (2007). Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity 26, 43–54.
Murali, R., Cheng, X., Berezov, A., Du, X., Schon, A., Freire, E., Xu, X., Chen, Y. H., and Greene, M. I. (2005). Disabling TNF receptor signaling by induced conformational perturbation of tryptophan-107. Proc. Natl. Acad. Sci. U.S.A. 102, 10970–10975.
Nika, K., Soldani, C., Salek, M., Paster, W., Gray, A., Etzensperger, R., Fugger, L., Polzella, P., Cerundolo, V., Dushek, O., Hofer, T., Viola, A., and Acuto, O. (2010). Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity 32, 766–77.
Remy, I., Wilson, I. A., and Michnick, S. W. (1999). Erythropoietin receptor activation by a ligand-induced conformation change. Science 283, 990–993.
Reth, M. (2002). Hydrogen peroxide as second messenger in lymphocyte activation. Nat. Immunol. 3, 1129–1134.
Risueno, R. M., Gil, D., Fernandez, E., Sanchez-Madrid, F., and Alarcon, B. (2005). Ligand-induced conformational change in the T-cell receptor associated with productive immune synapses. Blood 106, 601–608.
Risueno, R. M., Schamel, W. W., and Alarcon, B. (2008a). T cell receptor engagement triggers its CD3epsilon and CD3zeta subunits to adopt a compact, locked conformation. PLoS ONE 3, e1747. doi: 10.1371/journal.pone.0001747
Risueno, R. M., Ortiz, A. R., and Alarcon, B. (2008b). Conformational model. Adv. Exp. Med. Biol. 640, 103–112.
Rojo, J. M., and Janeway, C. A. Jr. (1988). The biologic activity of anti-T cell receptor V region monoclonal antibodies is determined by the epitope recognized. J. Immunol. 140, 1081–1088.
Rosette, C., Werlen, G., Daniels, M. A., Holman, P. O., Alam, S. M., Travers, P. J., Gascoigne, N. R., Palmer, E., and Jameson, S. C. (2001). The impact of duration versus extent of TCR occupancy on T cell activation: a revision of the kinetic proofreading model. Immunity 15, 59–70.
Rudolph, M. G., Stanfield, R. L., and Wilson, I. A. (2006). How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466.
Schamel, W. W., Arechaga, I., Risueno, R. M., van Santen, H. M., Cabezas, P., Risco, C., Valpuesta, J. M., and Alarcon, B. (2005). Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J. Exp. Med. 202, 493–503.
Schamel, W. W., Risueno, R. M., Minguet, S., Ortiz, A. R., and Alarcon, B. (2006). A conformation- and avidity-based proofreading mechanism for the TCR-CD3 complex. Trends Immunol. 27, 176–182.
Schrum, A. G., Gil, D., Turka, L. A., and Palmer, E. (2011). Physical and functional bivalency observed among TCR/CD3 complexes isolated from primary T cells. J. Immunol. 187, 870–878.
Sherman, E., Barr, V., Manley, S., Patterson, G., Balagopalan, L., Akpan, I., Regan, C. K., Merrill, R. K., Sommers, C. L., Lippincott-Schwartz, J., and Samelson, L. E. (2011). Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity 35, 705–720.
Siegel, R. M., Frederiksen, J. K., Zacharias, D. A., Chan, F. K., Johnson, M., Lynch, D., Tsien, R. Y., and Lenardo, M. J. (2000). Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 288, 2354–2357.
Strunk, J. J., Gregor, I., Becker, Y., Li, Z., Gavutis, M., Jaks, E., Lamken, P., Walz, T., Enderlein, J., and Piehler, J. (2008). Ligand binding induces a conformational change in ifnar1 that is propagated to its membrane-proximal domain. J. Mol. Biol. 377, 725–739.
Sun, Z. J., Kim, K. S., Wagner, G., and Reinherz, E. L. (2001). Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer. Cell 105, 913–923.
Sun, Z. Y., Kim, S. T., Kim, I. C., Fahmy, A., Reinherz, E. L., and Wagner, G. (2004). Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 16867–16872.
Tolar, P., Sohn, H. W., and Pierce, S. K. (2005). The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat. Immunol. 6, 1168–1176.
Valitutti, S., Dessing, M., Aktories, K., Gallati, H., and Lanzavecchia, A. (1995). Sustained signaling leading to T cell activation results from prolonged T cell receptor occupancy. Role of T cell actin cytoskeleton. J. Exp. Med. 181, 577–584.
van der Merwe, P. A., and Dushek, O. (2011). Mechanisms for T cell receptor triggering. Nat. Rev. Immunol. 11, 47–55.
Wade, F., Espagne, A., Persuy, M. A., Vidic, J., Monnerie, R., Merola, F., Pajot-Augy, E., and Sanz, G. (2011). Relationship between homo-oligomerization of a mammalian olfactory receptor and its activation state demonstrated by bioluminescence resonance energy transfer. J. Biol. Chem. 286, 15252–15259.
Walter, M. R., Windsor, W. T., Nagabhushan, T. L., Lundell, D. J., Lunn, C. A., Zauodny, P. J., and Narula, S. K. (1995). Crystal structure of a complex between interferon-gamma and its soluble high-affinity receptor. Nature 376, 230–235.
Wang, Y., Becker, D., Vass, T., White, J., Marrack, P., and Kappler, J. W. (2009). A conserved CXXC motif in CD3epsilon is critical for T cell development and TCR signaling. PLoS Biol. 7, e1000253. doi: 10.1371/journal.pbio.1000253
Wess, J. (1997). G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition. FASEB J. 11, 346–354.
Wess, J., Liu, J., Blin, N., Yun, J., Lerche, C., and Kostenis, E. (1997). Structural basis of receptor/G protein coupling selectivity studied with muscarinic receptors as model systems. Life Sci. 60, 1007–1014.
Xu, C., Gagnon, E., Call, M. E., Schnell, J. R., Schwieters, C. D., Carman, C. V., Chou, J. J., and Wucherpfennig, K. W. (2008). Regulation of T cell receptor activation by dynamic membrane binding of the CD3varepsilon cytoplasmic tyrosine-based motif. Cell 135, 702–713.
Yang, J., and Reth, M. (2010). Oligomeric organization of the B-cell antigen receptor on resting cells. Nature 467, 465–469.
Keywords: TCR, conformational changes, oligomers, nanoclusters
Citation: Blanco R and Alarcón B (2012) TCR nanoclusters as the framework for transmission of conformational changes and cooperativity. Front. Immun. 3:115. doi: 10.3389/fimmu.2012.00115
Received: 13 December 2011; Accepted: 22 April 2012;
Published online: 09 May 2012.
Edited by:
Michael Dustin, New York University School of Medicine, USAReviewed by:
Christopher E. Rudd, University of Cambridge, UKCopyright: © 2012 Blanco and Alarcón. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Balbino Alarcón, Centro de Biología Molecular Severo Ochoa, Nicolás Cabrera 1, Universidad Autónoma de Madrid, 28049 Madrid, Spain. e-mail:YmFsYXJjb25AY2JtLnVhbS5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.