- 1 Department of Immunobiology, Kings’ College London School of Medicine, Guy’s Campus, London, UK
- 2 Department of Pathology, School of Medicine, Cardiff University, Cardiff, UK
CD27 expression has been used to distinguish between memory and naive B cells in humans. However, low levels of mutated and isotype-switched CD27−IgD− cells are seen in healthy adults, and these are increased in some autoimmune diseases and in the elderly. Thus CD27 is not a universal marker of memory B cells in humans. Various hypotheses have been put forward as to the function of the CD27− memory population. Since we have previously found high-throughput IGHV repertoire analysis useful to distinguish “innate-like” memory B cells (CD27+IgD+), we have employed similar analyses to elucidate the relationship between CD27− and CD27+ memory B cells. IgM+IgD− memory cells in both the CD27+ and CD27− compartments share the unique characteristics of the “innate-like” IgM+IgD+CD27+ cells. The switched CD27+ and CD27− memory cells share a similar IGHV repertoire, having more in common with each other than with “innate-like” memory cells, although it is interesting that IgG2 and IgA2 subclasses of antibody in both switched memory populations have a more “innate-like” repertoire. Clonality analysis shows evidence of a close clonal relationship between the two populations in that both CD27− and CD27+ switched memory cells can be found in the same genealogical tree. The expression of CD27 does not appear to occur in a linear developmental fashion, since we see CD27− cells as precursors of CD27+ cells and vice versa. Despite the similarities, the CDR-H3 repertoire of the CD27− cells is significantly different from both the CD27+IgD+ and CD27+IgD− populations, indicating that perhaps the lack of CD27 might be related to binding properties of the Ig CDR-H3 region.
Introduction
Immunological memory in the humoral immune system is maintained by a very diverse pool of antibody-producing plasma cells (PCs) and memory B cells, which can respond more rapidly to challenge than do naive B cells of the same specificity (Blanchard-Rohner et al., 2009). In this way not only are antibodies available immediately for prevention of infection upon secondary challenge, but the improved B cell receptors that result from the costly and complex process of affinity maturation are preserved. Since naïve and memory B cells react very differently to exogenous stimuli (Agematsu et al., 1997; Tangye et al., 2003) and have a different repertoire of IGHV gene use (Wu et al., 2010), it is important to distinguish between the two in experiments. Morphologically, memory B cells are larger and of higher granule density than naïve B cells (Tangye et al., 1998; Ma et al., 2006). It is well documented that the cell surface phenotypes are distinct between naïve and memory B cells (Tangye et al., 1998; Wirths and Lanzavecchia, 2005). However, finding a precise and tractable method to identify memory B cells can be somewhat problematical.
Affinity maturation of B cells in a germinal center (GC) reaction results in cells carrying immunoglobulin (Ig) genes that have mutated variable regions as a result of the somatic hypermutation (SHM) process. Thus one-way in which memory and naïve cells can be distinguished is by the mutation status of the Ig genes, although the procedures required to determine this are not such that they can be used to sort cells. Since a large fraction of memory B cells also undergo class switch recombination (CSR) to switch their isotype from IgM and IgD to IgA, IgG, or IgE, it was once thought that the presence of IgM or IgD was a good marker of naïve cells. However, the discovery of a significant population of IgM+ IgD+ cells that have mutations in their Ig genes eliminated this option (Dunn-Walters et al., 1995; Klein et al., 1997). The alternative proposal was to use CD27 as a marker of memory B cells in humans on the basis that CD27 expression correlates with SHM in IgM+IgD+ cells (Klein et al., 1998). CD27 was found to be constitutively expressed in approximately 40% of peripheral blood B cells in humans (Klein et al., 1998). It is a member of TNF- receptor family and is an important marker of activation contributing to B cell expansion, differentiation, and antibody production (Kobata et al., 1995; Lens et al., 1996; Agematsu et al., 1997; Arens et al., 2004) via the interaction with its ligand, CD70, expressed on the surface of activated T cells (Hintzen et al., 1994). CD27–CD70 signaling is thought to orchestrate CD40–CD154 signaling in GCs to maintain long term immunological memory against T cell dependent (TD) antigens (Agematsu et al., 1997).
Although it was later found that memory cells can be distinguished from naïve cells by their absence of the ATP-binding cassette (ABCB1) transporter (Wirths and Lanzavecchia, 2005), the rhodamine staining protocol required for this is less tractable than simple surface staining protocols. Hence surface CD27 and IgD markers are still widely used to separate B cells into memory and naive subsets. The four main populations that are distinguished are: CD27− IgD+ antigen-inexperienced cells, two subsets of CD27+ memory cells (IgD+/IgD−) and CD27−IgD− memory cells. The existence of the latter population, containing B cells with class switched and mutated Ig genes that share similar morphology with CD27+ cells and have no ABCB1 transporter (Wirths and Lanzavecchia, 2005; Fecteau et al., 2006), calls into question the feasibility of using CD27 as a memory B cell marker. CD27− memory cells are present at birth (Van Gent et al., 2009), and in healthy adult subjects they account for approximately 5% of the healthy peripheral B cell population (Sanz et al., 2008). An alternative sorting strategy for separating B cells is to use CD24 and CD38, but the relationship with this strategy and that of using CD27/IgD is not clear cut and multiple subpopulations have differential expression of CD27, IgD, CD24, CD38, and B220 (Sanz et al., 2008). Recently, markers such as CD45RBMEM55 (Koethe et al., 2011) and CD330a (Silva et al., 2011) have been reported as alternative markers to help define human memory B cells. Whether they will prove more useful than current strategies remains to be determined.
The function of CD27− memory cells and their relationship to CD27+ cells is not clear. It is important to gain a better understanding of the CD27− cell population because they occur at greater frequency in immunosenescence, systemic lupus erythematosus (SLE), rotavirus infection, and human immunodeficiency virus (HIV) infection (Wei et al., 2007; Moir et al., 2008; Rojas et al., 2008; Colonna-Romano et al., 2009). Colonna-Romano et al. (2009) suggest they are exhausted memory cells that have downregulated CD27. However, if this were the case one might expect that they would carry equal or greater amounts of SHM in the Ig genes, while the CD27− IgG+ cells actually seem to have a lower level of mutation overall (Fecteau et al., 2006) which might suggest that they are precursors of memory cells that have not yet acquired CD27. It has also been noted that the proportions of IgG2 to IgG3, and IgA1 to IgA2 vary between CD27− and CD27+ populations (Fecteau et al., 2006; Berkowska et al., 2011), so the CD27− cells may be a different population of cells entirely, developed in response to different stimuli. In addition, analyses of replication history have shown differences between CD27− and CD27+ populations, leading to the hypothesis that a combination of CD27 expression and class switched status represents differences between T-independent (TI), primary TD responses and secondary TD responses (Berkowska et al., 2011).
We have previously used IGHV repertoire analysis to distinguish different populations of memory cells. The rationale being that if negative (tolerance) and positive (antigen activation) selection events result in the loss of some Ig genes and expansion of others, then a population of cells responding to different antigens will be distinguished by a different Ig gene repertoire. In this way we showed that the unswitched CD27+IgD+ repertoire could be distinguished from both CD27−IgD+ antigen-inexperienced cells and class switched CD27+IgD− memory cells by IGHV1 family and IGHV3-23 gene usage (Wu et al., 2010). The CD27+IgD+ population has been termed “IgM memory,” “Innate-like B cells” or “Natural Effector B cells” and it does not appear to require classical GCs or conventional T cell help for development, since it is present in individuals suffering from X-linked hyper-IgM syndrome where GCs and switched memory cells are absent due to the lack of CD40–CD154 signaling (Weller et al., 2004). It has been suggested that CD27+IgD+ cells represent an innate-like repertoire, equivalent to splenic marginal zone (MZ) cells that can be mobilized from the MZ into the peripheral blood in response to a TI challenge (Weller et al., 2004). In support of this, it has been shown that they play a key role in the protection against Streptococcus pneumoniae infections (Kelly et al., 2006; Langley et al., 2010). Hence the distinguishing IGHV1 and IGHV3 family characteristics of the CD27+IgD+ repertoire may reflect selection by a TI challenge.
In order to investigate the relationship between the CD27− and CD27+ memory B cell populations, we studied their Ig heavy chain (IgH) repertoire using high-throughput sequence analysis. We found a high degree of similarity between the two switched CD27+ and CD27− populations, both being distinct from the IgM memory cells. We also saw evidence of related B cells within clones that differed in their expression of CD27. These findings support the hypothesis that the two populations have a common origin, and that changes in CD27 expression are not always as a result of ordered progression through development. Crucially however, the present study also indicates heterogeneity of repertoire between different subclasses of switched memory cells, indicating that regardless of CD27 status there are different selection pressures on cells with different subclass-specific effector functions.
Materials and Methods
Separation and Analysis of B Cell Subsets
PBMCs were isolated using Ficoll-Paque Plus (GE Healthcare, UK) and Leucosep tubes (Greiner Bio-One Ltd., UK) from nine young healthy donors (21–45 years), after obtaining written consent as approved by the Guy’s Hospital Research ethics committee. Four B cell subsets were isolated as previously reported (Wu et al., 2010). Briefly, CD19+ B cells were isolated from PBMCs using the MACS B cell Isolation Kit (Miltenyi Biotec, Germany) and stained with CD27–FITC and IgD-PE (BD Biosciences Pharmingen, UK) before sorting into subsets (Figure 1A) on a FACSAria machine (BD) and being separately collected into 180 μL sort–lysis RT buffer (SLyRT).
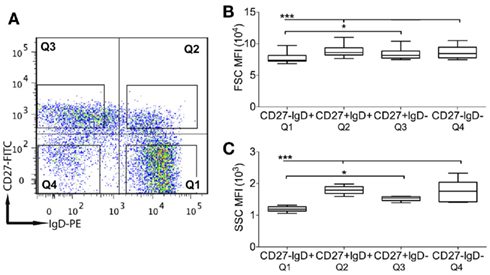
Figure 1. Identification of B cell subsets and analysis for cellular morphology. (A) Human peripheral blood CD19+ B cells were stained with CD27 and IgD and separated into four subsets: antigen-inexperienced (Q1: CD27−IgD−), IgM memory (Q2: CD27+IgD+), classical memory (Q3: CD27+IgD−), and CD27− (Q4: CD27−IgD−) cells, as indicated by four quadrants. The plot is one representative of nine. The mean fluorescence intensity (MFI) of (B) forward scatter (FSC) and (C) side scatter (SSC), measured by flow cytometry, were used to compare the cellular size and granularity, respectively, between cell subsets. Error bars indicate SEM. p Values for one-way ANOVA comparisons are indicated as *p < 0.05 and ***p < 0.0005 (n = 9).
High-Throughput Sequencing of Ig Genes
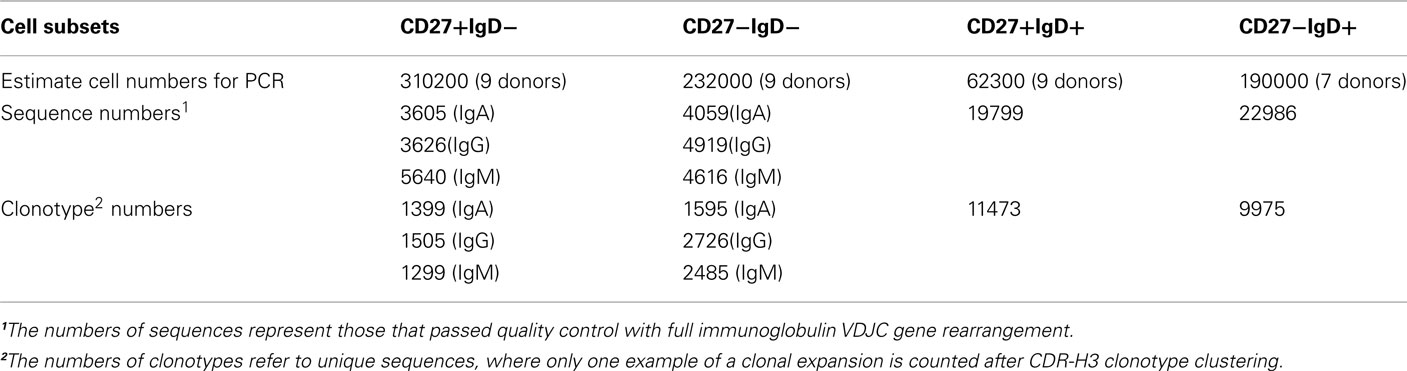
cDNA synthesis was performed by adding 500 U SuperScript III reverse transcriptase (Invitrogen, UK) in a volume of 20–180 μL cell sample, followed by the RT reaction at 42°C (10 min), 25°C (10 min), 50°C (60 min), and 72°C (15 min). The estimated numbers of cells used from each sample is given in Table 1. Ig genes were PCR amplified by a semi-nested, isotype-specific reaction as previously reported (Wu et al., 2010). Briefly, a 25-μL PCR1 reaction mix contained 6.25 μL of cDNA, 0.625 U Phusion DNA polymerase (NEB, UK), 200 μM each dNTP, 41.75 nM each of 5′ IGHV gene family primer and 250 nM constant region primer (for either IgA, IgG, or IgM). Twenty microliters of PCR2 reaction mix comprised 2 μL PCR1 products, 0.5 U Phusion DNA polymerase, 200 μM each dNTPs, 41.75 nM each of 5′ IGHV gene family, and 250 nM nested constant region primer where all primers contained matched multiplex identifiers (MIDS). PCR conditions are as follows: 98°C for (30 s), 15 (PCR1) or 20 (PCR2) cycles of 98°C (10 s); 58°C (15 s); 72°C (30 s), and 1 cycle of 72°C (5 min). Purification of PCR products in sufficient quantity for high-throughput sequencing on the Roche Titanium platform, and the downstream data processing are as previously published (Wu et al., 2010).
Sequence Analysis
Ig gene usage, CDR-H3 junction regions and degree of hypermutation (by percentage match to the equivalent germline IGHV gene) were determined using V-QUEST (Wilkins et al., 1999). Subclasses of sequences were further identified by internal motifs in the constant regions of each sequence. ProtParam (Brochet et al., 2008) was used to determine the physicochemical properties of the CDR-H3 peptide between the conserved first (cysteine) and last amino acid (tryptophan). Clonotype clustering was performed based on the DNA sequence of the CDR-H3, as previously reported (Ademokun et al., 2011). Genealogic trees of sequences were constructed after alignment of Ig sequences with putative germline genes using DNAstar alignment and editing software (Laser Gene).
Statistics
Statistics were performed with GraphPad Prism 4.0. Three methods were used: One-way ANOVA (parametric, with Bonferroni post-test), Kruskal–Wallis comparisons (non-parametric, with Dunn’s post-test), and chi-square (with Bonferroni post-test).
Results
Morphological Similarity between CD27− and CD27+ Memory Cells
Enriched CD19+ B cells from nine healthy donors were separated into four subsets based on their surface CD27 and IgD expression (Figure 1A): antigen-inexperienced transitional and naïve cells (CD27−IgD+; 61.0 ± 5.3%), classical memory (CD27+IgD−: 7.7 ± 2.3%), IgM memory (CD27+IgD+; 12.86 ± 2.8%), and CD27− memory cells (CD27−IgD−; 4.9 ± 1.6%). The cellular size and granularity were measured by FSC and SSC, respectively, using flow cytometry. CD27− cells were 10% larger and 40% more granular than antigen-inexperienced CD27−IgD+ cells (Figures 1B,C; p < 0.0005). However, neither the size nor granularity of CD27− cells showed a significant difference as compared with CD27+IgD− or CD27+IgD+ memory cells.
Ig Gene Hypermutation in CD27− Cells
Deep-sequencing resulted in a total of 69250 IGH sequences, which represented 32457 different clonotypes after clustering by CDR-H3 identity (Table 1). The extent of hypermutation is estimated by the “percentage similarity to germline” value returned from V-QUEST. This would include errors introduced during PCR and sequencing, and therefore we used the level of “mutation” in the antigen-inexperienced, CD27−IgD+, cells to estimate the background error rate (1.3 ± 0.02%, or 1.38 bp per IGHV gene) as in Figure 2A. The switched (IgA and IgG) CD27− cells had significantly more mutation in their IGHV genes (5.8 ± 0.06%, p < 0.0005) than CD27−IgD+ antigen-inexperienced and CD27+IgD+IgM+ memory (3.9 ± 0.32%) cells. As previously reported for IgG genes (Fecteau et al., 2006), the level of IGHV mutation in switched CD27− cells was significantly lower than in switched CD27+IgD− memory cells (p < 0.0005). This was seen in all classes and subclasses apart from IgA2 (Figure 2B). The lack of a significantly different mutation rate in IgA2 cells was due to skewing by one donor (data not shown). IgM memory mutation rates are lower than switched memory mutation rates in both populations. Higher average values of mutation load are generally due to greater numbers of cells with highly mutated Ig genes (Figures 2C–G). Further comparison of IGHV3-23 sequences showed there were no major differences in the types of mutation occurring in each population, with all showing the SHM characteristics of having more transition than transversion mutations and strand bias in the AT mutations (Table 2). Neither were there any differences in the targeting of mutations to hotspots (data not shown). Thus, the majority of switched CD27− cells appear to be memory cells that acquire mutations through a similar mechanism to the GC-derived switched CD27+IgD− memory cells.
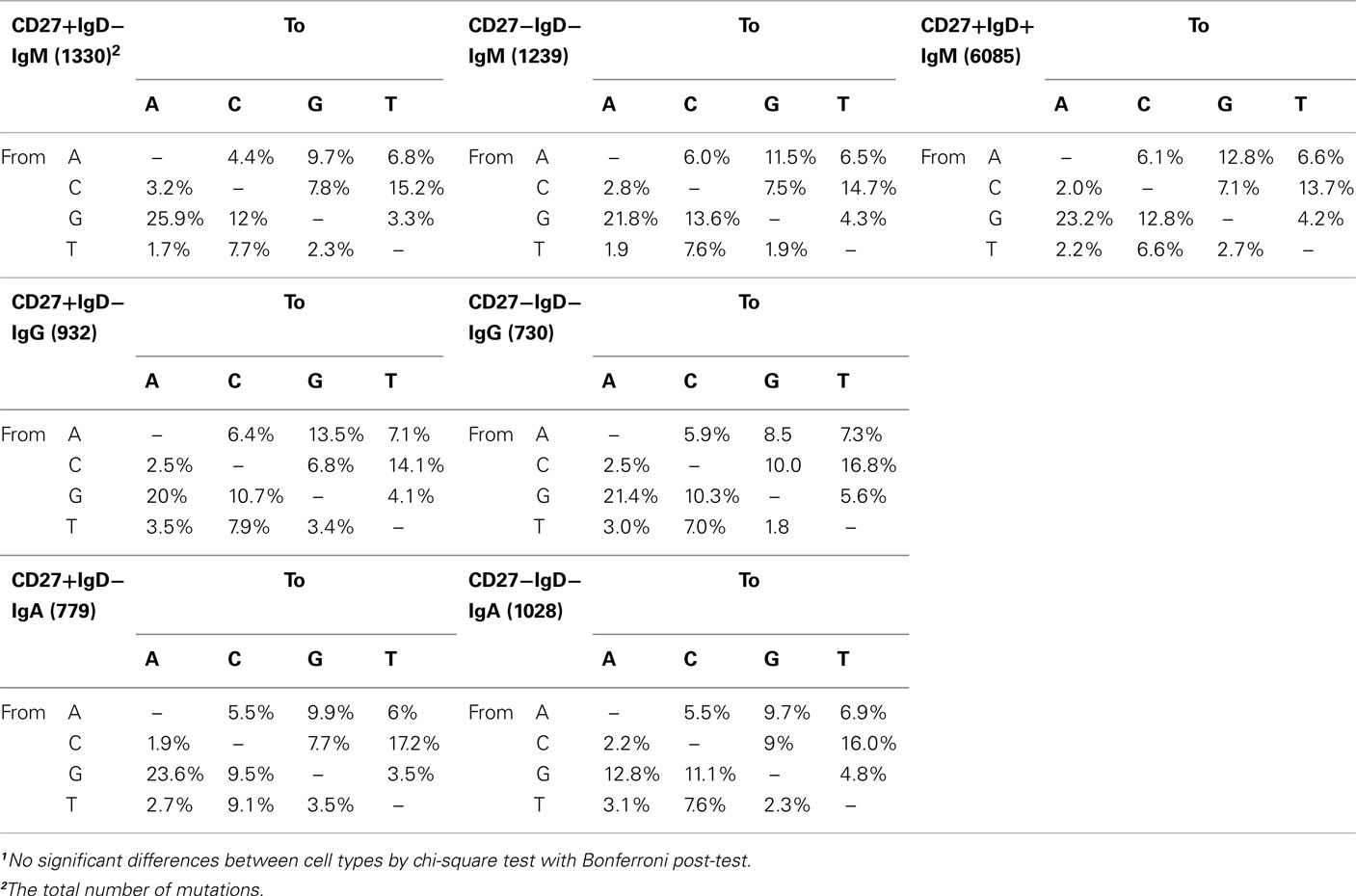
Table 2. Frequency (percent) of different types of mutations in the IGHV3-23 genes of switched IgD− memory cells1.
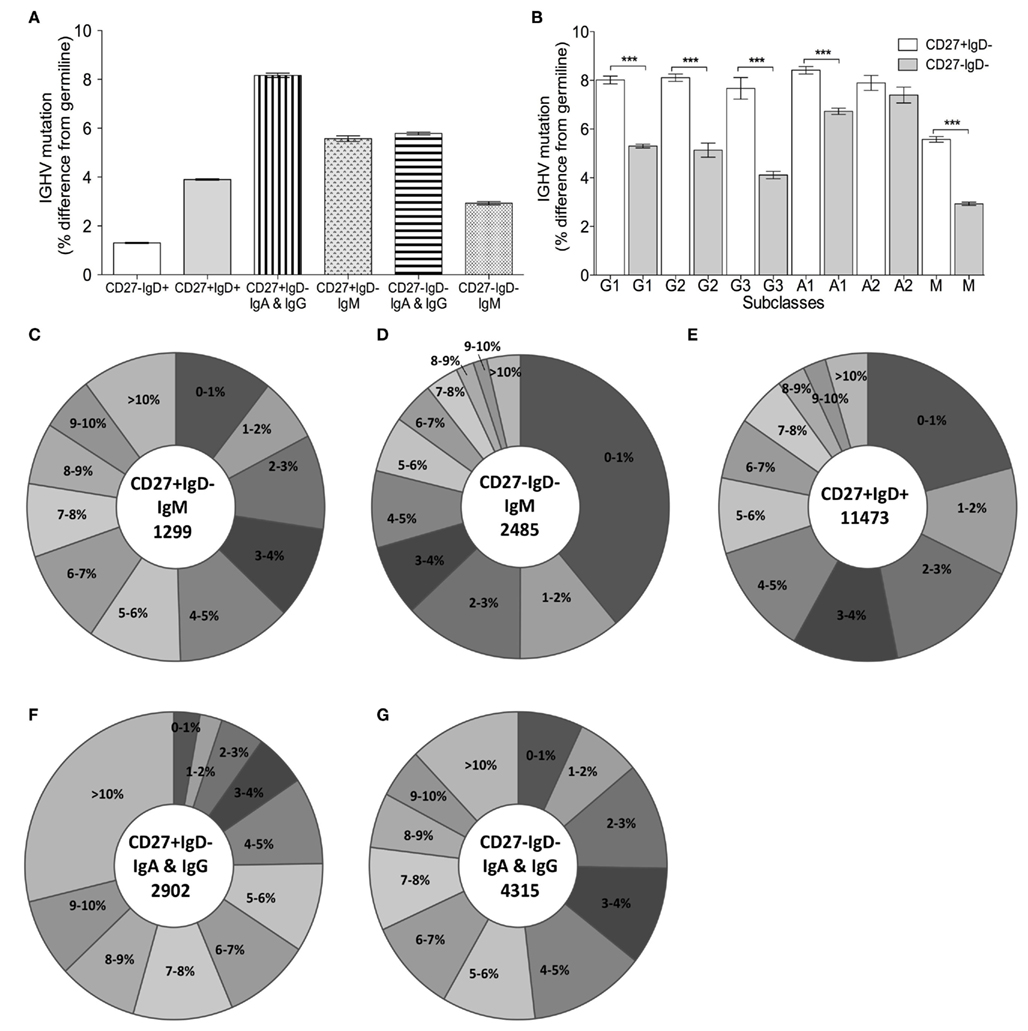
Figure 2. Somatic hypermutation in IGHV genes from different B cell subsets. (A) IGHV genes were identified from the Ig gene sequences by V-QUEST and the level of IGHV mutation is represented by the% difference from germline (y-axis) for CD27−IgD+ (n = 9975), CD27+IgD+ (n = 11473), switched CD27+IgD− (IgA+IgG; n = 2902), CD27+IgD−IgM+ (n = 1299) switched CD27−IgD− (IgA+IgG; n = 4315) and CD27−IgD−IgM+ (n = 2485) clonotypes from all donors combined. The low level of mutation in CD27−IgD+ was taken to be the background for the method. Kruskal–Wallis comparisons (non-parametric, with Dunn’s post-test) showed all comparisons were significant at p < 0.0001 except CD27+IgD−IgM+ compared with switched CD27−IgD−. (B) The mean IGHV mutation (y-axis;% difference from germline) is illustrated for CD27+IgD− (open bars) and CD27−IgD− (grey bars) clonotypes from all donors combined by different isotypes and subclasses, indicated as G for IgG, A for IgA, and M for IgM (G1: n = 764, G2: n = 764; G3: n = 132, A1: n = 1063, A2: n = 195, M: n = 1299 for CD27+IgD−; G1: n = 1777, G2: n = 120; G3: n = 132, A1: n = 1210, A2: n = 184, M: n = 2485 CD27−IgD−). Error bars indicate SEM. p Values for Kruskal–Wallis comparisons are indicated as ***p < 0.0005. Pie charts show the IGHV mutation levels, as indicated by the% difference from germline, for (C) CD27+IgD−IgM+, (D) CD27−IgD−IgM+. (E) CD27+IgD+IgM+ (F) switched CD27−IgD− and (G) switched CD27−IgD− clonotypes from all donors combined.
Clonally Related Memory Cells Independent of CD27 Expression
To determine whether CD27− cells are related to CD27+IgD− memory cells, we looked for evidence of clonally related cells, i.e., genes that shared unique CDR-H3 regions and mutation footprints, but were isolated from different populations. Out of 7225 IgA and IgG clones from nine donors, a total of 32 clones (0.44%) showed clonality between the two cell subsets. The sequences from these clones were used to generate genealogical trees (Figure 3). The majority of genealogical trees were of a single isotype: IgA (20 clones; e.g., Figures 3A–D) and IgG (7 clones; e.g., Figures 3E–F). There were also examples of clones containing different isotypes (four clones; Figures 3G–K,N,O) and subclasses (e.g., Figures 3F,G,L,M,O). We found four clones isolated from CD27+ and CD27− IgD− cells containing both IgM and IgG isotypes (Figures 3G,J,K,N). Class switching within a family was also seen in clones containing only CD27− cells (Figures 3L–O). In some lineage trees, sequences from CD27− and CD27+IgD− memory cells appeared on the same branch with no intermediate nodes, allowing us to determine the sequence of CD27 expression in the two cell subsets. While some trees indicated that CD27− cells were the progenitor of CD27+ cells (e.g., Figures 3F,J), other trees indicated that CD27+ cells were the progenitor of CD27− cells (e.g., Figures 3C,E,K). In other cases we could see examples of an identical clone, in terms of CDR-H3 and mutation footprint, but existing in two different populations (Figure 3D), or as a different class of antibody, or both (Figures 3G,H). We saw no relationships between CD27+IgD+IgM+ cells and switched IgD− cells, but we did find examples of related IgM clones in the three different memory populations (Figures 3P–U).
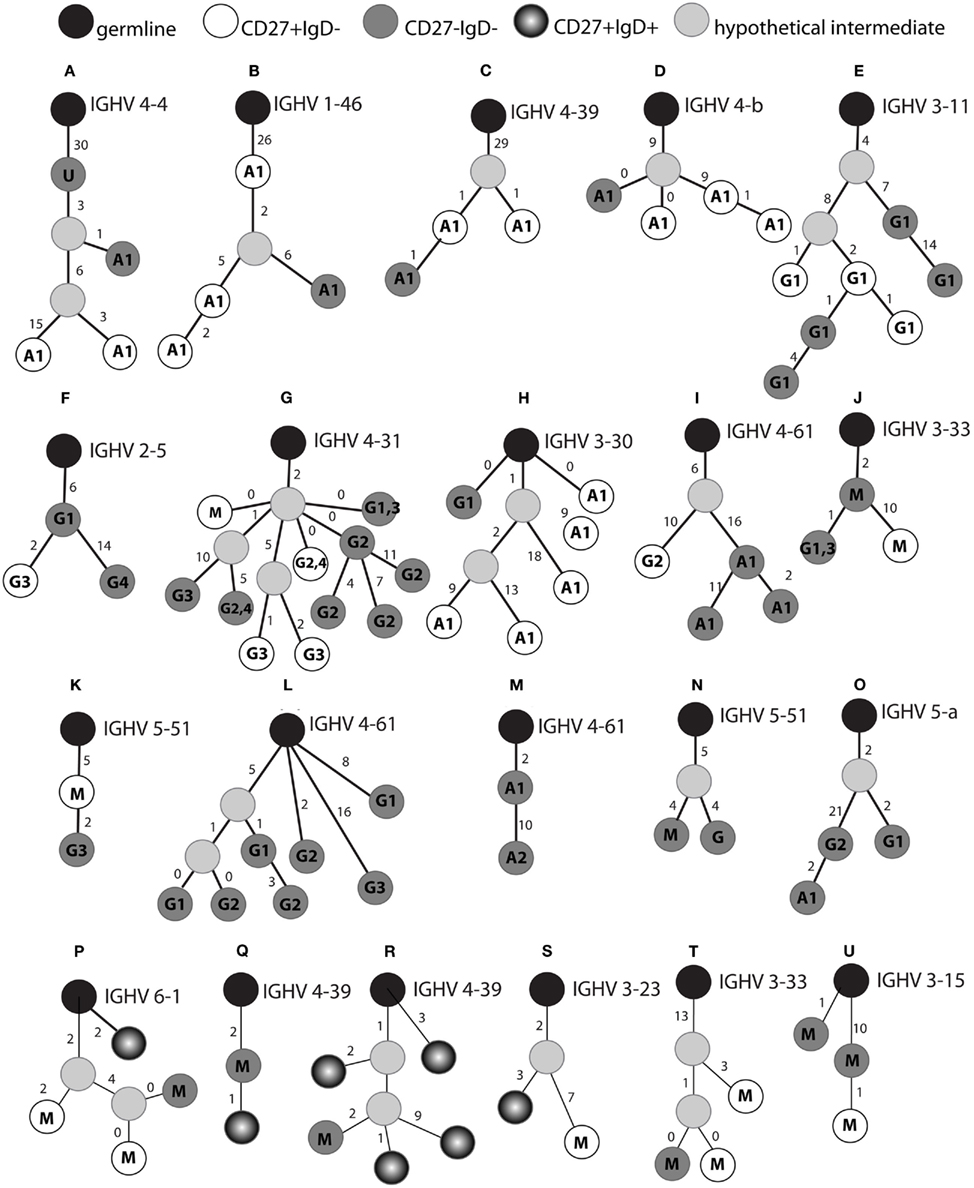
Figure 3. Genealogical relationships between class switched CD27+ and CD27− cells. The CDR-H3 sequence in each Ig gene sequence was identified by V-QUEST and used to cluster sequences sharing the same CDR-H3. Genealogical trees (A–U) were generated by aligning sequences with the germline sequences (black circles). Cells are represented as light grey (hypothetical intermediates), white (CD27+IgD−), dark grey (CD27−IgD−), and black gradient (CD27+IgD+). The class (G: IgG, A: IgA, M: IgM, U: unknown) or subclass (indicated by numbers) of the gene is indicated in each circle. Numbers besides the bars represent the number of IGHV mutations between two different sequences. If two sequences of different origins or different isotypes have exactly the same genotype this is indicated by a branch with zero beside the bar. Multiple identical sequences of the same origin and isotype did occur but are not indicated here.
CDR-H3 Repertoire in CD27− Cells
Mutated memory B cells generally use smaller, more hydrophilic and more basic CDR-H3 regions than naive cells (Meffre et al., 2001; Souto-Carneiro et al., 2004; Wu et al., 2010). In response to vaccination, B cells with these CDR-H3 properties were also preferentially expanded (Ademokun et al., 2011), indicating that the properties likely have some, as yet unknown, functional relevance. We analyzed the CDR-H3 size and peptide properties from clonotypes in the four different subsets. All groups showed a Gaussian distribution of CDR-H3 sizes, indicating a diversity of clonotypes with no preferential expansion of a particular CDR-H3 size (data not shown). Although the mean CDR-H3 size in CD27− memory cells was significantly smaller than antigen-inexperienced CD27−IgD+ cells, it was significantly bigger than CD27+IgD− and CD27+IgD+ memory cells (p < 0.0005; Figure 4A). The grand average hydropathicity (GRAVY) index, a positive indicator for peptide hydrophobicity, had a value in CD27− switched memory cells (−0.54 ± 0.01) that fell between that of antigen-inexperienced CD27−IgD+ cells (−0.49 ± 0.01; p < 0.0005) and the two CD27+ memory subsets (−0.57; p < 0.0005; Figure 4B), i.e., CDR-H3 from switched CD27− were more hydrophilic than antigen-inexperienced cells but more hydrophobic than CD27+ memory cells. CD27−IgD−IgM+ memory cells had a similar hydrophobicity to that of antigen-inexperienced CD27−IgD+ cells. The theoretical pI indicates the pH value at which a peptide carries no net charge. The CDR-H3 pI from switched CD27− cells (6.18 ± 0.32) significantly differed from those in the CD27+ memory cell subsets, with a value between CD27−IgD+ (5.98 ± 0.02, p < 0.0005) and the two CD27+ memory subsets (6.39–6.53; p < 0.05; Figure 4C). Therefore, CDR-H3 from switched CD27− cells would be more basic than antigen-inexperienced cells but more acidic than CD27+ memory cells in the peripheral blood. The CDR-H3 pI from the CD27− IgD− IgM+ memory cells was different again, showing the highest pI values of all subsets. To account for the possibility that the CD27− cells might be a mixed population, with some cells being directly related to CD27+ cells with a similar level of mutation (e.g., Figure 3) and others that may be less mutated of a different origin, we analyzed only the highly mutated genes in each group. However, this made no difference to the results (data not shown).
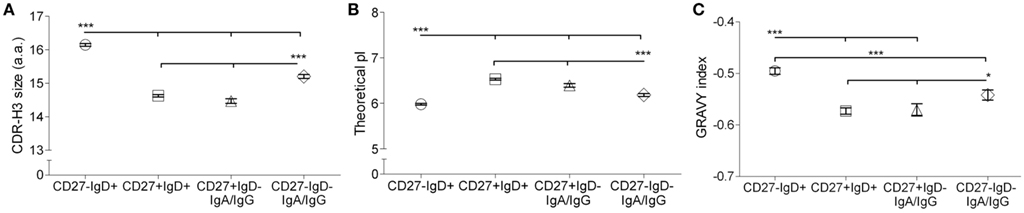
Figure 4. CDR-H3 properties in B cell subsets. The amino acids of the CDR-H3 region in the Ig sequences was determined from V-QUEST and subjected to ProtParam analysis to determine CDR-H3 peptide characteristics. (A) The size of CDR-H3 in amino acids, (B) the theoretical pI and (C) the GRAVY index for CD27−IgD+ (open circle; n = 9975), CD27+IgD+ (open square; n = 11473), switched CD27+IgD− (IgA+IgG; open triangle; n = 2902), CD27+IgD−IgM+ (grey triangle; n = 1299), switched CD27−IgD− (IgA+IgG; open diamond; n = 4315) and CD27−IgD−IgM+ (grey diamond; n = 2485) clonotypes with all ranges of IGHV mutations from all donors combined. Whiskers indicate 2.5–97.5%. p Values for Kruskal–Wallis comparisons are indicated as *p < 0.05 and ***p < 0.0005.
CD27− and CD27+ Cells Share IGHV Repertoire Characteristics
We previously reported that CD27+IgD+ memory cells use a very distinct IGHV repertoire from isotype-switched memory cells, and concluded that a large fraction of CD27+IgD+ memory cells are therefore not directly related to GC-derived, switched memory cells (Wu et al., 2010). Here we compared the IGH repertoire in CD27− clonotypes with that of the three other populations. In order to consider inter-individual variation, the frequency of IGH gene usage, returned by V-QUEST, was calculated for each individual donor before being used in grouped comparisons. The IGHJ gene usage was similar between CD27− and the two CD27+ memory subsets, all except the CD27−IgD−IgM+ cells having significantly higher IGHJ4 and lower IGHJ6 gene usage than antigen-inexperienced CD27−IgD+ cells (p < 0.0005; Figure 5A). The CD27−IgM+ memory cells had a significantly lower IGHJ6, but a similar IGHJ4 gene usage, compared to antigen-inexperienced CD27−IgD+ cells. As previously shown, there are differences in IGHV gene family usage between antigen-inexperienced CD27−IgD+ cells and memory subsets in IGHV1, IGHV3, and IGHV4 gene families (Figure 5B). All IgM+ cells from the three memory populations had the same unique characteristic of having a higher IGHV3 and lower IGHV1 usage (Figure 5B). When individual IGHV genes were considered the family differences were due to the same individual gene changes – IGHV3-23 in particular being higher in the IgM memory than the antigen-inexperienced cells (Figures 5D–F vs. 5C). There were no significant differences in IGHV gene family usage between CD27− and CD27+ switched cells, both populations contained a higher frequency of IGHV1 gene family at the expense of IGHV3 (p < 0.0005) in comparison with CD27+IgD+ memory cells. When the data was analyzed by individual IGHV genes rather than gene families a similar pattern emerged, with the repertoire of switched CD27− cells resembling that of switched CD27+ cells rather than any of the IgM+ populations. (Figures 5C–H; Table 3). Only three IGHV genes were found to be significantly different between the CD27− and CD27+ switched populations, whereas just two IGHV genes showed significant differences between the three IgM+ memory populations.
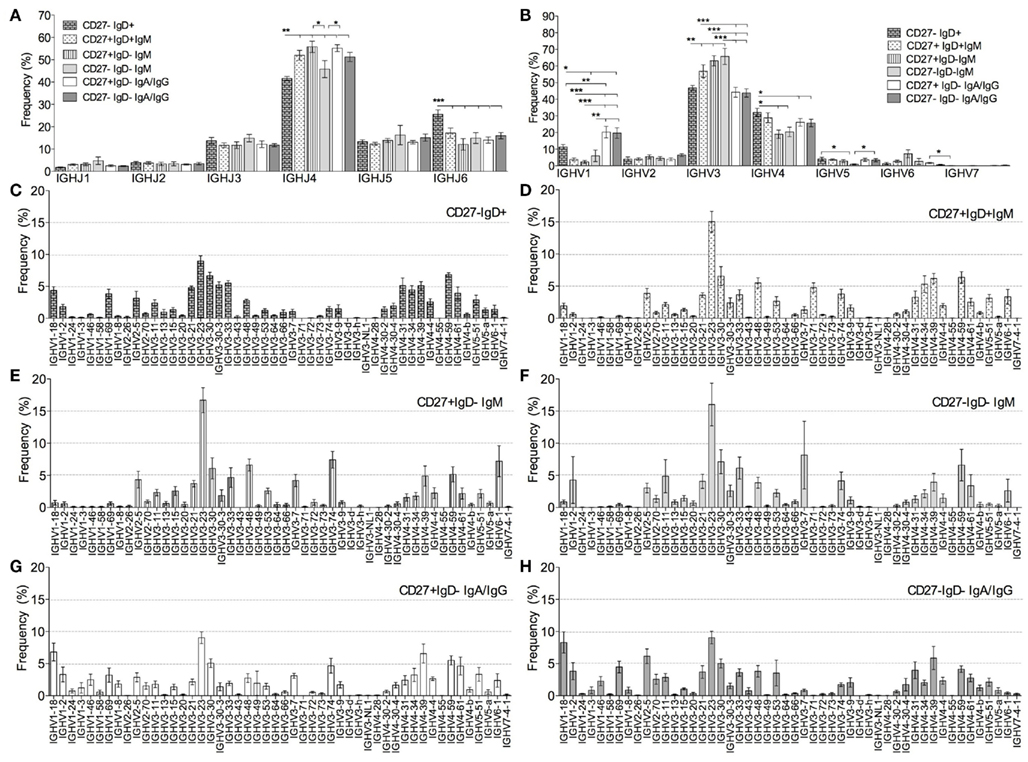
Figure 5. Ig gene usage in B cell subsets. IGHV and IGHJ genes were identified from the Ig gene sequences by V-QUEST. The relative frequency (% in y-axis) of (A) six IGHJ genes and (B) seven IGHV gene families was calculated for individual donors before being collectively compared between CD27−IgD+ (brick bars), CD27+IgD+IgM+ (dotted bars), CD27+IgD−IgM+ (stripe bars), CD27−IgD−IgM+ (light grey bars), switched CD27+IgD− (IgA+IgG; open bars) and switched CD27−IgD− (IgA+IgG; dark grey bars) clonotypes from all donors. p Values for one-way ANOVA comparisons are indicated as *p < 0.05, **p < 0.005, and ***p < 0.0005. IGHV gene families were further separated into individual IGHV genes and their relative frequencies were calculated for (C) CD27−IgD+, (D) CD27+IgD+, (E) IgM CD27+IgD−, (F) IgM CD27−IgD− (G) switched CD27+IgD− (IgA+IgG) and (H) switched CD27−IgD− (IgA+IgG) clonotypes. Genes showing a significant different in usage between different populations (one-way ANOVA) are shown in Table 3. Error bars indicate SEM.
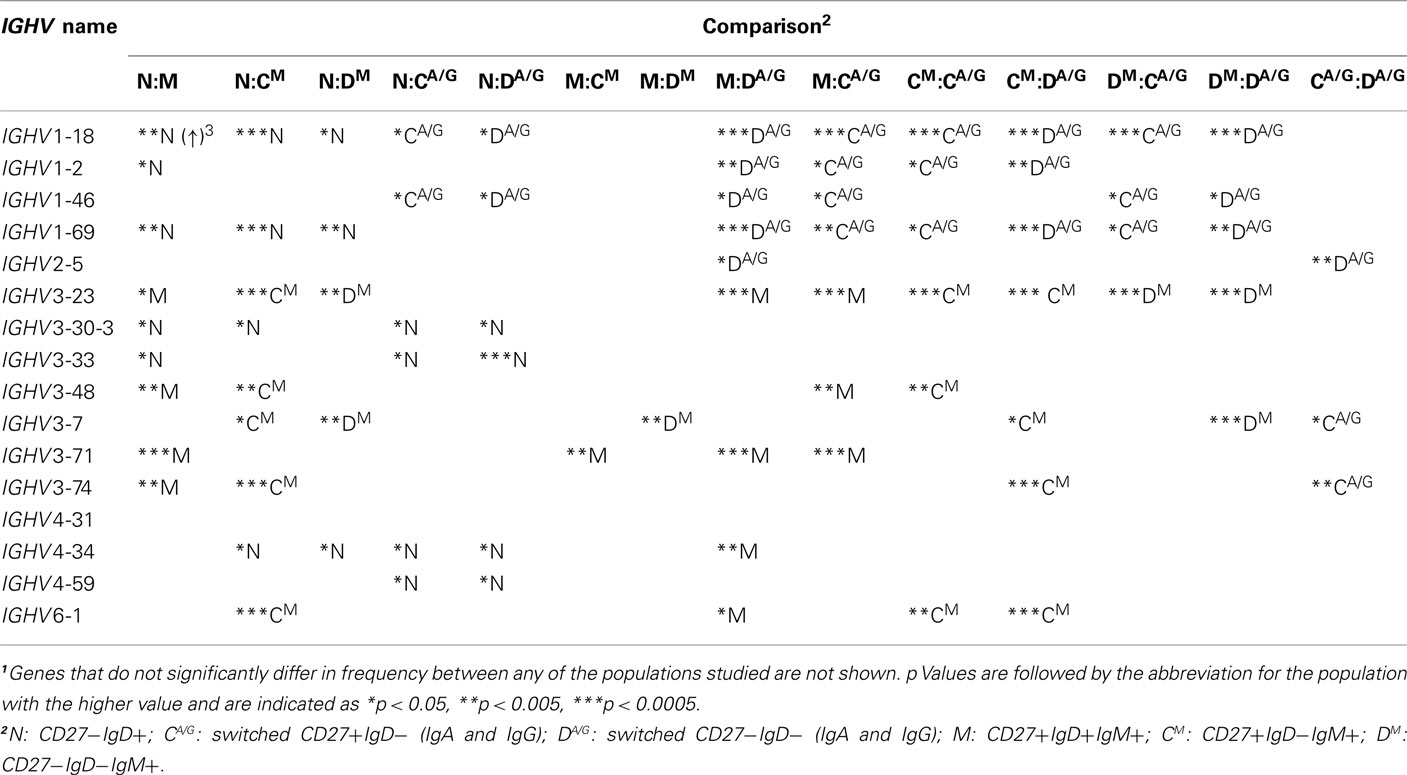
Table 3. One-way ANOVA comparisons of individual IGHV genes between different memory B cell subsets1.
Heterogeneity of IGHV Repertoires between Different Subclasses of Memory Cells
Memory B cells of different isotypes and subclasses have different functions and may display different IGHV repertoires as a result of selection by different type of antigens. When the data was split into subclasses the IGHJ repertoire in CD27− and CD27+IgD− switched memory cells were similar (data not shown). However, the IGHV repertoire varied between different classes and subclasses of genes (Figure 6). It can be seen that clonotypes of IgG1, IgG3, and IgA1 subclasses resembled one another in their IGHV gene family usage, with significantly higher levels of IGHV1 and lower levels of IGHV3 gene families compared to IgM+ memory (CD27+IgD+), as previously reported. Interestingly, there were also significant differences between these three subclasses and the IgG2, IgA2 subclasses, which had IGHV1 and IGHV3 family usage reminiscent of the IgM+ memory signature (Figure 6). IgG2 in particular had more in common with IgM memory cells (no significant difference in IGHV family usage) than with the other isotype-switched cells, having significantly lower use of IGHV1 and higher use of IGHV3 compared to the switched isotypes (Figure 6). This indicates that common selective influences shape the IgM and IgG2 memory repertoires.
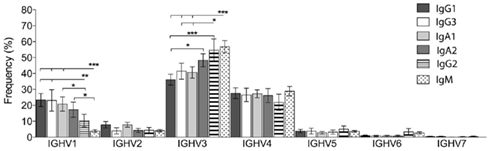
Figure 6. IGHV gene family usage between different subclasses of memory cells. IGHV gene families were identified from the Ig gene sequences by V-QUEST. Sequences from CD27+IgD− and CD27−IgD− clonotypes were combined and then grouped by different subclasses: IgG1, IgG2, IgG3, IgA1, and IgA2. IgM+ sequences were from the CD27+IgD+ cells. The relative frequency (% in y-axis) of seven IGHV gene families was calculated for individual donors before being collectively compared between groups. Error bars indicate SEM. p Values for one-way ANOVA comparisons are indicated as *p < 0.05, **p < 0.005, and ***p < 0.0005.
Discussion
We have shown that CD27− memory cells have properties in common with CD27+ memory. In agreement with previous reports (Fecteau et al., 2006), they are larger and more granular than CD27−IgD+ antigen-inexperienced cells, and are morphologically comparable with CD27+ memory cells (Figures 1B,C). As memory cells, CD27−IgD− cells contain class switched Ig genes that are mutated. Large-scale sequencing analysis has enabled the comparison of mutation characteristics within a single gene, the IGHV3-23 gene, which avoids comparison problems that can sometimes occur by comparing different IGHV genes. We showed that both populations have similar characteristics of SHM (Table 2). However, as previously reported, the level of mutation between the CD27− and CD27+ memory cells differs (Figure 2), with CD27− cells carrying more mutation than CD27+IgD+ memory cells but less than CD27+IgD− memory cells, a decrease not restricted to a particular class or subclass of antibody.
A decreased level of SHM could mean that the CD27− cells with lower mutation levels are related to the CD27+ cells but are at an earlier stage of development with regards to affinity maturation, perhaps representing early GC cells that fail to complete GC reactions (Wei et al., 2007). CD27− cells have significantly lower CD40 expression than CD27+IgD− memory cells (Colonna-Romano et al., 2009) so would be less able to receive T cell help. Whether decreased CD40 is a cause or consequence of reduced CD27 is not known, but loss of CD40 in a TD responding cell line would prevent it from progressing through the TD affinity maturation processes. It has also been suggested that CD27− cells are the memory cells from the primary immune responses, as opposed to the more extensively hypermutated CD27+ IgG memory cells from the secondary immune response, because they have been through less divisions (Berkowska et al., 2011), although these results could also be explained by a halting of a TD reaction due to downregulation of co-stimulatory molecules. Alternatively the CD27− cells may be unrelated to the CD27+ cells and are responding to a different set of stimuli which results in less mutation, perhaps TI stimuli as has been suggested for CD27+IgD+ cells.
Since we had previously distinguished “innate-like” memory B cells (CD27+IgD+) by their IGHV repertoire, we thought that any differences in functional selection would be noticeable in a comparison of switched Ig genes from CD27− and CD27+ cells. We found very few differences in repertoire; only three individual IGHV genes showed a significant differences and no differences in overall IGHV family usage (Figure 5; Table 3). The switched CD27− memory cell repertoire had more in common with that of switched CD27+ memory cells than it did with that of either CD27−IgD+ antigen-inexperienced B cells, or CD27+IgD+ innate-like B cells. The two IgD−IgM+ cell populations both had the same repertoire characteristics as the CD27+IgD+ innate-like B cells, indicating common influences on their development. Furthermore, using CDR-H3 clustering to group related cells, we found examples of clones with family members in both the CD27− and CD27+ switched populations (Figure 3). Although these were of a low frequency in our data (0.44% of clones were found to have mixed CD27 expression), it is clear evidence in favor of a common activation pathway for both CD27− and CD27+ switched memory cells. Since the number of cells sampled in this study only accounted for a small fraction of IgA and IgG B cells (approximately 0.04% in 5 L of blood), we estimate that there would be approximately 3 × 104 examples of clonotypes which comprised both CD27− and CD27+ switched memory cells in a healthy adult. That 1 out of 1000 switched CD27− clonotypes would have CD27+IgD− relatives in healthy blood or, conversely, 1 out of 1500 switched CD27+IgD− clones would have CD27− relatives. Therefore the degree of relatedness between switched CD27− and CD27+IgD− cells is relatively high, as compared with our estimation that 20,000 clonotypes of CD27−IgD+CD10− naïve cells are required to find one related to a CD27+IgD− memory cell (data not shown). We also found clonally related IgM cells from three different memory populations, additional evidence that the IgM memory cells share a common developmental pathway regardless of IgD and CD27 expression. Importantly, since we find both CD27− cells that are precursors of CD27+ cells and vice versa, or even clonotypes with exactly the same mutational footprint but different subclass and CD27 status (Figure 3), it would appear that CD27 expression is not an indicator of linear progression through B cell development, because it can be lost or gained within a clonal expansion of cells.
While it is clear from the above data that some of the cells in the CD27−IgD− gate are related to CD27+IgD− cells, there are differences at a population level that suggest there may be cells which are responding to a different type of antigen. The CDR-H3 region of the Ig gene is particularly important for antigen recognition. Ig genes from memory B cells generally contain CDR-H3 regions that are smaller, more hydrophilic and more basic in the peripheral blood than those from naive cells (Meffre et al., 2001; Souto-Carneiro et al., 2004; Wu et al., 2010). Preferential expansion of B cells using CDR-H3 with these properties was also observed in individuals challenged with vaccinations (Ademokun et al., 2011). Our results demonstrate that selection is imposed on switched CD27− cells favoring those with CDR-H3 regions that are shorter, more hydrophilic and more basic than those from antigen-inexperienced cells, i.e., they show characteristics of memory cells. However, despite having an IGHJ gene usage comparable to that of CD27+ memory cells (Figure 5), they do not have the same degree of “memory characteristics,” i.e., the composition of CDR-H3 regions from switched CD27− cells falls in between that of antigen-inexperienced CD27−IgD+ and memory CD27+ cells, being significantly different from both (Figure 4).
The relevance of these CDR-H3 differences is not known, since the CDR-H3 characteristics are also dissimilar to those of the “innate-like” CD27+IgD+ cells so they are unlikely to be due to a difference between TD and TI antigens. One hypothesis is that the CD27− memory compartment contains B cells that can be autoreactive, and hence have downregulated their activation molecules in an effort to limit autoreactivity. It is pertinent here that the population is increased in SLE, an antibody-mediated autoimmune disease (Wei et al., 2007). In addition, it has been reported that longer, more acidic CDR-H3 regions are associated with autoreactivity (Aguilera et al., 2001; Capello et al., 2004). The CD27− memory cells have decreased levels of stimulatory molecules (CD27; CD40; the α subunit of IL-6 receptor, CD126) and increased levels of inhibitory molecules (CD330a and FcRH4; Ehrhardt et al., 2005; Sanz et al., 2008; Silva et al., 2011) so would appear to be unable to continue development into antibody-secreting PCs. The occurrence of CD27− memory cells within lineage trees that are thought to be TD expansions of cells (Figure 3) could be explained by either gaining of autoreactivity through SHM, or by cross reactivity between activating- and auto-antigens.
A particularly interesting finding in this data was that, while the overall IGHV gene repertoire did not change between CD27− and CD27+ switched memory cells, and the IGHV gene repertoire matched by subclass also showed no differences, there were differences in IGHV repertoire between different subclasses of Ig genes. Previously we have shown that a decrease in IGHV1 family and increase in IGHV3 family distinguished CD27+IgD+ memory cells from switched CD27+ memory cells. We have shown here that IgG2, and to a lesser extent IgA2, share these characteristics (Figure 6). Thus on the basis of repertoire similarity we would group IgM, IgG2, and memory cells together, irrespective of CD27 status, under the heading of “innate-like” B cells that respond to antigen in a TI manner. This would be consistent with the finding that systemic vaccination against pneumococcal polysaccharides predominately induces antibody responses of IgG2 (Carson et al., 1995) and IgA2 (Lue et al., 1988) subclasses in humans. Since our “innate-like” repertoire characteristics are less obvious for IgA2 cells this population may contain a higher proportion of cells responding to TD antigens.
In conclusion, we have shown that switched CD27− cells are memory cells that in terms of IGHV gene usage are most closely related to switched CD27+ cells. IgM memory cells with or without IgD and CD27 are related to each other but have a different repertoire to the switched cells. Subclasses of antibody indicate that IgG2, and some IgA2, may be formed in response to TI antigens and the proportions of these subclasses are comparable between the two populations. Thus in this respect the lack of CD27 and lower mutational load does not appear to be a result of a difference between TI and TD antigen responders. However, in terms of CDR-H3 characteristics, there may be a tendency toward loss of CD27 (or failure to upregulate CD27) in cells with longer, more acidic and more hydrophobic antibody binding sites. The relevance of these characteristics remains to be determined and it will be interesting to compare whether these are qualities of autoreactive antibodies such as are found in SLE or in immunosenescence.
Author Contribution
Yu-Chang Bryan Wu designed and carried out experiments, analyzed data and wrote the manuscript; David Kipling designed and wrote data handling and analysis scripts and wrote the manuscript; Deborah K. Dunn-Walters, oversaw the project, designed experiments and analytical tools, carried out data analysis and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the Human Frontiers Science program and Research into Ageing, Age UK. The authors would like to thank all our volunteers for their blood donation.
References
Ademokun, A., Wu, Y. C., Martin, V., Mitra, R., Sack, U., Baxendale, H., Kipling, D., and Dunn-Walters, D. K. (2011). Vaccination-induced changes in human B-cell repertoire and pneumococcal IgM and IgA antibody at different ages. Aging Cell 10, 922–930.
Agematsu, K., Nagumo, H., Yang, F. C., Nakazawa, T., Fukushima, K., Ito, S., Sugita, K., Mori, T., Kobata, T., Morimoto, C., and Komiyama, A. (1997). B cell subpopulations separated by CD27 and crucial collaboration of CD27+B cells and helper T cells in immunoglobulin production. Eur. J. Immunol. 27, 2073–2079.
Aguilera, I., Melero, J., Nunez-Roldan, A., and Sanchez, B. (2001). Molecular structure of eight human autoreactive monoclonal antibodies. Immunology 102, 273–280.
Arens, R., Nolte, M. A., Tesselaar, K., Heemskerk, B., Reedquist, K. A., Van Lier, R. A., and Van Oers, M. H. (2004). Signaling through CD70 regulates B cell activation and IgG production. J. Immunol. 173, 3901–3908.
Berkowska, M. A., Driessen, G. J., Bikos, V., Grosserichter-Wagener, C., Stamatopoulos, K., Cerutti, A., He, B., Biermann, K., Lange, J. F., Van Der Burg, M., Van Dongen, J. J., and Van Zelm, M. C. (2011). Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood 118, 2150–2158.
Blanchard-Rohner, G., Pulickal, A. S., Jol-Van Der Zijde, C. M., Snape, M. D., and Pollard, A. J. (2009). Appearance of peripheral blood plasma cells and memory B cells in a primary and secondary immune response in humans. Blood 114, 4998–5002.
Brochet, X., Lefranc, M. P., and Giudicelli, V. (2008). IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–W508.
Capello, D., Guarini, A., Berra, E., Mauro, F. R., Rossi, D., Ghia, E., Cerri, M., Logan, J., Foa, R., and Gaidano, G. (2004). Evidence of biased immunoglobulin variable gene usage in highly stable B-cell chronic lymphocytic leukemia. Leukemia 18, 1941–1947.
Carson, P. J., Schut, R. L., Simpson, M. L., O’brien, J., and Janoff, E. N. (1995). Antibody class and subclass responses to pneumococcal polysaccharides following immunization of human immunodeficiency virus-infected patients. J. Infect. Dis. 172, 340–345.
Colonna-Romano, G., Bulati, M., Aquino, A., Pellicano, M., Vitello, S., Lio, D., Candore, G., and Caruso, C. (2009). A double-negative (IgD-CD27-) B cell population is increased in the peripheral blood of elderly people. Mech. Ageing Dev. 130, 681–690.
Dunn-Walters, D. K., Isaacson, P. G., and Spencer, J. (1995). Analysis of mutations in immunoglobulin heavy chain variable region genes of microdissected marginal zone (MGZ) B cells suggests that the MGZ of human spleen is a reservoir of memory B cells. J. Exp. Med. 182, 559–566.
Ehrhardt, G. R., Hsu, J. T., Gartland, L., Leu, C. M., Zhang, S., Davis, R. S., and Cooper, M. D. (2005). Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J. Exp. Med. 202, 783–791.
Fecteau, J. F., Cote, G., and Neron, S. (2006). A new memory CD27-IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. J. Immunol. 177, 3728–3736.
Hintzen, R. Q., Lens, S. M., Koopman, G., Pals, S. T., Spits, H., and Van Lier, R. A. (1994). CD70 represents the human ligand for CD27. Int. Immunol. 6, 477–480.
Kelly, D. F., Snape, M. D., Clutterbuck, E. A., Green, S., Snowden, C., Diggle, L., Yu, L. M., Borkowski, A., Moxon, E. R., and Pollard, A. J. (2006). CRM197-conjugated serogroup C meningococcal capsular polysaccharide, but not the native polysaccharide, induces persistent antigen-specific memory B cells. Blood 108, 2642–2647.
Klein, U., Kuppers, R., and Rajewsky, K. (1997). Evidence for a large compartment of IgM-expressing memory B cells in humans. Blood 89, 1288–1298.
Klein, U., Rajewsky, K., and Kuppers, R. (1998). Human immunoglobulin (Ig)M+ IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med. 188, 1679–1689.
Kobata, T., Jacquot, S., Kozlowski, S., Agematsu, K., Schlossman, S. F., and Morimoto, C. (1995). CD27-CD70 interactions regulate B-cell activation by T cells. Proc. Natl. Acad. Sci. U.S.A. 92, 11249–11253.
Koethe, S., Zander, L., Koster, S., Annan, A., Ebenfelt, A., Spencer, J., and Bemark, M. (2011). Pivotal advance: CD45RB glycosylation is specifically regulated during human peripheral B cell differentiation. J. Leukoc. Biol. 90, 5–19.
Langley, J. M., Dodds, L., Fell, D., and Langley, G. R. (2010). Pneumococcal and influenza immunization in asplenic persons: a retrospective population-based cohort study 1990–2002. BMC Infect. Dis. 10, 219. doi:10.1186/1471-2334-10-219
Lens, S. M., Keehnen, R. M., Van Oers, M. H., Van Lier, R. A., Pals, S. T., and Koopman, G. (1996). Identification of a novel subpopulation of germinal center B cells characterized by expression of IgD and CD70. Eur. J. Immunol. 26, 1007–1011.
Lue, C., Tarkowski, A., and Mestecky, J. (1988). Systemic immunization with pneumococcal polysaccharide vaccine induces a predominant IgA2 response of peripheral blood lymphocytes and increases of both serum and secretory anti-pneumococcal antibodies. J. Immunol. 140, 3793–3800.
Ma, C. S., Pittaluga, S., Avery, D. T., Hare, N. J., Maric, I., Klion, A. D., Nichols, K. E., and Tangye, S. G. (2006). Selective generation of functional somatically mutated IgM+ CD27+, but not Ig isotype-switched, memory B cells in X-linked lymphoproliferative disease. J. Clin. Invest. 116, 322–333.
Meffre, E., Milili, M., Blanco-Betancourt, C., Antunes, H., Nussenzweig, M. C., and Schiff, C. (2001). Immunoglobulin heavy chain expression shapes the B cell receptor repertoire in human B cell development. J. Clin. Invest. 108, 879–886.
Moir, S., Ho, J., Malaspina, A., Wang, W., Dipoto, A. C., O’shea, M. A., Roby, G., Kottilil, S., Arthos, J., Proschan, M. A., Chun, T. W., and Fauci, A. S. (2008). Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med. 205, 1797–1805.
Rojas, O. L., Narvaez, C. F., Greenberg, H. B., Angel, J., and Franco, M. A. (2008). Characterization of rotavirus specific B cells and their relation with serological memory. Virology 380, 234–242.
Sanz, I., Wei, C., Lee, F. E., and Anolik, J. (2008). Phenotypic and functional heterogeneity of human memory B cells. Semin. Immunol. 20, 67–82.
Silva, R., Moir, S., Kardava, L., Debell, K., Simhadri, V. R., Ferrando-Martinez, S., Leal, M., Pena, J., Coligan, J. E., and Borrego, F. (2011). CD300a is expressed on human B cells, modulates BCR-mediated signaling, and its expression is down-regulated in HIV infection. Blood 117, 5870–5880.
Souto-Carneiro, M. M., Longo, N. S., Russ, D. E., Sun, H. W., and Lipsky, P. E. (2004). Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J. Immunol. 172, 6790–6802.
Tangye, S. G., Avery, D. T., Deenick, E. K., and Hodgkin, P. D. (2003). Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J. Immunol. 170, 686–694.
Tangye, S. G., Liu, Y. J., Aversa, G., Phillips, J. H., and De Vries, J. E. (1998). Identification of functional human splenic memory B cells by expression of CD148 and CD27. J. Exp. Med. 188, 1691–1703.
Van Gent, R., Van Tilburg, C. M., Nibbelke, E. E., Otto, S. A., Gaiser, J. F., Janssens-Korpela, P. L., Sanders, E. A., Borghans, J. A., Wulffraat, N. M., Bierings, M. B., Bloem, A. C., and Tesselaar, K. (2009). Refined characterization and reference values of the pediatric T- and B-cell compartments. Clin. Immunol. 133, 95–107.
Wei, C., Anolik, J., Cappione, A., Zheng, B., Pugh-Bernard, A., Brooks, J., Lee, E. H., Milner, E. C., and Sanz, I. (2007). A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J. Immunol. 178, 6624–6633.
Weller, S., Braun, M. C., Tan, B. K., Rosenwald, A., Cordier, C., Conley, M. E., Plebani, A., Kumararatne, D. S., Bonnet, D., Tournilhac, O., Tchernia, G., Steiniger, B., Staudt, L. M., Casanova, J. L., Reynaud, C. A., and Weill, J. C. (2004). Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a pre diversified immunoglobulin repertoire. Blood 104, 3647–3654.
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., and Hochstrasser, D. F. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552.
Wirths, S., and Lanzavecchia, A. (2005). ABCB1 transporter discriminates human resting naive B cells from cycling transitional and memory B cells. Eur. J. Immunol. 35, 3433–3441.
Keywords: CD27, memory, Ig gene repertoire, subclass heterogeneity, CDR-H3
Citation: Wu Y-CB, Kipling D and Dunn-Walters DK (2011) The relationship between CD27 negative and positive B cell populations in human peripheral blood. Front. Immun. 2:81. doi: 10.3389/fimmu.2011.00081
Received: 13 October 2011;
Paper pending published: 16 November 2011;
Accepted: 05 December 2011;
Published online: 26 December 2011.
Edited by:
David Tarlinton, Walter and Eliza Hall Institute, AustraliaReviewed by:
Claudia Berek, Charite University Medicine Berlin, GermanyDinesh S. Rao, University of California Los Angeles, USA
Copyright: © 2011 Wu, Kipling and Dunn-Walters. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Deborah K. Dunn-Walters, Department of Immunobiology, King’s College London School of Medicine, Guy’s Campus, London SE1 9RT, UK. e-mail:ZGVib3JhaC5kdW5uLXdhbHRlcnNAa2NsLmFjLnVr

 David Kipling2
David Kipling2