
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 12 October 2011
Sec. Antigen Presenting Cell Biology
volume 2 - 2011 | https://doi.org/10.3389/fimmu.2011.00050
 Lucas Onder1
Lucas Onder1 Elke Scandella1
Elke Scandella1 Qian Chai1
Qian Chai1 Sonja Firner1
Sonja Firner1 Christian T. Mayer2
Christian T. Mayer2 Tim Sparwasser2
Tim Sparwasser2 Volker Thiel1
Volker Thiel1 Thomas Rülicke3
Thomas Rülicke3 Burkhard Ludewig1*
Burkhard Ludewig1*Stromal cells provide the structural foundation of secondary lymphoid organs (SLOs), and regulate leukocyte access and cell migration within the different compartments of spleen and lymph nodes (LNs). Furthermore, several stromal cell subsets have been implied in shaping of T cell responses through direct presentation of antigen. Despite significant gain of knowledge on the biology of different SLO-resident stromal cell subsets, their molecular and functional characterization has remained incomplete. To address this need, we have generated a bacterial artificial chromosome-transgenic mouse model that utilizes the podoplanin (pdpn) promoter to express the Cre-recombinase exclusively in stromal cells of SLOs. The characterization of the Pdpn–Cre mouse revealed transgene expression in subsets of fibroblastic reticular cells and lymphatic endothelial cells in LNs. Furthermore, the transgene facilitated the identification of a novel splenic perivascular stromal cell subpopulation that forms web-like structures around central arterioles. Assessment of the in vivo antigen expression in the genetically tagged stromal cells in Pdpn–Cre mice revealed activation of both MHC I and II-restricted TCR transgenic T cells. Taken together, stromal pdpn–Cre expression is well-suited to characterize the phenotype and to dissect the function of lymphoid organ stromal cells.
Secondary lymphoid organs (SLOs) are characterized by a complex architecture with specialized compartments that facilitate the efficient interaction between immune cells. The structural foundation of this compartmentalization is formed by elaborate frameworks of stromal cell elements (Junt et al., 2008; Mueller and Germain, 2009). These non-hematopoietic cells can be divided in subpopulations that are distinguished by their properties, functions, and localization. Endothelial cells are located at the interface between liquid-transporting compartments and the parenchyma. For example, the highly plastic system of lymphatic vessels that is formed by lymphatic endothelial cells (LECs), distributes lymph fluid through the lymphoid tissues and regulates lymphocyte trafficking (Pham et al., 2010). Likewise, blood endothelial cells (BECs) facilitate controlled access of leukocytes to SLOs by building the network of high endothelial venules (HEVs; Miyasaka and Tanaka, 2004; Kumar et al., 2010). A bridging function, in terms of junction formation between the lymphatic vasculature and the conduit network, is fulfilled by marginal reticular cells (MRCs) that form a dense web of stromal cells underlying the subcapsular endothelium (Katakai et al., 2008). Other stromal cells are critical for the formation and maintenance of distinct lymphocyte compartments. These include the CXCL13-producing follicular dendritic cells (FDCs) of the B cell zone (Gunn et al., 1998) and fibroblastic reticular cells (FRC) of the T cell zone. FRCs support T cell survival through expression of IL-7, and guide the migration of T cells and dendritic cells (DCs) via production of the constitutive chemokines CCL19 and CCL21 (Luther et al., 2000; Bajenoff et al., 2006; Link et al., 2007). Furthermore, FRCs form the microvascular conduit system that distributes small lymph-borne antigens within the lymphoid tissue (Sixt et al., 2005). Although significant insight into stromal cell biology has been generated over the recent years, the molecular and functional characterization of the above-described subsets has remained incomplete.
Phenotypical and functional characterization of cells in vivo can be achieved through the generation of transgenic mouse models expressing the bacteriophage P1 Cre-recombinase (Schmidt-Supprian and Rajewsky, 2007). Cell type-specific promoters can be utilized to express the Cre-recombinase in distinct cell subsets, and to activate markers for lineage tracing, to conditionally switch-off genes, or to express antigens in a cell type-restricted manner. The choice of a particular promoter for such studies depends on the pre-existing knowledge on the phenotypical diversification of the cell populations under investigation. The current distinction of SLO stromal cells largely relies on the differential expression of CD31 (Pecam-1) and podoplanin (pdpn, also known as glycoprotein 38). CD31 is a marker for endothelial cells and therefore is expressed on BECs and LECs. Pdpn is a mesenchymally expressed glycoprotein with distinct functions in the development of the lymphatic system such as the separation between the lymphatic and blood vascular system (Schacht et al., 2003). FRCs express pdpn, whereas LECs express both pdpn and CD31. Since pdpn is a major marker for stromal cells, we used a bacterial artificial chromosome (BAC)-encoded pdpn promoter to generate a Cre-recombinase-transgenic mouse model that could serve as a tool to facilitate in vivo characterization of SLO stromal subsets. We found that pdpn promoter-driven Cre-recombinase expression labels subsets of FRCs and LECs in lymph nodes (LNs). In addition, the analysis of Pdpn–Cre mice uncovered a novel splenic stromal cell subpopulation that forms web-like structures around central arterioles. Further immunological characterization revealed that antigen expression directed to pdpn–Cre-expressing cells led to activation of both MHC I and II-restricted TCR transgenic T cells, underscoring the potential of certain stromal cell subsets in antigen presentation and T cell activation.
Experiments were performed in accordance with federal and cantonal guidelines under permission numbers SG09/83 and SG09/87 following review and approval by the Cantonal Veterinary Office (St. Gallen, Switzerland).
Bacterial artificial chromosome transgenic Tg(Pdpn–Cre) mice were generated as described previously (Sparwasser et al., 2004; Sparwasser and Eberl, 2007). The coding sequence for the Cre-recombinase, including a stop codon, was inserted into pdpn exon 1 utilizing the endogenous ATG translation start codon on the 180-kb BAC RP23 146H1 (Invitrogen, Lucerne, Switzerland). The BAC carried at least 90 kb of sequence upstream of the pdpn transcription start site. Integration of the Cre-recombinase was analyzed by southern blot analysis using a digoxigenin-labeled riboprobe binding to the ATG region of the inserted Cre-recombinase according to the instructions of the manufacturer (Roche Diagnostics, Rotkreuz, Switzerland). Modified BACs were screened with 5′ATG PCR (forward 5′-tctcttgccgatacccactc-3′, reverse 5′-ctgcacacagacaggagcat-3′) and 3′polyA PCR (forward 5′-cgggtcagaaagaatggtgt-3′, reverse 5′-ccactccttcaccaggaaag-3′). Founder lines were genotyped by PCR using the following primers: forward 5′-atgctcctgtctgtgtgcag-3′, reverse 5′-tctctgcccagagtcatcct-3′. B6.129 × 1-Gt(ROSA)26Sortm1(EYFP)Cos/J (R26-EYFP) and B6.129S4-Gt(ROSA)26Sortm1Sor/J (R26-LacZ) were obtained from The Jackson Laboratory. To analyze Cre-recombinase expression, pdpn–Cre mice were crossed with R26-EYFP and R26-LacZ mice. Bg1 (Bolinger et al., 2008) and Bg2 (Tewalt et al., 2009) mice were generously provided by Nicolas P. Restifo (NCI, NIH, Bethesda, MD, USA). C57BL/6 mice were obtained from Charles River (Sulzfeld, Germany). All animals were kept under conventional conditions in individually ventilated cages.
Lymph nodes and spleens were dissected into small pieces and transferred into a well of a 24-well dish filled with RPMI 1640 medium containing 2% FCS, 20 mM Hepes (all from Lonza), 1 mg/ml Collagenase Type IV (Sigma), and 25 μg/ml DNaseI (Applichem). LN and spleen pieces were incubated at 37°C for 30 min. After enzymatic digestion, cell suspensions were passed through a 45-μm gauze filter and washed with phosphate-buffered saline (PBS) containing 0.5% FCS and 10 mM EDTA (Sigma). The cell pellet was resuspended in 5 ml MACS-buffer and cells were counted. To further enrich the stromal cell fraction, lymphocytes were depleted by incubating the cell suspension with MACS anti-CD45 microbeads and passing over a MACS LS column (Miltenyi Biotec). The cells in the flow-through were collected and analyzed by flow cytometry.
Stromal cell suspensions (106 cells per stain) were incubated for 20 min at 4°C in PBS containing 1% FCS and 10 mM EDTA with the following antibodies: PE-conjugated anti-gp38/podoplanin (clone8.1.1, BD Biosciences), Alexa647-conjugated anti-CD31, PE-conjugated anti-inter-cellular adhesion molecule-I (ICAM-1), PerCP-conjugated anti-vascular cell adhesion molecule-1 (VCAM-1; all from eBioscience). Cells were analyzed with a FACSCanto (BD Biosciences) and FlowJo software (Tree star Inc.). For peripheral blood lymphocyte (PBL) samples, erythrocytes were lysed with FACS Lysing Solution (BD Biosciences).
Lymphoid tissues were fixed over night in freshly prepared 4% paraformaldehyde (Sigma). Fixed tissues were embedded in 4% low melting agarose (Invitrogen) in PBS and sectioned with a vibratome (Leica VT-1200). Twenty to thirty micrometer thick sections were blocked in PBS containing 10% FCS, 1 mg/ml anti-Fcγ receptor (BD Biosciences) and 0.1% Triton X-100. Sections were incubated over night at 4°C with the following monoclonal antibodies: anti-gp38/podoplanin (clone8.1.1, BD Biosciences), Alexa647-conjugated anti-B220 (BD Biosciences), anti-EYFP (Clontech), Alexa647-conjugated anti-CD31 (eBioscience), anti-LyveI (eBioscience), or anti-βGal (Abcam). Unconjugated antibodies were detected with the following secondary antibodies: Dylight649-conjugated anti-rat-IgG, Alexa488-conjugated anti-rabbit-IgG, and Dylight549-conjugated anti-syrian hamster-IgG all purchased from Jackson Immunotools (Brunschwig AG). Microscopical analysis was done using a confocal microscope (Zeiss LSM-710, Carl Zeiss, Inc.) and images were processed with ZEN 2009 software (Carl Zeiss, Inc.).
Single-cell suspensions from the spleens of Bg1-Thy1.1 or Bg2-CD45.1 mice were subjected to hypotonic red blood cell lysis and stained with CFSE (Molecular Probes). A maximum concentration of 2.5 × 107 cells/ml were incubated in 5 μM CFSE in PBS for 10 min at 37°C. Cells were washed twice with ice-cold balanced salt solution (BSS) and resuspended in BSS. Pdpn-Cre, R26-LacZ or control mice were injected intravenously with 107 Bg1-Thy1.1 and/or Bg2-CD45.1 splenocytes in 200 μl BSS.
In order to target stromal cells in vivo, we used the BAC homologous recombination technology (Sparwasser et al., 2004; Sparwasser and Eberl, 2007). Here, a BAC containing approximately 190 kb of the mouse chromosome 4 with the pdpn locus was used to replace exon 1 of the pdpn gene with a codon-optimized Cre-recombinase sequence and a polyadenylization site (Figure 1A). The recombination resulted in the expected integration of the Cre-recombinase gene downstream of the pdpn start codon as shown by PCR reactions detecting the Cre-recombinase, the 5′ATG, and the 3′polyA regions (Figure 1B). Primer pairs of the 5′- and 3′-PCRs contained one primer within the transgene and one primer within the adjacent region, so that only properly modified BAC clones could be detected. Positive BAC clones were sequence-verified and the presence of the transgene in the correct location on the BAC was analyzed using a digoxigenin-labeled DNA probe in a southern blot (Figure 1C). Successfully recombined Pdpn–Cre BACs were linearized, purified, and injected into the pronucleus of C57BL/6 zygotes. The microinjection gave rise to 17 offsprings with three animals being PCR-positive for Cre-recombinase and the 5′ATG region (Figure 1D).
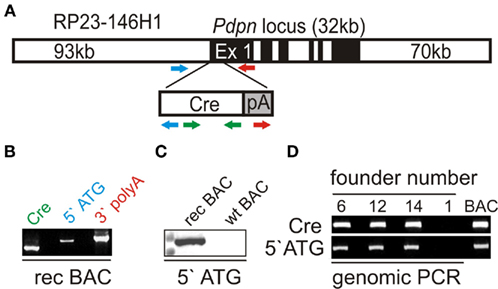
Figure 1. Generation of the Bacterial Artificial Chromosome (BAC) transgenic Pdpn–Cre mouse. (A) Schematic representation of BAC mutagenesis with introduction of the Cre-recombinase gene into the pdpn locus on BAC RP23 146H1. Colored arrows indicate primer location of Cre–PCR (green), 5′ATG PCR (blue) and 3′polyA PCR (red). (B) Initial screening of successfully recombined BAC by PCR for Cre-recombinase, 5′ATG region, and 3′polyA region. (C) PCR-positive BAC clones were examined by southern blot using a digoxigenin-labeled 5′ATG probe covering transgene and BAC sequences on the recombined (rec), but not the wild-type BAC (wt). (D) PCR analysis of purified genomic DNA from a cohort of potential founder mice. BAC indicates positive control DNA.
The founders C57BL/6N-Tg(Pdpn–Cre)406–408Biat , subsequently designated as line 6, 12, and 14 were crossed to R26-EYFP reporter mice in which a transcriptional stop-cassette can be excised in a Cre-recombinase-dependent manner (Srinivas et al., 2001). Initial examination of the resulting Pdpn–Cre,R26-EYFP mouse lines revealed that transgene expression could not be detected in mice derived from founder 12, suggesting that pdpn promoter-driven Cre expression was not sufficient for successful recombination. In mice derived from founder 6, some EYFP expression could be detected in the CD45+ non-stromal cell compartment (not shown). Founder line 6 was therefore excluded from further analysis. Examination of transgene activity in Pdpn–Cre,R26-EYFP offsprings from founder line 14 by flow cytometry revealed substantial transgene activity in CD45− LN cells and a complete absence of transgene activity in non-stromal cells (Figure 2A and Figure A1 in Appendix) with coexpression of the stromal cell markers ICAM-1 and VCAM-1 (Figure 2B). Further analysis of LN stromal cells revealed transgene activity in approximately 15% of cells in the LEC and FRC fractions (Figure 2C,D). Transgene expression in BECs was very low (Figure 2D, left histogram) or completely absent in the double-negative fraction (not shown). Assessment of transgene activity in splenic stromal cells showed that the Pdpn–Cre transgene is almost exclusively active in the pdpn+CD31− FRC fraction (Figure 2E,F). Taken together, these data indicate that the Pdpn–Cre transgene is well-suited to track particular stromal cell subsets within SLOs.
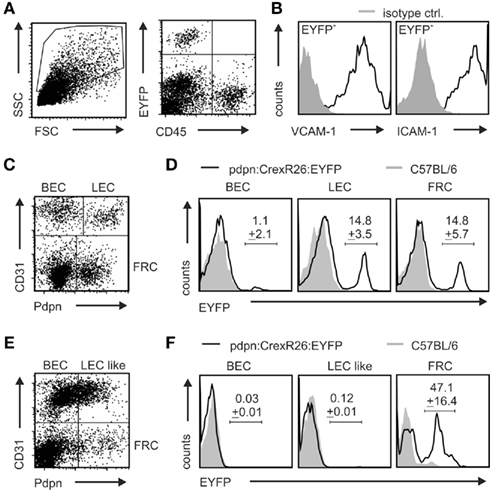
Figure 2. Validation of transgene expression and Cre-mediated recombination. Filial generation 1 of Pdpn–Cre founder 14 was crossed to R26-EYFP reporter mice. (A) Flow cytometric analysis of CD45-depleted cell suspensions utilized the distinct forward and side scatter properties (left dot plot) and lack of CD45-staining in EYFP+ population as found in right dot plot, upper left quadrant. (B) VCAM-1 (left histogram) and ICAM-1 (right histogram) expression on CD45−EYFP+ stromal cells from LNs of pdpn–Cre,R26-EYFP mice. (C–F) Stromal cell subsets in CD45-depleted cell preparations from LNs (C) and spleen (E) according to CD31 and pdpn expression. EYFP expression was determined on the indicated LN (D) or splenic (F) stromal cell subsets from Pdpn–Cre,R26-EYFP (black lines) and C57BL/6 control mice (gray shaded). Values in histograms in (D,F) indicate mean percentage ± SEM of EYFP-positive cells (n = 3 mice).
In order to localize transgene-expressing cells in spleen and LNs of Pdpn–Cre,R26-EYFP mice, we performed extensive in situ analysis using confocal laser scanning microscopy. LN sections were stained for Pdpn and B220 to visualize the Pdpn+ FRC network of the T cell zone, the Pdpn+ lymphatic endothelium, and B220+ B cell areas (Figure 3A). More detailed examination of transgene-expressing EYFP+ cells in the T cell zone showed that transgene-expressing cells were indeed part of the Pdpn+ FRC network (Figure 3B). In addition, pronounced transgene activity could be detected in lymphatic vessel endothelial hyaluron receptor I (LyveI)-positive cells with a particularly high accumulation of these cells in the subcapsular sinus of the LN (Figure 3C). Thus, the Pdpn–Cre transgene facilitates selective tagging of FRCs and LECs in LNs.
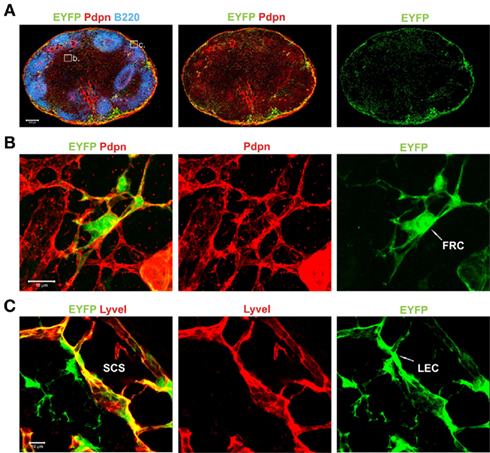
Figure 3. Localization and morphological phenotype of transgene-expressing stromal cells in LNs of Pdpn–Cre,R26-EYFP mice. (A) Histological section of inguinal LN from a Pdpn–Cre,R26-EYFP mouse stained for B cells (B220 in blue), Pdpn expressing stromal cells (red), and Cre-mediated transgene expression (EYFP, green). Inserts in the left panel mark the regions taken to acquire more detailed images in (B,C). Scale bar equals 200 μm. (B) Transgene-expressing stromal cells in the T cell zone with typical FRC structure marked with white arrow in the right panel. (C) Transgene-positive stromal cell in the subcapsular sinus (SCS) region visualized with anti-LyveI and anti-EYFP staining. Sinus-lining LEC is marked with white arrow in the right panel. Scale bars in (B,C) equal 10 μm.
Since the flow cytometric analysis of transgene activity in spleen revealed a more FRC-specific transgene expression pattern (Figure 2F), we wanted to confirm this finding using in situ analysis. Applying the identical immunofluorescent stainings as in the LN sections, we could detect transgene activity only within the Pdpn+ stromal cell network in the white pulp (Figure 4A). High resolution analysis revealed that EYFP+ cells accumulate in the central areas of the white pulp (Figure 4B). Co-staining with anti-CD31 showed that the Pdpn–Cre+ cells form a web-like structure around the central arteriole (Figure 4C). The EYFP+ perivascular stromal cells co-expressed the Pdpn protein, but were negative for CD31 (Figure 4D). It is noteworthy that Pdpn-expressing EYFP+ cells formed tube-like structures extending into the T cell zone (Figure 4D). In summary, transgene expression in spleens of Pdpn–Cre,R26-EYFP mice identified a novel stromal cell subset within the splenic white pulp that displays a FRC-signature (Pdpn+CD31−), but shows an exclusive location in the center of the white pulp.
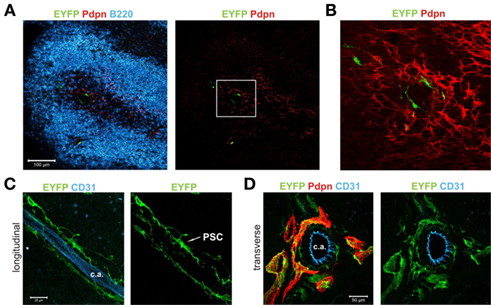
Figure 4. Distinct localization and morphology of Pdpn–Cre+ cells in spleen. (A) Splenic white pulp area of a Pdpn–Cre,R26-EYFP mouse stained for B cells (B220 in blue), Pdpn expressing stromal cells (red), and Cre-mediated transgene expression (EYFP, green). Insert in the right panel marks the region used for more detailed image in (B). Scale bar equals 100 μm. (B) Boxed area in (A) showing the central localization of transgene-expressing Pdpn+ cells. (C) Longitudinal section of the CD31+ central arteriole (c.a.) with surrounding transgene-expressing perivascular stromal cells (marked as PSC in right panel). Scale bar equals 20 μm. (D) 3D projection of transverse section of the CD31+ central arteriole (c.a.) and surrounding pdpn+EYFP+ perivascular stromal cells. Note the tube-like structures extending into the surrounding T cell zone. Scale bar equals 50 μm.
The organized structures of SLOs are critical for the activation of innate and adaptive immune responses (Junt et al., 2008). It is therefore intriguing that stromal cells can, at the same time, contribute to the tolerization of T cells. For example, it has been shown that direct presentation of self antigen by non-hematopoietic cells to autoreactive T cells results in activation, proliferation, and subsequent deletion of CD8+ T cells (Lee et al., 2007). Furthermore, a subpopulation of stromal cells in LNs and spleen was shown to express the transcription factor autoimmune regulator (AIRE) and was able to mediate the deletion of autoreactive CD8+ T cells (Gardner et al., 2008). Interestingly, it appears that different stromal cell subsets expressing peripheral tissue antigens may contribute to the maintenance of tolerance under distinct conditions (Cohen et al., 2010; Fletcher et al., 2010). To assess the impact of antigen expression by stromal cells in the Pdpn–Cre model, we crossed Pdpn–Cre to R26-LacZ mice that express the beta-galactosidase (βgal) antigen following Cre-recombinase-mediated removal of a transcriptional stop-cassette (Soriano, 1999). We have shown previously that βgal expression in non-hematopoietic stromal cells can be used to dissect mechanisms of peripheral tolerance (Bolinger et al., 2008, 2010). Histological analysis of LNs from Pdpn–Cre,R26-LacZ mice revealed an antigen expression pattern (Figure 5A) that was comparable to the EYFP expression in LNs of Pdpn–Cre,R26-EYFP mice (Figure 3A). To assess whether βgal expression in Pdpn–Cre+ stromal cells would suffice for activation of βgal-specific CD8+ and CD4+ T cells, we adoptively transferred Bg1 (Bolinger et al., 2008) and Bg2 (Tewalt et al., 2009) TCR transgenic cells into Pdpn–Cre,R26-LacZ mice. Single transfer of MHC I-restricted T cells did not result in significant activation (Figure 5B, left column), whereas transfer of Bg2 cells alone led to strong T cell proliferation as shown by substantial dilution of the intracellular dye CFSE (Figure 5B, right column). Co-transfer of Bg1 and Bg2 cells was accompanied by proliferation of both T cell populations (Figure 5B, middle columns) indicating that under these conditions, stromal cell-specific CD8+ T cells received help from CD4+ T cells and could therefore enter into proliferative cycles. Taken together, these data underscore the high versatility of the Pdpn–Cre model with successful and comparable Cre-mediated recombination in different reporter strains, and secondly indicate that Pdpn–Cre mice can be utilized to investigate the interplay of stromal cells during the activation and/or tolerization of T cells that encounter antigens exclusively expressed in these cells.
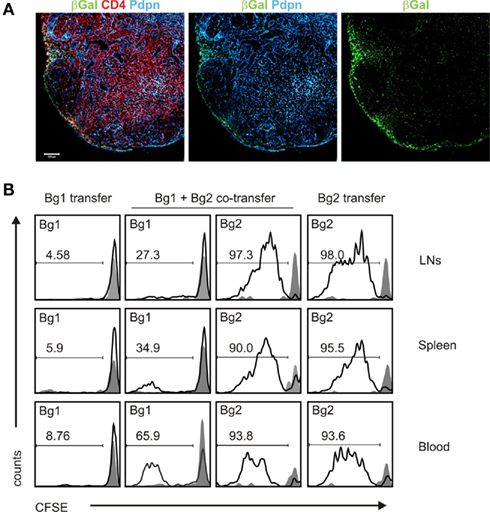
Figure 5. βgal expression in FRCs and LECs in Pdpn–Cre,R26-LacZ mice leads to activation of βgal-specific TCR transgenic T cells. (A) Pdpn–Cre mice were crossed to R26-LacZ reporter mice and βgal expression in LNs was detected by immunofluorescence analysis. Section of inguinal LN from Pdpn-Cre,R26-LacZ mouse stained for βgal (green), Pdpn (blue), and CD4 (red). Scale bar equals 100 μm. (B) Stimulation of βgal-specific TCR transgenic T cells by stromal βgal expression was determined by flow cytometry using CFSE-labeled congenic Bg1 and Bg2 cells. CFSE dilution in blood, spleen, and LNs was measured on day 5 after adoptive transfer in Pdpn–Cre,R26-LacZ mice (black lines) or Cre-negative littermates (gray shaded). Bg1 or Bg2 cells were adoptively transferred either alone or in combination. Values in the histograms indicate percentage of proliferated TCR transgenic cells; proliferation in Cre-negative littermates was always below 5%.
In this study, we describe the generation and first phenotypical analysis of a stromal cell-specific Pdpn–Cre BAC transgenic mouse. The importance of stromal cell-immune cell interaction has received increased attention over the recent years (Mebius, 2007; Mueller and Germain, 2009; Turley et al., 2010). However, the characterization of stromal cells within SLOs has relied largely on particular cell surface marker combinations. The present study has addressed the need to generate models that facilitate (i) genetic definition of distinct LN and splenic stromal cell populations, (ii) in vivo labeling and tracking of particular stromal cell subsets, and (iii) functional assessment of immune activation/tolerization processes through selective expression of model antigens. Furthermore, it will be possible to utilize this model to selectively delete molecularly defined target structures through crossing with conditionally gene-targeted mouse strains such as the LTβRfl/fl strain (Wang et al., 2010), or to transiently deplete stromal cells in the inducible diphtheria toxin receptor system (Buch et al., 2005).
Genomic integration of modified BACs is a widely used technology to generate transgenic mouse models. Comparable to other BAC transgenic mouse lines expressing the Cre-recombinase (Nishikawa et al., 2010), we observed varying levels of transgene expression and recombination frequencies in different founder lines. Phenotypic variability among several founder lines most likely resulted from different chromosomal integration sites and varying numbers of integrated BAC copies thereby determining the level of Cre-recombinase expression. Here, one of the three Pdpn–Cre founder lines displayed a suitable transgene expression pattern that facilitated the identification of novel stromal cell subpopulations in SLOs such as the splenic perivascular stromal cell. This finding implies that the currently used FRC phenotype description (Pdpn+CD31−) may obscure the true diversity of stromal cells in SLOs. This interpretation has been confirmed in a recent study that defines a particular FRC subpopulation through JAM-C controlled chemokine secretion (Frontera et al., 2011). Using genetic tools such as BAC transgenic mouse lines with stromal cell-specific promoters will help to define both function and phenotype of SLO stromal cells in future studies.
Antigen presentation and activation of T cells is a well-controlled process that relies on a division of labor between different myeloid cell subsets, mainly DCs (Turley et al., 2010). It is possible that a similar specialization in the display of self-antigens for the tolerization of autoreactive T cells is distributed between different stromal cell subsets. Indeed, recent studies suggest that diverse stromal cell subsets, in particular FRCs and endothelial cells, participate in presenting antigen via MHC class I and thereby critically contribute to the shaping of a T cell response (Cohen et al., 2010; Fletcher et al., 2010). The Pdpn–Cre mouse will facilitate future studies on the exact nature of direct or indirect stromal cell-T cell interaction. It will be important to determine whether the observed lack of βgal-specific CD8+ T cell activation in Pdpn–Cre,R26-LacZ mice is simply the lack of interaction, i.e., immunological ignorance (Bolinger et al., 2008) or an active process with down-tuning of T cell reactivity. Clearly, CD4+ T cells specific for a stromal cell-specific antigen were activated efficiently in the βgal system as presented here. Whether this activation was due to direct MHC II-mediated contact between stromal cells and CD4+ T cells or whether indirect activation through DCs had occurred, will require further investigations that exceed the scope of this initial characterization of this novel mouse strain.
In summary, stromal cell-specific Cre-recombinase expression is well-suited to better characterize SLO stromal cells through definition of functionally and phenotypically distinct subpopulations. In particular, the Pdpn–Cre mouse model can serve as a tool to dissect antigen presentation by FRCs and LECs in LNs and by the novel perivascular stromal cell population in spleen.
Burkhard Ludewig designed the study and wrote the paper; Lucas Onder and Elke Scandella performed research and wrote the paper; Qian Chai, Sonja Firner, Christian Mayer, and Thomas Rülicke performed research; Tim Sparwasser provided technical supervision and discussed data; Volker Thiel analyzed and discussed data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Rita de Giuli and Eva Allgäuer for technical support. This study received financial support from the Swiss National Science Foundation (CRSII3_125447 and 130823/1 to Burkhard Ludewig) and from the Austrian Genome Research Programme GEN-AU II and III (Austromouse) to Thomas Rülicke.
Bajenoff, M., Egen, J. G., Koo, L. Y., Laugier, J. P., Brau, F., Glaichenhaus, N., and Germain, R. N. (2006). Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity 25, 989–1001.
Bolinger, B., Engeler, D., Krebs, P., Miller, S., Firner, S., Hoffmann, M., Palmer, D. C., Restifo, N. P., Tian, Y., Clavien, P. A., and Ludewig, B. (2010). IFN-gamma-receptor signaling ameliorates transplant vasculopathy through attenuation of CD8+ T-cell-mediated injury of vascular endothelial cells. Eur. J. Immunol. 40, 733–743.
Bolinger, B., Krebs, P., Tian, Y., Engeler, D., Scandella, E., Miller, S., Palmer, D. C., Restifo, N. P., Clavien, P. A., and Ludewig, B. (2008). Immunologic ignorance of vascular endothelial cells expressing minor histocompatibility antigen. Blood 111, 4588–4595.
Buch, T., Heppner, F. L., Tertilt, C., Heinen, T. J., Kremer, M., Wunderlich, F. T., Jung, S., and Waisman, A. (2005). A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods 2, 419–426.
Cohen, J. N., Guidi, C. J., Tewalt, E. F., Qiao, H., Rouhani, S. J., Ruddell, A., Farr, A. G., Tung, K. S., and Engelhard, V. H. (2010). Lymph node-resident lymphatic endothelial cells mediate peripheral tolerance via Aire-independent direct antigen presentation. J. Exp. Med. 207, 681–688.
Fletcher, A. L., Lukacs-Kornek, V., Reynoso, E. D., Pinner, S. E., Bellemare-Pelletier, A., Curry, M. S., Collier, A. R., Boyd, R. L., and Turley, S. J. (2010). Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady-state and inflammatory conditions. J. Exp. Med. 207, 689–697.
Frontera, V., Arcangeli, M. L., Zimmerli, C., Bardin, F., Obrados, E., Audebert, S., Bajenoff, M., Borg, J. P., and Aurrand-Lions, M. (2011). Cutting edge: JAM-C controls homeostatic chemokine secretion in lymph node fibroblastic reticular cells expressing thrombomodulin. J. Immunol. 187, 603–607.
Gardner, J. M., Devoss, J. J., Friedman, R. S., Wong, D. J., Tan, Y. X., Zhou, X., Johannes, K. P., Su, M. A., Chang, H. Y., Krummel, M. F., and Anderson, M. S. (2008). Deletional tolerance mediated by extrathymic Aire-expressing cells. Science 321, 843–847.
Gunn, M. D., Ngo, V. N., Ansel, K. M., Ekland, E. H., Cyster, J. G., and Williams, L. T. (1998). A B-cell-homing chemokine made in lymphoid follicles activates Burkitt’s lymphoma receptor-1. Nature 391, 799–803.
Junt, T., Scandella, E., and Ludewig, B. (2008). Form follows function: lymphoid tissue microarchitecture in antimicrobial immune defence. Nat. Rev. Immunol. 8, 764–775.
Katakai, T., Suto, H., Sugai, M., Gonda, H., Togawa, A., Suematsu, S., Ebisuno, Y., Katagiri, K., Kinashi, T., and Shimizu, A. (2008). Organizer-like reticular stromal cell layer common to adult secondary lymphoid organs. J. Immunol. 181, 6189–6200.
Kumar, V., Scandella, E., Danuser, R., Onder, L., Nitschke, M., Fukui, Y., Halin, C., Ludewig, B., and Stein, J. V. (2010). Global lymphoid tissue remodeling during a viral infection is orchestrated by a B cell-lymphotoxin-dependent pathway. Blood 115, 4725–4733.
Lee, J. W., Epardaud, M., Sun, J., Becker, J. E., Cheng, A. C., Yonekura, A. R., Heath, J. K., and Turley, S. J. (2007). Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat. Immunol. 8, 181–190.
Link, A., Vogt, T. K., Favre, S., Britschgi, M. R., Acha-Orbea, H., Hinz, B., Cyster, J. G., and Luther, S. A. (2007). Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat. Immunol. 8, 1255–1265.
Luther, S. A., Tang, H. L., Hyman, P. L., Farr, A. G., and Cyster, J. G. (2000). Coexpression of the chemokines ELC and SLC by T zone stromal cells and deletion of the ELC gene in the plt/plt mouse. Proc. Natl. Acad. Sci. U.S.A. 97, 12694–12699.
Miyasaka, M., and Tanaka, T. (2004). Lymphocyte trafficking across high endothelial venules: dogmas and enigmas. Nat. Rev. Immunol. 4, 360–370.
Mueller, S. N., and Germain, R. N. (2009). Stromal cell contributions to the homeostasis and functionality of the immune system. Nat. Rev. Immunol. 9, 618–629.
Nishikawa, Y., Hirota, F., Yano, M., Kitajima, H., Miyazaki, J., Kawamoto, H., Mouri, Y., and Matsumoto, M. (2010). Biphasic Aire expression in early embryos and in medullary thymic epithelial cells before end-stage terminal differentiation. J. Exp. Med. 207, 963–971.
Pham, T. H., Baluk, P., Xu, Y., Grigorova, I., Bankovich, A. J., Pappu, R., Coughlin, S. R., McDonald, D. M., Schwab, S. R., and Cyster, J. G. (2010). Lymphatic endothelial cell sphingosine kinase activity is required for lymphocyte egress and lymphatic patterning. J. Exp. Med. 207, 17–27.
Schacht, V., Ramirez, M. I., Hong, Y. K., Hirakawa, S., Feng, D., Harvey, N., Williams, M., Dvorak, A. M., Dvorak, H. F., Oliver, G., and Detmar, M. (2003). T1alpha/podoplanin deficiency disrupts normal lymphatic vasculature formation and causes lymphedema. EMBO J. 22, 3546–3556.
Schmidt-Supprian, M., and Rajewsky, K. (2007). Vagaries of conditional gene targeting. Nat. Immunol. 8, 665–668.
Sixt, M., Kanazawa, N., Selg, M., Samson, T., Roos, G., Reinhardt, D. P., Pabst, R., Lutz, M. B., and Sorokin, L. (2005). The conduit system transports soluble antigens from the afferent lymph to resident dendritic cells in the T cell area of the lymph node. Immunity 22, 19–29.
Soriano, P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71.
Sparwasser, T., and Eberl, G. (2007). BAC to immunology – bacterial artificial chromosome-mediated transgenesis for targeting of immune cells. Immunology 121, 308–313.
Sparwasser, T., Gong, S., Li, J. Y., and Eberl, G. (2004). General method for the modification of different BAC types and the rapid generation of BAC transgenic mice. Genesis 38, 39–50.
Srinivas, S., Watanabe, T., Lin, C. S., William, C. M., Tanabe, Y., Jessell, T. M., and Costantini, F. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4. doi: 10.1186/1471-213X-1-4
Tewalt, E. F., Grant, J. M., Granger, E. L., Palmer, D. C., Heuss, N. D., Gregerson, D. S., Restifo, N. P., and Norbury, C. C. (2009). Viral sequestration of antigen subverts cross presentation to CD8(+) T cells. PLoS. Pathog. 5, e1000457. doi:10.1371/journal.ppat.1000457
Turley, S. J., Fletcher, A. L., and Elpek, K. G. (2010). The stromal and haematopoietic antigen-presenting cells that reside in secondary lymphoid organs. Nat. Rev. Immunol. 10, 813–825.
Wang, Y., Koroleva, E. P., Kruglov, A. A., Kuprash, D. V., Nedospasov, S. A., Fu, Y. X., and Tumanov, A. V. (2010). Lymphotoxin beta receptor signaling in intestinal epithelial cells orchestrates innate immune responses against mucosal bacterial infection. Immunity 32, 403–413.
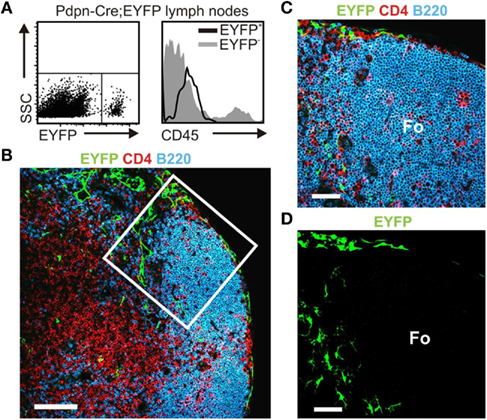
Figure A1. Pdpn–Cre transgene-expressing cells are non-hematopoietic stromal cells. (A) Flow cytometric analysis of Pdpn–Cre,R26-EYFP LN single-cell suspensions. Appropriate gates were established (boxed area) for subsequent analysis of EYFP+ and EYFP− populations. Histogram shows analysis of CD45 expression on EYFP+ (black line) and EYFP− (gray shaded) cells. (B) Confocal microscopy analysis of a Pdpn–Cre,R26-EYFP LN section stained for T lymphocytes (CD4 in red), B lymphocytes (B220 in blue), and transgene-expressing cells (EYFP in green), scale bar equals 50 μm, boxed area defines area for magnified view in (C) and three-dimensional rendering in (D), scale bars equal 20 μm, Fo: B cell follicle.
Keywords: stromal cells, podoplanin, fibroblastic reticular cells, lymphatic endothelial cells
Citation: Onder L, Scandella E, Chai Q, Firner S, Mayer CT, Sparwasser T, Thiel V, Rülicke T and Ludewig B (2011) A novel bacterial artificial chromosome-transgenic Podoplanin–Cre mouse targets lymphoid organ stromal cells in vivo. Front. Immun. 2:50. doi: 10.3389/fimmu.2011.00050
Received: 11 July 2011; Accepted: 19 September 2011;
Published online: 12 October 2011.
Edited by:
Christian Munz, University of Zurich, SwitzerlandReviewed by:
Christian Munz, University of Zurich, SwitzerlandCopyright: © 2011 Onder, Scandella, Chai, Firner, Mayer, Sparwasser, Thiel, Rülicke and Ludewig. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Burkhard Ludewig, Institute of Immunobiology, Cantonal Hospital St. Gallen, 9007 St. Gallen, Switzerland. e-mail:YnVya2hhcmQubHVkZXdpZ0Brc3NnLmNo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.