
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Imaging, 19 April 2024
Sec. Imaging Applications
Volume 3 - 2024 | https://doi.org/10.3389/fimag.2024.1373420
This article is part of the Research TopicHorizons in ImagingView all 6 articles
Efficient and accurate radiology reporting is critical in modern healthcare for timely diagnosis and patient care. In this paper, we present a novel deep learning approach that leverages BioGPT and co-attention mechanisms for automatic chest X-ray report generation. Our model, termed “ChestBioX-Gen” is designed to bridge the gap between medical images and textual reports. BioGPT, a biological language model, contributes its contextual understanding to the task, while the co-attention mechanism efficiently aligns relevant regions of the image with textual descriptions. This collaborative combination enables ChestBioX-Gen to generate coherent and contextually accurate reports that embed complex medical findings. Our model not only reduces the burden on radiologists but also enhances the consistency and quality of reports. By automating the report generation process, ChestBioX-Gen contributes to faster diagnoses and improved patient care. Quantitative evaluations, measured through BLEU-N and Rouge-L metrics, demonstrate the model's proficiency in producing clinically relevant reports with scores of 0.6685, 0.6247, 0.5689, 0.4806, and 0.7742 on BLUE 1, 2, 3, 4, and Rouge-L, respectively. In conclusion, the integration of BioGPT and co-attention mechanisms in ChestBioX-Gen represents an advancement in AI-driven medical image analysis. As radiology reporting plays a critical role in healthcare, our model holds the potential to revolutionize how medical insights are extracted and communicated, ultimately benefiting both radiologists and patients.
Medical imaging is an important aspect of modern healthcare, employing technologies like X-rays, CT scans, MRI, and ultrasound to non-invasively visualize internal body structures. These imaging modalities allow healthcare practitioners to observe and analyze anatomical details, detect abnormalities, and guide medical interventions. By offering a visual representation of the body structures, medical imaging aids in the early identification of diseases, informs treatment strategies, and enables ongoing monitoring of patient wellbeing. The continual advancements in medical imaging underscore the dedication of scientists and healthcare professionals to refining precision, minimizing confusion, and ultimately enhancing patient outcomes. In essence, medical imaging serves as a link between scientific understanding and practical clinical applications, providing essential insights into anatomical structures and pathological conditions for more effective and personalized healthcare.
The advent of automatic report generation for medical images, particularly in radiology, has emerged as a crucial area in the intersection of machine learning and healthcare. This task involves creating coherent and informative textual reports, with the potential to enhance clinical workflows and elevate the quality and standardization of care. Despite its potential benefits, this process presents significant challenges. Traditional image captioning approaches, designed for shorter and less complex reports, often fall short in addressing the highly templated nature of radiology reports. Generic natural language generation (NLG) methods, while prioritizing descriptive accuracy, may not align with the clinical priorities of accuracy and specificity in radiology reports.
In this work, we present a novel approach named ChestBioX-Gen for chest X-ray report generation, aimed at enhancing the precision of diagnostic outcomes and reducing the burden on radiologists. Leveraging advanced deep learning techniques, our methodology leverages an encoder-decoder architecture. The encoder utilizes the pretrained CheXNet model (Rajpurkar et al., 2017), based on the DenseNet121 backbone, to extract meaningful features from input chest X-ray images. Concurrently, we employ the BioGPT Tokenizer (Luo et al., 2022) to extract relevant embedding vectors from input captions, which are then combined with visual features Learned using CheXNet. Where the co-attention module computes attention weights, guiding the focus to the most pertinent information within the input image. This not only enhances the accuracy and informativeness of the generated reports but also reduces the burden on radiologists by highlighting the region of interest. The resulting vector serves as input to our RNN, contributing to the generation of a final sentence. Evaluation of the proposed model on the IU-X-Ray dataset (Demner-Fushman et al., 2016) highlights the effectiveness of our approach, notably demonstrated by its superior performance in the BLUE metric. This underscores its capacity to greatly assist radiologists in their diagnostic tasks.
In recent years, the field of chest X-ray image analysis has witnessed the emergence of various deep learning-based approaches. Liu et al. (2021) introduced an effective method that leverages the attention mechanism through contrastive learning. This model is specifically designed to enhance abnormal region detection in chest X-ray images. The approach incorporates known normal images, utilizing Aggregate and Differentiate Attention to prioritize images similar to the input. By extracting common features, the model augments abnormality detection. Evaluation on two datasets, MIMIC CXR (Johnson et al., 2016) and IU-X-ray (Demner-Fushman et al., 2016), demonstrates substantial performance improvements over baselines, as evident in both automated metrics and human assessments.
Another work was published by Liu et al. (2019), where the authors proposed a new framework. The study focuses on a domain-aware system employing a CNN-RNN-RNN architecture. The idea is to predict report topics and generates corresponding sentences, ensuring both readability and clinical accuracy. The training process involves reinforcement learning guided by the Clinically Coherent Reward. The image encoder CNN captures spatial features, while the sentence decoder RNN generates topics and stop signals. The word decoder RNN decodes words based on topics, all within a fully differentiable CNN-RNN-RNN architecture.
Kaur and Mittal (2023) presented a novel approach leveraging a deep neural network architecture enhanced with a multi-attention mechanism. The base model employs convolutional neural networks (CNNs) for feature extraction and multi-label classification, while attention mechanisms focus on salient image regions. Then, using LSTM networks, CheXPrune generates coherent reports from CXR images. Furthermore, the model uses pruning to reduce computational complexity, with experimental results suggesting significant pruning percentages without compromising accuracy.
Shetty et al. (2023) propose an encoder-decoder framework. The encoder, comprising the Unimodal Medical Visual Encoding Subnetwork (UM-VES) and the Unimodal Medical Text Embedding Subnetwork (UM-TES), processes images and corresponding reports during training. UM-VES extracts visual features from frontal and lateral CXR images using a depthwise separable convolutional neural network. UM-TES preprocesses radiology findings and learns word embeddings from medical terminology, combining glove word embeddings (Pennington et al., 2014) with those from a large knowledge base of Stanford reports (Zhang et al., 2018). The LSTM-based decoder generates reports by integrating visual and textual information aggregated by the encoder. Following the same idea, Akbar et al. (2023) utilized a DenseNet121 for image feature extraction and a GRU decoder for text generation, and categorical cross-entropy loss function is used for optimization.
Yang et al. (2023) introduces a novel approach to automatic chest X-ray radiology report generation. Their method features a self-updating Learned Knowledge Base, extracting medical knowledge from textual embeddings. The Multi-Modal Alignment module ensures consistency across textual, visual, and disease label modalities. The approach optimizes the bidirectional inter-relationship between image and report features and aligns visual features with disease labels. The training process involves balancing these alignments using a versatile loss function.
Recently, many works have focused on the use of graph neural networks (Wu et al., 2023), specifically, when the input data used for a specific task presents dependencies. Consequently, Li et al. (2023) introduced Dynamic Graph enhanced Contrastive Learning (DCL). This method addresses limitations in existing systems by incorporating a dynamic knowledge graph with contrastive learning. The dynamic graph construction involves both general and specific knowledge, enhancing relationships related to disease keywords. The dynamic graph encoder propagates information, and the graph attention module integrates knowledge with visual features. The paper also introduces image-report contrastive and image-report matching losses to improve both visual and textual representations.
In their recent work, Zhang et al. (2022) propose a framework based on knowledge graphs, for improving reasoning driven report generation. The framework is based on three main modules: Classifier, Generator, and Interpreter. The Classifier module integrates various submodules for disease topic extraction, multi-view image encoding, and text representation. By leveraging a Graph Convolutional Network, semantic information is extracted, followed by the integration of disease representation with prior knowledge. Subsequently, the Generator module, leveraging a Transformer decoder, generates reports based on the classifier's outputs. To ensure consistency, the Interpreter module, inspired by principles from CycleGan (Zhu et al., 2017), refines generated reports by comparing them with classifier outputs.
Li et al. (2023) presented a method for generating reports by utilizing a dynamic graph structure G that is enhanced with both specific and general knowledge. Qi et al. (2020), a Python natural language analysis package is used to extract anatomy and observation entities, and the graph is dynamically generated using insights from the topnT similar reports. Then, information flows through the dynamic graph while specific node attributes are learned with the help of a relational self-attention module and a cross-attention module that combines graph and visual encodings. Finally, the generated vector is fed into a Transformer decoder.
Kale et al. (2023) addresses the global challenge of timely generation of radiology reports and diagnoses due to a shortage of specialists. The proposed solution is a model called Knowledge Graph Augmented Vision Language BART (KGVL-BART). This model takes two chest X-ray images, along with diagnostic tags, and outputs patient-specific reports. The solution involves constructing a Chest X-ray knowledge graph (chestX-KG), extracting image features, and training the KGVL-BART model using visual, text, and KG data.
The advancement of deep learning models for automated X-ray report generation has been facilitated by the availability of comprehensive medical image datasets. Notable examples are illustrated in Figure 1, while Table 1 provides a detailed overview of the datasets utilized in the existing literature to support this task. Among these, the ChestX-ray14 dataset stands out, comprising 112,120 frontal-view X-ray images from 30,805 unique patients, each accompanied by corresponding reports. Additionally, datasets such as MIMIC-CXR, IU X-ray, NIH Chest X-ray, PadChest, and CheXpert have been used in automating X-ray report generation. Despite certain limitations, including abnormalities and class imbalances, these datasets, sourced from various institutions, have significantly contributed to the advancement of automatic report generation in the field. In the following, we present some of the common ones:
• ChestX-ray14 (Wang et al., 2017): this dataset contains 112,120 frontal-view X-ray images and reports from 30,805 patients. It includes additional thoracic diseases compared to the ChestX-ray8 dataset and is a collaboration between the National Institute of Health Clinical Center and the Indiana University School of Medicine.
• MIMIC-CXR (Johnson et al., 2016): with over 377,000 chest X-ray images and reports from 227,835 patients, this dataset, created by the Massachusetts Institute of Technology and Beth Israel Deaconess Medical Center, stands as one of the largest publicly available datasets for chest X-ray analysis.
• IU X-ray (Demner-Fushman et al., 2016): OpenI comprises 7,470 images (include both frontal and lateral views), accompanied by their corresponding 3,955 reports. The data was collected at various hospitals across the state of Indiana, USA.
• PadChest (Bustos et al., 2020): this dataset, originating from the University of Alicante in Spain, features 160,868 chest X-ray images and 109,931 Spanish reports, annotated with over 174 radiographic findings.
• CheXpert (Irvin et al., 2019): Stanford University's dataset includes 224,316 chest X-ray images and reports from 65,240 patients, annotated for 14 common thoracic pathologies.
• VinDr-CXR (Nguyen et al., 2022): comprised of over 100,000 DICOM-format images retrospectively gathered from Hospital 108 (H108) and Hanoi Medical University Hospital (HMUH), VinDr-CXR constitutes 18,000 postero-anterior (PA) view CXR scans. This dataset includes annotations for critical findings' localization and the classification of prevalent thoracic diseases. It encompasses 22 critical findings (local labels) and 6 diagnoses (global labels).
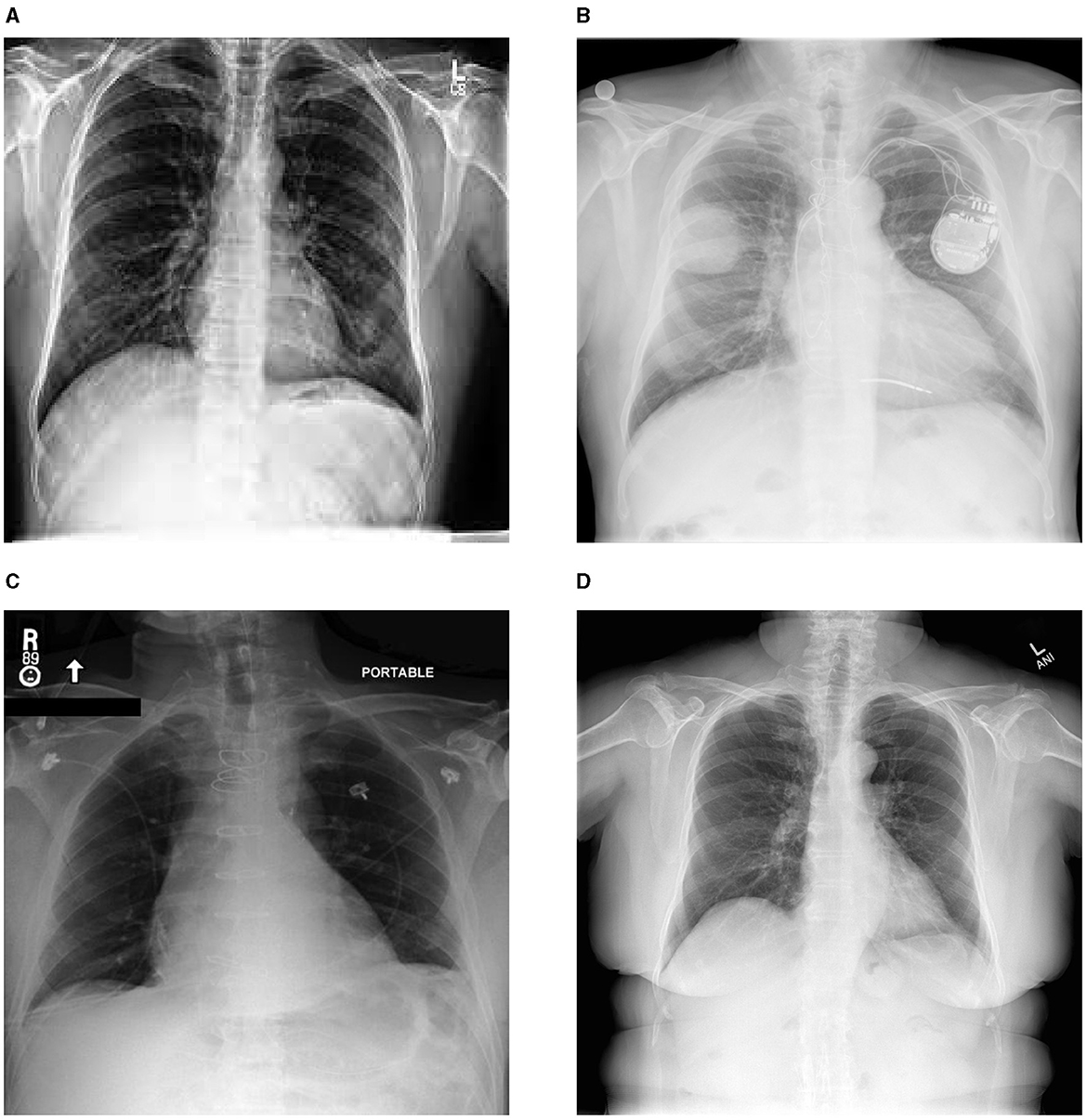
Figure 1. Sample images from diverse chest X-ray datasets. (A) chexpert, (B) padchest, (C) mimic, (D) Indiana University.
In our investigation, we exclusively used the IU X-ray (Demner-Fushman et al., 2016). This dataset, contains 7,470 images and 3,955 reports, providing an optimal size for efficient model training and testing. In our study, we focus on generating the impression section illustrated in Figure 2 as it provides a comprehensive summary of the findings, including the most significant observations and possible causes. Furthermore, in order to enhance data quality, we initiated a data preparation process. Initially, 7,460 images were obtained, but after removing records lacking impressions, we refined the dataset to 7,415 images. Subsequently, 500 images were randomly selected for testing, leaving the remainder for comprehensive model training.
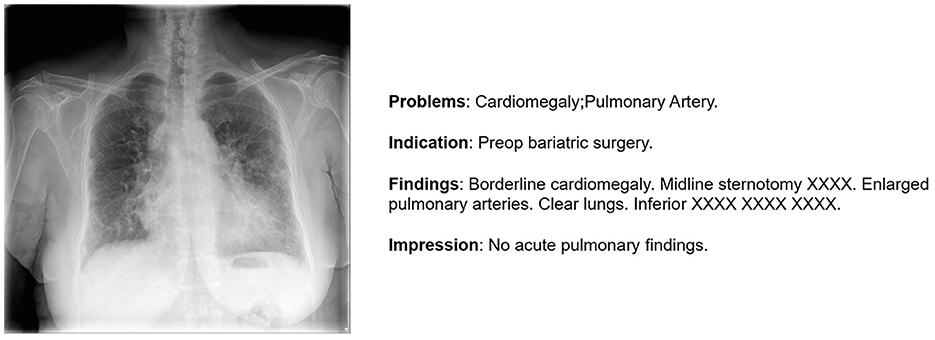
Figure 2. An illustration of one sample from IU-X-Ray (Demner-Fushman et al., 2016).
The assessment of machine learning models relies significantly on evaluation metrics. In the context of generating chest X-ray image reports, these metrics function as quantitative benchmarks, enabling the measurement of the quality and similarity of generated reports in comparison to reference reports. Their important role lies in objectively evaluating report generation models. In our work, we adopt several different metrics that focus on different aspects ranging from a natural language perspective to clinical adequacy.
• Bilingual evaluation understudy (BLEU) (Papineni et al., 2002): BLEU, a commonly employed metric, assesses the similarity between a generated report and a reference report by considering n-grams of different lengths (BLEU-1, BLEU-2, BLEU-3, and BLEU-4). It quantifies the overlap of n-grams to evaluate the quality of the generated text. BLEU is computed as follows:
where pn is the modified precision for n-gram, the base of log is the natural base e, wn is weight between 0 and 1 for logpn and .
where c is the number of unigrams (length) in all the candidate sentences, and r is the best match lengths for each candidate sentence in the corpus.
• Recall-Oriented Understudy for Gisting Evaluation-Longest Common Subsequence (ROUGE-L) (Lin, 2004): ROUGE-L is a metric designed to assess the similarity between a generated report and a reference report by examining the longest common subsequence (LCS) of words. It focuses on recall, effectively capturing essential information from the reference report. The mathematical formulation of Rouge-L precision, recall is shown below
• Consensus-based Image Description Evaluation (CIDEr) (Vedantam et al., 2015): CIDEr is a metric that evaluates the quality of generated text by comparing it to multiple reference reports. It measures the consensus among the reference reports and the generated report in terms of n-grams overlap and term frequency-inverse document frequency (TF-IDF) similarity.
where is a vector formed by gk(ci) corresponding to all n-grams of length n and is the magnitude of the vector . Similarly for .
• METEOR (Metric for Evaluation of Translation with Explicit ORdering) (Denkowski and Lavie, 2014): METEOR measures the similarity between the generated text and the reference text based on a combination of word matching and word order. Additionally, METEOR incorporates a set of pre-defined synonyms to further enhance the matching accuracy. The mathematical formulation is shown below:
Where P, R are the precision and recall correspondingly.
Our approach uses a CNN-RNN architecture illustrated in Figure 3, enhanced by the integration of a co-attention module (Lu et al., 2016) that jointly reasons about image and caption attention. This addition serves to focus on the most relevant regions within the image, enhancing the model's ability to extract and incorporate critical information for report generation. In the following sections, we explain our methodology by describing the different modules used in our architecture.
The co-attention mechanism is applied to both the image and caption as shown in Figure 4. We establish a connection between the image and caption by computing the similarity between their features across all pairs of image-locations and caption-locations. More precisely, having an image features vector V∈Rembed and a caption representation C∈RN×embed where Cn is the feature vector for the n−th word. The attention scores S∈RN×embed are determined through the calculation of
We then apply the softmax function to the affinity matrix values to transform them into a probability distribution, we call the result, attention weights.
Finally, in order to get the attended vector that represents our input image noted as AI∈Rembed
In the encoder section, the model, referred to as CheXNet, utilizes a pre-trained DenseNet121 (Huang et al., 2017) backbone. The process involves passing an input image, denoted as I, through the model layers to obtain a representative image features vector, V, of dimension Rembed. CheXNet, a 121-layer Dense Convolutional Network, is trained on the ChestX-ray 14 (Wang et al., 2017) dataset. It leverages DenseNets for efficient information flow and gradient optimization. The final layer is replaced with a single-output layer followed by sigmoid non linearity. Model weights are initialized from ImageNet (Deng et al., 2009) and trained end-to-end with the Adam optimizer using mini-batches of size 16. For pneumonia detection, images with pneumonia annotations are positive examples. The dataset is split into training, validation, and test sets. Images are downscaled to 224 × 224 and normalized using ImageNet statistics, with additional data augmentation of random horizontal flipping during training.
BioGPT stands out as a generative pre-trained Transformer language model designed and optimized for the specific purpose of generating and analyzing biomedical texts. It is based on the architecture of the GPT-2 model. BioGPT can be fine-tuned for various downstream tasks such as end-to-end relation extraction, question answering (QA), document classification, and text generation. The model was trained using a dataset comprising 15 million abstracts sourced from PubMed,1 with a vocabulary size of 42,384. The backbone network, GPT-2 model, has 24 layers, a hidden size of 1,024, and 16 attention heads, resulting in a total of 355 million parameters. In contrast, the BioGPT model comprises 347 million parameters, with differences from variations in embedding size and output projection size due to distinct vocabulary sizes. It has been scaled up to a larger size with the creation of the BioGPT-Large model using the GPT-2 XL architecture. The core component of the model is the multihead attention. Given the input, three linear transformations are applied to produce the query Q, the key K and the value V, and then the output is calculated as follows:
where h represents the number of heads, Q, K, and V are equally divided into Qi, Ki, and Vi along the feature dimension, denoted by i∈{1, 2, …, h}. The operation Concat signifies concatenating all inputs as a large tensor along the feature dimension, and W serves as the parameter for the affine transformation. The output of the multi-head attention layer is subsequently passed into a feed-forward layer to construct a Transformer layer (or Transformer block).
Pretrained BioGPT models are available in Huggingface directory.2 And for text generation, we fine-tune “microsoft/biogpt”3 on the IU-X-Ray dataset.
Given the attended vector AI, we used Long-Short Term Memory (LSTM) and model the hidden state as , where hi−1 and mi−1 are the hidden state vector and the memory vector for the previous sentence. The LSTM mechanism helps the model understand the order of information in the attended vector sequence. It learns subtle patterns and context. The hidden state hi−1 keeps track of the evolving understanding of the input, and the memory state mi−1 holds on to important context over time.
Finally, the model generates the final sentence. This generation process integrates the context and information Learned from the attended vector, facilitating the generation of coherent and contextually informed impression sections.
In our algorithm's implementation, we prioritize clinical interpretability by adopting an encoder-decoder architecture enhanced with attention mechanisms. This approach enables our model to focus on salient regions within input data, ensuring relevance to medical diagnosis and treatment. The validation of our results is improved through the utilization of pretrained models such as CheXNet and BioGPT, which have been extensively trained on diverse medical datasets.
Standardizing input images to a size of 512 × 512 pixels ensures consistency in the CheXNet (Rajpurkar et al., 2017) model by allowing the model to detect multi-scale features. The BioGPT (Luo et al., 2022) language model was fine-tuned with a vocabulary size of 42,384 to accommodate the terminology found in biomedical texts. Batch training was employed to enhance computational efficiency, and to handle varying sentence lengths, both < PAD> tokens for padding and < eos> tokens to denote sentence endings were incorporated. In our work, we used a batch size of 16 input images. The CheXNet model (Rajpurkar et al., 2017) served as the image encoder, generating a 1,024-dimensional output. The captioning component of BioGPT (Luo et al., 2022) produced text with a dimensionality of 1,024. Training parameters included a learning rate of 0.0001, and the model was trained for 100 epochs to facilitate learning and adaptation to the variations of biomedical data. During training, our model is applied specifically to the impression section only.
For the creation of our train and test subsets, we randomly selected 500 images from the IU-X-Ray dataset for testing, while the remaining images were used for training. Capturing a different set of cases for training and evaluation purposes. During evaluation, the model generates sentences, and once the predicted item token is < eos> or reaches a maximum length of 100 tokens, the model stops generating, thus providing coherent and complete outputs. Our proposed model demonstrates remarkable results in both quantitative and qualitative evaluations and surpasses many other works by enhancing the selection of relevant regions that contribute to generating the final output. The model provides challenging evaluation metrics as shown in Table 2, particularly the Blue 1 and Blue 2 metrics, in comparison to existing works in the literature. However, our model presents some limitations in terms of BLUE-3 and BLUE-4 metrics. We assume this limitation is due to the utilization of the LSTM architecture in our current model. Therefore, we suggest some potential solutions such as hierarchical LSTM architectures Zhang J. et al. (2023), or transformers Zhang H. et al. (2023) for the future. Furthermore, our qualitative analysis shown in Figure 5 highlights the model's capability to create different and contextually fitting responses, showcasing its adaptability in various situations.
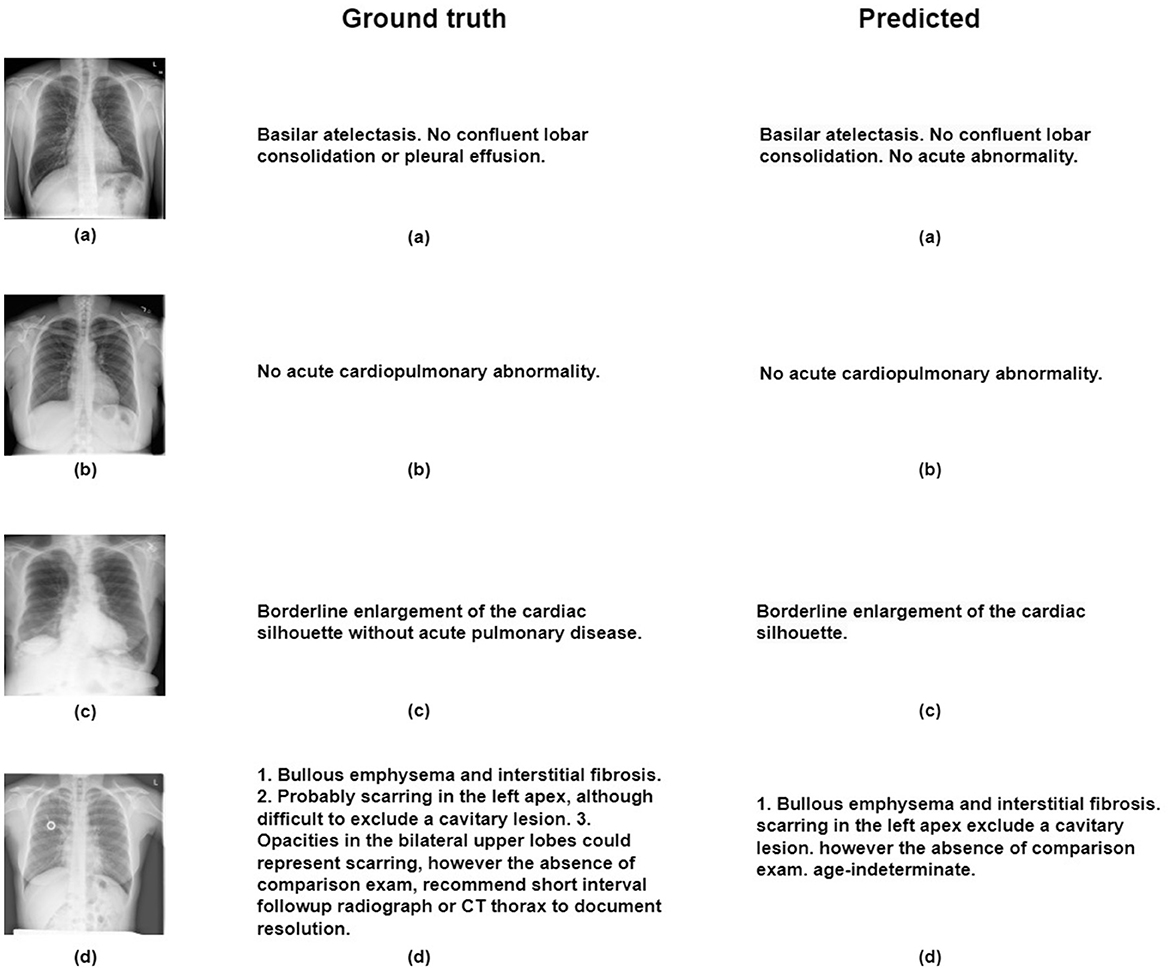
Figure 5. Figure illustrates qualitative results obtained through the proposed model on the IU-X-ray dataset, with reference images labeled as (A–D).
In our investigation, the integration of the co-attention module helped improve ChestBioX-Gen. Our model helps radiologists by reducing significantly time consuming burden during image interpretation. This module facilitates the extraction of contextual information from both image and text modalities, enabling a more effective cross-correlation. The integration of visual and textual information helps our model achieve a more sophisticated understanding of input images, allowing it to determine and prioritize key features essential for accurate and contextually rich medical report generation.
The utilization of BioGPT (Luo et al., 2022) as our language model proves to be a strategic choice, demonstrating significant success in generating medical sentences. In contrast to conventional text generators not pretrained on medical images, BioGPT shows its effectiveness in handling complex medical keywords, thereby enhancing the precision and relevance of the generated reports. This capability is crucial in the medical domain where accuracy and context are essential for effective reporting of diagnostic findings.
As we want to enhance our model capabilities, the exploration of alternative sentence generators beyond the conventional LSTM architecture such as hierarchical LSTM and transformers emerges as a promising path. This potential solution could help our model in generating longer, more detailed sentences, contributing to the overall quality of medical reports produced by ChestBioX-Gen.
However, it's important to acknowledge that AI-driven solutions have their limitations, especially in the sensitive medical domain where high precision and minimal error margins are important. Achieving such precision requires extensive training on larger datasets containing diverse real-world examples, such as MIMIC-CXR. Therefore, ongoing efforts to expand training datasets are essential to ensure the reliability and accuracy of AI-driven solutions like ChestBioX. This expanded training, covering a wider range of scenarios, will help validate the robustness and generalizability of ChestBioX-Gen, offering a more comprehensive understanding of its performance and highlighting specific areas for improvement. In enhancing AI-driven medical image analysis, improvements can target several key areas: integrating pretrained image encoders trained on larger datasets to capture crucial diagnostic information, diversifying datasets with images from various angles to enhance model robustness in detecting challenging anatomical structures, and incorporating multimodality inputs such as X-ray and ultrasound images to provide a comprehensive view of cases, potentially improving diagnostic accuracy and patient care reports.
Finally, the integration of BioGPT and the cross-attention mechanism in ChestBioX-Gen represents an interesting advancement in AI-driven medical image analysis. The model's success in generating clinically relevant and coherent chest X-ray reports demonstrates its potential to contribute to the field of radiology reporting, benefiting both healthcare professionals and patients.
In conclusion, our study presents a novel chest X-ray report generation model incorporating a cross-attention module that leverages information Learned from visual models and BioGPT, a language model used as a tokenizer, showcasing its proficiency in handling medical data. The collaborative functionality of these components plays a crucial role in extracting pertinent information from input X-ray images, consequently enhancing the coherence and contextual relevance of generated sentences. When applied to the IU-X-Ray dataset, the model demonstrates promising results in terms of BLUE-N and ROUGE-L metrics. To advance our work, it is crucial to test our model on larger datasets and explore alternative language models, especially for generating lengthy sentences. These efforts are directed toward further enhancing and validating the capabilities of the proposed model in the field of chest X-ray report generation. Our model represents a promising advancement in the field of medical image analysis, offering valuable insights into pathologies present in X-ray images and serving as a first-aided solution for radiologists. However, it is essential to recognize that while our model provides valuable support and guidance, the responsibility for diagnosis and treatment decisions lies with the radiologists. Our model complements the expertise and clinical judgment of radiologists, enhancing their workflow and potentially improving diagnostic accuracy. Moving forward, continued research and development in AI-driven medical image analysis should focus on further enhancing models to better assist healthcare professionals while ensuring that human expertise remains crucial to patient care.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
MO: Writing – review & editing, Writing – original draft. MA: Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work has been supported in part by the New Brunswick Health Research Foundation (NBHRF) and the New Brunswick Innovation Foundation (NBIF), New Brunswick Priority Occupation Student Support Fund (NBPOSS) POF2021-006.
Thanks to the support provided by Calcul Quebec (https://www.calculquebec.ca/) and the Digital Research Alliance of Canada (alliancecan.ca).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akbar, W., Haq, M. I. U., Soomro, A., Daudpota, S. M., Imran, A. S., Ullah, M., et al. (2023). “Automated report generation: a GRU based method for chest X-rays,” in 2023 4th International Conference on Computing, Mathematics and Engineering Technologies (iCoMET) (Sukkur: IEEE), 1–6. doi: 10.1109/iCoMET57998.2023.10099311
Bustos, A., Pertusa, A., Salinas, J.-M., and de la Iglesia-Vayá, M. (2020). Padchest: a large chest X-ray image dataset with multi-label annotated reports. Med. Image Anal. 66:101797. doi: 10.1016/j.media.2020.101797
Demner-Fushman, D., Kohli, M. D., Rosenman, M. B., Shooshan, S. E., Rodriguez, L., Antani, S., et al. (2016). Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 23, 304–310. doi: 10.1093/jamia/ocv080
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., Fei-Fei, L., et al. (2009). “Imagenet: a large-scale hierarchical image database,” in 2009 IEEE Conference on Computer vision And Pattern Recognition (Miami, FL: IEEE), 248–255. doi: 10.1109/CVPR.2009.5206848
Denkowski, M., and Lavie, A. (2014). “Meteor universal: language specific translation evaluation for any target language,” in Proceedings of the Ninth Workshop on statistical Machine Translation (Pittsburgh, PA), 376–380. doi: 10.3115/v1/W14-3348
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger, K. Q. (2017). “Densely connected convolutional networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (Honolulu, HI: IEEE), 4700–4708. doi: 10.1109/CVPR.2017.243
Irvin, J., Rajpurkar, P., Ko, M., Yu, Y., Ciurea-Ilcus, S., Chute, C., et al. (2019). “Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison,” in Proceedings of the AAAI Conference on Artificial Intelligence, Vol. 33 (Stanford, CA), 590–597. doi: 10.1609/aaai.v33i01.3301590
Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L.-w. H., Feng, M., Ghassemi, M., et al. (2016). Mimic-iii, a freely accessible critical care database. Sci. Data 3, 1–9. doi: 10.1038/sdata.2016.35
Kale, K., Bhattacharyya, P., Gune, M., Shetty, A., and Lawyer, R. (2023). “KGVL-BART: knowledge graph augmented visual language bart for radiology report generation,” in Proceedings of the 17th Conference of the European Chapter of the Association for Computational Linguistics (Mumbai), 3393–3403. doi: 10.18653/v1/2023.eacl-main.246
Kaur, N., and Mittal, A. (2023). Chexprune: sparse chest X-ray report generation model using multi-attention and one-shot global pruning. J. Ambient Intell. Humaniz. Comput. 14, 7485–7497. doi: 10.1007/s12652-022-04454-z
Li, M., Lin, B., Chen, Z., Lin, H., Liang, X., Chang, X., et al. (2023). “Dynamic graph enhanced contrastive learning for chest X-ray report generation,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Vancouver, BC: IEE), 3334–3343. doi: 10.1109/CVPR52729.2023.00325
Lin, C.-Y. (2004). “Rouge: a package for automatic evaluation of summaries,” in Text Summarization Branches Out (Marina del Rey, CA), 74–81.
Liu, F., Yin, C., Wu, X., Ge, S., Zhang, P., Sun, X., et al. (2021). “Contrastive attention for automatic chest X-ray report generation,” in Findings of the Association for Computational Linguistics: ACL-IJCNLP 2021 (Beijing), 269–280. doi: 10.18653/v1/2021.findings-acl.23
Liu, G., Hsu, T.-M. H., McDermott, M., Boag, W., Weng, W.-H., Szolovits, P., et al. (2019). “Clinically accurate chest X-ray report generation,” in Machine Learning for Healthcare Conference (Beijing: PMLR), 249–269.
Lu, J., Yang, J., Batra, D., and Parikh, D. (2016). Hierarchical question-image co-attention for visual question answering. Adv. Neural Inf. Process. Syst. 29.
Luo, R., Sun, L., Xia, Y., Qin, T., Zhang, S., Poon, H., et al. (2022). BioGPT: generative pre-trained transformer for biomedical text generation and mining. Brief. Bioinform. 23:bbac409. doi: 10.1093/bib/bbac409
Nguyen, H. Q., Lam, K., Le, L. T., Pham, H. H., Tran, D. Q., Nguyen, D. B., et al. (2022). VINDR-CXR: an open dataset of chest X-rays with radiologist's annotations. Sci. Data 9:429. doi: 10.1038/s41597-022-01498-w
Papineni, K., Roukos, S., Ward, T., and Zhu, W.-J. (2002). “Bleu: a method for automatic evaluation of machine translation,” in Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics (Yorktown Heights, NY), 311–318. doi: 10.3115/1073083.1073135
Pennington, J., Socher, R., and Manning, C. D. (2014). “Glove: global vectors for word representation,” in Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) (Stanford, CA), 1532–1543. doi: 10.3115/v1/D14-1162
Qi, P., Zhang, Y., Zhang, Y., Bolton, J., and Manning, C. D. (2020). “Stanza: a Python natural language processing toolkit for many human languages,” in Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics: System Demonstrations (Stanford, CA). doi: 10.18653/v1/2020.acl-demos.14
Rajpurkar, P., Irvin, J., Zhu, K., Yang, B., Mehta, H., Duan, T., et al. (2017). Chexnet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv [Preprint]. arXiv:1711.05225. doi: 10.48550/arXiv.1711.05225
Shetty, S., Ananthanarayana, V., and Mahale, A. (2023). Cross-modal deep learning-based clinical recommendation system for radiology report generation from chest X-rays. Int. J. Eng 36, 1569–1577. doi: 10.5829/IJE.2023.36.08B.16
Vedantam, R., Lawrence Zitnick, C., and Parikh, D. (2015). “Cider: consensus-based image description evaluation,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (Boston, MA: IEEE), 4566–4575. doi: 10.1109/CVPR.2015.7299087
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., Summers, R. M., et al. (2017). “ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (Honolulu, HI: IEEE), 2097–2106. doi: 10.1109/CVPR.2017.369
Wu, L., Chen, Y., Shen, K., Guo, X., Gao, H., Li, S., et al. (2023). Graph neural networks for natural language processing: asurvey. Found. Trends Mach. Learn 16, 119–328. doi: 10.1561/2200000096
Yang, S., Wu, X., Ge, S., Zheng, Z., Zhou, S. K., Xiao, L., et al. (2023). Radiology report generation with a learned knowledge base and multi-modal alignment. Med. Image Anal. 86:102798. doi: 10.1016/j.media.2023.102798
Zhang, D., Ren, A., Liang, J., Liu, Q., Wang, H., Ma, Y., et al. (2022). Improving medical X-ray report generation by using knowledge graph. Appl. Sci. 12:11111. doi: 10.3390/app122111111
Zhang, H., Song, H., Li, S., Zhou, M., and Song, D. (2023). A survey of controllable text generation using transformer-based pre-trained language models. ACM Comput. Surv. 56, 1–37.
Zhang, J., Xie, Y., Li, K., Wang, Z., and Du, W. (2023). Hierarchical decoding with latent context for image captioning. Neural Comput. Appl. 35, 2429–2442. doi: 10.1007/s00521-022-07726-z
Zhang, Y., Ding, D. Y., Qian, T., Manning, C. D., and Langlotz, C. P. (2018). Learning to summarize radiology findings. arXiv [Preprint]. arXiv:1809:04698. doi: 10.48550/arXiv.1809.04698
Keywords: radiology reporting, contextual understanding, deep learning, medical imaging, computer-aided diagnosis
Citation: Ouis MY and Akhloufi MA (2024) ChestBioX-Gen: contextual biomedical report generation from chest X-ray images using BioGPT and co-attention mechanism. Front. Imaging. 3:1373420. doi: 10.3389/fimag.2024.1373420
Received: 19 January 2024; Accepted: 22 March 2024;
Published: 19 April 2024.
Edited by:
Alessandro Piva, University of Florence, ItalyReviewed by:
Guokai Zhang, University of Shanghai for Science and Technology, ChinaCopyright © 2024 Ouis and Akhloufi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moulay A. Akhloufi, bW91bGF5LmFraGxvdWZpQHVtb25jdG9uLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.