- Department of Clinical Laboratory, The Second People’s Hospital of Jiangjin District, Chongqing, China
Thalassemia is one of the inherited hemoglobin disorders worldwide, resulting in ineffective erythropoiesis, chronic hemolytic anemia, compensatory hemopoietic expansion, hypercoagulability, etc., and when a mother carries the thalassemia gene, the child is more likely to have severe thalassemia. Furthermore, the economic and time costs of genetic testing for thalassemia prevent many thalassemia patients from being diagnosed in time. To solve this problem, we performed least absolute shrinkage and selection operator (LASSO) regression to analyze the correlation between thalassemia and blood routine indicators containing mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), and red blood cell (RBC). We then built a nomogram to predict the occurrence of thalassemia, and receiver operating characteristic (ROC) curve was used to verify the prediction efficiency of this model. In total, we obtained 7,621 cases, including 847 thalassemia patients and 6,774 non-thalassemia. Among the 847 thalassemia patients, with a positivity rate of 67.2%, 569 cases were positive for α-thalassemia, and with a rate of 31.5%, 267 cases were positive for β-thalassemia. The remaining 11 cases were positive for both α- and β-thalassemia. Based on machine learning algorithm, we screened four optimal indicators, namely, MCV, MCH, RBC, and MCHC. The AUC value of MCV, MCH, RBC, and MCHC were 0.907, 0.906, 0.796, and 0.795, respectively. Moreover, the AUC value of the prediction model was 0.911. In summary, a novel and effective machine learning model was built to predict thalassemia, which functioned accurately, and may provide new insights for the early screening of thalassemia in the future.
1 Introduction
Thalassemia, a prevalent monogenic disease worldwide, leads to hemolytic anemia due to impaired globin synthesis (1, 2). It is particularly prevalent in regions such as South Africa, the Middle East, and Southeast Asia, as well as in low- and middle-income areas like coastal cities in southern China and rural areas in western China. China bears the highest burden of thalassemia globally, with approximately 30 million individuals affected by thalassemia-related mutations and 3 million suffering from moderate to severe forms, posing significant challenges to families and society (3–5). Due to the autosomal recessive inheritance pattern of thalassemia, parents who are asymptomatic can still have children affected by thalassemia. When both parents carry the thalassemia gene, there is a substantial likelihood of their child developing severe thalassemia, which typically necessitates lifelong blood transfusions and is accompanied by various complications. This imposes a significant burden on affected children, their families, and society. Consequently, early detection of thalassemia during pregnancy assumes paramount importance. Existing approaches for thalassemia screening and diagnosis encompass osmotic fragility tests, assessment of red blood cell (RBC) smears, identification of inclusion bodies, evaluation of red blood cell indices, hemoglobin electrophoresis, high-performance liquid chromatography (HPLC), and genetic testing (6, 7). However, the cost and time associated with genetic testing often hinder timely diagnosis for many thalassemia patients. Conversely, blood routine indicators play a crucial role in the early identification of thalassemia due to the widespread availability of blood routine tests and the ability to distinguish different types of anemia based on red blood cell morphology (8, 9).
In this study, we analyzed the genetic test and blood routine results of 7,621 pregnant women being tested for thalassemia in the Jiangjin area of Chongqing from 2018 to 2022. Additionally, we employed a machine learning model to investigate the predictive value of blood routine indicators for thalassemia. The goal was to offer novel strategies for the early diagnosis, genetic counseling, and treatment of thalassemia in pregnant women within the Chongqing region.
2 Materials and methods
2.1 Patients
We conducted a retrospective study on thalassemia patients who were pregnant in the Jiangjin District, focusing on prenatal screening. From January 2018 to December 2022, blood routine and genetic tests for thalassemia were performed for the first time. After removing duplicates and cases with missing key information, a total of 7,621 cases were included in our analysis. This study received approval from the Medical Ethics Committee of The Second People’s Hospital of Jiangjin District, Chongqing.
2.2 Blood routine index test
The Sysmex XN-1000 automated blood cell analysis system along with its accompanying reagents were utilized to measure various hematological parameters, including red blood cell count (RBC), hemoglobin level (Hb), hematocrit level (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), and the standard deviation of red cell volume distribution width (RDW-SD).
2.3 Genetic testing for thalassemia
In our study, we employed polymerase chain reaction (PCR) in combination with diversion hybridization to detect various types of mutations and deletions associated with α-thalassemia and β-thalassemia. Specifically, we targeted three deletion types of α-thalassemia (i.e., –SEA, -α3.7 and -α4.2), three mutation types of α-thalassemia (CS, QS, and WS), as well as 17 mutation types of β-thalassemia [-28(A-G), -29(A-G), -30(T-C), -32(C-A), CD14/15(+G), CD17(A-T), CD27/28(+C), CD31(-C), CD41/42(-TCTT), CD43(G-T), CD71/72(+A), IVS-I-1(G-T,G-A), IVS-I-5(G-C), IVS-II-654(C-T), βE(G-A), CAP(A-C, A-AAAC), and Int(T-G)].
2.4 Model establishment
Data collation was conducted using Microsoft Excel (RRID : SCR_016137). The R Project for Statistical Computing (RRID : SCR_001905) was utilized for model establishment, training, verification of factors associated with hematological indicators, and thalassemia prediction. The samples were randomly divided into two parts: a training set and a validation set. The training set consisted of Type 1 [IVS-II-654 (C-T), 55 cases], Type 2 (–SEA/αα, 186 cases), Type 3 (-α3.7/αα, 287 cases), Type 4 [CD17 (A-T), 95 cases], and Type 5 [CD41-42 (-TCTT), 62 cases]. To identify the optimal indicators for the prediction model, least absolute shrinkage and selection operator (LASSO) regression was performed via the R package “glmnet”. The veen plot was visualized by the R package “ggVennDiagram”. The predictive accuracy of the model was verified using the receiver operating characteristic (ROC) curve by the R package “pROC”. Furthermore, a nomogram was constructed to establish the scoring criteria for the corresponding variables based on the coefficients of the LASSO regression model using the R package “rms”. The selected variables were plotted on the variable axis, and a straight line was drawn to determine the score for each variable value. Using the training set, the blood routine data were imported, and the scores corresponding to each variable were assigned. The total score was calculated by summing the scores of all variables. These data were then inputted into the linear predictor to predict the risk of thalassemia.
3 Results
3.1 Positive genetic types of thalassemia
Among the 7,621 cases analyzed, a total of 847 were identified as positive for thalassemia, resulting in a positivity rate of 11.11%. Specifically, 569 cases were positive for α-thalassemia, with a rate of 7.47%, while 267 cases were positive for β-thalassemia, with a rate of 3.50%. Furthermore, there were 11 cases that tested positive for both α- and β-thalassemia, as well as other combined genotypes, accounting for 0.14% of the cases. Compared to normal pregnant women, individuals with thalassemia exhibited decreased levels of hemoglobin (HGB), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), and increased red blood cell distribution width-standard deviation (RDW-SD) and red blood cell distribution width-coefficient of variation (RDW-CV). These findings are consistent with the clinical manifestations of small cell hypochromic anemia. Notably, the changes observed in individuals with β-thalassemia and α- combined β-thalassemia genotypes were more pronounced than those seen in individuals with α-thalassemia (as shown in Table 1).
3.2 Analysis of the relevant coefficient of thalassemia
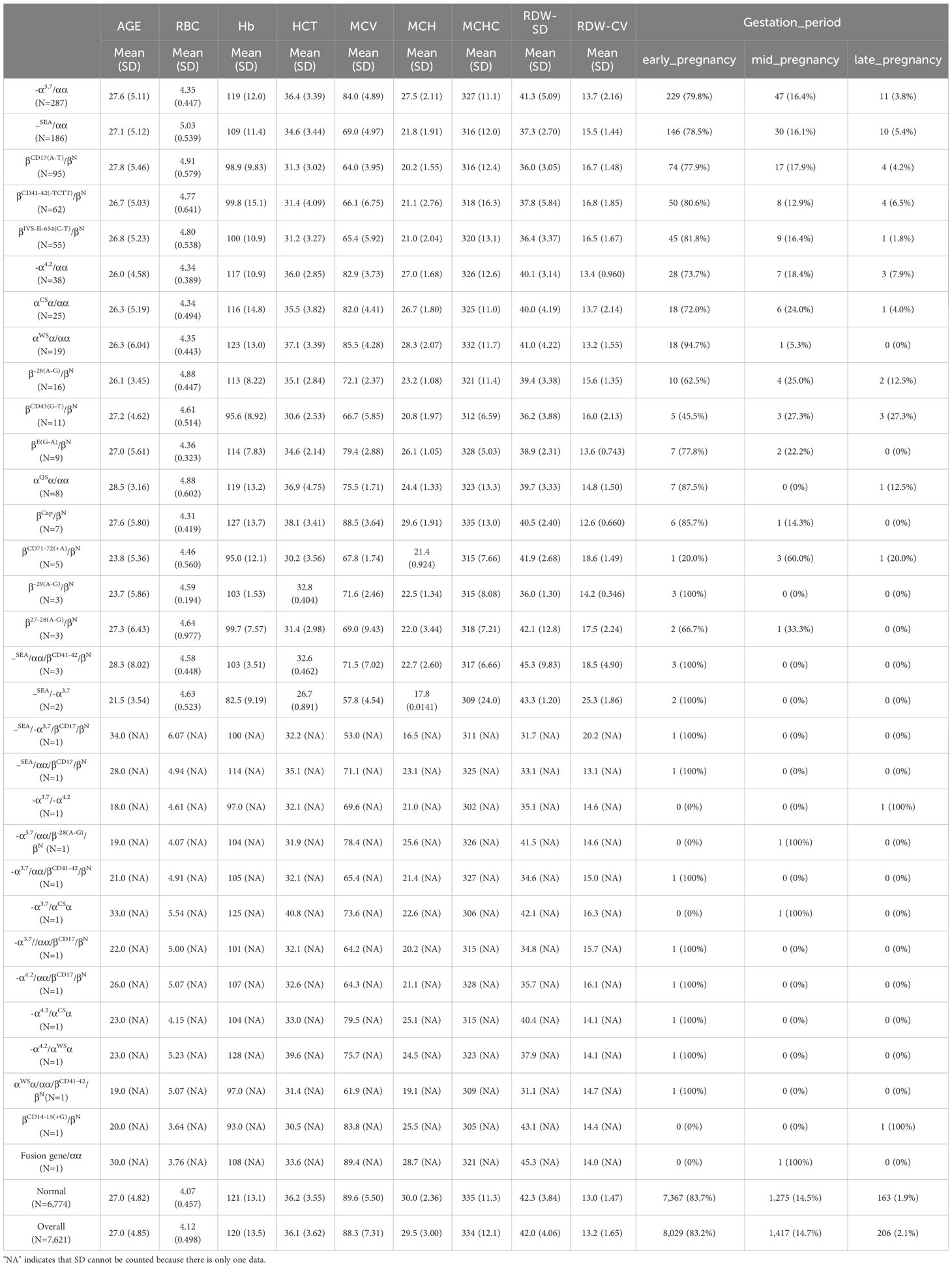
To identify the variables with the highest correlation to thalassemia, a LASSO prediction model was established using the data from the training set, which consisted of five thalassemia genotypes, namely, Type 1 [IVS-II-654 (C-T), 55 cases] (Figure 1A), Type 2 (–SEA/αα, 186 cases) (Figure 1B), Type 3 (-α3.7/αα, 287 cases) (Figure 1C), Type 4 [CD17 (A-T), 95 cases] (Figure 1D), and Type 5 [CD41-42 (-TCTT), 62 cases] (Figure 1E). Binomial deviation and Venn diagram analyses were employed to select the optimal variables (Figure 1F). As a result, four variables, namely, mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), and red blood cell count (RBC), were found to have the strongest correlation with thalassemia. To validate the predictive performance of these variables, receiver operating characteristic (ROC) curve analysis was conducted. The area under the curve (AUC) (Figure 1G) was used to interpret the results, with values greater than 0.5 and closer to 1 indicating higher effectiveness. It was observed that MCV and MCH exhibited a higher correlation with thalassemia compared to RBC and MCHC.
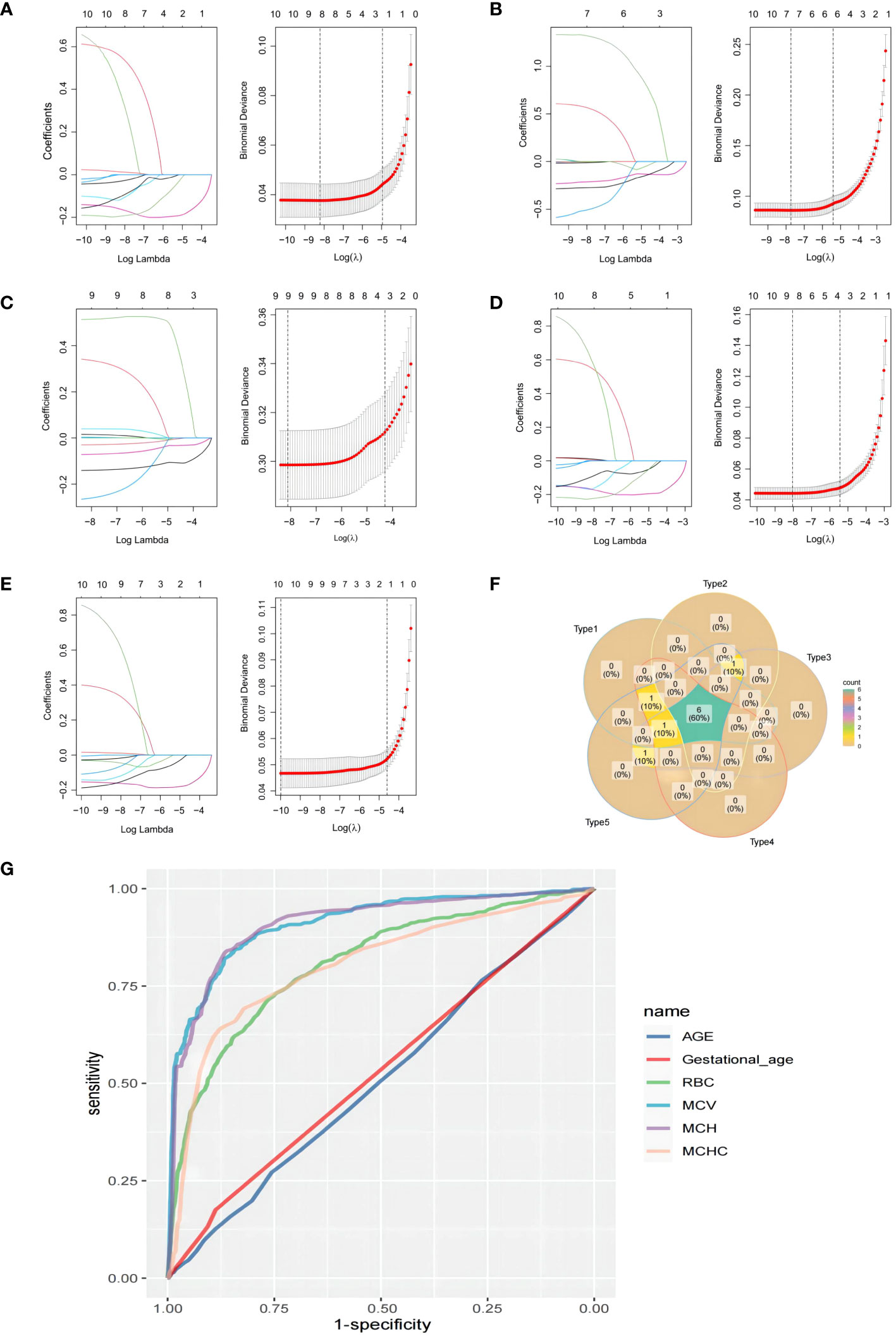
Figure 1 Least absolute shrinkage and selection operator (LASSO) prediction model was established using the data from the training set, which consisted of five thalassemia genotypes: (A) Type 1 [IVS-II-654(C-T), 55 cases], (B) Type 2 (–SEA/αα, 186 cases), (C) Type 3 (-α3.7/αα, 287 cases), (D) Type 4 [CD17(A-T), 95 cases], and (E) Type 5 [CD41-42(-TCTT), 62 cases]. (F) The veen plot of the LASSO result intersections among the five types. (G) The receiver operating characteristic (ROC) curve for the predictive value of each factor.
3.3 Establishment of a prediction model for thalassemia
Utilizing the correlation factors of MCV, MCH, RBC, and MCHC, a nomogram model (Figure 2A) was constructed, incorporating the scoring criteria for each variable derived from the coefficients of the LASSO regression model. The training set was selected to establish the scoring criteria for different variables. The receiver operating characteristic (ROC) curve analysis was performed to evaluate the predictive performance of MCV, MCH, RBC, and MCHC. The respective area under the curve (AUC) values were found to be 0.910, 0.909, 0.807, and 0.801 (Figures 2B–E). Furthermore, the AUC value of the overall model was determined to be 0.913 (Figure 2F). These results demonstrate that the prediction model effectively enhances the predictive power of these variables and reduces errors associated with using a single index alone.
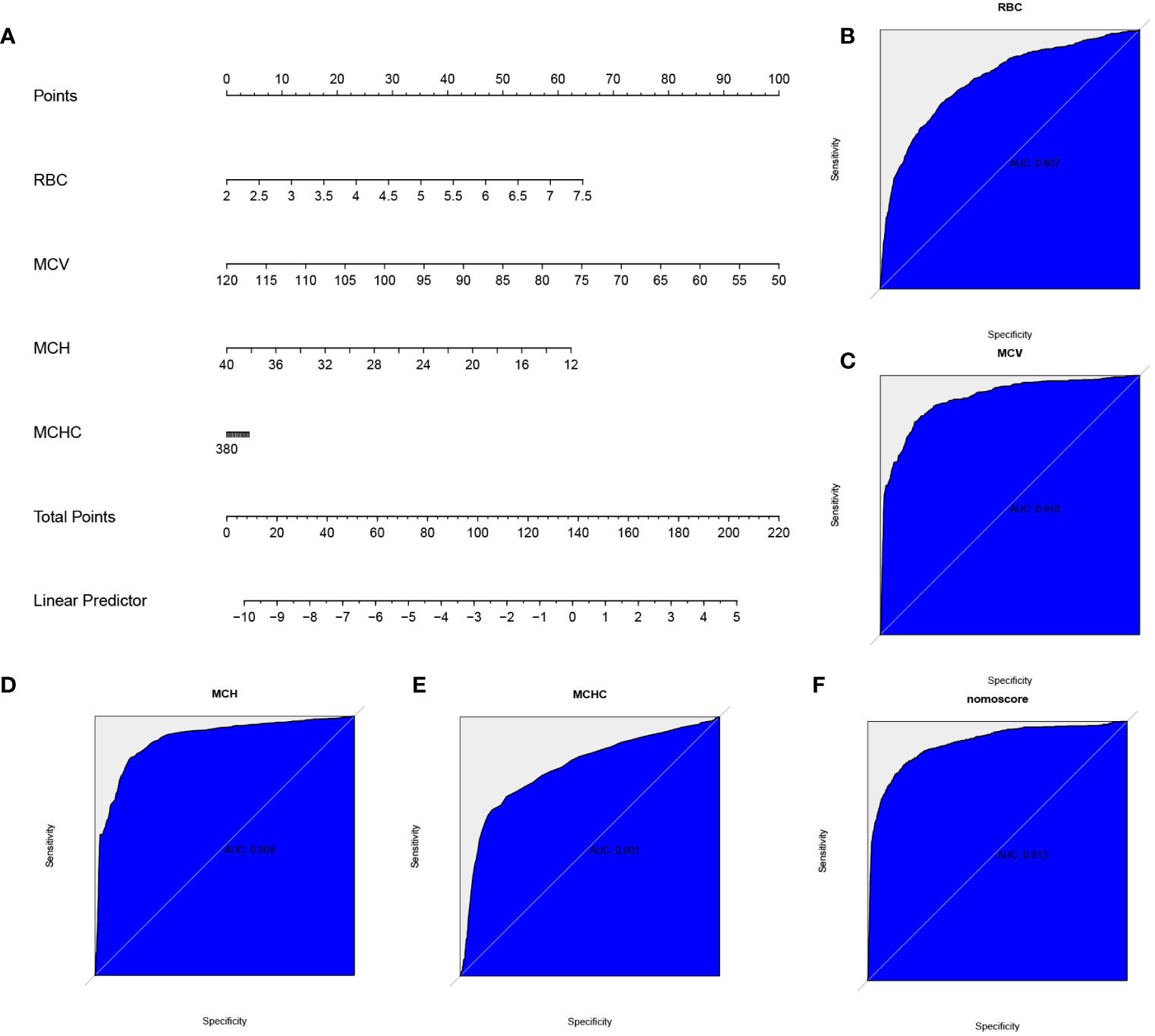
Figure 2 The nomogram and predictive efficiency of this model in the training set. (A) Construction of a predictive nomogram. The receiver operating characteristic (ROC) curve standing for the predictive efficiency of the correlation factors: (B) red blood cell (RBC), (C) mean corpuscular volume (MCV), (D) mean corpuscular hemoglobin (MCH), (E) mean corpuscular hemoglobin concentration (MCHC), and (F) the model.
3.4 Optimization of thalassemia prediction model
After observing the satisfactory performance of the model in the training set, we proceeded to fine-tune the variables to accommodate more complex data in the testing set (Figure 3A). As a result, the AUC values for MCV, MCH, RBC, and MCHC were found to be 0.907, 0.906, 0.796, and 0.795, respectively (Figures 3B–E). Notably, the overall model exhibited an AUC value of 0.911, indicating a strong predictive effect (Figure 3F). In the validation group, the addition of normal data for thalassemia and other genotypes led to a reduction in AUC. This adjustment was made to bring the model closer to real-world scenarios and enhance its reliability.

Figure 3 The nomogram and predictive efficiency of this model in the testing set. (A) Construction of a predictive nomogram. The ROC curve standing for the predictive efficiency of the correlation factors: (B) red blood cell (RBC), (C) mean corpuscular volume (MCV), (D) mean corpuscular hemoglobin (MCH), (E) mean corpuscular hemoglobin concentration (MCHC), and (F) the model.
4 Discussion
The rapid advancement of artificial intelligence (AI) technology has garnered significant attention worldwide, particularly in the realm of disease diagnosis. Machine learning and other related technologies have been widely explored to aid in this process. In comparison to manual diagnosis, computer-based diagnostic methods offer increased accuracy and efficiency, effectively reducing the misdiagnosis rate. Consequently, these methods enable more effective disease diagnoses at a lower cost (10–12). By harnessing the power of big data and clinical information, machine learning techniques have greatly improved the accuracy and efficiency of clinical diagnoses. This progress has propelled laboratory medicine toward precision medicine and intelligent testing (13, 14). Applying machine learning to the early diagnosis of thalassemia, for instance, allows for the prediction of thalassemia types with higher severity using a blood sample with high prevalence. This approach can effectively reduce the cost and time required for genetic identification of thalassemia, enabling healthcare providers to initiate early treatment measures and offer prompt genetic counseling to pregnant women with thalassemia. Such efforts are important in reducing the birth rate of children with moderate and severe thalassemia and conducting population health surveys in middle and low-income areas.
In this study, a total of 7,621 pregnant women underwent blood routine and thalassemia detection. Among them, 569 cases of α-thalassemia were identified, yielding a positive rate of 7.47%. Additionally, 267 cases of β-thalassemia were detected, with a positive rate of 3.5%. Furthermore, 11 cases of α- combined with β-thalassemia were observed, resulting in a positive rate of 0.14%. These prevalence rates were found to be lower compared to coastal cities such as Guangdong, particularly in relation to α thalassemia, indicating a significant difference as compared to previous studies (4, 15). To mitigate the cost and time associated with genetic diagnosis of thalassemia, a machine learning model was constructed in this study. The model employed routine blood test results for thalassemia prediction. Among the five genotypes with the highest incidence of thalassemia [α3.7/αα, –SEA/αα, CD17 (A-T), CD41-42 (-TCTT), and IVS-II-654 (C-T)], the four coefficients that exhibited the highest correlation with thalassemia were selected: MCV, MCH, RBC, and MCHC. Notably, the prediction performance of MCV and MCH demonstrated superior results. These findings align with previous studies, which have consistently reported reduced MCV and MCH levels across almost all types of thalassemia (15–17).
The prediction model was developed using four variables: MCV, MCH, RBC, and MCHC. The model’s predictive performance was assessed using ROC analysis, yielding AUC values of 0.910, 0.909, 0.807, and 0.801, respectively. These results indicate significant diagnostic value for thalassemia detection. The overall AUC value of the model was found to be 0.913, surpassing the individual variables in terms of prediction accuracy. This result demonstrates the successful construction of the model. To further optimize the model for complex clinical data, adjustments were made to each variable. Subsequently, the entire dataset was utilized for verification purposes. The obtained AUC values for MCV, MCH, RBC, and MCHC were 0.907, 0.906, 0.796, and 0.795, respectively. These values were slightly lower than those obtained during the training phase. However, the model still exhibited a good prediction effect, with an overall AUC value of 0.911. This minor decrease in AUC can be attributed to the inclusion of data from normal pregnant women. Their physiological anemia can interfere with the model, but incorporating such data ensures that the predictive performance of the model aligns more closely with clinical reality, thereby holding significant clinical significance.
It is worth mentioning that the main positive data in the model primarily consist of common thalassemia genotypes, such as -α3.7/αα, –SEA/αα, and CD17 (A-T). Consequently, the model’s predictions may be more accurate for these genotypes. Furthermore, it is crucial to note that unlike the findings of Khan et al. and El-Beshlawy et al. (18, 19), the current model’s predictive capability is limited when it comes to β-thalassemia in regions such as Africa, the Middle East, and other areas, as it primarily focuses on Asia, where α-thalassemia is prevalent. Additionally, since the study subjects predominantly consist of pregnant women, the model may face challenges in accurately predicting thalassemia in men and children. Another limitation of the model is its current inability to differentiate between varying degrees of thalassemia severity, such as severe, moderate, or characteristic types. Furthermore, research by Ferih et al. (20) provides us with a good idea that our model may face challenges in distinguishing between thalassemia and iron-deficiency anemia, as the diagnosis of iron-deficiency anemia often requires additional hematological indicators that may not be fully incorporated into the current model, which need further study.
5 Summary
This study aimed to predict thalassemia in pregnancy by means of economical and rapid blood routine detection by establishing a data prediction model. Through the analysis of 7,621 cases, MCV, MCH, RBC, and MCHC were selected as high correlation indicators. Subsequently, a machine learning prediction model was constructed, incorporating these four indicators as variables, and the results were verified using ROC analysis. The AUC values for MCV, MCH, RBC, and MCHC were 0.907, 0.906, 0.796, and 0.795, respectively. In particular, the prediction model achieved an AUC value of 0.911, demonstrating its effectiveness in thalassemia prediction and provided a novel strategy for the early screening of thalassemia.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Ethics Committee of The Second People’s Hospital of Jiangjin District, Chongqing. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of your previous study for which ethical approval was obtained. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
YL: Writing – original draft, Writing – review & editing. WB: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Science-Health Joint Medical Scientific Research Project of Chongqing (2022MSXM092).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Kattamis A, Kwiatkowski JL, Aydinok Y. thalassemia. Lancet. (2022) 399:2310–24. doi: 10.1016/S0140-6736(22)00536-0
2. Taher AT, Weatherall DJ, Cappellini MD. thalassemia. Lancet. (2018) 391:155–67. doi: 10.1016/S0140-6736(17)31822-6
3. Mo D, Zheng Q, Xiao B, Li L. Predicting thalassemia using deep neural network based on red blood cell indices. Clin Chim Acta. (2023) 543:117329. doi: 10.1016/j.cca.2023.117329
4. Wang WD, Hu F, Zhou DH, Gale RP, Lai YR, Yao HX, et al. thalassemia in China. Blood Rev. (2023) 60:101074. doi: 10.1016/j.blre.2023.101074
5. Weatherall DJ. The evolving spectrum of the epidemiology of thalassemia. Hematol Oncol Clin North Am. (2018) 32:165–75. doi: 10.1016/j.hoc.2017.11.008
6. Viprakasit V, Ekwattanakit S. Clinical classification, screening and diagnosis for thalassemia. Hematol Oncol Clin North Am. (2018) 32:193–211. doi: 10.1016/j.hoc.2017.11.006
7. Achour A, Koopmann TT, Baas F, Harteveld CL. The evolving role of next-generation sequencing in screening and diagnosis of hemoglobinopathies. Front Physiol. (2021) 12:686689. doi: 10.3389/fphys.2021.686689
8. Xu M, Lin G, Dong Z, Wang Q, Ma L, Su J. Logistic-Nomogram model based on red blood cell parameters to differentiate thalassemia trait and iron deficiency anemia in southern region of Fujian Province, China. J Clin Lab Anal. (2023) 37:e24940. doi: 10.1002/jcla.24940
9. Zheng S, Li Q, Ou T, Li Y, Wu S. Clinical performance study of a new fully automated red blood cell permeability fragility analyzer. J Healthc Eng. (2022) 2022:5642907. doi: 10.1155/2022/5642907
10. Yuan Y, Shi C, Zhao H. Machine learning-enabled genome mining and bioactivity prediction of natural products. ACS Synth Biol. (2023) 12(9):2650–62. doi: 10.1021/acssynbio.3c00234
11. Othman NA, Azhar MAAS, Damanhuri NS, Mahadi IA, Abbas MH, Shamsuddin SA, et al. Optimization of identifying insulinaemic pharmacokinetic parameters using artificial neural network. Comput Methods Programs Biomed. (2023) 236:107566. doi: 10.1016/j.cmpb.2023.107566
12. Bar-On M, Baharav S, Katzir Z, Mirelman A, Sosnik R, Maidan I. Task-related reorganization of cognitive network in Parkinson's disease using electrophysiology. Mov Disord. (2023) 38(11):2031–40. doi: 10.1002/mds.29571
13. Yuan W, Zhi W, Ma L, Hu X, Wang Q, Zou Y, et al. Neural oscillation disorder in the hippocampal CA1 region of different Alzheimer's disease mice. Curr Alzheimer Res. (2023) 20(5):350–9. doi: 10.2174/1567205020666230808122643
14. Rabbani N, Kim GYE, Suarez CJ, Chen JH. Applications of machine learning in routine laboratory medicine: Current state and future directions. Clin Biochem. (2022) 103:1–7. doi: 10.1016/j.clinbiochem.2022.02.011
15. Xian J, Wang Y, He J, Li S, He W, Ma X, et al. Molecular epidemiology and hematologic characterization of thalassemia in Guangdong Province, Southern China. Clin Appl Thromb Hemost. (2022) 28:10760296221119807. doi: 10.1177/10760296221119807
16. Zhuang J, Jiang Y, Wang Y, Zheng Y, Zhuang Q, Wang J, et al. Molecular analysis of α-thalassemia and β-thalassemia in Quanzhou region Southeast China. J Clin Pathol. (2020) 73:278–82. doi: 10.1136/jclinpath-2019-206179
17. Chen P, Lin WX, Li SQ. THALASSEMIA in ASIA 2021: thalassemia in Guangxi Province, People's Republic of China. Hemoglobin. (2022) 46:33–5. doi: 10.1080/03630269.2021.2008960
18. Khan AM, Al-Sulaiti AM, Younes S, Yassin M, Zayed H. The spectrum of beta-thalassemia mutations in the 22 Arab countries: a systematic review. Expert Rev Hematol. (2021) 14:109–22. doi: 10.1080/17474086.2021.1860003
19. El-Beshlawy A, Dewedar H, Hindawi S, Alkindi S, Tantawy AA, Yassin MA, et al. Management of transfusion-dependent β-thalassemia (TDT): Expert insights and practical overview from the Middle East. Blood Rev. (2024) 63:101138. doi: 10.1016/j.blre.2023.101138
Keywords: thalassemia, machine learning, blood routine indicators, pregnancy, prediction
Citation: Long Y and Bai W (2024) Constructing a novel clinical indicator model to predict the occurrence of thalassemia in pregnancy through machine learning algorithm. Front. Hematol. 3:1341225. doi: 10.3389/frhem.2024.1341225
Received: 28 November 2023; Accepted: 14 March 2024;
Published: 04 April 2024.
Edited by:
Mariasanta Napolitano, University of Palermo, ItalyReviewed by:
Richard Oscar Francis, Columbia University, United StatesMohamed A. Yassin, Qatar University, Qatar
Hakan Ayyildiz, Istanbul University, Türkiye
Copyright © 2024 Long and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxue Bai, NjY4MDgwNkBxcS5jb20=
†These authors have contributed equally to this work and share first authorship
 Yaoshui Long
Yaoshui Long Wenxue Bai
Wenxue Bai