
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Hematol., 16 November 2023
Sec. Blood Cancer
Volume 2 - 2023 | https://doi.org/10.3389/frhem.2023.1279863
This article is part of the Research TopicGlycobiology in Hematology: a Promising Avenue for Cancer Diagnostics and TherapeuticsView all 6 articles
Leukemic B-cells are lodged in the bone marrow [BM], a complex organ composed of many cell types and extracellular matrix. Determining how the reciprocal interactions between these components are regulated is critical to our understanding of the factors that allow leukemia cells to survive, multiply and withstand treatment. All cells in the bone marrow are surrounded by a glycocalyx, a glycan-rich layer of high complexity, which regulates such cell-cell and cell-matrix interactions. However, the structure and function of the glycan components of the biomolecules that constitute this layer have not been explored in much detail. Gaps are difficult to fill due to technical limitations as well as the fact that the composition of the BM in health, disease and aging is not static. This also applies to B-lineage malignancies that develop or persist in BM such as B-cell precursor acute lymphoblastic leukemia and Multiple Myeloma, and the effects of their treatment. In contrast, the proteomes and transcriptomes of different human bone marrow cells have been studied more extensively. A combination of technologies now increasingly allows correlations to be made between the expression of glycosyltransferases and glycan structures in cell lines, which could be extrapolated to RNAseq data from primary cells. Glycopeptide analysis will also be invaluable in providing details of specific glycan occupancy on glycoproteins, even if only as a snapshot in time. Functional studies on CD19, CD138/SDC1 and BCMA/TNFRSF17 have already demonstrated the importance of their glycosylation. Additional studies using such approaches are likely to find many more other instances in which malignant B-cell homeostasis is regulated by glycosylation, and lead to the identification of new targets to treat B-cell malignancies.
This review focuses only on human glycans and bone marrow B-cells. Mouse studies, which have provided substantial insight into the functional contribution of glycans to normal murine B-cells, were recently reviewed (1). Since there are no published studies that focus on the glycan component of human bone marrow, main sources of information are isolated primary cells or cells in tissue culture. Unfortunately, there are no glycomic studies on normal human bone marrow B-cell development, with older studies frequently using human leukemia cell lines such as NALM6 as a stand-in for normal early B-cells (2). Now that it has become more feasible to modify human hematopoietic cells genetically with Cas9/CRISPR technology, questions about the function of capping glycans can also be partly addressed, both in vitro as well as after transplant of glycosyltransferase-modified human B-lineage leukemia cells into immunocompromised mice (3). However, such in vivo experimentation introduces the variable of cross-species differences in glycosylation (4) and it is unclear if results from rodent cells can always be directly extrapolated to human cells (5, 6). Such limitations also apply to tissue culture. Nguyen et al. (7) showed that human leukemia cells take up N-glycolylneuraminic acid, a non-human glycan, as a component of the commonly used fetal bovine serum. Such incorporation clearly has functional consequences for immune cells (8–10). Additionally, the complexity of cell-cell and extracellular matrix [ECM]-cell interactions is only captured to a limited extent in cultured cells, and the culture of hematopoietic cell types at the non-natural oxygen conditions of tissue culture as compared to bone marrow also can induce changes (11). Thus, an optimal data set for human cells would include information from tissue culture or animal experimentation combined with primary cell analysis.
Glycan modifications add a staggering degree of diversity to biomolecules. Proteins, lipids and [more recently shown for] RNA (12) can all be modified by glycosylation, but the contributions of glycosylation to normal and abnormal B-cell development in the BM remain largely unknown. Glycosylation is regulated by glycosyltransferases, enzymes that are dedicated to one or more biosynthetic pathways of the glycosylation machinery. Its importance is illustrated by the fact that around 2% of all human protein-encoding genes are reserved for this activity: Growth et al. identified 403 human glyco-enzymes involved in biosynthesis of glycans (13).
Details of glycan biosynthesis can be found in reviews on glycosylation (14–17), their analysis (18–21) and in the Essentials of Glycobiology (22). The different forms of glycosylation that result in the formation of glycoconjugates are broadly classified based on the molecules to which they are attached, to include proteins [glycoproteins and glycophosphatidylinositol-linked proteins] and lipids [glycolipids]. Protein-linked glycans are also classified based on their attachment to asparagine [N-linked] or serine/threonine [O-linked, including Glycosaminoglycans, GAGs] residues, or on the type of initial sugar attachment such as mannose, fucose or N-acetylglucosamine (4). The contribution of monosaccharide N-acetyl glucosamine modification of serine/threonine residues on mostly intracellular proteins [termed O-GlcNAcylation (23)] in leukemias has been reviewed (24, 25). While biosynthetic steps involved in the different forms of glycosylation are frequently shared, many individual key steps are known to be unique to just one specific pathway (26).
The initial forms of glycan attachment to proteins, such as N-glycosylation, developed early on in the evolution of eukaryotes and remain highly conserved across all eukaryotic species studied to date (22). Individuality and diversification of glycosylation between species was largely introduced by the so-called glycan capping steps such as sialylation and fucosylation (4, 26). These terminal variations have produced highly antigenic epitopes and species-specific functions. In addition, post-glycosylation modifications such as acetylation of sialic acid attached to glycoproteins or glycolipids add further diversity and specificity (27, 28).
The “B” in B-cells designates the bone marrow [BM], the site at which immature B-cells are generated from hematopoietic stem cells. Early B-cell development proceeds from the common lymphoid progenitor, via pro-B1, pro-B2, pre-B1, pre-B2 and immature B cell stages (29). Immature B cells subsequently exit the bone marrow to undergo further differentiation and maturation (30, 31). This developmental process is regulated by changes in the expression of transcription factors (32) and by interactions within the niche of the hematopoietic precursors with bone marrow stromal cells and ECM (33).
The contribution of protein glycosylation to normal human B-cell development remains largely unexplored, although it seems certain that the glycan components of biomolecules are involved. For example, modification of the transcription factor c-Myc by O-GlcNAc in mice stimulates division of pre-B cells (34) suggesting a similar regulation of c-Myc in human B-cell development (35). Table 1 lists some of the important cell surface proteins involved in B-cell development/homeostasis and the contribution of glycosylation to their function, if any. For example, IL7 signaling is essential for early human B-cell development (76). In vitro, IL7 binds 300-fold more tightly to N-glycosylated IL7Rα than to the non-glycosylated protein (37). CRLF2, shown to be important for normal human B-cell development (77), needs N-linked glycosylation for cells surface expression and signal transduction after stimulation with TSLP (40).
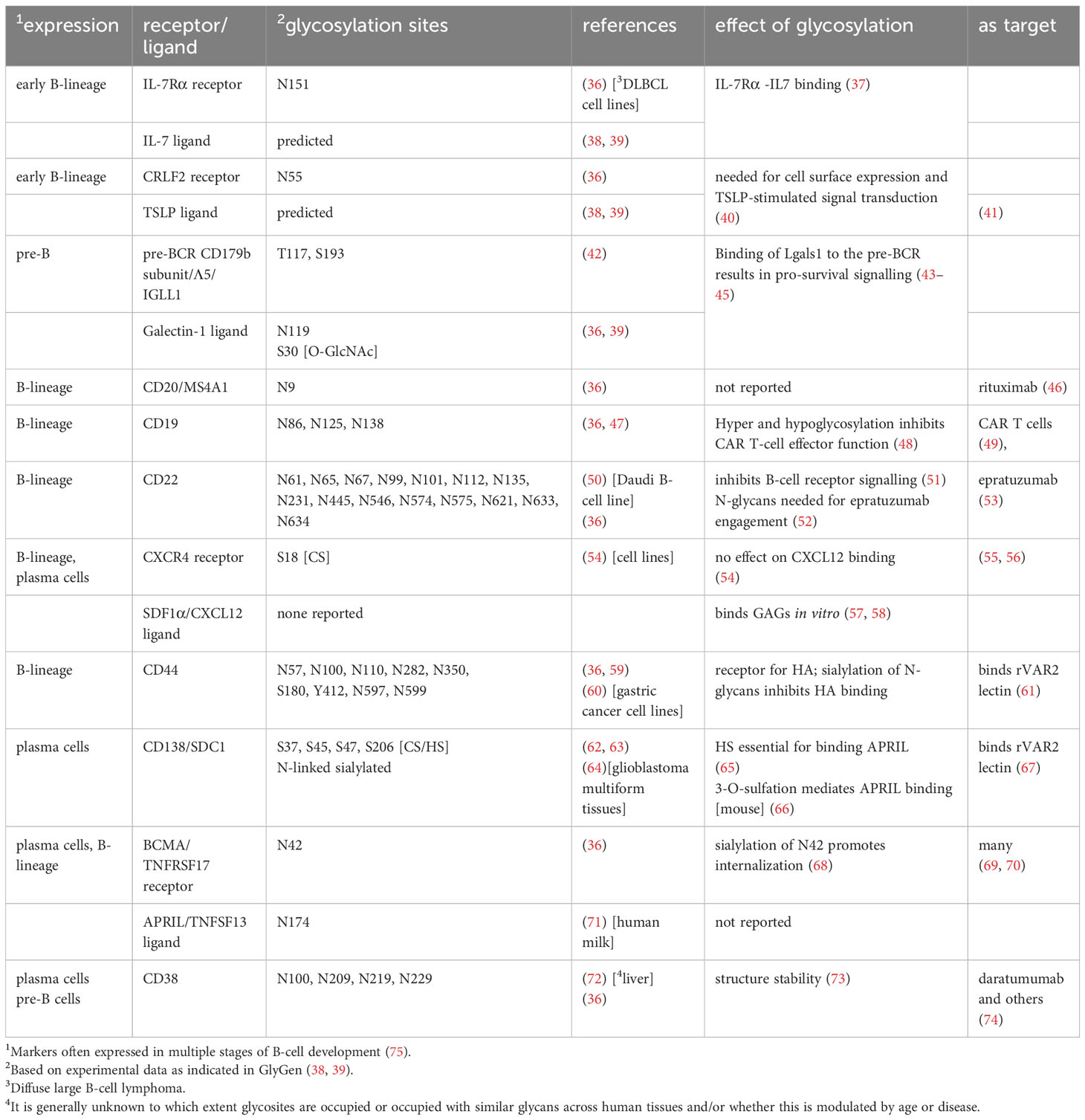
Table 1 Major glycoproteins important for normal and malignant human BM B-cell homeostasis and development.
Not surprisingly, the bone marrow is also the initiating site of precursor B-cell acute lymphoblastic leukemia [BCP-ALL], of which treatment has changed substantially over time (78). BCP-ALL has peaks of incidence in childhood and in older adults. It is the most common childhood malignancy but in recent years has become largely treatable, with survival in children of around 85-92% (79, 80). Older adults still have a much less favorable prognosis (81, 82).
All age groups of BCP-ALL can be divided into different subgroups and risk categories based on transcriptomes and genetic abnormalities (83). These leukemias represent different immature B-lineage precursor cells with a block in normal development and they differ in gene expression profiles to include glycosyltransferases and other glycan-relevant genes such as lectins, which are glycan-binding proteins. For example, the lectin Galectin-1 [Lgals1] is most highly expressed in one specific subgroup of BCP-ALL characterized by KMT2A rearrangements (84, 85), is implicated in other BCP-ALLs (43, 84, 86) and is overexpressed in the ECM of Multiple Myeloma [MM] (87).
Bone marrow is also a long-term repository for normal mature hematopoietic cells such as plasma cells, which confer immunological memory (88). Normal BM plasma cells are quiescent solitary cells that depend on, among others, SDF1α/CXCR4 and APRIL/BCMA (Table 1) signaling for retention in BM and survival (89–91). Based on single-cell RNA sequencing, different clusters of normal plasma cells co-exist in BM (92).
MM cells are the malignant counterparts of such plasma cells. However, the initiating mutations in MM may not have occurred in plasma cells located in the BM, but in those in germinal centers, where plasma cells develop (93, 94). MM is a lymphoid malignancy that is more prevalent in older adults and has an overall survival rate in the USA of 58% (79). MM is also subdivided into genetic subgroups with associated risk categories (95) and different treatments (96). Plasma cells and MM cells no longer express a B-cell receptor but can secrete copious amounts of antibodies [between ~10 and 10,000 Ab molecules per cell per second] (97). Since all immunoglobulins are glycosylated (98), this poses unique metabolic (99) and glycome-related challenges to these cells. Glycosylation has also been linked to specific aspects of MM pathology: osteolytic bone disease in patients correlated with reduced levels of galactose and sialic acid on the Fc part of the immunoglobulin and increased formation of immunoglobulin aggregates (100).
Whereas glycosylation is a wide-spanning modification, the environment of B-cells in the bone marrow is equally complex: BM is an organ consisting of bone as well as a large number of different hematopoietic cell types, stromal cells and extracellular matrix. These together regulate homeostasis and development of B-cells. Therefore, a comprehensive study of B-cell glycosylation would not only involve analysis of the B-cells themselves, but also incorporate glycan-based interactions and instructions from the cells and ECM that constitute the B-cell microenvironment.
Moreover, the hematopoietic compartment is a picture of constant change: development from hematopoietic stem cells into mature lineages such as the myeloid and lymphoid branches is not a stochastic process, but instead is represented by a continuum (101). For the B-lineage, this includes proliferation and resting phases which distinguish themselves by different metabolic states (102). Seeing that glycosylation is also regulated by metabolic flux (103, 104), one can anticipate that the different states will have different glycosylation as well. In addition, hematopoiesis is dynamically regulated by age (105–107) and inflammation (108, 109), as well as the presence of hematological malignancies (89, 110–112). Thus, glycan analysis of BM B-cells unavoidably represents a captured still image of what is essentially a motion picture.
Not surprisingly, analytical studies of human bone marrow in health and disease are less abundant than those of mice, because bone marrow aspiration and trephine biopsies are the only primary materials available for analysis. Generally, there are two approaches for analysis of BM glycans. One involves isolation and characterization of individual component cell types and ECM, which can provide significant detail. Although not glycan-focused, single-cell proteomic or transcriptomic analysis of bone marrow (Table 1) can generate subclassification of stromal and hematopoietic cell clusters in unprecedented detail. This provides some information on the potential or probable glycans expressed by these cells.
Imaging techniques, in contrast, allow visualization at less granularity but can capture potential glycan-based interactions of B-cells, if appropriate tools such as antibodies or lectins are available. Indeed, advances in microscopy techniques using immunostaining have greatly refined our view of the 3-dimensional structure of human bone marrow (111, 113), although this has not yet focused on the glycan component.
A first step towards analyzing contributions of glycans to normal and abnormal B-cell functioning in bone marrow would be an inventory of what is actually there, and where the glycans are located. Direct analysis of N-linked glycans has perhaps been the most widely used. An analysis of this type was performed on MM peripheral blood and bone marrow plasma using MALDI-TOF-MS for N-glycans. This technology employs enzymatic digestion to separate the N-glycans from the protein backbone, followed by mass spectrometry, which can yield the overall composition of the glycan and, through further analysis, glycosidic linkages. The study reported changes in all main glycosylation features in the MM samples compared to normal plasma (114).
Lectins and antibodies with specificity towards glycan and/or glycoconjugate structures have been widely employed to identify glycosylation traits in situ (115, 116). Indirect mass spectrometry imaging [MSI] approaches such as imaging CyTOF allow multiplexing of up to 40 different protein markers within a single experiment (117). Using a panel of scFv antibodies against heparin sulphate, Piszczatowski et al. (118) tracked differentiation from human CD34+ HSPCs into the erythroid and megakaryocyte lineages. Recently, lectins have also been used for cell identification in time-of-flight mass cytometry [CyTOF] to characterize human T-cell subsets (119). However, the vast majority of lectins employed for analytical purposes are of plant origin and unfortunately show a diverse range of affinities for specific glycan structures (120). The complex interaction preferences of the most widely applied lectins have just recently been critically re-evaluated (121), confirming that most lectins exhibit affinities to more than one specific glyco-epitope. While this does not preclude their use to visualize and study glycosylation, an observed change in lectin binding can be caused by more factors than just the simple presence/absence of one specific glyco-epitope. On the other hand, viral lectins and engineered sialyl-O-acetylesterases have been applied with exquisite specificity to allow discrimination of O-acetylation on the 9-, 7- or 4- position of sialic acid as biologically important post-glycosylation modifications (122, 123).
Direct MSI overcomes issues of multiplexing and probe specificity as it does not make use of lectins or antibodies (124). Using this technology, more than 100 biomolecules can be routinely imaged with a resolution down to 1-10 µm within a single experiment (125). Microdissection glycomics/glycoproteomics combines the benefits of microscopy and histopathology with the power of -omics technologies. In principle, cellular structures of interest can be isolated using microdissection and further processed for -omics analyses (126–128). Microdissection of areas of interest in formalin-fixed, paraffin-embedded tissue sections in combination with nano-scale liquid chromatography coupled to Mass Spectrometry [nanoLC] of N- and O-glycans was shown to be robustly possible down to around 1000 cells (129, 130), opening up novel opportunities to obtain detailed glycan structures and quantitative distribution information in a microenvironment context. To date no study has focused on normal or abnormal B-cells in human BM using any of the above-mentioned methods.
Glycosylation is a non-template driven process that is also regulated by factors beyond direct transcriptional control such as metabolomic flux, glycosyltransferase localization, intracellular transport, and complex enzymatic specificities. This poses specific and unique analytical challenges. However, some inferences can be drawn from transcriptome studies, which in depth and number vastly exceed the direct and indirect glycan analysis mentioned above. In particular, there are numerous transcriptomic studies on both stromal and non-stromal bone marrow cells which could be analyzed for glycosyltransferase transcripts and used to generate hypotheses concerning possible glycotraits expressed on these cells.
As noted, a significant limitation of this approach is that transcriptomics data are only partially indicative for the composition of the glycome: expression levels of glycosyltransferases do not necessarily reflect their enzymatic activity and the presence of final products (18). One example of an early study is that of Bret et al. (131) who analyzed Affymetrix-based signatures of 100 heparan sulphate and chondroitin sulphate-relevant genes in normal and abnormal plasma cells. Jöud et al. (132) also used glycosyltransferase gene expression data to predict carbohydrate blood group loci.
An inherent limitation of analysis of bulk RNA expression data is that signals from cell types which constitute only a minor percentage of the total number may be lost. For example, BM stromal stem/progenitor cells constitute only around 0.001–0.01% of the total amount of BM cells (133). Since glycosyltransferase RNAs are not always highly expressed (134), bulk RNA sequencing is likely to therefore miss their expression in cell types with a low representation. This can be mitigated to some extent by purification of specific cell types using antibody markers. More recently, single-cell RNA sequencing [scRNAseq] techniques, often with prior purification or selection of certain cell types, have been applied to generate unprecedented resolution of human bone marrow in normal and diseased states. Table 2 lists some of these studies [for BCP-ALL, also see (75)]. However, with the increased resolution on a cellular level, there is again potentially a loss of ability to detect glycosyltransferase transcripts due to their low expression combined with technical limitations of scRNAseq to detect all transcripts present in a cell (146). Such data, in combination with other data described below, could be used to generate a prediction of glycotraits expressed on cells that constitute only a minute fraction of the total bone marrow compartment including Minimal Residual Disease (MRD), the [frequently low percentage of] leukemia cells that can persist after chemotherapy.
One way to substantiate links between transcriptomes/proteomes and glycotraits is by multi-omics analysis of the same cells. In the only study of this type reported so far, analysis of proteins, RNA and glycans was performed on the same primary BCP-ALL and control samples (18). The study found increased sialylated N-glycans in the leukemia samples. A CRISPR survival screen on a leukemia cell line showed that MGAT1, the enzyme that initiates complex N-linked carbohydrate synthesis, is needed for survival of these cells in a tissue culture setting. Interestingly, compared to normal precursor B-cells, the leukemia cells showed a higher complexity of O-glycans, which could be a reflection of normal B-cell development (147–149). However, expression of mRNA and protein for GALNT7, an enzyme involved in O-glycan synthesis was also higher in the leukemia samples. This enzyme is one of the O−GalNAc glycosylation-initiating enzymes with a substrate preference for follow-up glycosylation of previously glycosylated regions (150–153). Glycopeptide studies with other cell types such as prostate cancer linked higher GALNT7 levels to the presence of the Tn antigen (152), but there is no evidence for Tn expression in BCP-ALL cells.
A landmark paper by Huang et al. (154) provided a critical framework in which RNAseq expression data for glycosyltransferases and the actual glycans expressed in HEK293, an immortalized human embryonic kidney cell line, were linked. The authors identified 38 N-glycan composition structures [complex 13.7%, hybrid 1.7%, oligomannose 84.7%] and 14 O-glycan compositions. Importantly, they validated these correlations by knocking out genes related to N-glycan processing and analysis of the remaining structures. Their GlycoMaple tool is available for analysis of other RNAseq data as well (155). Nielsen et al. (152) used a similar approach to correlate specific O-glycosyltransferases to O-glycan structures. Such studies and those reported by Zhu et al. (156) are likely to ultimately result in a degree of predictability, as reviewed (134) and could be applied to anticipate glycotraits of human BM B-cells based on transcriptome data (Table 2).
Other studies also combined different omics to investigate glycan traits in other cell types and organisms. A systems-based glycobiology approach integrating RNAseq and ChIP-Seq data was used to link transcription factor expression to that of glycogenes in breast cancer (157). Studying Alzheimer’s disease, Tang et al. (158) analyzed publicly available data sets for glycosyltransferase expression, with a confirmation through Q-PCR and subsequently N-glycan analysis of changes predicted by the transcriptome changes. Single-cell RNAseq has been combined with lectin-based glycan cell identification (159) for analysis of human induced pluripotent stem cells (160), which could be applied to human BM B-cells. Thus, a combination of many empirical and computational approaches is expected to lead to an increased ability to predict major glycotraits.
GAGs are a unique type of carbohydrate consisting of very long, pole-like linear carbohydrate chains that are built of repeated disaccharide units of varying repeat length. GAGs can be attached to proteins [proteoglycans] or secreted, like hyaluronic acid (HA). GAG chains are hydrophilic, with a strong negative charge. They also can be modified by the attachment of sulfur groups (sulfation). Heparin sulfate (HS) and chondroitin sulfate (CS) are two main distinct types of GAGs attached to cell surface proteins. GAGs regulate a variety of cellular interactions in the bone marrow because they are expressed by both the bone marrow stroma and by normal hematopoietic cells (161–163). To date, only 77-78 mammalian/human proteoglycan core proteins have been identified (61, 164).
Because core proteins of proteoglycans expressed in human bone marrow can reasonably be expected to have attached GAGs, their mRNA and protein expression can be used to extrapolate the presence of some GAGs. However, this data is limited in that it does not provide information about GAG modification. Main subclasses of proteoglycans include heparin-sulfate and chondroitin sulfate proteoglycans [HSPG and CSPG, respectively] and numerous proteins that interact with them have been identified (165, 166). In a study of the normal human bone marrow proteome, which included hematopoietic cells and mesenchymal stromal/stem cells (MSC), 12,000 proteins were identified. Interestingly, of the age-related changes in older MSC, abundance of proteins tightly linked to glycosaminoglycan metabolism were significantly changed (167).
Significant differences have been found in GAGs and proteoglycans in MM. For example, on a transcriptional level, increased expression of B4Galt7, involved in the synthesis of CS, was found in comparison with normal BM plasma cells (57, 131). Studying 56 trabecular bone tissue samples, Ho et al. found that ECM proteoglycans were higher in control and monoclonal gammopathy of undetermined significance samples, compared to smoldering and active MM (168). HSPG also have a specific significance to normal BM plasma cells and MM cells as reviewed in (161). In particular, HSPGs can bind and present chemokines and survival factors to hematopoietic cells in the BM. HSPGs were shown to bind SDF1α/CXCL12 (169). This is the key chemokine/receptor axis that attracts and anchors different hematopoietic cell types including normal progenitors, plasma cells as well as malignant hematopoietic cells [BCP-ALL, MM] in the bone marrow and mediates MM drug resistance (169).
APRIL [TNFSF13] is the key cytokine for maintaining viability of bone marrow plasma cells (170) and MM cells (171). APRIL is secreted by myeloid cells in bone marrow (172). It is sequestered by the proteoglycan CD138 [also known as Syndecan-1, SDC1] (173, 174) which is expressed on and is the defining hallmark of plasma cells. In this way, APRIL can be presented to its cell surface receptor BCMA [TNFRSF17], which can be located on the same cell or on other MM cells. Human CD138 exhibits a mixed glycosylation profile (62) (Table 1). Interestingly, using heparitinase and chondroitinase, Matthes et al. showed that the sequestration of APRIL by SDC1 is dependent only on the HS (65). Moreover, Baert et al. recently determined, using murine MM cells, that 3-O-sulfation of the HS on SDC1, as detected by a specific anti-HS antibody, mediates APRIL binding and was associated with increased drug resistance. Human diagnosis MM samples also contained cells reacting with this specific antibody (66). APRIL itself contains one N-linked glycosylation site, Asn174 that, in human milk (71), contains different complex sialylated and core fucosylated structures (38, 39) of unknown function.
BCMA is currently one of most well-studied targets for treatment of MM: clinically approved treatments that target BCMA include CAR-T cells, bispecific T-cell engagers and toxin conjugates (69, 175). Interestingly, glycosylation of BCMA (Table 1) is important for its function, although this does not appear to be studied in the context of therapies that target it, or resistance to or relapse on treatment. BCMA contains one N linked glycosylation site, N42, and sialylation of the glycan structure attached to this site promotes internalization of BCMA (68). The importance of sialic acids to cell surface expression of BCMA was recently confirmed in a Cas9/CRISPR screen, which also surprisingly showed SDC1 as important for BCMA cell surface expression (176).
GAGs and proteoglycans also are important to BCP-ALL, with the process of transformation to malignancy as well as chemotherapy modulating their representation. For example, based on transcriptomic data, it is likely that KMT2A-rearranged BCP-ALL cells undergo a shift from CS towards HS proteoglycans compared to normal BCP cells (18). Some proteoglycans and glycan structures have potential diagnostic and therapeutic applications. For example, CD44, the receptor for HA (177) in bone marrow, is widely expressed on BCP-ALL cells (178). It is a marker for MRD leukemia cells in both BCP-ALL (179, 180) and MM (181). Interestingly, CD44 is also functionally important: low molecular weight HA killed BCP-ALL cells and cell lines by a mechanism involving CD44 (182). CD44 is a complex glycoprotein (Table 1) with many isoforms generated through alternative splicing. Up to 9 potential N-linked glycosylation sites are present, and sialylation interferes with HA binding (183). A specific glycoform of CD44 that forms a ligand for L-selectin and is detected by the HECA-452 antibody (184) is expressed on human hematopoietic cells (185). In MM, relapsed samples had a high degree of HECA-452-positivity (186).
In addition, CD44 is a proteoglycan. A lectin called rVAR2 from P. falciparum detects an oncofetal CS structure on many cancer cell types. Candidate proteins expressed on malignant bone marrow B-cells that could bind this lectin include CD44 as well as CSPG4 and SDC1 (164, 187). Indeed, this special structure was detected on NALM-6, a BCP-ALL cell line, and MOLP-2, a MM cell line (164). Other proteoglycans could also present targets for immunotherapy: the proteoglycan CSPG4 is highly and specifically expressed in one of the subtypes of BCP-ALL (188).
Thus, glycosylation is strongly connected to normal and abnormal B-cell survival and development in bone marrow, among others through the regulation of adhesion and of cytokine/chemokine presentations. There are also therapeutic implications: immunotherapy targets are, for the most part, glycoproteins expressed on the cell surface (189). They could also be glycolipids (190) (191–193). Overall, immunotherapies based on antibodies and their derived products such as chimeric antigen receptors (CARs) have made a tremendous impact on treatment of human B-cell malignancies (82, 194–196) but are mainly targeted against CD19, CD22 or CD20 protein epitopes (78). However, there are also therapeutic antibodies specifically directed against glycan epitopes on other targets. It would be outside of the focus of this review to discuss this in-depth, and glycan-focused applications including targets of immunotherapy (164, 197–199) and drug delivery (200), have been recently reviewed, albeit not specifically for BCP-ALL or MM. Antibodies that detect acetylation of sialic acid on the 9-O position in glycoproteins and gangliosides could be the basis for immunological treatment in BCP-ALL (27, 28, 191, 201). Therapeutic applications related to proteoglycans and GAGs were already noted.
A relatively unexplored question is to which extent glycosylation of immunotherapy target proteins on the B-cell malignancies regulates the way the immune system could react with that target other than direct target recognition. For example, if and how glycosylation regulates the tertiary and quaternary structure of immunotherapy targets is mostly unknown. That glycosylation can regulate different aspects of immune responses is illustrated by the mAb camrelizumab, which interferes with binding of PD-L1 to its receptor PD-1 on T-cells. This approach represents an important breakthrough in treatment of solid tumors (202) although PD-1 blockade did not have a beneficial effect in MM (203). The crystal structure of PD-1 alone or in complex with blocking mAbs was also determined [for example (204–206)]. Since glycosylation was regarded as problematic for efficient crystal formation and structure analysis, crystallography of PD-1 and of almost all other cell surface immunotherapy targets has been traditionally performed on proteins expressed in bacteria, which lack eukaryotic glycosylation, or on proteins in which asparagine (Asn) residues, the sites of N-linked glycosylation, have been mutated. In the case of PD-1, this clearly has limited structure-function insights, seeing that glycosylation was found to regulate PD-1 interaction with PD-L1 as well as with some of the different therapeutic mAbs (207–209).
CD38 is an established treatment target for MM (Table 1). Its glycosylation was reported to be important for the assembly of homodimers and tetramers on the plasma membrane (73, 210) suggesting that glycosylation regulates features of the quaternary structure of this protein. Daly et al. (211) in fact reported that de-sialylation of CD38 increases NK-mediated antibody-dependent cytotoxicity of MM cell lines by daratumumab. A more direct line of evidence for the effect of glycosylation on immunotherapy was presented by Heard et al. (48). The authors performed a Cas9/CRISPR screen to identify factors that contribute to CART19 resistance. They reported that knockout of the Golgi protease SPPL3 led to branched N-glycan changes and increased glycosylation of CD19 in NALM6 BCP-ALL cells. Interestingly, these cells were also partly resistant to CART-19 mediated killing. Overexpression of SPPL3 reduced glycosylation of CD19 and its cell surface expression, also leading to CART19 treatment resistance.
The glycocalyx of B-cells and bone marrow stromal cells, as well as glycosylation of the ECM is ubiquitous, likely in constant flux, and diverse in composition. Although analysis of only the glycan component of this layer gives a global overview and inventory of what is present and what has changed, the ultimate goal would be to understand specific effects of specific glycans on the biology of these cells. To accomplish this, it will be necessary to analyze the composition of glycans in the context of glycopeptides or other glycoconjugates. Remarkably, this has become possible at the molecular level using electrospray deposition combined with scanning tunneling microscopy (212). However, sorting out which glycosylated sites on a protein are important from a functional point of view will be challenging, because those individual sites may also have variable site occupancy (152). Although it is unlikely that all glycotraits carry equal biological significance (213), constant features/trends do seem to exist: using a combination of analytical techniques, key glycan structures that regulate important biological functions are beginning to emerge. Glycoproteins such as CD44 and CD138 show how specific glycan components can play an essential functional role. Similar studies on other proteins will ultimately lead to a better understanding of how these highly diverse structures contribute to regulation of normal and abnormal B-cell homeostasis in the bone marrow.
NH: Conceptualization, Funding acquisition, Investigation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was partly supported by an award from the Judy and Bernard Briskin Center Multiple Myeloma Research Program to NH.
The authors of all the excellent reviews and above all primary research papers on this topic whose work could not be cited here are acknowledged. Daniel Kolarich is acknowledged for contributions to the draft of a related review.
The author declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The funding sponsor had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Vicente MM, Leite-Gomes E, Pinho SS. Glycome dynamics in T and B cell development: basic immunological mechanisms and clinical applications. Trends Immunol. (2023) 44(8):585–97. doi: 10.1016/j.it.2023.06.004
2. Engelmann S, Ebeling O, Schwartz-Albiez R. Modulated glycosylation of proteoglycans during differentiation of human B lymphocytes. Biochim. Biophys. Acta (1995) 1267(1):6–14. doi: 10.1016/0167-4889(95)00057-Y
3. Zhang M, Qi T, Yang L, Kolarich D, Heisterkamp N. Multi-faceted effects of ST6Gal1 expression on precursor B-lineage acute lymphoblastic leukemia. Front. Oncol. (2022) 12:828041. doi: 10.3389/fonc.2022.828041
4. Gagneux P, Panin V, Hennet T, Aebi M, Varki A. Evolution of glycan diversity. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al, editors. Essentials of Glycobiology, 4th ed. Cold Spring Harbor, NY (2022). p. 265–78.
5. Korzhenevich J, Janowska I, van der Burg M, Rizzi M. Human and mouse early B cell development: So similar but so different. Immunol. Lett. (2023) 261:1–12. doi: 10.1016/j.imlet.2023.07.004
6. Ichii M, Oritani K, Kanakura Y. Early B lymphocyte development: Similarities and differences in human and mouse. World J. Stem Cells (2014) 6(4):421–31. doi: 10.4252/wjsc.v6.i4.421
7. Nguyen DH, Tangvoranuntakul P, Varki A. Effects of natural human antibodies against a nonhuman sialic acid that metabolically incorporates into activated and Malignant immune cells. J. Immunol. (2005) 175(1):228–36. doi: 10.4049/jimmunol.175.1.228
8. Macauley MS, Kawasaki N, Peng W, Wang SH, He Y, Arlian BM, et al. Unmasking of CD22 co-receptor on germinal center B-cells occurs by alternative mechanisms in mouse and man. J. Biol. Chem. (2015) 290(50):30066–77. doi: 10.1074/jbc.M115.691337
9. Kawanishi K, Dhar C, Do R, Varki N, Gordts P, Varki A. Human species-specific loss of CMP-N-acetylneuraminic acid hydroxylase enhances atherosclerosis via intrinsic and extrinsic mechanisms. Proc. Natl. Acad. Sci. U S A. (2019) 116(32):16036–45. doi: 10.1073/pnas.1902902116
10. Brinkman-Van der Linden EC, Sjoberg ER, Juneja LR, Crocker PR, Varki N, Varki A. Loss of N-glycolylneuraminic acid in human evolution. Implications for sialic acid recognition by siglecs. J. Biol. Chem. (2000) 275(12):8633–40. doi: 10.1074/jbc.275.12.8633
11. Silva JC, Han X, Silva TP, Xia K, Mikael PE, Cabral JMS, et al. Glycosaminoglycan remodeling during chondrogenic differentiation of human bone marrow-/synovial-derived mesenchymal stem/stromal cells under normoxia and hypoxia. Glycoconj J. (2020) 37(3):345–60. doi: 10.1007/s10719-020-09911-5
12. Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG, et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell (2021) 184(12):3109–24 e22. doi: 10.1016/j.cell.2021.04.023
13. Groth T, Diehl AD, Gunawan R, Neelamegham S. GlycoEnzOnto: a GlycoEnzyme pathway and molecular function ontology. Bioinformatics (2022) 38(24):5413–20. doi: 10.1093/bioinformatics/btac704
14. Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. (2012) 13(7):448–62. doi: 10.1038/nrm3383
15. Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. (2020) 21(12):729–49. doi: 10.1038/s41580-020-00294-x
16. Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat. Rev. Nephrol. (2019) 15(6):346–66. doi: 10.1038/s41581-019-0129-4
17. Chin-Hun Kuo J, Gandhi JG, Zia RN, Paszek MJ. Physical biology of the cancer cell glycocalyx. Nat. Phys. (2018) 14(7):658–69. doi: 10.1038/s41567-018-0186-9
18. Oliveira T, Zhang M, Joo EJ, Abdel-Azim H, Chen CW, Yang L, et al. Glycoproteome remodeling in MLL-rearranged B-cell precursor acute lymphoblastic leukemia. Theranostics (2021) 11(19):9519–37. doi: 10.7150/thno.65398
19. Chernykh A, Kawahara R, Thaysen-Andersen M. Towards structure-focused glycoproteomics. Biochem. Soc. Trans. (2021) 49(1):161–86. doi: 10.1042/BST20200222
20. Kawahara R, Chernykh A, Alagesan K, Bern M, Cao W, Chalkley RJ, et al. Community evaluation of glycoproteomics informatics solutions reveals high-performance search strategies for serum glycopeptide analysis. Nat. Methods (2021) 18(11):1304–16. doi: 10.1038/s41592-021-01309-x
21. Karamanos NK, Piperigkou Z, Theocharis AD, Watanabe H, Franchi M, Baud S, et al. Proteoglycan chemical diversity drives multifunctional cell regulation and therapeutics. Chem. Rev. (2018) 118(18):9152–232. doi: 10.1021/acs.chemrev.8b00354
22. Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al. Essentials of Glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press (2015). The Consortium of Glycobiology Editors, La Jolla, California.
23. Mannino MP, Hart GW. The beginner's guide to O-glcNAc: from nutrient sensitive pathway regulation to its impact on the immune system. Front. Immunol. (2022) 13:828648. doi: 10.3389/fimmu.2022.828648
24. Spaner DE. O-glcNAcylation in chronic lymphocytic leukemia and other blood cancers. Front. Immunol. (2021) 12:772304. doi: 10.3389/fimmu.2021.772304
25. Nakajima H, Murakami K. O-GlcNAcylation: Implications in normal and Malignant hematopoiesis. Exp. Hematol. (2021) 101-102:16–24. doi: 10.1016/j.exphem.2021.07.003
26. Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, et al. An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol. Cell. (2019) 75(2):394–407.e5. doi: 10.1016/j.molcel.2019.05.017
27. Cavdarli S, Delannoy P. Groux-degroote S. O-acetylated gangliosides as targets for cancer immunotherapy. Cells (2020) 9(3):741. doi: 10.3390/cells9030741
28. Visser EA, Moons SJ, Timmermans S, de Jong H, Boltje TJ, Bull C. Sialic acid O-acetylation: From biosynthesis to roles in health and disease. J. Biol. Chem. (2021) 297(2):100906. doi: 10.1016/j.jbc.2021.100906
29. O'Byrne S, Elliott N, Rice S, Buck G, Fordham N, Garnett C, et al. Discovery of a CD10-negative B-progenitor in human fetal life identifies unique ontogeny-related developmental programs. Blood (2019) 134(13):1059–71. doi: 10.1182/blood.2019001289
30. Petkau G, Turner M. Signalling circuits that direct early B-cell development. Biochem. J. (2019) 476(5):769–78. doi: 10.1042/BCJ20180565
31. Tangye SG, Nguyen T, Deenick EK, Bryant VL, Ma CS. Inborn errors of human B cell development, differentiation, and function. J. Exp. Med. (2023) 220(7):e20221105. doi: 10.1084/jem.20221105
32. Sigvardsson M. Transcription factor networks link B-lymphocyte development and Malignant transformation in leukemia. Genes Dev. (2023) 37(15-16):703–23. doi: 10.1101/gad.349879.122
33. Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat. Rev. Mol. Cell Biol. (2019) 20(5):303–20. doi: 10.1038/s41580-019-0103-9
34. Lee DH, Kwon NE, Lee WJ, Lee MS, Kim DJ, Kim JH, et al. Increased O-glcNAcylation of c-myc promotes pre-B cell proliferation. Cells (2020) 9(1):158. doi: 10.3390/cells9010158
35. Yan Y, Park SS, Janz S, Eckhardt LA. In a model of immunoglobulin heavy-chain (IGH)/MYC translocation, the Igh 3' regulatory region induces MYC expression at the immature stage of B cell development. Genes Chromosomes Cancer. (2007) 46(10):950–9. doi: 10.1002/gcc.20480
36. Deeb SJ, Cox J, Schmidt-Supprian M, Mann M. N-linked glycosylation enrichment for in-depth cell surface proteomics of diffuse large B-cell lymphoma subtypes. Mol. Cell Proteomics. (2014) 13(1):240–51. doi: 10.1074/mcp.M113.033977
37. McElroy CA, Dohm JA, Walsh ST. Structural and biophysical studies of the human IL-7/IL-7Ralpha complex. Structure (2009) 17(1):54–65. doi: 10.1016/j.str.2008.10.019
38. York WS, Mazumder R, Ranzinger R, Edwards N, Kahsay R, Aoki-Kinoshita KF, et al. GlyGen: computational and informatics resources for glycoscience. Glycobiology (2020) 30(2):72–3. doi: 10.1093/glycob/cwz080
39. York WS, Mazumder R, Ranzinger R, Edwards N, Kahsay R, Aoki-Kinoshita KF, et al. GlyGen: Computational and informatics resources for glycoscience. Glygen, NIH Glycoscience Common Fund (2019). Available at: https://www.glygen.org/.
40. Yamamoto R, Segawa R, Liu J, Isaji T, Gu J, Hiratsuka M, et al. Effect of N-glycosylation on constitutive signal transduction by mutated cytokine receptor-like factor 2. Biochim. Biophys. Acta Gen. Subj. (2023) 1867(11):130465. doi: 10.1016/j.bbagen.2023.130465
41. Alkashgari HR, Ruiz-Jimenez C, Stoian C, Coats JS, Baez I, Chirshev E, et al. TSLP as a potential therapy in the treatment of CRLF2 B cell acute lymphoblastic leukemia. Int. J. Mol. Sci. (2022) 24(1):474. doi: 10.3390/ijms24010474
42. Yang W, Ao M, Hu Y, Li QK, Zhang H. Mapping the O-glycoproteome using site-specific extraction of O-linked glycopeptides (EXoO). Mol. Syst. Biol. (2018) 14(11):e8486. doi: 10.15252/msb.20188486
43. Pelletier J, Balzano M, Destin J, Montersino C, Delahaye MC, Marchand T, et al. Niche-expressed Galectin-1 is involved in pre-B acute lymphoblastic leukemia relapse through pre-B cell receptor activation. iScience (2023) 26(4):106385. doi: 10.1016/j.isci.2023.106385
44. Erasmus MF, Matlawska-Wasowska K, Kinjyo I, Mahajan A, Winter SS, Xu L, et al. Dynamic pre-BCR homodimers fine-tune autonomous survival signals in B cell precursor acute lymphoblastic leukemia. Sci. Signal. (2016) 9(456):ra116. doi: 10.1126/scisignal.aaf3949
45. Gauthier L, Rossi B, Roux F, Termine E, Schiff C. Galectin-1 is a stromal cell ligand of the pre-B cell receptor (BCR) implicated in synapse formation between pre-B and stromal cells and in pre-BCR triggering. Proc. Natl. Acad. Sci. U S A. (2002) 99(20):13014–9. doi: 10.1073/pnas.202323999
46. Hoshitsuki K, Zhou Y, Miller AM, Choi JK, Swanson HD, Bhakta NH, et al. Rituximab administration in pediatric patients with newly diagnosed acute lymphoblastic leukemia. Leukemia (2023) 37(9):1782–91. doi: 10.1038/s41375-023-01992-z
47. Teplyakov A, Obmolova G, Luo J, Gilliland GL. Crystal structure of B-cell co-receptor CD19 in complex with antibody B43 reveals an unexpected fold. Proteins (2018) 86(5):495–500. doi: 10.1002/prot.25485
48. Heard A, Landmann JH, Hansen AR, Papadopolou A, Hsu YS, Selli ME, et al. Antigen glycosylation regulates efficacy of CAR T cells targeting CD19. Nat. Commun. (2022) 13(1):3367. doi: 10.1038/s41467-022-31035-7
49. Chew S, Jammal N, Kantarjian H, Jabbour E. Monoclonal antibodies in frontline acute lymphoblastic leukemia. Best Pract. Res. Clin. Haematol. (2020) 33(4):101226. doi: 10.1016/j.beha.2020.101226
50. Wasim L, Buhari FHM, Yoganathan M, Sicard T, Ereno-Orbea J, Julien JP, et al. N-linked glycosylation regulates CD22 organization and function. Front. Immunol. (2019) 10:699. doi: 10.3389/fimmu.2019.00699
51. Enterina JR, Jung J, Macauley MS. Coordinated roles for glycans in regulating the inhibitory function of CD22 on B cells. BioMed. J. (2019) 42(4):218–32. doi: 10.1016/j.bj.2019.07.010
52. Ereno-Orbea J, Sicard T, Cui H, Mazhab-Jafari MT, Benlekbir S, Guarne A, et al. Molecular basis of human CD22 function and therapeutic targeting. Nat. Commun. (2017) 8(1):764. doi: 10.1038/s41467-017-00836-6
53. Valecha GK, Ibrahim U, Ghanem S, Asti D, Atallah JP, Terjanian T. Emerging role of immunotherapy in precursor B-cell acute lymphoblastic leukemia. Expert Rev. Hematol. (2017) 10(9):783–99. doi: 10.1080/17474086.2017.1350165
54. Farzan M, Babcock GJ, Vasilieva N, Wright PL, Kiprilov E, Mirzabekov T, et al. The role of post-translational modifications of the CXCR4 amino terminus in stromal-derived factor 1 alpha association and HIV-1 entry. J. Biol. Chem. (2002) 277(33):29484–9. doi: 10.1074/jbc.M203361200
55. Frede J, Anand P, Sotudeh N, Pinto RA, Nair MS, Stuart H, et al. Dynamic transcriptional reprogramming leads to immunotherapeutic vulnerabilities in myeloma. Nat. Cell Biol. (2021) 23(11):1199–211. doi: 10.1038/s41556-021-00766-y
56. Jonart LM, Ostergaard J, Brooks A, Fitzpatrick G, Chen L, Gordon PM. CXCR4 antagonists disrupt leukaemia-meningeal cell adhesion and attenuate chemoresistance. Br. J. Haematol. (2023) 201(3):459–69. doi: 10.1111/bjh.18607
57. Yao L, Wang JT, Jayasinghe RG, O'Neal J, Tsai CF, Rettig MP, et al. Single-cell discovery and multiomic characterization of therapeutic targets in multiple myeloma. Cancer Res. (2023) 83(8):1214–33. doi: 10.1158/0008-5472.CAN-22-1769
58. Kunze G, Huster D, Samsonov SA. Investigation of the structure of regulatory proteins interacting with glycosaminoglycans by combining NMR spectroscopy and molecular modeling - the beginning of a wonderful friendship. Biol. Chem. (2021) 402(11):1337–55. doi: 10.1515/hsz-2021-0119
59. Han H, Stapels M, Ying W, Yu Y, Tang L, Jia W, et al. Comprehensive characterization of the N-glycosylation status of CD44s by use of multiple mass spectrometry-based techniques. Anal. Bioanal Chem. (2012) 404(2):373–88. doi: 10.1007/s00216-012-6167-4
60. Campos D, Freitas D, Gomes J, Magalhaes A, Steentoft C, Gomes C, et al. Probing the O-glycoproteome of gastric cancer cell lines for biomarker discovery. Mol. Cell Proteomics. (2015) 14(6):1616–29. doi: 10.1074/mcp.M114.046862
61. Toledo AG, Pihl J, Spliid CB, Persson A, Nilsson J, Pereira MA, et al. An affinity chromatography and glycoproteomics workflow to profile the chondroitin sulfate proteoglycans that interact with malarial VAR2CSA in the placenta and in cancer. Glycobiology (2020) 30(12):989–1002. doi: 10.1093/glycob/cwaa039
62. Noborn F, Nilsson J, Larson G. Site-specific glycosylation of proteoglycans: A revisited frontier in proteoglycan research. Matrix Biol. (2022) 111:289–306. doi: 10.1016/j.matbio.2022.07.002
63. Kokenyesi R, Bernfield M. Core protein structure and sequence determine the site and presence of heparan sulfate and chondroitin sulfate on syndecan-1. J. Biol. Chem. (1994) 269(16):12304–9. doi: 10.1016/S0021-9258(17)32716-3
64. Autelitano F, Loyaux D, Roudieres S, Deon C, Guette F, Fabre P, et al. Identification of novel tumor-associated cell surface sialoglycoproteins in human glioblastoma tumors using quantitative proteomics. PloS One (2014) 9(10):e110316. doi: 10.1371/journal.pone.0110316
65. Matthes T, McKee T, Dunand-Sauthier I, Manfroi B, Park S, Passweg J, et al. Myelopoiesis dysregulation associated to sustained APRIL production in multiple myeloma-infiltrated bone marrow. Leukemia (2015) 29(9):1901–8. doi: 10.1038/leu.2015.68
66. Baert L, Manfroi B, Quintero M, Chavarria O, Barbon PV, Clement E, et al. 3-O sulfation of syndecan-1 mediated by the sulfotransferase HS3ST3a1 enhances myeloma aggressiveness. Matrix Biol. (2023) 120:60–75. doi: 10.1016/j.matbio.2023.05.005
67. Ayres Pereira M, Mandel Clausen T, Pehrson C, Mao Y, Resende M, Daugaard M, et al. Placental sequestration of plasmodium falciparum malaria parasites is mediated by the interaction between VAR2CSA and chondroitin sulfate A on syndecan-1. PloS Pathog. (2016) 12(8):e1005831. doi: 10.1371/journal.ppat.1005831
68. Huang HW, Chen CH, Lin CH, Wong CH, Lin KI. B-cell maturation antigen is modified by a single N-glycan chain that modulates ligand binding and surface retention. Proc. Natl. Acad. Sci. U S A. (2013) 110(27):10928–33. doi: 10.1073/pnas.1309417110
69. Holstein SA, Grant SJ, Wildes TM. Chimeric antigen receptor T-cell and bispecific antibody therapy in multiple myeloma: moving into the future. J. Clin. Oncol. (2023) 41(27):4416–29. doi: 10.1200/JCO.23.00512
70. Lee H, Ahn S, Maity R, Leblay N, Ziccheddu B, Truger M, et al. Mechanisms of antigen escape from BCMA- or GPRC5D-targeted immunotherapies in multiple myeloma. Nat. Med. (2023) 29(9):2295–306. doi: 10.1038/s41591-023-02491-5
71. Zhu J, Lin YH, Dingess KA, Mank M, Stahl B, Heck AJR. Quantitative longitudinal inventory of the N-glycoproteome of human milk from a single donor reveals the highly variable repertoire and dynamic site-specific changes. J. Proteome Res. (2020) 19(5):1941–52. doi: 10.1021/acs.jproteome.9b00753
72. Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, et al. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. (2009) 8(2):651–61. doi: 10.1021/pr8008012
73. Gao Y, Mehta K. N-linked glycosylation of CD38 is required for its structure stabilization but not for membrane localization. Mol. Cell Biochem. (2007) 295(1-2):1–7. doi: 10.1007/s11010-006-9265-9
74. van de Donk N, Usmani SZ, Yong K. CAR T-cell therapy for multiple myeloma: state of the art and prospects. Lancet Haematol. (2021) 8(6):e446–e61. doi: 10.1016/S2352-3026(21)00057-0
75. Morgan D, Tergaonkar V. Unraveling B cell trajectories at single cell resolution. Trends Immunol. (2022) 43(3):210–29. doi: 10.1016/j.it.2022.01.003
76. Kaiser FMP, Janowska I, Menafra R, de Gier M, Korzhenevich J, Pico-Knijnenburg I, et al. IL-7 receptor signaling drives human B-cell progenitor differentiation and expansion. Blood (2023) 142(13):1113–30. doi: 10.1182/blood.2023019721
77. Francis OL, Milford TA, Martinez SR, Baez I, Coats JS, Mayagoitia K, et al. A novel xenograft model to study the role of TSLP-induced CRLF2 signals in normal and Malignant human B lymphopoiesis. Haematologica (2016) 101(4):417–26. doi: 10.3324/haematol.2015.125336
78. Jabbour E, Short NJ, Jain N, Haddad FG, Welch MA, Ravandi F, et al. The evolution of acute lymphoblastic leukemia research and therapy at MD Anderson over four decades. J. Hematol. Oncol. (2023) 16(1):22. doi: 10.1186/s13045-023-01409-5
79. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J. Clin. (2023) 73(1):17–48. doi: 10.3322/caac.21763
80. Raetz EA, Bhojwani D, Devidas M, Gore L, Rabin KR, Tasian SK, et al. Children's Oncology Group blueprint for research: Acute lymphoblastic leukemia. Pediatr. Blood Cancer. (2023) 70 Suppl 6:e30585. doi: 10.1002/pbc.30585
81. Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J. (2017) 7(6):e577. doi: 10.1038/bcj.2017.53
82. Gavralidis A, Brunner AM. Novel therapies in the treatment of adult acute lymphoblastic leukemia. Curr. Hematol. Malig Rep. (2020) 15(4):294–304. doi: 10.1007/s11899-020-00591-4
83. Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. (2019) 51(2):296–307. doi: 10.1038/s41588-018-0315-5
84. Juszczynski P, Rodig SJ, Ouyang J, O'Donnell E, Takeyama K, Mlynarski W, et al. MLL-rearranged B lymphoblastic leukemias selectively express the immunoregulatory carbohydrate-binding protein galectin-1. Clin. Cancer Res. (2010) 16(7):2122–30. doi: 10.1158/1078-0432.CCR-09-2765
85. Paz H, Joo EJ, Chou CH, Fei F, Mayo KH, Abdel-Azim H, et al. Treatment of B-cell precursor acute lymphoblastic leukemia with the Galectin-1 inhibitor PTX008. J. Exp. Clin. Cancer Res. (2018) 37(1):67. doi: 10.1186/s13046-018-0721-7
86. Fei F, Zhang M, Tarighat SS, Joo EJ, Yang L, Heisterkamp N. Galectin-1 and galectin-3 in B-cell precursor acute lymphoblastic leukemia. Int. J. Mol. Sci. (2022) 23(22):14359. doi: 10.3390/ijms232214359
87. Glavey SV, Naba A, Manier S, Clauser K, Tahri S, Park J, et al. Proteomic characterization of human multiple myeloma bone marrow extracellular matrix. Leukemia (2017) 31(11):2426–34. doi: 10.1038/leu.2017.102
88. Slamanig SA, Nolte MA. The bone marrow as sanctuary for plasma cells and memory T-cells: implications for adaptive immunity and vaccinology. Cells (2021) 10(6):1508. doi: 10.3390/cells10061508
89. Miao R, Lim VY, Kothapalli N, Ma Y, Fossati J, Zehentmeier S, et al. Hematopoietic stem cell niches and signals controlling immune cell development and maintenance of immunological memory. Front. Immunol. (2020) 11:600127. doi: 10.3389/fimmu.2020.600127
90. Lightman SM, Utley A, Lee KP. Survival of long-lived plasma cells (LLPC): piecing together the puzzle. Front. Immunol. (2019) 10:965. doi: 10.3389/fimmu.2019.00965
91. Chang HD, Radbruch A. Maintenance of quiescent immune memory in the bone marrow. Eur. J. Immunol. (2021) 51(7):1592–601. doi: 10.1002/eji.202049012
92. Duan M, Nguyen DC, Joyner CJ, Saney CL, Tipton CM, Andrews J, et al. Understanding heterogeneity of human bone marrow plasma cell maturation and survival pathways by single-cell analyses. Cell Rep. (2023) 42(7):112682. doi: 10.1016/j.celrep.2023.112682
93. Rustad EH, Yellapantula V, Leongamornlert D, Bolli N, Ledergor G, Nadeu F, et al. Timing the initiation of multiple myeloma. Nat. Commun. (2020) 11(1):1917. doi: 10.1038/s41467-020-15740-9
94. Barwick BG, Gupta VA, Vertino PM, Boise LH. Cell of origin and genetic alterations in the pathogenesis of multiple myeloma. Front. Immunol. (2019) 10:1121. doi: 10.3389/fimmu.2019.01121
95. Rajkumar SV. Multiple myeloma: 2022 update on diagnosis, risk stratification, and management. Am. J. Hematol. (2022) 97(8):1086–107. doi: 10.1002/ajh.26590
96. Costello C, Davies FE, Cook G, Vela-Ojeda J, Omel J, Rifkin RM, et al. INSIGHT MM: a large, global, prospective, non-interventional, real-world study of patients with multiple myeloma. Future Oncol. (2019) 15(13):1411–28. doi: 10.2217/fon-2019-0013
97. Nguyen DC, Joyner CJ, Sanz I, Lee FE. Factors affecting early antibody secreting cell maturation into long-lived plasma cells. Front. Immunol. (2019) 10:2138. doi: 10.3389/fimmu.2019.02138
98. Zlatina K, Galuska SP. Immunoglobulin glycosylation - an unexploited potential for immunomodulatory strategies in farm animals. Front. Immunol. (2021) 12:753294. doi: 10.3389/fimmu.2021.753294
99. de Matos Simoes R, Shirasaki R, Downey-Kopyscinski SL, Matthews GM, Barwick BG, Gupta VA, et al. Genome-scale functional genomics identify genes preferentially essential for multiple myeloma cells compared to other neoplasias. Nat. Cancer. (2023) 4(5):754–73. doi: 10.1038/s43018-023-00550-x
100. Westhrin M, Kovcic V, Zhang Z, Moen SH, Nedal TMV, Bondt A, et al. Monoclonal immunoglobulins promote bone loss in multiple myeloma. Blood (2020) 136(23):2656–66. doi: 10.1182/blood.2020006045
101. Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. (2017) 19(4):271–81. doi: 10.1038/ncb3493
102. Akkaya M, Pierce SK. From zero to sixty and back to zero again: the metabolic life of B cells. Curr. Opin. Immunol. (2019) 57:1–7. doi: 10.1016/j.coi.2018.09.019
103. Scheper AF, Schofield J, Bohara R, Ritter T, Pandit A. Understanding glycosylation: Regulation through the metabolic flux of precursor pathways. Biotechnol. Adv. (2023) 67:108184. doi: 10.1016/j.biotechadv.2023.108184
104. McDonald AG, Hayes JM, Davey GP. Metabolic flux control in glycosylation. Curr. Opin. Struct. Biol. (2016) 40:97–103. doi: 10.1016/j.sbi.2016.08.007
105. Nikolich-Zugich J. The twilight of immunity: emerging concepts in aging of the immune system. Nat. Immunol. (2018) 19(1):10–9. doi: 10.1038/s41590-017-0006-x
106. Pang WW, Price EA, Sahoo D, Beerman I, Maloney WJ, Rossi DJ, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. U S A. (2011) 108(50):20012–7. doi: 10.1073/pnas.1116110108
107. Strzelecka PM, Damm F. Haematopoietic ageing through the lens of single-cell technologies. Dis. Model. Mech. (2021) 14(1):dmm047340. doi: 10.1242/dmm.047340
108. Xie X, Doody GM, Shuweihdi F, Conaghan PG, Ponchel F. B-cell capacity for expansion and differentiation into plasma cells are altered in osteoarthritis. Osteoarthritis Cartilage. (2023) 31(9):1176–88. doi: 10.1016/j.joca.2023.03.017
109. Takizawa H, Manz MG. Impact of inflammation on early hematopoiesis and the microenvironment. Int. J. Hematol. (2017) 106(1):27–33. doi: 10.1007/s12185-017-2266-5
110. Najman A, Kobari L, Khoury E, Baillou CL, Lemoine F, Guigon M. Suppression of normal hematopoiesis during acute leukemias. Ann. N Y Acad. Sci. (1991) 628:140–7. doi: 10.1111/j.1749-6632.1991.tb17231.x
111. Braunig S, Karmhag I, Li H, Enoksson J, Hultquist A, Scheding S. Three-dimensional spatial mapping of the human hematopoietic microenvironment in healthy and diseased bone marrow. Cytometry A. (2023) 103(100):763–76. doi: 10.1002/cyto.a.24775
112. Batsivari A, Grey W, Bonnet D. Understanding of the crosstalk between normal residual hematopoietic stem cells and the leukemic niche in acute myeloid leukemia. Exp. Hematol. (2021) 95:23–30. doi: 10.1016/j.exphem.2021.01.004
113. Takaku T, Malide D, Chen J, Calado RT, Kajigaya S, Young NS. Hematopoiesis in 3 dimensions: human and murine bone marrow architecture visualized by confocal microscopy. Blood (2010) 116(15):e41–55. doi: 10.1182/blood-2010-02-268466
114. Zhang Z, Westhrin M, Bondt A, Wuhrer M, Standal T, Holst S. Serum protein N-glycosylation changes in multiple myeloma. Biochim. Biophys. Acta Gen. Subj. (2019) 1863(5):960–70. doi: 10.1016/j.bbagen.2019.03.001
115. Walker RA. The use of lectins in histopathology. Pathol. Res. Pract. (1989) 185(6):826–35. doi: 10.1016/S0344-0338(89)80282-1
116. Cummings RD, Darvill AG, Etzler ME, Hahn MG. Glycan-recognizing probes as tools. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, et al, editors. Essentials of Glycobiology. Cold Spring Harbor, NY (2015). p. 611–25.
117. Elaldi R, Hemon P, Petti L, Cosson E, Desrues B, Sudaka A, et al. High dimensional imaging mass cytometry panel to visualize the tumor immune microenvironment contexture. Front. Immunol. (2021) 12:666233. doi: 10.3389/fimmu.2021.666233
118. Piszczatowski RT, Schwenger E, Sundaravel S, Stein CM, Liu Y, Stanley P, et al. A glycan-based approach to cell characterization and isolation: Hematopoiesis as a paradigm. J. Exp. Med. (2022) 219(11):e20212552. doi: 10.1084/jem.20212552
119. Ma T, McGregor M, Giron L, Xie G, George AF, Abdel-Mohsen M, et al. Single-cell glycomics analysis by CyTOF-Lec reveals glycan features defining cells differentially susceptible to HIV. Elife (2022) 11:e78870. doi: 10.7554/eLife.78870.sa2
120. Gao C, Hanes MS, Byrd-Leotis LA, Wei M, Jia N, Kardish RJ, et al. Unique binding specificities of proteins toward isomeric asparagine-linked glycans. Cell Chem. Biol. (2019) 26(4):535–47 e4. doi: 10.1016/j.chembiol.2019.01.002
121. Bojar D, Meche L, Meng G, Eng W, Smith DF, Cummings RD, et al. A useful guide to lectin binding: machine-learning directed annotation of 57 unique lectin specificities. ACS Chem. Biol. (2022) 17(11):2993–3012. doi: 10.1101/2021.08.31.458439
122. Albers M, Schroter L, Belousov S, Hartmann M, Grove M, Abeln M, et al. The sialyl-O-acetylesterase NanS of Tannerella forsythia encompasses two catalytic modules with different regiospecificity for O7 and O9 of sialic acid. Glycobiology (2021) 31(9):1176–91. doi: 10.1093/glycob/cwab034
123. Langereis MA, Bakkers MJ, Deng L, Padler-Karavani V, Vervoort SJ, Hulswit RJ, et al. Complexity and diversity of the mammalian sialome revealed by nidovirus virolectins. Cell Rep. (2015) 11(12):1966–78. doi: 10.1016/j.celrep.2015.05.044
124. Drake RR, Powers TW, Jones EE, Bruner E, Mehta AS, Angel PM. MALDI mass spectrometry imaging of N-linked glycans in cancer tissues. Adv. Cancer Res. (2017) 134:85–116. doi: 10.1016/bs.acr.2016.11.009
125. Unsihuay D, Mesa Sanchez D, Laskin J. Quantitative mass spectrometry imaging of biological systems. Annu. Rev. Phys. Chem. (2021) 72:307–29. doi: 10.1146/annurev-physchem-061020-053416
126. Ezzoukhry Z, Henriet E, Cordelieres FP, Dupuy JW, Maitre M, Gay N, et al. Combining laser capture microdissection and proteomics reveals an active translation machinery controlling invadosome formation. Nat. Commun. (2018) 9(1):2031. doi: 10.1038/s41467-018-04461-9
127. Herrera JA, Mallikarjun V, Rosini S, Montero MA, Lawless C, Warwood S, et al. Laser capture microdissection coupled mass spectrometry (LCM-MS) for spatially resolved analysis of formalin-fixed and stained human lung tissues. Clin. Proteomics. (2020) 17:24. doi: 10.1186/s12014-020-09287-6
128. Zhu Y, Dou M, Piehowski PD, Liang Y, Wang F, Chu RK, et al. Spatially resolved proteome mapping of laser capture microdissected tissue with automated sample transfer to nanodroplets. Mol. Cell Proteomics. (2018) 17(9):1864–74. doi: 10.1074/mcp.TIR118.000686
129. Hinneburg H, Korac P, Schirmeister F, Gasparov S, Seeberger PH, Zoldos V, et al. Unlocking cancer glycomes from histopathological formalin-fixed and paraffin-embedded (FFPE) tissue microdissections. Mol. Cell Proteomics. (2017) 16(4):524–36. doi: 10.1074/mcp.M116.062414
130. Hinneburg H, Schirmeister F, Korać P, Kolarich D. N- and O-glycomics from minor amounts of formalin-fixed, paraffin-embedded tissue samples. In: Lauc G, Wuhrer M, editors. High-Throughput Glycomics and Glycoproteomics: Methods and Protocols. New York, NY: Springer New York (2017). p. 131–45.
131. Bret C, Hose D, Reme T, Sprynski AC, Mahtouk K, Schved JF, et al. Expression of genes encoding for proteins involved in heparan sulphate and chondroitin sulphate chain synthesis and modification in normal and Malignant plasma cells. Br. J. Haematol. (2009) 145(3):350–68. doi: 10.1111/j.1365-2141.2009.07633.x
132. Joud M, Moller M, Olsson ML. Identification of human glycosyltransferase genes expressed in erythroid cells predicts potential carbohydrate blood group loci. Sci. Rep. (2018) 8(1):6040. doi: 10.1038/s41598-018-24445-5
133. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science (1999) 284(5411):143–7. doi: 10.1126/science.284.5411.143
134. Stewart N, Wisnovsky S. Bridging glycomics and genomics: new uses of functional genetics in the study of cellular glycosylation. Front. Mol. Biosci. (2022) 9:934584. doi: 10.3389/fmolb.2022.934584
135. Li H, Braunig S, Dhapolar P, Karlsson G, Lang S, Scheding S. Identification of phenotypically, functionally, and anatomically distinct stromal niche populations in human bone marrow based on single-cell RNA sequencing. Elife (2023) 12:e81656. doi: 10.7554/eLife.81656
136. Hay SB, Ferchen K, ChEtal K, Grimes HL, Salomonis N. The Human Cell Atlas bone marrow single-cell interactive web portal. Exp. Hematol. (2018) 68:51–61. doi: 10.1016/j.exphem.2018.09.004
137. Lee NYS, Li M, Ang KS, Chen J. Establishing a human bone marrow single cell reference atlas to study ageing and diseases. Front. Immunol. (2023) 14:1127879. doi: 10.3389/fimmu.2023.1127879
138. Oetjen KA, Lindblad KE, Goswami M, Gui G, Dagur PK, Lai C, et al. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight (2018) 3(23):e124928. doi: 10.1172/jci.insight.124928
139. Triana S, Vonficht D, Jopp-Saile L, Raffel S, Lutz R, Leonce D, et al. Single-cell proteo-genomic reference maps of the hematopoietic system enable the purification and massive profiling of precisely defined cell states. Nat. Immunol. (2021) 22(12):1577–89. doi: 10.1038/s41590-021-01059-0
140. Qin P, Pang Y, Hou W, Fu R, Zhang Y, Wang X, et al. Integrated decoding hematopoiesis and leukemogenesis using single-cell sequencing and its medical implication. Cell Discovery (2021) 7(1):2. doi: 10.1038/s41421-020-00223-4
141. Jung SH, Park SS, Lim JY, Sohn SY, Kim NY, Kim D, et al. Single-cell analysis of multiple myelomas refines the molecular features of bortezomib treatment responsiveness. Exp. Mol. Med. (2022) 54(11):1967–78. doi: 10.1038/s12276-022-00884-z
142. Ledergor G, Weiner A, Zada M, Wang SY, Cohen YC, Gatt ME, et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat. Med. (2018) 24(12):1867–76. doi: 10.1038/s41591-018-0269-2
143. Iacobucci I, Witkowski MT, Mullighan CG. Single-cell analysis of acute lymphoblastic and lineage-ambiguous leukemia: approaches and molecular insights. Blood (2023) 141(4):356–68. doi: 10.1182/blood.2022016954
144. Caron M, St-Onge P, Sontag T, Wang YC, Richer C, Ragoussis I, et al. Single-cell analysis of childhood leukemia reveals a link between developmental states and ribosomal protein expression as a source of intra-individual heterogeneity. Sci. Rep. (2020) 10(1):8079. doi: 10.1038/s41598-020-64929-x
145. Shahi P, Kim SC, Haliburton JR, Gartner ZJ, Abate AR. Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Sci. Rep. (2017) 7:44447. doi: 10.1038/srep44447
146. Guruprasad P, Lee YG, Kim KH, Ruella M. The current landscape of single-cell transcriptomics for cancer immunotherapy. J. Exp. Med. (2021) 218(1):e20201574. doi: 10.1084/jem.20201574
147. Chakraborty A, Staudinger C, King SL, Erickson FC, Lau LS, Bernasconi A, et al. Galectin-9 bridges human B cells to vascular endothelium while programming regulatory pathways. J. Autoimmun. (2021) 117:102575. doi: 10.1016/j.jaut.2020.102575
148. Giovannone N, Antonopoulos A, Liang J, Geddes Sweeney J, Kudelka MR, King SL, et al. Human B cell differentiation is characterized by progressive remodeling of O-linked glycans. Front. Immunol. (2018) 9:2857. doi: 10.3389/fimmu.2018.02857
149. Giovannone N, Liang J, Antonopoulos A, Geddes Sweeney J, King SL, Pochebit SM, et al. Galectin-9 suppresses B cell receptor signaling and is regulated by I-branching of N-glycans. Nat. Commun. (2018) 9(1):3287. doi: 10.1038/s41467-018-05770-9
150. Bennett EP, Hassan H, Hollingsworth MA, Clausen H. A novel human UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc-glycosylated acceptor substrates. FEBS Lett. (1999) 460(2):226–30. doi: 10.1016/S0014-5793(99)01268-5
151. Kong Y, Joshi HJ, Schjoldager KT, Madsen TD, Gerken TA, Vester-Christensen MB, et al. Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology (2015) 25(1):55–65. doi: 10.1093/glycob/cwu089
152. Nielsen MI, de Haan N, Kightlinger W, Ye Z, Dabelsteen S, Li M, et al. Global mapping of GalNAc-T isoform-specificities and O-glycosylation site-occupancy in a tissue-forming human cell line. Nat. Commun. (2022) 13(1):6257. doi: 10.1038/s41467-022-33806-8
153. Scott E, Hodgson K, Calle B, Turner H, Cheung K, Bermudez A, et al. Upregulation of GALNT7 in prostate cancer modifies O-glycosylation and promotes tumour growth. Oncogene (2023) 42(12):926–37. doi: 10.1038/s41388-023-02604-x
154. Huang YF, Aoki K, Akase S, Ishihara M, Liu YS, Yang G, et al. Global mapping of glycosylation pathways in human-derived cells. Dev. Cell. (2021) 56(8):1195–209 e7. doi: 10.1016/j.devcel.2021.02.023
155. Glycomaple. Available at: https://glycosmos.org/glycomaple/index.
156. Zhu Y, Groth T, Kelkar A, Zhou Y, Neelamegham S. A GlycoGene CRISPR-Cas9 lentiviral library to study lectin binding and human glycan biosynthesis pathways. Glycobiology (2021) 31(3):173–80. doi: 10.1093/glycob/cwaa074
157. Groth T, Gunawan R, Neelamegham S. A systems-based framework to computationally describe putative transcription factors and signaling pathways regulating glycan biosynthesis. Beilstein J. Org Chem. (2021) 17:1712–24. doi: 10.3762/bjoc.17.119
158. Tang X, Tena J, Di Lucente J, Maezawa I, Harvey DJ, Jin LW, et al. Transcriptomic and glycomic analyses highlight pathway-specific glycosylation alterations unique to Alzheimer's disease. Sci. Rep. (2023) 13(1):7816. doi: 10.1038/s41598-023-34787-4
159. Kearney CJ, Vervoort SJ, Ramsbottom KM, Todorovski I, Lelliott EJ, Zethoven M, et al. SUGAR-seq enables simultaneous detection of glycans, epitopes, and the transcriptome in single cells. Sci. Adv. (2021) 7(8):eabe3610. doi: 10.1126/sciadv.abe3610
160. Minoshima F, Ozaki H, Odaka H, Tateno H. Integrated analysis of glycan and RNA in single cells. iScience (2021) 24(8):102882. doi: 10.1016/j.isci.2021.102882
161. Ren Z, Spaargaren M, Pals ST. Syndecan-1 and stromal heparan sulfate proteoglycans: key moderators of plasma cell biology and myeloma pathogenesis. Blood (2021) 137(13):1713–8. doi: 10.1182/blood.2020008188
162. Papy-Garcia D, Albanese P. Heparan sulfate proteoglycans as key regulators of the mesenchymal niche of hematopoietic stem cells. Glycoconj J. (2017) 34(3):377–91. doi: 10.1007/s10719-017-9773-8
163. Morris AJ, Turnbull JE, Riley GP, Gordon MY, Gallagher JT. Production of heparan sulphate proteoglycans by human bone marrow stromal cells. J. Cell Sci. (1991) 99(Pt 1):149–56. doi: 10.1242/jcs.99.1.149
164. Salanti A, Clausen TM, Agerbaek MO, Al Nakouzi N, Dahlback M, Oo HZ, et al. Targeting human cancer by a glycosaminoglycan binding malaria protein. Cancer Cell. (2015) 28(4):500–14. doi: 10.1016/j.ccell.2015.09.003
165. Gomez Toledo A, Sorrentino JT, Sandoval DR, Malmstrom J, Lewis NE, Esko JD. A systems view of the heparan sulfate interactome. J. Histochem Cytochem. (2021) 69(2):105–19. doi: 10.1369/0022155420988661
166. Ding Xu JHP, Linhardt RJ, Esko. JD. Chapter 38 proteins that bind sulfated glycosaminoglycans. In: Essentials of glycobiology, 4th edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press (2022). Available at: https://www.ncbi.nlm.nih.gov/books/NBK579914/.
167. Hennrich ML, Romanov N, Horn P, Jaeger S, Eckstein V, Steeples V, et al. Cell-specific proteome analyses of human bone marrow reveal molecular features of age-dependent functional decline. Nat. Commun. (2018) 9(1):4004. doi: 10.1038/s41467-018-06353-4
168. Ho M, Dasari S, Visram A, Drake MT, Charlesworth MC, Johnson KL, et al. An atlas of the bone marrow bone proteome in patients with dysproteinemias. Blood Cancer J. (2023) 13(1):63. doi: 10.1038/s41408-023-00840-8
169. Ren Z, Lantermans H, Kuil A, Kraan W, Arenzana-Seisdedos F, Kersten MJ, et al. The CXCL12gamma chemokine immobilized by heparan sulfate on stromal niche cells controls adhesion and mediates drug resistance in multiple myeloma. J. Hematol. Oncol. (2021) 14(1):11. doi: 10.1186/s13045-021-01031-3
170. Nguyen DC, Garimalla S, Xiao H, Kyu S, Albizua I, Galipeau J, et al. Factors of the bone marrow microniche that support human plasma cell survival and immunoglobulin secretion. Nat. Commun. (2018) 9(1):3698. doi: 10.1038/s41467-018-05853-7
171. Quinn J, Glassford J, Percy L, Munson P, Marafioti T, Rodriguez-Justo M, et al. APRIL promotes cell-cycle progression in primary multiple myeloma cells: influence of D-type cyclin group and translocation status. Blood (2011) 117(3):890–901. doi: 10.1182/blood-2010-01-264424
172. Matthes T, Dunand-Sauthier I, Santiago-Raber ML, Krause KH, Donze O, Passweg J, et al. Production of the plasma-cell survival factor a proliferation-inducing ligand (APRIL) peaks in myeloid precursor cells from human bone marrow. Blood (2011) 118(7):1838–44. doi: 10.1182/blood-2011-01-332940
173. Moreaux J, Sprynski AC, Dillon SR, Mahtouk K, Jourdan M, Ythier A, et al. APRIL and TACI interact with syndecan-1 on the surface of multiple myeloma cells to form an essential survival loop. Eur. J. Haematol. (2009) 83(2):119–29. doi: 10.1111/j.1600-0609.2009.01262.x
174. Hendriks J, Planelles L, de Jong-Odding J, Hardenberg G, Pals ST, Hahne M, et al. Heparan sulfate proteoglycan binding promotes APRIL-induced tumor cell proliferation. Cell Death Differ. (2005) 12(6):637–48. doi: 10.1038/sj.cdd.4401647
175. More S, Offidani M, Corvatta L, Petrucci MT, Fazio F. Belantamab mafodotin: from clinical trials data to real-life experiences. Cancers (Basel). (2023) 15(11):2948. doi: 10.3390/cancers15112948
176. Ramkumar P, Abarientos AB, Tian R, Seyler M, Leong JT, Chen M, et al. CRISPR-based screens uncover determinants of immunotherapy response in multiple myeloma. Blood Adv. (2020) 4(13):2899–911. doi: 10.1182/bloodadvances.2019001346
177. Sutherland TE, Dyer DP, Allen JE. The extracellular matrix and the immune system: A mutually dependent relationship. Science (2023) 379(6633):eabp8964. doi: 10.1126/science.abp8964
178. Horst E, Meijer CJ, Radaskiewicz T, van Dongen JJ, Pieters R, Figdor CG, et al. Expression of a human homing receptor (CD44) in lymphoid Malignancies and related stages of lymphoid development. Leukemia (1990) 4(5):383–9.
179. Coustan-Smith E, Song G, Clark C, Key L, Liu P, Mehrpooya M, et al. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood (2011) 117(23):6267–76. doi: 10.1182/blood-2010-12-324004
180. Tembhare PR, Ghogale S, Ghatwai N, Badrinath Y, Kunder N, Patkar NV, et al. Evaluation of new markers for minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia: CD73 and CD86 are the most relevant new markers to increase the efficacy of MRD 2016; 00B: 000-000. Cytometry B Clin. Cytom. (2018) 94(1):100–11. doi: 10.1002/cyto.b.21486
181. Paiva B, Corchete LA, Vidriales MB, Puig N, Maiso P, Rodriguez I, et al. Phenotypic and genomic analysis of multiple myeloma minimal residual disease tumor cells: a new model to understand chemoresistance. Blood (2016) 127(15):1896–906. doi: 10.1182/blood-2015-08-665679
182. Kasai S, Furuichi Y, Ando N, Kagami K, Abe M, Nakane T, et al. Inflammatory mediator ultra-low-molecular-weight hyaluronan triggers necrosis of B-precursor leukemia cells with high surface CD44 expression. Cell Death Dis. (2017) 8(6):e2857. doi: 10.1038/cddis.2017.249
183. Faller CE, Guvench O. Terminal sialic acids on CD44 N-glycans can block hyaluronan binding by forming competing intramolecular contacts with arginine sidechains. Proteins (2014) 82(11):3079–89. doi: 10.1002/prot.24668
184. Duijvestijn AM, Horst E, Pals ST, Rouse BN, Steere AC, Picker LJ, et al. High endothelial differentiation in human lymphoid and inflammatory tissues defined by monoclonal antibody HECA-452. Am. J. Pathol. (1988) 130(1):147–55.
185. Dimitroff CJ, Lee JY, Fuhlbrigge RC, Sackstein R. A distinct glycoform of CD44 is an L-selectin ligand on human hematopoietic cells. Proc. Natl. Acad. Sci. U S A. (2000) 97(25):13841–6. doi: 10.1073/pnas.250484797
186. Natoni A, Smith TAG, Keane N, McEllistrim C, Connolly C, Jha A, et al. E-selectin ligands recognised by HECA452 induce drug resistance in myeloma, which is overcome by the E-selectin antagonist, GMI-1271. Leukemia (2017) 31(12):2642–51. doi: 10.1038/leu.2017.123
187. Spliid CB, Toledo AG, Sanderson P, Mao Y, Gatto F, Gustavsson T, et al. The specificity of the malarial VAR2CSA protein for chondroitin sulfate depends on 4-O-sulfation and ligand accessibility. J. Biol. Chem. (2021) 297(6):101391. doi: 10.1016/j.jbc.2021.101391
188. Harrer DC, Schuler G, Dorrie J, Schaft N. CSPG4-specific CAR T cells for high-risk childhood B cell precursor leukemia. Int. J. Mol. Sci. (2019) 20(11):2764. doi: 10.3390/ijms20112764
189. Xiao H, Wu R. Quantitative investigation of human cell surface N-glycoprotein dynamics. Chem. Sci. (2017) 8(1):268–77. doi: 10.1039/C6SC01814A
190. Soliman C, Chua JX, Vankemmelbeke M, McIntosh RS, Guy AJ, Spendlove I, et al. The terminal sialic acid of stage-specific embryonic antigen-4 has a crucial role in binding to a cancer-targeting antibody. J. Biol. Chem. (2020) 295(4):1009–20. doi: 10.1016/S0021-9258(17)49911-X
191. Parameswaran R, Lim M, Arutyunyan A, Abdel-Azim H, Hurtz C, Lau K, et al. O-acetylated N-acetylneuraminic acid as a novel target for therapy in human pre-B acute lymphoblastic leukemia. J. Exp. Med. (2013) 210(4):805–19. doi: 10.1084/jem.20121482
192. Pick J, Anh-Tuan N, Mod A, Hollan S. Ganglioside composition in common acute lymphoblastic leukaemia. Haematologia (Budap). (1986) 19(1):33–9.
193. Merritt WD, Sztein MB, Taylor B, Reaman GH. Immunoreactivity of leukemic lymphoblasts of T-cell and B-cell precursor origin with monoclonal anti-GD3 and anti-GM3 antibodies. Leukemia (1991) 5(12):1087–91.
194. Visweshwar N, Rico JF, Killeen R, Manoharan A. Harnessing the immune system: an effective way to manage diffuse large B-cell lymphoma. J. Hematol. (2023) 12(4):145–60. doi: 10.14740/jh1112
195. Tapia-Galisteo A, Alvarez-Vallina L, Sanz L. Bi- and trispecific immune cell engagers for immunotherapy of hematological Malignancies. J. Hematol. Oncol. (2023) 16(1):83. doi: 10.1186/s13045-023-01482-w
196. Glasser CL, Chen J. Harnessing the immune system: current and emerging immunotherapy strategies for pediatric acute lymphoblastic leukemia. Biomedicines (2023) 11(7):1886. doi: 10.3390/biomedicines11071886
197. Buettner MJ, Shah SR, Saeui CT, Ariss R, Yarema KJ. Improving immunotherapy through glycodesign. Front. Immunol. (2018) 9:2485. doi: 10.3389/fimmu.2018.02485
198. RodrIguez E, Schetters STT, van Kooyk Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. (2018) 18(3):204–11. doi: 10.1038/nri.2018.3
199. Steentoft C, Migliorini D, King TR, Mandel U, June CH, Posey AD Jr. Glycan-directed CAR-T cells. Glycobiology (2018) 28(9):656–69. doi: 10.1093/glycob/cwy008
200. Diniz F, Coelho P, Duarte HO, Sarmento B, Reis CA, Gomes J. Glycans as targets for drug delivery in cancer. Cancers (Basel). (2022) 14(4):911. doi: 10.3390/cancers14040911
201. Pal S, Ghosh S, Bandyopadhyay S, Mandal C, Bandhyopadhyay S, Kumar Bhattacharya D, et al. Differential expression of 9-O-acetylated sialoglycoconjugates on leukemic blasts: a potential tool for long-term monitoring of children with acute lymphoblastic leukemia. Int. J. Cancer. (2004) 111(2):270–7. doi: 10.1002/ijc.20246
202. Kraehenbuehl L, Weng CH, Eghbali S, Wolchok JD, Merghoub T. Enhancing immunotherapy in cancer by targeting emerging immunomodulatory pathways. Nat. Rev. Clin. Oncol. (2022) 19(1):37–50. doi: 10.1038/s41571-021-00552-7
203. Salik B, Smyth MJ, Nakamura K. Targeting immune checkpoints in hematological Malignancies. J. Hematol. Oncol. (2020) 13(1):111. doi: 10.1186/s13045-020-00947-6
204. Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci. Rep. (2016) 6:35297.
205. Lee SH, Lee HT, Lim H, Kim Y, Park UB, Heo YS. Crystal structure of PD-1 in complex with an antibody-drug tislelizumab used in tumor immune checkpoint therapy. Biochem. Biophys. Res. Commun. (2020) 527(1):226–31. doi: 10.1016/j.bbrc.2020.04.121
206. Zhang F, Qi X, Wang X, Wei D, Wu J, Feng L, et al. Structural basis of the therapeutic anti-PD-L1 antibody atezolizumab. Oncotarget (2017) 8(52):90215–24. doi: 10.18632/oncotarget.21652
207. Liu K, Tan S, Jin W, Guan J, Wang Q, Sun H, et al. N-glycosylation of PD-1 promotes binding of camrelizumab. EMBO Rep. (2020) 21(12):e51444. doi: 10.15252/embr.202051444
208. Lu D, Xu Z, Zhang D, Jiang M, Liu K, He J, et al. PD-1 N58-glycosylation-dependent binding of monoclonal antibody cemiplimab for immune checkpoint therapy. Front. Immunol. (2022) 13:826045.
209. Wang M, Wang J, Wang R, Jiao S, Wang S, Zhang J, et al. Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation. Commun. Biol. (2019) 2:392. doi: 10.1038/s42003-019-0642-9
210. Hara-Yokoyama M, Kukimoto-Niino M, Terasawa K, Harumiya S, Podyma-Inoue KA, Hino N, et al. Tetrameric interaction of the ectoenzyme CD38 on the cell surface enables its catalytic and raft-association activities. Structure (2012) 20(9):1585–95. doi: 10.1016/j.str.2012.06.017
211. Daly J, Sarkar S, Natoni A, Stark JC, Riley NM, Bertozzi CR, et al. Targeting hypersialylation in multiple myeloma represents a novel approach to enhance NK cell-mediated tumor responses. Blood Adv. (2022) 6(11):3352–66. doi: 10.1182/bloodadvances.2021006805
212. Anggara K, Srsan L, Jaroentomeechai T, Wu X, Rauschenbach S, Narimatsu Y, et al. Direct observation of glycans bonded to proteins and lipids at the single-molecule level. Science (2023) 382(6667):219–23. doi: 10.1126/science.adh3856
Keywords: B-cell precursor acute lymphoblastic leukemia, multiple myeloma, bone marrow, scRNAseq, glycans, glycosaminoglycans, proteoglycans
Citation: Heisterkamp N (2023) Glycosylation as regulator of human B-cell leukaemias in bone marrow. Front. Hematol. 2:1279863. doi: 10.3389/frhem.2023.1279863
Received: 18 August 2023; Accepted: 31 October 2023;
Published: 16 November 2023.
Edited by:
Valerie R. Wiersma, University Medical Center Groningen, NetherlandsReviewed by:
Karen-Sue Barker Carlson, Medical College of Wisconsin, United StatesCopyright © 2023 Heisterkamp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nora Heisterkamp, ZWhlaXN0ZXJrYW1wQGNvaC5vcmc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.