
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genome Ed., 04 April 2025
Sec. Genome Editing in Human Health and Disease
Volume 7 - 2025 | https://doi.org/10.3389/fgeed.2025.1558432
This article is part of the Research TopicApplication and Advantages of Genome Editing Technology in Anti-AgingView all articles
CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-associated protein 9) has emerged as a transformative genome-editing tool with significant therapeutic potential for age-related diseases, including Alzheimer’s disease, Parkinson’s disease, cardiovascular disorders, and osteoporosis. This study presents a bibliometric analysis of CRISPR-Cas9 research in age-related diseases, identifying key contributors, major research hotspots, and critical technological advancements. While promising applications have been demonstrated in gene repair, functional regulation, and molecular interventions, significant barriers persist, including off-target effects, low delivery efficiency, and limited editing in non-dividing cells. Ethical concerns over germline editing and gaps in long-term safety data further complicate clinical translation. Future directions emphasize the development of high-precision Cas9 variants, homology-directed repair-independent tools, and efficient delivery systems, alongside the establishment of international regulatory frameworks and multicenter clinical trials. These efforts are essential to fully realize the potential of CRISPR-Cas9 in addressing the global health challenges of aging.
The global aging population has led to a sharp rise in age-related diseases, including Alzheimer’s (Hou et al., 2019), Parkinson’s (Lopes et al., 2022), cardiovascular disorders (Liang et al., 2024a), and cancer (Berben et al., 2021), placing considerable strain on healthcare systems due to their high prevalence and prolonged care requirements. Aging is a multifaceted biological process shaped by genetic (Maldonado et al., 2023), epigenetic (Liang et al., 2024b), and environmental factors (Dodig et al., 2019), characterized by key hallmarks such as genomic instability, aberrant epigenetic regulation, telomere attrition, mitochondrial dysfunction, and chronic low-grade inflammation (López-Otín et al., 2023).
CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-associated protein 9), a transformative genome-editing tool (Figure 1), enables precise targeting of genes and epigenetic regulators implicated in aging (Doudna and Charpentier, 2014). It has demonstrated potential in mitigating cellular senescence, genomic instability, and inflammatory processes (Hazrati et al., 2022). Applications in age-related diseases include correcting pathogenic mutations in Alzheimer’s and Parkinson’s, modulating metabolic pathways in cardiovascular disorders, and slowing the progression of osteoporosis and metabolic dysfunctions (Caobi et al., 2020). With superior precision and efficiency compared to conventional methods, CRISPR-Cas9 represents a powerful approach to unraveling and addressing the molecular mechanisms of aging (Zhang et al., 2024).
Despite significant advancements, research on the therapeutic applications of CRISPR-Cas9 in age-related diseases remains fragmented, with limited understanding of its mechanistic roles, long-term effects, and safety profiles (Caobi et al., 2020). Clinical translation faces major hurdles, including off-target effects leading to genomic instability, low tissue-specific delivery efficiency, and reduced efficacy in non-dividing cells (Dai et al., 2016). Ethical concerns surrounding germline editing and uncertainties about long-term impacts further constrain widespread adoption (Cwik, 2020). Bibliometric analysis offers a valuable framework to evaluate global research trends, identify key contributors, and map critical gaps, providing essential insights to guide future research and advance the field (Moed, 2009).
This study aims to construct a comprehensive landscape of CRISPR-Cas9’s therapeutic applications in age-related diseases, highlighting current progress, technological advancements, and translational barriers. Through the integration of bibliometric analysis and an evaluation of its therapeutic potential and challenges, this work offers insights to accelerate the clinical translation of CRISPR-Cas9 in addressing the health challenges of aging.
This study systematically retrieved CRISPR-Cas9 research related to age-associated diseases from PubMed, Embase, Cochrane Library, Scopus, and Web of Science, covering the period from January 1, 2005, to December 31, 2024. To ensure consistency and reliability, two authors independently conducted searches on the same day. Detailed search strategies and database-specific results are provided in Supplementary Table S1.
The inclusion criteria were as follows: 1) studies investigating CRISPR-Cas9 interventions targeting molecular pathways involved in age-associated diseases; 2) experimental validation through in vitro, in vivo, or clinical studies; and 3) provision of primary data analysis. Exclusion criteria included: 1) duplicate publications; 2) meeting abstracts; 3) editorial materials; 4) letters; 5) book chapters; 6) notes; 7) studies on unrelated diseases; 8) studies involving unrelated technologies; 9) articles with unavailable full texts; and 10) non-English language articles. Title, abstract and full-text screenings were independently conducted by two authors using pre-specified eligibility criteria and a piloted data extraction form. Discrepancies were resolved through iterative discussion, and unresolved conflicts were adjudicated by a third reviewer according to predefined rules, which included: 1) focusing on age-associated diseases as classified by the WHO; 2) mandatory re-examination of full texts for studies with ambiguous CRISPR-Cas9 applications; and 3) documenting all decisions with cross-references to the eligibility criteria. Ultimately, 923 original articles met the inclusion criteria and were included in the final analysis (Figure 2).
Bibliometric information, including publication and citation counts, countries, institutions, authors, journals, co-cited references, funding agencies, and keywords, was exported for analysis. Following bibliometric principles, co-occurrence analysis of countries, authors, institutions, and co-cited references was performed using CiteSpace to create visual atlases (Donthu et al., 2021). CiteSpace also facilitated cluster analysis and keyword burst detection. Cluster analysis, an unsupervised learning algorithm widely applied in machine learning and information recognition, was employed to uncover patterns and groupings within the data (Shirkhorshidi et al., 2014). A global distribution and collaboration map of CRISPR-Cas9 research in age-related diseases was generated using Python (v3.10.12), with Geopandas (v0.13.2) for geospatial data processing and Matplotlib (v3.7.1) for visualization. Additionally, the publication growth trend was modeled in Excel through a fitting formula, offering insights into research trajectories.
The growth of annual publications and citations reflects the dynamic expansion of the field. Total citations have surged to 23,835 (Figure 3A), with an average of 26.03 citations per article. The H-index, a robust metric of research impact, stands at 73, signifying that 73 articles have been cited at least 73 times (Lü et al., 2016). To illustrate the relationship between publication year and research output, a fitting formula was applied, where “x” represents the year and “y” denotes the annual publication count (y = 0.6073x2 - 4.449x + 5.6798, R2 = 0.9068) (Figure 3B). This exponential growth pattern correlates with critical therapeutic milestones in the field. The acceleration phase (2018–2022) coincided with the first CRISPR-based therapies entering clinical trials for age-related conditions, including CTX001 for β-thalassemia and sickle cell disease (Frangoul et al., 2021).
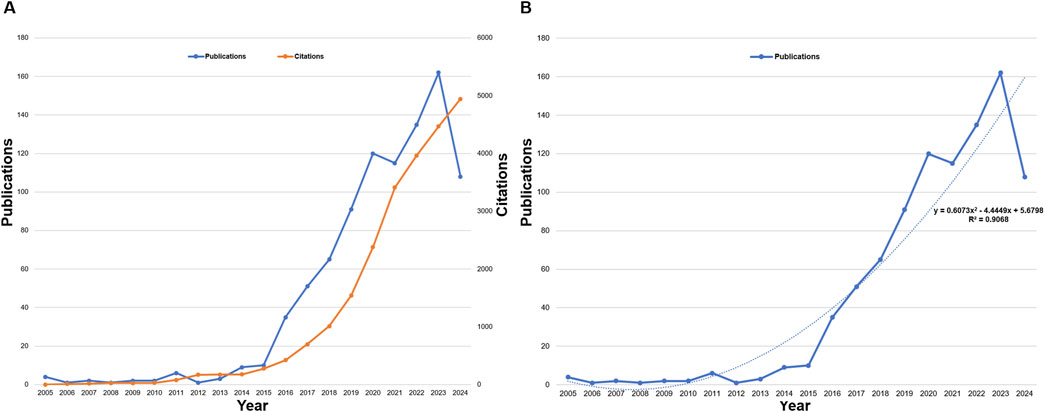
Figure 3. (A) Annual trends in publications and citations, and (B) curve fitting of annual growth trends in publications on CRISPR-Cas9 research.
Research on CRISPR-Cas9 in age-related diseases is distributed across 73 countries and regions, with the United States and China dominating the field, contributing 44.78% of the total publications (Figure 4A). Germany, England, and France also feature prominently, as reflected by their significant nodes in the country co-occurrence network, underscoring their substantial influence in this domain (Figure 4B). The international collaboration network comprises 135 interconnections among participating countries, resulting in a network density of 0.051. Although this indicates a relatively well-established framework for global cooperation, opportunities for strengthening cross-border partnerships remain (Figure 4B). These insights underscore both the leadership of key nations and the potential for fostering deeper international engagement.
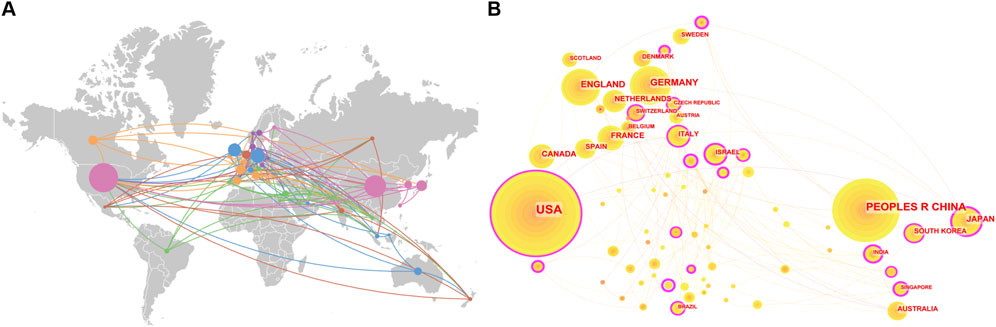
Figure 4. (A) Global distribution, and (B) country co-occurrence network map of CRISPR-Cas9 research and international collaborations in age-related diseases.
Our analysis also reveals national differences in CRISPR-Cas9 research on age-related diseases. The United States focuses on aging mechanisms and neurodegenerative disease models (Marschallinger et al., 2020), while China leads in gene screening, particularly whole-genome functional screening, and clinical translation (Li and Huang, 2018). Germany and the United Kingdom emphasize multi-center studies on neurodegenerative diseases (Barazesh et al., 2021), and France pioneers gene-editing therapies for cardiovascular diseases (Vermersch et al., 2020). These variations reflect distinct national strategies for advancing CRISPR-Cas9 in age-related diseases. Future studies could explore the underlying factors, such as demographic needs, funding priorities, and available expertise.
The top ten authors and institutions (Supplementary Tables S2, S3) primarily originate from the United States and China, highlighting the leading role of these two countries in the field. Several collaborative networks have emerged, with strong ties within groups but limited connections between them (Figure 5). Liu Guang-Hui and Qu Jing, both from the Chinese Academy of Sciences, are among the most prolific authors. Other notable contributors include Belmonte Juan Carlos Izpisua (Salk Institute) and Brunet Anne (Stanford University). Harvard University and the Chinese Academy of Sciences are the most productive institutions, with 87 and 77 publications, respectively, followed by the University of California system, Institut National de la Santé et de la Recherche Médicale, and the University of London, which also serve as key hubs for collaboration.
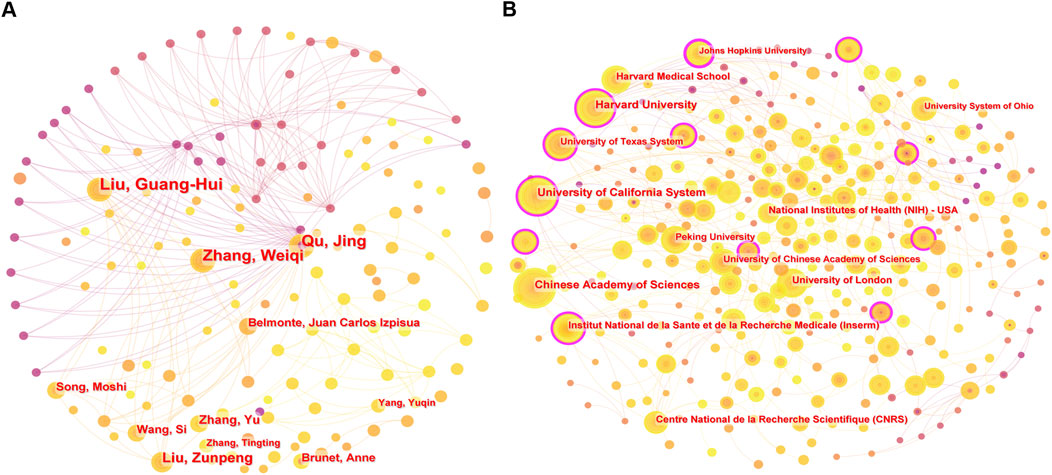
Figure 5. (A) Author co-occurrence network map, and (B) institution co-occurrence network map in CRISPR-Cas9 research on age-related diseases.
Despite robust partnerships within clusters, intergroup collaboration remains limited, reflecting potential regional or disciplinary barriers. Enhancing these connections could accelerate advancements in CRISPR-Cas9 applications for age-related diseases.
A total of 414 funding agencies were identified. The top ten funding bodies subsidized 399 studies, accounting for 43.23% of the literature in this field. The top three donors were the National Institutes of Health (77), the United States Department of Health and Human Services (57), and the National Natural Science Foundation of China (51). These results indicate that the United States and Chinese governments have provided great support and funding for CRISPR-Cas9 research on age-related diseases (Supplementary Table S4.
The top ten journals contributing to CRISPR-Cas9 research on age-related diseases are recognized as core outlets in the field, collectively accounting for 18.42% of total publications (Supplementary Table S5). All are classified within Q1 and Q2 categories of the Journal Citation Reports (2024) and specialize in molecular biology, genetics, and aging research. This reflects the interdisciplinary scope of CRISPR-Cas9 applications in tackling complex biological processes and age-related disorders. Given their prominence and focus, these journals are poised to remain pivotal in disseminating future breakthroughs, continuing to shape progress in this rapidly advancing domain.
Besides, the evolution of publication patterns across these journals tracks the therapeutic maturation of the field. Early publications predominantly appeared in basic science journals focusing on CRISPR mechanisms and tool development. By 2018–2020, a notable shift occurred toward disease-specific journals, coinciding with proof-of-concept studies demonstrating therapeutic efficacy in models of age-related diseases. Most recently (2021-present), clinical and translational journals have featured prominently, reflecting the advancement of CRISPR therapies toward patient applications. This progression through journal categories illustrates the field’s transition from technical innovation to therapeutic implementation, with recent publications increasingly addressing clinical considerations such as delivery optimization, safety profiling, and patient selection criteria.
Co-citation analysis, which identifies studies frequently cited together, provides insights into the intellectual framework of a research field (Small and Griffith, 1974). Using CiteSpace, a co-citation network comprising 716 nodes and 2,216 connections with a topological density of 0.0087 was constructed (Figure 6A), revealing key articles and their interrelationships. A timeline visualization (Figure 6B) illustrates the field’s progression, from the foundational discoveries of Jinek et al. (2012), and Cong et al. (2013) on the CRISPR-Cas9 system to advancements in precision editing techniques by Gaudelli et al. (2017), and Anzalone et al. (2019).
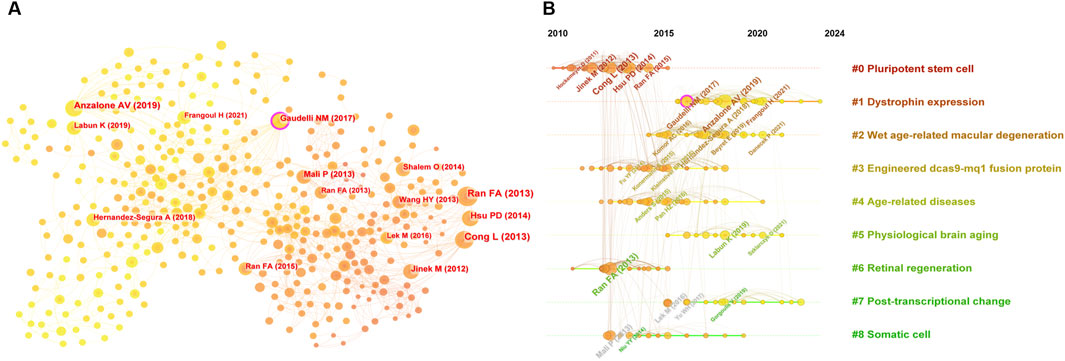
Figure 6. (A) Knowledge map of co-citation literature, and (B) timeline visualization of co-cited studies on CRISPR-Cas9 for combating age-related diseases.
Highly co-cited and cited works (Table 1) offer a framework for understanding the application of CRISPR-Cas9 in addressing age-related diseases. Foundational studies, including Ran et al. (2013) and Hsu et al. (2014), laid the groundwork for therapeutic innovations, driving significant progress in the use of genome editing technologies to combat aging-related conditions.
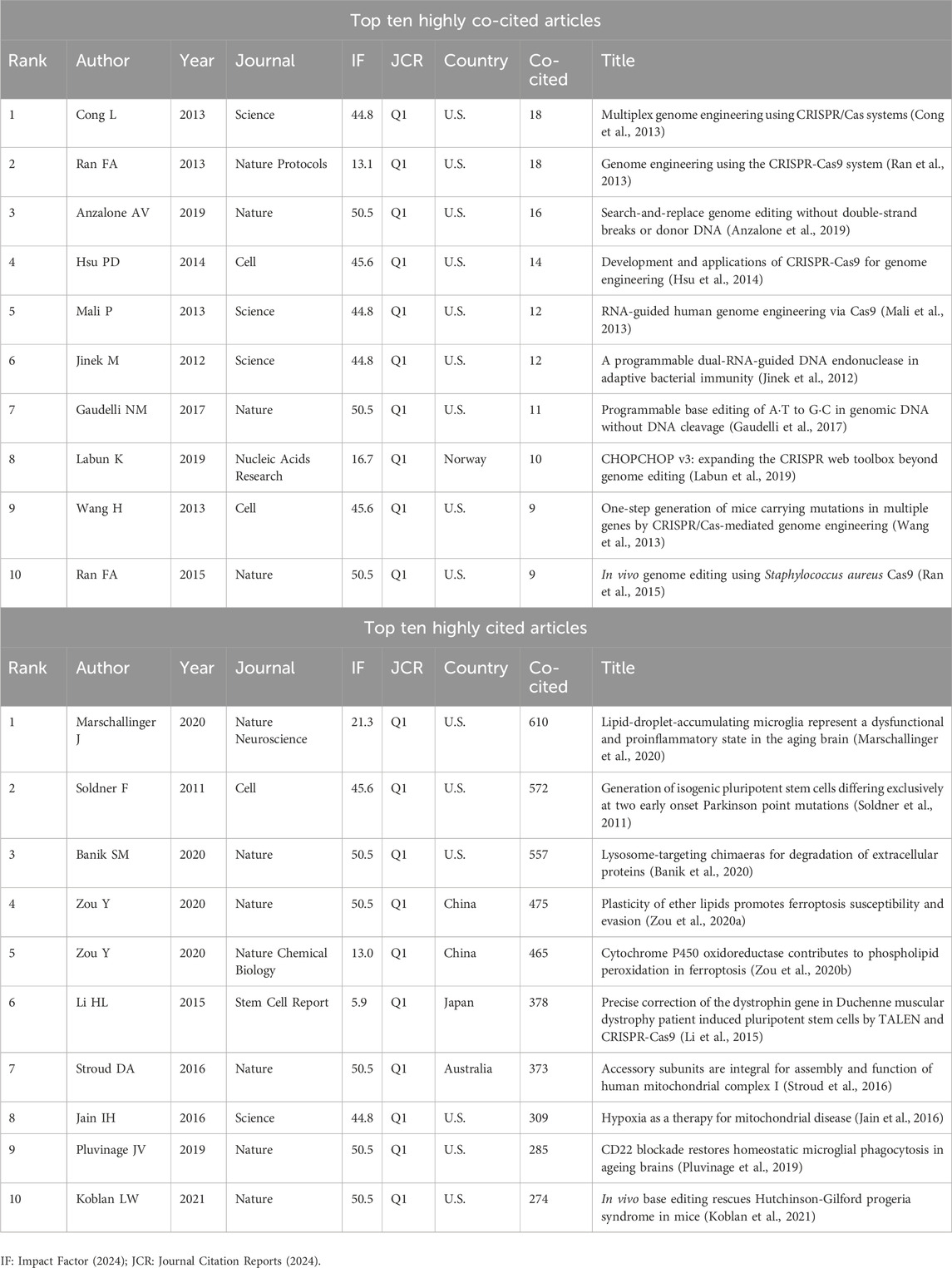
Table 1. Top ten highly co-cited and cited articles on CRISPR-Cas9 research in age-related diseases.
Keyword clustering analysis revealed the top ten clusters in CRISPR-Cas9 research on age-related diseases, including (0) Alzheimer’s disease, (1) Pathway, (2) DNA damage, (3) Expression, (4) Genome-wide association, (5) Life span, (6) Parkinson’s disease, (7) Genome editing, (8) Gene, and (9) Cellular senescence (Figure 7A). The modularity (Q) value of 0.8174 indicates a well-defined cluster structure (Q > 0.3), while the mean silhouette (S) value of 0.9004 confirms high reliability of the clustering (S > 0.7) (Yu and La, 2015). These clusters reflect the thematic focus and interdisciplinary nature of CRISPR-Cas9 applications in addressing key biological processes and diseases associated with aging. For instance, recent advances in Alzheimer’s disease (Cluster 0) highlight the translational potential of CRISPR-Cas9. Duan et al. utilized the blood-brain-barrier-penetrant AAV-PHP.eB vector to systemically deliver CRISPR-Cas9 in 5XFAD and APP/PS1 transgenic mice, achieving allele-specific editing of the human APPswe gene (Duan et al., 2022). This intervention reduced Aβ pathology, mitigated microgliosis and neuritic dystrophy, and rescued cognitive deficits. Similarly, Park et al. demonstrated that amphiphilic R7L10 nanoparticle-mediated hippocampal delivery of CRISPR-Cas9 targeting Bace1 in APPNL-G-F/NL-G-F and 5XFAD models attenuated BACE1 expression and ameliorated cognitive dysfunction5 (Park et al., 2019). These studies exemplify the growing emphasis on in vivo therapeutic strategies (Burst Keyword: In vivo, Strength 3.63) within Cluster 0. The Parkinson’s disease cluster (Cluster 6) further underscores innovations in delivery systems. Lipid nanoparticles (LNPs) encapsulating Cas9 protein and guide RNA (gRNA) have emerged as promising tools to protect CRISPR components from enzymatic degradation and enhance cellular uptake (Yadav et al., 2025). Surface functionalization of LNPs with targeting ligands, such as dopamine neuron-specific markers, enables precise delivery to degenerating neurons in Parkinson’s disease models (Wang et al., 2024). Polymer-based nanoparticles with tailored surface modifications similarly facilitate CRISPR/Cas9 targeting to dopamine pathways, illustrating the convergence of genome editing (Cluster 7) and translational neurotherapeutics.
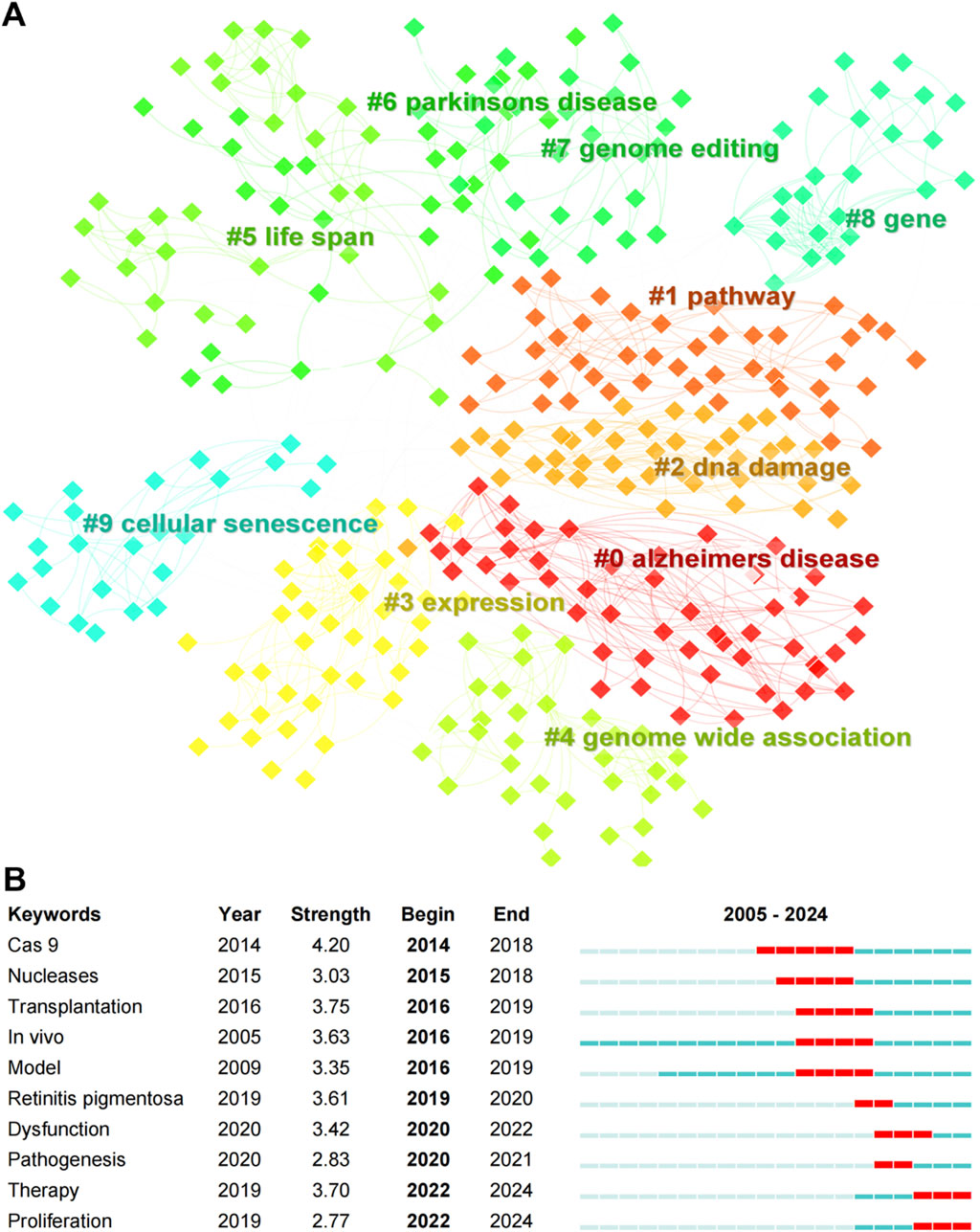
Figure 7. (A) Clustering map of keywords, and (B) top ten keywords with the strongest citation bursts (2005–2024) in CRISPR-Cas9 research on age-related diseases.
Burst detection analysis identified the top ten keywords with the strongest citation bursts from 2005 to 2024 (Figure 7B). Among these, “Cas9” exhibited the highest burst strength (4.20) and the longest burst period, signifying its foundational role in the field. Other keywords with notable burst strengths include “Transplantation” (3.75) and “In vivo” (3.63), indicating their early significance in CRISPR-Cas9 research. The therapeutic focus is further emphasized by recent bursts in “Therapy” and “Dysfunction,” paralleling breakthroughs in retinal degeneration research. For example, Latella et al. demonstrated allele-specific disruption of the dominant S334ter mutation in the RHO gene via CRISPR/Cas9 in autosomal dominant retinitis pigmentosa (adRP) models (Latella et al., 2016). Subretinal delivery of gRNA/Cas9 plasmids prevented photoreceptor degeneration and restored visual function, validating its efficacy in gene editing (Cluster 7) for inherited retinal disorders (Cai et al., 2018). Such advances align with the rising prominence of therapy-oriented research (Burst Keyword: “Therapy”) in age-related disease models. These findings underscore the ongoing evolution of CRISPR-Cas9 research, with a shift toward translational and clinical advancements for combating age-related diseases.
This study reveals a global research landscape on CRISPR-Cas9 in age-related diseases, with the United States and China serving as central contributors, driven by institutions such as Harvard University and the Chinese Academy of Sciences. Keyword clustering and burst analyses highlight research hotspots in Alzheimer’s disease, Parkinson’s disease, gene editing, and cellular senescence, reflecting a transition from fundamental mechanisms to clinical applications. By identifying key contributors, major research focuses, and the phased characteristics of technological development, this study provides a comprehensive overview and valuable data to inform future research directions.
CRISPR-Cas9 has demonstrated significant therapeutic potential in age-related diseases, particularly in neurodegenerative disorders, metabolic diseases, and osteoporosis (Table 2). In Alzheimer’s disease, CRISPR-Cas9 reduces β-amyloid deposition by knocking out APP and PSEN1 genes, alleviating neuroinflammation and improving cognitive function (Yoon et al., 2024). In Parkinson’s disease, it decreases α-synuclein aggregation by targeting the SNCA gene and restores mitochondrial function by correcting mutations in LRRK2 and PINK1 (Mansour and El-Khatib, 2023). Similarly, in cardiovascular diseases, targeting PCSK9 lowers cholesterol levels, while NLRP3 knockout reduces myocardial inflammation, improving atherosclerosis and decreasing cardiovascular event risk (Makhmudova et al., 2024). For osteoporosis, modulating the SOST gene enhances bone formation, while inhibiting RANKL reduces bone resorption, thereby increasing bone density and preventing fractures (Lin et al., 2024). These findings provide a foundation for direct molecular interventions in disease mechanisms. Recent research highlights the role of PANoptosis in immune infiltration during Alzheimer’s disease and the anti-aging effects of Panax notoginseng saponins through apoptosis and neurodegeneration pathways, offering novel therapeutic insights (Mei et al., 2024; Zhao et al., 2024).
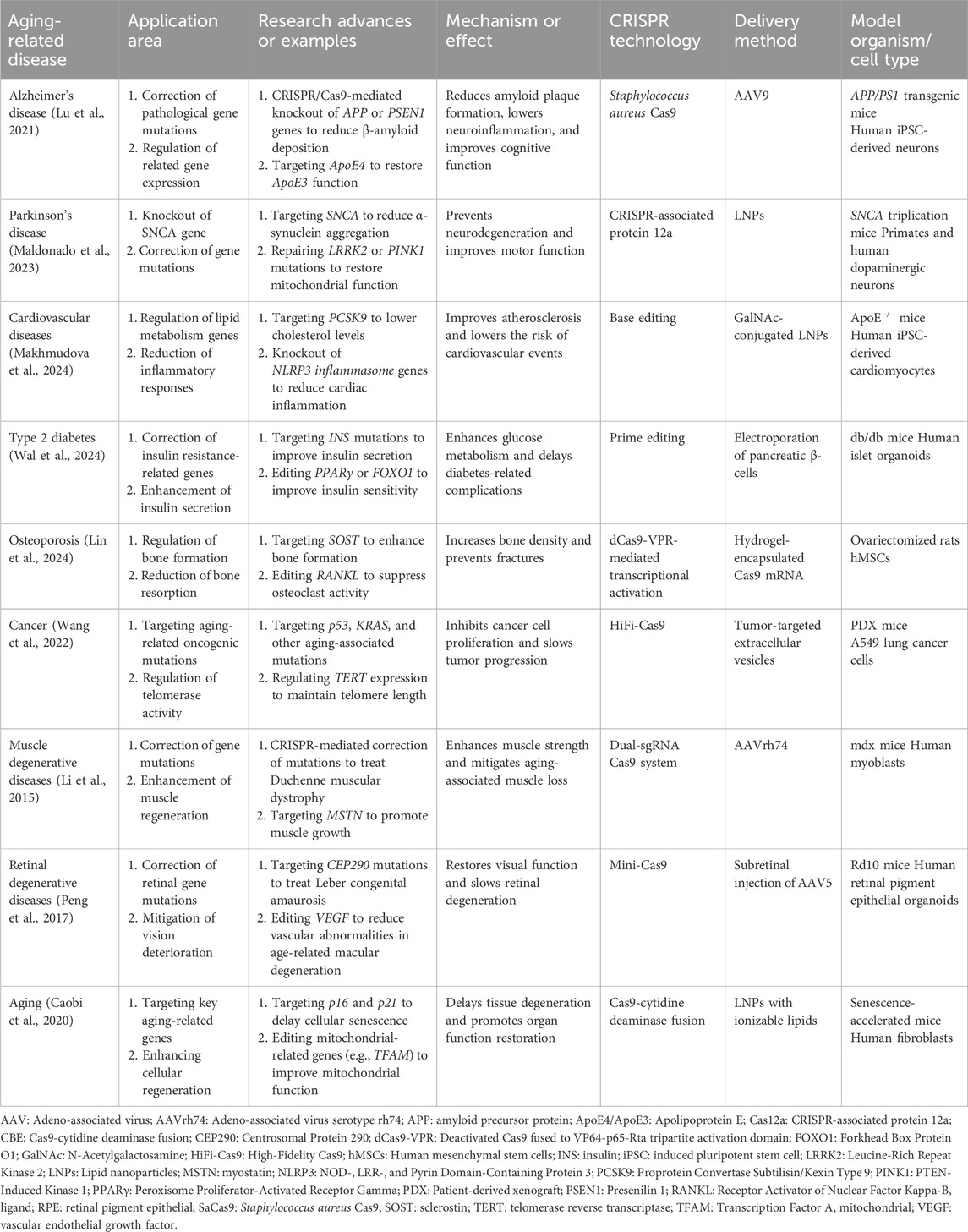
Table 2. CRISPR-based therapeutic innovations for aging-related disorders: technological platforms, delivery systems, and preclinical models.
On the technical front, high-fidelity (HiFi) Cas9 variants, such as eSpCas9 and HiFi Cas9, have significantly minimized off-target effects (Chen et al., 2017; Vakulskas et al., 2018), while HDR-independent tools like base editing and prime editing have improved gene editing efficiency in non-dividing cells (Kantor et al., 2020). Advances in non-viral delivery systems and low-immunogenicity Cas9 variants address challenges of delivery inefficiency and high immune risks (Li et al., 2018; Kenjo et al., 2021). These innovations enhance the precision, safety, and feasibility of CRISPR-Cas9 in both research and potential clinical applications for age-related diseases.
Co-citation analyses and preclinical validation studies consistently identify off-target effects as the primary technical constraint limiting clinical translation of CRISPR-Cas9 systems (Yuan, 2025). Emerging data demonstrate that nonspecific Cas9 activity not only induces genomic instability but also disrupts epigenetic regulatory networks, presenting particular risks in polygenic age-related disorders where pathway interdependencies magnify biological consequences (Table 3). These challenges manifest most prominently within two interconnected research domains: DNA damage repair mechanisms associated with error-prone nonhomologous end joining, and gene expression dysregulation stemming from unintended editing of regulatory elements. Technological innovations are addressing these concerns, as evidenced by high-fidelity editors such as HiFi-Cas9 achieving 20-fold reductions in off-target activity within human hematopoietic stem cells (Vakulskas et al., 2018). Concurrently, artificial intelligence platforms exemplified by DeepCRISPR provide robust prediction of guide RNA specificity, attaining 0.981 ROC-AUC values in cross-validated datasets (Chuai et al., 2018). However, maintaining functional equilibrium in aging-related gene networks requires further refinement of these precision tools, particularly when targeting genomic regions containing homologous pseudogenes or repetitive sequences.
Delivery limitations, prominently identified through citation burst dynamics of keywords such as “Transplantation” and “In vivo,” persist as critical translational barriers. Current strategies increasingly adopt multifunctional integration paradigms to simultaneously address tissue selectivity, delivery efficiency, and immune compatibility. Viral vector optimization efforts demonstrate particular promise, exemplified by AAV-mediated delivery of miniaturized dystrophin constructs achieving functional recovery in canine muscular dystrophy models with negligible adverse events (Birch et al., 2023). Conversely, non-viral approaches are advancing through ligand-directed targeting, as evidenced by epidermal growth factor receptor functionalized lipid nanoparticles delivering CRISPR-SOX2 therapeutics that achieved 90% tumor growth suppression and extended survival beyond 84 days in head-and-neck squamous cell carcinoma xenografts (Masarwy et al., 2025). These case studies underscore the potential of hybrid delivery architectures combining viral precision with non-viral safety profiles. Nevertheless, critical gaps remain: AAV systems face payload capacity limitations for larger Cas9 variants, while nanoparticle formulations exhibit suboptimal penetration through fibrotic microenvironments characteristic of aged tissues.
Ethical considerations and longitudinal safety uncertainties remain inextricably linked to CRISPR-Cas9 clinical development. Germline editing controversies, particularly regarding irreversible hereditary modifications and transgenerational epigenetic effects, have catalyzed international scientific consensus-building efforts. The 2018 gene-edited infant incident precipitated rigorous institutional responses, including the Chinese Academy of Sciences’ seminal position statement advocating for stringent oversight frameworks and restricted therapeutic applications (Wang et al., 2019).
To address current technical and ethical challenges, the future development of CRISPR-Cas9 should focus on optimizing precision, enhancing delivery strategies, and establishing robust regulatory frameworks. For off-target effects, high-precision sgRNA design platforms and single-base editors offer improved specificity and safety in polygenic environments (Doench et al., 2016; Hryhorowicz et al., 2023). Tools independent of HDR, such as prime editors, could further expand applicability by overcoming low editing efficiency in non-dividing cells (Kantor et al., 2020). In terms of delivery, ligand-modified lipid nanoparticles show promise for improving tissue specificity, while combining viral and non-viral delivery methods could enhance efficiency and mitigate immunogenic risks (Li et al., 2018). To address long-term safety concerns, developing low-immunogenic Cas9 variants and incorporating controllable gene-editing switches could enable reversible and precise interventions.
On the ethical and regulatory front, dynamic ethical oversight and international collaboration are essential for establishing unified frameworks to govern the use of germline editing. Multicenter clinical trial networks should prioritize gathering long-term safety and efficacy data to ensure the responsible application of CRISPR-Cas9. These strategies aim to seamlessly bridge technological advancements and clinical translation, accelerating its application in treating age-related diseases.
This study has certain limitations, primarily in the scope of data coverage and the depth of technical analysis. While we retrieved a substantial number of publications from major databases such as Web of Science, the exclusion of regional studies and non-English literature may result in incomplete global coverage. To minimize this bias, we employed a multi-database integration approach and a rigorous three-reviewer screening process. However, future studies should incorporate regional databases (e.g., CNKI) and open-access resources (e.g., bioRxiv) to enhance data comprehensiveness. Additionally, although this study systematically identified the technical challenges of CRISPR-Cas9, it provides limited exploration of the molecular mechanisms underlying these issues and cutting-edge solutions. Future work should integrate experimental data and disease-specific models to validate the biological efficacy of proposed strategies, offering more concrete guidance for advancing the technology.
This study provides a comprehensive overview of the global development, therapeutic potential, and technical challenges of CRISPR-Cas9 in age-related disease research. The findings highlight its promising applications in gene repair and functional regulation for diseases such as Alzheimer’s and Parkinson’s. However, significant barriers remain, including off-target effects, low delivery efficiency, and challenges in editing non-dividing cells, alongside ethical concerns over germline editing and long-term safety. Future efforts should prioritize the development of high-precision Cas9 variants, HDR-independent tools, and efficient delivery systems. Integrating multicenter clinical trials with an international ethical framework will be critical to unlocking the transformative potential of CRISPR-Cas9 for treating age-related diseases.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
QH: Conceptualization, Data curation, Formal Analysis, Writing–original draft. YW: Data curation, Formal Analysis, Methodology, Writing–original draft. ZT: Conceptualization, Methodology, Software, Writing–original draft. XZ: Writing–review and editing. CY: Supervision, Writing–review and editing. XJ: Supervision, Writing–review and editing.
The author(s) declare that no financial support was received for the research and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2025.1558432/full#supplementary-material
APP, amyloid precursor protein; CNKI, China National Knowledge Infrastructure; CRISPR-Cas9, clustered regularly interspaced short palindromic repeats-associated protein 9; eSpCas9, enhanced specificity Cas9; HDR, homology-directed repair; HiFi Cas9, high-fidelity Cas9; LRRK2, leucine-rich repeat kinase 2; NHEJ, non-homologous end joining; NLRP3, NOD-, LRR-, and pyrin domain-containing protein 3; PCSK9, proprotein convertase subtilisin/kexin type 9; PINK1, PTEN-induced kinase 1; PSEN1, presenilin 1; RANKL, receptor activator of nuclear factor kappa-Β ligand; sgRNA, single-guide RNA; SNCA, synuclein alpha.
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 (7785), 149–157. doi:10.1038/s41586-019-1711-4
Banik, S. M., Pedram, K., Wisnovsky, S., Ahn, G., Riley, N. M., and Bertozzi, C. R. (2020). Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 584 (7820), 291–297. doi:10.1038/s41586-020-2545-9
Barazesh, M., Mohammadi, S., Bahrami, Y., Mokarram, P., Morowvat, M. H., Saidijam, M., et al. (2021). CRISPR/Cas9 technology as a modern genetic manipulation tool for recapitulating of neurodegenerative disorders in large animal models. Curr. Gene Ther. 21 (2), 130–148. doi:10.2174/1566523220666201214115024
Berben, L., Floris, G., Wildiers, H., and Hatse, S. (2021). Cancer and aging: two tightly interconnected biological processes. Cancers 13 (6), 1400. doi:10.3390/cancers13061400
Birch, S. M., Lawlor, M. W., Conlon, T. J., Guo, L. J., Crudele, J. M., Hawkins, E. C., et al. (2023). Assessment of systemic AAV-microdystrophin gene therapy in the GRMD model of Duchenne muscular dystrophy. Sci. Transl. Med. 15 (677), eabo1815. doi:10.1126/scitranslmed.abo1815
Cai, B., Sun, S., Li, Z., Zhang, X., Ke, Y., Yang, J., et al. (2018). Application of CRISPR/Cas9 technologies combined with iPSCs in the study and treatment of retinal degenerative diseases. Hum. Genet. 137 (9), 679–688. doi:10.1007/s00439-018-1933-9
Caobi, A., Dutta, R. K., Garbinski, L. D., Esteban-Lopez, M., Ceyhan, Y., Andre, M., et al. (2020). The impact of CRISPR-Cas9 on age-related disorders: from pathology to therapy. Aging Dis. 11 (4), 895–915. doi:10.14336/AD.2019.0927
Chen, J. S., Dagdas, Y. S., Kleinstiver, B. P., Welch, M. M., Sousa, A. A., Harrington, L. B., et al. (2017). Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550 (7676), 407–410. doi:10.1038/nature24268
Chuai, G., Ma, H., Yan, J., Chen, M., Hong, N., Xue, D., et al. (2018). DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 19 (1), 80. doi:10.1186/s13059-018-1459-4
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121), 819–823. doi:10.1126/science.1231143
Cwik, B. (2020). Revising, correcting, and transferring genes. Am. J. Bioeth. 20 (8), 7–18. doi:10.1080/15265161.2020.1783024
Dai, W. J., Zhu, L. Y., Yan, Z. Y., Xu, Y., Wang, Q. L., and Lu, X. J. (2016). CRISPR-Cas9 for in vivo gene therapy: promise and hurdles. Mol. Ther. Nucleic Acids 5 (8), e349. doi:10.1038/mtna.2016.58
Dodig, S., Čepelak, I., and Pavić, I. (2019). Hallmarks of senescence and aging. Biochem. Med. 29 (3), 030501. doi:10.11613/BM.2019.030501
Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W., Donovan, K. F., et al. (2016). Optimized sgrna design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34 (2), 184–191. doi:10.1038/nbt.3437
Doman, J. L., Pandey, S., Neugebauer, M. E., An, M., Davis, J. R., Randolph, P. B., et al. (2023). Phage-assisted evolution and protein engineering yield compact, efficient prime editors. Cell 186 (18), 3983–4002.e26. doi:10.1016/j.cell.2023.07.039
Donthu, N., Kumar, S., Mukherjee, D., Pandey, N., and Lim, W. M. (2021). How to conduct a bibliometric analysis: an overview and guidelines. J. Bus. Res. 133, 285–296. doi:10.1016/j.jbusres.2021.04.070
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346 (6213), 1258096. doi:10.1126/science.1258096
Duan, Y., Ye, T., Qu, Z., Chen, Y., Miranda, A., Zhou, X., et al. (2022). Brain-wide Cas9-mediated cleavage of a gene causing familial Alzheimer's disease alleviates amyloid-related pathologies in mice. Nat. Biomed. Eng. 6 (2), 168–180. doi:10.1038/s41551-021-00759-0
Frangoul, H., Altshuler, D., Cappellini, M. D., Chen, Y. S., Domm, J., Eustace, B. K., et al. (2021). CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 384 (3), 252–260. doi:10.1056/NEJMoa2031054
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature 551 (7681), 464–471. doi:10.1038/nature24644
Gheibi Hayat, S. M., Bianconi, V., Pirro, M., and Sahebkar, A. (2019). Stealth functionalization of biomaterials and nanoparticles by CD47 mimicry. Int. J. Pharm. 569, 118628. doi:10.1016/j.ijpharm.2019.118628
Hazrati, A., Malekpour, K., Soudi, S., and Hashemi, S. M. (2022). CRISPR/Cas9-engineered mesenchymal stromal/stem cells and their extracellular vesicles: a new approach to overcoming cell therapy limitations. Biomed. Pharmacother. 156, 113943. doi:10.1016/j.biopha.2022.113943
Hou, Y., Dan, X., Babbar, M., Wei, Y., Hasselbalch, S. G., Croteau, D. L., et al. (2019). Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15 (10), 565–581. doi:10.1038/s41582-019-0244-7
Hryhorowicz, M., Lipiński, D., and Zeyland, J. (2023). Evolution of CRISPR/Cas systems for precise genome editing. Int. J. Mol. Sci. 24 (18), 14233. doi:10.3390/ijms241814233
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157 (6), 1262–1278. doi:10.1016/j.cell.2014.05.010
Hu, J. H., Miller, S. M., Geurts, M. H., Tang, W., Chen, L., Sun, N., et al. (2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. doi:10.1038/nature26155
Jain, I. H., Zazzeron, L., Goli, R., Alexa, K., Schatzman-Bone, S., Dhillon, H., et al. (2016). Hypoxia as a therapy for mitochondrial disease. Science 352 (6281), 54–61. doi:10.1126/science.aad9642
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. doi:10.1126/science.1225829
Kantor, A., McClements, M. E., and MacLaren, R. E. (2020). CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 21 (17), 6240. doi:10.3390/ijms21176240
Kenjo, E., Hozumi, H., Makita, Y., Iwabuchi, K. A., Fujimoto, N., Matsumoto, S., et al. (2021). Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice. Nat. Commun. 12 (1), 7101. doi:10.1038/s41467-021-26714-w
Koblan, L. W., Erdos, M. R., Wilson, C., Cabral, W. A., Levy, J. M., Xiong, Z. M., et al. (2021). In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature 589 (7843), 608–614. doi:10.1038/s41586-020-03086-7
Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H., and Valen, E. (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47 (W1), W171–W174. doi:10.1093/nar/gkz365
Latella, M. C., Di Salvo, M. T., Cocchiarella, F., Benati, D., Grisendi, G., Comitato, A., et al. (2016). In vivo editing of the human mutant rhodopsin gene by electroporation of plasmid-based CRISPR/Cas9 in the mouse retina. Mol. Ther. Nucleic Acids 5 (11), e389. doi:10.1038/mtna.2016.92
Li, H., and Huang, C. (2018). Functional genetic screening using CRISPR-Cas9 system. Sheng Wu Gong Cheng Xue Bao 34 (4), 461–472. doi:10.13345/j.cjb.170348
Li, H. L., Fujimoto, N., Sasakawa, N., Shirai, S., Ohkame, T., Sakuma, T., et al. (2015). Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep. 4 (1), 143–154. doi:10.1016/j.stemcr.2014.10.013
Li, L., Hu, S., and Chen, X. (2018). Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials 171, 207–218. doi:10.1016/j.biomaterials.2018.04.031
Liang, R., Qi, X., Cai, Q., Niu, L., Huang, X., Zhang, D., et al. (2024a). The role of NLRP3 inflammasome in aging and age-related diseases. Immun. Ageing 21 (1), 14. doi:10.1186/s12979-023-00395-z
Liang, R., Tang, Q., Chen, J., and Zhu, L. (2024b). Epigenetic clocks: beyond biological age, using the past to predict the present and future. Aging Dis., 0. Online ahead of print. doi:10.14336/AD.2024.1495
Lin, W., Hu, S., Li, K., Shi, Y., Pan, C., Xu, Z., et al. (2024). Breaking osteoclast-acid vicious cycle to rescue osteoporosis via an acid responsive organic framework-based neutralizing and gene editing platform. Small 20 (22), e2307595. doi:10.1002/smll.202307595
Lopes, D. M., Llewellyn, S. K., and Harrison, I. F. (2022). Propagation of tau and α-synuclein in the brain: therapeutic potential of the glymphatic system. Transl. Neurodegener. 11 (1), 19. doi:10.1186/s40035-022-00293-2
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., and Kroemer, G. (2023). Hallmarks of aging: an expanding universe. Cell 186 (2), 243–278. doi:10.1016/j.cell.2022.11.001
Lu, L., Yu, X., Cai, Y., Sun, M., and Yang, H. (2021). Application of CRISPR/Cas9 in Alzheimer's disease. Front. Neurosci. 15, 803894. doi:10.3389/fnins.2021.803894
Lü, L., Zhou, T., Zhang, Q. M., and Stanley, H. E. (2016). The H-index of a network node and its relation to degree and coreness. Nat. Commun. 7, 10168. doi:10.1038/ncomms10168
Makhmudova, U., Steinhagen-Thiessen, E., Volpe, M., and Landmesser, U. (2024). Advances in nucleic acid-targeted therapies for cardiovascular disease prevention. Cardiovasc Res. 120 (10), 1107–1125. doi:10.1093/cvr/cvae136
Maldonado, E., Morales-Pison, S., Urbina, F., and Solari, A. (2023). Aging hallmarks and the role of oxidative stress. Antioxidants 12 (3), 651. doi:10.3390/antiox12030651
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339 (6121), 823–826. doi:10.1126/science.1232033
Mansour, H. M., and El-Khatib, A. S. (2023). Exploring Parkinson-associated kinases for CRISPR/Cas9-based gene editing: beyond alpha-synuclein. Ageing Res. Rev. 92, 102114. doi:10.1016/j.arr.2023.102114
Marschallinger, J., Iram, T., Zardeneta, M., Lee, S. E., Lehallier, B., Haney, M. S., et al. (2020). Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23 (2), 194–208. doi:10.1038/s41593-019-0566-1
Masarwy, R., Breier, D., Stotsky-Oterin, L., Ad-El, N., Qassem, S., Naidu, G. S., et al. (2025). Targeted CRISPR/Cas9 lipid nanoparticles elicits therapeutic genome editing in head and neck cancer. Adv. Sci. (Weinh) 12 (7), e2411032. doi:10.1002/advs.202411032
Mei, J. L., Wang, S. F., Zhao, Y. Y., Xu, T., Luo, Y., and Xiong, L. L. (2024). Identification of immune infiltration and PANoptosis-related molecular clusters and predictive model in Alzheimer's disease based on transcriptome analysis. Ibrain 10 (3), 323–344. doi:10.1002/ibra.12179
Moed, H. F. (2009). New developments in the use of citation analysis in research evaluation. Arch. Immunol. Ther. Exp. 57 (1), 13–18. doi:10.1007/s00005-009-0001-5
Park, H., Oh, J., Shim, G., Cho, B., Chang, Y., Kim, S., et al. (2019). In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer's disease. Nat. Neurosci. 22 (4), 524–528. doi:10.1038/s41593-019-0352-0
Peng, Y. Q., Tang, L. S., Yoshida, S., and Zhou, Y. D. (2017). Applications of CRISPR/Cas9 in retinal degenerative diseases. Int. J. Ophthalmol. 10 (4), 646–651. doi:10.18240/ijo.2017.04.23
Pluvinage, J. V., Haney, M. S., Smith, B. A. H., Sun, J., Iram, T., Bonanno, L., et al. (2019). CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 568 (7751), 187–192. doi:10.1038/s41586-019-1088-4
Ran, F. A., Cong, L., Yan, W. X., Scott, D. A., Gootenberg, J. S., Kriz, A. J., et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520 (7546), 186–191. doi:10.1038/nature14299
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8 (11), 2281–2308. doi:10.1038/nprot.2013.143
Shen, X., Lin, Q., Liang, Z., Wang, J., Yang, X., Liang, Y., et al. (2022). Reduction of pre-existing adaptive immune responses against SaCas9 in humans using epitope mapping and identification. CRISPR J. 5 (3), 445–456. doi:10.1089/crispr.2021.0142
Shirkhorshidi, A. S., Aghabozorgi, Sr., Wah, T., and Herawan, T. (2014). Big data clustering: a review. Comput. Sci. Its Appl. - ICCSA 2014 8583, 707–720. doi:10.1007/978-3-319-09156-3_49
Small, H., and Griffith, B. C. (1974). The structure of scientific literatures I: identifying andgraphing specialties. Sci. Stud. 4 (1), 17–40. doi:10.1177/030631277400400102
Soldner, F., Laganière, J., Cheng, A. W., Hockemeyer, D., Gao, Q., Alagappan, R., et al. (2011). Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 146 (2), 318–331. doi:10.1016/j.cell.2011.06.019
Stroud, D. A., Surgenor, E. E., Formosa, L. E., Reljic, B., Frazier, A. E., Dibley, M. G., et al. (2016). Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538 (7623), 123–126. doi:10.1038/nature19754
Vakulskas, C. A., Dever, D. P., Rettig, G. R., Turk, R., Jacobi, A. M., Collingwood, M. A., et al. (2018). A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 24 (8), 1216–1224. doi:10.1038/s41591-018-0137-0
Vermersch, E., Jouve, C., and Hulot, J. S. (2020). CRISPR/Cas9 gene-editing strategies in cardiovascular cells. Cardiovasc Res. 116 (5), 894–907. doi:10.1093/cvr/cvz250
Wal, P., Aziz, N., Prajapati, H., Soni, S., and Wal, A. (2024). Current landscape of various techniques and methods of gene therapy through CRISPR Cas9 along with its pharmacological and interventional therapies in the treatment of type 2 diabetes mellitus. Curr. Diabetes Rev. 20 (6), e201023222414. doi:10.2174/0115733998263079231011073803
Wang, C., Zhai, X., Zhang, X., Li, L., Wang, J., Liu, D. P., et al. (2019). Gene-edited babies: Chinese Academy of medical sciences' response and action. Lancet 393 (10166), 25–26. doi:10.1016/S0140-6736(18)33080-0
Wang, H., and Yang, H. (2019). Gene-edited babies: what went wrong and what could go wrong. PLoS Biol. 17 (4), e3000224. doi:10.1371/journal.pbio.3000224
Wang, H., Yang, H., Shivalila, C. S., Dawlaty, M. M., Cheng, A. W., Zhang, F., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153 (4), 910–918. doi:10.1016/j.cell.2013.04.025
Wang, S. W., Gao, C., Zheng, Y. M., Yi, L., Lu, J. C., Huang, X. Y., et al. (2022). Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol. Cancer 21 (1), 57. doi:10.1186/s12943-022-01518-8
Wang, X., Cai, C., Lv, W., Chen, K., Li, J., Liao, K., et al. (2024). Short cell-penetration peptide conjugated bioreducible polymer enhances gene editing of CRISPR system. J. Nanobiotechnology 22 (1), 284. doi:10.1186/s12951-024-02554-w
Yadav, V. K., Dhanasekaran, S., Choudhary, N., Nathiya, D., Thakur, V., Gupta, R., et al. (2025). Recent advances in nanotechnology for Parkinson's disease: diagnosis, treatment, and future perspectives. Front. Med. (Lausanne) 12, 1535682. doi:10.3389/fmed.2025.1535682
Yoon, J., Monarch, K., Kiesewetter, E., Rodriguez-Lukey, K., Koh, S., Uh, K., et al. (2024). Development of a novel swine model carrying humanized APP and PSEN1. Alzheimers Dement. 20 (Suppl. 1), e089607. doi:10.1002/alz.089607
Yu, C., and La, W. (2015). The methodology function of Cite Space mapping knowledge domains. Stud. Sci. Sci. doi:10.1515/jdis-2017-0006
Yuan, Z. (2025). From origin to the present: establishment, mechanism, evolutions and biomedical applications of the CRISPR/Cas-based macromolecular system in brief. Molecules 30 (4), 947. doi:10.3390/molecules30040947
Zhang, J., Zhou, Y., Qiao, J., and Liu, Y. (2024). Recent advances in spatiotemporal control of the CRISPR/Cas9 system. Colloids Surf. B Biointerfaces 248, 114474. doi:10.1016/j.colsurfb.2024.114474
Zhang, J. P., Yang, Z. X., Zhang, F., Fu, Y. W., Dai, X. Y., Wen, W., et al. (2021). HDAC inhibitors improve CRISPR-mediated HDR editing efficiency in iPSCs. Sci. China Life Sci. 64 (9), 1449–1462. doi:10.1007/s11427-020-1855-4
Zhao, Y. Y., Yang, L. X., Que, S. Y., An, L. X., Teeti, A. A., and Xiao, S. W. (2024). Systemic mechanism of Panax noteginseng saponins in antiaging based on network pharmacology combined with experimental validation. Ibrain 10 (4), 519–535. doi:10.1002/ibra.12165
Zou, Y., Henry, W. S., Ricq, E. L., Graham, E. T., Phadnis, V. V., Maretich, P., et al. (2020a). Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature 585 (7826), 603–608. doi:10.1038/s41586-020-2732-8
Keywords: age-related diseases, bibliometric analysis, CRISPR-Cas9, genome editing, gene therapy
Citation: He Q, Wang Y, Tan Z, Zhang X, Yu C and Jiang X (2025) Mapping the therapeutic landscape of CRISPR-Cas9 for combating age-related diseases. Front. Genome Ed. 7:1558432. doi: 10.3389/fgeed.2025.1558432
Received: 10 January 2025; Accepted: 19 March 2025;
Published: 04 April 2025.
Edited by:
Wenchuan Qi, Chengdu University of Traditional Chinese Medicine, ChinaReviewed by:
Dhruva Katrekar, Arc Institute, United StatesCopyright © 2025 He, Wang, Tan, Zhang, Yu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Yu, Nzc5MDg4NDFAcXEuY29t; Xiaoqin Jiang, MTU5ODg2MjY1N2p4cUBzY3UuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.